Abstract
The mRNA vaccine route from injection site to critical immunologic tissues, as well as the localization of protein antigen following intramuscular (i.m.) administration, is crucial to generating an effective immune response. Here, we quantified mRNA at the injection site, lymph nodes, and in select tissues. mRNA was primarily present 24 h after administration and then rapidly degraded from local and systemic tissues. Histological analyses of mRNA and expressed protein at the site of administration and in the lymph nodes following i.m. administration of our vaccine in rodents and nonhuman primates (NHPs) were completed, and mRNA and protein expression were detected in tissue resident and infiltrating immune cells at the injection site. In addition, high levels of protein expression were observed within subcapsular and medullary sinus macrophages in draining lymph nodes. More important, results were similar between rodents and NHPs, indicating cross-species similarities.
Keywords: MT: Delivery Strategies, lipid nanoparticles, mRNA, vaccine, intramuscular, delivery, trafficking
Graphical abstract
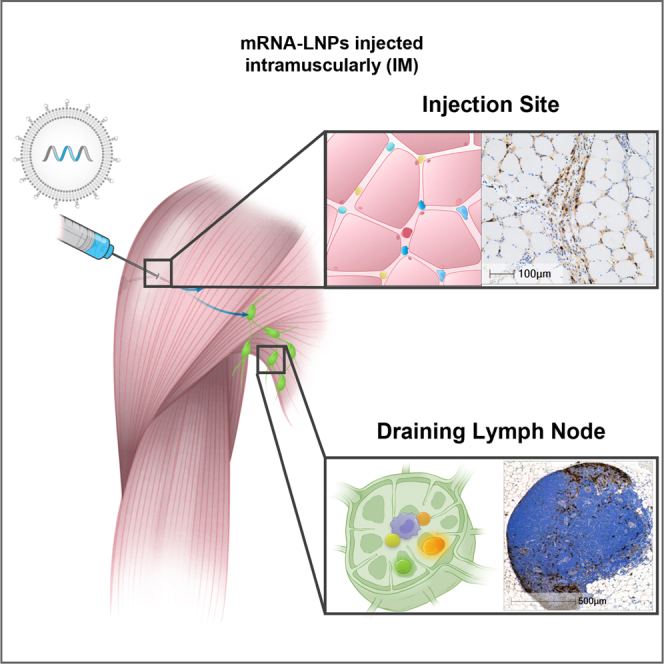
Burke and colleagues demonstrated that following intramuscular administration of mRNA-lipid nanoparticles, mRNA was present for 24 h and then rapidly degraded. mRNA and expressed protein were detected in injection site tissue-resident and infiltrating immune cells and draining lymph nodes. mRNA-lipid nanoparticle vaccine trafficking and resulting protein expression are crucial for effective immune response.
Introduction
A greater understanding of vaccine delivery system dynamics and resulting protein expression in target tissues and specific immune cells over time could inform future mRNA vaccine design, because questions remain regarding the trafficking of mRNA formulated in lipid nanoparticles (mRNA-LNPs).1,2,3,4
Naked mRNA is unstable in biological fluids and will rapidly degrade, due to the abundance of RNases. In addition, the large size, negative charge, and hydrophilic nature of mRNA prevents it from crossing cell membranes.5 LNPs are an ideal delivery system because they encapsulate mRNA, protecting it from degradation and allowing for transport from the injection site to target tissues. Once at the intended tissue, LNPs facilitate rapid cellular uptake and endosomal escape, enabling the entry of mRNA into the cytosol, where it can engage with ribosomes and be translated into protein.6
A successful mRNA vaccine must elicit a strong immune response to confer protection, which requires distribution to critical tissues.5 LNPs are sized (<200 nm) such that they can engage with cells at the injection site or drain through the lymphatic system to lymph nodes (LNs).7,8 We believe that there are two pathways by which the mRNA-encoded protein can reach the LNs. First, delivery of mRNA-LNPs may facilitate the rapid recruitment of immune cells to the injection site, triggering an initial innate immune response. Antigen-presenting cells (APCs), including dendritic cells (DCs) and macrophages, internalize mRNA-LNPs at the injection site9; then, the APCs transport the expressed antigen to draining LNs (dLNs), which triggers an adaptive immune response. Second, mRNA-LNPs may drain to the dLNs, engaging with APCs directly. Regardless, transient innate immune activation ultimately results in B and T cell priming in the dLNs, which provides protection against the pathogen.5,6,10,11
Intramuscular (i.m.) administration is the most common vaccine delivery route. Previously, we screened a biodegradable ionizable lipid library to identify a lead to include in mRNA-LNPs optimized for i.m. vaccine delivery. We demonstrated that this LNP formulation improves local protein expression and tolerability without affecting immunogenicity compared to legacy LNP formulations.12 Earlier work demonstrated that mRNA-LNPs delivered i.m. can express protein in tissues critical for immune response generation.11,13 Although our mRNA-LNP vaccine delivery system has been proven to induce robust clinical responses, additional data are required to determine vaccine trafficking after i.m. delivery and the specific cell types engaged.14,15,16,17,18
To better understand the delivery path of our mRNA-LNP vaccine, we conducted a trafficking study in select tissues evaluating levels of mRNA and expressed protein over time in rodents and nonhuman primates (NHPs) following i.m. administration. We also identified specific cell types critical to creating a protective immune response. Although the previous literature has investigated resulting protein expression after administration, no other studies have looked at mRNA-LNP vaccine trafficking over an extended time course.
Results
Formulation of mRNA-LNPs
To study both the delivery system trafficking and visualize mRNA and expressed protein, two reporter mRNA constructs were used. Using a modified firefly luciferase mRNA-LNP, we measured the concentration of mRNA in select NHP tissues over time. Using enhanced green flourescent protein (EGFP) mRNA-LNPs, we visualized mRNA and expressed protein at the injection site and dLNs of rodents and NHPs. EGFP also allowed us to analyze specific immune cells expressing protein within the dLNs in NHPs through flow cytometry. By using two distinct mRNA constructs, we collected more samples using fewer animals. In each NHP, we used multiple limbs: one injected with modified firefly luciferase mRNA-LNPs and another injected with EGFP mRNA-LNPs. Because each injection contained different mRNA constructs, cross-body drainage should not affect results.
The biophysical properties of all mRNA-LNP formulations used are consistent with our vaccine drug product and did not differ based on the mRNA construct encapsulated within the mRNA-LNP. The mRNA-LNP particle size used in these studies is 100 ± 20 nm, with a high level (>95%) of mRNA entrapment.
mRNA and protein expression visualization in rodents
Because rodents are commonly used in vaccine preclinical studies, we conducted a mouse study to visualize both mRNA and expressed protein after i.m. administration of our mRNA-LNP vaccine. Immunohistochemistry (IHC) was used to evaluate protein expression at both the injection site and LNs in mice administered mRNA-LNPs containing 3 μg of EGFP mRNA. At the injection site, the intermuscular adipose and connective tissue were infiltrated by numerous intact and degenerated neutrophils, macrophages, and few lymphocytes.
In mice, both the mRNA and EGFP protein were observed at the injection site in macrophages, fibroblasts, and adipocytes in the vicinity of the inflammatory cell infiltrates in the intermuscular connective and adipose tissue, with the 6- and 24-h time points showing comparable signals upon visual assessment of the in situ hybridization (ISH) and IHC staining (Figure 1A). The distribution of mRNA is observed in some cells that do not express protein, indicating mRNA uptake but no translation. To assess staining in muscle cells, we used both H&E and 3,3′-diaminobenzidine (DAB) chromogenic staining to identify the nucleus in blue and cytoplasm in red for both visual assessment and morphological analysis. The DAB brown stained mRNA (RNAScope) or protein (IHC) in muscle fibers. At both time points, no mRNA or protein expression was observed in muscle fibers.
