Summary
Background
Pancreatic Duct Adenocarcinoma (PDAC) screening can enable early-stage disease detection and long-term survival. Current guidelines use inherited predisposition, with about 10% of PDAC cases eligible for screening. Using Electronic Health Record (EHR) data from a multi-institutional federated network, we developed and validated a PDAC RISk Model (Prism) for the general US population to extend early PDAC detection.
Methods
Neural Network (PrismNN) and Logistic Regression (PrismLR) were developed using EHR data from 55 US Health Care Organisations (HCOs) to predict PDAC risk 6–18 months before diagnosis for patients 40 years or older. Model performance was assessed using Area Under the Curve (AUC) and calibration plots. Models were internal-externally validated by geographic location, race, and time. Simulated model deployment evaluated Standardised Incidence Ratio (SIR) and other metrics.
Findings
With 35,387 PDAC cases, 1,500,081 controls, and 87 features per patient, PrismNN obtained a test AUC of 0.826 (95% CI: 0.824–0.828) (PrismLR: 0.800 (95% CI: 0.798–0.802)). PrismNN's average internal-external validation AUCs were 0.740 for locations, 0.828 for races, and 0.789 (95% CI: 0.762–0.816) for time. At SIR = 5.10 (exceeding the current screening inclusion threshold) in simulated model deployment, PrismNN sensitivity was 35.9% (specificity 95.3%).
Interpretation
Prism models demonstrated good accuracy and generalizability across diverse populations. PrismNN could find 3.5 times more cases at comparable risk than current screening guidelines. The small number of features provided a basis for model interpretation. Integration with the federated network provided data from a large, heterogeneous patient population and a pathway to future clinical deployment.
Funding
Prevent Cancer Foundation, TriNetX, Boeing, DARPA, NSF, and Aarno Labs.
Keywords: Pancreatic cancer, Risk prediction, Machine learning, Electronic health records, Federated data
Research in context.
Evidence before this study
We searched PubMed for publications on pancreatic cancer risk prediction models for the general population. We focused on articles published between 2013 and 2023, using the search terms “pancreatic cancer”, “risk prediction models”, and “general population”. Previous studies have developed and validated Pancreatic Duct Adenocarcinoma (PDAC) risk models on large populations. However, they are limited by their lack of racial and geographic diversity, external validation, and a clear pathway to clinical implementation. Moreover, while other models use standard classification metrics such as AUC for performance evaluation, they provide little insight into the comparison with currently utilised PDAC screening inclusion criteria for high-risk individuals with an inherited predisposition.
Added value of this study
We used Electronic Health Record (EHR) data from 55 Health Care Organisations (HCOs) across the US within a federated network platform including over 1.5 million PDAC cases and controls. We developed, internally and internal-externally validated, and simulated the deployment of PDAC risk models for early prediction of 6–18 months before diagnosis. Our PDAC RISk Model (Prism) uses 87 features derived from EHR diagnosis, medication, lab, and demographic data from a racially and geographically diverse population. Prism maintained its accuracy across internal-external validation and simulated deployment within the network platform and can now be tested prospectively on multiple institutional data within the network. The model captured 3.5 times more patients than the current inclusion criteria used to identify patients for PDAC screening programs at similar risk levels.
Implications of all the available evidence
Prism can potentially help primary care providers nationwide noninvasively identify high-risk individuals for PDAC screening or serve as a first filter before subsequent biomarker testing. Prism sets the stage for model deployment within a federated network to identify high-risk patients at multiple institutions participating in the network.
Introduction
Most cases of Pancreatic Duct Adenocarcinoma (PDAC) are diagnosed as advanced-stage disease, leading to a five-year relative survival rate of only 11%.1 Expanding the population currently being screened is crucial for increasing early detection and improving survival. Current screening guidelines2, 3, 4 targeting stage I cancers and high-grade PDAC precursors have significantly improved long-term survival.5,6 Current guidelines target patients with a family history or genetic predisposition to PDAC,7,8 with screening eligibility based on estimated absolute (5%) and relative (five times) risk compared to the general population.6 These patients comprise only about 10% of all PDAC cases. No consensus or guidelines exist for PDAC screening in the general population,9 where the majority of PDAC cases are found.
Several groups have developed PDAC risk models for the general population using various data sources.10, 11, 12 Most such models aim for integration with Electronic Health Record (EHR) systems for clinical implementation. One effort used EHR data from an aggregated multi-institutional database.13 Their evaluation focused on risk prediction up to one month before diagnosis without evaluating generalizability across locations or races. Several other efforts using EHR data had limited validation across locations and races.14, 15, 16 Other efforts worked with small sample sizes10,17 and internal validation only.12,17
We used EHR data from 55 US Health Care Organisations (HCOs) from a federated data network to develop and validate PDAC risk prediction models for the general population. Our models enable identifying individuals at high risk for PDAC from the general population, so they can be offered early screening or referred for lower overhead testing such as biomarker testing.
The data network provides access to harmonised, de-identified EHR data of over 89 million patients for model development and testing. Because the network is connected to the EHR systems of the participating HCOs, it provides a pathway to model deployment in a clinical setting, a critical step in the progression toward successful clinical adoption.18
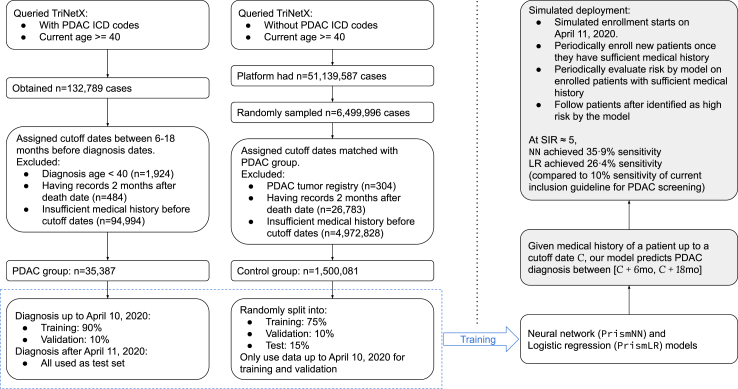
We developed a methodology to train PDAC RISk prediction Model (Prism) on federated network EHR data. We worked with two classes of models: neural networks (PrismNN) and logistic regression (PrismLR). Prism models identify high-risk patients 6–18 months before an initial PDAC diagnosis. Our evaluation reports Area Under the Curve (AUC) and risk calibration. We also conducted three types of internal-external validation: location-based, race-based, and temporal. Furthermore, we simulated the deployment of Prism models with temporally separate training/test data to evaluate their performance in a more realistic setting. Evaluation metrics include sensitivity and Standardised Risk Ratio (SIR). SIR was based on demographically matched PDAC incidence rates of the general US population from the SEER database.19 Fig. 1 summarises the process of our model development.
