Abstract
We have constructed an Escherichia coli strain lacking the small heat shock proteins IbpA and IbpB and compared its growth and viability at high temperatures to those of isogenic cells containing null mutations in the clpA, clpB, or htpG gene. All mutants exhibited growth defects at 46°C, but not at lower temperatures. However, the clpA, htpG, and ibp null mutations did not reduce cell viability at 50°C. When cultures were allowed to recover from transient exposure to 50°C, all mutations except Δibp led to suboptimal growth as the recovery temperature was raised. Deletion of the heat shock genes clpB and htpG resulted in growth defects at 42°C when combined with the dnaK756 or groES30 alleles, while the Δibp mutation had a detrimental effect only on the growth of dnaK756 mutants. Neither the overexpression of these heat shock proteins nor that of ClpA could restore the growth of dnaK756 or groES30 cells at high temperatures. Whereas increased levels of host protein aggregation were observed in dnaK756 and groES30 mutants at 46°C compared to wild-type cells, none of the null mutations had a similar effect. These results show that the highly conserved E. coli small heat shock proteins are dispensable and that their deletion results in only modest effects on growth and viability at high temperatures. Our data also suggest that ClpB, HtpG, and IbpA and -B cooperate with the major E. coli chaperone systems in vivo.
Living organisms respond to stressful environmental conditions by increasing the production of specific proteins which alleviate or reduce damage incurred by the cell. In Escherichia coli, temperature increase upregulates two groups of heat-shock proteins (Hsps) that are transcribed by the Eς32 and EςE holoenzymes. The ς32 regulon is implicated in the management of cellular stress in the cytoplasm, whereas EςE-transcribed proteins are specifically upregulated in response to stress in the periplasm or the cell envelope (reviewed in reference 13). While the identity and cellular function of most members of the ςE regulon remain unclear, a great deal of effort has been directed at understanding the regulation of ς32 and the role of Eς32-transcribed Hsps in the maintenance of heat-shocked cells. Most members of the ς32 regulon have been classified as either molecular chaperones or ATP-dependent proteases (13). Molecular chaperones, which include the DnaK-DnaJ-GrpE and GroEL-GroES systems, facilitate the proper folding of newly synthesized polypeptides and help thermally damaged proteins regain a biologically active conformation (14). Heat shock proteases, such as ClpP, Lon, and HflB, apparently degrade misfolded proteins that cannot be rescued by chaperone action (10). The signal responsible for induction of the heat shock response is believed to be an increase in the intracellular concentration of unfolded and misfolded proteins (13). This can be caused by temperature increase or other stresses, including phage infection; the presence of organic solvents, heavy metals, and certain antibiotics; and the production of aggregation-prone proteins (25, 27, 37).
The DnaK-DnaJ-GrpE and GroEL-GroES systems are the best-characterized molecular chaperones of E. coli. Based on in vitro studies and homology to eukaryotic proteins, other members of the ς32 regulon are also believed to perform a molecular chaperone function in vivo. These include the Clp ATPases (ClpB, ClpX, and ClpY), the Hsp90 homolog HtpG, and the small Hsps (sHsps) IbpA and IbpB (13, 30). To date, relatively little is known about the cellular functions of these “minor” chaperones. It has been shown that clpB null mutants exhibit growth defects at 44°C and undergo a higher rate of killing at 50°C than the wild type (31). Nevertheless, ClpB overexpression does not enhance the viability of wild-type E. coli at 55°C (8). While htpG null mutants also display growth defects at 44°C (4), little is known about the in vivo function of HtpG, except for a possible involvement in secretion (29, 36). IbpA and IbpB have been found in association with thermally aggregated host proteins (20) and recombinant protein inclusion bodies (2), but knowledge of their in vivo function is limited, since the construction and characterization of an ibp mutant have not been previously reported. Despite the fact that the Clp ATPase ClpA is not itself an Hsp, it displays molecular chaperone activity in vitro (40) and provides substrate specificity to the heat shock protease ClpP (17). While a clpA null mutation was previously found to have no effect on growth at 42°C (16), other aspects of the role of ClpA in thermal stress management have not been examined.
In this report, we have characterized the effects of deletion or overexpression of the sHsps IbpA and -B on the growth and viability of heat-shocked E. coli cells. Results were compared to the effects of deletion or overproduction of ClpA, ClpB, or HtpG. We further investigated the influence of manipulation of the intracellular concentration of these minor chaperones in dnaK756 and groES30 genetic backgrounds. We show that IbpA and -B are dispensable in E. coli and that ClpB, HtpG, and IbpA and -B cooperate with the major chaperone systems in the management of thermal stress in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, and routine growth conditions.
Relevant characteristics of the bacterial strains and plasmids used in this study are described in Tables 1 and 2. Top10 and XL1-blue cells were transformed with plasmid DNA by electroporation; all other transformations were performed by the RbCl method (Promega). Routine growth was carried out at 30°C in Luria-Bertani (LB) medium supplemented with the appropriate antibiotics at the following concentrations: chloramphenicol, 34 μg/ml; carbenicillin, 100 μg/ml; neomycin or kanamycin, 50 μg/ml; and streptomycin, 50 μg/ml.
TABLE 1.
