Abstract
Enzymes from psychrophilic (cold-loving) organisms have attracted considerable interest over the past decades for their potential in various low-temperature industrial processes. However, we still lack large-scale commercialization of their activities. Here, we review their properties, limitations and potential. Our review is structured around answers to 5 central questions: 1. How do cold-active enzymes achieve high catalytic rates at low temperatures? 2. How is protein flexibility connected to cold-activity? 3. What are the sequence-based and structural determinants for cold-activity? 4. How does the thermodynamic stability of psychrophilic enzymes reflect their cold-active capabilities? 5. How do we effectively identify novel cold-active enzymes, and can we apply them in an industrial context? We conclude that emerging screening technologies combined with big-data handling and analysis make it reasonable to expect a bright future for our understanding and exploitation of cold-active enzymes.
Keywords: Psychrophilicity, Enzyme catalysis, Thermodynamic stability, Protein dynamics, Bioprospecting
Abbreviations
- ΔCp
heat capacity
- ΔGD-N
free energy of unfolding
- ΔG‡
free energy of activation
- ΔH‡
enthalpy and of activation
- ΔS‡
entropy of activation
- Ea
activation barrier
- kB
Boltzmann's constant
- kcat
catalytic reaction rate
- Km
Michaelis constant
- T
temperature in Kelvin
- t
temperature in oC
- Tc
cold unfolding midpoint
- tm
melting temperature
- topt
optimal temperature
- TS
transition state
Introduction to psychrophilicity
Our planet harbors a great diversity of habitats in which organisms have adapted to live and thrive within an enormous range of temperature, salinity, pH, humidity and pressure. Of these, temperature adaptation is perhaps the most fascinating phenomenon. Hyperthermophiles grow optimally at up to 105°C, e.g. in deep-sea hydrothermal vents [1]; in contrast, psychrophiles are adapted to temperatures well below 20°C. Since the average temperature on Earth is around 14°C, psychrophiles are not niche species but make up a significant part of biomass, in particular polar regions, mountains and the deep ocean [2]. They also occur in air samples, permafrost and freshwater regions [3]. Two methanogenic archaea have been isolated from the Antarctic Ace Lake [4], while the few known eukaryotic psychrophiles include yeast [5], lichen [6], ciliates [7] and an Antarctic midge [8]. However, most psychrophiles are bacteria. They are now divided into “narrow” or stenopsychrophiles (formerly true psychrophiles, restricted to < 20°C, often optimal at 5-15°C [9]) and “broad” or eurypsychrophiles (psychrotolerants, optimal growth > 20°C but able to grow at temperatures as low as -15°C). Both groups can be considered cryophiles, i.e. able to grow < 0°C.
Psychrophiles are less studied than other extremophiles, partly due to the more obvious applications of temperature- or acid-resistant enzymes, but also because of their general fragility under normal working conditions. However, their unique survival strategies may provide insight into extraterrestrial life, e.g. habitability of Mars, not least because they sometimes combine cold tolerance with ability to withstand high pressure (deep sea sediments) or high salinity (Arctic permafrost) [9]. Low temperatures affect many aspects of life to which psychrophiles have to adjust. The cell membrane has adapted to the reduction in dynamics, permeability and tensile strength by using more branched, unsaturated and short-length lipids [10] as well as modified pigment molecules [11], while the peptidoglycan can increase in thickness and tensor strength to protect against ice formation and osmotic pressure [12]. Other tricks include cryoprotection through accumulation of soluble osmolytes (glycerol, betaine, trehalose) [13] and reduction of ice nucleation through secretion of complex carbohydrates as a protective matrix [14] or antifreeze proteins [15]. Cold shock proteins can be produced to increase translation efficiency [16] or counter cold-denaturation which would otherwise compromise protein folding [17]. Perhaps counterintuitively, the production of toxic reactive oxygen species increases at low temperatures due to the rise in oxygen solubility; this is countered by downregulation of ROS-producing pathways such as glycolysis and the tricarboxylic acid cycle [18].
Irrespective of all these measures, a fundamental challenge remains: how to maintain a metabolically viable level of enzyme activity at these low temperatures? This challenge can be split up into five critical questions:
-
1.
How do cold-active enzymes achieve high catalytic rates at low temperatures?
-
2.
How is protein flexibility connected to cold-activity?
-
3.
What are the sequence-based and structural determinants for cold-activity?
-
4.
How does the thermodynamic stability of psychrophilic enzymes reflect their cold-active capabilities?
-
5.
How do we effectively identify novel cold-active enzymes, and can we apply them in an industrial context?
We will address each of these in turn in the next sections.
1. How do cold-active enzymes achieve high catalytic rates at low temperatures?By switching from enthalpic to entropic activation, involving a more dynamic active site and decrease in substrate specificity
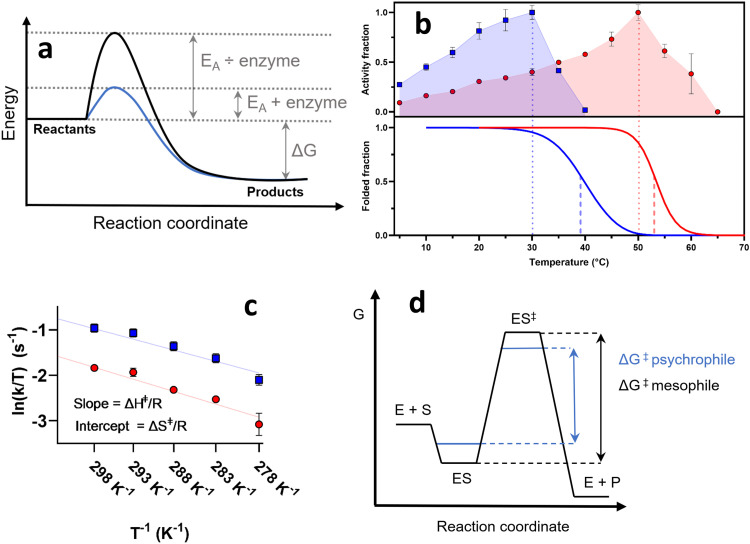
The cold activity of enzymes is critical for psychrophiles to maintain sufficiently high chemical reaction rates at low temperatures. Enzymes catalyze chemical reactions primarily by lowering the activation barrier Ea, that is, by stabilizing the transition state (TS) separating substrate and product [19] (Fig. 1a). Empirically, the relationship between temperature T and the catalytic reaction rate (kcat) can be described through the Arrhenius equation [20]:
| (1) |
where A is an empirical pre-exponential factor. Lowering T lowers kcat, which for mesophilic enzymes typically falls 20- to 250-fold between 37°C and 0°C, effectively preventing metabolic activity at low temperatures [21,22]. Psychrophiles have two options to counteract this: either upregulate the expression of the insufficiently active enzymes or adapt the enzymes towards more efficient catalysis at lower temperatures. Continuous upregulation is energetically costly and appears to be rare [23,24]. Instead, psychrophiles lower Ea, leading to a characteristic decrease in optimal temperature (topt) that is most often accompanied by lower thermal stability, i.e. melting temperature (tm), compared to their meso- and thermophilic homologues [21] (Fig. 1b). (Note that T is in Kelvin and t in °C). Notably, psychrophilic enzymes show a characteristic temperature difference between topt and tm [25], [26], [27]. A more sophisticated approach is provided by TS theory, in which the enzyme E in complex with the substrate S (ES) is in equilibrium with the TS (ES‡), separated by a free energy of activation ΔG‡. Breakdown of ES‡ leads to the formation of product (P). ΔG‡ can be split up into the enthalpy and entropy of activation (ΔH‡ and ΔS‡), i.e. ΔG‡ = ΔH‡ - TΔS‡, so that the Arrhenius equation can be rewritten as:
| (2) |
Here kB is Boltzmann's constant. While TΔS‡ tends to be an order of magnitude smaller than ΔH‡, it plays an important role in psychrophilicity: cold-adapted enzymes have redistributed contributions to ΔG‡ from ΔH‡ to ΔS‡ (reducing the magnitude of the positive value of ΔH‡ and correspondingly increasing the positive value of -TΔS‡, i.e. a more negative value of ΔS‡), which allows maintaining catalytic activity despite a decrease in temperature [28]. This is because the ΔS‡/R component is temperature independent, in contrast to ΔH‡/RT (Fig. 1c); it is the latter which reduces kcat exponentially with temperature. The upshot of all this is that psychrophiles’ rates do not decline so much at low temperatures as mesophiles’ rates.
Fig. 1.
Thermodynamic aspects of enzyme catalysis. (a) Schematic of activation energy (EA) as a barrier between the reactant and the products. Enzymes catalyze reactions by lowering EA. (b) Activity versus temperature (upper box) of two structurally homologous psychrophilic (blue) and mesophilic (red) enzymes and a comparison with structural unfolding (lower box) (J.S.N. and D.E.O., unpublished data). topt is represented by dotted lines, while tm is shown as a stippled line. (c) Eyring plot of the catalytic rates in the range 5-25°C of structurally homologous psychrophilic (blue) and mesophilic (red) enzymes from panel b. The slope of the mesophilic enzyme is 13% steeper than the psychrophilic, while the secondary axis intercept is 7% higher for the psychrophilic enzyme. (d) Transition state diagram according to Eyring-Polanyi. A reduction of the stability of ES (and possibly an increase in the stability of ES‡) in a psychrophile can lead to a lower ΔG‡.