Fig. 1.
Flowchart of our study, including data collection and training/evaluation of simulated deployment.
Methods
Data source and setting
This is an observational retrospective study with both a case–control and cohort design. Our goal is to develop machine learning models to predict risk of PDAC diagnosis 6–18 months in the future based on existing EHRs of a patient. We used data from the federated EHR database platform of TriNetX.20 TriNetX is global research network that specialises in EHR data collection and distribution. The network consists of mostly large, academic medical centres, community hospitals, and outpatient clinics. TriNetX supports pulling data from any EHR systems used by HCOs.
We used retrospective de-identified EHR data from 55 HCOs across the United States. On average, each HCO provides approximately 13 years of historical data. Data include values from structured EHR fields (e.g., demographics, date-indexed encounters, diagnoses, procedures, labs, and medications) as well as facts and narratives from free text (e.g., medications identified through Natural Language Processing (NLP)). TriNetX harmonises all data from each HCO's EHR to the TriNetX standard data model and a common set of controlled terminologies.
Study population
We worked with a PDAC case group and a control group. We obtained all data during Nov and Dec, 2022. We obtained the PDAC group by querying TriNetX to obtain EHR data for patients currently 40 years old or older with one of the following ICD-10/ICD-9 codes: C25.0, C25.1, C25.2, C25.3, C25.7, C25.8, C25.9, and 157. We obtained n = 132,789 PDAC cases. We excluded patients who were diagnosed before 40 years of age (n = 1924), patients with entries two months after their death record (n = 484, likely due to mixed entries of different patients), and patients with insufficient medical history (n = 94,994, defined in Section 2.3), to obtain a PDAC group with n = 35,387 cases.
For the control group, we queried TriNetX for patients at least 40 years old without any of the above ICD-10 or ICD-9 codes. The query matched n = 51,139,587 patients. From them, we uniformly sampled n = 6,499,996 patients. We excluded patients with a PDAC tumour registry entry but no PDAC diagnosis entries (n = 304), patients with records two months after their death record (n = 26,783), and patients with insufficient medical history (n = 4,972,828, defined in Section 2.3) to obtain a control group with n = 1,500,081 cases.
Model development
We trained and evaluated two model classes, neural networks (PrismNN) and logistic regression (PrismLR). Data were randomly partitioned into training (75%), validation (10%), and test (15%) sets. Note that the validation set refers to the dataset for model hyperparameter selection and should not be confused with our internal-external validation that accesses model performance.
Our training and testing procedures work with a cutoff date C for every patient. We derived features from entries available before C to predict PDAC risk between C +6 months and C +18 months. We sampled C uniformly between 6 and 18 months before diagnosis for each PDAC case and matched the cutoff dates for control cases. Patient age was assessed at the cutoff date. Given a cutoff date C, we empirically defined a patient to have sufficient medical history if their total number of diagnosis, medication, or lab entries within 2 years before C is at least 16, their first entry is at least 3 months earlier than their last entry before C, and their age at C is at least 38.5 years (40 years minus 18 months). We removed patients without sufficient medical history as described in Section 2.2.
We derived four classes of features from EHR: basic features, diagnosis features, medication features, and lab features. Basic features have six values, four for demographic information (age and sex, and their existence bits) and two for frequencies of clinical encounters. We counted the number of diagnosis, medication, or lab entries greater than 18 months before the cutoff date as the number of early records, and entries within 18 months as the number of recent records. The numbers of records serve as a proxy of clinical encounter frequencies, which are correlated with cancer diagnosis.21 Other features encode information about the existence, frequency, time span, and lab results (if applicable) of the corresponding type of EHR entries. We ignored EHR entry types appearing in less than 1% of the PDAC cases in the training set.
For each EHR entry type, we included an existence bit feature (0 or 1) indicating if the patient's EHR contains such entries. This encoding accounts for the additional effect of the healthcare process on EHR data.22 The encoding also enables model predictions based on whether a feature is present or missing. Because PrismNN can use sophisticated nonlinear reasoning, data imputation provides little to no useful additional information for these models. Therefore, we did not use any imputation.
The above process generates over five thousand features. To improve interpretability, we automatically selected fewer features by L0 regularisation on a binary input mask23 and iterative feature removal. We will present all the selected features ranked by their univariate predictive power with the PrismNN model, calculated as the AUC achieved by using each type of feature while zeroing all other features.
Model was calibrated with a variant of the Platt calibration.24 Calibration was evaluated with calibration plots on the test set. We also calculated the Geometric Mean of Over Estimation (GMOE), the geometric mean of the ratios of predicted risks to the actual risks, for quantitative calibration evaluation.
We evaluated the mean AUC and GMOE on test sets in nine independent runs with different random seeds for dataset split and model initialisation. We also evaluated model AUC and GMOE for a few age subgroups: 40–49, 50–69, 70+, and 50+ years old. Since bootstrapping has shown more accurate performance estimations in certain settings,25 we further evaluated the model AUC using optimism-corrected bootstrapping.
The Supplemental Material (Sections A.2 and A.3) provides more details on model development, including details on feature selection, model training/calibration, and model evaluation.
Internal-external validation
Our use of a large federated network enabled internal-external validation by splitting data within the network.26,27 Our internal-external validation considered three attributes: geographic location of the HCO (or its headquarter if an HCO covers multiple locations), patient race, and training data time. For each attribute, we split the dataset accordingly, trained models on one split, and tested on the other. We repeated all model development steps, including the automatic feature selection, on the training set for each internal-external validation.
