Abstract
Background
Penile cancer is a rare tumor and few studies have focused on the prognosis of M0 penile squamous cell carcinoma (PSCC). This retrospective study aimed to identify independent prognostic factors and construct predictive models for the overall survival (OS) and cancer-specific survival (CSS) of patients with M0 PSCC.
Methods
Data was extracted from the Surveillance, Epidemiology, and End Results database for patients diagnosed with malignant penile cancer. Eligible patients with M0 PSCC were selected according to predetermined inclusion and exclusion criteria. These patients were then divided into a training set, a validation set, and a test set. Univariate and multivariate COX regression analyses were initially performed to identify independent prognostic factors for OS and CSS in M0 PSCC patients. Subsequently, traditional and machine learning prognostic models, including random survival forest (RSF), COX, gradient boosting, and component-wise gradient boosting modelling, were constructed using the scikit-survival framework. The performance of each model was assessed by calculating time-dependent area under curve (AUC), C-index, and integrated Brier score (IBS), ultimately identifying the model with the highest performance. Finally, the Shapley additive explanation (SHAP) value, feature importance, and cumulative rates analyses were used to further estimate the selected model.
Results
A total of 2, 446 patients were included in our study. Cox regression analyses demonstrated that age, N stage, and tumor size were predictors of OS, while the N stage, tumor size, surgery, and residential area were predictors of CSS. The RSF and COX models had a higher time-independent AUC and C-index, and lower IBS value than other models in OS and CSS prediction. Feature importance analysis revealed the N stage as a common significant feature for predicting M0 PSCC patients’ survival. The SHAP and cumulative rate analyses demonstrated that the selected models can effectively evaluate the prognosis of M0 PSCC patients.
Conclusion
In M0 PSCC patients, age, N stage, and tumor size were predictors of OS. In addition, the N stage, tumor size, surgery, and residential area were predictors of CSS. The machine learning-based RSF and COX models effectively predicted the prognosis of M0 PSCC patients.
Keywords: Penile cancer, Penile squamous cell carcinoma, Survival, Machine learning, Random survival forest
1. Introduction
Penile cancer is a relatively rare tumor of the male urogenital system. Based on a 2022 statistical research, 1,583 new penile cancer cases may be diagnosed, and 435 patients may die of penile cancer in the USA [1]. Squamous cell carcinomas account for about 95 % of all penile cancers [2]. Despite the low incidence rate in developed countries, penile squamous cell carcinoma (PSCC) constitutes up to 10 % of male malignancies in certain parts of Africa, South America and Asia [3]. Hence, PSCC remains a serious worldwide male health problem and requires further research.
The prognosis of rate tumors such as PSCC can be analyzed by predictors and constructing predictive models. Lymph node metastasis is one of the main factors affecting penile cancer prognosis [4]. Previous studies reported that a five-year survival rate as low as 10–20 % in patients with inguinal lymph node metastasis, and a 96 % five-year survival rate in patients without lymph node metastasis [5,6]. Furthermore, distant metastasis is rare in penile cancer, and significantly affected survival. Therefore, a prognostic study including only penile cancer patients without distant metastases may provide a better understanding of this rare tumor. Li et al. performed a retrospective study and found that the primary site was a prognostic factor in M0 PSCC [7]. However, this study included a small number of patients and combined primary site data. Currently, only a few large-sample studies have investigated M0 PSCC with machine learning.
The Surveillance, Epidemiology, and End Results (SEER) database is a systematic population-based cancer database that provides the largest cohort of PSCC patients [8]. The Cox regression model, commonly utilized in prognostic analysis studies, assumes linear correlations between predictors and survival outcomes. However, this model is constrained by its inability to account for non-linear associations. In contrast, machine learning, a subfield of artificial intelligence, can effectively capture non-linear associations between variables and prognosis by applying large training datasets and adjustable algorithm parameters [9,10]. Machine-learning algorithms offer an alternative approach to traditional prediction models and may address their limitations to improve model accuracy [11]. Therefore, the current research had two objectives: (1) identifying the prognostic factors for overall survival (OS) and cancer-specific survival (CSS) in M0 PSCC patients; (2) constructing machine learning models to predict OS and CSS in M0 PSCC patients.
2. Methods
2.1. Patient population and study design
Penile cancer patient data were obtained from the SEER database by using the SEER*Stat software (Version 8.4.0.1, ID:14120-nov2021). The data were screened against the inclusion and exclusion criteria. Patients were enrolled if the following inclusion criteria were met: (1) The pathological diagnosis was penile squamous cell carcinoma; (2) Penile cancer was the first primary tumor; (3) T stage was available; (4) M0 stage patients. Exclusion criteria: (1) patients were diagnosed only by autopsy or death certificate; (2) T stage of Tx, T0, Cx, Px, or 88; (3) N stage of Nx, Cx, or Px; (4) Unknown race; (5) Unknown marital status; (6) Unkown residential information; (7) The value of radiation therapy was ‘recommended, unknown if administered’; (8) Unclear tumor size (code: 990, 994,999); (9) Unclear surgery (code:99). In addition, patients were excluded from CSS analysis when cancer-specific death was unknown.