E. coli strains used in this study
| Strain | Genotype and/or descriptiona | Source or referenceb |
|---|---|---|
| Top10 | F′ λ−endA1 recA1 hsdR17 (rK− mK+) supE44 thi1 gyrA96 relA1 φ80ΔlacZΔM15Δ(lacZYA-argF) U169 deoR | Invitrogen |
| XL1-blue | endA1 hsdR17 (rK− mK+) supE44 thi1 recA1 gyrA96 relA1 lac [F′ proAB lacIqlacZΔM15 Tn10] (Tetr) | Stratagene |
| JCB495 | MC1000 recD | C. Manoil |
| CC160 | MC1000 dam | C. Manoil |
| MC4100 | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | Laboratory stock |
| SG1126 | SG1101 clpA::kan | 11 |
| JGT4 | MC4100 clpA::kan | P1(SG1126) × MC4100→Neor Strr Lac− |
| MC1000ΔclpB | MC1000 ΔclpB::kan | 31 |
| JGT3 | MC4100 ΔclpB::kan | P1(MC1000ΔclpB) × MC4100→Neor Strr Lac− |
| JCB23 | MC1655 F−lacZ::Tn10 zba315::kan ΔhtpG1::lacZ | 4 |
| JGT11 | MC4100 zba315::kan ΔhtpG1::lacZ | P1(JCB23) × MC4100→Neor Strr Lac+ |
| JGT1 | JCB495 Δibp1::kan | LinTr(pBRΔibp::kan 14-3) × JCB495→Neor Amps |
| JGT17 | MC4100 Δibp1::kan | P1(JGT1) × MC4100→Neor Strr Lac− |
| CG800 | C600 leu dnaK756 thr::Tn10 | S. van der Vies |
| JGT20 | MC4100 dnaK756 thr::Tn10 | P1(CG800) × MC4100→Tetr Strr Ts44 Lac− |
| CG712 | B178 zjd::Tn10 groES30 | 34 |
| JGT6 | MC4100 zjd::Tn10 groES30 | P1(CG712) × MC4100→Tetr Strr Ts44 Lac− |
| JGT44 | MC4100 zjd::Tn10 | P1(CG712) × MC4100→Tetr Strr Lac− |
| SG1162a | SG1101 clpP::cat | 11 |
| JGT19 | MC4100 clpP::cat | P1(SG1162a) × MC4100→Chlr Strr Lac− |
| JGT31 | MC4100 dnaK756 thr::Tn10 clpA::kan | P1(SG1126) × JGT20→Neor Lac− |
| JGT32 | MC4100 dnaK756 thr::Tn10 ΔclpB::kan | P1(MC1000ΔclpB) × JGT20→Neor Lac− |
| JGT30 | MC4100 dnaK756 thr::Tn10 zba315::kan ΔhtpG1::lacZ | P1(JCB23) × JGT20→Neor Lac+ |
| JGT34 | MC4100 dnaK756 thr::Tn10 Δibp1::kan | P1(JGT1) × JGT20→Neor Lac− |
| JGT27 | MC4100 zjd::Tn10 groES30 clpA::kan | P1(SG1126) × JGT6→Neor Lac− |
| JGT38 | MC4100 zjd::Tn10 groES30 ΔclpB::kan | P1(CG712) × JGT3→Neor Ts44 Lac− |
| JGT39 | MC4100 zjd::Tn10 groES30 zba315::kan ΔhtpG1::lacZ | P1(CG712) × JGT11→Neor Ts44 Lac+ |
| JGT28 | MC4100 zjd::Tn10 groES30 Δibp1::kan | P1(JGT1) × JGT6→Neor Lac− |
| BT66 | MC4100 malG(Am) malTc-Tn10 secB::Tn5 (F′ lacpro lacIq) | B. Traxler |
| JGT43 | MC4100 clpP::cat (F′ lacpro lacIq) | F′(BT66) × JGT19→Chlr Lac+ Neos Tets |
| JGT47 | MC4100 zjd::Tn10 (F′ lacpro lacIq) | F′(JGT43) × JGT44→Tetr Lac+ Chls |
| JGT61 | MC4100 dnaK756 thr::Tn10 (F′ lacpro lacIq) | F′(JGT43 × JGT20→Tetr Lac+ Chls |
| JGT49 | MC4100 zjd::Tn10 groES30 (F′ lacpro lacIq) | F′(JGT43 × JGT6→Tetr Lac+ Chls |
Amp, ampicillin; Neo, neomycin or kanamycin; Chl, chloramphenicol; Tet, tetracycline; Str, streptomycin. Superscripts r and s indicate resistance and sensitivity, respectively. Lac refers to the formation of blue colonies on M9 plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG.
P1 transductions are represented as P1(donor) × recipient→phenotypes used for selection and screening. Linear transformations (LinTr) are represented as LinTr(plasmid) × recipient→phenotypes used for selection and screening. F′ mating is represented as F′(donor) × recipient→phenotypes used for selection and screening. Ts44 indicates that the selected cells were unable to form colonies at 44°C.
TABLE 2.
E. coli plasmids used in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pMON18003 | pUC19 derivative carrying a 5.2-kbp fragment of the E. coli chromosome containing the ibp operon (Ampr) | 2 |
| pBRibp | pBR322 derivative encoding the ibp operon and surrounding genomic DNA (Ampr) | This study |
| pBSL14 | Encodes the gene for neomycin phosophotransferase flanked by multiple restriction sites (Neor) | 1 |
| pBRΔibp::kan14-3 | pBRibp derivative with most of the ibp operon removed by BstXI digestion and a Neor cartridge from pBSL14 inserted at this location (Ampr Neor) | This study |
| pWPC3 | pUC19 derivative encoding the clpA gene (Ampr) | 11 |
| pclpB | pBR322 derivative encoding the clpB gene (Ampr) | 31 |
| pBJ5 | pBR322 derivative encoding the htpG gene (Ampr) | 4 |
| pT7blue | pUC19-derived cloning vector (Ampr) | Novagen |
| pTb-ibp | pT7blue derivative encoding a PCR-amplified ibp operon (Ampr) | This study |
| pTG10 | pACYC184-derived cloning vector (Chlr) | 9 |
| pDnaK/J | pTG10 derivative encoding the dnaKJ operon under control of the native promoter (Chlr) | A. A. Gatenby |
| pGroESL | pTG10 derivative encoding the groE operon under control of the lac and native promoters (Chlr) | 9 |
| pς32 | pTG10 derivative encoding the rpoH gene under control of the lac promoter only (Chlr) | 33 |
| pClpA | pTG10 derivative encoding the clpA gene under control of the native promoter (Chlr) | This study |
| pClpB | pTG10 derivative encoding the clpB gene under control of the native promoter (Chlr) | This study |
| pHtpG | pTG10 derivative encoding the htpG gene under control of the native promoter (Chlr) | This study |
| pIbp | pTG10 derivative encoding the ibp operon under control of the lac and native promoters (Chlr) | This study |
Amp, ampicillin; Neo, neomycin or kanamycin; Chl, chloramphenicol. Superscript r indicates resistance.