For location and race-based validations, we assessed model generalizability by comparing the AUCs of internal-external-validation models with corresponding control models. Control models used the same sizes of training and test sets as the attribute-based split but used random splitting that ignores attribute values. We also calculated the gap between AUCs on the validation and test sets as an extra generalizability assessment. We further calculated the I2 index of the AUCs of geographic or racial subgroups to assess the heterogeneity of model performance. Table 1 provides the location and race distributions. We excluded patients with unknown HCO locations or races in the internal-external validation.
Table 1.
Demographics of our dataset.
| Attribute | Cancer group (n = 35,387) |
Control group (n = 1,500,081) |
||
|---|---|---|---|---|
| N | (%) | N | (%) | |
| Sex | ||||
| Female | 18,341 | (51.83) | 841,042 | (56.07) |
| Male | 17,045 | (48.17) | 637,674 | (42.51) |
| Unknown | 1 | (0.00) | 21,365 | (1.42) |
| Age at cutoff | ||||
| Mean (SD) | 67.55 | (10.61) | 59.50 | (12.87) |
| <40 | 135 | (0.38) | 103,566 | (6.90) |
| 40–50 | 2052 | (5.80) | 285,492 | (19.03) |
| 50–60 | 5762 | (16.28) | 342,037 | (22.80) |
| 60–70 | 10,727 | (30.31) | 354,907 | (23.66) |
| 70–80 | 10,531 | (29.76) | 251,333 | (16.75) |
| >80 | 4175 | (11.80) | 86,841 | (5.79) |
| Unknown | 2005 | (5.67) | 75,905 | (5.06) |
| Race | ||||
| AIAN | 93 | (0.26) | 5527 | (0.37) |
| Asian | 504 | (1.42) | 32,998 | (2.20) |
| Black | 5315 | (15.02) | 228,256 | (15.22) |
| NHPI | 21 | (0.06) | 1883 | (0.13) |
| White | 25,634 | (72.44) | 1,046,240 | (69.75) |
| Unknown | 3820 | (10.79) | 185,177 | (12.34) |
| HCO location | ||||
| Midwest | 8371 | (23.66) | 230,088 | (15.34) |
| Northeast | 11,831 | (33.43) | 426,469 | (28.43) |
| South | 12,246 | (34.61) | 682,417 | (45.49) |
| West | 2595 | (7.33) | 120,961 | (8.06) |
| Unknown | 344 | (0.97) | 40,146 | (2.68) |
| No. medical records | ||||
| Mean (SD) | 854.06 | (1501.89) | 440.23 | (939.50) |
Race abbreviations: AIAN: American Indian or Alaska Native; Black: Black or African American; NHPI: Native Hawaiian or Other Pacific Islander. The numbers in brackets indicate percentages of the corresponding category, except for the two rows with Mean (SD) where the bracketed numbers are standard deviations.
For temporal validation, we selected the 50%, 60%,...,90% percentile of diagnosis dates as split dates. We trained models on data available prior to those split dates. We used the data available after Oct 10, 2021 (the 90% percentile) as the test set for all models. Since the split date also impacts the size of the training set, we trained control models with the same number of randomly sampled cases (i.e., the control models used data available before each split dates but sampled only N PDAC cases from them, where N is half the total number of PDAC cases, corresponding to the number of cases with 50% percentile).
Simulated deployment
We estimated the model performance in a clinical setting by simulating model deployment using a prospective cohort study design. We trained the model on data available before Apr 11, 2020 (70% percentile of diagnosis dates) as the above temporal validation. We then simulated a clinical study. We periodically enrolled new patients into the study when they first satisfied the age and sufficient medical history requirements (see Section 2.3) after Apr 11, 2020. For each enrolled patient, we evaluated their PDAC risk using our models every 90 days until there was no more sufficient data (i.e., 18 months before dataset query date, or no more sufficient medical history on future dates) or the patient got a PDAC diagnosis. We followed up each enrolled patient starting 6 months after their first risk evaluation until 18 months after their last risk evaluation to see if they were diagnosed with PDAC during the follow up period. The selection of enrolment, risk evaluation, and follow up dates was independent of the model. We computed model performance statistics, including sensitivity, specificity, Positive Predictive Value (PPV), and Standardised Incidence Ratio (SIR), based on whether a patient ever received a high-risk prediction between 6 and 18 months before their PDAC diagnosis.
We chose multiple high-risk thresholds according to the 89.00%, 92.00%, 96.60%, 97.80%, 99.70%, and 99.95% specificity levels on the validation set. Overall, our design simulated the anticipated clinical application of our models to periodically evaluating every patient's PDAC risk. Supplemental Material (Section A.4) provides more details on the study design.
We accounted for unbalanced data sampling (we used all PDAC cases but a subset of the control cases) to estimate the PPV and SIR that would be obtained if we had evaluated the model on the entire TriNetX population. SIR is the ratio of the observed PDAC cases (true positives) in the high-risk group to the expected number of PDAC cases of that group. To calculate the expected number of cases, we used the SEER database,19 matched with age, sex, race, and calendar year for each individual in the high-risk group, as done by Porter, Laheru, Lau, He, Zheng, Narang et al.28
Ethics
All EHR data were obtained through TriNetX and de-identified by TriNetX. We accessed the data under a no-cost collaboration agreement. Some EHR entries were obfuscated by TriNetX to protect patient privacy. No human subjects were directly involved in this study. No patient could be re-identified from the data used in this study. Because this study used only de-identified patient records and did not involve the collection, use, or transmittal of individually identifiable data, this study was exempted from Institutional Review Board approval, as determined by the Western IRB.
Statistics
The PDAC group used all available cases on TriNetX. The sample size of control group (6,499,996) was determined by the size that can be effectively handled by our storage capacity and computational resources.
Our dataset took up 734 GiB of storage. We aimed to include everyone satisfying our age and medical history sufficiency requirements, with minimum exclusion to improve data quality.
Bootstrapping was performed with 16 repetitions; each repetition included all model development steps including automatic feature selection and took about an hour. We followed the algorithm for optimism-corrected bootstrapping described by Steyerberg, Section 5.3.4.29
Confidence intervals of AUCs were calculated with an optimised version of the DeLong's algorithm.30,31 Confidence intervals of binomial proportions (e.g., sensitivity, specificity) were calculated with the exact Clopper-Pearson method.32 Confidence intervals of the AUC mean in internal-external validation were calculated assuming a Gaussian mixture model to avoid the assumption of a global mean. The Supplemental Material (Section A.4) provides more details for calculation in simulated deployment.
