ABSTRACT
ERM proteins are conserved regulators of cortical membrane specialization that function as membrane-actin linkers and molecular hubs. The activity of ERM proteins requires a conformational switch from an inactive cytoplasmic form into an active membrane- and actin-bound form, which is thought to be mediated by sequential PIP2 binding and phosphorylation of a conserved C-terminal threonine residue. Here, we use the single Caenorhabditis elegans ERM ortholog, ERM-1, to study the contribution of these regulatory events to ERM activity and tissue formation in vivo. Using CRISPR/Cas9-generated erm-1 mutant alleles, we demonstrate that a PIP2-binding site is crucially required for ERM-1 function. By contrast, dynamic regulation of C-terminal T544 phosphorylation is not essential but modulates ERM-1 apical localization and dynamics in a tissue-specific manner, to control cortical actin organization and support lumen formation in epithelial tubes. Our work highlights the dynamic nature of ERM protein regulation during tissue morphogenesis and the importance of C-terminal phosphorylation in fine-tuning ERM activity in a tissue-specific context.
Keywords: Caenorhabditis elegans, ERM-1, Ezrin, Moesin, Phosphorylation, Radixin
Summary: CRISPR/Cas9-generated erm-1 mutant alleles reveal that a PIP2-binding site is crucial for ERM-1 function, and phosphorylation of a C-terminal regulatory threonine modulates its localization and dynamics to control actin organization and support lumen formation in development.
INTRODUCTION
Morphological and molecular specialization of defined regions at the cell cortex is crucial for the development and function of most animal cell types. The formation of specialized cortical domains relies on local reorganization of membrane composition and the cortical cytoskeleton. Ezrin/Radixin/Moesin (ERM) proteins form an evolutionarily conserved family that plays a major role in organizing the cell cortex and signaling (Fehon et al., 2010; McClatchey, 2014; Neisch and Fehon, 2011). For example, ERM family members promote the formation of microvilli at the apical surface of epithelial tissues, are required for lumen formation in tubular epithelia, and control the mechanical properties of the cell cortex in processes such as mitosis, cell migration, and immunological synapse formation in B and T cells (Kunda et al., 2008; McClatchey, 2014; Parameswaran and Gupta, 2013; Pelaseyed and Bretscher, 2018). To perform this wide range of functions, ERM proteins can link the plasma membrane physically with the actin cytoskeleton and orchestrate the assembly of a broad array of multiprotein complexes at the cell surface.
ERM proteins consist of an N-terminal band Four-point-one/Ezrin/Radixin/Moesin (FERM) domain that mediates binding to transmembrane and membrane-associated proteins, a C-terminal tail that mediates actin binding, and a central α-helical linker region (Fehon et al., 2010; McClatchey, 2014). The activity of ERM proteins is regulated by a reversible intramolecular interaction. In the inactive closed conformation, extensive interactions between the FERM domain and the C-terminal tail mask the actin-binding site, membrane-binding sites and protein interaction sites (Gary and Bretscher, 1995; Li et al., 2007; Magendantz et al., 1995; Pearson et al., 2000). The transition to an open and active conformation involves binding to the plasma membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2) and phosphorylation of a specific C-terminal threonine residue (T567 in ezrin, T564 in radixin and T558 in moesin) (Simons et al., 1998; Nakamura et al., 1999; Barret et al., 2000; Fievet et al., 2004; Coscoy et al., 2002; Yonemura et al., 2002; Hao et al., 2009; Roch et al., 2010). This transition is thought to occur in a multistep process, in which binding to PIP2 induces a partial conformational change that enables binding of a kinase and phosphorylation of the C-terminal threonine (Fievet et al., 2004; Hao et al., 2009; Bosk et al., 2011; Pelaseyed et al., 2017).
C-terminal phosphorylation is often considered to be essential for the activity of ERM proteins (Matsui et al., 1998; Oshiro et al., 1998; Simons et al., 1998; Gautreau et al., 2000; Kunda et al., 2008; Nakamura et al., 1995; Parameswaran et al., 2011; Viswanatha et al., 2012). However, several studies indicate that phosphorylation might not be a universal requirement. For example, the phosphorylation of ERM proteins appears dispensable for the formation of microvilli-like structures in A431 and MDCK II cells (Yonemura et al., 2002). Likewise, although some studies have reported that phosphorylation is essential for the activity of Drosophila Moesin (Karagiosis and Ready, 2004; Polesello et al., 2002), significant rescuing capacity for a non-phosphorylatable variant has been reported (Roch et al., 2010). Thus, ERM proteins might be activated without phosphorylation, at least in some cell types or biological conditions.
Here, we make use of the nematode Caenorhabditis elegans to gain a better understanding of the contributions of PIP2-binding and C-terminal threonine phosphorylation to the functioning of ERM proteins in vivo. Caenorhabditis elegans expresses a single ERM protein, termed ERM-1, and the corresponding gene can be engineered endogenously to express mutant variants. Thus, the role of ERM-1 can be studied without the confounding presence of additional family members or non-mutated protein. ERM-1 is highly similar in sequence and domain composition to other ERM proteins, and the residues crucial for PIP2 binding and the C-terminal threonine residue are fully conserved (Fig. 1A). ERM-1 localizes to the apical surface of most polarized tissue types and is essential for apical membrane morphogenesis (Göbel et al., 2004; van Fürden et al., 2004). Loss of erm-1 causes early larval lethality, with severe cystic defects in the intestine and excretory canals (Göbel et al., 2004). In the intestine, loss of erm-1 causes constrictions, loss of microvilli, severe reduction in the levels of apical actin, and defects in the accumulation of junctional proteins (Bernadskaya et al., 2011; Göbel et al., 2004; van Fürden et al., 2004). In the excretory canals, ERM-1 controls the extension of the canal lumen in a dose-dependent manner (Göbel et al., 2004; Khan et al., 2013).
Fig. 1.
ERM-1 activity and efficient membrane targeting require the PIP2-binding domain. (A) Conservation and domain organization of ERM-1. AB is the actin-binding domain. (B) Differential interference contrast microscopy images of an N2 animal and erm-1 mutants. Arrowheads point to the excretory canal and arrows to the intestinal lumen. (C) Intestinal lumen discontinuities (arrowheads) and widened intestinal lumen in erm-1[4KN]::GFP second generation homozygous 2.5-fold embryos and L1 larvae. ERM-1 localization in the schematic diagram is in green. Dashed line indicates the boundary between the pharynx and intestine. (D,E) Apical levels of GFP-tagged ERM-1 variants in the intestine of L1 larvae with indicated genotypes. Images were acquired and displayed with the same settings, except +/+ in E. In this and all other figures, imaging was done with spinning disk confocal microscopy unless otherwise indicated. PM, plasma membrane. Data shown are means±s.d. of six measurements per animal for the apical membrane, normalized to the mean intensity of GFP in erm-1::GFP controls (n=10, 7, 6 and 8 in D; and n=24, 40 and 28 in E). Error bars are mean±s.d. Tests of significance: Dunnett's T3 multiple comparisons test. ns, not significant. ***P≤0.001, ****P≤0.0001. Unless indicated otherwise, statistical comparisons are with erm-1::GFP.
We used CRISPR/Cas9 to engineer the erm-1 locus endogenously to express ERM-1 variants with a mutated PIP2-binding site, or with a mutated threonine residue that cannot be phosphorylated or that mimics phosphorylation. As in other systems, the PIP2-binding site was essential for the functioning of ERM-1. By contrast, animals expressing only non-phosphorylatable or phosphomimetic ERM-1 protein were viable, demonstrating that C-terminal threonine phosphorylation is not a strict requirement for ERM-1 activity in C. elegans. Nevertheless, phosphorylation contributed to multiple aspects of ERM-1 function, including localization, mobility, and the ability to organize an apical actin network. Effects caused by the phosphorylation mutants were often tissue specific, highlighting versatility for C-terminal phosphorylation in controlling ERM-1. Finally, in support of an essential role for phosphorylation cycling, most of the defects we observed were highly similar between non-phosphorylatable and phosphomimetic variants.
RESULTS
A lipid binding site is essential for the activity of ERM-1
Binding to PIP2 has been shown to be essential for the membrane localization and activity of ERM proteins, but has not been investigated in C. elegans (Nakamura et al., 1999; Barret et al., 2000; Coscoy et al., 2002; Yonemura et al., 2002; Fievet et al., 2004; Hao et al., 2009; Roch et al., 2010). We used CRISPR/Cas9 to mutate two pairs of lysine residues within a plasma-binding region to asparagines (K254/255N and K263/264N, hereafter referred to as 4KN) (Fig. 1A). This change has been shown virtually to abolish membrane localization of mammalian ezrin and fly Moesin (Barret et al., 2000; Fievet et al., 2004; Ben-Aissa et al., 2012; Roch et al., 2010). The first homozygous generation of erm-1(mib11[4KN]) is viable, whereas >95% of their offspring arrest during larval development, mostly as L1, with cysts in the lumens of the intestine and excretory canals (Fig. 1B,C). Both the maternal effect lethality and the cystic luminal phenotypes have been described for the putative erm-1(tm667) null allele (Göbel et al., 2004). In addition, first generation homozygous erm-1(mib11[4KN]) animals die prematurely in adulthood and have a greatly reduced brood size, as has been observed for erm-1(tm677). Thus, erm-1(mib11[4KN]) behaves in a similar manner to a putative null allele.
To examine whether the ERM-1[4KN] mutation prevents membrane association, we generated endogenous COOH-terminal fusions of eGFP (referred to as GFP) with wild-type ERM-1 and ERM-1[4KN]. In early embryogenesis, ERM-1::GFP localized to the entire plasma membrane in addition to the cytoplasm (Fig. S1A). As morphogenesis initiated, ERM-1::GFP was primarily detected at the apical surface of epithelial tissues and in primordial germ cells (PGCs) (Fig. S1A). In larval stages, we observed apical localization of ERM-1::GFP in epithelial tissues including the intestine, seam cells and excretory canals (Fig. 1D; Fig. S1B). In the syncytial germline, ERM-1 was associated with the entire plasma membrane but enriched at the apical domain (Fig. S1B).
Addition of N- or C-terminal tags to ERM proteins has been suggested to interfere with the intramolecular interaction between the FERM domain and C-terminus (Chambers and Bretscher, 2005; Viswanatha et al., 2012). Consistent with this, erm-1::GFP animals had a reduced brood size and incomplete outgrowth of the excretory canals, but showed no other developmental or morphological abnormalities (Fig. S1C-E). These defects are not likely to be attributable to constitutive ERM-1 activity, because excretory canal defects were absent in erm-1::GFP/+ heterozygotes (Fig. S1D,E). C-terminal GFP fusions have been used extensively to characterize the distribution of ERM proteins (Coscoy et al., 2002; Göbel et al., 2004; Roch et al., 2010; Viswanatha et al., 2012; Garbett and Bretscher, 2012; Khan et al., 2013; Babich and Di Sole, 2015), and the localization of our endogenous ERM-1::GFP fusion mimicked previous reports of ERM-1 localization in C. elegans (Göbel et al., 2004; van Fürden et al., 2004; Bernadskaya et al., 2011; Khan et al., 2013). ERM-1::GFP therefore appears to reflect the localization of the endogenous protein accurately.
ERM-1[4KN]::GFP failed to localize to the plasma membrane in PGCs and seam cells of second generation homozygous erm-1[4KN]::GFP L1 larvae (Fig. S1B), and levels in the intestine were severely reduced (Fig. 1D,E). Surprisingly, ERM-1[4KN]::GFP appeared still to be restricted to the apical domain of the excretory canal (Fig. S1B).