Plasmid constructions.
All enzymes and kits were used according to the manufacturer’s recommendations. PCR amplifications were performed with the Expand high-fidelity kit (Boehringer Mannheim) and primers purchased from Gibco BRL. PCR products from genomic DNA were purified with the Qiaquick PCR purification kit (Qiagen). All other PCR products and DNA restriction fragments were purified after agarose gel electrophoresis by using the Qiaquick gel extraction kit (Qiagen). Plasmid DNA was purified using the QIAprep spin miniprep kit (Qiagen). Ligations were carried out with the Rapid DNA ligation system (Boehringer Mannheim) and verified by restriction analysis or sequencing with the PRISM ready reaction dyedeoxy terminator cycle sequencing kit (Applied Biosystems). Ligation products were maintained in either XL1-blue or Top10, and CC160 was used for purification of plasmids requiring restriction with enzymes blocked by dam methylation.
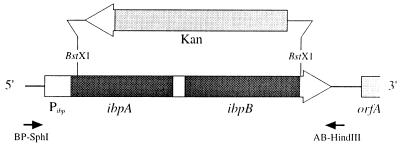
Plasmid pBRibp was constructed by ligation of a 2.9-kbp HindIII-PvuII fragment from pMON18003 to a pBR322 backbone digested with the same enzymes. This plasmid encodes the entire ibp operon surrounded by 500 to 1,000 bp of flanking genomic DNA. The majority of the operon was removed by digestion of pBRibp with BstXI (Fig. 1). The large fragment was blunted with T4 polymerase, dephosphorylated with shrimp alkaline phosphatase (Boehringer Mannheim), and ligated to a neomycin phosphotransferase gene isolated on an SmaI fragment from pBSL14. Plasmid pBRΔibp::kan14-3, which carries the kanamycin resistance cassette transcribed in the opposite orientation of the ibp operon, was selected for further manipulations.
FIG. 1.
Structure of the ibp operon. An ibp null mutant was constructed by insertion of a kanamycin resistance cartridge (Kan) between the indicated BstXI sites. This operation removes about 60% of the ibpA open reading frame and the entire ibpB gene. The presence of the Δibp1::kan mutation in E. coli JGT1 and JGT17 was confirmed by PCR analysis of chromosomal DNA with the primer set shown. The same primers were used for PCR amplification of the ibp operon. The figure is not drawn to scale.
The cloning vector pTG10 was used as a backbone to generate a series of expression plasmids similar to the previously described plasmids pDnaK/J, pGroESL, and pς32 (Table 2). Plasmid pClpA was generated by insertion of a BamHI-PstI fragment from pWPC3 into the BclI-PstI backbone of pTG10. Plasmid pClpB was constructed by ligation of a BamHI-SphI fragment from pclpB into the same sites of pTG10. Plasmid pHtpG was constructed by ligation of a BclI-SalI fragment from pBJ5 to the BamHI-SalI backbone of pTG10. The ibp operon was amplified by PCR with plasmid pMON18003 as a template and the primer BP-SphI (5′-GCCCCCTCAGTGCATGCAATAGACC), which hybridizes before the promoter region, and primer AB-HindIII (5′-ATCGGTGAAGAAGCTTTGCCTT), which binds between the putative transcription terminator and orfA (Fig. 1). The PCR-amplified ibp operon was cloned into the pT7blue blunt vector system (Novagen), verified by restriction digests and DNA sequencing, and inserted into pTG10 as a HindIII fragment. Since the lac promoter is very weak compared to heat shock promoters at high temperatures, increased chaperone expression from these plasmids can be considered to mostly result from an increase in gene dosage. The sole exception is pς32, which contains the rpoH gene under control of the lac promoter only in order to avoid known stability issues (35). Overexpression of all proteins was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis and/or Western blotting (data not shown and reference 33).
Strain constructions.
An ibp null mutant was created by digestion of pBRΔibp::kan14-3 with ScaI and transformation of the linear DNA into the recD mutant strain JCB495. Putative recombinants were selected on the basis of kanamycin resistance and ampicillin sensitivity. The presence of the mutation was confirmed by PCR analysis of chromosomal DNA, which was isolated by the method of Marmur (21) followed by phenol extraction. JGT1, a homologous recombinant containing the Δibp1::kan mutation, was used for further studies.
Strains were constructed by standard techniques (23). P1 transduction was used to create a panel of chaperone mutants isogenic to E. coli MC4100 (Table 1). Transductants were selected at 30°C on LB plates containing the appropriate antibiotics. An F′ episome bearing the lacIq allele was conjugated into several strains with selection on M9 lactose agar plates containing the appropriate antibiotics and no amino acids, except for 100 μg of threonine per ml in the case of cells carrying the thr::Tn10 mutation (Table 1).
Growth studies and viability measurements.
