Abstract
Objective
Pediatric acute respiratory distress syndrome (PARDS) is a source of substantial morbidity and mortality in the pediatric ICU (PICU), and different plasma biomarkers have identified different PARDS and ARDS subgroups. We have a poor understanding of how these biomarkers change over time and with changing lung injury. We sought to determine how biomarker levels change over PARDS course, whether they are correlated, and whether they are different in critically ill non-PARDS patients.
Design
Two center prospective observational study.
Setting
Two quaternary care academic children’s hospitals
Patients
Subjects under 18 years of age admitted to the PICU who were intubated and met PALICC-2 PARDS diagnostic criteria and non-intubated critically ill subjects without apparent lung disease.
Interventions
None
Measurements
Plasma samples were obtained on study days 1, 3, 7, and 14. The levels of 16 biomarkers were measured using a fluorometric bead-based assay.
Main Results
Compared to non-PARDS subjects, on Day 1 PARDS subjects had increased concentrations of TNFα, IL8, IFNγ, IL17, GrB, sICAM1, SPD, and IL18 but reduced MMP-9 concentrations (all p<0.05). Day 1 biomarker concentrations and PARDS severity were not correlated. Over PARDS course, changes in 11 of the 16 biomarkers positively correlated with changing lung injury with sICAM1 (R=0.69, p=2.2×10−16) having the strongest correlation. By Spearman Rank correlation of biomarker concentrations in PARDS subjects, we identified two patterns. One had elevations of PAI1, MMP-9, and MPO and the other had higher inflammatory cytokines.
Conclusions
sICAM1 had the strongest positive correlation with worsening lung injury across all study time points suggesting that it is perhaps the most biologically relevant of the 16 analytes. There was no correlation between biomarker concentration on day 1 and day 1 PARDS severity; however, changes in most biomarkers over time positively correlated with changing lung injury. Finally, in day 1 samples, 7 of the 16 biomarkers were not significantly different between PARDS and critically ill non-PARDS subjects. These data highlight the difficulty of using plasma biomarkers to identify organ specific pathology in critically ill patients.
Keywords: pediatrics, critical care, PARDS, plasma biomarkers, immunology
Introduction
Pediatric acute respiratory distress syndrome (PARDS) is a biologically and clinically heterogeneous condition that is a major cause of morbidity and mortality in the pediatric ICU (PICU) (1, 2). PARDS has traditionally been classified as arising from a direct lung injury such as pneumonia or smoke inhalation or from an indirect lung injury such as sepsis or trauma. The most recent PALICC definition stratifies PARDS severity as mild/moderate or severe based on degree of oxygenation impairment (2). Many studies have used plasma biomarkers for classification and prognostication (3–10). In adult ARDS hyper- and hypo-inflammatory subclasses were found using latent class analysis and were associated with differential responses to fluid administration and high PEEP (5, 6). In the pediatric population, several biomarkers including Receptor for Advanced Glycation End Products (RAGE), Tumor Necrosis Factor-α (TNFα), matrix metalloproteinases (MMPs), and Surfactant Protein D (SPD) were elevated in PARDS.(7, 8) In addition, PARDS subjects with elevated levels of Angiopoietin 2 (Ang2) and inflammatory cytokines interleukin-1, (IL1), IL6, IL8, IL10, and IL18 have worse outcomes (7, 9–11). Most recently, a latent class analysis PARDS subjects found both a hyper- and hypo-inflammatory subclasses similar to the adult data mentioned above. These subclasses differed in ventilator free days and overall mortality (12). Despite data indicating differential outcomes and responses to therapy (5, 6), biomarker-directed management of ARDS and PARDS is not yet the standard of care. We chose our 16 biomarkers based on this previous body of research and their potential to reflect underlying pathophysiologic processes known to occur in PARDS, including endothelial and alveolar epithelial injury, activation of inflammation, dysregulation of hemostasis, and extracellular matrix degradation.
Understanding biomarker change over time is important because in some cases (e.g., viral pneumonia) the cause of PARDS was present days before meeting PARDS criteria while in others (e.g. trauma) onset is abrupt. Likewise, with PARDS resolution, identifying biomarkers that change in tandem with improving lung status could help identify those with the greatest biological relevance. Few studies have compared plasma biomarker values of PARDS subjects to critically ill children without PARDS. Biomarkers that lack specificity in this regard would likely be less reliable in patients with multiple concomitant acute conditions. Here, we describe both how the concentration of 16 plasma biomarkers differs between PARDS and critically ill non-PARDS subjects and how the concentrations change over time. We hypothesized that some but not all plasma biomarkers would correlate with changing levels of lung injury and that these previously described biomarkers would be significantly more elevated in the plasma of PARDS subjects compared to critically ill subjects without PARDS.
Methods and Materials:
Human Subject Research:
The study was conducted at two quaternary PICUs. The study (PARDS Subtypes) was approved by the Cincinnati Children’s Hospital Institutional Review Board (IRB 2015–8514 & 2017–1345) on November 28, 2017, and the Children’s Hospital of Philadelphia (CHOP) Institutional Review Board (CHOP IRB 20–017205). The trial was registered with ClinicalTrial.gov (NCT#03539783). All procedures and protocols were in accordance with the ethical standards set forth by the Cincinnati Children’s Hospital IRB, CHOP IRB, and with the Helsinki Declaration.