We also analyzed the distribution of ERM-1[4KN]::GFP in heterozygous erm-1[4KN]::GFP/+ animals. Although ERM-1[4KN]::GFP still failed to localize to the membrane in PGCs and seam cells, in the intestine, excretory canal and larval germline we observed clear apical localization of ERM-1[4KN]::GFP at the apical membrane domain (Fig. 1D; Fig. S1B). Interestingly, in first generation homozygous animals, apical levels of ERM-1[4KN]::GFP were comparable to wild-type ERM-1::GFP (Fig. 1D,E). First generation homozygous animals had low levels of maternally contributed wild-type ERM-1 (Fig. 1E). Thus, it appears that wild-type ERM-1 can support apical recruitment of ERM-1[4KN], albeit not in all tissues. These results contrast with observations in Drosophila and mammalian tissue culture cells, where C-terminally tagged ERM 4KN mutants do not localize to the plasma membrane, even in the presence of wild-type ezrin (Babich and Di Sole, 2015; Barret et al., 2000; Fievet et al., 2004; Hao et al., 2009; Roch et al., 2010).
Based on these observations, we conclude that the N-terminal PIP2-binding domain is crucial for activity of C. elegans ERM-1, as it is for ERM proteins in other organisms. However, the presence of wild-type ERM-1 protein can compensate for the loss of membrane-binding activity of the ERM-1[4KN] mutant in at least some tissues.
Phosphorylation of T544 is not essential for ERM-1 functioning
We next investigated the contribution of phosphorylation of the conserved C-terminal threonine residue to ERM-1 functioning. We used CRISPR/Cas9 to generate erm-1 mutants in which T544 was replaced with an alanine (T544A) or aspartic acid (T544D), to mimic the non-phosphorylated and phosphorylated states, respectively (Fig. 1A). Using an antibody that specifically recognizes C-terminally phosphorylated ERM proteins (pERM), we observed extensive pERM staining at apical membranes of the intestine, seam cells and excretory canals of N2 embryos and larvae, in addition to the larval germline (Fig. 2A,B). No staining was observed in erm-1[T544A] mutants, indicating that the pERM antibody was specific for the phosphorylated form of ERM-1 (Fig. S2). Thus, ERM-1 is broadly phosphorylated on the conserved C-terminal threonine residue.
Fig. 2.
T544 phosphorylation is not essential for ERM-1 activity. (A) Immunostaining of phosphorylated ERM-1 (pERM) and the junctional marker AJM-1 in N2 embryos. Arrows point to the intestinal lumen or seam cells and arrowheads to excretory canals. Images are maximum intensity projections. (B) Immunostaining of pERM in adults. In the graphical representations in A and B, red indicates junctional structures and green areas of ERM-1 localization. Dashed rectangles indicate areas imaged in B. The first two panels in B were imaged on an epifluorescence microscope. (C-E) Quantifications of brood size, larval lethality and embryonic lethality. Each symbol represents the progeny of an individual animal (n=12, 11, 8, 14 and 7 in C; n=8, 5 and 7 in D; and n=7, 5 and 7 in E). Error bars are the mean±s.d. Tests of significance: Dunnet's multiple comparisons test for C; Tukey's multiple comparisons test for D and E. ns, not significant. *P≤0.05, ***P≤0.001, ****P≤0.0001. Unless indicated otherwise, comparisons are with N2.
Surprisingly, both homozygous erm-1[T544A] and erm-1[T544D] mutants were viable, demonstrating that T544 phosphorylation is not essential for ERM-1 activity in C. elegans. Nevertheless, both mutants had a reduced brood size and increased embryonic and larval lethality compared with wild-type animals (Fig. 2C-E). These defects were not observed in heterozygous erm-1[T544A]/+ and erm-1[T544D]/+ animals, indicating that neither mutation exerts a dominant effect. Finally, consistent with the broad expression pattern of ERM-1, we observed a variety of partially penetrant phenotypes, including small, dumpy, tail defects, protruding vulva, exploded through vulva, uncoordinated and clear. Together, these results demonstrate that the C-terminal phosphorylation site is important, but not essential, for ERM-1 function in C. elegans.
T544 phosphorylation contributes to lumen formation in tubular epithelia
To gain a better understanding of the defects caused by mutation of T544, we investigated the effects on the intestine and excretory canal. We examined the appearance and extension of the canal tubes using a COOH-terminal VHA-5∷GFP fusion protein, which localizes to the apical membrane and canaliculi (Liégeois et al., 2006, 2007). In both erm-1[T544A] and erm-1[T544D] animals, we observed severe canal extension defects, in addition to cystic canals and widened canal lumens (Fig. 3). In accordance with a non-dominant effect of both mutations, canal defects were absent from heterozygous erm-1[T544A]/+ and erm-1[T544D]/+ animals (Fig. 3B,E). We also examined canal morphology by transmission electron microscopy (TEM) and observed widened lumens and canals in erm-1[T544A] and erm-1[T544D] animals (Fig. 3D). Overexpression of ERM-1 leads to lumen formation defects in the excretory canal, which can be rescued by depletion of the ERM-1 binding partner aquaporin/AQP-8 (Khan et al., 2013). Given that phosphorylation-mimicking ERM variants are widely assumed to have constitutive activity, we assessed whether cyst formation in T544 mutants also required AQP-8 activity. However, depletion of aqp-8 by RNA interference (RNAi) enhanced the severity of canal defects (Fig. S3), further demonstrating that defects in erm-1[T544D] are not attributable to increased ERM-1 activity.
Fig. 3.
ERM-1 T544 phosphorylation is required for excretory canal lumenogenesis. (A) Schematic drawing of the excretory canal system in an L4 animal. Red dashed rectangle and blue dashed circle indicate areas shown in D by fluorescence and electron microscopy, respectively. (B) Quantification of excretory canal outgrowth in L4 animals. All four canal branches were measured per animal, and each data point represents one branch (n=25, 25, 22, 25 and 14 animals from top to bottom). Error bars are the median±95% CI. (C) Frequency of anterior and posterior canals from B extending <35% of the distance between cell body and tips. (D) Lumen morphology in L4 animals visualized by using a VHA-5::GFP transgene (top panels) or in cross-section by transmission electron microscopy (bottom panels). (E) Quantification of canal width in L4 animals. Each data point represents the average of three measurements at the three widest points in a single posterior canal (n=15, 17, 13, 18 and 13). Error bars are the median±95% CI. Tests of significance: Dunn's multiple comparisons test for B and E; Fisher's exact test for C. ns, not significant. *P≤0.05, **P≤0.01, ****P≤0.0001. Outgrowth and width were measured by fluorescence microscopy using VHA-5::GFP as a marker. Unless indicated otherwise, statistical comparisons are with the VHA-5::GFP control line.
We next investigated intestinal lumen formation by staining T544 mutant embryos with an antibody directed against the junctional protein AJM-1 (Fig. 4A). In wild-type embryos, separation of junctions on opposing sides of the lumen was visible from the bean stage. By contrast, in both erm-1[T544A] and erm-1[T544D] mutants we did not detect clear separation of opposing junctions until the comma stage (Fig. 4A). Between comma and 2-fold stages, junction separation distance was reduced, and constrictions were visible along the length of the intestinal epithelium. At the 2-fold stage, erm-1[T544D] animals showed an almost complete separation of junctions throughout the epithelium. In addition, we observed an apparent expansion or ectopic accumulation of junctional material. By contrast, in erm-1[T544A] embryos, junction constrictions were still often detected at the 2-fold stage.
Fig. 4.
Positioning of cell junctions in intestinal cells requires dynamic regulation of ERM-1 T544 phosphorylation. (A) Junction organization visualized by AJM-1 staining of fixed embryos at different stages. Insets to the right and bottom are enlarged views of the regions indicated by dashed rectangles. Red brackets indicate junction separation in bean stage. Blue arrows point to expansion or ectopic accumulation of junction material, and red arrows point to partial and full constrictions. Images are maximum intensity projections. In all panels, developmental times are the approximate time after fertilization at 20°C. (B) Junction organization in live L1 larvae expressing DLG-1::mCherry and HMR-1::GFP. Blue and red arrows are as in A. (C) Transmission electron microscopy images of intestinal microvilli in L4 animals. (D) Quantification of the length of microvilli (total microvilli quantified: wild type, 88 from five animals; erm-1[T544A], 168 from four animals; erm-1[T544D], 135 from five animals). Error bars are the median±95% CI. Dunn's multiple comparisons test of significance, ****P≤0.0001. Unless indicated otherwise, statistical comparisons are with N2.
We characterized the junction defects of T544 ERM-1 mutants further in living animals during larval stages, using a strain that expresses endogenously tagged DLG-1::mCherry/Discs large and HMR-1::GFP/E-cadherin to mark the C. elegans apical junctions (CeAJ). We rarely observed full constrictions in erm-1[T544D] at the first larval stage (L1), although junctions had a wavy appearance and formed occasional aggregates and ectopic ring-like structures (Fig. 4B). Junctions in erm-1[T544A] animals had a higher frequency of partial or full constrictions (Fig. 4B). However, we did not detect obvious junction defects in either mutant at subsequent larval stages (data not shown).
The lumen formation defects we observed in early intestinal development are similar to those reported for knockdown of erm-1 by RNAi (van Fürden et al., 2004). The mild defects in intestines of L1 larvae and the observed recovery at later stages are, however, in stark contrast to previous descriptions of erm-1 mutants or RNAi (Göbel et al., 2004; van Fürden et al., 2004), suggesting that T544 phosphorylation is important for ERM-1 activity during intestinal lumen formation, but is not strictly required.
ERM proteins in mammals, Drosophila and C. elegans are all crucial for the formation of microvilli (Bonilha et al., 1999, 2006; Speck et al., 2003; Göbel et al., 2004; Saotome et al., 2004; Karagiosis and Ready, 2004; Casaletto et al., 2011), and C-terminal phosphorylation is thought to be essential for ERM proteins to support the formation of microvilli (Chen et al., 1995; Kondo et al., 1997; Gautreau et al., 2000; Pelaseyed et al., 2017; Viswanatha et al., 2012). We therefore examined the formation of intestinal microvilli by electron microscopy. Surprisingly, microvilli still formed in erm-1[T544A] and erm-1[T544D] mutants (Fig. 4C), although their length was reduced (Fig. 4D). Collectively, our results show that ERM-1 phosphorylation plays a major role in lumen formation during early embryogenesis but is not essential for formation of a functional intestine or formation of microvilli.
Dynamic ERM-1 T544 phosphorylation contributes to molecular specialization of the apical domain
We next addressed whether ERM-1 T544 mutations perturb the functional specialization of the intestinal apical membrane. We introduced the erm-1 T544 alleles in a strain overexpressing reporters for the peptide transporter PEPT-1::DsRed, which marks the apical membrane, and the small GTPase GFP::RAB-11, which marks apically enriched recycling endosomes (Winter et al., 2012). Both proteins have important functions in intestinal development and homeostasis across species (Spanier, 2014; Welz et al., 2014). In L1 larvae, we observed a dramatic reduction in the levels of PEPT-1::DsRed at the apical membrane in both erm-1[T544A] and erm-1[T544D] mutants (Fig. 5A,B,D). Both mutants also showed a reduction in GFP::RAB-11 enrichment near the apical plasma, although the effect was more pronounced in erm-1[T544A] (Fig. 5C,E). Thus, the balance between phosphorylated and non-phosphorylated ERM-1 forms contributes to the molecular specialization of the apical domain.
Fig. 5.
![Fig. 5. Dynamic T544 phosphorylation is important for molecular specialization of the apical domain. (A) Graphical representation of the area imaged in B and C (dashed red rectangle). (B,C) Intestinal distribution of PEPT-1::DsRed and GFP::RAB-11 transgenes in L1 larvae. Images were acquired and displayed with the same settings for comparison. Images are maximum intensity projections. Arrow in B indicates a small patch of apical membrane observed at the lateral domain, which occurs with low frequency in ERM-1 T544D mutant animals. (D,E) Distribution plots of the mean±s.d. fluorescence intensity of PEPT-1::DsRed and GFP::RAB-11 along the apical-basolateral axis in intestinal cells of L1 larvae. Two measurements per animal were plotted separately (n=27 animals for control, n=22 for erm-1[T544A] and n=19 for erm-1[T544D]).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/ae42/10755404/5b8fd70d0cbd/develop-147-188011-g5.jpg)
Dynamic T544 phosphorylation is important for molecular specialization of the apical domain. (A) Graphical representation of the area imaged in B and C (dashed red rectangle). (B,C) Intestinal distribution of PEPT-1::DsRed and GFP::RAB-11 transgenes in L1 larvae. Images were acquired and displayed with the same settings for comparison. Images are maximum intensity projections. Arrow in B indicates a small patch of apical membrane observed at the lateral domain, which occurs with low frequency in ERM-1 T544D mutant animals. (D,E) Distribution plots of the mean±s.d. fluorescence intensity of PEPT-1::DsRed and GFP::RAB-11 along the apical-basolateral axis in intestinal cells of L1 larvae. Two measurements per animal were plotted separately (n=27 animals for control, n=22 for erm-1[T544A] and n=19 for erm-1[T544D]).