Growth studies were performed with 125-ml shake flasks containing 25 ml of LB medium supplemented with 0.2% glucose and the appropriate antibiotics. A New Brunswick G76 water bath was used for culturing cells at 44°C and above in order to maintain precise temperature control (±0.2°C). Growth rates were determined by recording the culture A600 at various time points. Cultures were inoculated at the ratios indicated below to obtain A600 readings below 0.1 at the initial time point. For typical growth experiments, seed cultures were grown for 20 h at 30°C and diluted 50-fold into supplemented medium prewarmed to the indicated temperatures. To examine the effect of chaperone overexpression on the growth of dnaK756 and groES30 mutants, the cells were grown at 30°C to mid-exponential phase and induced by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and temperature shift to 37°C for 1 h. The cells were diluted 25-fold into fresh medium held at 44 or 46°C, and growth was monitored for up to 24 h. For spot test experiments, 10-μl samples of cultures grown overnight at 30°C were aliquoted onto LB agar plates supplemented with chloramphenicol and IPTG. The plates were incubated for 24 h at 30, 37, 42, or 44°C, and growth was estimated by visual inspection. The ability of the cells to recover from heat shock at 50°C was examined by transfer of cultures grown to mid-exponential phase at 30 to 50°C for 1 h. Thereafter, culture aliquots were diluted 10-fold into fresh medium prewarmed to the indicated temperatures. The data presented were obtained from simultaneous cultures of the various strains. All experiments were repeated on two or more separate occasions to confirm the results. The data shown are the averages from duplicate cultures when indicated.
Thermotolerance at 50°C was determined by measuring CFU. For experiments involving chaperone mutants, cells growing exponentially at 30°C were transferred to 50°C. Plasmid-bearing cells were grown at 30°C to mid-exponential phase, induced by the addition of 1 mM IPTG and temperature upshift to 37°C for 1 h, and transferred to 50°C. In all cases, serial dilutions of cells were spread onto agar plates containing the appropriate antibiotics. The numbers of colonies formed after overnight incubation at 30°C were obtained from duplicate cultures. Experiments were repeated on at least two separate occasions.
SDS-PAGE fractionation analysis.
To examine the effects of chaperone mutations on the aggregation of host proteins, cultures growing in mid-exponential phase at 30°C were transferred to 42, 46, or 50°C for 1 h, and culture samples were collected. SDS-PAGE analysis of the cellular fractions was performed as described previously (33). Briefly, culture samples were passaged through a French pressure cell at 10,000 lb/in2, and soluble and insoluble materials were separated by centrifugation at 30,000 × g for 12 min. Soluble proteins were concentrated by methanol-chloroform precipitation (39). Aliquots of soluble and insoluble cellular fractions corresponding to identical absorbance units were fractionated by reducing SDS-PAGE and visualized by Coomassie brilliant blue staining.
RESULTS
Construction of an ibp knockout strain and isogenic Hsp mutants.
IbpA and IbpB are highly homologous 16-kDa proteins displaying more than 50% identity at the amino acid level (2). The ibp operon, which lies at 82.5 min on the E. coli genetic map, contains a ς32-regulated promoter followed by the ibpA and ibpB genes in succession (Fig. 1) (2, 5). Directly after the transcription terminator lies an open reading frame (orfA) of unknown function. To gain further insight into the cellular function of IbpA and -B, we constructed an ibp null mutant by inserting a kanamycin resistance cartridge between the two BstXI restriction sites located within the operon (Fig. 1). The resulting deletion, Δibp1::kan, removes most of the ibpA gene and all of the ibpB gene without disturbing orfA. Since the kanamycin resistance gene is transcribed in the opposite orientation of the ibp operon, polar effects are not expected. P1 transduction was next used to create a panel of MC4100 derivatives containing mutations in the genes encoding DnaK (dnaK756), GroES (groES30), ClpA (clpA::kan), ClpB (ΔclpB::kan), ClpP (clpP::cat), HtpG (ΔhtpG1::lacZ), and IbpA and -B (Δibp1::kan) (see Table 1 for further details).
The E. coli sHsps are dispensable for growth at high temperatures.
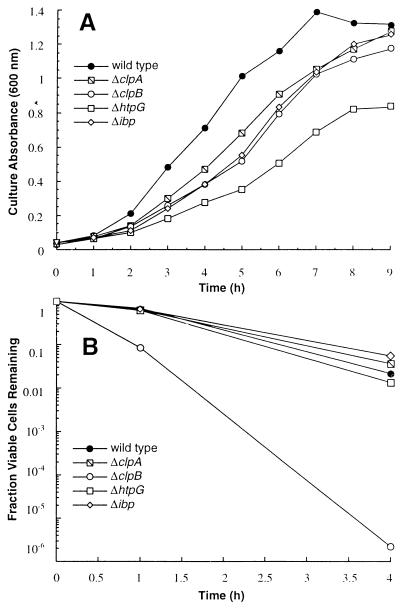
We first compared the growth of the ibp null mutant to that of isogenic ΔclpA, ΔclpB, and ΔhtpG mutants over the 30 to 46°C temperature range. Fresh medium prewarmed to various temperatures was inoculated with cells grown overnight at 30°C. While the growth of all mutants was indistinguishable from that of the wild type at temperatures up to 45°C (data not shown), growth deficiencies were readily apparent when the temperature was raised to 46°C (Fig. 2A). Under these conditions, the specific growth rates of the clpB and htpG mutants were 30 to 40% lower than that of the wild type. Although it was previously found that the growth of ΔclpB and ΔhtpG mutants is impaired at 44°C (4, 31), the 2°C difference in temperature for full expression of the growth-deficient phenotype in these genetic backgrounds is likely explained by variations in host strains, growth medium, or other experimental conditions. The Δibp cells grew at a rate similar to that of the ΔclpB cells at 46°C. More surprisingly, we reproducibly found that the specific growth rate of the clpA mutant was 80% that of the wild type, despite the fact that ClpA is not an Hsp.
FIG. 2.
Growth and viability of chaperone mutants at high temperatures. (A) Stationary-phase cells grown at 30°C were diluted directly into fresh medium held at 46°C. Average variation (coefficient of variation [CV]) between duplicate cultures was 6.2%. (B) Cells containing the indicated mutations were grown to mid-exponential phase at 30°C and shifted to 50°C. Data shown represent the ratio of viable cells after high-temperature incubation to that of viable cells before the shift to 50°C. The average CV for duplicate cultures was 23%.
Differential influence of chaperones on basal thermotolerance.