The patients diagnosed from 2004 to 2015 were randomly divided into a training set and a validation set following a 7:3 ratio. The dataset for 2016–2017 and 2018–2019 were combined as the test set. To account for the differences in TNM staging systems, the TNM stage in the test set was re-staged according to the AJCC 6th edition.
2.2. Features and outcomes
A total of 12 features were selected, including age, race, primary site, T stage, N stage, tumor size, surgery, chemotherapy, radiation therapy, marital status, residential area, and median household income. Age was divided into three subgroups: <50,50–69, and 70 above. Race was categorized into white, black, and other. Tumor size was divided into <3 cm, 3–5 cm, and >5 cm. According to the surgical site and types, surgery was categorized as none, local tumor (local tumor destruction/excision, code:10–27), local excision of primary sit, total excision of primary site, and others (debulking, radical surgery and NOS-surgery, code: 50–90). The patients were further categorized by whether they underwent chemotherapy and radiation or not. Finally, the outcomes included OS status, survival time and CSS status.
2.3. Model building
Univariate COX regression analysis was performed on the training set to identify features correlating with OS and CSS. Subsequently, multivariate COX analyses were performed to identify survival-related prognostic features. Based on these features, RSF (random survival forest), COX, gradient boosting (GB), and component-wise gradient boosting (CGB) models were constructed using scikit-survival (version 0.20.0). These models are based on different algorithms. The RSF model fits a set of survival trees independently and then averages their predictions, while the GB model is constructed sequentially in a greedy stagewise fashion. CGB uses component-wise least squares as base learner. Usually, estimators have hyper-parameters that are optimize by the operator. Therefore, scikit-learn's GridSearchCV was used to determine the most effective hyper-parameter configuration on average. The hyper-parameters (n_estimators, max_depth) between the RSF and GB models were searched by GridSearchCV. In the CGB model, n_estimators were obtained by GridSearchCV, whereas other parameters were fixed (learn_rate = 1, random_state = 0).
2.4. Model evaluation, selection, and explanation
Time-dependent area under curve (AUC), C-index, and integrated Brier score (IBS) were used to evaluate the performance of each model in data sets. Based on these performances, an effective and accurate model was selected for further analysis. The importance and weight of features in the selected model were implemented using scikit-learn's permutation_importance function. Furthermore, Shapley Additive Explanations (SHAP) were drawn based on training set to explain the selected model [12]. The cumulative survival and risk rate of partial validation set patients were calculated using the selected model.
2.5. Operation and analysis
Data processing, grouping, and COX-regression survival analysis were performed using R version 4.2.2. Model building, evaluation, and explanation were carried out with Python version 3.9.13. The R package “survival” version 3.4-0 and Python package scikit-learn version 1.2.2 were used. Categorical variables were presented as numbers (percentages). In this study, P < 0.05 represented a statistically significant difference.
3. Results
3.1. Baseline characteristics
In total, the data of 6,520 penile cancer patients were initially obtained from the SEER database. After strict screening, 2,446 M0 PSCC patients were enrolled in the study, including 1,214 in the training set, 521 in the validation set, and 711 in the test set. CSS data was available in 2,426 patients, with 1,206 patients included in the training set, 515 in the validation set, and 705 in the test set. The patient screening and overall analysis sub-flows are displayed in Fig. 1A and B respectively. Table 1 shows the distribution of each feature in the OS analysis. Approximately 40 % of the patients were 70 or older, 85 % were white, 60 % were married, 50 % lived in large metropolitan areas, and 48 % had an annual income of $50,000–69,999. In terms of tumor-related information, 40 % of patients had unclear penis primary sits, 47 % of patients had a tumor size < 3 cm, 50 % demonstrated T1 stage cancer, and 80 % were N0 stage. Regarding treatment, 55 % of patients underwent local excision of the primary site, 90 % of the patients received no chemotherapy, and 91 % of patients had no radiotherapy. The OS analysis revealed that 1,095 patients died during the follow-up, including 654 in the training set, 290 in the validation set, and 151 in the test set. Furthermore, the CSS analysis indicated that 477 patients died due to PSCC, including 260 in the training set, 126 in the validation set, and 91 in the test set.
Fig. 1.
Flow chart of the patient selection process (A) and analysis process (B).
Table 1.