Calculation of I2 for geographic/racial subgroups assumes a random effects model yi = μ + ϵi + Ei where μ is assumed as the “true” AUC, yi is the measurement of μ based on subgroup i, ϵi models the measurement uncertainty, and Ei ∼ N (0, τ2) models the heterogeneity between subgroups.33 We used the definition I2 = (Q − (k − 1))/Q.34 We calculated the Q statistics using the log-odds of AUCs to better match the normal assumption.35 We estimated Q's confidence interval using Monte Carlo simulation with 3 × 106 samples.
Role of funders
TriNetX provided cloud computing resources and access to the TriNetX data platform. A few TriNetX employees, listed as coauthors, made direct contributions to this research (see the Contributors section). Other funders did not have any role in the study design, data collection, data analyses, interpretation, or writing.
Results
Model evaluation
Both PrismNN and PrismLR used 35,387 patients with cancer and 1,500,081 controls up to 98.1 years old. Table 1 shows demographics, including sex, age, race, and HCO location. Fig. 1 presents a flowchart of dataset creation. The Supplemental Material (Table A1) has more demographic details.
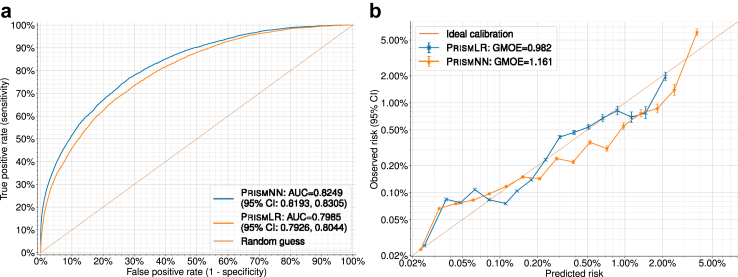
The average AUCs of PrismNN and PrismLR on nine random runs were 0.826 (95% CI: 0.824–0.828) and 0.800 (95% CI: 0.798–0.802), respectively. With bootstrapping, the optimism-corrected AUCs of. PrismNN and PrismLR were 0.825 (95% CI: 0.823–0.827) and 0.801 (95% CI: 0.799–0.804), respectively. Because our models incorporate the presence or absence of features, each feature is a predictor and we have no participants with missing predictors.22 The average GMOE on nine random runs was 1.169 (95% CI: 1.145–1.192) and 0.969 (95% CI: 0.945–0.993) for PrismNN and PrismLR, respectively.
Fig. 2a shows the ROC curve of one of the nine random runs, with AUCs being 0.825 (95% CI: 0.819–0.830) (PrismNN) and 0.798 (95% CI: 0.793–0.804) (PrismLR). Fig. 2b shows the corresponding log-scale calibration plots on the test set. GMOE was 1.161 (PrismNN) and 0.982 (PrismLR).
Fig. 2.
Model evaluation results. (a) ROC (Receiver Operating Characteristic) curves on the test set. (b) Log-scale calibration plots on the test set.
The PrismNN AUCs for different age groups were 0.847 (95% CI: 0.826–0.869), 0.796 (95% CI: 0.787–0.806), 0.775 (95% CI: 0.765–0.785), and 0.797 (95% CI: 0.790–0.804) for 40–49, 50–69, 70+, and 50+ years old, respectively. The corresponding GMOEs were 11.277, 1.057, 1.400, and 1.201. The PrismLR AUCs were 0.822 (95% CI: 0.799–0.846), 0.767 (95% CI: 0.757–0.777), 0.741 (95% CI: 0.730–0.752), and 0.766 (95% CI: 0.759–0.773) and GMOEs were 90.107, 0.804, 1.253, and 1.068. Fig. 3 presents all the selected features ranked by feature predictive power with PrismNN. Model features include known PDAC risk factors such as age, sex, diabetes mellitus, pancreatitis, pancreatic cysts, and abdominal pain; other features include hypertension, hypercholesterolemia, kidney function, and frequency of clinical visits preceding PDAC diagnosis. Figure A1 in the Supplemental Material presents how model performance varies with different numbers of selected features.
Fig. 3.
List of selected features ranked by univariate AUC of PrismNN. The label diag refers to diagnosis features, med to medication features, and lab to lab features. Letters in the brackets indicate the types of derived features: e for existence, fd for first date, ld for last date, p for time span, f for frequency, v for latest lab value, ve for whether a valid lab value exists, s for lab value slope, and se for whether lab value slope can be computed.
Internal-external validation results
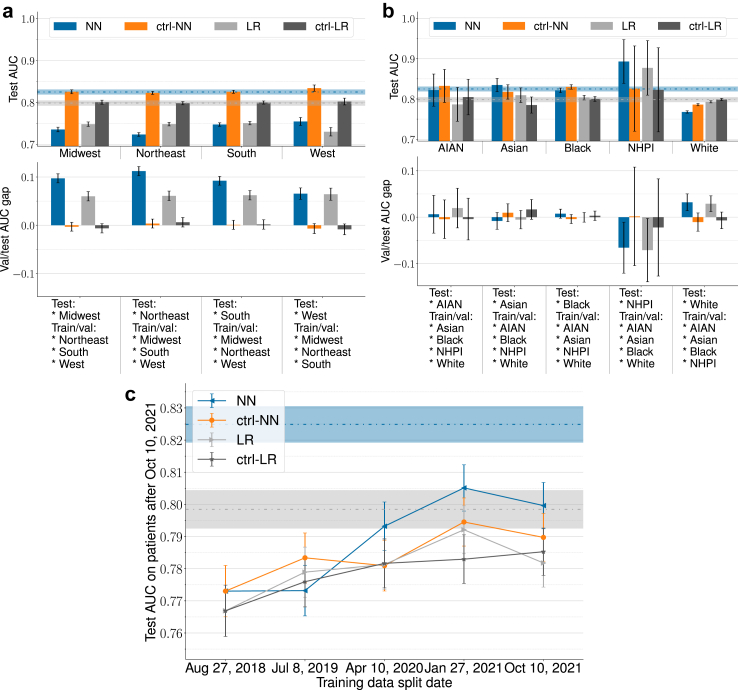
Fig. 4 shows the results for location-based, race-based, and temporal internal-external validation. The number of patients of each HCO location or racial group can be seen in Table 1. Note that as stated in Section 2.4, the results were obtained by training models on non-random data splits according to race/location/time. Fig. 4a presents location-based internal-external validation results. PrismNN AUCs on the test sets were 0.735 (95% CI: 0.730–0.741), 0.723 (95% CI: 0.719–0.728), 0.747 (95% CI: 0.743–0.751), and 0.754 (95% CI: 0.745–0.764) for the Midwest, Northeast, South, and West, respectively. PrismLR AUCs were 0.748 (95% CI: 0.743–0.753), 0.748 (95% CI: 0.744–0.753), 0.751 (95% CI: 0.746–0.755), and 0.730 (95% CI: 0.720–0.740). AUC drop between test and control models was between 0.078 and 0.099 for PrismNN, and between 0.049 and 0.072 for PrismLR. The average test AUCs on the four locations were 0.740 (95% CI: 0.716–0.764) and 0.744 (95% CI: 0.727–0.762) for PrismNN and PrismLR, respectively. The I2 indexes of PrismNN and PrismLR were 99.2% (95% CI: 86.7%–99.8%) and 95.9% (95% CI: 33.5%–98.8%), respectively.