This redistribution has been reported for many psychrophilic enzymes and provides a quantifiable explanation of how psychrophiles reduce the exponential dampening of activity at low temperatures [29], [30], [31]. Although the structural origin of this phenomenon is not yet fully understood, in broad terms, it is believed to be achieved by a more flexible protein structure - in many cases combined with a larger and more dynamic and accessible active site [30,32]. Increased flexibility leads to a more dynamic or “softer” catalytic site where a smaller amount of bonds between enzyme and substrate must be formed and broken during the formation of the TS, thereby lowering ΔH‡ and effectively reducing ΔG‡ (Fig. 1d) [32]. Nevertheless, a more negative ΔS‡ compensates for this to some extent, as a consequence of increased structural disorder in the ground state (ES) that may also explain ES’ relative loss of specificity (increased Km, see below) and remarkable heat lability [33]. In addition, the TS (ES‡) itself has also been postulated to be stabilized by a comparatively greater disorder or enhanced water displacement; some recent computer simulations show how delocalization of charge during bond hydrolysis also could lead to favorable entropy of activation [28]. All of this contributes to the further lowering of ΔG‡.
Notably, psychrophilic enzymes tend to have a higher Michaelis constant (Km) than their mesophilic counterparts. This reflects their looser structure, which can also lead to either broader specificity [34] or faster product release [35,36]. To complicate matters, there are also examples of lower Km values for psychrophiles [37,38]. It is challenging to prove the relationship between KM values among individual psychrophilic enzymes and their mesophilic counterparts. Indeed, even closely related proteins may be optimized to bind substrates with different affinities according to their availability in natural environments [39,40]. To tackle these issues, some researchers have selected highly conserved house-keeping enzymes that are not likely to differ in substrate specificity or catalytic mechanism, such as triosephosphate isomerases or lactate dehydrogenases, and combined molecular dynamic (MD) simulations with laboratory experiments [30,41]. As MD is a powerful tool for understanding the thermodynamics of enzyme catalysis, this approach could be extended by performing MD simulations as a function of temperature combined with systematic mutagenesis of individual psychrophilic enzymes’ temperature-dependent activity. Such work could truly help to map out the molecular and mechanistic basis for psychrophilicity.
2. How is flexibility connected to activity? The active site is more flexible and hydrophilic than in meso/thermophiles; local and sometimes global flexibility is also increased by chain loop modifications
2.1. Flexibility around the active site
The notion of increased disorder during substrate binding and formation of the TS suggests that mainly the active site of psychrophilic enzymes should be the object of adaptive changes. In support of this, psychrophilic enzymes often lose activity at temperatures well below global unfolding (Fig. 1b), suggesting exceptional heat-lability of psychrophilic active sites. However, the large number of structures of cold-adapted enzymes reveals strict conservation of the architecture of the active sites [42], and the increased dynamics of catalytic residues of cold-adapted enzymes have been linked to the global flexibility of the protein structure [30,43]. Nevertheless, the heat-labile activity of psychrophilic enzymes suggests dynamics-inducing structural changes that promote diffusion in and out of the site and the changes in the ES and ES‡ complexes mentioned earlier [34]. Examples include removal of bulky hydrophobic residues resulting in a more spacious catalytic cleft [44], changes or deletions in loops near the active site leading to increased dynamics [45,46], and alterations in metal binding sites, inducing enlargement of the active site opening [47]. Especially polysaccharide-binding enzymes, such as α-amylases, have been shown to contain highly dynamic active sites that exhibit broad substrate specifity [25,26,48]. Another feature of psychrophilic enzyme structures is increased electrostatic potential in and around the active site. Increased hydrophilicity at the catalytic cleft may be an adaptive feature to optimize Km to low temperature as the strength of hydrophobic interactions (driven by the release of water molecules) falls drastically at lower temperatures [49].
These examples show that local regions with increased flexibility, which are responsible for the high catalytic rates at low temperatures, are particularly heat-labile compared to the rest of the structure and this results in the characteristic loss of activity far below the observable tm. Nevertheless, other explanations are also possible. Recent computational studies of a psychrophilic α-amylase suggested early thermal inactivation to be caused either by a formation of a dead-end substrate-enzyme complex, whose formation is more favourable at high temperatures, or a difference in heat capacity between the ground and TS (ΔCp‡) [50]. Common for both explanations is that the TS is more favored at lower temperatures than at higher temperatures, rather than local unfolding.
2.2. How do we measure protein flexibility?
Intuitively one might expect that a combination of low stability and high heat sensitivity implies high flexibility, which is induced either locally or globally by structural adaptations to psychrophilic conditions. However, flexibility is a complex structural phenomenon that requires a direct comparison of closely related proteins within well-defined conditions to draw concrete conclusions. To complicate matters further, flexibility can either be static or dynamic depending on which technique is chosen to measure this phenomenon.
Static flexibility accounts for the structural degree of freedom, i.e., how many different separate and stable states in which the protein can exist. This is most directly measured by hydrogen-deuterium exchange (HDX) which determines how quickly amide protons can be exchanged with solvent. This is normally measured under conditions (neutral pH or EX2 exchange) which report on the equilibrium distribution between states rather than their rate of interconversion [51]. HDX is normally measured by mass spectrometry (MS) [52] or (for smaller proteins) nuclear magnetic resonance spectroscopy (NMR), though Fourier-transformed infrared (FTIR) spectroscopy can also be used [53]. Structural techniques such as X-ray crystallography or cryogenic electron microscopy (cryo-EM) may also provide information about the number of different conformations through the atomic B-factors, also known as the temperature factors or atomic displacement factors. Although the origin of B-factors of the respective techniques is fundamentally different, they both express the function of atom placement uncertainty, thereby indicating static flexibility within a protein structure. However, biases caused by differences in experimental setups and data processing suggest that B-factors should be interpreted within a structure rather than by comparing different proteins [54,55]. For example, a cold-adapted malate dehydrogenase displayed two-fold higher B-factor values in the binding cavity than its mesophilic counterpart despite having a lower overall B-factor [56]. Nevertheless, many psychrophilic enzymes show significantly higher B-factors than their mesophilic homologs, especially in specific secondary structures such as long turns and β-strands [57].
Dynamic flexibility is time-resolved and describes how fast the different conformations exchange. This can be studied experimentally by Trp fluorescence quenching or proteolytic digestion [58] or in silico by molecular dynamics (MD) simulations [30,43]. Small quenchers such as acrylamide or iodide penetrate a protein structure over time; the more flexible the overall structure is, the faster the quencher will infiltrate the interior of a protein and collide with the Trp residue, leading to greater overall quenching [59]. Comparative studies of homologous α-amylases show that quenching by acrylamide correlates to their melting points, indicating that low thermal stability can be linked to an increase in overall flexibility [25]. Proteolytic nicking by broad-spectrum proteases (e.g. proteinase K or pepsin) identifies local regions of protein flexibility [32], which can be combined with MS to identify sites [29]. When proteolysis-MS is done in parallel with HDX, it can powerfully differentiate between protons in folded and disordered regions [60,61]. Finally, MD simulations follow the movements of individual atoms on ns time scales. The strength of the MD approach lies in its versatility; the researcher can relatively easily define the temperature, solvent contents, protein sequence, and even restrain certain parts of the structure [30,62]. MD simulations are a powerful means to understand the concept of flexibility, especially when combined with experimental results, as shown by Åqvist and colleagues [28].
2.3. Evidence for the role of local and global flexibility
In many psychrophilic enzymes, the increase in flexibility is separated from the active site, nevertheless leading to an entropic shift in TS formation [32,63]. This raises the question of whether specific regions enhance flexibility (local flexibility) or whether overall flexibility resonates throughout the structure (global flexibility).
Local flexibility is supported by the identification of specific regions directly responsible for the increased dynamics though often separate from the active site. For example, psychrophilic cellulase from P. haloplanktis has unusually long linkers that lead to a larger number of steric conformations [64] while a cold-adapted xylanase has two particularly long surface-loops thought to allow an increase in movement of the neighbouring secondary structures [65]. Dynamic surface loops seem crucial for the increased flexibility of many psychrophilic enzymes, which tend toward increased length and comparatively high Gly and low Pro content [45,62,[66], [67], [68]]. Inserting Pro residues into an extended loop of the cold-adapted subtilisin S41 substantially reduced its conformational mobility and increased its thermostability [68]. Similarly, shortening a uniquely long surface loop L3 in a psychrophilic β-Glucosidase decreased its kcat at low temperatures [69], just as deletion of a whole surface loop in a Streptomyces phospholipase reduced kinetics and increased stability [70]. Interestingly, shifting mesophilic enzymes towards psychrophilic behaviour is rarely reported. However, insertion of a prolonged loop into mesophilic subtilisin from B. lentus (to mimic its psychrophilic homologue from Bacillus TA39) led to a hybrid enzyme with increased flexibility and more cold-adapted behavior [71].
Global flexibility: Flexibility can be more subtly distributed over the whole protein structure, as seen in the thoroughly investigated psychrophilic α-amylase from P. haloplanktis [25,72]. MD simulations of cold-adapted salmon trypsin have shown that its temperature dependence of catalysis is defined mainly by surface flexibility [63]. In this elegant study, the movement of the outer atoms was systematically restrained, which caused the psychrophilic enzyme to approach the catalytic parameters of the mesophilic enzyme. In an interesting twist, MD simulations of the aforementioned hybrid subtilisin S41 showed that insertion of the prolonged loop induces increased global flexibility [71].
To summarize, flexibility is undeniably essential for high catalytic rates at low temperatures, whether global or local, and described from either a static or dynamic point of view. However, we have not yet seen the full picture of flexibility in connection to catalysis, structure and stability of psychrophilic enzymes. To disentangle these phenomena, simulations of molecular motion (MD) and catalysis (quantum mechanics coupled with molecular mechanics, i.e. QM/MM) should be combined with experimental catalysis and structural analysis, as exemplified by a study of a cold-adapted lipase and its mutants [73].