Demographic characteristics of M0 penile squamous cell carcinoma patients.
| Features | Training Set (N = 1214) | Validation Set (N = 521) | Test Set (N = 711) |
|---|---|---|---|
| Age, years | |||
| <50 | 187 (15.4 %) | 77 (14.8 %) | 108 (15.2 %) |
| 50–59 | 223 (18.4 %) | 103 (19.8 %) | 148 (20.8 %) |
| 60–69 | 324 (26.7 %) | 131 (25.1 %) | 181 (25.5 %) |
| ≥70 | 480 (39.5 %) | 210 (40.3 %) | 274 (38.5 %) |
| Race | |||
| White | 1035 (85.3 %) | 438 (84.1 %) | 620 (87.2 %) |
| Black | 104 (8.6 %) | 54 (10.4 %) | 52 (7.3 %) |
| Others | 75 (6.1 %) | 29 (5.5 %) | 39 (5.5 %) |
| Primary Site | |||
| Prepuce | 160 (13.2 %) | 61 (11.7 %) | 68 (9.7 %) |
| Glans | 447 (36.8 %) | 204 (39.1 %) | 271 (38.1 %) |
| Body | 63 (5.2 %) | 26 (5.0 %) | 36 (5.0 %) |
| Overlapping lesion | 61 (5.0 %) | 27 (5.2 %) | 40 (5.6 %) |
| Penis, NOS | 483 (39.8 %) | 203 (39.0 %) | 296 (41.6 %) |
| T | |||
| Ta | 9 (0.8 %) | 3 (0.6) | 4 (0.5 %) |
| T1 | 652 (53.7 %) | 286 (54.9 %) | 346 (48.7 %) |
| T2 | 334 (27.5 %) | 129 (24.8 %) | 189 (26.6 %) |
| T3 | 198 (16.3 %) | 93 (17.8 %) | 156 (21.9 %) |
| T4 | 21 (1.7 %) | 10 (1.9 %) | 16 (2.3 %) |
| N | |||
| N0 | 994 (81.9 %) | 424 (81.4 %) | 575 (80.9 %) |
| N1 | 79 (6.5 %) | 38 (7.3 %) | 41 (5.7 %) |
| N2 | 83 (6.8 %) | 40 (7.7 %) | 43 (6.1 %) |
| N3 | 58 (4.8 %) | 19 (3.6 %) | 52 (7.3 %) |
| Tumor size, cm | |||
| <3 | 597 (49.2 %) | 245 (47.0 %) | 325 (45.7 %) |
| 3–5 | 432 (35.6 %) | 180 (34.6 %) | 238 (33.5 %) |
| >5 | 185 (15.2 %) | 96 (17.4 %) | 148 (20.8 %) |
| Surgery | |||
| None | 28 (2.3 %) | 13 (2.5 %) | 13 (1.8 %) |
| Local tumor | 321 (26.4 %) | 139 (26.7 %) | 174 (24.4 %) |
| Local excision | 676 (55.7 %) | 284 (54.5 %) | 390 (54.9 %) |
| Total excision | 141 (11.6 %) | 63 (12.1 %) | 100 (14.1 %) |
| Others | 48 (4.0 %) | 22 (4.2 %) | 34 (4.8 %) |
| Chemotherapy | |||
| No | 1094 (90.0 %) | 472 (89.4 %) | 618 (86.9 %) |
| Yes | 120 (10.0 %) | 49 (10.6 %) | 93 (13.1 %) |
| Radiotherapy | |||
| No | 1113 (91.7 %) | 480 (92.1 %) | 660 (92.8 %) |
| Yes | 101 (8.3 %) | 41 (7.9 %) | 51 (7.3 %) |
| Marital Status | |||
| Married | 735 (60.5 %) | 310 (59.5 %) | 426 (60.0 %) |
| Single | 232 (19.1 %) | 94 (18.0 %) | 151 (21.2 %) |
| Divorced | 142 (11.7 %) | 53 (10.2 %) | 72 (10.1 %) |
| Widowed | 105 (8.7 %) | 64 (12.3 %) | 62 (8.7 %) |
| Residential Area | |||
| Large metropolitan | 619 (51.0 %) | 295 (56.7 %) | 368 (51.7 %) |
| Middle metropolitan | 275 (22.7 %) | 95 (18.2 %) | 168 (23.6 %) |
| Small metropolitan | 103 (8.5 %) | 46 (8.8 %) | 59 (8.3 %) |
| Contiguous nonmetropolitan | 116 (9.5 %) | 52 (10.0 %) | 63 (8.9 %) |
| Remote nonmetropolitan | 101 (8.3 %) | 33 (6.3 %) | 53 (7.5 %) |
| Median Household Income | |||
| < $50,000 | 261 (21.5 %) | 96 (18.4 %) | 134 (18.8 %) |
| $50,000–69,999 | 613 (50.5 %) | 244 (46.8 %) | 326 (45.9 %) |
| ≥$70,000 | 340 (28.0 %) | 181 (34.8 %) | 251 (35.3 %) |
3.2. Model construction
Univariate COX regression analysis showed that OS was associated with age, T stage, N stage, tumor size, surgery, chemotherapy, radiotherapy, and marital status (Table 2). Subsequently, the multivariate COX regression analysis based on the above features revealed that age, N stage, and tumor size were independent prognostic factors for OS. The RSF, COX, GB, and CGB prognostic models were then constructed with the prognostic factors. The optimal hyper-parameters were 3 max depth and 88 estimators for the RSF model, 2 max depth and 161 estimators for the GB model, and 43 estimators for the CGB model.