Eligibility Criteria:
Subjects were eligible if they were admitted to the PICU and were less than 18 years old. Neonates and any subjects with neonatal lung disease were excluded. PARDS subjects were mechanically ventilated and met PALICC PARDS criteria for hypoxemia and chest radiograph findings.(13) Critically ill comparison subjects were eligible if admitted to the PICU with no apparent lung disease and expected hospital stay of at least 7 days. Most critically ill subjects without PARDS were post operative subjects. We chose this population as a comparison with the goal of determining which PARDS biomarkers are most confounded by conditions that are commonly encountered in the PICU. The most common surgeries these subjects had included neurosurgical procedures (craniotomy for tumor removal or seizure focus removal and cervical or lumbar laminectomy), airway reconstruction (slide tracheoplasty or laryngotracheoplasty), and total pancreatectomy with auto-islet transplant. Exclusion criteria included order of limited resuscitation or baseline oxygen requirement of greater than 2 liters per minute. The exclusion of subjects with high baseline oxygen requirement was to prevent confounding. Written, informed consent was obtained from parents or legal guardians, and subject assent was obtained as appropriate.
Protocol and Procedure:
PARDS subjects were enrolled within 24 hours of meeting PARDS criteria and non-PARDS subjects were enrolled within 24 hours of admission to the PICU. Blood samples were collected on study days 1, 3, 7, and 14. Samples were collected in subjects with vascular access that drew or during clinically indicated lab draw. Sample collection continued until subjects were discharged from the hospital or completed the study. We had twelve PARDS patients discharge prior to day 14, two PARDS patients die prior to day 14, and 5 control patients discharge prior to day 14. Plasma was stored at −80°C until analysis.
Plasma samples were analyzed for Angiopoietin-2 (ANG2), Granzyme B (GrB), Soluble Intercellular Adhesion Molecule-1 (sICAM1), Interferon-γ (IFNγ), lnterleukin-6 (IL6), IL8, IL10, IL17, IL18, Matrix Metalloproteinase 9 (MMP-9), Plasminogen Activator Inhibitor-1 (PAI1), Surfactant Protein D (SPD), Tumor Necrosis Factor alpha (TNFα), TNF Receptor Soluble Factor 1A (TNFRSF1A), Myeloperoxidase (MPO), and Receptor for Advanced Glycation End Products (RAGE) using proprietary bead-based assays from R&D Systems, a Luminex 200 instrument (Luminex Corp, Austin, TX), and manufacturer recommended protocols. Recognizing the potential for technical differences between batches to influence results, we chose to analyze coefficients of variation (CV) for each sample. We decided on an acceptable CV value of less than 20%. If we found higher CV values, batch correction would be performed.
Batch Correction
During data analysis, we discovered that one of our four batches (batch one) had significantly different measurements than other batches. Details on the coefficients of variation are provided in (Supplemental Figure 1). The observed differences between batch one and all others was likely due to a manufacturing change in the standards provided in the kit. Using replicates between batch 1 and batches 2, 3, and 4, we performed analyte-specific correction of batch 1 fluorescent intensity values (Supplemental Figure 1 and 2) and used these values to calculate analyte concentrations used in downstream analysis.
Data Collection:
PARDS was classified as mild/moderate or severe (2) using oxygenation index or oxygen saturation index (Masimo pediatric pulse oximeter). The OSI was measured as close to 4 AM on each of the specimen collection days as possible. As PARDS subjects improved and PARDS resolved, their PARDS was reclassified to “none”. Clinical and demographic information was collected at enrollment and on specimen collection days and was entered into a RedCap Database.
Data Analysis:
All data were analyzed using the R statistical package (version 4.1.0)(14), rstatix (version 0.7.0) (15), corrplot (version 0.91) (16), and ggpubr (version 0.4.0) (17). Wilcoxon-Rank Sum and Kruskal-Wallis with Bonferroni post hoc tests were used for group comparisons and Spearman rank test with Bonferroni correction was used for correlative analyses. p-values <0.05 were considered significant. For longitudinal analyses, change in OSI was calculated as ΔOSI = (OSIn+1 – OSIn)/(dayn+1 – dayn), with n being the sample collection day. Similarly change in biomarker concentration was calculated as ΔConcentration = (Concentrationn+1 – Concentrationn)/(dayn+1 – dayn), with n denoting sample collection day. We then determined ΔConcentraiton/ΔOSI which was moded using generalized linear modeling in R. OSI data was recorded only on sample collection days (i.e. 1, 3, 7, of 14). Since the calculation accounts for differences in time between datapoints, missing values were not imputed.
Racial Differences in Pulse Oximetry:
Recent studies in both adult and pediatric patients have demonstrated that pulse oximeters over-estimate the true arterial oxygen saturation in non-white patients. (18, 19). Given our use of OSI in this study, we conducted a sensitivity analysis. Since pulse oximetry overestimated true arterial oxygen saturation by a mean of 4.3% in non-white children (18), we subtracted five percentage points from the measured pulse oximetry saturation from each non-white subject in our cohort. We then re-calculated the OSI and re-analyzed the data.
Results
Demographics
We analyzed 138 specimens from 50 PARDS subjects and 13 critically ill subjects without PARDS. The demographic and clinical characteristics are shown in Table 1. As expected, PARDS subjects had significantly higher OSI and PELOD 2 score. PARDS subjects were intubated for longer, had fewer ventilator free days, and were admitted to the PICU and hospital for longer than non-PARDS subjects. Mortality was 14% for PARDS subjects compared to 0% for non-PARDS subjects.
