Abstract
Endochondral ossification (ECO) is a form of bone formation whereby the newly deposited bone replaces the cartilage template. A decellularized artificial cartilage graft (dLhCG), which is composed of hyaline cartilage matrixes, has been developed in our previous study. Herein, the osteogenesis of bone marrow-derived MSCs in the dLhCG through chondrogenic differentiation, chondrocyte hypertrophy, and subsequent transdifferentiation induction has been investigated by simulating the physiological processes of ECO for repairing critical-sized bone defects. The MSCs were recellularized into dLhCGs and subsequently allowed to undergo a 14-day proliferation period (mrLhCG). Following this, the mrLhCG constructs were subjected to two distinct differentiation induction protocols to achieve osteogenic differentiation: chondrogenic medium followed by chondrocytes culture medium with a high concentration of fetal bovine serum (CGCC group) and canonical osteogenesis inducing medium (OI group). The formation of a newly developed artificial bone graft, ossified dLhCG (OsLhCG), as well as its capability of aiding bone defect reconstruction were characterized by in vitro and in vivo trials, such as mRNA sequencing, quantitative real-time PCR (qPCR), immunohistochemistry, the greater omentum implantation in nude mice, and repair for the critical-sized femoral defects in rats. The results reveal that the differentiation induction of MSCs in the CGCC group can realize in vitro ECO through chondrogenic differentiation, hypertrophy, and transdifferentiation, while the MSCs in the OI group, as expected, realize ossification through direct osteogenic differentiation. The angiogenesis and osteogenesis of OsLhCG were proved by being implanted into the greater omentum of nude mice. Besides, the OsLhCG exhibits the capability to achieve the repair of critical-size femoral defects.
Keywords: Tissue engineering, Regeneration medicine, Endochondral ossification, Hypertrophic chondrocytes, Transdifferentiation, Biomaterials, Scaffold
Graphical abstract
Endochondral ossification (ECO) is a form of bone formation whereby the newly deposited bone replaces the cartilage template. A decellularized artificial cartilage graft (dLhCG), which is composed of hyaline cartilage matrixes, has been developed in our previous study. Herein, the osteogenesis of MSCs in the dLhCG through chondrogenic differentiation and subsequent transdifferentiation induction has been investigated by simulating the physiological processes of ECO for repairing critical-sized bone defects. The MSCs were recellularized into dLhCGs (mrLhCGs) and then subjected to two distinct differentiation induction protocols to achieve osteogenic differentiation: chondrogenic medium followed by chondrocytes culture medium with a high concentration of fetal bovine serum (CGCC group) and canonical osteogenesis inducing medium (OI group). The formation of a newly developed artificial bone graft, ossified dLhCG (OsLhCG), as well as its capability of aiding bone defect reconstruction were characterized by in vitro and in vivo trials, such as mRNA sequencing, quantitative real-time PCR (qPCR), immunohistochemistry, the greater omentum implantation in nude mice, and repair for the critical-sized femoral defects in rats. The results reveal that the differentiation induction of MSCs in the CGCC group can realize in vitro ECO through chondrogenic differentiation, hypertrophy, and transdifferentiation, while the MSCs in the OI group, as expected, realize ossification through direct osteogenic differentiation. The angiogenesis and osteogenesis of OsLhCG were proved by being implanted into the greater omentum of nude mice. Besides, the OsLhCG exhibits the capability to achieve the repair of critical-size femoral defects.
1. Introduction
Bone defects occur occasionally and usually have a higher incidence in the older population. Apart from old-age factors, various inducements, such as trauma, tumors, and infection, can also cause bone defects. Autologous bone grafts (autografts) are regarded as the gold standard for treating bone defects in clinical practice [1]. Nevertheless, the shortage of autografts remains unneglectable. As the autograft is obtained from the patient, a second surgery must be conducted, and postoperative complications may occur at the donor site [2,3]. Autografts are not applicable when there is a significant bone defect, especially for children and the elderly. Therefore, engineering live bone grafts by using the patient's own cells via a tissue engineering approach should be an alternative method to establish biomimetic autografts for the treatment of bone defects.