Phosphorylation of T544 controls subcellular localization of ERM-1
Replacing the C-terminal threonine of mammalian ezrin or fly Moesin with a phosphomimetic aspartic acid or glutamic acid causes relocalization to the entire plasma membrane, whereas reported effects of alanine substitutions vary from reduced apical enrichment to loss of membrane localization (Coscoy et al., 2002; Karagiosis and Ready, 2004; Roch et al., 2010; Viswanatha et al., 2012; Babich and Di Sole, 2015). We therefore investigated the localization of the phosphorylation mutants using an antibody directed against C. elegans ERM-1. In the embryonic intestine, ERM-1[T544A] failed to accumulate at the apical membrane (Fig. S4A). ERM-1[T544D] also failed to accumulate during early stages of intestinal development, but apical enrichment was evident by the 2-fold stage (Fig. S4A). Nevertheless, we observed persistent basolateral localization of both ERM-1[T544A] and ERM-1[T544D] throughout embryogenesis, which was not observed for wild-type ERM-1 (Fig. S4A). We were not able to assess ERM-1 distribution accurately in larval stages by antibody staining, owing to limitations in the quality of antibody staining.
To analyze the distribution of ERM-1 T544 mutants in live animals, we engineered erm-1[T544A] and erm-1[T544D] alleles carrying a COOH-terminal GFP fusion. In the developing embryonic intestine, the GFP-tagged T544 mutants also showed a delay in apical enrichment (Fig. 6A). However, we did observe apical enrichment of ERM-1[T544A]::GFP starting from the 2-fold stage (Fig. 6A). Strikingly, basolateral localization of ERM-1 persisted throughout development in both mutant variants, albeit more readily visible in erm-1[T544A] (Fig. 6A-C). In the larval intestine, apical ERM-1[T544A] levels were sharply reduced at the L1 stage, but largely recovered by the L4 stage (Fig. 6E; Fig. S7A). In the excretory canal, ERM-1[T544A]::GFP and ERM-1[T544D]::GFP were detected exclusively at the apical plasma membrane (Fig. 6D). In the germline, ERM-1[T544A]::GFP levels were lower than those of non-phosphomutant ERM-1::GFP, but still showed apical enrichment (Fig. S5). By contrast, ERM-1[T544D]::GFP showed increased accumulation at the apical domain (Fig. S5). Thus, C-terminal phosphorylation dynamics modulate ERM-1 distribution in a tissue-specific manner.
Fig. 6.
T544 phosphorylation cycling supports apical recruitment of ERM-1 and intestinal lumen formation. (A) Distribution of ERM-1::GFP variants in different embryonic stages. Blue arrows indicate lumen discontinuities, and red arrowheads indicate examples of ectopic basolateral ERM-1 localization. (B) Basolateral localization (red arrowheads) of ERM-1::GFP variants in the intestine of L1 larvae. Images are overexposed for clarity. (C) Frequency of animals with ERM-1::GFP detectable at the basolateral membrane (n=58, 39 and 26). Statistical comparisons are with erm-1::GFP. (D) Apical localization of ERM-1::GFP variants in the excretory canal of L4 larvae. Red dashed rectangles in B and D indicate the area imaged. (E) Intensity of ERM-1::GFP variants at the apical intestinal membrane of L1 larvae. Each symbol represents an individual animal (n=10 for L1, and n=22, 19 and 17 for L4). Error bars are the median±95% CI. Statistical comparisons are with erm-1::GFP unless indicated otherwise. (F) Number of intestinal lumen discontinuities (median±95% CI) and fraction of animals with discontinuities in late embryos and L1 larvae (n=34, 41, 18 and 16 from top to bottom). Tests of significance: Fisher's exact test for D and bar graph in F; Tukey's multiple comparisons test for E; and Dunn's multiple comparisons test for dot plot in F. ns, not significant. **P≤0.01, ****P≤0.0001.
We also used the erm-1[T544A]::GFP and erm-1[T544D]::GFP alleles to examine intestinal lumen morphology. We observed discontinuities in the GFP signal along the lumen of erm-1[T544A]::GFP and erm-1[T544D]::GFP embryos, which largely resolved by the L1 stage (Fig. 6F). These presumably correspond to the junction constrictions shown in Fig. 4. The penetrance of intestinal phenotypes was slightly higher than in untagged T544 mutants (Fig. S4B), presumably owing to a detrimental influence of the COOH-terminal GFP tag. These results further demonstrate the importance of ERM-1 phosphorylation in the early stages of intestinal lumenogenesis.
Mobility of ERM-1 at the membrane is modulated by T544 phosphorylation status
The altered distribution of ERM-1 T544 mutants might be attributable to changes in the dynamics of ERM-1 association with the membrane. To investigate this, we performed fluorescence recovery after photobleaching (FRAP) experiments using GFP tagged wild-type and T544 mutant strains. In the intestine, ERM-1::GFP was extraordinarily stable, with recovery of bleached areas averaging 15% after 45 min (Fig. 7A,B). However, the fraction and speed of recovery were dramatically increased for the ERM-1[T544A]::GFP and ERM-1[T544D]::GFP mutant forms (Fig. 7A,B). Recovery profiles for wild type and T544 mutants were best fitted with a two-component curve, suggesting the presence of two distinct populations (Table S1). We did not observe a noticeable difference in recovery of ERM-1[T544A]::GFP at the middle versus the borders of the bleached region (Fig. S6A,B), which suggests that the increased recovery resulted from faster association/dissociation rates, rather than a change in lateral diffusion rate at the plasma membrane.
Fig. 7.
T544 phosphorylation cycling is required for stable ERM-1 localization in the intestine. Stills from time-lapse movies (A,C,E) and FRAP curves (B,D,F) of GFP-tagged ERM-1 variants at the apical membrane of the intestine of L1 larvae (n=7 for ERM-1::GFP, n=8 for ERM-1[T544A] and n=7 for ERM-1[T544D]), the apical membrane of the excretory canal of L4 larvae (n=5 for ERM-1::GFP, n=4 for ERM-1[T544A] and n=4 for ERM-1[T544D]), or the basolateral membrane of the germline of L4 larvae (n=10 for ERM-1::GFP, n=7 for ERM-1[T544A] and n=7 for ERM-1[T544D]). Thin lines and shading represent the mean±s.d., and thick lines represent curve fitting of averaged FRAP data with a double exponential equation.
Recovery rates of wild-type ERM-1::GFP in the excretory canal were similar to those in the intestine (Fig. 7C,D). However, in this tissue no changes in recovery rates were detected for T544 mutant forms (Fig. 7C,D). To rule out the possibility that the disrupted canal morphology resulting from the COOH-terminal GFP tag affected the recovery of ERM-1, we also analyzed ERM-1::GFP fusions in heterozygous animals, which have morphologically normal canals. We observed no difference in recovery (Fig. S6C). Finally, we analyzed the recovery of ERM-1::GFP in the adult germline. Challenges related to imaging depth and tissue movement prevented us from accurate prolonged analysis of recovery at the apical membrane, thus we focused on the more easily imaged basolateral membrane. ERM-1::GFP was much more dynamic in this tissue, with bleached areas recovering ∼70% after 10 min (Fig. 7E,F). Surprisingly, both ERM-1[T544A]::GFP and ERM-1[T544D]::GFP had slower recovery profiles (Fig. 7E,F; Table S1), suggesting that these forms are more stably associated with the basolateral membrane than ERM-1::GFP. Our data show that ERM-1 dynamics are modulated by C-terminal phosphorylation and tissue-specific properties, potentially including different binding partners or differences in lipid membrane composition.
Phosphorylation of T544 controls apical actin enrichment and dynamics
C-terminal phosphorylation of ERM proteins has been reported to be required for apical actin enrichment in epithelial cells (Hipfner, 2004; Roch et al., 2010; Abbattiscianni et al., 2016). In C. elegans, loss of erm-1 function results in reduced apical actin levels in the intestine and in the excretory canal, whereas overexpression of ERM-1 in the excretory canal leads to excessive accumulation of apical actin (Göbel et al., 2004; Bernadskaya et al., 2011; Khan et al., 2013). The apical actin network of tubular epithelia in C. elegans is mostly composed of the specialized actin ACT-5 (MacQueen et al., 2005). To analyze ACT-5 in the canal, we generated a transgenic line expressing mCherry::ACT-5 from the sulp-4 promoter. Consistent with our results using VHA-5::GFP, the excretory canals showed variable, but fully penetrant, morphological defects in both ERM-1 T544 mutants (Fig. 8A). In canal regions with a widened lumen, apical ACT-5 coating was sparser and circumferential bundles were visible. The apical-to-cytoplasmic ratio of mCherry::ACT-5 was decreased in both erm-1[T544A] and erm-1[T544D] mutants, relative to control animals expressing wild-type erm-1 (Fig. 8A). This suggests that, in the excretory canal, cycling of T544 phosphorylation is important for recruitment of actin to the apical membrane.
Fig. 8.
Phosphorylation of T544 controls apical actin recruitment and dynamics. (A) Lumen morphology and distribution of mCherry::ACT-5 in the excretory canal of L4 larvae. Canal widths vary (see Fig. 3E), and examples of severely widened and less severely widened canals are shown. Graph shows apical-to-cytoplasmic ratio of mean intensity of mCherry::ACT-5. Each data point represents the average of three measurements in a single animal (n=15, 9 and 14). Error bars are the mean±s.d. (B,C) Intestinal distribution of YFP::ACT-5. Arrows indicate discontinuities. Arrowheads in comma stage indicate basolateral YFP::ACT-5, and arrowheads in 2.5-fold stage indicate ectopic expansions of the cortical actin network. (D) Quantification of YFP::ACT-5 levels at the intestinal apical membrane. Each symbol represents a single animal (n=23, 26 and 19 for L1 and n=24, 21 and 22 for L4). Error bars are the median±95% CI. Tests of significance: Dunnett's T3 multiple comparisons test for A; Dunn's multiple comparisons test for D. ns, not significant. **P≤0.01, ***P≤0.001, ****P≤0.0001. Statistical comparisons are with control unless indicated otherwise. (E) FRAP curves of apical mCherry::ACT-5 in the excretory canal of L4 larvae (n=14 for control, n=6 for erm-1[T544A] and n=7 for erm-1[T544D]) and apical YFP::ACT-5 in the intestine of L1 larvae (n=8 for control, n=13 for erm-1[T544A] and n=8 for erm-1[T544D]). Thin lines and shading represent the mean±s.d., and thick lines were obtained by curve fitting averaged FRAP data with a double exponential equation.
To analyze ACT-5 distribution in intestinal cells, we used an integrated YFP::ACT-5 transgene (Bossinger et al., 2004). In intestines of wild-type embryos, we detected a strong enrichment of ACT-5 at the apical domain of intestinal cells soon after polarization (Fig. 8B). By contrast, apical enrichment of YFP::ACT-5 in comma-stage embryos of erm-1[T544A] and erm-1[T544D] mutants was not readily detected (Fig. 8B), and YFP::ACT-5 was clearly detectable in the cytoplasm and along the basolateral membrane (Fig. 8B). Similar to the distribution of ERM-1 T544 mutants during embryogenesis, we did observe apical enrichment of YFP::ACT-5 at later embryonic stages (Fig. 8B,C). In larval stages, YFP::ACT-5 levels also followed the pattern we observed for ERM-1 in the T544 mutants (Fig. 8D; compare with Fig. 6E; Fig. S7A). In erm-1[T544D] mutants, apical YFP::ACT-5 levels were normal throughout larval development. In erm-1[T544A] mutants, apical levels were reduced in L1 larvae, but partially recovered by the L4 stage.