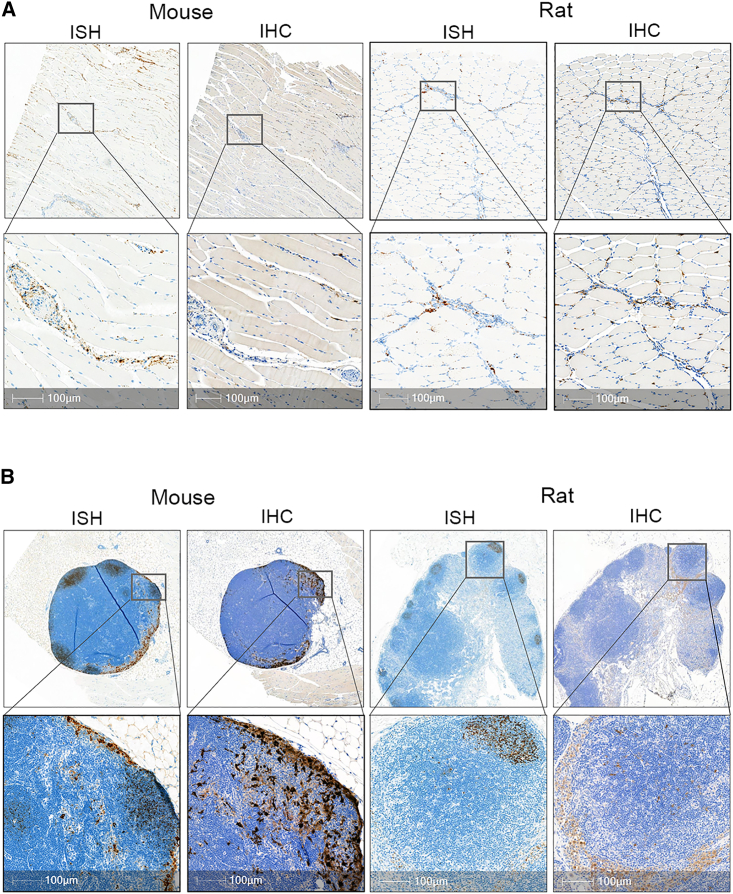
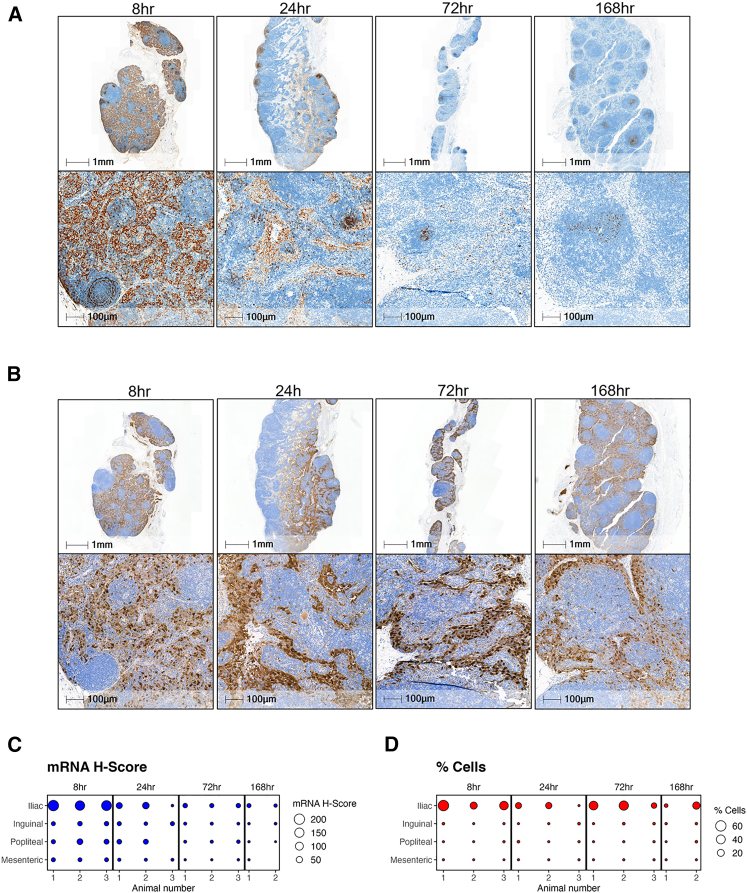
Figure 1.
Activation of immune cells and recruitment of innate immune response to site of administration in rodents
(A) ISH staining of EGFP mRNA (brown) was detected in mouse and rat muscle 24 h post-mRNA-LNP administration using RNAScope assay. IHC staining of muscle with anti-EGFP antibody (brown) showed positive expression in tissue resident and infiltrating immune cells. No protein expression or mRNA observed in the muscle fiber. (B) EGFP positive mRNA and protein in mouse and rat dLN post-i.m. injection. Majority of mRNA signal is cleared from the LN and residual mRNA signal is localized to B cell follicles and interfollicular regions by 24 h EGFP expression was primarily in macrophages located within the subcapsular and medullary sinus of the draining LNs. Scale bar, 100 μm.
Positive EGFP signal by IHC was quantified for both the popliteal and inguinal LNs in mice. The left popliteal LN showed ∼5% positivity compared with minimal positivity (<0.1%) in either the right popliteal and right or left inguinal LN. Expression was observed primarily in macrophages located within the subcapsular sinus (SCS) and medullary sinus of the left-sided dLNs at both time points (Figure 1B), with the highest expression at 6 h. Very few macrophages and reticular cells displayed EGFP protein expression in the medullary region of the LNs. The small sample size (N = 3 per group) and inconsistent LN collection (6/24 LNs missing) may be contributing factors to the observed variability and low expression in the inguinal LNs.
To address the variability of expressed EGFP observed in mouse LNs, we conducted a rat study to further evaluate the lymphatic drainage patterns. The objective of this study was to (1) compare protein expression in three different LN locations, (2) examine two different LN collection techniques, and (3) use the “bread loaf” sectioning technique to analyze EGFP protein expression changes throughout the entire LN tissue. In rats, the iliac, inguinal, and popliteal LNs were collected 24 h post-i.m. injection, and protein expression was confirmed by IHC. Animals were placed into 2 groups (N = 10 per group), and 2 different histology collection methods were conducted (“standard” versus “large collection”). In the large collection group, LNs were collected with intact surrounding outer fat, connective tissue, and blood vessels, potentially achieving better orientation/embedding of the LNs in the paraffin block, allowing for more standardized sections. In the standard collection groups, LNs were collected without accessory fat, tissue, or blood vessels.