3. What are the sequence-based and structural determinants of cold-activity? Psychrophiles increase flexibility and reduce stability via more Gly clusters and Met residues, fewer disulfide bridges and Pro residues, weaker metal ion binding, more aromatic residues on the surface, a destabilized hydrophobic core, helix kinks, weakened dipoles and long loops and turns; an altered level of oligomerization may also contribute
It can be difficult to identify psychrophilicity-promoting adaptations in enzyme sequences, as they may hide among changes induced by genetic drift or combine with adaptations towards other extreme conditions, such as high salinity or pressure. Traditionally, most insight has been obtained by comparing closely related protein homologues from different temperature optima. More recently, comprehensive genomic, transcriptomic, and proteomic studies of psychrophilic organisms have expanded our insight further. Here we summarize different molecular adaptations found in the structures of psychrophilic enzymes.
3.1. Global differences in sequence content
-
•
Increased Met content: This may be rationalized by Met's high mobility and lack of charge [58]. Met biosynthesis is also upregulated in cryophilic organisms at low temperatures [74].
-
•
Decrease in disulfide bridges: Disulfide bridges are generally linked to thermostability [32], though specifically located disulfide bridges in a psychrophilic α-amylase have counterintuitively induced local destabilization of the active site [75].
-
•
Increased Gly clusters: Gly residues have high intrinsic flexibility [33]. Gly clusters, which consist of at least two Gly residues in succession, contribute to the lowering of EA in an Antarctic alkaline phosphatase [76].
-
•
Low Pro content [21,77]: Introduction of Pro residues generally increases rigidity in otherwise flexible regions [78]. In addition, cis-trans isomerization of Pro's peptidyl-prolyl bond is often rate-limiting for protein folding. This particularly impacts psychrophilic proteins due to a decrease in the rate of isomerization as the temperature falls [42].
3.2. The protein surface
The surface of psychrophilic enzymes often differs from their warm-adapted homologues through mutations that increase their flexibility. This is critical for low-temperature activity [63].
-
•
Reduced Arg/Lys ratio: This is especially seen on the surface [54,56,64,79]. Arg can increase thermostability, as its guanidino group can form more electrostatic interactions than Lys’ amino group [33].
-
•
Increased (negative) surface charge: This is thought to counteract the increase in the dielectric constant of water at low temperatures [32] and increase flexibility by the repulsion of surface charges [80].
-
•
More aromatic surface residues [69,71,79,80]: Water forms clathrates (crystalline-like structures) around these aromatic residues due to the entropically driven hydrophobic effect, whose strength decreases at low temperatures [49,81]. Increased water mobility around the aromatic residues at low temperatures leads to a more flexible surface [4].
-
•
Fewer salt bridges: An absence of critical salt bridges can contribute to the protein's thermolability [45,79]. However, examples of the contrary are also known [4].
-
•
Weaker metal ion binding: Binding of metal ions such as Ca2+ and Mg2+ will stabilize the native state [33]. Several psychrophilic enzymes show lower binding affinity for, and thus lower stabilization by, these ions [26,82].
3.3. Protein core
Buried hydrophobic residues in the core of a protein are essential to its stability but have to be carefully chosen to fit sterically. The incorporation of either smaller or larger residues may lead to destabilization or misfolding, respectively [83]. Destabilization can however be important to increase general flexibility and the tendency to unfold.
-
•
Smaller hydrophobic residues in the core: Psychrophilic enzymes tend to incorporate smaller and branched aliphatic internal residues (e.g., Ala or Ile) instead of large and bulky residues (e.g., Phe or Trp), leading to a less dense hydrophobic core [45,55,84].
-
•
Larger internal cavities: Less dense hydrophobic core packing leads to an increase in volume and number of solvent-accessible cavities leading to an increase in internal motions of the cold-adapted proteins [57].
3.4. Secondary structure
α-helices and β-sheets in psychrophilic enzymes exhibit increased static flexibility, heterogeneously distributed throughout the structure, compared to their mesophilic counterparts [57,85]. This can be caused by the following:
-
•
α-helix breaks: Central Pro residues in α-helices can destabilize psychrophilic enzymes by inducing a kink in the helix (the helix breaker effect) due to the distinctive cyclic structure of the pyrrolidine group [86] and the loss of H-bonding. Note that introducing Pro into flexible regions has the opposite effect [45].
-
•
Charged helix caps: α-helices tend to have a cumulatively positive charge towards the N-terminal part of the helix (N-cap) and negatively charged at the C-terminal part (C-cap), leading to a charge-dipole that stabilizes the secondary structure [87]. This is weakened in psychrophilic enzymes, decreasing their stability [88].
-
•
Long and flexible loops and turns: Lowered Pro and increased Gly content in loops, both increasing degrees of freedom, are common to many cold-adapted enzymes [26,56,82,84]. Loops and turns are often extraordinarily long, resulting in large, flexible surface regions [64,89] which contribute to psychrophilic flexibility. This increased flexibility increases the contribution of ΔS‡ to ΔG‡ [30,90].
3.5. Oligomeric assembly
Changes in subunit interfaces and oligomeric assembly have been proposed as an adaptive mechanism towards cold activity; however, it is not universally accepted. A recent MD study of a psychrophilic short-chain dehydrogenase showed that thermodynamic activation parameters were insensitive to the oligomeric state of the protein, despite increased mobility of the solvent-exposed interface residues [91]. Nevertheless, in many other cases, the level of oligomerization may be important:
-
•
Weaker subunit interfaces: Interfaces of oligomeric assemblies in psychrophilic enzymes are significantly different from their meso- and thermophilic homologs, e.g. fewer H-bonds and fewer hydrophobic interactions, but an increase in ion-pairs [92]. This also leads to increased thermolability of cold-adapted enzymes [45].
-
•
Changes in oligomeric assembly: Increased oligomerization (more subunits per oligomer) in thermophilic proteins is linked to thermostabilization [93,94]. Analogously, reduced oligomerization is often seen in psychrophilic enzymes [95,96]. Interestingly, increased oligomerization could also be a cold-adaptive strategy when multimerization increases flexibility at surface-exposed patches around the active site [97] or an increase in the number of internal cavities [98].
Our own investigation of a psychrophilic β-galactosidase shows a clear link between multimeric assembly and stability (J.S.N. and D.E.O., unpublished); however, there is as yet no obvious connection to low temperature optimum. An obvious next step is to disassemble the oligomer by mutations in the inter-chain interface and investigate the impact of this disassembly on the temperature profile.
4. How does the thermodynamic stability of psychrophilic enzymes reflect their cold-active profile? They have a narrow window of thermal stability and need to adapt to avoid cold denaturation
Although both kcat and KM tend to increase in psychrophilic enzymes, their kcat/KM values (the catalytic efficiency constant) remain similar to mesophilic homologs near their respective topt, prompting the now widely accepted activity-stability trade-off axiom. That is, the shift in the thermodynamic parameters of activation (eq. 2) leads to enzymes with higher turnover rates through an increase in entropy of activation but with a cost in stability. Although the inverse activity-stability relationship is somewhat reductionistic and cannot explain the molecular mechanisms of adaptation, it can help us understand the fundamental principles that drive the evolution of psychrophilic enzymes.
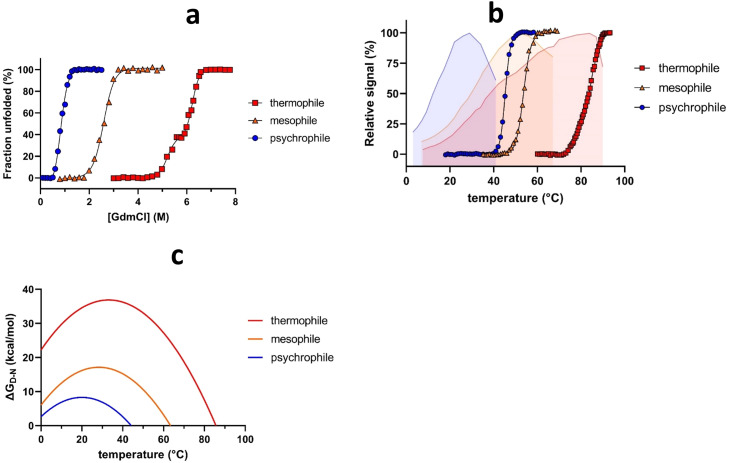
Protein folding is mainly driven by hydrophobic interactions forming the hydrophobic core, together with contributions from van der Waals forces, H-bonds, and to some degree, salt bridges, aromatic interactions, disulfide bridges, and an array of higher-order structural factors [99]. Unfolding curves of a psychrophilic α-amylase from P. haloplanktis and its meso- and thermophilic structural homologs (Fig. 2a) reveal that this cold-active enzyme has a comparatively lower midpoint of denaturation ([den]50%) and higher slope of denaturation (mD-N), suggesting lower stability and increased cooperativity of unfolding, respectively [25]. Similar relative destabilization in chemical denaturants is seen for other psychrophilic enzymes, including a superoxide dismutase from P. haloplanktis unfolding with a [den]50% of 0.9 M GdmCl compared to the 2.6 M GdmCl in the porcine homologue [100]; also, two variants of γS-Crystallin protein from the Antarctic Nototheniid Toothfish unfold at 3.8 and 5.6 M urea while the human homolog unfolds at 6.3 M urea [101].
Fig. 2.
Stability profiles of different temperature classes of enzymes. (a) Chemical unfolding of a psychro-, meso- and thermophilic amylase using the denaturant guanidinium chloride (GdmCl). Data adapted from ref. 24 and fit to a two-state unfolding model (psychro- and mesophile) and a three-state model (thermophile). (b) - Thermal unfolding of a psychro- (blue circles), meso- (orange triangles), and thermophilic (red squares) α-amylase measured by a shift in Trp fluorescence. The relative activity of the enzymes is shown as semitransparent fill, colour-coded in the same way as unfolding data. Data adapted from ref. 24 and fit to a two-state thermal unfolding model. (c) Conformational stability curves of a psychro- (blue), meso- (orange), and thermophilic (red) α-amylase measured by DSC. All 3 panels based on data from ref. 25 using digitization, normalization and replotting in GraphPad Prism 9.