We next investigated ACT-5 dynamics by FRAP. The effects on ACT-5 recovery by erm-1[T544A] and erm-1[T544D] mutations were consistent with the effects we observed on ACT-5 levels. In the excretory canal, recovery of mCherry::ACT-5 was slightly faster in both mutants than in erm-1 wild-type animals, with the fastest recovery observed in the erm-1[T544A] background (Fig. 8E). In the intestine, recovery of YFP::ACT-5 in control animals was slow (∼23% in 15 min) and similar to that of ERM-1::GFP (Fig. 7A; Fig. 8E). Recovery of apical YFP::ACT-5 in the intestine of erm-1[T544A] mutants was faster than in control animals (∼41% in 15 min), whereas no difference was seen in erm-1[T544D] (Fig. 8E; Fig. S7B).
Collectively, our results show that T544 phosphorylation is important for cortical actin organization by ERM-1 in tubular epithelia. The similar delay in apical enrichment of ERM-1 and ACT-5 in the embryonic intestine, and the similar defects in ACT-5 and ERM-1 levels in larval intestines of erm-1[T544A] mutants, suggest that the presence of ERM-1 at the apical membrane is a major factor in ACT-5 recruitment.
DISCUSSION
ERM proteins drive morphological specialization events required for the function of numerous cell types across animal species. Here, we characterized the contribution of the major conserved regulatory sites important for the activity of ERM proteins in vivo in C. elegans. The use of CRISPR/Cas9 allowed us to analyze the effects of PIP2-binding and C-terminal phosphorylation mutants in the absence of wild-type product, without addition of any tags and at endogenous expression levels. This is especially relevant because ERM proteins are affected by fluorescent protein tags and often have dose-dependent effects (Berryman, 1995; Gautreau et al., 2000; Cao et al., 2005; Chambers and Bretscher, 2005; Khan et al., 2013). In agreement with studies in Drosophila and mammalian cultured cells (Yonemura et al., 2002; Fievet et al., 2004; Hao et al., 2009; Roch et al., 2010), we showed that the PIP2-binding site is essential for ERM-1 activity. By contrast, our phosphorylation mutants demonstrate that phosphorylation is not essential for ERM activity in C. elegans, despite contributing to multiple aspects of ERM-1 function, often in a tissue-specific manner.
The similarity in phenotypes caused by the erm-1[4KN] mutation and the putative erm-1(tm667) null allele (Göbel et al., 2004) demonstrates the essentiality of the PIP2-binding site. Effects on protein localization were also largely consistent with an essential role for the PIP2-binding site in localizing ERM-1 to the plasma membrane. Unexpectedly, however, we detected substantial amounts of ERM-1 at the plasma membrane of intestinal cells and the excretory canal in heterozygous erm-1[4KN]::GFP/+ animals and in first generation homozygous animals. Given that first generation erm-1[4KN]::GFP homozygous animals contain very low levels of wild-type ERM-1, it is unlikely that dimerization with wild-type ERM-1 can account for the significant plasma membrane localization of ERM-1[4KN]::GFP. An alternative model is the formation of higher order ERM-1 complexes, in which the presence of a small fraction of wild-type ERM-1 can mediate membrane localization of ERM-1[4KN]. Finally, wild-type ERM-1 might enact apical membrane modifications that promote ERM-1[4KN] recruitment.
To investigate the contribution of T544 phosphorylation, we mutated the endogenous erm-1 locus to express either a non-phosphorylatable T544A variant or a phosphomimetic T544D variant. Our results show that non-phosphorylatable ERM-1 can support C. elegans development. Although T544 mutant embryos display intestinal lumenogenesis defects that resemble those we observed in erm-1[4KN] mutants and that have been described for erm-1(RNAi) (van Fürden et al., 2004), these defects largely resolve by the L1 stage, and both erm-1(T544A) and erm-1(T544D) mutants are homozygous viable. Perhaps most strikingly, both ERM-1 T544A and T544D animals develop intestines with a normal appearance and only a mildly reduced length of microvilli. These results contrast with the findings of studies in mammalian cell culture and Drosophila that non-phosphorylatable and phosphomimetic ERM variants cannot substitute for wild-type ERM proteins (Viswanatha et al., 2012; Parameswaran et al., 2011; Polesello et al., 2002; Karagiosis and Ready, 2004). However, one study in Drosophila showed that expression of a Moesin-T559A transgene (but not Moesin-T559D) could significantly rescue viability of a strong moesin allele (Roch et al., 2010), and two others demonstrated rescuing activity for Moesin-T559D, but not for Moesin-T559A (Hipfner, 2004; Speck et al., 2003). Moreover, phosphorylation of ERM proteins is not required for the formation of microvillus-like structures in A431 and MDCK II cells (Yonemura et al., 2002). Thus, non-phosphorylated ERM proteins can be active, and the importance of C-terminal phosphorylation is likely to depend on the biological setting and experimental system used.
One of the main functions of ERM proteins is to link the plasma membrane and the actin cytoskeleton. Binding to PIP2 and C-terminal phosphorylation have been shown cooperatively to promote the actin-binding activity of ERM proteins (Matsui et al., 1998; Bosk et al., 2011; Braunger et al., 2014; Huang et al., 1999; Nakamura et al., 1999). In this process, phosphorylation does not appear to regulate the F-actin binding strength of individual ERM proteins directly, but instead increases the pool of ERM protein available for interaction with actin by promoting the open conformation (Bosk et al., 2011; Braunger et al., 2014). Our results are consistent with this model because the levels of apical actin in the intestine and canal were correlated with the levels of apical ERM-1, and not with the phosphorylation state. The fact that microvilli are formed in erm-1[T544A] mutants also indicates that ERM-1[T544A] can be active and capable of organizing apical actin. The close correlation between ERM-1 and ACT-5 levels in the intestine also highlights the important role of ERM-1 in organizing apical actin in this tissue.
Multiple aspects of the defects in erm-1[T544A] and erm-1[T544D] mutants were strikingly similar. This includes the lumen formation defects in the excretory canal and early intestine, the delayed apical accumulation in embryonic development, and the increased mobility in the larval intestine. These results are consistent with a model in which cycling between phosphorylated and non-phosphorylated states is crucial to controlling the activity of ERM-1, as has been proposed to account for ERM protein activity in activation of B and T cells, in secretion of gastric parietal cells, and in formation of microvilli in epithelial cells (Zhu et al., 2007; Parameswaran et al., 2011; Viswanatha et al., 2012). This interpretation does rely on the assumption that phosphomimetic mutations mimic the phosphorylated state, which is supported by extensive characterization of phosphomimetic ERM mutants in other systems (Huang et al., 1999; Matsui et al., 1998; Nakamura et al., 1999; Oshiro et al., 1998; Simons et al., 1998; Gautreau et al., 2000; Polesello et al., 2002; Speck et al., 2003; Chambers and Bretscher, 2005; Charras et al., 2006; Carreno et al., 2008; Kunda et al., 2008; Bosk et al., 2011).
How does the dynamic turnover of C-terminal phosphorylation regulate ERM activity? A previous study in cultured epithelial cells found that local phosphocycling by apical kinases restricts ezrin localization and activity to microvilli (Viswanatha et al., 2012). Similar polarized kinase activity was shown to restrict Moesin activity to the apical cortex of tracheal cells in Drosophila (Ukken et al., 2014). During embryonic intestinal development, we observed a delay in the apical enrichment of ERM-1 in erm-1[T544A] and erm-1[T544D] mutants, and in the larval intestine, both T544A and T544D substitutions greatly increased the mobility of ERM-1. These results indicate that in C. elegans, local phosphorylation cycling promotes the apical enrichment and basolateral exclusion of ERM-1. How phosphorylation cycling promotes the stable apical localization of ERM-1 is not clear. One possibility is that ERM-1 needs to cycle between different conformations in order to cope with a changing microenvironment, which presumably involves actin treadmilling and local changes in the concentration or availability of PIP2 and protein binding partners. ERM-1 stability in the excretory canal was unaffected by the phosphorylation mutants, perhaps reflecting a difference in the dynamics of apical membrane components between tissues. Regardless of the underlying reason, ERM-1 turnover dynamics can vary greatly depending on the biological context and are subject to tissue-specific regulation. Finally, we note that although both mutant forms behave in a similar manner in many respects, apical ERM-1[T544A]::GFP levels are significantly lower than either ERM-1::GFP or ERM-1[T544D]::GFP levels in the L1 intestine and never become completely normal. Thus, cycling-independent aspects of T544 phosphorylation presumably do play a role in regulating ERM-1.
Previous FRAP studies in microvilli of LLC-PK1 cells and in blebbing M2 melanoma cells indicate the presence of three pools of ezrin with recovery half-times in the range of seconds to minutes (Coscoy et al., 2002; Fritzsche et al., 2014). Our analysis of ERM-1::GFP protein dynamics by FRAP also supports the presence of at least two pools of ERM-1 in C. elegans, but reveals an unexpected stability for ERM-1 in the intestine and excretory canal. Similar results were obtained by A. Bidaud Meynard and G. Michaux (personal communication). Slow recovery of ERM-1 is consistent with a recent study which found that the ezrin FERM domain binds phosphoinositide-rich membranes with very high affinity and slow dissociation dynamics (Senju et al., 2017), whereas ERM-1::GFP levels at the basolateral domain of the gonad recovered more rapidly. Our results in the intestine might therefore reflect an inherent physiological difference from cells in culture and between tissues.
In contrast to the intestine, lumen formation and outgrowth of the excretory canal were severely affected in both erm-1[T544A] and erm-1[T544D] mutant animals throughout their lifespan. Surprisingly, however, neither localization nor mobility of ERM-1 in the canal was affected by altered T544 regulation. ERM proteins interact with and regulate the function of numerous proteins, including trafficking components, transmembrane channels or pumps, polarity determinants and junction proteins (Médina et al., 2002; Deretic et al., 2004; Pilot et al., 2006; Zhu et al., 2007; Chirivino et al., 2011; Kvalvaag et al., 2013; Khan et al., 2013; Bryant et al., 2014). Moreover, several interaction partners have been shown to interact selectively with either non-phosphorylated or phosphorylated forms of ezrin (Viswanatha et al., 2013). It is possible that a difference in the subset of ERM-1 interaction partners involved in each tissue explains why effects on ERM-1 localization and mobility are not strictly correlated with the phenotypic defects at the tissue level. The absence of changes in ERM-1 protein behavior in the canal further indicate that C-terminal threonine phosphorylation is not the only mechanism that controls ERM-1 localization and stability. Binding to specific interaction partners might contribute to ERM-1 localization, but alternative mechanisms of regulation are also possible. For example, a recent study found that in breast epithelial cells, ezrin membrane association is regulated by acetylation of ezrin (Song et al., 2019). Overall, these results demonstrate that C-terminal phosphorylation is a versatile regulatory modification that modulates ERM-1 function in a context-dependent manner and underscore the importance of studying ERM regulation in different biological scenarios.
MATERIALS AND METHODS
Caenorhabditis elegans strains and culture conditions
Caenorhabditis elegans strains were cultured in standard conditions (Brenner, 1974). Only hermaphrodites were used, and all experiments were performed with animals grown at 20°C on nematode growth medium (NGM) agar plates. Table S2 contains a list of all the strains used.