Table 2.
Univariate and multivariate Cox regression analysis of M0 squamous cell penile carcinoma.
| Univariate Analysis |
Multivariate Analysis |
|||||||
|---|---|---|---|---|---|---|---|---|
| OS |
CSS |
OS |
CSS |
|||||
| HR | P | HR | P | HR | P | HR | P | |
| Age, years | ||||||||
| <50 | Reference | Reference | – | |||||
| 50-59 | 1.063 | 0.717 | 0.905 | 0.651 | 1.124 | 0.495 | ||
| 60-69 | 1.465 | 0.012 | 0.973 | 0.893 | 1.465 | 0.013 | ||
| ≥70 | 3.059 | <0.001 | 1.300 | 0.161 | 3.172 | <0.001 | ||
| Race | ||||||||
| Black | Reference | Reference | – | – | ||||
| Others | 0.688 | 0.078 | 0.548 | 0.081 | ||||
| White | 0.853 | 0.232 | 0.746 | 0.144 | ||||
| Primary Site | ||||||||
| Body | Reference | Reference | – | |||||
| Glans | 0.867 | 0.413 | 0.914 | 0.737 | 1.353 | 0.295 | ||
| Prepuce | 0.715 | 0.089 | 0.502 | 0.036 | 1.235 | 0.549 | ||
| Overlapping lesion | 0.806 | 0.373 | 0.934 | 0.853 | 1.315 | 0.285 | ||
| Penis, NOS | 0.882 | 0.570 | 0.851 | 0.548 | 1.355 | 0.285 | ||
| T | ||||||||
| T1 | Reference | Reference | – | – | ||||
| Ta | 0.955 | 0.927 | 1.449 | 0.604 | 0.964 | 0.941 | 1.849 | 0.393 |
| T2 | 1.380 | <0.001 | 1.776 | <0.001 | 1.050 | 0.636 | 1.014 | 0.935 |
| T3 | 1.519 | <0.001 | 2.549 | <0.001 | 1.069 | 0.589 | 1.130 | 0.513 |
| T4 | 2.304 | 0.002 | 4.348 | <0.001 | 1.344 | 0.302 | 1.547 | 0.263 |
| N | ||||||||
| N0 | Reference | Reference | – | – | ||||
| N1 | 1.612 | 0.002 | 2.820 | <0.001 | 1.602 | 0.003 | 2.575 | <0.001 |
| N2 | 2.139 | <0.001 | 4.438 | <0.001 | 2.100 | <0.001 | 3.409 | <0.001 |
| N3 | 2.633 | <0.001 | 5.907 | <0.001 | 2.755 | <0.001 | 4.344 | <0.001 |
| Tumor size, cm | ||||||||
| <3 | Reference | Reference | – | – | ||||
| 3-5 | 1.346 | <0.001 | 1.670 | <0.001 | 1.189 | 0.060 | 1.349 | 0.045 |
| >5 | 1.565 | <0.001 | 2.536 | <0.001 | 1.330 | 0.020 | 1.757 | 0.002 |
| Surgery | ||||||||
| Local excision | Reference | Reference | – | – | ||||
| Local tumor | 0.700 | <0.001 | 0.450 | <0.001 | 0.936 | 0.550 | 0.665 | 0.044 |
| None | 1.449 | 0.106 | 1.741 | 0.089 | 0.963 | 0.879 | 0.934 | 0.846 |
| Others | 1.316 | 0.131 | 1.718 | 0.034 | 1.289 | 0.181 | 1.313 | 0.313 |
| Total excision | 1.133 | 0.311 | 1.433 | 0.039 | 0.988 | 0.925 | 1.048 | 0.801 |
| Chemotherapy | ||||||||
| No | Reference | Reference | – | – | ||||
| Yes | 1.486 | 0.001 | 2.858 | <0.001 | 0.997 | 0.984 | 1.015 | 0.939 |
| Radiotherapy | ||||||||
| No | Reference | Reference | – | – | ||||
| Yes | 1.665 | <0.001 | 2.814 | <0.001 | 1.133 | 0.389 | 1.436 | 0.059 |
| Marital Status | ||||||||
| Divorced | Reference | Reference | – | |||||
| Married | 0.786 | 0.050 | 0.843 | 0.373 | 0.799 | 0.071 | ||
| Single | 0.870 | 0.333 | 1.012 | 0.957 | 1.039 | 0.797 | ||
| Widowed | 1.645 | 0.001 | 0.893 | 0.693 | 1.226 | 0.208 | ||
| Residential Area | ||||||||
| Contiguous nonmetropolitan | Reference | Reference | – | |||||
| Large metropolitan | 0.816 | 0.123 | 0.619 | 0.012 | 0.634 | 0.052 | ||
| Middle metropolitan | 0.861 | 0.304 | 0.611 | 0.023 | 0.623 | 0.056 | ||
| Remote nonmetropolitan | 1.210 | 0.266 | 1.068 | 0.789 | 1.059 | 0.818 | ||