There is a clear trend that cold adaptation involves lower tm-values (Fig. 2b). Furthermore, psychrophilic enzymes show a significant temperature gap between topt and tm [42]. Like chemical denaturation, the thermal unfolding of psychrophilic proteins is more cooperative than their warm-adapted homologs [77]. Hydrophobic interactions, which typically lead to misfolding and aggregation upon renaturation [42], increase with temperature, so psychrophilic unfolding could be expected to be more reversible. However, several psychrophilic enzymes show irreversible unfolding, e.g. ornithine carbamoyltransferase from Moritellaabyssi [102] and the alkaline phosphatase from Vibrio splendidus [103]. Obviously, irreversible unfolding compromises thermodynamic analysis which assumes reversibility. Strictly speaking, irreversibility only allows for the Eyring formalism, where the energy of activation (ΔG‡) for the transition can be studied through the kinetics of folding. Nevertheless, thermodynamic equations are often applied for convenience. Lack of reversibility should always be investigated and clearly stated in such studies. However, the assumption of reversibility allows us to calculate the temperature dependence of the free energy of unfolding ΔGD-N, which results in a hyperbolic curve (Fig. 2c). Positive ΔGD-N values, implying native state stability, reveal the stability window of protein driven by enthalpic contributions. Intercepts with the x-axis provide the hot and cold unfolding midpoints Tm and Tc. Tc can be predicted by extrapolation of the bell-shaped curvature; it can be difficult to measure directly due to the freezing at subzero temperatures and may require the use of alcohols or salts (with concomitant consequences for stability) to avoid ice formation in the sample [104].
Despite psychrophiles’ lower thermodynamic stability, most enzymes have the same maximal ΔGD-N, typically near room temperature [25]. This has been attributed to the fact that the hydrophobic effect, which is a major driver of protein folding, peaks around 20°C [105,106]. Clearly psychrophilic enzymes have a smaller stability window than their meso- and thermophilic homologs, implying that cold adaptation involves both heat- and cold-lability. In evolutionary terms, this means that while hot environments push thermophiles to adapt against protein unfolding at high temperatures, psychrophiles inhabiting subzero environments may primarily experience selective pressure away from cold denaturation - at least in theory. However, due to the symmetrical the bell-shaped curvature of ΔGD-N’s temperature dependence, lowering of TC would involve increasing ΔGD-N, or decreasing ΔCp, to broaden the stability window. This would inherently lead to a higher Tm as well, given that the stability maximum remains the same. Cold denaturation of psychrophilic enzymes is rarely studied; however, investigating it more closely may unveil the mechanisms underlying the functioning of these enzymes at low temperatures.
The kinetics of folding and unfolding of psychrophilic proteins, which provide insight into global conformational dynamics, have received less attention than their stability. However, we have some indications. Protein folding rates for a psychrophilic transcription factor (TF) from. P. haloplanktis and a tryptophan synthase α subunit are somewhat lower than their mesophilic counterparts [107,108] but cannot account for the huge difference in stability. Moreover, there were no unusual kinetic adapations; the TF's temperature dependence of folding was similar to its meso- and thermophilic homologues. Slow folding kinetics at low temperatures may result in protein misfolding and aggregation, and psychrophiles are likely to possess some other type of adaptations to overcome this. Also, folding can be limited by the peptidyl-prolyl cis/trans isomerization around Pro residues, which is slow and temperature-dependent [109]. Psychrophilic enzymes typically have a lower Pro content, which may benefit folding rates. Furthermore, several psychrophilic organisms overexpress prolyl isomerases (PPI), which accelerate isomerization [110]. Notably, PPI expression is also upregulated at low temperatures in the case of the aforementioned TF from P. haloplanktis [111]. Similarly, a transcriptomic study showed that two molecular chaperones responsible for correct assembly of disulfide bridges are the most upregulated genes in the cold response of Polaromonas [112], emphasizing the importance of chaperones in translation processes at low temperatures. Differences in unfolding between psychrophilic proteins and their meso- and thermophilic counterparts are even more dramatic. While thermophilic proteins unfold unusually slowly, psychrophiles such as the tryptophan synthase α subunit unfold very rapidly [107,108,111]. Thus the narrow thermodynamic stability window of psychrophilic enzymes is constrained more by fast unfolding than by slow refolding.
5. How do we effectively identify novel cold-active enzymes and apply them in an industrial context? Solutions involve smart combinations of bioprospecting, shotgun expression, genome mining and ultra-high throughput screening/engineering approaches
Although some psychrophilic enzymes exhibit higher kcat values at their topt than their warm-adapted homologues [82,113], generally meso- and thermophilic enzymes achieve the highest maximal turnover rates due to their high unfolding temperatures [69,79]. Indeed, the lack of selective pressure for thermal stability in cold environments might be responsible for the low tm of psychrophilic enzymes; this remains to be investigated in more detail. The possibility that many known psychrophilic enzymes may have experienced genetic drift towards low stability in turn opens up the possibility of improvement, that is to engineer enzymes to combine low-temperature activity with high-temperature stability [99]. Search towards such optimized “super-cold-active enzymes” has already been achieved by methods such as directed evolution [114,115], rational [116,117] or random mutagenesis [118], and chemical modifications [119]. Optimized variants of closely related enzymes that are both more thermostable and active cold-active are also found in nature, such as a highly active xylanase from Scopulariopsis sp., which seems to combine local flexibility at the active site with global rigidity [120]. Eurypsychrophilic organisms are promising candidates for discovering such highly optimized enzymes, as their environments require them to maintain metabolic activity at low temperatures, but also survive relatively high temperatures. Despite the seemingly inherent reduction in cold activity as a consequence of enhanced stability, the examples of the thermostable cold-active enzymes mentioned above suggest that the search for novel psychrophilic enzymes in permafrost environments may reveal unique adaptation strategies that challenge this apparent activity/stability trade-off.
5.1. Search for novel enzymes
Enzymes with high catalytic rates at low temperatures are an attractive and sustainable alternative to meso- and thermophilic enzymes in biotechnology-based industries which operate at ambient temperatures or lower, e.g. within food (processing and conservation), medicinal (pharmaceuticals and diagnostics), chemistry (organic synthesis and biopolymers), textile (fabric processing and washing), paper (wood fiber processing), and environmental (wastewater treatment and bioremediation) industries. However, only a small number of psychrophilic enzymes have been commercialized due to their inherently low stability [29]. Many cold-adapted enzymes described in the literature originate from the narrowly-adapted stenopsychrophiles. Enzymes in these organisms are exceptionally active at low temperatures but are highly heat-labile. As a result, these enzymes often lose their activity at characteristically low temperatures [9,25]. This property makes them generally less attractive in biotech industries, as stable shelf life at room temperatures is crucial for commercial use [121]. In the absence of a robust psychrophilic enzyme, it is simpler and cheaper to use larger doses of less active but more stable mesophilic enzymes. However, eurypsychrophilic organisms have the potential to address the issue of instability by being capable of functioning in both moderate and extremely cold temperatures [122,123], provided that the molecular adaptations of the enzymes reflect the broad adaptation of the host. Future detailed comparative studies of enzymes from eurypsychrophilic organisms may uncover molecular adaptations to both cold and warm temperatures. In general, psychrophilic enzymes pose a novel point of origin for optimization, which may prove to result in better final products than their mesophilic counterparts. Furthermore, the otherwise undesirable heat lability of psychrophilic enzymes may pose a key solution to fast and efficient processing at low temperatures followed by a fast and unintrusive deactivation of the reaction by subsequent high-temperature processing steps, thereby turning a weakness into a strength.
5.2. Isolation of enzymes from microbiomes and metagenomes
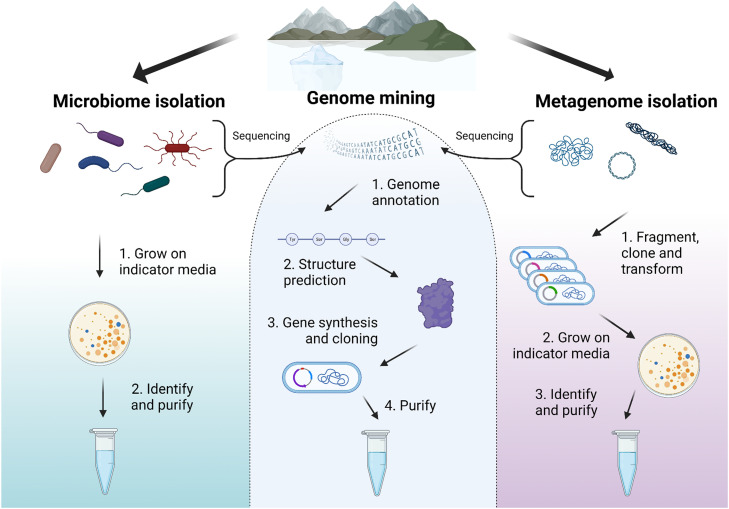
Given the vast size of the low-temperature habitats on our planet, there are many opportunities for bioprospecting (summarized in Fig. 3). Isolation of microbiomes from e.g. permafrost allows for direct identification of novel psychrophilic enzymes by utilizing indicator media that contain color- or fluorometric substrate analogs, e.g., 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) for detection of β-galactosidase activity, or fluorogenic triglyceride analogs for identification of lipases [124]. Furthermore, use of selective conditions, such as temperature, salinity, or even substrate availability can narrow down the range of microorganism growth to the desired characteristics. A drawback of the isolation of enzymes from microbiomes is that <1% of microorganisms can be grown in the lab. This excludes many potentially interesting hosts and their enzymes. A potential solution is provided by functional metagenomics, which screens total DNA from environmental samples by cloning (random) fragments of the DNA into an easily culturable host. Following bulk DNA extraction, it is fragmented (typically sheared by sonication or partially digested by restriction endonucleases), cloned into vectors (e.g., plasmids or cosmids), and transfected into a host optimized for construction of gene banks [125,126]. From this stage, it becomes possible to identify the enzymes of interest by cultivating the gene bank host on indicator media, similar to the process of identifying microbiomes through isolation techniques. It may also be possible to screen for activity at low temperatures by growing the host bacteria under mesophilic conditions and carrying out the assays at lower temperatures. Although functional metagenomics expands the potential for discovering novel enzymes compared to conventional microbial screening, its success relies on the likelihood of capturing the whole reading frame of a protein in an insert and the ability of the host organism to decipher the genetic material encoded by the gene banks. It is estimated that ∼ 40 % of coding sequences can be expressed in E. coli this way [127].