CRISPR/Cas9 genome engineering
Endogenous eGFP protein fusions and point mutations were generated by homology-directed repair of CRISPR/Cas9-induced DNA double-strand breaks. erm-1[T544A] and eGFP protein fusions were generated in an N2 background, with the exception of erm-1[4KN]::GFP, in which erm-1[KN]/+ was used as the starting genetic background; remaining point mutations were generated in a pha-1(e2123ts) background. In all cases, two single guide RNA (sgRNA) plasmids targeting each locus were used. The sgRNA plasmids were generated by ligation of annealed oligo pairs into the pU6::sgRNA expression vectors pMB70 (Addgene 47943) or pJJR50 (Addgene 75026) as previously described (Waaijers et al., 2013, 2016). To introduce point mutations, synthesized single-stranded oligodeoxynucleotides (ssODNs) with 33-45 bp homology arms were used as a repair template, and integration events were selected using either dpy-10 (Arribere et al., 2014) or pha-1 (Ward, 2015) co-CRISPR approaches. eGFP knock-ins were introduced using a plasmid-based repair template with 450-600 bp homology arms and containing a self-excising cassette (SEC) for selection, as previously described (Dickinson et al., 2015). To introduce eGFP, we created a custom SEC vector, pJJR82 (Addgene 75027), by replacing a fragment of pDD282 (Addgene 66823) comprising the GFP sequence with a similar fragment comprising a codon-optimized and synthetic intron-containing eGFP sequence using the flanking Bsu36I and BglII restriction sites. In all cases, correct genome editing was confirmed by Sanger sequencing (Macrogen Europe) of PCR amplicons encompassing the edited genomic region. The maternal-effect lethal erm-1[KN] and erm-1[KN]::GFP alleles were balanced with dpy-5(e61); unc-29(e403) I (strain DR102). In all cases, edited strains were backcrossed twice with N2 to eliminate any non-linked unspecific editing events, and one additional round of backcrossing was done for strains generated in a pha-1(e2123ts) background. The sequences of all oligonucleotides used (synthesized by Integrated DNA Technologies) are listed in Table S3.
Microscopy
Imaging of C. elegans was done by mounting embryos or larvae on a 5% agarose pad in a 10 mM Tetramisole solution in M9 buffer (0.22 M KH2PO4, 0.42 M Na2HPO4, 0.85 M NaCl and 0.001 M MgSO4) to induce paralysis. Spinning disk confocal imaging was performed using a Nikon Ti-U microscope equipped with a Yokogawa CSU-X1 spinning disk and an Andor iXon+ EMCCD camera, using ×60 and ×100 1.4 NA objectives. Time-lapse imaging for FRAP experiments was performed on a Nikon Eclipse-Ti microscope equipped with a Yokogawa CSU-X1-A1 spinning disk and a Photometrics Evolve 512 EMCCD camera, using a ×100 1.4 NA objective. Targeted photobleaching was done using an ILas system (Roper Scientific France/PICT-IBiSA, Institut Curie). Two epifluorescence microscopy set-ups were used. Digital interference contrast (DIC) imaging was done in an upright Zeiss Axioplan2 microscope using a ×63 1.4 NA objective, and imaging to quantify excretory canal outgrowth was done in an upright Zeiss Axio Imager 2 microscope using a ×20 0.5 NA objective. Microscopy data were acquired using MetaMorph Microscopy Automation & Image Analysis Software (spinning disk), Zeiss AxioVision (DIC) and Zeiss Zen (epifluorescence). All stacks along the z-axis were obtained at 0.25 μm intervals, and all images were analyzed and processed using ImageJ and Adobe Photoshop. For quantifications, the same laser power and exposure times were used within experiments.
Quantitative image analysis
Quantitative analysis of spinning disk images was done in ImageJ. In all quantifications, the mean background intensity was quantified by drawing a circular region of diameter 50 pixels in areas within the field of view that did not contain any animals, and values were normalized using the mean intensity of eGFP, YFP or mCherry at the apical membrane of the corresponding tissue in control animals. In the intestine, all measurements were done in cells forming int2 through int6, and the intensity of fluorescence at the apical membrane was quantified in regions where opposing apical membranes could be seen clearly as two lines. To quantify the intensity of fluorescence of ERM-1[4KN]::GFP in intestinal cells and mCherry::ACT-5 in the excretory canal, measurements were performed in maximum intensity projections of three consecutive z-slices showing the highest intensity at the apical membrane. The peak intensity at the apical membrane was calculated by averaging the peak values of intensity profiles from multiple 40-pixel-wide line scans perpendicular to the membrane per animal. The mean cytoplasmic intensity was obtained by averaging the mean intensity values of multiple elliptical regions within the cytoplasm. Each measurement was corrected for background noise and normalized as described above. Averaged apical and cytoplasmic intensity values were used to calculate the apical-to-cytoplasmic intensity ratio per animal. Distribution plots of the fluorescence intensity of GFP-tagged ERM-1 variants in the larval germline, and for PEPT-1::DsRed and GFP::RAB-11 in the intestine, were obtained using the same method. For GFP-tagged ERM-1 variants in the germline, three measurements were done per animal in single frames where both apical and basal membranes were clearly visible. For animals expressing both PEPT-1::DsRed and GFP::RAB-11 in the intestine, two measurements were done per animal in maximum intensity projections of the three consecutive z-slices with the highest intensity of PEPT-1::DsRed at the apical membrane. Measurements from single animals were not averaged in either case. Intensity distribution profiles were obtained by tracing line scans (15 pixels wide for the germline and 40 pixels wide for the intestine) encompassing the entire germ cell compartments or intestinal cells along the apical-basal axis. Intensity profiles were trimmed manually to exclude values outside the cells/compartments of interest, and each value was corrected for background noise and normalized as described above. To make direct comparisons and plot intensity profiles despite differences in the distance between basal and apical membranes, a custom R script was made to interpolate intensity values linearly on the y-axis to a fixed distance along the x-axis for each intensity profile defined by the average apical-basal distance in control animals (script is available upon request). For the germline, apical-to-basal intensity ratios were calculated using the peak intensity values at the apical and basal membranes per intensity profile. To quantify the intensity of fluorescence of eGFP-tagged ERM-1 variants and YFP::ACT-5 in intestinal cells, a free-hand region was drawn either surrounding the apical membrane or in the cytoplasm, and mean intensity values were extracted for all z-slices in which apical membrane was visible. The background was subtracted per frame, and each value was normalized as described above. The mean intensity at the apical membrane and cytoplasm were calculated by averaging measurements through the z-axis of two intestinal cells per animal. Averaged intensity values were used to calculate the apical-to-cytoplasmic ratio per animal.
Brood size and lethality
Starting at the L4 stage, individual P0 animals were cultured at 20°C and transferred to a fresh plate every 24 h for 6 days. Hatched and unhatched progeny were scored 24 h after removal of the P0, and larval lethality was scored 48 h after removal of the P0.
Relative excretory canal outgrowth and canal width
To quantify relative canal outgrowth in the excretory canal cell, F1 progeny of L4 animals expressing the VHA-5::GFP transgene grown in standard or RNAi culture plates were scored at the L4 stage. The distance between the cell body and either the anterior or posterior distal body tips was determined by tracing a segmented line along the center of the animal. The length of each individual canal was measured with a segmented line from the anterior-posterior bifurcation points close to the cell body until the canal tip. Relative outgrowth was calculated as the fraction of canal length over the distance between the cell body and distal tips. Severe outgrowth defects were defined as canals that extend ≤35% of the distance between the excretory canal cell body and either anterior or posterior tips. The frequency was calculated by the sum of both anterior and posterior canals with severe defects over total canals quantified per genotype. For canal width, we measured the width at the three widest points, including at any cysts present, in the most severely affected posterior canal of an animal. The average measurement of these three points represents one data point.
FRAP experiments and analysis
For FRAP assays, the laser power was adjusted in each experiment to avoid complete photobleaching of the selected area. Photobleaching was performed on a circular region with a diameter of 30–100 pixels at the cortex, and recovery was followed at 5 s intervals for 15-45 min depending on the tissue. Time-lapse movies were analyzed in ImageJ. The size of the area for FRAP analysis was defined by the full width at half-maximum of an intensity plot across the bleached region in the first post-bleach frame. For each time-lapse frame, the mean intensity value within the bleached region was determined, and the background, defined as the mean intensity of a non-bleached region outside the animal, was subtracted. The mean intensities within the bleached region were corrected for acquisition photobleaching per frame using the background-subtracted mean intensity of a similar non-bleached region at the cortex, which was normalized to the corresponding pre-bleach mean intensity. FRAP recovery was calculated as the change in corrected intensity values within the bleach region from the first frame after bleach (set to zero) normalized to the mean intensity of the 10 frames before bleaching. Curve fitting was done on averaged recovery data per sample using the non-linear regression analysis in GraphPad. One- and two-phase association were tested, and in all cases the data were best fitted with a two-phase curve. Intensity distribution plots were obtained by performing a three-pixel-wide line scan perpendicular to the apical membrane.
Immunohistochemistry
Antibody stainings were performed in mixed-stage (anti-pERM) or synchronous (anti-ERM-1) populations. Synchronized animals were obtained from gravid adult animals by bleaching and allowed to develop in M9 for 4-6 h for embryonic stages, or in standard culture plates for 10 h for early larval stages. Mixed-stage or synchronous larval populations were collected from plates and were washed three times in M9 (for stainings of post-embryonic stages, samples were incubated for 30 min with gentle shaking before the last wash), washed once in water, and transferred to a poly-l-lysine-coated slide. Samples were permeabilized by freeze-cracking and fixed at −20°C with methanol for 5 min and acetone for 10 min for staining with the ERM-1 antibody, or with P buffer (3.7% formaldehyde, 75% methanol, 250 µM EDTA and 50 mM NaF) for 15 min and methanol for 5 min for the pERM antibody. Samples were rehydrated in an ethanol series (90, 60 and 30%, for 10 min each at −20°C), rinsed three times in PBS-Tween (PBST; 1.35 M NaCl, 27 mM KCl, 100 mM Na2HPO4, 18 mM KH2PO4 and 0.05% Tween-20), and blocked for 1 h with 1% bovine serum albumin and 10% serum in PBST at room temperature (RT). Samples were incubated with primary antibodies in blocking solution overnight at 4°C, washed four times for 15 min in PBST, and secondary antibodies in blocking solution were incubated for 2 h at RT. Finally, samples were washed three times in PBST and once in PBS, for 10 min each, and mounted with Prolong Gold Antifade (Thermofisher). The following antibodies and dilutions were used: MH27 mouse monoclonal (Developmental Studies Hybridoma Bank, validated to have no immunoreactivity in mutant embryos), 1:20; ERM-1 rabbit polyclonal [gift from O. Bossinger, validated to have no immunoreactivity in erm-1(RNAi) embryos (van Fürden et al., 2004)], 1:100; pERM rabbit polyclonal (Cell Signaling Technology #3144, verified here to not stain ERM-1[T544A]), 1:100; Alexa-Fluor 488 goat anti-rabbit and Alexa-Fluor 568 goat anti-mouse (Life Technologies, A-11008 and A11004, respectively) 1:500.
Feeding RNAi
The aqp-8 RNAi clone was obtained from the genome-wide Vidal full-length HT115 RNAi feeding library derived from the ORFeome v.3.1 collection (Rual et al., 2004). An HT115 bacterial clone expressing the L4440 vector lacking an insert was used as a control. For feeding RNAi experiments, bacteria were precultured in 2 ml lysogeny broth (LB) supplemented with 100 µg/ml ampicillin (Amp) and 2.5 µg/ml tetracyclin (Tet) at 37°C in an incubator rotating at 200 rpm for 6-8 h, then transferred to new tubes with a total volume of 10 ml for overnight culturing. To induce production of double-stranded RNA, cultures were incubated for 90 min in the presence of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Bacterial cultures were pelleted by centrifugation at 4000 g for 15 min and concentrated five times. NGM agar plates supplemented with 100 μg/ml Amp and 1 mM IPTG were seeded with 250 μl of bacterial suspension and kept at RT for 48 h in the dark. Six to eight L4 hermaphrodites per strain were transferred to individual NGM-RNAi plates against target genes, and phenotypes were analyzed in the F1 generation.