In nature, bone formation typically follows two physiological processes, namely endochondral ossification (ECO) and intramembranous ossification (IMO). IMO often occurs during the formation of parietal, frontal, and clavicle bones. The connective tissue membrane, periosteum, which is derived from mesenchyme, first creates a region in which the bone formation process happens. The long bone originates from mesoderm-derived mesenchymal tissues, transiting a series of primary stages of chondrogenesis, chondrocyte hypertrophy, and osteogenesis, ultimately developing into a delicate and complex assembly of highly mineralized and vascularized solid connective tissues that construct the axial skeleton of the human body. This developmental process is also known as endochondral ossification [4,5]. In the middle of this process, a subpopulation of chondrocytes stops proliferation and undergoes hypertrophy; the hypertrophic chondrocytes (HCCs) play a central role in signaling for initiation of osteogenesis and induction of vascularization in the bulk of the preestablished cartilage template. Recent studies have discovered that at least a certain subpopulation of HCCs commits transdifferentiation instead of apoptosis. Various in vivo tracking experiments performed in genetically recombinant animal models through embryonic, fetal, and postnatal periods of development have confirmed that HCCs, or a certain subpopulation of HCCs, can directly transdifferentiate into osteoblasts for osteogenesis [[6], [7], [8]].
In our previous work, we developed a hyaline cartilage graft named living hyaline-like cartilage graft (LhCG) through a tissue engineering approach. LhCG is a pure cartilaginous tissue comprising only living chondrocytes and their endogenous extracellular matrix (ECM), which provides an ideal template for studying the mechanisms involved in ECO [9,10]. The decellularized LhCG (dLhCG) can be obtained by decellularizing LhCG with deoxyribonuclease and ribonuclease. As a pure hyaline cartilaginous ECM, dLhCG inherits all cartilage-specific features from LhCG; therefore, it can act as a cartilaginous template (scaffold) for investigating ECO, and the engineered dLhCG can act as a bone graft for repair of bone defects.
In this study, the ECO of human bone marrow-derived mesenchymal stem cells (MSCs) in the dLhCG was studied for preparing engineered bone grafts. We hypothesize that the MSCs in the dLhCG will suffer chondrogenic differentiation, hypertrophy, and transdifferentiation to osteoblasts, as well as ossification. To test this hypothesis, the MSCs-laden dLhCG (mrLhCG) constructs were established by seeding MSCs into dLhCG, which were cultured in the chondrogenic medium containing TGF-β3 and then in the chondrocytes culture medium with a high concentration of fetal bovine serum (CGCC group). As the control group, the osteogenic differentiation of the mrLhCG constructs was performed in the osteogenesis-inducing medium containing dexamethasone (OI group). To evaluate the differentiation processes, the expressions of marker genes at mRNA and protein levels were characterized by qPCR, mRNA-seq, and immunohistochemical staining. Ossification and vascularization of the ossified mrLhCG constructs (OsLhCGs) were assessed by the in vitro greater omentum implantation trials. Besides, the OsLhCG constructs undergoing decellularization treatments were explored to reconstruct critical-sized bone defects in rats.
2. Methods
2.1. Materials and reagents
Unless otherwise mentioned, all chemical reagents were purchased from Sigma-Aldrich through a local distributor. The osteopontin ELISA kit was purchased from Thermo Fisher Scientific. Mesenchymal stem cells were purchased from ATCC through a local distributor. All cell culture medium and related supplements were purchased from Thermo Fisher Scientific except ITS+, which was purchased from Corning. Antibodies used for immunohistochemical staining were purchased from Abcam unless otherwise mentioned. All reagents were used as received. The decellularized living hyaline cartilage grafts (dLhCGs) were prepared by following the protocol reported in our previous study [11].