Fig. 3.
Potential ways to isolate and identify novel psychrophilic enzymes. Created with BioRender.com
5.3. Genome mining
The emergence of next-generation sequencing (NGS) during the last two decades has led to the discovery and characterization of a vast number of microorganisms, including many eurypsychrophiles [128,129]. These organisms may possess enzymes with novel molecular adaptations that allow them to maintain high enzymatic activity at low temperatures while resisting denaturation by heat at ambient temperatures. Genomic databases arising from these findings constitute a possibility of bioinformatic exploration leading to the discovery of novel enzymes using an array of annotation and prediction tools - an approach termed genome mining [130]. Thanks to large databases of previously characterized and structurally resolved enzymes, it is possible to pinpoint enzymes in large genomes or metagenomes with high accuracy and predict a significant amount of information on structure and activity before taking the sequences to the lab. However, the commonly used databases, which allow easy searching, make up <1% of all publicly available sequenced DNA and computational expertise is required for more thorough analysis [131]. Once this is in place, utilization of genome annotation tools, such as Rapid Annotations using Subsystems Technology (RAST), allows for relatively quick prediction of gene encoding sequences and their putative function [132]. This approach significantly simplifies identification of novel enzymes, as it already at this point allows for the synthesis and cloning of the gene of interest, followed by transfection into a host organism optimized for psychrophilic protein expression, such as the commercially available ArcticExpress E. coli strain. Furthermore, sequences of interest can be utilized in structure prediction through homology prediction methods like SWISS-MODEL [133], as well as advanced AI tools such as AlphaFold [134]. Both prediction approaches allow for preliminary structural analysis of the putative enzymes and may even elucidate substrate specificity by comparison to related enzymes described in specific classification databases, e.g. CAZy in the case for carbohydrate-active enzymes [135] or the recently established plastic-active enzymes database, PAZy [136]. Although this approach enables rapid and exclusively in silico discovery of enzymes, it relies heavily on accurate genome annotation tools that are built upon databases of previously characterized proteins. As a result, its applicability may be limited when it comes to identifying enzymes with exotic or novel activities. However, the search for specific activity of methyl halide transferases, which was not evident in the genome annotations of various organisms, yielded positive results through the expression of candidates annotated as methyl transferases [137]. This demonstrates that it is possible to successfully identify specific enzymes by expressing candidates that are predicted to be only weakly related in activity. Present and future activity screenings of libraries of enzyme sequences discovered through a broad genome mining search will hopefully combine the efficiency and expediency of bioinformatic exploration with the extensive scope of functional metagenomics.
5.4. Enzyme engineering
Once a suitable candidate has been found, it is invariably necessary to improve it further. In addition to rational protein engineering, recent advances in ultra-high throughput screening of protein variants may combine with directed evolution to provide a much-needed tool. A thorough description of this progress is outside our scope, but we refer to excellent recent reviews [138,139] and an inspiring example (to generate thermophilic variants of a plastic-degrading enzyme) based on ultra performance liquid chromatography [140]. In closing, we would like to highlight the emerging microdroplet technology. Here picoliter-size droplets are generated so rapidly that it is possible to screen thousands of individual randomly generated variants per second and select positive variants based on colorimetric or fluorimetric readouts [141]. Provided the enzymes in the individual droplets can be identified through their encoding DNA in the same droplet, it is possible to select promising candidates and sequence them to identify promising mutations. The approach is limited by the need for a fluorescent read-out which often requires adjustments to the enzyme assay and may bias the selection of variants. However, they can provide the first step in a more elaborate screening pipeline. Microfluidics are sufficiently versatile that each droplet can sequentially be used to produce the protein under a certain set of conditions, add substrate and incubate at another (psychrophilic) set of conditions and then select for the right hits. These approaches have only recently been applied systematically [142] but may have a bright future ahead of them.
6. Conclusion
It is important to point out that none of the approaches described above are sure to deliver enzymes with the desired psychrophilic features. The host may have adapted towards low temperatures in other ways than changing the protein in question. Despite the recent advances in precision and convenience of genome annotation and structure prediction, the sheer multitude of sequence-based and structural determinants of cold-activity makes it difficult to predict temperature dependency on the structure-function relationship of novel enzymes. Ultimately, in-house purification and characterization will be necessary to identify enzymes with psychrophilic characteristics, but given the emerging techniques and opportunities for massive data generation and analysis, there is every reason to expect a bright future for our understanding and exploitation of cold-active enzymes.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful for support from Independent Research Foundation Denmark | Technology and Production (grant 9041-00123B) to D.E.O and for inspiring discussions on psychrophilicity with collaoborators Mariane Thøgersen and Peter Stougaard and impressive inputs from the many hard-working, talented and committed students who have worked with psychrophilic enzymes in our group (Malthe Kjær Bendtsen, Elia Viezzi, Nikoline Kruuse, Julie Astono, Rune Efferbach Toft and Alexander Højlund Toftgaard).
Data availability
Data will be made available on request.
References
- 1.Canganella F., Wiegel J. Extremophiles: from abyssal to terrestrial ecosystems and possibly beyond. Naturwissenschaften. 2011;98:253–279. doi: 10.1007/s00114-011-0775-2. [DOI] [PubMed] [Google Scholar]
- 2.Siddiqui K.S., et al. Psychrophiles. Annu. Rev. Earth Planet. Sci. 2013;41:87–115. doi: 10.1146/annurev-earth-040610-133514. [DOI] [Google Scholar]
- 3.De Maayer P., Anderson D., Cary C., Cowan D.A. Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 2014;15:508–517. doi: 10.1002/embr.201338170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders N.F., et al. Mechanisms of thermal adaptation revealed from the genomes of the Antarctic Archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 2003;13:1580–1588. doi: 10.1101/gr.1180903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turchetti B., et al. Psychrophilic yeasts in glacial environments of Alpine glaciers. FEMS Microbiol. Ecol. 2008;63:73–83. doi: 10.1111/j.1574-6941.2007.00409.x. [DOI] [PubMed] [Google Scholar]
- 6.Barták M., Váczi P., Hájek J., Smykla J. Low-temperature limitation of primary photosynthetic processes in Antarctic lichens Umbilicaria antarctica and Xanthoria elegans. Polar Biol. 2007;31:47–51. doi: 10.1007/s00300-007-0331-x. [DOI] [Google Scholar]
- 7.Pucciarelli S., et al. Molecular cold-adaptation of protein function and gene regulation: The case for comparative genomic analyses in marine ciliated protozoa. Mar. Geonomics. 2009;2:57–66. doi: 10.1016/j.margen.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Kozeretska I., Serga S., Kovalenko P., Gorobchyshyn V., Convey P. Belgica antarctica (Diptera: Chironomidae): a natural model organism for extreme environments. Insect Sci. 2022;29:2–20. doi: 10.1111/1744-7917.12925. [DOI] [PubMed] [Google Scholar]
- 9.Feller G., Gerday C. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Micro. 2003;1:200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- 10.Hassan N., et al. Temperature driven membrane lipid adaptation in glacial psychrophilic bacteria. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey N., Jain R., Pandey A., Tamta S. Optimisation and characterisation of the orange pigment produced by a cold adapted strain of Penicillium sp. (GBPI_P155) isolated from mountain ecosystem. Mycology. 2018;9:81–92. doi: 10.1080/21501203.2017.1423127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mykytczuk N.C., et al. Bacterial growth at -15 degrees C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J. 2013;7:1211–1226. doi: 10.1038/ismej.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonseca F., Meneghel J., Cenard S., Passot S., Morris G. Determination of intracellular vitrification temperatures for unicellular micro organisms under conditions relevant for cryopreservation. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casillo A., et al. Structure-activity relationship of the exopolysaccharide from a psychrophilic bacterium: a strategy for cryoprotection. Carbohydr. Polym. 2017;156:364–371. doi: 10.1016/j.carbpol.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorv J., Rose D.R., Glick B.R. Bacterial ice crystal controlling proteins. Scientifica. 2014;2014 doi: 10.1155/2014/976895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodrigues D.F., et al. Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: a genome and transcriptome approach. Bmc Genomics [Electronic Resource] 2008;9:547. doi: 10.1186/1471-2164-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strocchi M., Ferrer M., Timmis K.N., Golyshin P.N. Low temperature-induced systems failure in Escherichia coli: insights from rescue by cold-adapted chaperones. Proteomics. 2006;6:193–206. doi: 10.1002/pmic.200500031. [DOI] [PubMed] [Google Scholar]