Transmission electron microscopy
For TEM, L4 animals were fixed by high-pressure freezing with the EMPACT-2 system (Leica Microsystems). Freeze-substitution (FS) was done in anhydrous acetone containing 1% OsO4, 0.5% glutaraldehyde and 0.25% uranyl acetate for 60 h in an FS system (AFS-2; Leica Microsystems). Larvae were embedded in an Epon-Araldite mix (hard formula; EMS). Adhesive frames were used (Gene Frame 65 μl; ThermoFisher) for flat-embedding, as previously described (Kolotuev et al., 2012), to facilitate anterior-posterior orientation and sectioning. Ultrathin sections were cut on an ultramicrotome (UC7; Leica Microsystems) and collected on formvar-coated slot grids (FCF2010-CU; EMS). Each larva was sectioned in five different places, with ≥10 μm between each grid to ensure that different cells were observed. Each grid contained ≥10 consecutive sections of 70 nm thickness. The TEM grids were observed using a JEM-1400 transmission electron microscope (JEOL) operated at 120 kV, equipped with a Gatan Orius SC200 camera (Gatan) and piloted by the Digital Micrograph program. The length of the microvilli was quantified using Fiji on TEM pictures of at least five sections per worm.
Excretory canal-specific mCherry::ACT-5 reporter
The Psulp-4::mCherry::ACT-5 construct was cloned into the pBSK vector using Gibson Assembly (Gibson et al., 2009). A fragment containing 2.3 kb immediately upstream of the sulp-4 coding sequence, and a fragment of 1.7 kb containing the entire genomic sequence of act-5 and 215 bp of the 3′ untranslated region, were amplified from C. elegans genomic DNA. The codon-optimized mCherry sequence with synthetic introns and a C-terminal linker was amplified from pJJR83 (Addgene 75028). Correct amplification and assembly were confirmed by Sanger sequencing. Primers used are listed in Table S3. Several stable transgenic lines were generated by microinjection of N2 young adult animals with 5 ng/µl Psulp-4::mCherry::ACT-5 and 10 ng/µl Plin-48::GFP as a co-injection marker, which did not affect excretory canal development, and one was selected for further analysis.
Statistical analysis
All statistical analyses were performed using GraphPad Prism v.8. For population comparisons, a D'Agostino & Pearson test of normality was first performed to determine whether the data were sampled from a Gaussian distribution. For data drawn from a Gaussian distribution, comparisons between two populations were made using Student's unpaired t-test, with Welch's correction if the standard deviations of the populations differed significantly, and comparisons between more than two populations were made using a one-way ANOVA, or Welch's ANOVA if the standard deviations of the populations differed significantly. For data not drawn from a Gaussian distribution, a non-parametric test was used (Mann–Whitney U-test for two populations and Kruskal–Wallis test for more than two populations). ANOVA and non-parametric tests were followed up with multiple comparison tests of significance (Dunnett's, Tukey's, Dunnett's T3 or Dunn's). The tests of significance used and the sample sizes are indicated in the figure legends. No statistical method was used to predetermine sample sizes. No samples or animals were excluded from analysis. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary Material
Acknowledgements
We thank J. Kerver-Stumpfova for technical assistance, H. R. Pires for the dlg-1::mCherry knock-in, O. Bossinger for providing the ERM-1 antibody and the BJ49 strain, M. Labouesse for sharing the ML846 strain, and M. Zerial for sharing the MZE1 strain. We also thank WormBase, the Biology Imaging Center Faculty of Sciences, Department of Biology, Utrecht University, and the electron microscopy facility of the Microscopy Rennes Imaging Center (MRIC). Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the US National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). The MH27 monoclonal antibody developed by R. Francis and R. H. Waterston was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the NIH and maintained at The University of Iowa. We thank S. van den Heuvel, M. Harterink and members of the S. van den Heuvel, M.B. and R. Korswagen groups for helpful discussions.
Footnotes
Author contributions
Conceptualization: J.J.R., M.B.; Methodology: J.J.R., J.J.S., O.N., R.S., J.C.; Formal analysis: J.J.R., J.J.S., O.N., R.S.; Investigation: J.J.R., J.J.S., O.N., R.S.; Writing - original draft: J.J.R., M.B.; Writing - review & editing: J.J.S., R.S., G.M., M.B.; Visualization: J.J.R., J.J.S., O.N., R.S., M.B.; Supervision: G.M., M.B.; Project administration: M.B.; Funding acquisition: G.M., M.B.
Funding
This work was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) (CW ECHO 711.014.005 and NWO-VICI 016.VICI.170.165 to M.B.), by funding from the Ligue Contre le Cancer Grand-Ouest/22-29, by the Centre National de la Recherche Scientifique and Université de Rennes (G.M.), and by the European Union Horizon 2020 research and innovation program under Marie Skłodowska-Curie Actions (675407 – PolarNet).
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.188011.reviewer-comments.pdf
References
- Abbattiscianni, A. C., Favia, M., Mancini, M. T., Cardone, R. A., Guerra, L., Monterisi, S., Castellani, S., Laselva, O., Di Sole, F., Conese, M., et al. (2016). Correctors of mutant CFTR enhance subcortical cAMP–PKA signaling through modulating ezrin phosphorylation and cytoskeleton organization. J. Cell Sci. 129, 1128-1140. 10.1242/jcs.177907 [DOI] [PubMed] [Google Scholar]
- Arribere, J. A., Bell, R. T., Fu, B. X. H., Artiles, K. L., Hartman, P. S. and Fire, A. Z. (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837-846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babich, V. and Di Sole, F. (2015). The Na+/H+ exchanger-3 (NHE3) activity requires ezrin binding to phosphoinositide and its phosphorylation. PLOS ONE 10, e0129306. 10.1371/journal.pone.0129306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret, C., Roy, C., Montcourrier, P., Mangeat, P. and Niggli, V. (2000). Mutagenesis of the phosphatidylinositol 4,5-bisphosphate (Pip2) binding site in the Nh2 -terminal domain of ezrin correlates with its altered cellular distribution. J. Cell Biol. 151, 1067-1080. 10.1083/jcb.151.5.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aissa, K., Patino-Lopez, G., Belkina, N. V., Maniti, O., Rosales, T., Hao, J.-J., Kruhlak, M. J., Knutson, J. R., Picart, C. and Shaw, S. (2012). Activation of moesin, a protein that links actin cytoskeleton to the plasma membrane, occurs by phosphatidylinositol 4,5–bisphosphate (PIP2) binding sequentially to two sites and releasing an autoinhibitory linker. J. Biol. Chem. 287, 16311-16323. 10.1074/jbc.M111.304881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadskaya, Y. Y., Patel, F. B., Hsu, H.-T. and Soto, M. C. (2011). Arp2/3 promotes junction formation and maintenance in the Caenorhabditis elegans intestine by regulating membrane association of apical proteins. Mol. Biol. Cell 22, 2886-2899. 10.1091/mbc.e10-10-0862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryman, M. (1995). Ezrin oligomers are major cytoskeletal components of placental microvilli: a proposal for their involvement in cortical morphogenesis. J. Cell Biol. 131, 1231-1242. 10.1083/jcb.131.5.1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha, V. L., Finnemann, S. C. and Rodriguez-Boulan, E. (1999). Ezrin promotes morphogenesis of apical microvilli and basal infoldings in retinal pigment epithelium. J. Cell Biol. 147, 1533-1548. 10.1083/jcb.147.7.1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha, V. L., Rayborn, M. E., Saotome, I., McClatchey, A. I. and Hollyfield, J. G. (2006). Microvilli defects in retinas of ezrin knockout mice. Exp. Eye Res. 82, 720-729. 10.1016/j.exer.2005.09.013 [DOI] [PubMed] [Google Scholar]
- Bosk, S., Braunger, J. A., Gerke, V. and Steinem, C. (2011). Activation of F-actin binding capacity of ezrin: synergism of PIP2 interaction and phosphorylation. Biophys. J. 100, 1708-1717. 10.1016/j.bpj.2011.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossinger, O., Fukushige, T., Claeys, M., Borgonie, G. and McGhee, J. D. (2004). The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev. Biol. 268, 448-456. 10.1016/j.ydbio.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Braunger, J. A., Brückner, B. R., Nehls, S., Pietuch, A., Gerke, V., Mey, I., Janshoff, A. and Steinem, C. (2014). Phosphatidylinositol 4,5-bisphosphate alters the number of attachment sites between ezrin and actin filaments: a colloidal probe study. J. Biol. Chem. 289, 9833-9843. 10.1074/jbc.M113.530659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, D. M., Roignot, J., Datta, A., Overeem, A. W., Kim, M., Yu, W., Peng, X., Eastburn, D. J., Ewald, A. J., Werb, Z., et al. (2014). A molecular switch for the orientation of epithelial cell polarization. Dev. Cell 31, 171-187. 10.1016/j.devcel.2014.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., Ding, X., Guo, Z., Zhou, R., Wang, F., Long, F., Wu, F., Bi, F., Wang, Q., Fan, D., et al. (2005). PALS1 specifies the localization of ezrin to the apical membrane of gastric parietal cells. J. Biol. Chem. 280, 13584-13592. 10.1074/jbc.M411941200 [DOI] [PubMed] [Google Scholar]
- Carreno, S., Kouranti, I., Glusman, E. S., Fuller, M. T., Echard, A. and Payre, F. (2008). Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J. Cell Biol. 180, 739. 10.1083/jcb.200709161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaletto, J. B., Saotome, I., Curto, M. and McClatchey, A. I. (2011). Ezrin-mediated apical integrity is required for intestinal homeostasis. Proc. Natl. Acad. Sci. USA 108, 11924-11929. 10.1073/pnas.1103418108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, D. N. and Bretscher, A. (2005). Ezrin mutants affecting dimerization and activation. Biochemistry 44, 3926-3932. 10.1021/bi0480382 [DOI] [PubMed] [Google Scholar]