| Small metropolitan | 0.731 | 0.087 | 0.439 | 0.006 | 0.420 | 0.006 | ||
| Median Household Income | ||||||||
| $50,000–69,999 | Reference | Reference | – | |||||
| < $50,000 | 1.051 | 0.617 | 1.352 | 0.045 | 0.995 | 0.981 | ||
| ≥$70,000 | 0.837 | 0.059 | 0.954 | 0.757 | 0.953 | 0.764 | ||
In CSS, the univariate COX regression analysis followed by multivariate COX regression analysis found that the N stage, tumor size, surgery, and residential area were independent prognostic factors. Similarly, the RSF, COX, GB, and CGB prognostic models were constructed based on the prognostic factors. The optimal hyperparameters included 1 max depth and 135 estimators for the RSF model, 1 max depth and 259 estimators for the GB model, and 113 estimators for the CGB model.
3.3. Model evaluation and selection
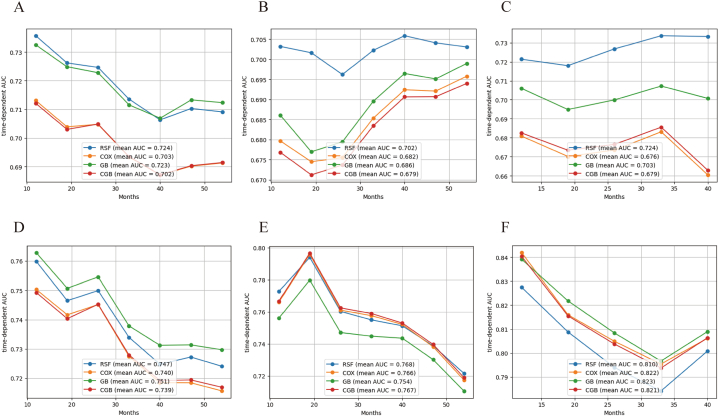
In the OS analysis, the RSF model resulted in a mean time-dependent AUC of 0.724, which was superior to that of other models in the training set (Fig. 2 A). Furthermore, the mean time-dependent AUC in the validation set was: 0.702 (RSF),0.682 (COX), 0.663 (GB), and 0.672 (CGB) (Fig. 2 B). In addition, the C-index and IBS of the RSF model were superior to that of the other models in the validation and test set, as displayed in Table 3. In the test set, the RSF model reached a mean time-dependent AUC of 0.724, which was significantly higher than that of other models (Fig. 2C). Hence, the RSF model outperformed other models in predicting the OS of M0 PSCC patients and was selected for further analysis.
Fig. 2.
Time-dependent AUC in models and datasets. (A) Time-dependent AUC of overall survival in training set; (B) Time-dependent AUC of overall survival in validation set; (C) Time-dependent AUC of overall survival in test set; (D)Time-dependent AUC of cancer specific survival in training set; (E.) Time-dependent AUC of cancer specific survival in validation set; (F) Time-dependent AUC of cancer specific survival in test set.
Table 3.
Model performance summary.
| Validation Set |
Test Set |
|||
|---|---|---|---|---|
| OS Model | C-index | IBS | C-index | IBS |
| RSF | 0.662 | 0.206 | 0.675 | 0.094 |
| COX | 0.662 | 0.203 | 0.664 | 0.096 |
| GB | 0.659 | 0.204 | 0.681 | 0.095 |
| CGB | 0.661 | 0.205 | 0.666 | 0.098 |
| Random | 0.500 | 0.250 | 0.500 | 0.146 |
| Kaplan-Meier | – | 0.230 | – | 0.107 |
| CSS Model | ||||
| RSF | 0.719 | 0.168 | 0.774 | 0.074 |
| COX | 0.715 | 0.160 | 0.785 | 0.065 |
| GB | 0.709 | 0.160 | 0.783 | 0.067 |
| CGB | 0.716 | 0.160 | 0.784 | 0.066 |
| Random | 0.500 | 0.246 | 0.500 | 0.141 |
| Kaplan-Meier | – | 0.182 | – | 0.081 |
In the CSS analysis, the GB model exhibited the best time-dependent AUC in the training set (Fig. 2 D). However, no significant difference in predicting CSS was observed among the RSF, COX, and CGB models in the validation set (Fig. 2 E). In the test set, the COX model exhibited a high mean AUC (0.822), C-index (0.785) and low IBS (0.065), which was superior to that of other models (Fig. 2 F, Table 3). Considering the stability and accuracy of results, the COX model was identified as the optimal model to predict the CSS of M0 PSCC patients.