- 18.Piette F., Leprince P., Feller G. Is there a cold shock response in the Antarctic psychrophile Pseudoalteromonas haloplanktis? Extremophiles: Life Under Extreme Conditions. 2012;16:681–683. doi: 10.1007/s00792-012-0456-x. [DOI] [PubMed] [Google Scholar]
- 19.Knowles J.R. Enzyme catalysis: not different, just better. Nature. 1991;350:121–124. doi: 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- 20.Arrhenius S. Über die Dissociationswärme und den Einfluss der Temperatur auf den Dissociationsgrad der Elektrolyte. Zeitschrift für Physikalische Chemie. 1889;4U:96–116. doi: 10.1515/zpch-1889-0408. [DOI] [Google Scholar]
- 21.Feller G., Gerday C. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 2003;1:200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- 22.Collins T., Margesin R. Psychrophilic lifestyles: mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol. 2019;103:2857–2871. doi: 10.1007/s00253-019-09659-5. [DOI] [PubMed] [Google Scholar]
- 23.Miyake R., et al. Construction of a low-temperature protein expression system using a cold-adapted bacterium, Shewanella sp. strain Ac10, as the host. Appl. Environ. Microbiol. 2007;73:4849–4856. doi: 10.1128/AEM.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oswald V.F., Chen W., Harvilla P.B., Magyar J.S. Overexpression, purification, and enthalpy of unfolding of ferricytochrome c552 from a psychrophilic microorganism. J. Inorg. Biochem. 2014;131:76–78. doi: 10.1016/j.jinorgbio.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Amico S., Marx J.C., Gerday C., Feller G. Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 2003;278:7891–7896. doi: 10.1074/jbc.M212508200. [DOI] [PubMed] [Google Scholar]
- 26.Feller G., d'Amic D., Gerday C. Thermodynamic stability of a cold-active α-amylase from the antarctic bacterium Alteromonas haloplanctis. Biochemistry. 1999;38:4613–4619. doi: 10.1021/bi982650+. [DOI] [PubMed] [Google Scholar]
- 27.Gerday C., et al. Psychrophilic enzymes: a thermodynamic challenge. Biochim. Biophy. Acta. Prot. Struct. Mol. Enzymol. 1997;1342:119–131. doi: 10.1016/S0167-4838(97)00093-9. [DOI] [PubMed] [Google Scholar]
- 28.Aqvist J., Kazemi M., Isaksen G.V., Brandsdal B.O.Entropy, Catalysis Enzyme. Acc. Chem. Res. 2017;50:199–207. doi: 10.1021/acs.accounts.6b00321. [DOI] [PubMed] [Google Scholar]
- 29.Santiago M., Ramirez-Sarmiento C.A., Zamora R.A., Parra L.P.Discovery. Molecular mechanisms, and industrial applications of cold-active enzymes. Front. Microbiol. 2016;7:1408. doi: 10.3389/fmicb.2016.01408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Åqvist J. Cold adaptation of triosephosphate isomerase. Biochemistry. 2017;56:4169–4176. doi: 10.1021/acs.biochem.7b00523. [DOI] [PubMed] [Google Scholar]
- 31.Bjelic S., Brandsdal B.O., Åqvist J. Cold adaptation of enzyme reaction rates. Biochemistry. 2008;47:10049–10057. doi: 10.1021/bi801177k. [DOI] [PubMed] [Google Scholar]
- 32.Siddiqui K.S., Cavicchioli R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 33.Feller G., Gerday C. Psychrophilic enzymes: molecular basis of cold adaptation. Cell. Mol. Life Sci. 1997;53:830–841. doi: 10.1007/s000180050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerday, C. in Cold-adapted Yeasts: Biodiversity, Adaptation Strategies and Biotechnological Significance (eds Pietro Buzzini & Rosa Margesin) 325-350 (Springer Berlin Heidelberg, 2014).
- 35.De Vos D., et al. Oligosaccharide binding in family 8 glycosidases: crystal structures of active-site mutants of the β-1, 4-xylanase pXyl from Pseudoaltermonas haloplanktis TAH3a in complex with substrate and product. Biochemistry. 2006;45:4797–4807. doi: 10.1021/bi052193e. [DOI] [PubMed] [Google Scholar]
- 36.De Vos D., Xu Y., Hulpiau P., Vergauwen B., Van Beeumen J.J. Structural investigation of cold activity and regulation of aspartate carbamoyltransferase from the extreme psychrophilic bacterium Moritella profunda. J. Mol. Biol. 2007;365:379–395. doi: 10.1016/j.jmb.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 37.Hoyoux A., et al. Cold-adapted beta-galactosidase from the Antarctic psychrophile Pseudoalteromonas haloplanktis. Appl. Environ. Microb. 2001;67:1529–1535. doi: 10.1128/aem.67.4.1529-1535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi S., Satyanarayana T. Biotechnology of cold-active proteases. Biology. 2013;2:755–783. doi: 10.3390/biology2020755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Amico S., et al. Molecular basis of cold adaptation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:917–925. doi: 10.1098/rstb.2002.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Amico S., Collins T., Marx J.C., Feller G., Gerday C. Psychrophilic microorganisms: challenges for life. EMBO Rep. 2006;7:385–389. doi: 10.1038/sj.embor.7400662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koenekoop L., Åqvist J. Principles of cold adaptation of fish lactate dehydrogenases revealed by computer simulations of the catalytic reaction. Mol. Biol. Evol. 2023;40 doi: 10.1093/molbev/msad099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feller G. Psychrophilic enzymes: from folding to function and biotechnology. Scientifica. 2013;2013:1–28. doi: 10.1155/2013/512840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sočan J., Purg M., Åqvist J. Computer simulations explain the anomalous temperature optimum in a cold-adapted enzyme. Nat. Commun. 2020;11:2644. doi: 10.1038/s41467-020-16341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung S.K., et al. Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum. Proteins Struct. Funct. Bioinf. 2008;71:476–484. doi: 10.1002/prot.21884. [DOI] [PubMed] [Google Scholar]
- 45.Russell R.J., Gerike U., Danson M.J., Hough D.W., Taylor G.L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure. 1998;6:351–361. doi: 10.1016/s0969-2126(98)00037-9. [DOI] [PubMed] [Google Scholar]
- 46.Rutkiewicz M., Bujacz A., Bujacz G. Structural features of cold-adapted dimeric GH2 β-D-galactosidase from Arthrobacter sp. 32cB. Biochim. Biophys. Acta Proteins Proteom. 2019;1867:776–786. doi: 10.1016/j.bbapap.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Aghajari N., et al. Crystal structures of a psychrophilic metalloprotease reveal new insights into catalysis by cold-adapted proteases. Proteins Struct. Funct. Bioinf. 2003;50:636–647. doi: 10.1002/prot.10264. [DOI] [PubMed] [Google Scholar]
- 48.Cipolla A., D'Amico S., Barumandzadeh R., Matagne A., Feller G. Stepwise adaptations to low temperature as revealed by multiple mutants of psychrophilic α-amylase from Antarctic Bacterium. J. Biol. Chem. 2011;286:38348–38355. doi: 10.1074/jbc.M111.274423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schellman J.A. Temperature, stability, and the hydrophobic interaction. Biophys. J. 1997;73:2960–2964. doi: 10.1016/s0006-3495(97)78324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sočan J., Purg M., Åqvist J. Computer simulations explain the anomalous temperature optimum in a cold-adapted enzyme. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-16341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fersht A.R. Freeman & Co.; 1999. Structure and mechanism in protein science. A guide to enzyme catalysis and protein folding. [Google Scholar]
- 52.Ozohanics O., Ambrus A. Hydrogen-deuterium exchange mass spectrometry: a novel structural biology approach to structure, dynamics and interactions of proteins and their complexes. Life (Basel) 2020;10 doi: 10.3390/life10110286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berthomieu C., Hienerwadel R. Fourier transform infrared (FTIR) spectroscopy. Photosynth. Res. 2009;101:157–170. doi: 10.1007/s11120-009-9439-x. [DOI] [PubMed] [Google Scholar]
- 54.Van Petegem F., et al. The structure of a cold-adapted family 8 xylanase at 1.3 A resolution. Structural adaptations to cold and investgation of the active site. J. Biol. Chem. 2003;278:7531–7539. doi: 10.1074/jbc.M206862200. [DOI] [PubMed] [Google Scholar]
- 55.Aghajari N., Feller G., Gerday C., Haser R. Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 1998;7:564–572. doi: 10.1002/pro.5560070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S.Y., et al. Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum. J. Biol. Chem. 1999;274:11761–11767. doi: 10.1074/jbc.274.17.11761. [DOI] [PubMed] [Google Scholar]
- 57.Paredes D.I., Watters K., Pitman D.J., Bystroff C., Dordick J.S. Comparative void-volume analysis of psychrophilic and mesophilic enzymes: Structural bioinformatics of psychrophilic enzymes reveals sources of core flexibility. BMC Struct. Biol. 2011;11:42. doi: 10.1186/1472-6807-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cavicchioli, R. & Siddiqui, K. S. in Enzyme Technology (eds Ashok Pandey, Colin Webb, Carlos Ricardo Soccol, & Christian Larroche) 615-638 (Springer New York, 2006).