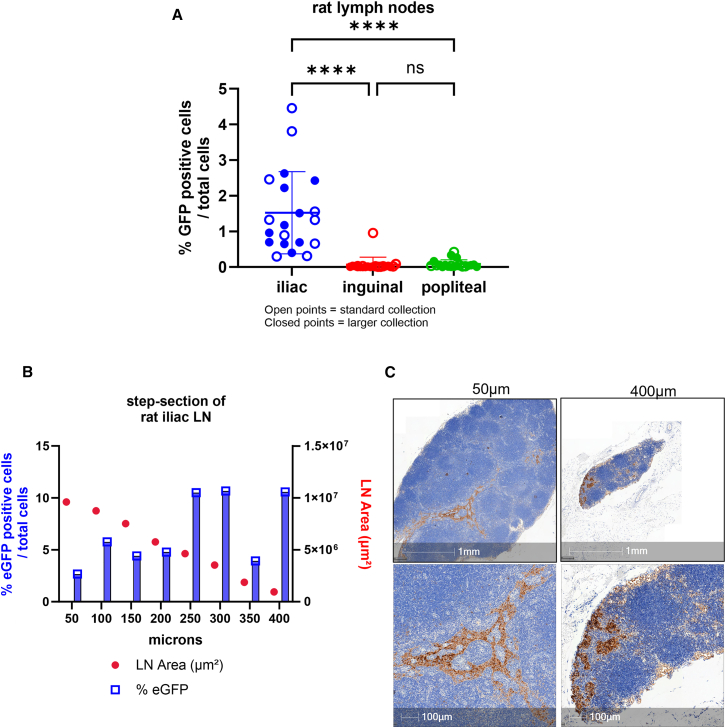
Expression in the iliac LN was significantly higher (1.5%) compared to the inguinal or popliteal LNs (<0.1%) (Figure 2A). Expression in the ipsilateral iliac is comparable to what has been previously reported.19 Overall, there was no difference in expression using the two different collection methods within all LN expression. Next, a bread loaf or step-sectioning technique was performed on a subset of iliac LNs. Each LN was cut ∼50 μM in depth throughout the entire tissue block. At each 50-μM depth, 3 serial sections were obtained and placed on 3 different slides; each slide had 3 sections 50 μM apart per slide (Figure S1). Quantitative analysis of each serial section per LN revealed that protein expression can vary across depth. For example, in one animal, the iliac LN was sectioned from 50 to 400 μm and showed a range of <0.1%–10% positivity (Figure 2B). An LN from another rat showed a range from 0.7% to 28% positivity. Most staining was localized to the medullary sinuses (Figure 2C). In summary, the mRNA-LNP vaccine drained primarily to the iliac LNs, with variable drainage to the inguinal and popliteal LNs. Even within the iliac LNs, there are variable levels of protein expression throughout the entire LN tissue.
Figure 2.
Evaluation of draining LN protein expression throughout entire LN
(A) Values represent mean positive EGFP staining across all rat LNs represented by 3 sections 50 μm apart. Expression in iliac is significant (∗∗∗∗p < 0.0001) compared to inguinal and popliteal LNs by 1-way analysis of variance followed by Tukey’s multiple comparison test. (B) Scatterplot of percentage of EGFP positivity across series of step-sections of a single draining iliac LN in rat. (C) Immunostaining of rat LN. Images represent EGFP expression at 50 μm section and 400 μm depth to demonstrate varied expression observed. Scale bars, 1 mm and 100 μm.
During the follow-up rat study, the injection site was also collected 24 h post-i.m. dosing, and EGFP protein expression was confirmed by IHC. The EGFP positivity seen was similar to what was observed in mice.
Delivery system trafficking in NHP
Upon confirming EGFP protein expression at the injection site and dLNs in rodents, we evaluated vaccine trafficking and protein expression in NHPs. NHP models possess characteristics similar to those of the human immune system and thus represent a favorable model to study vaccine delivery.11,20 After i.m. administration, the mRNA-LNP was tracked by measuring the mRNA level in select tissues through the branched DNA (bDNA) assay. Tissues were collected at 8 (N = 3), 24 (N = 3), 72 (N = 3), and 168 (N = 2) h and serum was collected at 0 (N = 11), 2 (N = 11), 4 (N = 11), 6 (N = 11), 8 (N = 11), 16 (N = 8), 24 (N = 8), 72 (N = 5), 96 (N = 2), and 168 (n = 2) h postdosing to gain an understanding of the time course.
Due to the nature of sparse sampling, to capture the full concentration profile over 168 h, a naive pooled data approach was used in which concentrations were averaged at each time point for both plasma and tissues (Figure 3A). The averaged concentration profiles were further used to evaluate pharmacokinetic (PK) parameters. The naive pooled data approach limited our ability to perform statistical analysis on individual PK parameters. PK parameters were calculated using Phoenix 64 software by Certara (version 8.3.4.295) and summarized (Figure 3B).
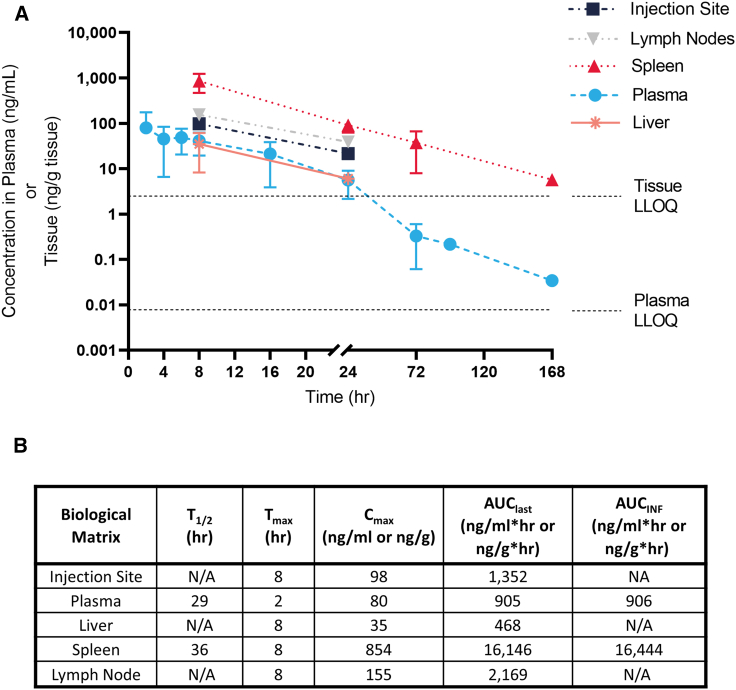
Figure 3.
mRNA concentration found at the injection site, LNs, spleen, liver, and plasma up to 1 week after IM administration of mRNA-LNPs in NHP
(A) mRNA concentration and (B) corresponding PK parameters. Data points below the tissue lower limit of quantitation (LLOQ) are not displayed. Error bars represent the standard deviation. AUCINF, the area under the concentration versus time from the start of dose administration to extrapolated infinite time; AUClast, the area under the concentration versus time curve from the start of dose administration to the last observed quantifiable concentration calculated using the linear trapezoidal method; Cmax, the maximum observed concentration measured after dosing; Tmax, the time after dosing at which the maximum concentration was observed; T1/2, the apparent terminal elimination half-life.
At the first tissue harvest (8 h) or plasma collection (2 h), the maximum mRNA concentration (Cmax) was observed in all of the samples evaluated. Cmax was the highest in the spleen (854 ng/g), followed by LNs (155 ng/g), the injection site (98 ng/g), plasma (80 ng/mL), and liver (35 ng/g). mRNA exposure was calculated as the area under the mRNA concentration curve from the start of dose administration to the last observed quantifiable concentration (AUClast) and from the start of dose administration to infinity (AUCinf). In dLN, AUClast (2,169 ng/g × h) was similar to the exposure at the injection site (1,352 ng/g × h) and was higher than systemic exposure (plasma levels [905 ng/mL × h]), suggesting that LNPs traffic from the injection site to LNs, plasma, and other tissues. The highest levels of mRNA were observed in the spleen, most likely from infiltrating cells (AUClast was 16,146 ng/g × h).
mRNA was detected up to 24 h at the injection site, LNs, and liver. Plasma and spleen mRNA levels were detected over 7 days (168 h) (Figure 3A). mRNA half-life (T1/2) could not be calculated at the injection site, LN, or liver tissues because only 2 time points were above the limit of mRNA detection: the T1/2 was 29 and 36 h in plasma and spleen, respectively. The observed variability in tissues was high and was driven by observed high mRNA concentrations in one animal; however, the same animal showed consistently high concentration in all tissues, which provides confidence that observed trends are consistent, regardless of high variability. Our analyses demonstrated that mRNA degrades rapidly from the injection site as well as from systemic compartments (plasma and tissues). The presented mRNA distribution in selected tissues represents the true trafficking of intact mRNA-LNPs, because any free mRNA will be degraded quickly.
mRNA and protein expression visualization in NHPs
We then looked at the time course of mRNA and expressed protein at the injection site out to 1 week (8, 24, 72, and 168 h) after i.m. administration using RNAscope (Figure 4A) and IHC staining (Figure 4B). Peak mRNA was detected at the first collection time point of 8 h, with the majority of mRNA gone by 72 h (Figure 4C). Analysis of the time course revealed EGFP protein expression out to 1 week postinjection, with peak protein expression at 24 h postinjection, which matched the rodent data (Figure 4D).