Table 1:
Comparison of demographic and clinical variables between PARDS subjects and critically ill subjects without PARDS.
| Age | Median (IQR) | 6 (3.2 to 6.8) | 10 (4.3 to 15.2) |
| Sex | Female Male |
22 (44) 28 (56) |
4 (31) 9 (69) |
| Race | White Non-white |
36 (72) 14 (28) |
11 (85) 2 (15) |
| OSI | Median (IQR) | 9 (6.91 to 11.1) | 0.1 (0.1 to 3) |
| PELOD 2 | Median (IQR) | 9 (6 to 11) | 6 (6 to 10) |
| Ventilator Free Days | Median (IQR) | 19 (10.5 to 22) | 28 (27 to 28) |
| Days Intubated | Median (IQR) | 10 (6 to 17.5) | 0 (0 to 1) |
| Principle Comorbidity | Airway/Lung Disorder Baseline Genetic Condition Developmental Delay Bone Marrow Transplant Solid Organ Transplant Hematologic/Oncologic Disorder Chronic Pancreatitis AKI None |
15 (30) 15 (30) 13 (26) 9 (18) 2 (4) 1 (2) 0 (0) 0 (0) 8 (16) |
3 (23) 1 (8) 1 (8) 2 (15) 1 (11) 1 (8) 2 (15) 2(15) 2 (15) |
| PARDS Acute Trigger | Pneumonia Sepsis Aspiration Unknown Trauma Pulmonary Hemorrhage Cardiac Arrest None |
35 (70) 10 (20) 3 (6) 3 (6) 1 (2) 1 (2) 1 (2) 3 (6) |
0 (0) 3 (23) 0 (0) 0 (0) 1 (8) 0 (0) 0 (0) 9 (69) |
| PICU Days | Median (IQR) | 16 (10 to 25) | 6 (2 to 7) |
| Hospital Days | Median (IQR) | 28 (13.25 to 48.75) | 17 (9 to 19) |
| Mortality | Percent | 14% | 0% |
Changing PARDS Biomarker Concentrations Correlate Inversely with Changes in Oxygenation Saturation Index
To identify associations between biomarkers and lung injury severity, we compared change in OSI to change in biomarker concentration. This was calculated as current collection day biomarker and OSI values minus the previous collection day values as described above. With the exception of RAGE, IL6, IL8, IFNγ, and MMP-9, all biomarkers were positively correlated with changing lung injury (Supplemental Figure 3). sICAM1 had the strongest positive correlation (R=0.69, p =2.2 × 10−16, Figure 1). We then conducted a sensitivity analysis of the data to account for possible confounding due to the potential for falsely elevated pulse oximetry readings in non-white subjects but found no substantial change in findings (Supplemental Figure 4).
Figure 1:
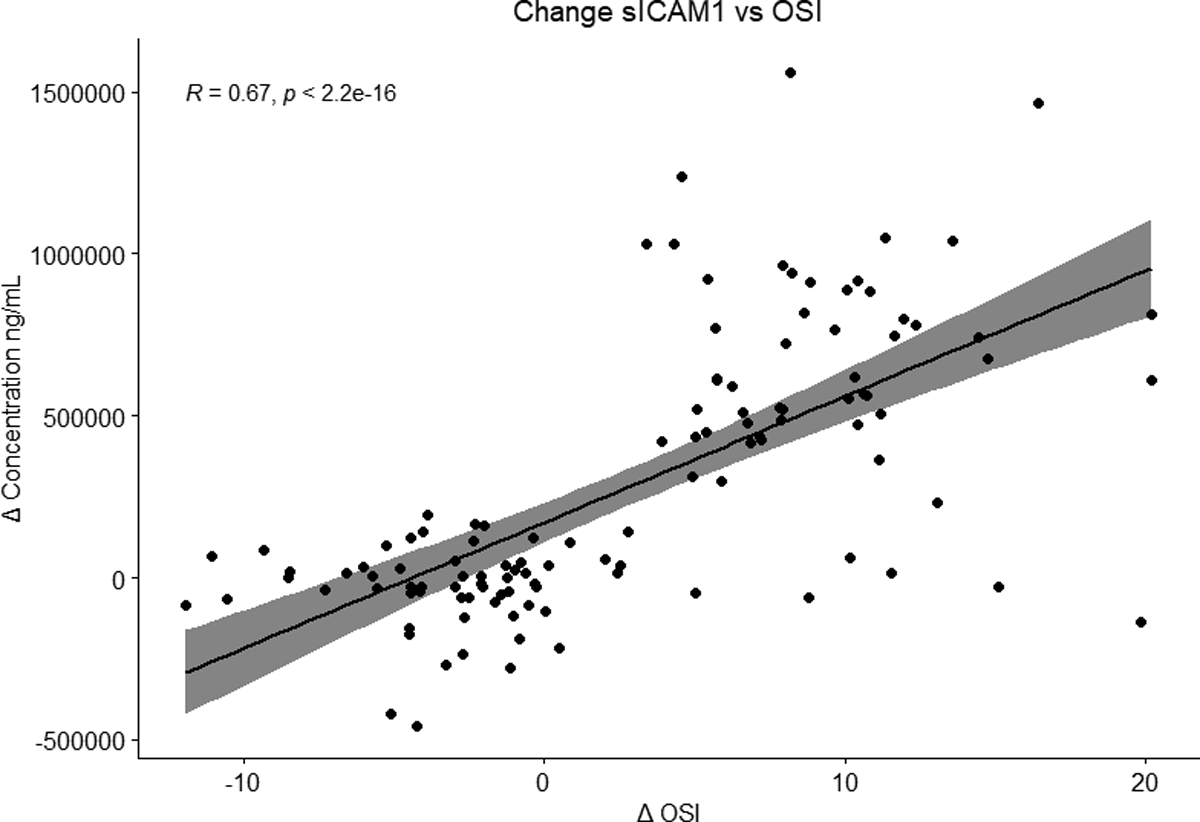
Changing Soluble Intercellular Adhesion Molecule-1 (sICAM1) concentration over time with changing lung injury as measured by change in OSI. The number of PARDS samples from each day are as follows day 1 (n = 46), day 3 (n = 33), day 7 (n = 25), and day 14 (n = 13).