We next examined the viability of the various mutants following incubation at 50°C. For these experiments, cells growing exponentially at 30°C were shifted to 50°C, and the number of viable cells remaining after 1 and 4 h was determined by plating culture aliquots at 30°C. In agreement with previous studies (31), the death rate of the ΔclpB strain was about 3.5-fold higher than that of the wild type and was comparable to that of an isogenic dnaK756 mutant (data not shown). In contrast, cellular viability was not affected by the presence of the ΔclpA, ΔhtpG, Δibp mutation (Fig. 2B). We further observed that colonies formed overnight by ΔclpB cells exposed to 50°C for 1 h were smaller and heterogeneous compared to those formed by other minor chaperone mutants. From these observations, it is clear that individual ΔclpB cells which remain viable after incubation at 50°C are severely compromised.
We also investigated whether overproduction of various chaperones would improve the viability of E. coli cells exposed to lethal temperatures. For these experiments, E. coli JGT47, an MC4100 derivative containing an F′ episome encoding the lacIq gene, was transformed with a series of pTG10-derived plasmids encoding DnaK-DnaJ, GroEL-GroES, ς32, ClpA, ClpB, HtpG, or IbpA and -B (see Table 2 and Materials and Methods for details). Basal thermotolerance at 50°C was determined as described in Materials and Methods. Whereas GroEL-GroES overexpression had a 7-fold beneficial effect, higher intracellular concentrations of DnaK-DnaJ, ClpA, ClpB, or IbpA and -B led to 5- to 10-fold reductions in viability (data not shown). To examine the rapid loss of viability of ClpB cells at 50°C in more detail, the experiments described above were repeated with JGT51, an MC4100 ΔclpB derivative carrying an F′ lacIq episome. In this genetic background, only pClpB was able to restore viability to control levels. Cell viabilities in all other strains were 4 to 6 orders of magnitude less than those in pClpB transformants (data not shown).
ClpA, ClpB, and HtpG—but not IbpA and -B—are required for optimal recovery from exposure to high temperatures.
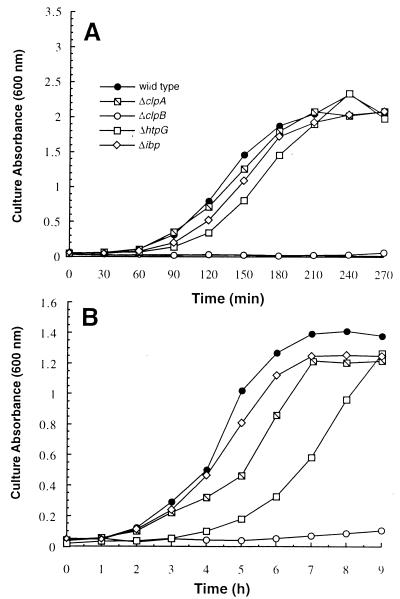
Because growth and survival at high temperatures may involve different pathways from that of recovery following exposure to extreme stress, we examined how chaperone mutations influenced the ability of E. coli to recover from transient incubation at 50°C. For this purpose, the various mutants were grown to mid-exponential phase at 30°C, shifted to 50°C for 1 h, and diluted into fresh medium held at 37, 42, or 45°C. At 37°C, all strains except for the ΔclpB mutant recovered similarly to the wild type (data not shown). When cultures were transferred to 42°C, ΔclpB cells were unable to recover for several hours, and an increased growth lag was observed in ΔhtpG cells (Fig. 3A). The latter effect was much more obvious when the temperature was raised to 45°C (Fig. 3B). Under these conditions, the ΔclpA mutant also displayed a pronounced lag in recovery. However, the Δibp strain behaved comparably to the wild type except for a lower final culture density. It should finally be noted that ΔclpB cultures began to grow slowly after 4 to 6 h of incubation, depending on recovery temperature, and reached a density similar to that of the wild type after 24 h (data not shown). Since the viability of clpB mutants is greatly reduced after 1 h of incubation at 50°C (Fig. 2B) and viable cells exhibit morphological changes, both processes are likely to account for the extreme behavior of this strain.
FIG. 3.
Recovery of chaperone mutants following transient incubation at 50°C. Cells grown to mid-exponential phase at 30°C were shifted to 50°C for 1 h and diluted into fresh medium held at 42°C (A) or 45°C (B). The average coefficient of variation for duplicate cultures was 7.2%. Note the difference in scales for culture absorbance and incubation time.
The Hsps ClpB, HtpG, and IbpA and -B cooperate with the DnaK-DnaJ-GrpE and GroEL-GroES systems in stress management.
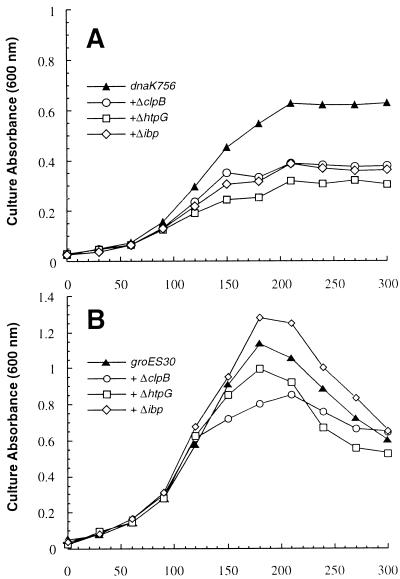
To investigate the possibility that ClpA, ClpB, HtpG, or IbpA and -B cooperate with DnaK-DnaJ-GrpE and/or GroEL-GroES at high temperatures, the various null mutations were introduced into MC4100 derivatives carrying either the dnaK756 or groES30 allele, and the growth of the double mutants was characterized at 42°C. Whereas the clpA deletion had no detrimental effect on the growth of dnaK756 or groES30 cells at 42°C (data not shown), the combination of the dnaK756 allele with the ΔclpB, ΔhtpG, or Δibp mutations exerted a clear deleterious effect on cell growth, since all double mutants only reached half of the maximum turbidity of dnaK756 control cells (Fig. 4A). The simplest explanation for this behavior is that the double mutants experience a higher degree of cellular damage and lose the ability to replicate more rapidly.