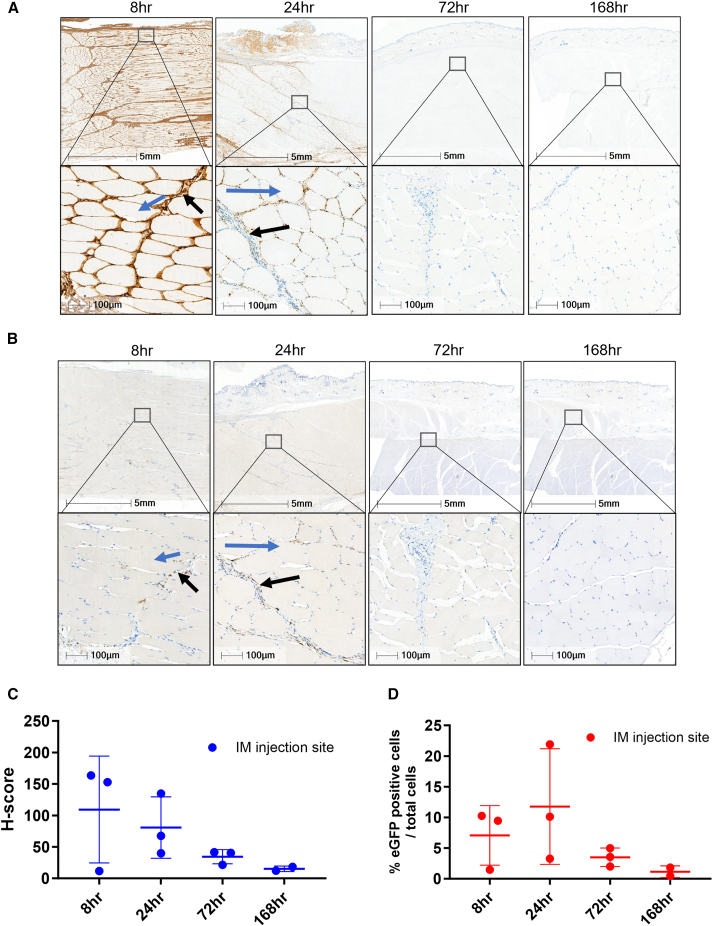
Figure 4.
Localization of EGFP mRNA at the NHP injection site
(A) RNAscope EGFP labeling of NHP muscle at the injection site. Highest levels of mRNA were detected at the 8 h time point and mRNA levels depleted by 72 h. Muscle fibers are absent of EGFP mRNA (red arrows), with the majority of mRNA distributing to the connective tissue with uptake in macrophages from the inflammatory infiltrates, fibroblasts/fibrocytes, and randomly distributed adipocytes. (B) IHC images of the NHP injection site tissue. Cells at the NHP injection sites expressing EGFP protein were primarily macrophages, fibroblasts/fibrocytes, and adipocytes. Protein expression also observed in endothelial cells. No EGFP expression in muscle fibers (blue arrows), with some positivity in connective tissue (black arrows). (C) H-score analysis of mRNA. (D) Plot of percentage of EGFP positivity from 8 to 168 h. Error bars represent the standard deviation. Area shown in insets is indicated in the low-magnification images. Scale bars, 5 mm and 100 μm.
At the i.m. injection site, the intermuscular fascia was expanded by edema and an inflammatory infiltrate was composed of numerous intact and degenerated neutrophils, macrophages, and a few lymphocytes; there were multifocal hemorrhages. The inflammatory infiltrate was extending and invading the perimysium of adjacent muscles. As in the rodent studies, the cells at the NHP injection sites expressing EGFP protein were macrophages from the inflammatory infiltrate, fibroblasts/fibrocytes, and randomly distributed adipocytes. In addition, EGFP protein expression was observed in endothelial cells lining the lumen of small, usually dilated capillaries, considered to be of lymphatic origin. Visual assessment of EGFP immunostaining by a pathologist confirmed that there was no expression in the muscle fibers.
Overall, at the injection site, macrophage, fibrocytes/fibroblast, and adipocyte cells displaying EGFP protein expression were similar between the mouse and NHPs. However, for the tissue samples examined for the mouse, no protein expression was observed in endothelial cells.
We also conducted RNAscope (Figure 5A) and IHC (Figure 5B) staining on the LNs using the same time points as the injection site (8, 24, 72, and 168 h). The EGFP mRNA was present out to 168 h in the iliac, popliteal, and inguinal LNs. A bubble plot of mRNA H-score reveals that the highest mRNA levels were found in iliac LN (Figure 5C). The iliac LNs showed the highest mRNA levels and were primarily detected throughout the medullary and SCS at the first collection time point (8 h). At later time points, the mRNA was detected in lower quantities and localized to the B cell follicular regions. EGFP protein expression was seen out to day 7 in the iliac LN. Positive EGFP expression was observed in histiocytes, reticular cells, and DCs in the medulla region. In addition, EGFP expression was also observed in the medullar sinus and SCSs in histiocytes, reticular cells, and endothelial cells lining the sinuses. Few histiocytes and reticular cells were displaying EGFP expression in the marginal zone of the follicles.
Figure 5.
Time course of mRNA and protein distribution in NHP dLN
(A) RNAscope EGFP labeling of NHP iliac LN out to 7 days post-mRNA-LNP i.m. injection. (B) EGFP staining of the iliac LN at 8, 24, 72, and 169 h post-i.m. injection. (C and D) Bubble plot of mRNA H-score (C) and percentage of EGFP positivity (D) comparing dLN versus non-dLN (mesenteric). Area shown in insets is indicated in the low-magnification images. Scale bars, 1 mm and 100 μm.
In the dLNs, EGFP protein expression was similar in mouse and NHP, in the macrophages of the subcapsular region in medullary sinuses and in the medullary region within macrophages and reticular cells. However, there was no evidence of protein expression in the endothelial cells in the LNs from the mouse.
The iliac LN showed the highest percentage positivity, with 10 of 11 total samples across all time points collected showing EGFP expression, with a mean positivity of 40%. At the 24-h time point, the iliac LN mean decreased to 10%; the 72- and 96-h time points were 32% and 19%, respectively. There was little to no expression in the inguinal and popliteal LNs at all time points (Figure 5D). Although there was a small amount of mRNA in the mesenteric follicle zones, as expected of a non-dLN, the mesenteric LN showed no EGFP protein expression. The low levels of mRNA in the mesenteric LNs could be indicative of mRNA clearance. For all of the RNAScope assays, positive and negative control probes were used as technical controls to assess sample and RNA quality. Similarly, for all of the IHC assays performed, a positive control sample and an immunoglobulin G isotype control was stained to validated antibody-specific and nonspecific binding (Figures S2A, S2B, S3A, and S3B).