Plasma levels of TNFα, IL8, IFNγ, IL17, GrB, sICAM1, SPD, and IL18, are elevated in subjects with PARDS
We found that concentrations of TNFα, IL8, IL18, IFNγ, IL17, GrB, sICAM1, SPD were significantly increased in the plasma of PARDS subjects compared to non-PARDS subjects on day 1 samples (Supplemental Figure 5). Interestingly, the concentration of MMP-9 was significantly increased in our non PARDS subjects (Figure 2). In a sensitivity analysis to account skin pigmentation-related inaccuracies in pulse oximetry data in we again found no substantial difference (Supplemental Figure 6).
Figure 2:
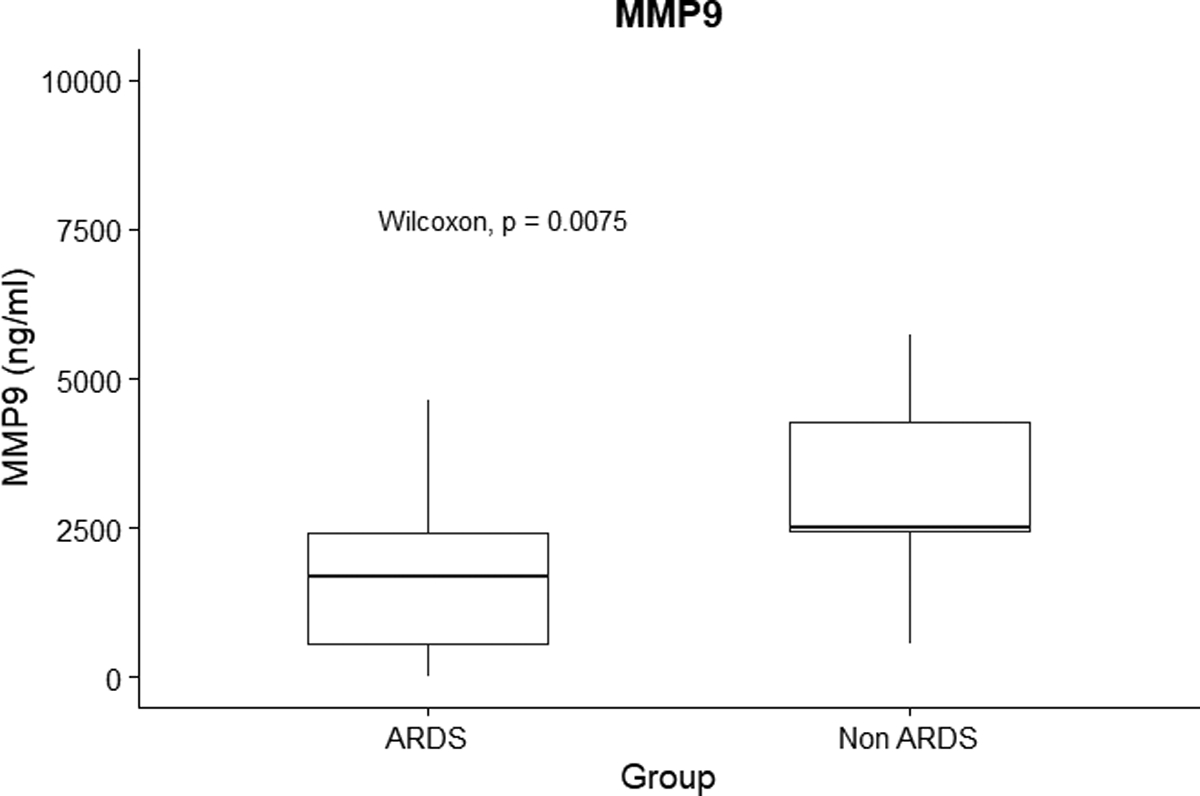
Concentration of matrix metalloproteinase 9 (MMP-9) in ng/mL on day 1 samples between critically ill non-PARDS (n = 13) subjects and PARDS (n = 50) subjects. P value is noted on the plot.
There were outliers with very elevated levels of several biomarkers among the PARDS group. To determine if this represented a subgroup of PARDS subjects with very elevated levels of every biomarker, we visualized biomarker levels of PARDS day 1 specimens using a heatmap (Supplemental Figure 7). One subject (ARDS 29) showed elevated levels of most biomarkers assayed. Most other outlying points were scattered among the remainder of the PARDS subjects.