3.4. Feature importance of the model
Table 4 shows the importance and weight of each prognostic features. N stage was the most important common predictor between the OS and CSS analysis. In contrast, age was the most significant prognostic feature in OS, whereas the N stage was the most significant prognostic factor in CSS. Compared with the COX model, the RSF model exhibited a lower weight for the N stage and age in OS analysis (0.041 ± 0.006,0.109 ± 0.010). However, the importance and weight of tumor size were higher in COX model predicting CSS.
Table 4.
Feature importance calculated based on permutation importance.
| RSF |
COX |
||||||
|---|---|---|---|---|---|---|---|
| OS |
CSS |
OS |
CSS |
||||
| Feature | Weight | Feature | Weight | Feature | Weight | Feature | Weight |
| Age | 0.109 ± 0.010 | N | 0.091 ± 0.010 | Age | 0.111 ± 0.011 | N | 0.082 ± 0.009 |
| N | 0.041 ± 0.006 | Surgery | 0.028 ± 0.005 | N | 0.043 ± 0.007 | Tumor Size | 0.024 ± 0.008 |
| Tumor Size | 0.021 ± 0.004 | Tumor Size | 0.016 ± 0.006 | Tumor Size | 0.014 ± 0.003 | Surgery | 0.013 ± 0.003 |
| Residential Area | 0.005 ± 0.001 | Residential Area | 0.009 ± 0.005 | ||||
Fig. 3 illustrates the performance of the RSF model and COX model in the training set. Advanced age, higher N stage, and larger tumor size contributed to higher risk scores in OS (Fig. 3 A). Conversely, higher N stage, larger tumor size, living in remote nonmetropolitan areas, and not undergoing surgery were associated with high-risk scores in CSS (Fig. 3 B). Three patients were selected to show detailed SHAP values in OS and CSS risk (Fig. 3C, D).
Fig. 3.
SHAP plot of overall survival and cancer specific survival. (A) SHAP summary plot of overall survival; (B) SHAP summary plot of cancer specific survival; (C) SHAP value plot in three overall survival analysis patients. (D) SHAP value plot in three cancer specific survival analysis patients.
3.5. Model demonstration on patients
Six patients were selected from the validation set to evaluated the performance of the RSF and COX models. The table in Fig. 4 shows the detailed features and survival information of the selected patients. The survival probability over time in terms of OS and CSS are displayed in Fig. 4 A and C, respectively. Moreover, the cumulative mortality risk for all-cause and cancer-specific death are presented in Fig. 4 B and D, respectively. According to the prediction model, patients with higher predictive scores have a lower survival rate and a higher risk rate. In the CSS analysis, patients with ID 0 and ID 2 had overlapping survival and risk rate curves due to the same COX predictive score.
Fig. 4.
Six patients' survival functions and cumulative curve functions were predicted by the selected models. (A) cumulative survival rate curve of overall survival. (B) cumulative hazard rate curve of overall survival. (C) cumulative survival rate curve of cancer-specific survival. (D) cumulative hazard rate curve of cancer-specific survival.
4. Discussion
Our study found that age, N stage, and tumor size were prognostic factors for OS in M0 PSCC patients. Additionally, N stage, tumor size, surgery, and residential area were prognostic factors for CSS in M0 PSCC patients. Based on the above prognostic factors, the performance of the RSF model and COX model were found to be superior to other models. A high events-to-predictor ratio confirmed the reliability of the results. In each set, the number of deaths was more than 50, whereas the number of predictors was at most four, yielding an events-to-predictor of ratio of >10. Owing to the short clinical follow-up of the patients, the time-dependent AUC of the test set was different from that of the training set and validation set.