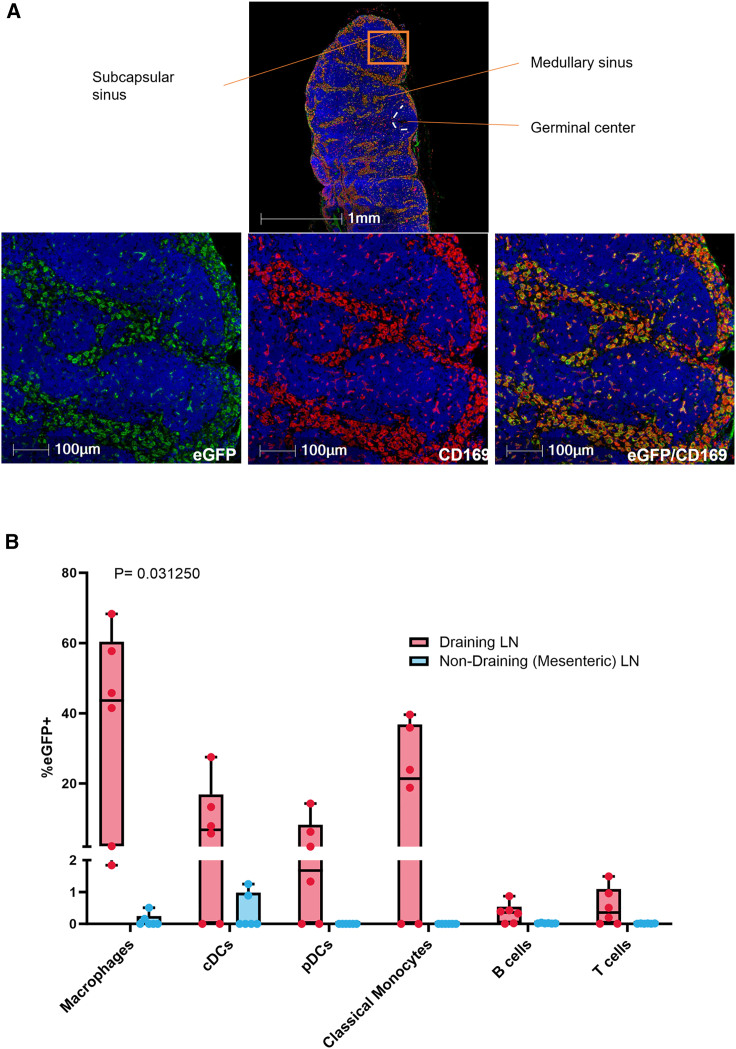
Colocalization of expressed protein and immune cells in dLNs
A separate NHP study was conducted to determine specific immune cells expressing protein within the dLNs 24 h post-i.m. administration. The classification of macrophages is based on macrophage polarization, and macrophages have traditionally been divided into two subsets. M1 macrophages are classically activated and M2 are the alternatively activated macrophages. Other subpopulations of macrophages have been categorized, including CD169+ macrophages found in lymphoid organs. CD169, a member of the Siglec family, is a cell surface molecule primarily expressed on a specific subset of macrophages and can also be induced on some monocytes and DCs. CD169+ macrophages are localized in the SCS and medullar sinus macrophages (MSMs) and are involved in immune system regulation.21 Cell-specific staining revealed the CD169 macrophage marker colocalizing with EGFP protein expression within both SCS and MSMs of the draining LNs (Figure 6A). Flow cytometry further confirmed high levels of EGFP positivity in macrophages with an average positivity of 36.2%. In addition, EGFP positivity was also seen in monocytes (19.7%) and conventional DC (cDC) and plasmacytoid DC (pDC) populations, 9.1 and 4.0% respectively. Low levels of EGFP positivity (<1%) were seen in B and T cells (Figure 6B). Although EGFP positivity increased in the dLN compared to the mesenteric (non-DLN) LN across all cell types analyzed, statistical analyses revealed a significant increase in positivity in macrophages (p = 0.031250) based on a Wilcoxon matched-pairs signed rank test.
Figure 6.
Co-localization of expressed protein and macrophages in dLN-specific cells in NHPs
(A) Fluorescent microscopic image of NHP popliteal LN 24 h post-i.m. injection of mRNA-LNP. LN was stained with anti-CD169 (red) and anti-GFP (green) antibodies. The majority of EGFP expression is detected in the SCS and medullary sinus. (B) Graph depicting flow cytometry analysis of EGFP-expressing immune cells in the dLNs and non-dLNs postinjection. Non-dLN tissue (mesenteric) and dLN tissue (axillary [left and right], popliteal [left and right], and iliac LNs [left and right]) were collected at 24 h (N = 6) postdosing. Box plot whiskers are set to contain the min and max data points. LN tissue was processed to cells, which were then stained with fluorescently labeled antibodies to delineate macrophages, cDCs, pDCs, classical monocytes, B cells, and T cells. Graph shows the level of EGFP positivity in the dLN compared to the non-dLN across all of the cell types analyzed. Area shown in insets is indicated in the low-magnification images. Scale bars, 1 mm and 100 μm.
Discussion
The efficiency of an mRNA vaccine delivery system and subsequent protein expression in tissues is critical to generating robust innate and adaptive immune responses, both of which contribute to efficacy. mRNA-LNPs possess favorable physiochemical properties to facilitate vaccine delivery.6 Previous work demonstrated that our ionizable lipid designed for i.m. vaccines improves immunogenicity and local expression compared to legacy mRNA-LNPs.12 Further tailoring of this delivery system requires a greater understanding of mRNA-LNP trafficking and protein expression following vaccine administration.
We sought to identify the path of our mRNA-LNPs following i.m. administration in tissues that we believe are critical to generating a protective immune response. After i.m. administration and drainage through the lymphatic system, mRNA-LNPs must pass through the architecture of the dLN, including the medullary sinus, SCS, and germinal center.22,23 It was previously reported that vaccine antigen drainage upon i.m. injection to the quadriceps will drain primarily to iliac LNs and subsequently to the inguinal LNs, similar to our findings.19 There are two potential ways for an antigen to reach the LN: through active transport by APC and through passive transport through the lymphatic system to dLNs. Therefore, expressed protein can be detected from the APC traveling from the injection site or from mRNA-LNP transfection of cells within the LN.24
Vaccine trafficking studies were conducted in both rodents and NHPs. Histological analyses were performed on samples taken from the site of administration and dLNs, because previous studies have shown that mRNA distributes to these tissues.13,24 Two different mRNA reporters (a modified firefly luciferase and EGFP) were used to quantify and visualize mRNA and expressed protein in select tissues, as well as analyze specific cell types that expressed protein within the dLNs.
At the injection site in rodents, we found that protein is readily detectable in the infiltrating cells, connective tissue, and adipose tissue. We did not detect expression in the muscle fibers. The LN contains three main areas: the follicular B cell zone; the paracortex, which is rich in T cells; and the medulla, which includes plasma and macrophage cells. The plane of section is critical for the histopathology evaluation of LNs.25 In the Results section, we describe a bread loaf technique to evaluate protein expression in a step-order fashion throughout the LNs and found that protein expression can vary based on depth of dLN. The subcapsular and medullary sinus of the dLNs also showed both mRNA and expressed protein. We hypothesize that the staining pattern between RNA and expressed protein differs due to the difference in endosomal escape and therefore protein expression between cell types.