All nude mice were provided by City University of Hong Kong. All rats were provided by the Chinese University of Hong Kong. The experiments were approved and conducted under the regulations and guidelines of the Animal Research Ethics Sub-Committee of City University of Hong Kong for laboratory animals. (AEC, # AN-STA-00000070, A0697 and A0649)
2.2. Recellularization of dLhCG
Human mesenchymal stem cells (hMSCs) were subcultured to passage 8 with MSC expansion medium (Dulbecco's Modified Eagle's Medium F-12 (DMEM/F-12), 10 % fetal bovine serum (FBS), 1 % Penicillin-Streptomycin, 10 ng/mL bFGF, and 20 μg/mL ascorbic acid) in a CO2 incubator (37 °C, 5 % CO2). When the confluence reached 70 %, the P8 MSCs were dissociated with 0.05 % trypsin-EDTA and transferred to a centrifuge tube. Cell numbers were determined with trypan blue and a cell counter (Thermo Fisher Scientific). The cell suspension was centrifuged at a speed of 1200 rpm for 5 min. After centrifugation, the MSC pellet was resuspended in the MSC expansion medium and recullularized to dLhCG at a cell density of 25k per construct. Briefly, dLhCGs were placed on tissue paper to blot out the moisture and sterilized with UV light for 2 h in a biosafety cabinet (BSC) before transferring to a 24-well cell culture plate. Fifty microliters of MSC expansion medium containing 25k MSCs were gently seeded onto dLhCG and then transferred the plate to the incubator, allowing cells to attach to the surface of dLhCG. After 4 h of incubation, 1.5 mL MSC expansion medium was added to each well. The dLhCG recellularized with MSCs (mrLhCG) was immersed in an MSC expansion medium for 14 days at 37 °C in a 5 % CO2 incubator (Fig. 1). The cell culture medium was changed every three days.
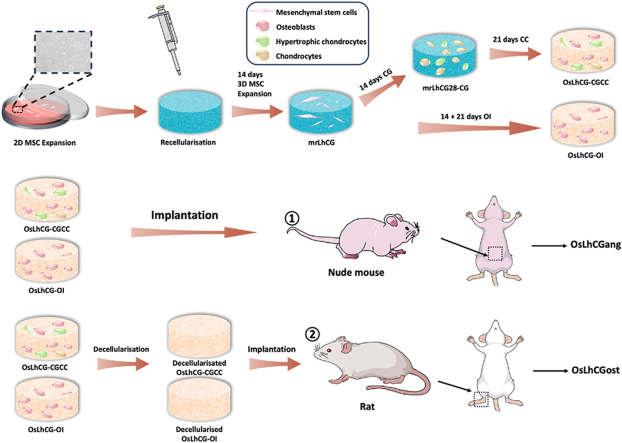
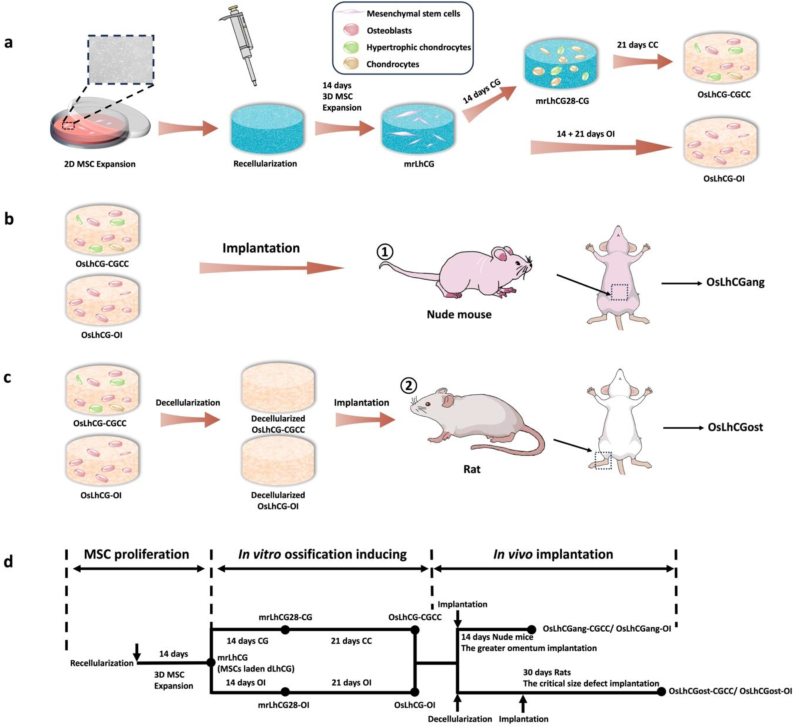
Fig. 1.