- Charras, G. T., Hu, C.-K., Coughlin, M. and Mitchison, T. J. (2006). Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175, 477. 10.1083/jcb.200602085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Cohn, J. A. and Mandel, L. J. (1995). Dephosphorylation of ezrin as an early event in renal microvillar breakdown and anoxic injury. Proc. Natl. Acad. Sci. USA 92, 7495-7499. 10.1073/pnas.92.16.7495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirivino, D., Del Maestro, L., Formstecher, E., Hupé, P., Raposo, G., Louvard, D. and Arpin, M. (2011). The ERM proteins interact with the HOPS complex to regulate the maturation of endosomes. Mol. Biol. Cell 22, 375-385. 10.1091/mbc.e10-09-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy, S., Waharte, F., Gautreau, A., Martin, M., Louvard, D., Mangeat, P., Arpin, M. and Amblard, F. (2002). Molecular analysis of microscopic ezrin dynamics by two-photon FRAP. Proc. Natl. Acad. Sci. USA 99, 12813-12818. 10.1073/pnas.192084599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic, D., Traverso, V., Parkins, N., Jackson, F., de Turco, E. B. R. and Ransom, N. (2004). Phosphoinositides, Ezrin/Moesin, and rac1 regulate fusion of rhodopsin transport carriers in retinal photoreceptors. Mol. Biol. Cell 15, 359-370. 10.1091/mbc.e03-04-0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D. J., Pani, A. M., Heppert, J. K., Higgins, C. D. and Goldstein, B. (2015). Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200, 1035-1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon, R. G., McClatchey, A. I. and Bretscher, A. (2010). Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11, 276-287. 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fievet, B. T., Gautreau, A., Roy, C., Del Maestro, L., Mangeat, P., Louvard, D. and Arpin, M. (2004). Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J. Cell Biol. 164, 653-659. 10.1083/jcb.200307032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche, M., Thorogate, R. and Charras, G. (2014). Quantitative analysis of ezrin turnover dynamics in the actin cortex. Biophys. J. 106, 343-353. 10.1016/j.bpj.2013.11.4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett, D. and Bretscher, A. (2012). PDZ interactions regulate rapid turnover of the scaffolding protein EBP50 in microvilli. J. Cell Biol. 198, 195-203. 10.1083/jcb.201204008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary, R. and Bretscher, A. (1995). Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol. Biol. Cell 6, 1061-1075. 10.1091/mbc.6.8.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau, A., Louvard, D. and Arpin, M. (2000). Morphogenic effects of ezrin require a phosphorylation-induced transition from oligomers to monomers at the plasma membrane. J. Cell Biol. 150, 193-204. 10.1083/jcb.150.1.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A. and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343-345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Göbel, V., Barrett, P. L., Hall, D. H. and Fleming, J. T. (2004). Lumen Morphogenesis in C. elegans Requires the Membrane-Cytoskeleton Linker erm-1. Dev. Cell 6, 865-873. 10.1016/j.devcel.2004.05.018 [DOI] [PubMed] [Google Scholar]
- Hao, J.-J., Liu, Y., Kruhlak, M., Debell, K. E., Rellahan, B. L. and Shaw, S. (2009). Phospholipase C–mediated hydrolysis of PIP2 releases ERM proteins from lymphocyte membrane. J. Cell Biol. 184, 451-462. 10.1083/jcb.200807047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipfner, D. R. (2004). Slik Sterile-20 kinase regulates Moesin activity to promote epithelial integrity during tissue growth. Genes Dev. 18, 2243-2248. 10.1101/gad.303304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., Wong, T. Y. W., Lin, R. C. C. and Furthmayr, H. (1999). Replacement of threonine 558, a critical site of phosphorylation of moesin in Vivo, with aspartate activates f-actin binding of moesin: regulation by conformational change. J. Biol. Chem. 274, 12803-12810. 10.1074/jbc.274.18.12803 [DOI] [PubMed] [Google Scholar]
- Karagiosis, S. A. and Ready, D. H. (2004). Moesin contributes an essential structural role in Drosophila photoreceptor morphogenesis. Development 131, 725-732. 10.1242/dev.00976 [DOI] [PubMed] [Google Scholar]
- Khan, L. A., Zhang, H., Abraham, N., Sun, L., Fleming, J. T., Buechner, M., Hall, D. H. and Gobel, V. (2013). Intracellular lumen extension requires ERM-1-dependent apical membrane expansion and AQP-8-mediated flux. Nat. Cell Biol. 15, 143-156. 10.1038/ncb2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolotuev, I., Bumbarger, D. J., Labouesse, M. and Schwab, Y. (2012). Targeted ultramicrotomy. In Methods in Cell Biology (ed. T. Müller-Reichert and P. Verkade), pp. 203-222. Elsevier. [DOI] [PubMed] [Google Scholar]
- Kondo, T., Takeuchi, K., Doi, Y., Yonemura, S., Nagata, S., Tsukita, S. and Tsukita, S. (1997). ERM (Ezrin/Radixin/Moesin)-based molecular mechanism of microvillar breakdown at an early stage of apoptosis. J. Cell Biol. 139, 749-758. 10.1083/jcb.139.3.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda, P., Pelling, A. E., Liu, T. and Baum, B. (2008). Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr. Biol. 18, 91-101. 10.1016/j.cub.2007.12.051 [DOI] [PubMed] [Google Scholar]
- Kvalvaag, A. S., Pust, S., Sundet, K. I., Engedal, N., Simm, R. and Sandvig, K. (2013). The ERM proteins ezrin and moesin regulate retrograde shiga toxin transport: the ERM proteins regulate retrograde transport. Traffic 14, 839-852. 10.1111/tra.12077 [DOI] [PubMed] [Google Scholar]
- Li, Q., Nance, M. R., Kulikauskas, R., Nyberg, K., Fehon, R., Karplus, P. A., Bretscher, A. and Tesmer, J. J. G. (2007). Self-masking in an intact ERM-merlin protein: an active role for the central α-helical domain. J. Mol. Biol. 365, 1446-1459. 10.1016/j.jmb.2006.10.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liégeois, S., Benedetto, A., Garnier, J.-M., Schwab, Y. and Labouesse, M. (2006). The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol. 173, 949-961. 10.1083/jcb.200511072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liégeois, S., Benedetto, A., Michaux, G., Belliard, G. and Labouesse, M. (2007). Genes required for osmoregulation and apical secretion in Caenorhabditis elegans. Genetics 175, 709-724. 10.1534/genetics.106.066035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen, A. J., Baggett, J. J., Perumov, N., Bauer, R. A., Januszewski, T., Schriefer, L. and Waddle, J. A. (2005). ACT-5 is an essential caenorhabditis elegans actin required for intestinal microvilli Formation. Mol. Biol. Cell 16, 13. 10.1091/mbc.e04-12-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magendantz, M., Henry, M. D., Lander, A. and Solomon, F. (1995). Interdomain interactions of radixin in Vitro. J. Biol. Chem. 270, 25324-25327. 10.1074/jbc.270.43.25324 [DOI] [PubMed] [Google Scholar]
- Matsui, T., Maeda, M., Doi, Y., Yonemura, S., Amano, M., Kaibuchi, K., Tsukita, S. and Tsukita, S. (1998). Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J. Cell Biol. 140, 647-657. 10.1083/jcb.140.3.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey, A. I. (2014). ERM proteins at a glance. J. Cell Sci. 127, 3199-3204. 10.1242/jcs.098343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médina, E., Williams, J., Klipfell, E., Zarnescu, D., Thomas, G. and Le Bivic, A. (2002). Crumbs interacts with moesin and β Heavy -spectrin in the apical membrane skeleton of Drosophila. J. Cell Biol. 158, 941-951. 10.1083/jcb.200203080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, F., Amieva, M. R. and Furthmayr, H. (1995). Phosphorylation of threonine 558 in the carboxyl-terminal actin-binding domain of moesin by thrombin activation of human platelets. J. Biol. Chem. 270, 31377-31385. 10.1074/jbc.270.52.31377 [DOI] [PubMed] [Google Scholar]
- Nakamura, F., Huang, L., Pestonjamasp, K., Luna, E. J. and Furthmayr, H. (1999). Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphatidylinositides. Mol. Biol. Cell 10, 2669-2685. 10.1091/mbc.10.8.2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch, A. L. and Fehon, R. G. (2011). Ezrin, Radixin and Moesin: key regulators of membrane–cortex interactions and signaling. Curr. Opin. Cell Biol. 23, 377-382. 10.1016/j.ceb.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro, N., Fukata, Y. and Kaibuchi, K. (1998). Phosphorylation of moesin by Rho-associated kinase (Rho-kinase) plays a crucial role in the formation of microvilli-like structures. J. Biol. Chem. 273, 34663-34666. 10.1074/jbc.273.52.34663 [DOI] [PubMed] [Google Scholar]
- Parameswaran, N. and Gupta, N. (2013). Re-defining ERM function in lymphocyte activation and migration. Immunol. Rev. 256, 63-79. 10.1111/imr.12104 [DOI] [PubMed] [Google Scholar]
- Parameswaran, N., Matsui, K. and Gupta, N. (2011). Conformational switching in ezrin regulates morphological and cytoskeletal changes required for B cell chemotaxis. J. Immunol. 186, 4088-4097. 10.4049/jimmunol.1001139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, M. A., Reczek, D., Bretscher, A. and Karplus, P. A. (2000). Structure of the ERM protein moesin reveals the ferm domain fold masked by an extended actin binding tail domain. Cell 101, 259-270. 10.1016/S0092-8674(00)80836-3 [DOI] [PubMed] [Google Scholar]
- Pelaseyed, T. and Bretscher, A. (2018). Regulation of actin-based apical structures on epithelial cells. J. Cell Sci. 131, jcs221853. 10.1242/jcs.221853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaseyed, T., Viswanatha, R., Sauvanet, C., Filter, J. J., Goldberg, M. L. and Bretscher, A. (2017). Ezrin activation by LOK phosphorylation involves a PIP2-dependent wedge mechanism. eLife 6, e22759. 10.7554/eLife.22759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot, F., Philippe, J.-M., Lemmers, C. and Lecuit, T. (2006). Spatial control of actin organization at adherens junctions by a synaptotagmin-like protein. Nature 442, 580-584. 10.1038/nature04935 [DOI] [PubMed] [Google Scholar]
- Polesello, C., Delon, I., Valenti, P., Ferrer, P. and Payre, F. (2002). Dmoesin controls actin-based cell shape and polarity during Drosophila melanogaster oogenesis. Nat. Cell Biol. 4, 782-789. 10.1038/ncb856 [DOI] [PubMed] [Google Scholar]
- Roch, F., Polesello, C., Roubinet, C., Martin, M., Roy, C., Valenti, P., Carreno, S., Mangeat, P. and Payre, F. (2010). Differential roles of PtdIns(4,5)P2 and phosphorylation in moesin activation during Drosophila development. J. Cell Sci. 123, 2058-2067. 10.1242/jcs.064550 [DOI] [PubMed] [Google Scholar]