Additional studies were conducted in NHPs to expand upon the rodent findings and analyze immune cell populations. For these studies, the dose level was increased 100-fold, from 3 to 300 μg to account for the difference in body weight between rodents and NHPs. The NHP study was designed to capture a detailed time course up to 1 week after i.m. administration. Like the rodent studies, we detected mRNA and expressed protein at both the injection site and dLNs. At the injection site, peak levels of mRNA were found 8 h postadministration (the first tissue collection), and peak protein expression occurred at 24 h. After the 24 h time point, mRNA was not detectable at the injection site, LNs, or liver and was significantly reduced in plasma and spleen (as measured by the bDNA assay). Histology results demonstrated comparable results, with the level of mRNA detected being significantly reduced after the 24 h time point. Together, the bDNA and histology results indicate rapid degradation of the delivery system. At the iliac LN, mRNA was detected throughout the medullary and SCS at 8 h, with protein expression seen out to the last tissue collection (168 h). Expressed protein was detected longer than mRNA, because its time course is dependent on the protein half-life, which is much longer than that of mRNA.26 Peak expression occurred earlier at the injection site than draining through lymphatics, indicating the time course of LNP trafficking from injection site to dLN. As expected, we did not detect expressed protein in the non-dLN (mesenteric). More important, EGFP protein positivity showed the same staining pattern in the medullary sinus and SCS of LNs in both rodents and NHPs. These results demonstrate cross-species confirmation of immune response following i.m. administration.
Further analysis using high-throughput, multiparameter flow cytometry interrogated which immune cells were positive for expressed protein within the dLNs at 24 h after administration. Expressed protein is primarily found within APCs, specifically macrophages. Macrophages are critical in generating a strong immune response through interactions with T and B cells, leading to protection against a pathogen.5,6,10,11 These data support that antigen capture and processing by APCs, specifically macrophages, helps trigger an innate immune response, whereas antigen presentation and innate immune signaling lead to adaptive immune responses following i.m. administration of our mRNA-LNP vaccine delivery system.
Our study design, which includes the use of two different mRNA reporters, provides a favorable framework for time course analysis of mRNA-LNPs, allowing for the ability to monitor the delivery system through mRNA, while also detecting protein expression in tissues important for creating an immune response at the injection site and dLNs. However, there are important limitations to note. We performed a limited distribution study in which mRNA was measured only in select tissues, and other components of the delivery system were not measured. Also, our first tissue collection occurred at 8 h, limiting our knowledge of the early path and time course of our vaccine mRNA-LNP delivery system components. Additional data from a detailed biodistribution study are being analyzed and will shed further light on mRNA, lipid, and protein expression across several tissues following i.m. administration of our vaccine mRNA-LNP. We also did not investigate the mechanism of mRNA-LNP clearance; however, we expect that degradation of the ionizable lipid occurs through cleavage of the ester bond by esterases naturally found throughout the body and that degradation of mRNA occurs through endogenous RNases once the mRNA is no longer protected by the LNP.27
Although our vaccines are usually given in a two-dose series (a priming dose followed by a booster) several weeks apart, here, we conducted a single-dose analysis.14,28 Our results are therefore limited in their ability to capture subsequent changes in distribution and time course following a booster dose. In addition, the dose level chosen for NHP was also increased from a typical dose used in humans (50–100 μg) to allow for better detection of mRNA-LNP trafficking.
The results of this study provide essential information on the path that our mRNA-LNP vaccine takes following i.m. administration and the time course of detection in select tissues critical to generating an immune response. After i.m. delivery of mRNA-LNPs, we demonstrated that mRNA distribution and protein expression are seen primarily in infiltrating immune cells in both NHPs and rodents at the injection site, followed by expression in the dLNs. Within the dLN, macrophages exhibit the greatest level of protein expression. These data will help us optimize our mRNA-LNP system for future vaccines and provide important guidance on developing mRNA vaccines.
Materials and methods
mRNA and mRNA-LNP production
All mRNA constructs were manufactured in vitro by T7 RNA polymerase-mediated transcription, with complete replacement of uridine by N1-methyl-pseudouridine, as previously described.29 Briefly, the DNA template used in the in vitro reaction contained the immunogen open reading frame flanked by 5′ UTR and 3′ UTR sequences and was terminated by a polyA tail. After transcription, the pre-mRNA was purified by oligo-dT affinity, and the cap 1 structure was added to the 5′ end using Vaccinia capping enzyme (New England Biolabs, Ipswich, MA) and Vaccinia 2′O-methyltransferase (New England Biolabs, Ipswich, MA). The capped mRNA was then purified by reverse-phase purification; buffer exchanged by tangential flow filtration into sodium citrate, pH 6.5, sterile filtered, and kept frozen at −80°C until further use.
mRNA-LNPs were manufactured via nanoprecipitation by mixing the ionizable lipid (heptadecan-9-yl-8-((2-hydroxyethyl) (6-oxo-6-(undecyloxy)hexyl)amino)octanoate), distearoylphosphatidylcholine, cholesterol, and PEG2k-DMG lipids dissolved in ethanol with mRNA diluted in sodium acetate buffer (pH 5.0).12 mRNA-LNPs were then buffer exchanged into a physiologically relevant buffer system and sterile filtered before storage. All of the formulations were tested for particle size, mRNA encapsulation, and endotoxin levels and were deemed acceptable for in vivo study.
Rodent in vivo studies
Study procedures involving the care and use of rodents were approved by the Institutional Animal Care and Use Committee (IACUC) at Charles River Laboratories (CRL) Discovery Services (Shrewsbury, MA). Female BALB/c mice 8–9 weeks old (n = 3 per group; CRL) were dosed by i.m. injection in the left quadricep with a fixed volume of 50 μL of mRNA-LNPs containing 3 μg EGFP mRNA. Following perfusion, left and right popliteal and inguinal LNs along with injection site (quadriceps muscle) were collected at 6 and 24 h postdosing.
An additional study was conducted at Moderna using male Sprague-Dawley rats 8–9 weeks old obtained from CRL. The animal study was approved by the IACUC at Moderna. Animals were housed in microisolator cages in a BSL-2 facility and provided water and food ad libitum. Rats (n = 10 per group) were given two 50-μL i.m. injections of mRNA-LNPs containing 3 μg EGFP into the left and right quadriceps, respectively. Left and right popliteal, inguinal, and iliac LN tissues were collected at 24 h postdosing.
NHP in vivo studies
Study procedures involving the care and use of animals were approved by the IACUC of CRL Preclinical Services (Sherbrooke, QC, Canada). NHP studies were conducted using non-naive cynomolgus male monkeys 2–5 years old weighing 2–5 kg. Animals were housed in stainless-steel, perforated-floor cages in a temperature- and humidity-controlled environment (20°C–26°C and 30%–70%, respectively) with an automatic 12-h/12-h dark/light cycle. Animals were fed PMI Nutrition (St. Louis, MO) Certified Primate Chow No. 5048 twice daily.
A single-dose study was conducted in cynomolgus monkeys to evaluate mRNA-LNP trafficking following i.m.injection into the lateral compartment of the quadriceps. In the right quadriceps, modified firefly luciferase mRNA-LNPs were injected and to monitor the trafficking of mRNA. In the left quadriceps, EGFP mRNA-LNPs were injected and used for histology endpoints. Each injection site received a 0.5-mL injection of mRNA-LNPs containing a fixed dose of 300 μg mRNA. Plasma samples were collected at 0, 2, 4, 6, 8, 16, 24, 72, 96, and 168 h postdosing. LNs (iliac, inguinal, popliteal, and mesenteric), spleen, liver, and injection site muscle were collected at 8, 24, 72, and 168 h postdosing (N = 3, except N = 2 for the 168-h time point).