We were most interested in sICAM1 since this showed the strongest correlation with OSI and MMP-9 since this biomarker was unexpectedly more elevated in non-PARDS subjects on day 1, and thus chose to examine these biomarkers in more temporal detail. We examined the mean level of MMP-9 and sICAM1 between groups on each collection day (Supplemental Figure 8 and 9). ICAM 1 was significantly elevated in PARDS compared to non-PARDS subjects on each collection day; although, no non-PARDS samples were collected on day 14. sICAM1 levels peaked in PARDS subjects on day 7 and started to decline by day 14. Interestingly, MMP-9 was significantly higher in non-PARDS subjects on day 1, but there was no significant difference in mean MMP-9 levels between PARDS and non-PARDS subjects on days 3 or 7.
Comparison of Biomarker Level and Severity of Lung Injury
We examined the correlation between day 1 biomarker level and day 1 PARDS severity using OSI (Supplemental Figure 10) based upon the 2023 PALICC-2 recommendations (2). There was no correlation between day 1 biomarker concentration and day 1 PARDS severity for any of the biomarkers we examined. We then conducted a sensitivity analysis to account for racial differences in the accuracy of pulse oximetry and found no substantial differences (Supplemental Figure 11).
Correlation Of Plasma Biomarkers With Each Other:
We then used a correlation analysis to examine how biomarker concentrations correlated with each other across all study days. We found that PAI1, MMP-9, and MPO were positively correlated with each other and negatively correlated with other measured biomarkers. Levels of assayed interleukins, IFNγ, sICAM1, ANG2, RAGE, GrB, TNFα, and TNFRSF1A were positively correlated with each other (Figure 3). We noted that the neutrophil marker GrB did not group with the other two neutrophil markers (MPO and MMP-9). We conducted a sub-analysis to investigate this. We hypothesized that the presence of sepsis in some patients might be influencing these results. We examined the concentrations of MMP9, MPO, and GrB in PARDS patients with and without sepsis at all study time points. We found no statistically significant difference in the concentrations of any of these three markers between subjects with sepsis and those without sepsis (Supplemental Figure 12).
Figure 3:
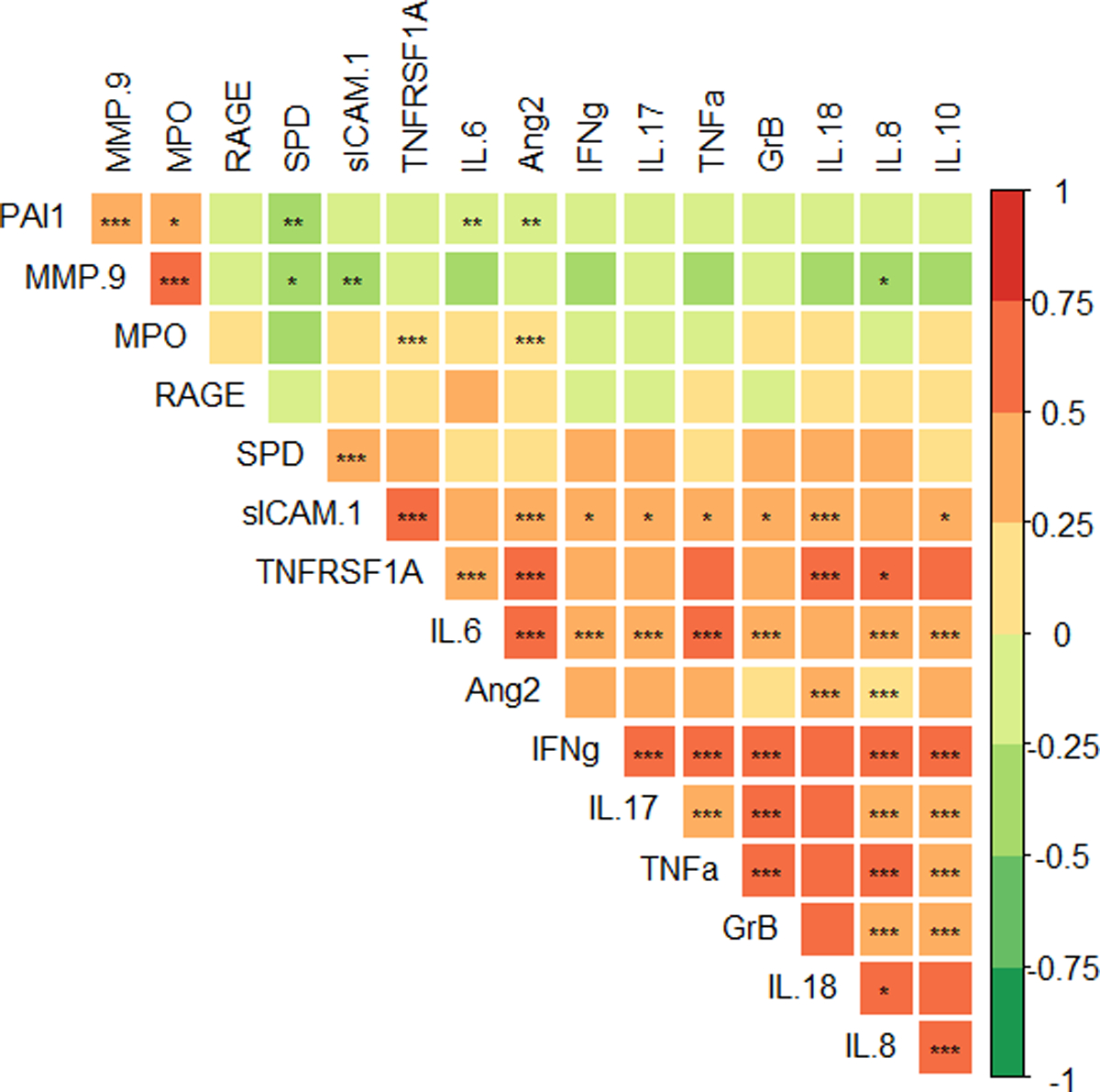
Heatmap of Spearman correlations between biomarkers in PARDS subjects. In the color scale, red is positively and green negatively correlated. *p<0.05, **p<0.01, *** p<0.001 with Bonferroni correction.