Schematic illustrations. a) In vitro experiment of OsLhCG preparation, including MSC expansion, recellularization, and differentiation induction. b) The greater omentum implantation. c) The critical size defect implantation. d) Flow chart of in vitro and in vivo experiments.
2.3. 3D cell culture of mrLhCG
As shown in Fig. 1, mrLhCG constructs were randomly separated into two groups. In the CGCC group, the mrLhCG constructs were cultured in the chondrogenic medium (CG medium) for 14 days (mrLhCG28-CG) and then in the chondrocytes culture medium (CC medium) for 21 days (OsLhCG-CGCC); in the OI group, the mrLhCG constructs are cultured in the osteogenesis inducing medium (OI medium) for 14 days (mrLhCG28-OI) and further 21 days (OsLhCG-OI). Briefly, the MSC expansion medium (as mentioned above) of each well was aspirated out, and then 1.5 mL of the corresponding medium was added. The cell culture plate was transferred back to the incubator. The cell culture medium was changed every three days.
The CG medium is the high glucose DMEM supplemented with 100 nM dexamethasone; 50 μg/mL ascorbic acid; 50 μg/mL proline; 100 μg/mL sodium pyruvate; 1 % ITS+; 1 % Penicillin-Streptomycin; 10 ng/mL BMP6; 10 ng/mL TGF-β3. The CC medium is the high glucose DMEM supplemented with 20 % (v/v) FBS; 0.1 mM nonessential amino acids (NEAA); 0.01 M 4-(2-hydroxyethyl)-piperazine-1-ethanesulfonic acid (HEPES); 0.05 mg/ml ascorbic acid; 0.4 mM proline; 1 % Penicillin-Streptomycin. The OI medium is the high glucose DMEM supplemented with 20 % (v/v) FBS; 0.1 mM NEAA; 0.01 M HEPES; 0.05 mg/ml ascorbic acid; 0.4 mM proline; 1 % Penicillin-Streptomycin; 100 nM dexamethasone; 50 μM ascorbic acid 2-phosphate; 10 mM β-glycerophosphate.
2.4. Animal study
2.4.1. The great omentum implantation
OsLhCG-CGCC and OsLhCG-OI were implanted into the great omentum of 4- to 6-week-old Balb/c nude mice. Briefly, nude mice were anesthetized with isoflurane. After sterilization with povidone-iodine, a 1.5 cm longitudinal incision was created on the abdomen to expose the great omentum. Then, one sample was rolled into the great omentum and placed back into the abdominal cavity. Nude mice were euthanized after 14 days of implantation, and the samples (named OsLhCGang-CGCC and OsLhCGang-OI for the OsLhCG-CGCC and OsLhCG-OI, respectively) were retrieved for histological evaluation.