An additional NHP study was conducted to determine which immune cells in the dLNs were expressing the protein of interest. Cynomolgus monkeys were concurrently vaccinated in both the left and right deltoids and left and right vastus lateralis muscles. The same mRNA-LNP formulation was injected at one of the four sites in each of the six animals to account for possible biodistribution differences. Each injection site received a 0.15-mL injection of mRNA-LNPs containing a fixed dose of 300 μg EGFP mRNA. LN tissues (mesenteric, axillary [left and right], popliteal [left and right], and iliac LNs [left and right]) were collected at 24 h (N = 6) postdosing.
mRNA quantification by bDNA assay
The plate-based hybridization assay measures mRNA levels of in vivo samples. This assay is based on a QuantiGene Singleplex kit (Thermo Fisher Scientific, Waltham, MA), which uses bDNA and a customized probe set containing capture, label, and blocking probes to identify the target mRNA. Briefly, calibrators, quality controls, and samples are diluted 1:40 in lysis buffer with 0.5 mg/mL Proteinase K and heated to 65°C for 30 min. After heating, calibrators, quality controls, and samples are cooled to room temperature. A capture probe mixture is then created using lysis buffer, blocking reagents, and a capture probe set. A total of 20 μL of the capture probe mixture is added to a precoated plate, and then 80 μL of room temperature calibrators, quality controls, and samples are added for a final volume of 100 μL. The plate is then sealed and incubated at 55°C for 18–20 h. The following day, the plate is washed 3 times and 100 μL of preamplification mixture (37°C) is added. The plate is sealed and then incubated at 55°C for 60 min.
After incubation, the plate is washed 3 times with wash buffer, and 100 μL of amplification mixture at 37°C is added. The plate is sealed and then incubated at 55°C for an additional 60 min. After incubation, the plate is washed 3 times and 100 μL of the label probe working reagent is added. The plate is sealed and then incubated at 50°C for 60 min. After incubation, the plate is washed 3 times, and 100 μL of luminescent substrate is added. After adding the substrate, the plate is analyzed using a luminometer, and a luminescent signal is generated in proportion to the amount of mRNA present in the sample.
IHC
IHC, conducted to visual EGFP protein expression in injection site and LN tissues, was performed on formalin-fixed paraffin embedded sections using the Leica Bond RX autostainer (Leica Microsystems, Buffalo Grove, IL). Sections were baked and deparaffinized on the instrument, followed by an epitope retrieval for 10 min using Leica Epitope Retrieval Buffer 1 (catalog no. AR9961). Dako serum-free protein block (catalog no. X090930-2, Agilent Dako, Santa Clara, CA) was incubated on the slides for 15 min at room temperature. Anti-EGFP antibody (catalog no. ab6673, or ab290 Abcam, Cambridge, UK) was used at 2.4 or 0.3 μg/mL dilution at room temperature for 30 min. Secondary antibody and detection were performed using the Goat-on-Rodent HRP-Polymer (catalog no. GHP516, BioCare Medical, Pacheco, CA) and/or the Bond Polymer Refine Detection Kit (catalog no. DS9800, Leica Microsystems). Bond DAB Enhancer (catalog no. AR9432, Leica Microsystems) and bluing reagent (catalog no. 3802918, Leica Microsystems) were used to enhance the color.
RNAScope (ISH assay)
ISH was performed using RNAscope 2.5 LS Reagent Kit-BROWN (catalog no. 322100, Advanced Cell Diagnostics (ACD), Hayward, CA) for use with Leica Biosystems’ BOND RX System, according to the manufacturer’s instructions. An exclusive target probe with proprietary sequences was designed by ACD to target EGFP mRNA. Control probes to the housekeeping gene Macaca fascicularis peptidylprolyl isomerase B (cyclophilin B) (catalog no. 424148, ACD) were used as a positive control; the bacterial gene DapB (catalog no. 312038, ACD) was used a negative control. Briefly, slides were baked offline for 30 min at 60°C before use. Slides were then placed on a Leica Bond RX autostainer (Leica Microsystems), baked, and dewaxed. Next, slides were processed using a Leica staining protocol according to the ACD user manual (document no. 322100-USM).
Image acquisition and image analysis
Whole-tissue slides were scanned at 20× magnification with the Pannoramic 250 Flash III (3DHISTECH, Budapest, Hungary) digital slide scanner. Digital images were analyzed for GFP quantification using Halo (Indica Labs, Albuquerque, NM) software. First, manual annotations were created to identify individual tissue regions per slides and next to detect positive eGFP cells. For RNAScope staining, the Halo ISH module was performed and the output parameter for H-score was generated. The H-score is the score to indicate the amount of expression for probe. The H-score is calculated with the following equation: H-Score = (1 × % Probe 1 + Cells) + (2 × % Probe 2 + Cells) + (3 × % Probe 3 + Cells) + (4 × % Probe 4 + Cells). For IHC staining the HALO Multiplex IHC module was optimized to detect nuclear stain and anti-EGFP immunolabeling. The results are expressed as the percentage of EGFP cell positivity = EGFP positive cells/total number of cells and was analyzed using GraphPad Prism 10, with ordinary one-way ANOVA test followed by Tukey’s multiple comparisons test.
Flow cytometry
Frozen LN samples were thawed at 37°C and washed twice in warm RPMI 1640 + 10% fetal bovine serum. Samples were then stained with Live/Dead (L/D) FVD780 in PBS for 15 min at 4°C. All of the other staining steps were conducted in fluorescence-activated cell sorting buffer containing PBS (Gibco, Waltham, MA), 0.1% BSA (Sigma-Aldrich, St. Louis, MO), and 2 mM EDTA (Invitrogen, Carlsbad, CA). Cells were washed twice, resuspended in Fc block (Human TruStain FcX, BioLegend, San Diego, CA), and incubated for 15 min at 4°C. After blocking, cells were stained with the surface antibodies listed in Table S1. Samples were analyzed on an Attune NxT multicolor flow cytometer (Thermo Fisher Scientific). Data were analyzed using FlowJo version X (Tree Star, Ashland, OR).
A Wilcoxon matched-pairs signed rank test was performed to determine significant differences between percentage of EGFP positivity of our mRNA-LNP in the dLN compared with the mesenteric LN across cell types, including macrophages, cDCs, pDCs, classical monocytes, and B and T cells. Gates were set using mesenteric LNs (% EGFP+ cells) samples and fluorescence minus one. Table S2 lists the primary markers for all of the NHP cell populations characterized.
Data and code availability
All of the data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We acknowledge Luis Brito, Kristen Hopson, and Maija Garnaas for input on study design; Jaclyn Higgins, Ben Geilich, and Sean Severt for providing mRNA-LNP formulations; Elena Shevtsova, Edward Acosta, and the Moderna In Vivo Pharmacology team for coordinating and conducting rodent studies; John Wickwire and Andy Lynn for planning and coordinating NHP studies; Erica Regan and Brandon Jones for mRNA quantification; Molly Bell for staining of tissues; Chinmayi Bankar for generating plots; Doug Burdette for input on PK analysis; and Loren DeVito for writing support. This work was funded entirely by Moderna, Inc.
Author contributions
Conceptualization: K.J.H., I.L.R., K.B., and K.E.B. Methodology: K.E.B. Investigation: R.W. and K.C. Formal analysis: I.L.R., K.C., and E.J. Writing – original draft: K.J.H., I.L.R., K.B., and K.E.B. Writing – review & editing: K.J.H., I.L.R., K.B., R.W., K.C., E.J., and K.E.B. Visualization: R.W. and K.E.B. Supervision: K.J.H., I.L.R., K.E.B.