FIG. 4.
Effect of double chaperone mutations on growth at 42°C. (A) dnaK756 single or double mutants grown overnight at 30°C were diluted into fresh medium held at 42°C. The average coefficient of variation (CV) for duplicate cultures was 5.9%. (B) groES30 single or double mutants were grown overnight at 30°C and diluted into fresh medium held at 42°C. The average CV for duplicate cultures was 6.8%. Note the difference in scale for culture absorbances.
The effect of minor chaperone deletions was less dramatic in the groES30 background. The absorbance of groES30 control cultures declined 3 h after transfer to 42°C, and the Δibp and ΔhtpG double mutants displayed similar growth patterns (Fig. 4B). However, we reproducibly found in several independent experiments that the Δibp groES30 double mutant grew to a slightly higher turbidity than the control, while ΔhtpG groES30 cells exhibited the opposite behavior (Fig. 4B). The ΔclpB mutation had the most detrimental effect in the groES30 background. This double mutant only reached 70% of the maximal turbidity of groES30 control cultures before gradual lysis occurred. We additionally found that the basal thermotolerance levels of groES30 cells at 50°C were identical to that of the wild type and that the ΔclpB mutation was the only null mutation to reduce the viability of groES30 cells held at 50°C for 1 h (data not shown). Similar experiments were not performed with dnaK756 cells, since the death rate of this mutant is similar to that of ΔclpB cells.
Overexpression of the minor chaperones does not restore the growth of dnaK756 or groES30 mutants at high temperatures.
To determine whether an increase in the intracellular concentration of ClpB, HtpG, or IbpA and -B could compensate for the deleterious effects of the dnaK756 or groES30 mutations on growth at high temperature, the pTG10-derived series of chaperone expression plasmids were introduced into MC4100 dnaK756 (JGT61) or groES30 (JGT49) derivatives conjugated with an F′ episome carrying the lacIq allele (Tables 1 and 2). Transformants were tested for growth at high temperatures either in liquid culture or by their ability to grow on LB agar plates, as described in Materials and Methods. Under both sets of experimental conditions, the only plasmids capable of restoring growth of dnaK756 or groES30 cells at lethal temperatures (i.e., above 42°C) were pDnaK/J and pGroESL, respectively (data not shown).
Mutations in the minor chaperones do not result in wholesale aggregation of host proteins at high temperature.
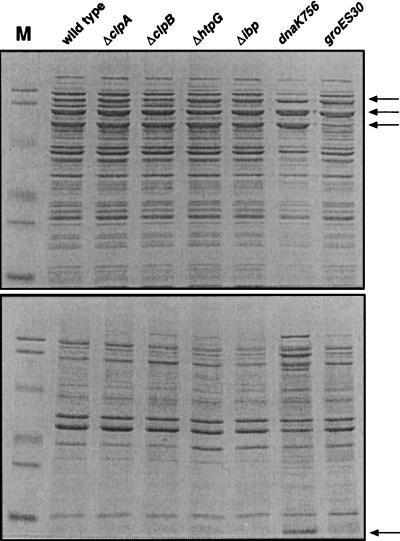
To directly test the in vivo chaperone activity of ClpA, ClpB, HtpG, and IbpA and -B, we examined whether their deletion would affect the aggregation of host proteins in heat-shocked cells. Single chaperone mutants grown to mid-exponential phase at 30°C were transferred to 46°C for 1 h, and culture samples were fractionated by SDS-PAGE following separation into soluble and insoluble fractions. Although wholesale aggregation of host proteins does not take place in dnaK756 or groES30 mutants at 42°C (12), an obvious increase in the amount of insoluble proteins was seen in dnaK756 cells and, to a lesser extent, in groES30 cells at 46°C (Fig. 5). Overproduction of Hsps was readily apparent in the dnaK756 mutant, in which gross aggregation occurred at the expense of the soluble protein. A similar phenotype was not observed in any of the minor chaperone mutants following incubation at 46°C (Fig. 5) or 50°C (data not shown). Finally, obvious synergistic effects between the dnaK756 and groES30 mutations and the various minor chaperone deletions could not be detected by SDS-PAGE when these experiments were repeated with the double mutants (data not shown).
FIG. 5.
Effect of chaperone mutations on host protein aggregation at 46°C. Cells containing the indicated mutations were grown to mid-exponential phase at 30°C and shifted to 46°C for 1 h. Soluble (upper gel) and insoluble (lower gel) cellular fractions from identical absorbance units (A600) of culture were separated by reducing SDS-PAGE (12.5% polyacrylamide). Insoluble fractions were loaded at three times the absorbance units of soluble fractions. Markers (M [Bio-Rad]) correspond to the molecular masses 104, 81, 47.7, 34.6, 28.3, and 19.2 kDa. The positions of the following proteins are indicated by arrows: upper gel from top to bottom, ClpA and ClpB, DnaK and HtpG, and GroEL; lower gel, GroES, IbpA, and IbpB. Gels were digitized with a Sharp JX-325 high-resolution scanner and the NIH Image 1.60 software for PowerPC.
DISCUSSION
In this study, we show that the sHsps IbpA and IbpB are dispensable for normal E. coli growth at temperatures as high as 45°C, but that their absence leads to growth defects at 46°C (Fig. 2A). Although the cellular roles of IbpA and -B are unknown, it has been shown in vitro that sHsps and Hsp90s can maintain partially folded proteins in a conformation that can be reactivated through interactions with Hsp70s (6, 7, 15, 38). These findings have led to the proposal that sHsps and Hsp90s act as reservoirs of thermally denatured or otherwise stress-damaged proteins, thereby facilitating the reactivation of misfolded proteins upon removal of the insult. The results shown in Fig. 3 are consistent with the idea that the E. coli Hsp90 homolog HtpG plays an important role in cell recovery following exposure to lethal temperatures, but also indicate that IbpA and -B are not absolutely required for this process. Thus, it appears that the role of IbpA and -B in protein reactivation following stress is relatively minor or that alternative cellular pathways can compensate for the absence of sHsps. We also found that overproduction of the ibp operon did not improve cell viability at 50°C, although overexpression of sHsps from other organisms can improve E. coli thermotolerance (24, 28). Taken together, these results suggest that the mode of action of the bacterial sHsps may differ from that of their eukaryotic homologs.