2.4.2. Critical-size defect implantation
To evaluate the capability of OsLhCG-CGCC and OsLhCG-OI in reconstructing bone defects, they underwent decellularization before implantation to reduce their immunogenicity. In short, the OsLhCG samples, including OsLhCG-CGCC and OsLhCG-OI, were washed twice with PBS, and decellularization was initiated by cycling the freezing and thawing process for three times (−80 °C for 3 h followed by room temperature for 4 h). Samples were soaked in Tris hypertonic solution for 24 h after the freeze-thaw cycles, and then Tris hypertonic containing 1 % Triton X-100 was added to each sample for 48 h [[11], [12], [13], [14]]. Subsequently, the samples were soaked in 0.5 mg/mL DNase and 50 μg/mL RNase solution for 3 h to remove DNA and RNA in a 37 °C, 150 rpm shaking incubator, followed by washing with PBS for 24 h and DI water for 6 h. The decellularized OsLhCG was sterilized under UV light for 2 h in BSC. Seven-to nine-week-old rats were used to establish bone defect models. Briefly, rats were anesthetized with isoflurane, followed by sterilization with povidone-iodine and hair removal. A 2 cm longitudinal incision was created on each leg of the rat to expose the condyle of the femur. A cylindrical bone defect measuring 5 mm in diameter and 3 mm in depth was surgically generated with a drill on each femur, and the decellularized OsLhCG was implanted in the defect of the left femur, leaving the right femur untreated as a control group. Rats were euthanized after 30 days of feeding, and samples (i.e., OsLhCGost-CGCC and OsLhCGost-OI) were retrieved for micro-CT scanning and histological evaluation.
2.5. Analytical method
2.5.1. Quantitative polymerase chain reaction (qPCR)
Samples (n = 3) were collected at each time point and washed with PBS before RNA extraction. RNA samples were extracted using an RNA extraction kit (Norgen Biotek Thorold, Canada) under the extraction protocol. RNA samples were converted to cDNA by reverse transcription. qPCR was performed by a StepOnePlus Real-Time PCR System (Applied Biosystems). Gene expression was determined with the comparative 2- Δ Δ CT. GAPDH was used as a housekeeping gene to normalize the value. All primers used in qPCR were engineered and prepared by BGI Group (China). The primer sequences are presented in Table 1.
Table 1.
Primer sequences.
| Primer name | Base-pair order |
|---|---|
| β-actin-F | CTTCGCGGGCGACGAT |
| β-actin-R | CCACATAGGAATCCTTCTGACC |
| SPP1–F | AAACGCCGACCAAGGAAAAC |
| SPP1-R | GCCACAGCATCTGGGTATTTG |
| SPARC-F | TGAGAATGAGAAGCGCCTGG |
| SPARC-R | TGGGAGAGGTACCCGTCAAT |
| ALP-F | GGGAACGAGGTCACCTCCAT |
| ALP-R | TGGTCACAATGCCCACAGAT |
| Col1A2-F | ACAAGGCATTCGTGGCGATA |
| Col1A2-R | ACCATGGTGACCAGCGATAC |
| CBFA1-F | TCTCCAGGAGGACAGCAAGA |
| CBFA1-R | CTGCTTGCAGCCTTAAATGACT |
| BGLAP-F | CACCGAGACACCATGAGAGC |
| BGLAP-R | CTGCTTGGACACAAAGGCTGC |
| Col10A1-F | CATAAAAGGCCCACTACCCAAC |
| Col10A1-R | ACCTTGCTCTCCTCTTACTGC |
| Col2A1-F | AGACTTGCGTCTACCCCAATC |
| Col2A1-R | GCAGGCGTAGGAAGGTCATC |
| SOX9-F | GGCAAGCTCTGGAGACTTCTG |
| SOX9-R | CCCGTTCTTCACCGACTTCC |
| MMP13-F | CCAGACTTCACGATGGCATTG |
| MMP13-R | GGCATCTCCTCCATAATTTGGC |
| IBSP-F | GAACCTCGTGGGGACAATTAC |
| IBSP-R | CATCATAGCCATCGTAGCCTTG |
| GAPDH-F | GTCTCCTCTGACTTCAACAGCG |
| GAPDH-R | ACCACCCTGTTGCTGTAGCCAA |
2.5.2. mRNA sequencing (mRNA-seq)
OsLhCG-CGCC and OsLhCG-OI were collected (n = 3) for mRNA-seq. Samples were homogenized manually with a tissue homogenizer before RNA extraction. The RNA extraction procedure followed the manufacturer's instructions for the RNA extraction kit (Norgen Biotek Thorold, Canada). The Agilent 2100 system was used to determine RNA concentration and RIN/RQN. Six samples with high RNA integrity number values (RIN>7) were used to prepare the library [15]. The sequencing process was performed through the Illumina platform by BGI Health (HK) Company Limited.