Discussion
The use of biomarkers for predictive and prognostic enrichment in both pediatric and adult ARDS subjects has shown promise (5, 6, 12); however, there are limitations to their use. This appears to be especially true in patients with concomitant processes such as sepsis. For example, ANG2 has been shown to be elevated in pediatric subjects with sepsis (20) and MMP-9 is known to be elevated in adult sepsis (21, 22) and burn subjects (23). Despite previously promising results, biomarkers of epithelial and endothelial damage did not perform well in our cohort, and this is likely due to our selection of other critically ill patients as a comparator population. These results highlight the potential for other pathophysiologic conditions commonly encountered in the PICU to confound plasma biomarker interpretation, the best approach to prognostic and predictive enrichment is likely to involve the combination of several biomarkers (12), clinical parameters, and even genomics data as these technologies continue to rapidly evolve. In our study, we found that sICAM1 was the most correlated with changing levels of lung injury, that there was no correlation between day 1 biomarker concentration and day 1 PARDS severity, that 7 previously described PARDS biomarkers were not elevated in PARDS subjects compared to critically ill non-PARDS patients on day 1, and that there were largely two groups of biomarkers apparent on correlation analysis.
In our cohort, sICAM1 showed the strongest correlation with changing oxygenation (R = 0.69). sICAM1 is an adhesion molecule that is involved in neutrophil recruitment and is expressed on many cell types including endothelial cells, alveolar type 1 cells, and several different immune cells.(24) Adult subjects with ARDS that have increased sICAM1 levels have fewer ventilator free days and are at an increased risk of death (25, 26). In pediatric subjects, early elevated sICAM1 is associated longer duration of mechanical ventilation and higher risk of death (24). Most existing studies of ICAM have focused on the levels of sICAM1 early in the disease course (24, 25). We examined sICAM1 levels over 14 days and found that in PARDS subjects, levels peaked on day 7 and were decreasing by day 14. We also examined the relationship between biomarker level and PARDS classification. We found no correlation between day 1 biomarker concentration and day 1 PARDS severity. This suggests that while changing biomarker concentrations from an individual baseline might provide information about evolving lung injury, comparison of point-in-time between subjects is confounded by inter-subject differences.
In contrast to sICAM1, we found that MMP-9 did not have a statistically significant correlation with worsening lung injury. MMP-9 was also higher in critically ill non-PARDS compared to PARDS subjects on day 1. MMP-9 is an enzyme released from activated neutrophils and is involved in degradation of extracellular matrix proteins as well as cleavage of membrane bound cytokines (27). Our study included critically ill non-PARDS subjects as a comparison population. Most of our subjects were post operative after major surgeries and thus were likely experiencing varying degrees of inflammation and SIRS that could contribute to elevated MMP-9. Several previous studies have shown that MMP-9 is elevated in non-PARDS processes commonly encountered in the PICU including sepsis (21, 22) and burns (23). The inclusion of these critically ill non-PARDS subjects thus could have resulted in a comparison population with elevated levels of MMP-9.
Another potential explanation for our MMP-9 results is the existence of MMP-9 subclasses amongst PARDS subjects. In one PARDS study specifically looking at MMP profiles, latent class analysis found two classes that were different in MMP profile (27). The class the authors termed MMP Profile 1 had relatively lower levels of MMP-9 compared to MMP Profile 2. Outcomes including mortality were better in the MMP Profile 2 group compared to the MMP Profile 1 group. It is possible given our small sample size, that we had a disproportionate number of PARDS subjects with relatively lower levels of MMP-9 (MMP profile 1). Overall, in our population of critically ill non-PARDS subjects, MMP-9 was not a good marker for PARDS and did not correlate with changing lung injury. Our data suggest MMP-9 should be used with caution in subjects with other disease processes that are known to affect MMP-9 levels.
We examined the correlation between biomarkers and observed two patterns in PARDS subjects. The first pattern included all assayed biomarkers except MMP-9, PAI1, and MPO and likely reflects overall inflammatory state. The second showed a positive correlation between MMP-9, PAI1, and MPO. The anticorrelation of neutrophil markers MPO and MMP-9 with GrB is interesting and could be consistent with the importance of different neutrophil populations in inflammatory processes such as lung injury and sepsis (28, 29). Overall, these findings are consistent with a recent latent class analysis of PARDS subjects that found two different groups with one group being characterized by a hyper-inflammatory state and the other by a hypo-inflammatory state (12).