Declaration of interests
All of the authors are current employees of Moderna, Inc., and own shares in the company.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.102083.
Supplemental information
References
- 1.Kirschman J.L., Bhosle S., Vanover D., Blanchard E.L., Loomis K.H., Zurla C., Murray K., Lam B.C., Santangelo P.J. Characterizing exogenous mRNA delivery, trafficking, cytoplasmic release, and RNA-protein correlations at the level of single cells. Nucleic Acids Res. 2017;45:e113. doi: 10.1093/nar/gkx290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupuis M., Denis-Mize K., Woo C., Goldbeck C., Selby M.J., Chen M., Otten G.R., Ulmer J.B., Donnelly J.J., Ott G., McDonald D.M. Distribution of DNA vaccines determines their immunogenicity after intramuscular injection in mice. J. Immunol. 2000;165:2850–2858. doi: 10.4049/jimmunol.165.5.2850. [DOI] [PubMed] [Google Scholar]
- 3.Liang F., Loré K. Local innate immune responses in the vaccine adjuvant-injected muscle. Clin. Transl. Immunology. 2016;5:e74. doi: 10.1038/cti.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards D.K., Carfi A. Messenger ribonucleic acid vaccines against infectious diseases: current concepts and future prospects. Curr. Opin. Immunol. 2022;77:102214. doi: 10.1016/j.coi.2022.102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iavarone C., O’hagan D.T., Yu D., Delahaye N.F., Ulmer J.B. Mechanism of action of mRNA-based vaccines. Expert Rev. Vaccines. 2017;16:871–881. doi: 10.1080/14760584.2017.1355245. [DOI] [PubMed] [Google Scholar]
- 6.Cagigi A., Loré K. Immune responses induced by mRNA vaccination in mice, monkeys and humans. Vaccines. 2021;9:61. doi: 10.3390/vaccines9010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassett K.J., Higgins J., Woods A., Levy B., Xia Y., Hsiao C.J., Acosta E., Almarsson Ö., Moore M.J., Brito L.A. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Control. Release. 2021;335:237–246. doi: 10.1016/j.jconrel.2021.05.021. [DOI] [PubMed] [Google Scholar]
- 8.Howard G.P., Verma G., Ke X., Thayer W.M., Hamerly T., Baxter V.K., Lee J.E., Dinglasan R.R., Mao H.Q. Critical size limit of biodegradable nanoparticles for enhanced lymph node trafficking and paracortex penetration. Nano Res. 2019;12:837–844. doi: 10.1007/s12274-019-2301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindsay K.E., Bhosle S.M., Zurla C., Beyersdorf J., Rogers K.A., Vanover D., Xiao P., Araínga M., Shirreff L.M., Pitard B., et al. Visualization of early events in mRNA vaccine delivery in non-human primates via PET-CT and near-infrared imaging. Nat. Biomed. Eng. 2019;3:371–380. doi: 10.1038/s41551-019-0378-3. [DOI] [PubMed] [Google Scholar]
- 10.Ols S., Yang L., Thompson E.A., Pushparaj P., Tran K., Liang F., Lin A., Eriksson B., Karlsson Hedestam G.B., Wyatt R.T., Loré K. Route of vaccine administration alters antigen trafficking but not innate or adaptive immunity. Cell Rep. 2020;30:3964–3971.e7. doi: 10.1016/j.celrep.2020.02.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Röhss J., John S., Hassett K., Yuzhakov O., Bahl K., et al. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in Rhesus Macaques. Mol. Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J., et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.August A., Shaw C.A., Lee H., Knightly C., Kalidindia S., Chu L., Essink B.J., Seger W., Zaks T., Smolenov I., Panther L. Safety and immunogenicity of an mRNA-based human metapneumovirus and parainfluenza virus type 3 combined vaccine in healthy adults. Open Forum Infect. Dis. 2022;9:ofac206. doi: 10.1093/ofid/ofac206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu L., Vrbicky K., Montefiori D., Huang W., Nestorova B., Chang Y., Carfi A., Edwards D.K., Oestreicher J., Legault H., et al. Immune response to SARS-CoV-2 after a booster of mRNA-1273: an open-label phase 2 trial. Nat. Med. 2022;28:1042–1049. doi: 10.1038/s41591-022-01739-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panther L., Fierro C., Brune D., Leggett R., Peterson J., Pickrell P., Lin J., Wu K., Lee H., Hasselbeck R., et al. Interim results from a Phase 2, randomized, observer-blind, placebo-controlled, dose-finding trial of an mRNA-based cytomegalovirus vaccine in healthy adults. Open Forum Infect. Dis. 2022;9:ofac492. [Google Scholar]
- 18.Chalkias S., Eder F., Essink B., Khetan S., Nestorova B., Feng J., Chen X., Chang Y., Zhou H., Montefiori D., et al. Safety, immunogenicity and antibody persistence of a bivalent Beta-containing booster vaccine against COVID-19: a phase 2/3 trial. Nat. Med. 2022;28:2388–2397. doi: 10.1038/s41591-022-02031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barber-Axthelm I.M., Kelly H.G., Esterbauer R., Wragg K.M., Gibbon A.M., Lee W.S., Wheatley A.K., Kent S.J., Tan H.X., Juno J.A. Coformulation with tattoo ink for immunological assessment of vaccine immunogenicity in the draining lymph node. J. Immunol. 2021;207:735–744. doi: 10.4049/jimmunol.2001299. [DOI] [PubMed] [Google Scholar]
- 20.Rivera-Hernandez T., Carnathan D.G., Moyle P.M., Toth I., West N.P., Young P.R., Silvestri G., Walker M.J. The contribution of non-human primate models to the development of human vaccines. Discov. Med. 2014;18:313–322. [PMC free article] [PubMed] [Google Scholar]
- 21.Chávez-Galán L., Olleros M.L., Vesin D., Garcia I. Much more than M1 and M2 macrophages, there are also CD169(+) and TCR(+) macrophages. Front. Immunol. 2015;6:263. doi: 10.3389/fimmu.2015.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schudel A., Francis D.M., Thomas S.N. Material design for lymph node drug delivery. Nat. Rev. Mater. 2019;4:415–428. doi: 10.1038/s41578-019-0110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havenar-Daughton C., Carnathan D.G., Boopathy A.V., Upadhyay A.A., Murrell B., Reiss S.M., Enemuo C.A., Gebru E.H., Choe Y., Dhadvai P., et al. Rapid germinal center and antibody responses in non-human primates after a single nanoparticle vaccine immunization. Cell Rep. 2019;29:1756–1766.e8. doi: 10.1016/j.celrep.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutz J., Lazzaro S., Habbeddine M., Schmidt K.E., Baumhof P., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Heidenreich R., Fotin-Mleczek M. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmore S.A. Enhanced histopathology of the lymph nodes. Toxicol. Pathol. 2006;34:634–647. doi: 10.1080/01926230600939997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corish P., Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- 27.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Sahly H.M., Baden L.R., Essink B., Doblecki-Lewis S., Martin J.M., Anderson E.J., Campbell T.B., Clark J., Jackson L.A., Fichtenbaum C.J., et al. Efficacy of the mRNA-1273 SARS-CoV-2 vaccine at completion of blinded phase. N. Engl. J. Med. 2021;385:1774–1785. doi: 10.1056/NEJMoa2113017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson J., Sorensen E.W., Mintri S., Rabideau A.E., Zheng W., Besin G., Khatwani N., Su S.V., Miracco E.J., Issa W.J., et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6:eaaz6893. doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available from the corresponding author upon reasonable request.