- 59.Phillips S.R., Wilson L.J., Borkman R.F. Acrylamide and iodide fluorescence quenching as a structural probe of tryptophan microenvironment in bovine lens crystallins. Curr. Eye Res. 1986;5:611–619. doi: 10.3109/02713688609015126. [DOI] [PubMed] [Google Scholar]
- 60.Balasubramaniam D., Komives E.A. Hydrogen-exchange mass spectrometry for the study of intrinsic disorder in proteins. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2013;1834:1202–1209. doi: 10.1016/j.bbapap.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramírez-Sarmiento César A, et al. Observation of Solvent Penetration during Cold Denaturation of E. coli Phosphofructokinase-2. Biophys. J. 2013;104:2254–2263. doi: 10.1016/j.bpj.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parveen T., Kamran M., Fatmi M.Q. Structural and dynamical thermostability of psychrophilic enzyme at various temperatures: molecular dynamics simulations of tryptophan synthase. Arch. Biochem. Biophys. 2019;663:297–305. doi: 10.1016/j.abb.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 63.Isaksen G.V., Åqvist J., Brandsdal B.O. Enzyme surface rigidity tunes the temperature dependence of catalytic rates. Proc. Natl. Acad. Sci. 2016;113:7822–7827. doi: 10.1073/pnas.1605237113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Violot S., et al. Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering. J. Mol. Biol. 2005;348:1211–1224. doi: 10.1016/j.jmb.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 65.Zheng Y., et al. Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-β-1,4-xylanase. J. Struct. Biol. 2016;193:206–211. doi: 10.1016/j.jsb.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 66.Gerike U., Danson M.J., Russell N.J., Hough D.W. Sequencing and expression of the gene encoding a cold-active citrate synthase from an Antarctic bacterium, strain DS2-3R. Eur. J. Biochem. 1997;248:49–57. doi: 10.1111/j.1432-1033.1997.00049.x. [DOI] [PubMed] [Google Scholar]
- 67.Isaksen G.V., Åqvist J., Brandsdal B.O. Protein surface softness is the origin of enzyme cold-adaptation of trypsin. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arnold F.H., Wintrode P.L., Miyazaki K., Gershenson A. How enzymes adapt: lessons from directed evolution. Trends Biochem. Sci. 2001;26:100–106. doi: 10.1016/s0968-0004(00)01755-2. [DOI] [PubMed] [Google Scholar]
- 69.Miao L.L., et al. Molecular structural basis for the cold adaptedness of the Psychrophilic β-Glucosidase BglU in Micrococcus antarcticus. Appl. Environ. Microb. 2016;82:2021–2030. doi: 10.1128/aem.03158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Damnjanović J., Nakano H., Iwasaki Y. Deletion of a dynamic surface loop improves stability and changes kinetic behavior of phosphatidylinositol-synthesizingStreptomycesphospholipase D. Biotechnol. Bioeng. 2014;111:674–682. doi: 10.1002/bit.25149. [DOI] [PubMed] [Google Scholar]
- 71.Tindbaek N., Svendsen A., Oestergaard P.R., Draborg H. Engineering a substrate-specific cold-adapted subtilisin. Protein Eng. Des. Sel. 2004;17:149–156. doi: 10.1093/protein/gzh019. [DOI] [PubMed] [Google Scholar]
- 72.Georlette D., et al. Some like it cold: biocatalysis at low temperatures. FEMS Microbiol. Rev. 2004;28:25–42. doi: 10.1016/j.femsre.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 73.van der Ent F., et al. Structure and mechanism of a cold-adapted bacterial lipase. Biochemistry. 2022;61:933–942. doi: 10.1021/acs.biochem.2c00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raymond-Bouchard I., Tremblay J., Altshuler I., Greer C.W., Whyte L.G. Comparative transcriptomics of cold growth and adaptive features of a eury- and steno-psychrophile. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siddiqui K.S., et al. Role of disulfide bridges in the activity and stability of a cold-active alpha-amylase. J. Bacteriol. 2005;187:6206–6212. doi: 10.1128/jb.187.17.6206-6212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mavromatis K., Tsigos I., Tzanodaskalaki M., Kokkinidis M., Bouriotis V. Exploring the role of a glycine cluster in cold adaptation of an alkaline phosphatase. Eur. J. Biochem. 2002;269:2330–2335. doi: 10.1046/j.1432-1033.2002.02895.x. [DOI] [PubMed] [Google Scholar]
- 77.Feller G. Protein stability and enzyme activity at extreme biological temperatures. J. Phys. Condens. Matter. 2010;22 doi: 10.1088/0953-8984/22/32/323101. [DOI] [PubMed] [Google Scholar]
- 78.Yu H., Zhao Y., Guo C., Gan Y., Huang H. The role of proline substitutions within flexible regions on thermostability of luciferase. Biochimica et Biophysica Acta (BBA) - Protein. Proteomics. 2015;1854:65–72. doi: 10.1016/j.bbapap.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 79.Feller G., et al. Purification, characterization, and nucleotide sequence of the thermolabile alpha-amylase from the antarctic psychrotroph Alteromonas haloplanctis A23. J. Biol. Chem. 1992;267:5217–5221. [PubMed] [Google Scholar]
- 80.Violot S., et al. Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering. J. Mol. Biol. 2005;348:1211–1224. doi: 10.1016/j.jmb.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 81.Head-Gordon T. Is water structure around hydrophobic groups clathrate-like? Proc. Natl. Acad. Sci. U.S.A. 1995;92:8308–8312. doi: 10.1073/pnas.92.18.8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davail S., Feller G., Narinx E., Gerday C. Cold adaptation of proteins. Purification, characterization, and sequence of the heat-labile subtilisin from the antarctic psychrophile Bacillus TA41. J. Biol. Chem. 1994;269:17448–17453. [PubMed] [Google Scholar]
- 83.Munson M., et al. What makes a protein a protein? Hydrophobic core designs that specify stability and structural properties. Protein Sci. 1996;5:1584–1593. doi: 10.1002/pro.5560050813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leiros H.K., Willassen N.P., Smalås A.O. Residue determinants and sequence analysis of cold-adapted trypsins. Extremophiles. 1999;3:205–219. doi: 10.1007/s007920050118. [DOI] [PubMed] [Google Scholar]
- 85.Siglioccolo A., Gerace R., Pascarella S. Cold spots” in protein cold adaptation: Insights from normalized atomic displacement parameters (B′-factors) Biophys. Chem. 2010;153:104–114. doi: 10.1016/j.bpc.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 86.Kim M.K., Kang Y.K. Positional preference of proline in alpha-helices. Protein Sci. 1999;8:1492–1499. doi: 10.1110/ps.8.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sinden, R. R. in DNA Structure and Function (ed Richard R. Sinden) 287-325 (Academic Press, 1994).
- 88.Rentier-Delrue F., et al. Cloning and overexpression of the triosephosphate isomerase genes from psychrophilic and thermophilic bacteria. Structural comparison of the predicted protein sequences. J. Mol. Biol. 1993;229:85–93. doi: 10.1006/jmbi.1993.1010. [DOI] [PubMed] [Google Scholar]
- 89.Gerike U., Danson M.J., Russell N.J., Hough D.W. Sequencing and expression of the gene encoding a cold-active citrate synthase from an Antarctic bacterium, strain DS2-3R. Eur. J. Biochem. 1997;248:49–57. doi: 10.1111/j.1432-1033.1997.00049.x. [DOI] [PubMed] [Google Scholar]
- 90.Sočan J., Isaksen G.V., Brandsdal B.O., Åqvist J. Towards rational computational engineering of psychrophilic enzymes. Sci. Rep. 2019;9:19147. doi: 10.1038/s41598-019-55697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koenekoop L., van der Ent F., Purg M., Åqvist J. The activation parameters of a cold-adapted short chain dehydrogenase are insensitive to enzyme oligomerization. Biochemistry. 2022;61:514–522. doi: 10.1021/acs.biochem.2c00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tronelli D., Maugini E., Bossa F., Pascarella S. Structural adaptation to low temperatures−analysis of the subunit interface of oligomeric psychrophilic enzymes. FEBS J. 2007;274:4595–4608. doi: 10.1111/j.1742-4658.2007.05988.x. [DOI] [PubMed] [Google Scholar]
- 93.Míguez Amil S., et al. The cryo-EM structure of thermotoga maritima β-galactosidase: quaternary structure guides protein engineering. ACS Chem. Biol. 2020;15:179–188. doi: 10.1021/acschembio.9b00752. [DOI] [PubMed] [Google Scholar]
- 94.Walden H., et al. Tiny TIM: a small, tetrameric, hyperthermostable triosephosphate isomerase 11. Huber R., editor. Tiny TIM: a small, tetrameric, hyperthermostable triosephosphate isomerase 11J. Mol. Biol. 2001;306:745–757. doi: 10.1006/jmbi.2000.4433. Edited by. [DOI] [PubMed] [Google Scholar]
- 95.Hildebrandt P., Wanarska M., Kur J. A new cold-adapted β-D-galactosidase from the Antarctic Arthrobacter sp. 32c – gene cloning, overexpression, purification and properties. BMC Microbiol. 2009;9:151. doi: 10.1186/1471-2180-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robin S., Togashi D.M., Ryder A.G., Wall J.G. Trigger factor from the psychrophilic bacterium Psychrobacter frigidicola is a monomeric chaperone. J. Bacteriol. 2009;191:1162–1168. doi: 10.1128/jb.01137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zanphorlin L.M., et al. Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 β-glucosidase from Exiguobacterium antarcticum B7. Sci. Rep. 2016;6:23776. doi: 10.1038/srep23776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Skálová T., et al. Cold-active β-Galactosidase from Arthrobacter sp. C2-2 forms compact 660kDa hexamers: crystal structure at 1.9Å resolution. J. Mol. Biol. 2005;353:282–294. doi: 10.1016/j.jmb.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 99.Cornely K.P. Wiley; 2004. Essential Biochemistry; pp. 96–107. C. [Google Scholar]