Our study supports the idea that future strategies for prognostic and predictive enrichment should not rely solely on plasma biomarkers and highlights several of the associated challenges in interpreting them. For example, the lack of a dose response relationship between PARDS severity and biomarker concentration as well as the high inter-subject variability makes interpretation of any biomarker concentration at a single time point challenging. The incorporation of genomic information in the form of transcriptomics and epigenomics among other techniques into future prognostic and predictive enrichment tools will be valuable. One such example of the potential of these techniques is the ability of mRNA seq and scRNA seq to identify important pathobiologic differences in COVID and non-COVID PARDS and correctly identify the most appropriate treatment (30).
This study has several limitations. First, we have a small sample size which may limit the ability to detect differences between groups. Second, PARDS and critically ill non-PARDS subjects had plasma samples drawn only when they had existing vascular access from which blood could be drawn. This likely limited sampling at less ill timepoints. Third, our critically ill non-PARDS subjects were largely drawn from subjects who underwent a major elective surgery requiring prolonged hospitalization. Finally, recent studies have shown that pulse oximetry has the potential to underestimate true arterial oxygen saturation in non-white individuals. This is problematic for our study given our use of OSI which was necessary because few subjects had arterial lines for the duration of the study; however, in sensitivity analysis there was no substantial impact of race on the findings.
Conclusion
We found that several ARDS-associated biomarkers correlate with changing lung injury; although, most correlations were weak. The strongest correlation was with sICAM1 (R = 0.69) suggesting potential pathophysiologic importance. Additionally, we found that the day 1 concentration of 7 previously described PARDS biomarkers were no different between PARDS patients and critically ill non-PARDS patients indicating a lack of specificity. Furthermore, there was no correlation between day 1 biomarker concentration and day 1 PARDS severity consistent with substantial intersubject variability is these measures. Our study adds to the growing body of evidence that future prognostic and predictive enrichment strategies will need to include clinical, plasma biomarker, and even genomic data to be most effective.
Supplementary Material
RESEARCH IN CONTEXT:
Intrinsic heterogeneity in PARDS has made conducting successful clinical trials difficult.
One proposed strategy to overcome this is plasma-based biomarkers for predictive and prognostic enrichment.
There is little longitudinal data about which plasma biomarkers are most correlated with changing lung injury and thus might be most biologically significant.
AT THE BEDSIDE:
In our study most plasma biomarkers correlated only weakly with changing lung injury.
Plasma markers may be confounded by other systemic processes commonly encountered in the PICU as demonstrated by our non-PARDS patients having higher levels of MMP9.
Future studies should seek to determine which plasma biomarkers are most biologically meaningful and least susceptible to confounding by other systemic processes.
Acknowledgments:
We would like to thank Toni Yunger, Erin Stoneman, and Stephen Famularo. Grant support SCCM award 2020 (Varisco). NHLBI R01HL141229 (Varisco). NHLBI 1R01HL148054-01 (Yehya). 5T32GM008478-30 (Williams). We would also like to thank the Cincinnati Children’s Hospital Center for Pediatric Genomics. Finally, we acknowledge the contributions of Dr. Hector Wong in the conception of this study. Due to Dr. Wong’s untimely passing, he was unable to review the final manuscript.
Copyright Form Disclosure:
Drs. Williams, Lahni, and Yehya’s institutions received funding from the institution received funding from the National Institutes of Health (NIH). Dr. Williams’ institution received funding from the National Heart, Lung, and Blood Institute (NHLBI) (5T32GM008478-30). Drs. Williams, Lahni, Yehya, and Varisco received support for article research from the NIH. Dr. Yehya’s institution received funding from the NHLBI (1R01HL148054-01) and Pfizer. Dr. Varisco’s institution received funding from NHLBI (R01HL141229); he disclosed that he received the Society of Critical Care Medicine Award 2020. The remaining authors have disclosed that they do not have any potential conflicts of interest.
References:
- 1.Khemani RG, Smith L, Lopez-Fernandez YM, et al. : Pediatric Acute Respiratory Distress Syndrome Incidence and Epidemiology (PARDIE): an international observational study. Lancet Respir Med 2019; 7:115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yehya N, Smith L, Thomas NJ, et al. : Definition, Incidence, and Epidemiology of Pediatric Acute Respiratory Distress Syndrome: From the Second Pediatric Acute Lung Injury Consensus Conference. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc 2023; 24:S87–S98 [DOI] [PubMed] [Google Scholar]
- 3.Dahmer MK, Flori H, Sapru A, et al. : Surfactant Protein D Is Associated With Severe Pediatric Acute Respiratory Distress Syndrome, Prolonged Ventilation, and Death in Children With Acute Respiratory Failure [Internet]. Chest 2020; [cited 2020 May 8] Available from: http://www.sciencedirect.com/science/article/pii/S0012369220305730