Interestingly, we observed that clpA null mutants are defective for growth at 46°C despite the fact that ClpA is not an Hsp. This protein also appears to play a role in cellular recovery from transient incubation at 50°C (Fig. 3). Since the clpP and clpX genes are part of an Eς32-transcribed operon (11), it would be reasonable to assume that the ClpXP protease plays a more essential role in E. coli survival at high temperatures than ClpAP (10). Nevertheless, the results in Fig. 2 and 3 suggest that ClpAP, or ClpA itself, is also important in heat-shocked cells.
Although a role of ClpB in thermotolerance was reported by Squires et al. (31), the mechanisms responsible for the rapid death of ΔclpB cells at 50°C remain unclear. The unique role of ClpB in thermotolerance is further highlighted by our observations that (i) high intracellular concentrations of other molecular chaperones or all Hsps cannot compensate for the deleterious effect of the ΔclpB mutation at 50°C (data not shown) and (ii), unlike dnaK756 cells, ΔclpB mutants do not show an obvious increase in host protein aggregation upon incubation at 50°C, even though both mutants exhibit nearly equivalent death rates at this temperature. ClpB and its yeast homolog, Hsp104, have been implicated in the clearance of thermally denatured proteins in E. coli (19, 26). It is possible that E. coli ClpB plays a vital role in stress recovery through this mechanism.
Among the chaperone mutants tested, groES30 cells exhibited a unique and somewhat paradoxical behavior. In contrast to all other strains, the turbidity of groES30 cultures declined after prolonged incubation at 42 or 46°C (Fig. 4 and data not shown), suggesting that the GroE chaperonins are required to maintain cellular integrity and/or cell viability at these temperatures. However, the same mutation did not have an adverse effect on viability or turbidity when the incubation temperature was raised to 50°C, a temperature at which the growth of groES30 cells is halted (data not shown). A possible explanation for these results is that the GroE chaperonins play a key role in cell division. This hypothesis is in agreement with a recent report suggesting that GroEL and GroES are important in the folding of DapA, an enzyme involved in the synthesis of the cell wall precursor diaminopimelic acid (22). It should finally be noted that the intracellular levels of GroEL were greatly reduced in groES30 cells (Fig. 5). Interestingly, groES619 cells, but not groEL140 mutants, exhibit a similar behavior (32). Although the GroEL140 protein is known to interact suboptimally with GroES, it is still able to associate with the cochaperonin (3). Since the mutations in both GroES30 and GroES619 map in the mobile loop region, which plays a key role in the formation of GroEL-GroES hetero-oligomers (18), complex formation between GroEL and GroES may be severely reduced or completely abolished in groES30 and groES619 strains. Thus, it is possible that interactions with GroES are required to confer stability to GroEL.
To investigate the possible interplay between the Clp ATPases, HtpG, IbpA and -B, and the DnaK-DnaJ-GrpE and GroEL-GroES systems, we characterized the growth of isogenic double chaperone mutants at 42°C. Although the clpB, htpG, and ibp deletions did not affect cell growth at this temperature, all mutations exerted a deleterious effect in the dnaK756 background. These data suggest that minor heat shock chaperones cooperate with the DnaK-DnaJ-GrpE system in thermal stress management. The fact that the ΔclpB and ΔhtpG mutations, but not the Δibp deletion, affected the growth of groES30 cells further suggests that while ClpB and HtpG interact with the GroEL-GroES system, the bacterial sHsps do not. These results are in agreement with recent biochemical data showing that IbpB-bound malate dehydrogenase and lactate dehydrogenase are specifically transferred to the DnaK-DnaJ-GrpE system but that the GroEL-GroES chaperonins do not interact directly with IbpB-released proteins (38). Overall, our findings are consistent with the idea that ClpB, HtpG, and IbpA and -B function as molecular chaperones in vivo. However, their overexpression could not restore the growth of dnaK756 or groES30 mutants at or above 44°C. While it remains possible that minor chaperone overexpression may partially suppress other phenotypes of dnaK or groES mutants, it is obvious that they are not interchangeable with DnaK-DnaJ-GrpE or GroEL-GroES. Thus, the putative chaperone activities of ClpB, HtpG, and IbpA and -B are likely to be of a specialized nature in heat-shocked cells. A more precise examination of the roles of ClpA, ClpB, HtpG, and IbpA and -B in cellular protein folding is in progress.
ACKNOWLEDGMENTS
We thank Mikhail Alexeyev, Elizabeth Craig, Alan Easton, Anthony Gatenby, Costa Georgopoulos, Susan Gottesman, Colin Manoil, Catherine Squires, Beth Traxler, and Saskia van der Vies for their generous gifts of bacterial strains, plasmids, and P1 phage. We thank Andria Costello, Colin Manoil, and Beth Traxler for technical advice and Jeff Shearstone for assistance with DNA sequencing. We are grateful to Tom Horbett and Mary Lidstrom for comments on early versions of the manuscript.
This work was supported by NSF award BES-9501212.