2.5.3. Calcium quantification
Calcium quantification was conducted with a calcium colorimetric assay kit. Calcium ions formed a chromogenic complex with o-cresol phthalein, which can be measured at 575 nm using a microplate reader. The collected mrLhCG and OsLhCG samples were washed twice with DI water before freeze-drying and weighting. Subsequently, the freeze-dried samples were immersed in the hydrochloric acid (3 M) solution overnight. Then, after centrifugation, the supernatant was transferred to a 96-well plate for calcium quantification based on the kit's protocol.
2.5.4. DNA assay
The collected samples were washed with DI water, freeze-dried overnight, and weighed, respectively. Papain solution was added to each sample-containing tube and left in a 60 °C water bath overnight. Salmon sperm DNA (1 mg/mL) was prepared and serially diluted to 500 μg/mL, 250 μg/mL, 125 μg/mL, 62.5 μg/mL, 31.25 μg/mL, 15.625 μg/mL and 7.8125 μg/mL. Hoechst 33258 (1 μg/mL) was added to each papain-digested sample or salmon sperm DNA solution, mixed thoroughly with a pipette, and allowed to equilibrate for 5 min. For each sample, 150 μL of the solution was transferred to a 96-well plate and measured with a microplate reader (excitation: 350 nm; emission: 450 nm). The cell number equals the total DNA weight (pg) divided by 7.7 pg/cell.
2.5.5. Glycosaminoglycan (GAG) assay
The dimethylmethylene blue (DMMB, 0.016 mg/mL) solution was freshly prepared. The standard solutions were prepared by dissolving chondroitin sulfate in DI water by performing serial dilution. 40 μL of DMMB solution was added into 1 mL of sample solution, and 150 μL of the resultant solution was transferred to a 96-well plate. The absorbance at 525 nm was measured with a microplate reader to calculate glycosaminoglycan content.
2.5.6. Enzyme-linked immunosorbent assay (ELISA)
To quantify the concentration of osteopontin (OPN), the cell culture medium was removed and refilled with 1.5 mL fresh medium 1 day before quantification. After 24 h of cell culture, the medium was collected and analyzed by following the protocol of the ELISA kit.
2.5.7. MSC viability testing
The cell viability of MSCs within mrLhCG constructs was assessed through live/dead staining and CCK-8 assay at different time points. Briefly, after recellularizing the MSCs onto dLhCG, the constructs were placed in an incubator and cultured in the MSC expansion medium for three-dimensional cultivation. The day of dLhCG recellularization was considered Day 0, and subsequently, samples were retrieved on Day 2, Day 6, Day 8, Day 10, and Day 14. For live/dead staining, the samples were thoroughly rinsed with PBS after retrieval and stained using Calcein AM and Propidium Iodide (PI) according to the protocol of the Live/Dead staining kit. After staining, confocal microscopy was utilized to capture images, enabling the visualization of the distribution of live and dead cells within the construct. For the CCK-8 assay, three parallel samples were retrieved at corresponding time points, rinsed with PBS, and then, following the protocol of the CCK-8 assay, 10 % CCK-8 solution was added to the fresh culture medium and incubated for an additional 4 h in the incubator. After 4 h, absorbance readings at 450 nm were measured using a microplate reader, and a comparative analysis was performed at different time points to obtain a comprehensive value considering the combined effect of cell viability and cell number. To eliminate systematic errors, CCK-8 cell viability testing of the recellularized constructs was performed normalization at each time point using a certain number of MSC cells from the same passage. Following the measurement of the absorbance values, normalization of the measured values was achieved by dividing the absorbance obtained from the construct measurements by the absorbance of the cells. After the completion of the experiment, a graph depicting the relative cell viability was generated.