- Rual, J.-F., Ceron, J., Koreth, J., Hao, T., Nicot, A.-S., Hirozane-Kishikawa, T., Vandenhaute, J., Orkin, S. H., Hill, D. E., van den Heuvel, S., et al. (2004). toward improving caenorhabditis elegans phenome mapping with an ORFeome-based RNAi Library. Genome Res. 14, 2162-2168. 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome, I., Curto, M. and McClatchey, A. I. (2004). Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev. Cell 6, 855-864. 10.1016/j.devcel.2004.05.007 [DOI] [PubMed] [Google Scholar]
- Senju, Y., Kalimeri, M., Koskela, E. V., Somerharju, P., Zhao, H., Vattulainen, I. and Lappalainen, P. (2017). Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc. Natl. Acad. Sci. USA 114, E8977-E8986. 10.1073/pnas.1705032114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, P. C., Pietromonaco, S. F., Reczek, D., Bretscher, A. and Elias, L. (1998). C-terminal threonine phosphorylation activates ERM proteins to link the cell's cortical lipid bilayer to the cytoskeleton. Biochem. Biophys. Res. Commun. 253, 561-565. 10.1006/bbrc.1998.9823 [DOI] [PubMed] [Google Scholar]
- Song, X., Wang, W., Wang, H., Yuan, X., Yang, F., Zhao, L., Mullen, M., Du, S., Zohbi, N., Muthusamy, S., et al. (2019). Acetylation of ezrin regulates membrane-cytoskeletal interaction underlying CCL18-elicited cell migration. J. Mol. Cell Biol., mjz099. 10.1093/jmcb/mjz099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanier, B. (2014). Transcriptional and functional regulation of the intestinal peptide transporter PEPT1. J. Physiol. 592, 871-879. 10.1113/jphysiol.2013.258889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck, O., Hughes, S. C., Noren, N. K., Kulikauskas, R. M. and Fehon, R. G. (2003). Moesin functions antagonistically to the Rho pathway to maintain epithelial integrity. Nature 421, 83-87. 10.1038/nature01295 [DOI] [PubMed] [Google Scholar]
- Ukken, F. P., Aprill, I., JayaNandanan, N. and Leptin, M. (2014). Slik and the receptor tyrosine kinase breathless mediate localized activation of moesin in terminal tracheal cells. PLoS ONE 9, e103323. 10.1371/journal.pone.0103323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Fürden, D., Johnson, K., Segbert, C. and Bossinger, O. (2004). The C. elegans ezrin-radixin-moesin protein ERM-1 is necessary for apical junction remodelling and tubulogenesis in the intestine. Dev. Biol. 272, 262-276. 10.1016/j.ydbio.2004.05.012 [DOI] [PubMed] [Google Scholar]
- Viswanatha, R., Ohouo, P. Y., Smolka, M. B. and Bretscher, A. (2012). Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J. Cell Biol. 199, 969-984. 10.1083/jcb.201207047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha, R., Wayt, J., Ohouo, P. Y., Smolka, M. B. and Bretscher, A. (2013). Interactome analysis reveals ezrin can adopt multiple conformational states. J. Biol. Chem. 288, 35437-35451. 10.1074/jbc.M113.505669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers, S., Portegijs, V., Kerver, J., Lemmens, B. B. L. G., Tijsterman, M., van den Heuvel, S. and Boxem, M. (2013). CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195, 1187-1191. 10.1534/genetics.113.156299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers, S., Muñoz, J., Berends, C., Ramalho, J. J., Goerdayal, S. S., Low, T. Y., Zoumaro-Djayoon, A. D., Hoffmann, M., Koorman, T., Tas, R. P., et al. (2016). A tissue-specific protein purification approach in Caenorhabditis elegans identifies novel interaction partners of DLG-1/Discs large. BMC Biol. 14, 66. 10.1186/s12915-016-0286-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, J. D. (2015). Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199, 363-377. 10.1534/genetics.114.172361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz, T., Wellbourne-Wood, J. and Kerkhoff, E. (2014). Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 24, 407-415. 10.1016/j.tcb.2014.02.004 [DOI] [PubMed] [Google Scholar]
- Winter, J. F., Höpfner, S., Korn, K., Farnung, B. O., Bradshaw, C. R., Marsico, G., Volkmer, M., Habermann, B. and Zerial, M. (2012). Caenorhabditis elegans screen reveals role of PAR-5 in RAB-11-recycling endosome positioning and apicobasal cell polarity. Nat. Cell Biol. 14, 666-676. 10.1038/ncb2508 [DOI] [PubMed] [Google Scholar]
- Yonemura, S., Matsui, T., Tsukita, S. and Tsukita, S. (2002). Rho-dependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. J. Cell Sci. 115, 2569. [DOI] [PubMed] [Google Scholar]
- Zhu, L., Zhou, R., Mettler, S., Wu, T., Abbas, A., Delaney, J. and Forte, J. G. (2007). High turnover of ezrin T567 phosphorylation: conformation, activity, and cellular function. Am. J. Physiol. Cell Physiol. 293, C874-C884. 10.1152/ajpcell.00111.2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Fig. 1. ERM-1 activity and efficient membrane targeting require the PIP2-binding domain. (A) Conservation and domain organization of ERM-1. AB is the actin-binding domain. (B) Differential interference contrast microscopy images of an N2 animal and erm-1 mutants. Arrowheads point to the excretory canal and arrows to the intestinal lumen. (C) Intestinal lumen discontinuities (arrowheads) and widened intestinal lumen in erm-1[4KN]::GFP second generation homozygous 2.5-fold embryos and L1 larvae. ERM-1 localization in the schematic diagram is in green. Dashed line indicates the boundary between the pharynx and intestine. (D,E) Apical levels of GFP-tagged ERM-1 variants in the intestine of L1 larvae with indicated genotypes. Images were acquired and displayed with the same settings, except +/+ in E. In this and all other figures, imaging was done with spinning disk confocal microscopy unless otherwise indicated. PM, plasma membrane. Data shown are means±s.d. of six measurements per animal for the apical membrane, normalized to the mean intensity of GFP in erm-1::GFP controls (n=10, 7, 6 and 8 in D; and n=24, 40 and 28 in E). Error bars are mean±s.d. Tests of significance: Dunnett's T3 multiple comparisons test. ns, not significant. ***P≤0.001, ****P≤0.0001. Unless indicated otherwise, statistical comparisons are with erm-1::GFP.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/ae42/10755404/22a004a4f02c/develop-147-188011-g1.jpg)
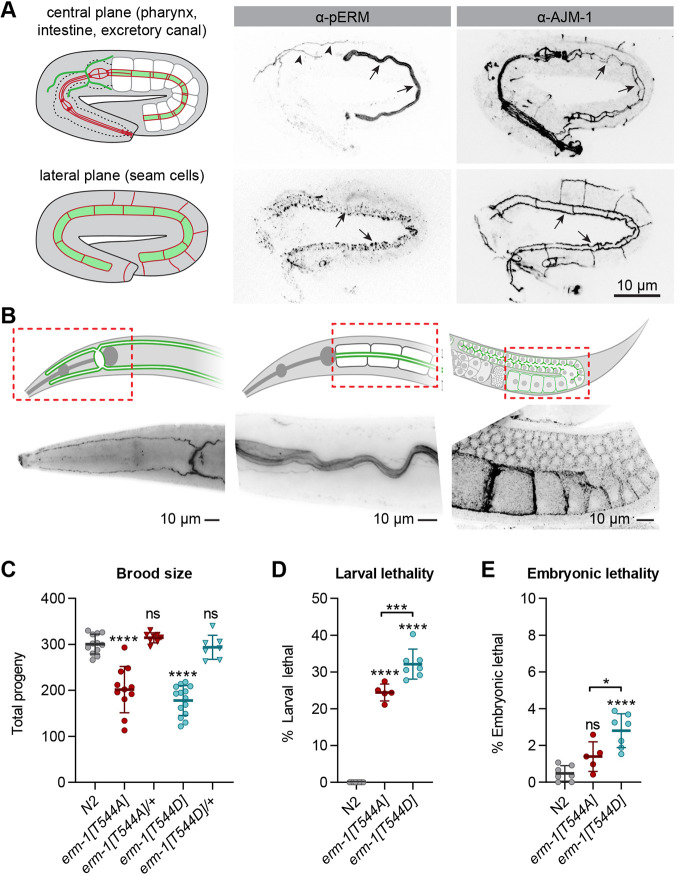
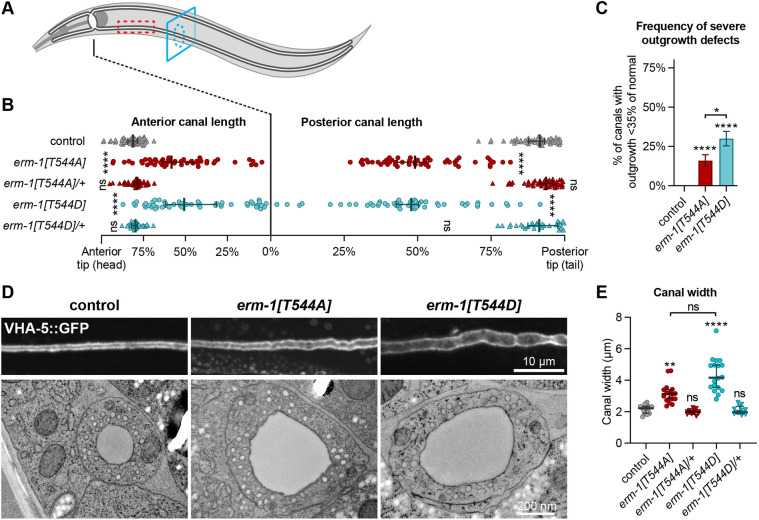
![Fig. 4. Positioning of cell junctions in intestinal cells requires dynamic regulation of ERM-1 T544 phosphorylation. (A) Junction organization visualized by AJM-1 staining of fixed embryos at different stages. Insets to the right and bottom are enlarged views of the regions indicated by dashed rectangles. Red brackets indicate junction separation in bean stage. Blue arrows point to expansion or ectopic accumulation of junction material, and red arrows point to partial and full constrictions. Images are maximum intensity projections. In all panels, developmental times are the approximate time after fertilization at 20°C. (B) Junction organization in live L1 larvae expressing DLG-1::mCherry and HMR-1::GFP. Blue and red arrows are as in A. (C) Transmission electron microscopy images of intestinal microvilli in L4 animals. (D) Quantification of the length of microvilli (total microvilli quantified: wild type, 88 from five animals; erm-1[T544A], 168 from four animals; erm-1[T544D], 135 from five animals). Error bars are the median±95% CI. Dunn's multiple comparisons test of significance, ****P≤0.0001. Unless indicated otherwise, statistical comparisons are with N2.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/ae42/10755404/4e4f346cc2af/develop-147-188011-g4.jpg)
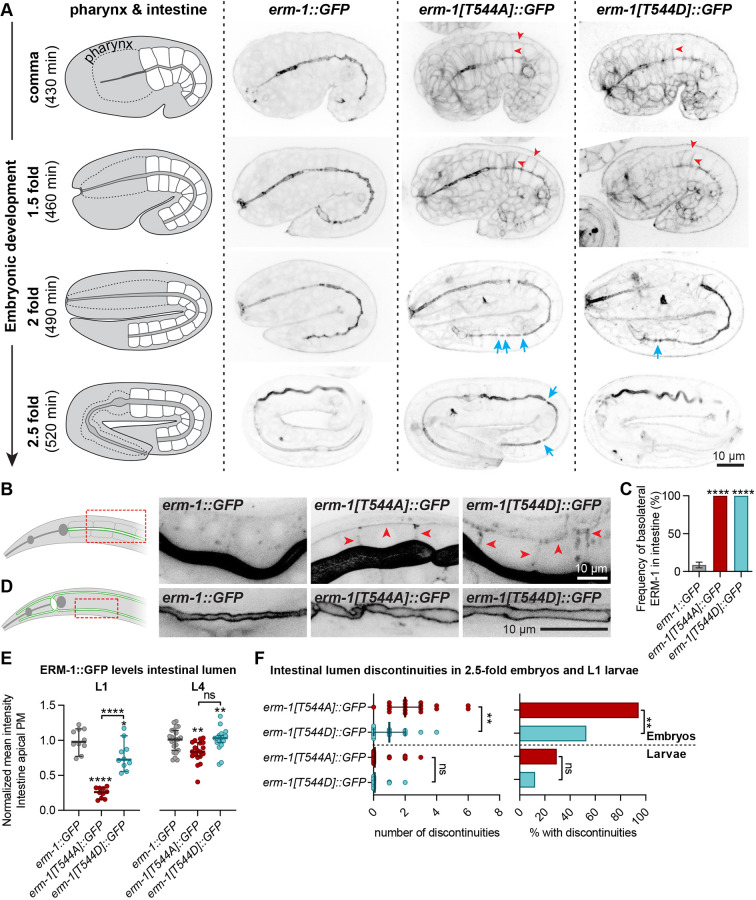
![Fig. 7. T544 phosphorylation cycling is required for stable ERM-1 localization in the intestine. Stills from time-lapse movies (A,C,E) and FRAP curves (B,D,F) of GFP-tagged ERM-1 variants at the apical membrane of the intestine of L1 larvae (n=7 for ERM-1::GFP, n=8 for ERM-1[T544A] and n=7 for ERM-1[T544D]), the apical membrane of the excretory canal of L4 larvae (n=5 for ERM-1::GFP, n=4 for ERM-1[T544A] and n=4 for ERM-1[T544D]), or the basolateral membrane of the germline of L4 larvae (n=10 for ERM-1::GFP, n=7 for ERM-1[T544A] and n=7 for ERM-1[T544D]). Thin lines and shading represent the mean±s.d., and thick lines represent curve fitting of averaged FRAP data with a double exponential equation.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/ae42/10755404/46fe9bd88da1/develop-147-188011-g7.jpg)
![Fig. 8. Phosphorylation of T544 controls apical actin recruitment and dynamics. (A) Lumen morphology and distribution of mCherry::ACT-5 in the excretory canal of L4 larvae. Canal widths vary (see Fig. 3E), and examples of severely widened and less severely widened canals are shown. Graph shows apical-to-cytoplasmic ratio of mean intensity of mCherry::ACT-5. Each data point represents the average of three measurements in a single animal (n=15, 9 and 14). Error bars are the mean±s.d. (B,C) Intestinal distribution of YFP::ACT-5. Arrows indicate discontinuities. Arrowheads in comma stage indicate basolateral YFP::ACT-5, and arrowheads in 2.5-fold stage indicate ectopic expansions of the cortical actin network. (D) Quantification of YFP::ACT-5 levels at the intestinal apical membrane. Each symbol represents a single animal (n=23, 26 and 19 for L1 and n=24, 21 and 22 for L4). Error bars are the median±95% CI. Tests of significance: Dunnett's T3 multiple comparisons test for A; Dunn's multiple comparisons test for D. ns, not significant. **P≤0.01, ***P≤0.001, ****P≤0.0001. Statistical comparisons are with control unless indicated otherwise. (E) FRAP curves of apical mCherry::ACT-5 in the excretory canal of L4 larvae (n=14 for control, n=6 for erm-1[T544A] and n=7 for erm-1[T544D]) and apical YFP::ACT-5 in the intestine of L1 larvae (n=8 for control, n=13 for erm-1[T544A] and n=8 for erm-1[T544D]). Thin lines and shading represent the mean±s.d., and thick lines were obtained by curve fitting averaged FRAP data with a double exponential equation.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/ae42/10755404/8aa9e5061a9a/develop-147-188011-g8.jpg)