Fig. 4.
Internal-external validation results. Error bars indicate 95% CI. Dashed horizontal lines and the surrounding regions indicate the original NN/LR test AUCs and 95% CI without attribute-based splitting. NN is short for PrismNN, LR short PrismLR, and ctrl- for control models that use random data split with matched training set size for race/location validation and use matched training size to separate the effect of data size increase for temporal validation. (a) Location-based internal-external validation. (b) Race-based internal-external validation. See notes under Table 1 for race abbreviations. (c) Model performance over time in temporal validation.
Fig. 4b presents race-based internal-external validation results. PrismNN AUCs on the test sets were 0.822 (95% CI: 0.782–0.862), 0.835 (95% CI: 0.818–0.851), 0.821 (95% CI: 0.816–0.827), 0.893 (95% CI: 0.839–0.947), and 0.768 (95% CI: 0.765–0.771) for AIAN, Asian, Black, NHPI, and White, respectively. The PrismLR AUCs were 0.787 (95% CI: 0.745–0.829), 0.809 (95% CI: 0.791–0.828), 0.803 (95% CI: 0.798–0.809), 0.877 (95% CI: 0.809–0.945), and 0.793 (95% CI: 0.790–0.796). AUC drop between test and control models was between −0.067 and 0.018 for PrismNN, and between −0.054 and 0.018 for PrismLR. The average test AUCs on the five races were 0.828 (95% CI: 0.744–0.912) and 0.814 (95% CI: 0.740–0.888) for PrismNN and PrismLR, respectively. The I2 indexes of PrismNN and PrismLR were 99.8% (95% CI: 92.9%–100.0%) and 96.4% (95% CI: 2.9%–99.2%), respectively.
Fig. 4c present temporal validation results. Models achieved average test AUCs 0.789 (95% CI: 0.762–0.816) (PrismNN) and 0.780 (95% CI: 0.763–0.798) (PrismLR). Performance tends to become better with more recent training data and larger training sets, but the change is not statistically significant.
Simulated deployment results
We simulated model deployment on 185,932 patients (with 7095 PDAC cases) in the test set, with enrolment from Apr 11, 2020 to Apr 6, 2021. Mean age at enrolment was 61.62 (SD 11.98). Mean age at PDAC diagnosis was 69.75 (SD 10.37). Each patient was followed up for 1.82 (SD 0.31) years (Table 2).
Table 2.
Simulated deployment results.
| Model | Risk level | Sensitivity | Specificity | PPV (TrxPop. Est.) | SIR (TrxPop. Est.) |
|---|---|---|---|---|---|
| PrismNN | 1 | 54.6% (53.4–55.8) | 85.3% (85.1–85.5) | 0.28% (0.27–0.29) | 2.38 (2.34–2.41) |
| 2 | 48.7% (47.5–49.9) | 89.1% (89.0–89.3) | 0.34% (0.33–0.35) | 2.87 (2.82–2.91) | |
| 3 | 35.9% (34.8–37.1) | 95.3% (95.2–95.4) | 0.58% (0.56–0.60) | 5.10 (5.02–5.18) | |
| 4 | 31.4% (30.4–32.5) | 96.9% (96.9–97.0) | 0.77% (0.74–0.80) | 7.07 (6.96–7.17) | |
| 5 | 17.3% (16.4–18.2) | 99.5% (99.5–99.6) | 2.81% (2.59–3.06) | 29.0 (28.6–29.5) | |
| 6 | 11.9% (11.2–12.7) | 99.9% (99.9–99.9) | 8.62% (7.42–10.0) | 96.0 (94.3–97.6) | |
| PrismLR | 1 | 52.3% (51.1–53.4) | 86.2% (86.1–86.4) | 0.29% (0.28–0.29) | 2.22 (2.19–2.25) |
| 2 | 46.4% (45.2–47.6) | 89.9% (89.7–90.0) | 0.35% (0.34–0.36) | 2.66 (2.61–2.70) | |
| 3 | 31.8% (30.7–32.9) | 95.4% (95.3–95.5) | 0.53% (0.50–0.55) | 4.06 (3.99–4.12) | |
| 4 | 26.4% (25.3–27.4) | 97.0% (96.9–97.1) | 0.66% (0.63–0.69) | 5.17 (5.09–5.25) | |
| 5 | 8.95% (8.30–9.64) | 99.6% (99.5–99.6) | 1.51% (1.36–1.67) | 13.1 (12.9–13.3) | |
| 6 | 2.93% (2.55–3.35) | 99.9% (99.9–99.9) | 2.88% (2.32–3.56) | 24.2 (23.7–24.7) |
Numbers in brackets are 95% CI.
PPV: Positive Predictive Value.
SIR: Standardised Incidence Ratio.
TrxPop. Est.: Estimation on the whole TriNetX population that accounts for unbalanced sampling.
All calculations were based on the outcome during the followup period of individual patients. Followup period was determined by the age and EHR data availability of each patient. More details can be found in Section 2.5 and in the Supplemental Material.