In OS analysis, age was the prognostic factor with the largest weight. Compared with age <60 patients, the patients in the 60–69 and 70 or higher age groups had higher overall death risk (HR:1.465, HR:3.059). Similar results have been reported in previous penile cancer research and other tumor studies [12]. However, no significant difference in overall death risk was observed between patients aged <50 and 50–59. N stage were second most important feature in predicting OS. Furthermore, the SHAP value showed that the overall death risk score of N3 patients was significantly higher than that of N2, N1, and N0 patients. Previous studies also found that the N stage associated with overall survival. Soodana-Prakash et al. performed a COX regression analysis involving 364 PSCC patients and found that the positive N stage was an independent predictor of poor OS [13]. Xu et al. Reported similar results [14].Therefore, the N stage and intraoperative lymph node examination play an important role and correlate directly with the survival of the M0 PSCC patients. Consistent with previous studies, tumor size was also identified as an OS prognostic factor [15,16]. Larger tumors are associated with poorer OS prognosis. The HR of patients with >5 cm tumors size was significantly higher than those with 3–5 cm tumors size and <3 tumors size. Nevertheless, whether marital status and race are predictors of OS in penile cancer remains controversial. Chen et al. evaluated 1,643 penile cancer patients and performed a multivariate COX analysis, revealing that marital status and race were independent prognostic risk factors associated with OS. However, univariate COX analyses found no significant difference in race. Our univariate analysis found that widowed patients have a higher risk of overall death risk (HR:1.645), while no significant difference in overall death was observed in the multivariate COX analysis. Similarly, Zheng et al. found that age and N stage were prognostic factor of OS in PSCC. Notably, the P and HR values of patients with a household income of more than $70,000 were 0.059 and 0.837, respectively. Although no significant difference was observed in the median household income subgroups, a higher-income M0 PSCC patients might have a better OS prognosis (HR:0.837, P:0.059).
Moreover, the N stage was also significantly associated with CSS in M0 PSCC patients. Our multivariate COX and SHAP analysis demonstrated that N3-stage patients exhibited higher HR and risk scores. Several studies reported the importance of the N stage in predicting the CSS in M0 PSCC. Kawase et al. performed a retrospective study, reporting that the N stage was a vital prognostic indicator for CSS in PSCC patients [17]. In addition, AI-Najar et al. included 89 PSCC patients and reported a similar conclusion [18]. Tumor size was secondary most important variable. In our multivariate COX analysis, the HR of patients with tumors larger than 5 cm was higher than in patients with 3–5 cm tumors. Similarly, Zhu et al. reported that tumor size was related to CSS in penile cancer [19]. However, few studies reported the effect of the residential area and surgery on CSS. This study found an association between metropolitan patients and lower cancer-specific death risk (HR < 1). This finding may be related to higher health awareness and access to medical resources in large metropolitan areas.
Machine learning is widely used to aid clinical decision-making due to its advanced data analysis and model fitting. Currently, no machine learning-based prognostic study has investigated M0 PSCC patients. Li et al. performed a Kaplan-Meier analysis study and found that the tumor site affected survival prognosis in M0 PSCC patients [7]. Our study subdivided the tumor site and yielded a different conclusion. Furthermore, our models suggested that the N stage and advanced age significantly affected prognosis. Lymph node biopsy and imaging in patients with PSCC should be performed thoroughly. Moreover, online predictive models based on RSF and COX can offer a convenient and effective tool in the clinical management of M0 PSCC patients.
Nevertheless, the limitations of the current study should be acknowledged. First, due to the limitation of the scikit-survival function, we selected only four machine-learning algorithms, namely RSF, COX, GB, and CGB. Evaluating additional machine learning may improve the results. Secondly, to ensure an appropriate sample size, some important information, such as lymph-vascular invasion and HPV infection, was not be included. These important variables also influence the prognosis of patients and should be considered in future studies. Thirdly, no external validation was performed duo to the rarity of penile cancer. External validation sets from other clinical centers would provide a better assessment of the performance of the model. Still, the SEER database remains a valuable source for studying such rare cancers.
5. Conclusion
Age, N stage, and tumor size were independent risk factors affecting the OS of M0 PSCC patients, while N stage, tumor size, surgery and residential area were independent risk factors for CSS. Furthermore, the RSF and COX models effectively predicted the OS and CSS, respectively.
Funding
None
Data availability statement
All data of this study can be obtained from the Surveillance, Epidemiology, and End Results (SEER) database (www.seer.cancer.gov): incidence-SEER Research Plus Data, 18 Registries, Nov 2020 Sub. The login account was 14120-nov2021.
CRediT authorship contribution statement
Di Chen: Writing – original draft. Shengsheng Liang: Conceptualization, Data curation, Formal analysis. Jinji Chen: Conceptualization, Data curation, Formal analysis. Kezhen Li: Visualization. Hua Mi: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Home for Researchers editorial team (www.home-for-researchers.com) for language editing service.