REFERENCES
- 1.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–56. [PubMed] [Google Scholar]
- 2.Allen S P, Polazzi J O, Gierse J K, Easton A M. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol. 1992;174:6938–6947. doi: 10.1128/jb.174.21.6938-6947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baneyx F, Gatenby A A. A mutation in GroEL interferes with protein folding by reducing the rate of discharge of sequestered polypeptides. J Biol Chem. 1992;267:11637–11644. [PubMed] [Google Scholar]
- 4.Bardwell J C A, Craig E A. Ancient heat shock gene is dispensable. J Bacteriol. 1988;170:2977–2983. doi: 10.1128/jb.170.7.2977-2983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuang S-E, Burland V, Plunkett III G, Daniels D L, Blattner F R. Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coli. Gene. 1993;134:1–6. doi: 10.1016/0378-1119(93)90167-2. [DOI] [PubMed] [Google Scholar]
- 6.Ehrnsperger M, Gräber S, Gaestel M, Buchner J. Binding of non-native protein to hsp25 during heat-shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman B C, Morimoto R I. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 8.Fuge E K, Farr S B. AppppA-binding protein E89 is the Escherichia coli heat shock protein ClpB. J Bacteriol. 1993;175:2321–2326. doi: 10.1128/jb.175.8.2321-2326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goloubinoff P, Gatenby A A, Lorimer G H. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman S, Clark W P, de Crecy Lagard V, Maurizi M R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 12.Gragerov A L, Nudler E, Komissarova E, Gaitanaris G A, Gottesman M E, Nikiforov V G. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:10341–10344. doi: 10.1073/pnas.89.21.10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 14.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 15.Jakob U, Lilie H, Meyer I, Buchner J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J Biol Chem. 1995;270:7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- 16.Katayama Y, Gottesman S, Pumphrey J, Ridikoff S, Clark W P, Maurizi M R. The two-component, ATP-dependent Clp protease of Escherichia coli: purification, cloning, and mutational analysis of the ATP-binding component. J Biol Chem. 1988;263:15226–15236. [PubMed] [Google Scholar]
- 17.Katayama Y, Kasahara A, Kuraishi H, Amano F. Regulation of activity of an ATP-dependent protease, Clp, by the amount of a subunit, ClpA, in the growth of Escherichia coli cells. J Biochem. 1990;108:37–41. doi: 10.1093/oxfordjournals.jbchem.a123158. [DOI] [PubMed] [Google Scholar]
- 18.Landry S J, Zeilstra-Ryalls J, Fayet O, Georgopoulos C, Gierasch L M. Characterization of a functionally important mobile domain of GroES. Nature. 1993;364:255–258. doi: 10.1038/364255a0. [DOI] [PubMed] [Google Scholar]
- 19.Laskowska E, Kuczynska-Wisnik D, Skorko-Glonek J, Taylor A. Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol Microbiol. 1996;22:555–571. doi: 10.1046/j.1365-2958.1996.1231493.x. [DOI] [PubMed] [Google Scholar]
- 20.Laskowska E, Wawryznów A, Taylor A. IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie. 1996;78:117–122. doi: 10.1016/0300-9084(96)82643-5. [DOI] [PubMed] [Google Scholar]
- 21.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 22.McLennan N, Masters M. GroE is vital for cell-wall synthesis. Nature. 1988;392:139. doi: 10.1038/32317. [DOI] [PubMed] [Google Scholar]
- 23.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 24.Muchowski P J, Clark J I. ATP-enhanced molecular chaperone functions of the small heat shock protein human αB crystallin. Proc Natl Acad Sci USA. 1998;95:1004–1009. doi: 10.1073/pnas.95.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neidhardt F C, VanBogelen R A. Heat shock response. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1334–1345. [Google Scholar]
- 26.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 27.Parsell D A, Sauer R T. Induction of a heat shock-like response by unfolded protein in Escherichia coli: dependence on protein level not protein degradation. Genes Dev. 1989;3:1226–1232. doi: 10.1101/gad.3.8.1226. [DOI] [PubMed] [Google Scholar]
- 28.Plater M L, Goode D, Crabbe M J. Effects of site-directed mutations on the chaperone-like activity of alphaB-crystallin. J Biol Chem. 1996;271:28558–28566. doi: 10.1074/jbc.271.45.28558. [DOI] [PubMed] [Google Scholar]
- 29.Shirai Y, Akiyama Y, Ito K. Suppression of ftsH mutant phenotypes by overproduction of molecular chaperones. J Bacteriol. 1996;178:1141–1145. doi: 10.1128/jb.178.4.1141-1145.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Squires C, Squires C L. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992;174:1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Squires C L, Pedersen S, Ross B M, Squires C. ClpB is the Escherichia coli heat shock protein F84. 1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas J G, Baneyx F. Protein folding in the cytoplasm of Escherichia coli: requirements for the DnaK-DnaJ-GrpE and GroEL-GroES molecular chaperone machines. Mol Microbiol. 1996;21:1185–1196. doi: 10.1046/j.1365-2958.1996.651436.x. [DOI] [PubMed] [Google Scholar]
- 33.Thomas J G, Baneyx F. Protein misfolding and inclusion body formation in recombinant Escherichia coli cells overproducing heat-shock proteins. J Biol Chem. 1996;271:11141–11147. doi: 10.1074/jbc.271.19.11141. [DOI] [PubMed] [Google Scholar]
- 34.Tilly K, McKittrick N, Georgopoulos C, Murialdo H. Studies on Escherichia coli mutants which block bacteriophage morphogenesis. Prog Clin Biol Res. 1981;64:35–45. [PubMed] [Google Scholar]
- 35.Tilly K, Spence J, Georgopoulos C. Modulation of stability of the Escherichia coli heat shock regulatory factor ς32. J Bacteriol. 1989;171:1585–1589. doi: 10.1128/jb.171.3.1585-1589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueguchi C, Ito K. Multicopy suppression: an approach to understanding intracellular functioning of the protein export system. J Bacteriol. 1992;174:1454–1461. doi: 10.1128/jb.174.5.1454-1461.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VanBogelen R A, Kelley P M, Neidhardt F C. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J Bacteriol. 1987;169:26–32. doi: 10.1128/jb.169.1.26-32.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1988;273:11032–11037. doi: 10.1074/jbc.273.18.11032. [DOI] [PubMed] [Google Scholar]
- 39.Wessel D, Flügge U I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 40.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]