The estimated model PPV range on the whole TriNetX population was 0.28%–8.62% for PrismNN and 0.29%–2.88% for PrismLR. PrismNN and PrismLR SIR ranges were 2.38–96.0 and 2.22–24.2, respectively. The SIR of all the enrolled patients during the follow-up period was 1.00 (95% CI: 0.98–1.01). A SIR close to one indicates that our TriNetX test population with patient exclusion has similar PDAC incidence as the general US population.
We determined the high-risk group to be individuals with a SIR of 5.10 or above, based on PrismNN, which is correlated with a 35.9% sensitivity and 95.3% specificity. This SIR threshold is similar to the current eligibility cutoff for inclusion into screening programs.2
The Supplemental Material has more comprehensive simulated deployment results with more risk levels (Tables A2 and A3) and breakdowns of final model performance within different race/location/age/sex subgroups (Tables A4–A9).
Discussion
Our study leveraged routinely collected EHR data from a federated network including 55 US HCOs to develop and validate two families of models (PrismNN and PrismLR) for identifying patients in the general population at high PDAC risk, 6–18 months before the first PDAC diagnosis. Both models were trained on 35,387 PDAC cases and 1,500,081 controls with features derived from demographics, diagnosis, medication, and lab entries in EHR. Both models used 87 features (Fig. 3) automatically selected using the training data. PrismNN achieved better AUC than PrismLR on the test set, delivering test AUCs of 0.826 (95% CI: 0.824–0.828) and 0.800 (95% CI: 0.798–0.802), respectively. Bootstrapping gave similar AUC estimations. Both models showed worse performance for patients over 50 years old compared to over 40 years old, while PrismNN maintained better performance than PrismLR for different age groups; the deviation of GMOE from 1 indicates recalibration is needed if an age-based subgroup of patients is targeted. PrismNN showed worse average AUC in location-based validation than PrismLR, but performed favourably compared to PrismLR in other validations. The large capacity and flexibility of neural networks make them a good choice for modelling complex relationships in EHR data, but such capacity may hinder generalizability compared to simpler models. Although interpretation of neural networks is more challenging, our automatic feature selection provides insight into the reasoning process of the models.
We anticipate two potential clinical use cases for Prism. The first is to expand the eligibility for current screening programs that utilise imaging modalities such as Endoscopic UltraSound (EUS) and MRI/MRCP.6 Current eligibility criteria are based on familial PDAC or a known germline mutation syndrome (e.g., Lynch, Peutz-Jeghers).6 The identified population have a minimum lifetime SIR of 5 and includes only about 10% of PDAC cases.7,8 Depending on the chosen high-risk threshold, PrismNN exhibited a two-year SIR of 2.38–96.0. At a SIR of 5.10, PrismNN identified 35.9% of the PDAC cases as high risk 6–18 months before diagnosis, a significant improvement over current eligibility criteria.
The second use case is to identify an enriched group for lower overhead testing (such as biomarker testing) followed by screening based on the lower overhead test. In this use case, the model could be deployed at a higher sensitivity than in our first use case. For example, at 85.3% specificity, PrismNN exhibited 54.6% sensitivity.
To facilitate the integration of this model into clinical practice, a prospective study that examines the impact of implementing a PDAC risk model in real-time on screening behaviour is needed. This study would involve deploying the Prism models on patient EHR data and screening the high-risk group. The aim would be to evaluate both quantitative clinical outcome measures, such as the identification of a higher number of early-stage PDAC cases compared to typical hospital figures, and qualitative measures of model adoption by practitioners such as end-user (primary care provider) satisfaction.
We considered three types of internal-external validation to evaluate model generalizability. We split the dataset according to some attribute, trained models on one part, and tested the models on the other. Results of race-based validation highlight the generalizability across diverse racial populations. When tested on White, there was a small AUC drop, which we attribute to the fact that the White group constituted about 70% of the dataset while training was performed on the remaining 30% of data. Location-based validation showed modest AUC drops, implying potentially systematic differences between EHR data from geographically different HCOs. Additional validation is urged for model deployment to HCOs outside of the network. The high values of I2 indicate that model performance differences between locations or races were more likely due to intrinsic heterogeneity instead of randomness, while PrismLR exhibited slightly less heterogeneity. Temporal validation results showed good performance across time and suggested (with insufficient statistical significance) that model performance may improve with more recent training data and/or larger training data.
We evaluated the effectiveness of Prism in clinical implementation by simulated deployment. A key aspect is training models on data available before a simulated enrolment date to identify high-risk individuals after that date. We periodically evaluated PDAC risk for each individual and followed the identified high-risk individuals over time to evaluate model performance. This simulated deployment methodology contrasts previous methodologies that do not temporally separate the training and test data or test each individual at multiple cutoff dates.10,13 By tracking the envisioned deployment scenario more closely, we eliminated a potential source of inaccuracy and obtained a potentially more accurate prediction of model performance in clinical use. Simulated deployment also reveals model performance differences overlooked by traditional evaluation; although PrismNN and PrismLR have numerically close AUCs, the gap between their sensitivities in simulated deployment with SIR ≥5 is large.
A significant strength of our work is using a federated EHR network that ingests EHR data from multiple HCOs and presents data as a single format. This network enabled our three types of external validation and simulated deployment.
Integration with EHR systems is crucial in clinical deployment of risk prediction models. Without proper integration, clinicians must manually enter information into a program, which forms a significant barrier to model adoption.18 By contrast, federated networks allow seamless model integration due to their close interaction with existing HCO EHR systems. Federated networks provide a clear pathway for integrated model development, validation, and clinical deployment, all within a single platform.36
Other researchers have used EHR data to develop PDAC risk prediction models for the general population.10,13, 14, 15, 16 Data set sizes ranged from 1792 PDAC cases/1.8 M controls15 to 24,000 PDAC cases/6.2 M controls.14 Some studies lacked an external validation,13 completed the external validation/evaluated model generalizability only with data from a single geographic area,14,16 or validated only on one sex (male)15 or race.11 While some studies worked with data obtained from multiple organisations,13, 14, 15 none worked with a federated network that harmonises and standardises the data, none provided a clear path to clinical deployment, and none supported seamlessly deploying the model to new HCOs joining the federated network. Some previous studies evaluated the ability of their models to identify high-risk individuals either until or shortly before PDAC diagnosis,13, 14, 15 when the clinical benefit is improbable. By contrast, our evaluation focused on risk identification at least six months before diagnosis, when early-stage disease detection and potential cure are more likely.