References
- 1.Xia C., Dong X., Li H., Cao M., Sun D., He S., Yang F., Yan X., Zhang S., Li N., et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin. Med. J. 2022;135(5):584–590. doi: 10.1097/CM9.0000000000002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang T., Cheng X., Chahoud J., Sarhan A., Tamboli P., Rao P., Guo M., Manyam G., Zhang L., Xiang Y., et al. Effective combinatorial immunotherapy for penile squamous cell carcinoma. Nat. Commun. 2020;11(1):2124. doi: 10.1038/s41467-020-15980-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downes M.R. Review of in situ and invasive penile squamous cell carcinoma and associated non-neoplastic dermatological conditions. J. Clin. Pathol. 2015;68(5):333–340. doi: 10.1136/jclinpath-2015-202911. [DOI] [PubMed] [Google Scholar]
- 4.Yu J., Long Q., Zhang Z., Liao S., Zheng F. The prognostic value of lymph node ratio in comparison to positive lymph node count in penile squamous cell carcinoma. Int. Urol. Nephrol. 2021;53(12):2527–2540. doi: 10.1007/s11255-021-02996-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ottenhof S.R., Djajadiningrat R.S., Thygesen H.H., Jakobs P.J., Jozwiak K., Heeren A.M., de Jong J., Sanders J., Horenblas S., Jordanova E.S. The prognostic value of immune factors in the tumor microenvironment of penile squamous cell carcinoma. Front. Immunol. 2018;9:1253. doi: 10.3389/fimmu.2018.01253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagliaro L.C., Crook J. Multimodality therapy in penile cancer: when and which treatments? World J. Urol. 2009;27(2):221–225. doi: 10.1007/s00345-008-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K., Le X., Wang J., Fan C., Sun J. Tumor Location may independently predict survival in patients with M0 squamous cell carcinoma of the penis. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.927088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourlon M.T., Verduzco-Aguirre H., Molina E., Meyer E., Kessler E., Kim S.P., Spiess P.E., Flaig T. Patterns of treatment and outcomes in older men with penile cancer: a SEER dataset analysis. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.926692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Zhao Y., Liu X., Huang J. Artificial intelligence for prediction of response to cancer immunotherapy. Semin. Cancer Biol. 2022;87:137–147. doi: 10.1016/j.semcancer.2022.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., Wei X. Artificial intelligence-assisted selection and efficacy prediction of antineoplastic strategies for precision cancer therapy. Semin. Cancer Biol. 2023;90:57–72. doi: 10.1016/j.semcancer.2023.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Uddin S., Khan A., Hossain M.E., Moni M.A. Comparing different supervised machine learning algorithms for disease prediction. BMC Med. Inf. Decis. Making. 2019;19(1):281. doi: 10.1186/s12911-019-1004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scavuzzo C.M., Scavuzzo J.M., Campero M.N., Anegagrie M., Aramendia A.A., Benito A., Periago V. Feature importance: opening a soil-transmitted helminth machine learning model via SHAP. Infect Dis Model. 2022;7(1):262–276. doi: 10.1016/j.idm.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soodana-Prakash N., Koru-Sengul T., Miao F., Lopategui D.M., Savio L.F., Moore K.J., Johnson T.A., Alameddine M., Barboza M.P., Parekh D.J., et al. Lymph node yield as a predictor of overall survival following inguinal lymphadenectomy for penile cancer. Urologic Oncology-Seminars and Original Investigations. 2018;36(10) doi: 10.1016/j.urolonc.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Xu W.B., Qi F., Liu Y., Zheng L.Z., Kang Z.J. Nomograms to predict overall and cancer-specific survival in patients with penile cancer. Transl. Cancer Res. 2020;9(4):2326–2339. doi: 10.21037/tcr.2020.03.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li K., Wu G., Fan C., Yuan H. The prognostic significance of primary tumor size in squamous cell carcinoma of the penis. Discov Oncol. 2021;12(1):22. doi: 10.1007/s12672-021-00416-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez D.F., Soares F., Alvarado-Cabrero I., Canete S., Fernandez-Nestosa M.J., Rodriguez I.M., Barreto J., Cubilla A.L. Pathological factors, behavior, and histological prognostic risk groups in subtypes of penile squamous cell carcinomas (SCC) Semin. Diagn. Pathol. 2015;32(3):222–231. doi: 10.1053/j.semdp.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Kawase M., Takagi K., Kawada K., Ishida T., Tomioka M., Enomoto T., Fujimoto S., Taniguchi T., Ito H., Kameyama K., et al. Clinical lymph node involvement as a predictor for cancer-specific survival in patients with penile squamous cell cancer. Curr. Oncol. 2022;29(8):5466–5474. doi: 10.3390/curroncol29080432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Najar A., Alkatout I., Al-Sanabani S., Korda J.B., Hegele A., Bolenz C., Junemann K.P., Naumann C.M. External validation of the proposed T and N categories of squamous cell carcinoma of the penis. Int. J. Urol. 2011;18(4):312–316. doi: 10.1111/j.1442-2042.2011.02722.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Y., Gu W.J., Wang H.K., Gu C.Y., Ye D.W. Surgical treatment of primary disease for penile squamous cell carcinoma: a Surveillance, Epidemiology, and End Results database analysis. Oncol. Lett. 2015;10(1):85–92. doi: 10.3892/ol.2015.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data of this study can be obtained from the Surveillance, Epidemiology, and End Results (SEER) database (www.seer.cancer.gov): incidence-SEER Research Plus Data, 18 Registries, Nov 2020 Sub. The login account was 14120-nov2021.