- 4.Lim MJ, Zinter MS, Chen L, et al. : Beyond the Alveolar Epithelium: Plasma Soluble Receptor for Advanced Glycation End Products Is Associated With Oxygenation Impairment, Mortality, and Extrapulmonary Organ Failure in Children With Acute Respiratory Distress Syndrome. Crit Care Med 2022; 50: 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Famous KR, Delucchi K, Ware LB, et al. : Acute Respiratory Distress Syndrome Subphenotypes Respond Differently to Randomized Fluid Management Strategy. Am J Respir Crit Care Med 2017; 195:331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calfee CS, Delucchi K, Parsons PE, et al. : Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2014; 2:611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yehya N, Thomas NJ, Meyer NJ, et al. : Circulating markers of endothelial and alveolar epithelial dysfunction are associated with mortality in pediatric acute respiratory distress syndrome. Intensive Care Med 2016; 42:1137–45 [DOI] [PubMed] [Google Scholar]
- 8.Gallagher DC, Parikh SM, Balonov K, et al. : Circulating angiopoietin 2 correlates with mortality in a surgical population with acute lung injury/adult respiratory distress syndrome. Shock 2008; 29:656–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahmer MK, Quasney MW, Sapru A, et al. : Interleukin-1 Receptor Antagonist Is Associated With Pediatric Acute Respiratory Distress Syndrome and Worse Outcomes in Children With Acute Respiratory Failure. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc 2018; 19:930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flori H, Sapru A, Quasney MW, et al. : A prospective investigation of interleukin-8 levels in pediatric acute respiratory failure and acute respiratory distress syndrome. Crit Care Lond Engl 2019; 23:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zinter MS, Orwoll BE, Spicer AC, et al. : Incorporating Inflammation into Mortality Risk in Pediatric Acute Respiratory Distress Syndrome. Crit Care Med 2017; 45:858–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahmer MK, Yang G, Zhang M, et al. : Identification of phenotypes in paediatric patients with acute respiratory distress syndrome: a latent class analysis. Lancet Respir Med 2022; 10:289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khemani RG, Smith LS, Zimmerman JJ, et al. : Pediatric acute respiratory distress syndrome: definition, incidence, and epidemiology: proceedings from the Pediatric Acute Lung Injury Consensus Conference. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc 2015; 16:S23–40 [DOI] [PubMed] [Google Scholar]
- 14.R Core Team: R: A Language and Environment for Statistical Computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2019. Available from: https://www.R-project.org/ [Google Scholar]
- 15.Kassambara A: rstatix: Pipe-Friendly Framework for Basic Statistical Tests [Internet]. 2020; [cited 2020 Aug 20] Available from: https://CRAN.R-project.org/package=rstatix [Google Scholar]
- 16.Wei T, Simko V: R package “corrplot”: Visualization of a Correlation Matrix [Internet]. 2017. Available from: https://github.com/taiyun/corrplot
- 17.ggpubr: Publication Ready Plots - Articles - STHDA [Internet]. 2020; [cited 2020 May 15] Available from: http://www.sthda.com/english/articles/24-ggpubr-publication-ready-plots/
- 18.Andrist E, Nuppnau M, Barbaro RP, et al. : Association of Race With Pulse Oximetry Accuracy in Hospitalized Children. JAMA Netw Open 2022; 5:e224584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seitz KP, Wang L, Casey JD, et al. : Pulse Oximetry and Race in Critically Ill Adults. Crit Care Explor 2022; 4:e0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter RP, Zheng L, Ashtekar AR, et al. : Associations of Plasma Angiopoietins-1 and −2 and Angiopoietin-2/−1 Ratios With Measures of Organ Injury and Clinical Outcomes in Children With Sepsis: A Preliminary Report. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc 2020; 21:e874–e878 [DOI] [PubMed] [Google Scholar]
- 21.Mühl D, Nagy B, Woth G, et al. : Dynamic changes of matrix metalloproteinases and their tissue inhibitors in severe sepsis. J Crit Care 2011; 26:550–555 [DOI] [PubMed] [Google Scholar]
- 22.Wang M, Zhang Q, Zhao X, et al. : Diagnostic and prognostic value of neutrophil gelatinase-associated lipocalin, matrix metalloproteinase-9, and tissue inhibitor of matrix metalloproteinases-1 for sepsis in the Emergency Department: an observational study. Crit Care Lond Engl 2014; 18:634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy B, Szélig L, Rendeki S, et al. : Dynamic changes of matrix metalloproteinase 9 and tissue inhibitor of metalloproteinase 1 after burn injury. J Crit Care 2015; 30:162–166 [DOI] [PubMed] [Google Scholar]
- 24.Flori HR, Ware LB, Glidden D, et al. : Early elevation of plasma soluble intercellular adhesion molecule-1 in pediatric acute lung injury identifies patients at increased risk of death and prolonged mechanical ventilation. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc 2003; 4:315–321 [DOI] [PubMed] [Google Scholar]
- 25.Calfee CS, Eisner MD, Parsons PE, et al. : Soluble intercellular adhesion molecule-1 and clinical outcomes in patients with acute lung injury. Intensive Care Med 2009; 35:248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boldt J, Wollbrück M, Kuhn D, et al. : Do plasma levels of circulating soluble adhesion molecules differ between surviving and nonsurviving critically ill patients? Chest 1995; 107:787–792 [DOI] [PubMed] [Google Scholar]
- 27.Zinter MS, Delucchi KL, Kong MY, et al. : Early Plasma Matrix Metalloproteinase Profiles. A Novel Pathway in Pediatric Acute Respiratory Distress Syndrome. Am J Respir Crit Care Med 2019; 199:181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alder MN, Opoka AM, Lahni P, et al. : Olfactomedin-4 Is a Candidate Marker for a Pathogenic Neutrophil Subset in Septic Shock. Crit Care Med 2017; 45:e426–e432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kangelaris KN, Clemens R, Fang X, et al. : A neutrophil subset defined by intracellular olfactomedin 4 is associated with mortality in sepsis. Am J Physiol Lung Cell Mol Physiol 2021; 320:L892–L902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarma A, Christenson SA, Byrne A, et al. : Tracheal aspirate RNA sequencing identifies distinct immunological features of COVID-19 ARDS. Nat Commun 2021; 12:5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