Our study has a few limitations: (i) Model development and validation were retrospective. Prospective studies are needed to evaluate the efficacy of clinical detection of early-stage disease; (ii) Despite the favourable generalizability across racial groups demonstrated in our internal-external validation, certain racial groups may have biased presentation in our data because their socioeconomic status limits their access to the healthcare system. Future research should further evaluate the fairness of Prism, particularly concerning underrepresented groups; (iii) Although TriNetX incorporates a diverse set of US HCOs, they are still a small portion on the global scale; future work should evaluate Prism on more geographically diverse data. The lack of standardisation in data collection and the heterogeneity of EHR systems may have impacted model generalizability, as hinted by the PDAC prevalence differences and the performance drop in location-based validation; and (iv) Our study does not try to interpret the model reasoning process or extract clinical knowledge from models. Future work should improve model interpretability to make the decision process more reliable and transparent.
In conclusion, we have built, validated, and simulated the deployment of PDAC risk prediction models for the general population on multi-institutional EHR data from a federated network. Prism models can be used to help primary care providers across the country identify high-risk individuals for PDAC screening or used as a first filter before subsequent biomarker testing. Both PrismNN and PrismLR maintained their accuracy across diverse racial groups and geographic regions in the US and over time, outperforming widely-used clinical guideline criteria2,3 for PDAC screening inclusion.
Our approach enables potential expansion of the population targeted for screening beyond the traditionally screened minority with an inherited predisposition. Prism models set the stage for model deployment within the network to identify high-risk patients at multiple institutions within the network. The next step is a prospective study to validate the models before full clinical deployment.
Contributors
Conceptualisation: LA, MR, KJ, SK. Data acquisition: KH, JW, KJ. Data curation: KJ, MR, LA. Data verification: KJ, MR, LA. Data interpretation: KJ, MR, LA, MP, IDK. Project administration: LA, MR, KH. Supervision: MR, LA, SK, MP. ALL writing review and editing. ALL approved published version and agreed to be accountable for all aspects of the work.
Data sharing statement
The de-identified data in TriNetX federated network database can only be accessed by researchers that are either part of the network or have a collaboration agreement with TriNetX. We used data from the TriNetX database under a no-cost collaboration agreement between BIDMC, MIT, and TriNetX. Under this agreement, we accessed de-identified data under the agreements and institutional approvals already in place between TriNetX and their partner institutions. The data used in this study are stored on Amazon S3 storage under a TriNetX account and could be shared with future researchers who establish a collaboration with TriNetX.
Declaration of interests
KJ and MR are not aware of any payments or services, paid to themselves or MIT, that could be perceived to influence the submitted work. IK and LA are not aware of any payments or services, paid to themselves or BIDMC, that could be perceived to influence the submitted work. During the time the research was performed MR received consulting fees and payment for expert testimony for Comcast, Google, Motorola, Qualcomm, and IBM, is a member of the scientific advisory board and owns stock at Vali Cyber, and acknowledges support from Boeing, DARPA, and the NSF for salary and research support including meeting attendance and travel. MR has the following patents: United States Patent 10,539,419. Method and apparatus for reducing sensor power dissipation. Phillip Stanley- Marbell, Martin Rinard. United States Patent 10,135,471. System, method, and apparatus for reducing power dissipation of sensor data on bit-serial communication interfaces. Phillip Stanley-Marbell, Martin Rinard. United States Patent 9,189,254. Translating text to, merging, and optimizing graphical user interface tasks. Nathaniel Kushman, Regina Barzilay, Satchuthananthavale Branavan, Dina Katabi, Martin Rinard. United States Patent 8,839,221. Automatic acquisition and installation of software upgrades for collections of virtual machines. Constantine Sapuntzakis, Martin Rinard, Gautam Kachroo. United States Patent 8,788,884. Automatic correction of program logic. Jeff Perkins, Stylianos Sidiroglou, Martin Rinard, Eric Lahtinen, Paolo Piselli, Basil Krikeles, Timothy Anderson, Greg Sullivan. United States Patent 7,260,746. Specification based detection and repair of errors in data structures. Brian Demsky, Martin Rinard. JW is a TriNetX employee and owns TriNetX stock. The remaining authors declare no competing interests.
Acknowledgements
We are grateful to Gadi Lachman and TriNetX for providing support and resources for this work, and to Lydia González for her help with identifying and mitigating data quality issues. MR, LA, KJ acknowledge the contribution of resources by TriNetX, including secured laptop computers, access to the TriNetX EHR database, and clinical, technical, legal, and administrative assistance from the TriNetX team of clinical informaticists, engineers, and technical staff. LA acknowledges support from the Prevent Cancer Foundation for this work. MR and KJ received funding from DARPA and Boeing. MR also received funding from the NSF and Aarno Labs. SK, MP, JW, and KH are employees of TriNetX.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104888.
Contributor Information
Kai Jia, Email: jiakai@mit.edu.
Steven Kundrot, Email: steve.kundrot@trinetx.com.
Matvey B. Palchuk, Email: matvey.palchuk@trinetx.com.
Jeff Warnick, Email: jeff.warnick@trinetx.com.
Kathryn Haapala, Email: kathryn.haapala@trinetx.com.
Irving D. Kaplan, Email: ikaplan@bidmc.harvard.edu.
Martin Rinard, Email: rinard@csail.mit.edu.
Limor Appelbaum, Email: lappelb1@bidmc.harvard.edu.