- 100.Merlino A., et al. Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis. J. Struct. Biol. 2010;172:343–352. doi: 10.1016/j.jsb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 101.Kingsley C.N., Bierma J.C., Pham V., Martin R.W. γS-Crystallin Proteins from the Antarctic Nototheniid Toothfish: A Model System for Investigating Differential Resistance to Chemical and Thermal Denaturation. J. Phys. Chem. B. 2014;118:13544–13553. doi: 10.1021/jp509134d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu Y., Feller G., Gerday C., Glansdorff N. Metabolic enzymes from psychrophilic bacteria: challenge of adaptation to low temperatures in ornithine carbamoyltransferase from Moritellaabyssi. J. Bacteriol. 2003;185:2161–2168. doi: 10.1128/JB.185.7.2161-2168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hjörleifsson J.G., Ásgeirsson B. Cold-active alkaline phosphatase is irreversibly transformed into an inactive dimer by low urea concentrations. Biochimica et Biophysica Acta (BBA) - Protein. Proteomics. 2016;1864:755–765. doi: 10.1016/j.bbapap.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 104.Sanfelice D., Temussi P.A. Cold denaturation as a tool to measure protein stability. Biophys. Chem. 2016;208:4–8. doi: 10.1016/j.bpc.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kumar S., Tsai C.-J., Nussinov R. Maximal stabilities of reversible two-state proteins. Biochemistry. 2002;41:5359–5374. doi: 10.1021/bi012154c. [DOI] [PubMed] [Google Scholar]
- 106.Schellman J.A. Temperature, stability, and the hydrophobic interaction. Biophys. J. 1997;73:2960–2964. doi: 10.1016/S0006-3495(97)78324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mitsuya D., et al. Strategy for cold adaptation of the tryptophan synthase α subunit from the psychrophile Shewanella frigidimarina K14-2: crystal structure and physicochemical properties. J. Biochem. 2013;155:73–82. doi: 10.1093/jb/mvt098. [DOI] [PubMed] [Google Scholar]
- 108.Struvay C., Negro S., Matagne A., Feller G. Energetics of protein stability at extreme environmental temperatures in bacterial trigger factors. Biochemistry. 2013;52:2982–2990. doi: 10.1021/bi4002387. [DOI] [PubMed] [Google Scholar]
- 109.Baldwin R.L. The search for folding intermediates and the mechanism of protein folding. Annu. Rev. Biophys. 2008;37:1–21. doi: 10.1146/annurev.biophys.37.032807.125948. [DOI] [PubMed] [Google Scholar]
- 110.Piette F., Struvay C., Feller G. The protein folding challenge in psychrophiles: facts and current issues. Environ. Microbiol. 2011;13:1924–1933. doi: 10.1111/j.1462-2920.2011.02436.x. [DOI] [PubMed] [Google Scholar]
- 111.Piette F., et al. Proteomics of life at low temperatures: trigger factor is the primary chaperone in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Mol. Microbiol. 2010;76:120–132. doi: 10.1111/j.1365-2958.2010.07084.x. [DOI] [PubMed] [Google Scholar]
- 112.Raymond-Bouchard I., Tremblay J., Altshuler I., Greer C.W., Whyte L.G. Comparative transcriptomics of cold growth and adaptive features of a eury- and steno-psychrophile. Front. Microbiol. 2018;9:1565. doi: 10.3389/fmicb.2018.01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Georlette D., et al. Structural and functional adaptations to extreme temperatures in psychrophilic, mesophilic, and thermophilic DNA ligases. J. Biol. Chem. 2003;278:37015–37023. doi: 10.1074/jbc.M305142200. [DOI] [PubMed] [Google Scholar]
- 114.Miyazaki K., Wintrode P.L., Grayling R.A., Rubingh D.N., Arnold F.H. Directed evolution study of temperature adaptation in a psychrophilic enzyme. J. Mol. Biol. 2000;297:1015–1026. doi: 10.1006/jmbi.2000.3612. [DOI] [PubMed] [Google Scholar]
- 115.Zhang N., et al. Improving tolerance of Candida antarctica lipase B towards irreversible thermal inactivation through directed evolution. Protein. Eng. 2003;16:599–605. doi: 10.1093/protein/gzg074. [DOI] [PubMed] [Google Scholar]
- 116.Deng Z., Yang H., Shin H.-d., Li J., Liu L. Structure-based rational design and introduction of arginines on the surface of an alkaline α-amylase from Alkalimonas amylolytica for improved thermostability. Appl. Microbiol. Biotechnol. 2014;98:8937–8945. doi: 10.1007/s00253-014-5790-8. [DOI] [PubMed] [Google Scholar]
- 117.Asghari S.M., et al. Remarkable improvements of a neutral protease activity and stability share the same structural origins. Protein Eng. Des. Sel. 2010;23:599–606. doi: 10.1093/protein/gzq031. [DOI] [PubMed] [Google Scholar]
- 118.Rha E., et al. Simultaneous improvement of catalytic activity and thermal stability of tyrosine phenol-lyase by directed evolution. FEBS J. 2009;276:6187–6194. doi: 10.1111/j.1742-4658.2009.07322.x. [DOI] [PubMed] [Google Scholar]
- 119.Siddiqui K., Poljak A., Cavicchioli R. Improved activity and stability of alkaline phosphatases from psychrophilic and mesophilic organisms by chemically modifying aliphatic or amino groups using tetracarboxy-benzophenone derivatives. Cell. Mol. Biol. (Noisy-le-grand) 2004;50:657–667. [PubMed] [Google Scholar]
- 120.Afzal A.J., Ali S., Latif F., Rajoka M.I., Siddiqui K.S. Innovative kinetic and thermodynamic analysis of a purified superactive xylanase from Scopulariopsis sp. Appl. Biochem. Biotechnol. 2005;120:51–70. doi: 10.1385/abab:120:1:51. [DOI] [PubMed] [Google Scholar]
- 121.Al-Ghanayem A.A., Joseph B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 2020;104:2871–2882. doi: 10.1007/s00253-020-10429-x. [DOI] [PubMed] [Google Scholar]
- 122.Dopson M., González-Rosales C., Holmes D.S., Mykytczuk N. Eurypsychrophilic acidophiles: from (meta)genomes to low-temperature biotechnologies. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1149903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Goordial J., et al. Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 2015;92 doi: 10.1093/femsec/fiv154. [DOI] [PubMed] [Google Scholar]
- 124.Duque M., et al. New fluorogenic triacylglycerol analogs as substrates for the determination and chiral discrimination of lipase activities. J. Lipid Res. 1996;37:868–876. doi: 10.1016/S0022-2275(20)37584-2. [DOI] [PubMed] [Google Scholar]
- 125.McGarvey K.M., Queitsch K., Fields S. Wide variation in antibiotic resistance proteins identified by functional metagenomic screening of a soil DNA library. Appl. Environ. Microb. 2012;78:1708–1714. doi: 10.1128/aem.06759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Biver S., Portetelle D., Vandenbol M. Characterization of a new oxidant-stable serine protease isolated by functional metagenomics. Springerplus. 2013;2:410. doi: 10.1186/2193-1801-2-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gabor E.M., Alkema W.B., Janssen D.B. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ. Microbiol. 2004;6:879–886. doi: 10.1111/j.1462-2920.2004.00640.x. [DOI] [PubMed] [Google Scholar]
- 128.Goordial J., et al. Improved-high-quality draft genome sequence of Rhodococcus sp. JG-3, a eurypsychrophilic Actinobacteria from Antarctic Dry Valley permafrost. Standard. Genomic Sci. 2015;10 doi: 10.1186/s40793-015-0043-8. 61-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mykytczuk N.C.S., Wilhelm R.C., Whyte L.G. Planococcus halocryophilus sp. nov., an extreme sub-zero species from high Arctic permafrost. Int. J. Syst. Evol. Microbiol. 2012;62:1937–1944. doi: 10.1099/ijs.0.035782-0. [DOI] [PubMed] [Google Scholar]
- 130.Zaparucha A., de Berardinis V., Vaxelaire-Vergne C. The Royal Society of Chemistry; 2018. Modern Biocatalysis: Advances Towards Synthetic Biological Systems 1-27. [Google Scholar]
- 131.Heenan, J., Eiermann, A. & Kabisch, J. Proteineer GmbH, https://www.proteineer.com/ (2023).
- 132.Aziz R.K., et al. The RAST Server: rapid annotations using subsystems technology. Bmc Genomics [Electronic Resource] 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Waterhouse A., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic. Acids. Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jumper J., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cantarel B.L., et al. The carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic. Acids. Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Buchholz P.C.F., et al. Plastics degradation by hydrolytic enzymes: The plastics-active enzymes database—PAZy. Proteins Struct. Funct. Bioinf. 2022;90:1443–1456. doi: 10.1002/prot.26325. [DOI] [PubMed] [Google Scholar]
- 137.Bayer T.S., et al. Synthesis of methyl halides from biomass using engineered microbes. J. Am. Chem. Soc. 2009;131:6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 138.Markel U., et al. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem. Soc. Rev. 2020;49:233–262. doi: 10.1039/c8cs00981c. [DOI] [PubMed] [Google Scholar]
- 139.Pfaff L., Breite D., Badenhorst C.P.S., Bornscheuer U.T., Wei R. Weber Gert, Bornscheuer Uwe T., Wei Ren., editors. Methods Enzymol. 2021;648:253–270. doi: 10.1016/bs.mie.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 140.Bell E.L., et al. Directed evolution of an efficient and thermostable PET depolymerase. Nature Catal. 2022;5:673–681. doi: 10.1038/s41929-022-00821-3. [DOI] [Google Scholar]
- 141.Zurek P.J., Hours R., Schell U., Pushpanath A., Hollfelder F. Growth amplification in ultrahigh-throughput microdroplet screening increases sensitivity of clonal enzyme assays and minimizes phenotypic variation. Lab Chip. 2021;21:163–173. doi: 10.1039/D0LC00830C. [DOI] [PubMed] [Google Scholar]
- 142.Holstein J.M., Gylstorff C., Hollfelder F. Cell-free directed evolution of a protease in microdroplets at ultrahigh throughput. ACS Synth. Biol. 2021;10:252–257. doi: 10.1021/acssynbio.0c00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.