Appendix A. Supplementary data
Material
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Goggins M., Overbeek K.A., Brand R., et al. Management of patients with increased risk for familial pancreatic cancer: updated recommendations from the International Cancer of the Pancreas Screening (CAPS) consortium. Gut. 2020;69:7–17. doi: 10.1136/gutjnl-2019-319352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daly M.B., Pal T., AlHilli Z., et al. 2023. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic.https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf [Google Scholar]
- 4.Aslanian H.R., Lee J.H., Canto M.I. AGA clinical practice update on pancreas cancer screening in high-risk individuals: expert review. Gastroenterology. 2020;159:358–362. doi: 10.1053/j.gastro.2020.03.088. [DOI] [PubMed] [Google Scholar]
- 5.Lu C., Xu C.F., Wan X.Y., Zhu H.T., Yu C.H., Li Y.M. Screening for pancreatic cancer in familial high-risk individuals: a systematic review. World J Gastroenterol. 2015;21:8678. doi: 10.3748/wjg.v21.i28.8678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canto M.I., Harinck F., Hruban R.H., et al. International cancer of the pancreas screening (CAPS) consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut. 2013;62:339–347. doi: 10.1136/gutjnl-2012-303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphris J.L., Johns A.L., Simpson S.H., et al. Clinical and pathologic features of familial pancreatic cancer. Cancer. 2014;120:3669–3675. doi: 10.1002/cncr.28863. [DOI] [PubMed] [Google Scholar]
- 8.Petersen G.M. Familial pancreatic cancer. In: Semin Oncol. Vol. 43. Elsevier; 2016. pp. 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owens D.K., Davidson K.W., Krist A.H., et al. Screening for pancreatic cancer: US preventive services task force reaffirmation recommendation statement. JAMA. 2019;322:438–444. doi: 10.1001/jama.2019.10232. [DOI] [PubMed] [Google Scholar]
- 10.Baecker A., Kim S., Risch H.A., et al. Do changes in health reveal the possibility of undiagnosed pancreatic cancer? development of a risk-prediction model based on healthcare claims data. PLoS One. 2019;14 doi: 10.1371/journal.pone.0218580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J., Yuan C., Babic A., et al. Genetic and circulating biomarker data improve risk prediction for pancreatic cancer in the general population. Cancer Epidemiol Biomarkers Prev. 2020;29:999–1008. doi: 10.1158/1055-9965.EPI-19-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein A.P., Lindström S., Mendelsohn J.B., et al. An absolute risk model to identify individuals at elevated risk for pancreatic cancer in the general population. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q., Cherry D.R., Nalawade V., et al. Clinical data prediction model to identify patients with early-stage pancreatic cancer. JCO Clin Cancer Inform. 2021;5:279–287. doi: 10.1200/CCI.20.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Placido D., Yuan B., Hjaltelin J.X., et al. A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories. Nat Med. 2023;29:1–10. doi: 10.1038/s41591-023-02332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W., Zhou Y., Xie F., et al. Derivation and external validation of machine learning-based model for detection of pancreatic cancer. Am J Gastroenterol. 2022;118(1):157–167. doi: 10.14309/ajg.0000000000002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Appelbaum L., Cambronero J.P., Stevens J.P., et al. Development and validation of a pancreatic cancer risk model for the general population using electronic health records: an observational study. Eur J Cancer. 2021;143:19–30. doi: 10.1016/j.ejca.2020.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Muhammad W., Hart G.R., Nartowt B., et al. Pancreatic cancer prediction through an artificial neural network. Front Artif Intell. 2019;2:2. doi: 10.3389/frai.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Videha Sharma I.A., van der Veer S., Martin G., Ainsworth J., Augustine T. Adoption of clinical risk prediction tools is limited by a lack of integration with electronic health records. BMJ Health Care Inform. 2021;28 doi: 10.1136/bmjhci-2020-100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Surveillance, epidemiology, and end results (SEER) program SEER∗stat database: incidence SEER research limited-field data, 22 registries, Nov 2021 sub (2000-2019) - linked to county attributes time dependent (1990-2019) income/rurality, 1969-2020 counties, National Cancer Institute, DCCPS, Surveillance Research Program, Released April 2022, based on the November 2021 submission. 2022. [Google Scholar]
- 20.Topaloglu U., Palchuk M.B. Using a federated network of real-world data to optimize clinical trials operations. JCO Clin Cancer Inform. 2018;2:1–10. doi: 10.1200/CCI.17.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen H., Vedsted P., Møller H. Consultation frequency in general practice before cancer diagnosis in relation to the patient's usual consultation pattern: a population-based study. Cancer Epidemiol. 2018;55:142–148. doi: 10.1016/j.canep.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Agniel D., Kohane I.S., Weber G.M. Biases in electronic health record data due to processes within the healthcare system: retrospective observational study. BMJ. 2018;361 doi: 10.1136/bmj.k1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia K., Rinard M. Effective neural network L0 regularization with binmask. arXiv. 2023 [Google Scholar]
- 24.Platt J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Adv Large Margin Classifiers. 1999;10:61–74. [Google Scholar]
- 25.Steyerberg E.W., Harrell F.E. Prediction models need appropriate internal, internal–external, and external validation. J Clin Epidemiol. 2016;69:245–247. doi: 10.1016/j.jclinepi.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riley R.D., Ensor J., Snell K.I., et al. External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ. 2016;353 doi: 10.1136/bmj.i3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sperrin M., Riley R.D., Collins G.S., Martin G.P. Targeted validation: validating clinical prediction models in their intended population and setting. Diagn Progn Res. 2022;6:24. doi: 10.1186/s41512-022-00136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porter N., Laheru D., Lau B., et al. Risk of pancreatic cancer in the long-term prospective follow-up of familial pancreatic cancer kindreds. J Natl Cancer Inst. 2022;114:1681–1688. doi: 10.1093/jnci/djac167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steyerberg E. Statistics for biology and Health. 2nd ed. Springer International Publishing; 2019. Clinical prediction models: a practical approach to development, validation, and updating. [Google Scholar]
- 30.Sun X., Xu W. Fast implementation of DeLong's algorithm for comparing the areas under correlated receiver operating characteristic curves. IEEE Signal Process Lett. 2014;21:1389–1393. [Google Scholar]
- 31.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 32.Clopper C.J., Pearson E.S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- 33.Biggerstaff B., Tweedie R. Incorporating variability in estimates of heterogeneity in the random effects model in meta-analysis. Stat Med. 1997;16:753–768. doi: 10.1002/(sici)1097-0258(19970415)16:7<753::aid-sim494>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 34.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 35.Klaveren D van, Steyerberg E.W., Perel P., Vergouwe Y. Assessing discriminative ability of risk models in clustered data. BMC Med Res Methodol. 2014;14:1–10. doi: 10.1186/1471-2288-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rieke N., Hancox J., Li W., et al. The future of digital health with federated learning. NPJ Digit Med. 2020;3:1–7. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Material