2.5.8. Surface morphology characterization
The mrLhCG28-CG, mrLhCG28-OI, OsLhCG-CGCC, and OsLhCG-OI samples were fixed in at 4 °C for 12 h after thorough rinsing. Subsequently, dehydration was carried out using a graded ethanol series, followed by complete drying using a critical point dryer (CPD) for further processing. The dried samples were then subjected to surface sputter coating and affixed to the sample stage using conductive adhesive tape for surface morphological characterization using a scanning electron microscope (SEM) (FEI Quanta 250 e-SEM). The SEM electron beam accelerating voltage utilized was 15 kV.
2.5.9. Histology
The collected samples were fixed with 4 % (w/v) paraformaldehyde for 24 h, embedded in paraffin, and sectioned with a fully motorized rotary microtome to a thickness of 7 μm. Slides were used for immunohistochemical staining (OCN, OPN, SPARC, COL1, COL2, VEGFA, CD31, HIF1-α, VWF), Hematoxylin-eosin (HE) staining, Von Kossa staining, and Alizarin red S staining. The staining procedure followed the protocol of the staining kit. For HE staining, slides underwent deparaffinization, and the nuclei were stained with hematoxylin solution for 3 min, followed by differentiation with differentiation solution for 1 min. The samples were restained with Eosin Y Aqueous Solution for 1 min. The sections were dehydrated in alcohol and cleared with xylene before mounting. For von Kossa staining, slides were deparaffinized and hydrated before incubation under UV for 60 min. The slides were incubated in sodium thiosulfate solution (5 %) for 3 min and rinsed in tap water. A nuclear fast red solution was used for staining the nucleus for 5 min before dehydrating and mounting. For Alizarin Red staining, the rehydrated samples were stained with Alizarin Red staining solution for 5 min and then dehydrated and mounted. For immunohistochemical staining, the pepsin was added to each slide and incubated in a 37 °C oven for 25 min to retrieve the antigen. Then, 3 % H2O2 and 10 % goat serum were successively added to the samples and incubated for 1.5 h and 40 min, respectively. After washing with PBS, the primary antibodies were diluted (ColI 1:200; ColII 1:200; OCN 1:250; OPN 1:200; SPARC 1:200; VEGF 1:500; CD31 1:50; HIF1-α 1:200; VWF 1:200) and added to each sample, and then incubated at 4 °C overnight. Subsequently, a secondary antibody was added to each slide and incubated for 30 min at room temperature, followed by adding a DAB working solution for coloration. Slides were then dehydrated and mounted for further observation. The percentage of positive IHC staining of the in vitro sample is obtained through Image J.
2.5.10. Micro-CT scanning
To evaluate the repair of bone defect, rat condyles of the femur were scanned by micro-CT after 48 h of fixation using a Scanco Xtreme II Micro CT Scanner. The scans were reconstructed in order to obtain 3D representations. Ten researchers unrelated to the project were asked to go through the 3D reconstructed photo of bone tissue to determine the extent of bone reparation. The researchers were asked to rate the degree of bone reparation on a scale of 0–10, with 0 indicating no bridging and 10 indicating complete bridging or full recovery. The average score was calculated to reflect the extent of reparation. Further analysis was conducted to obtain data that indicate bone repair efficacy, including bone volume/tissue volume ratio (BV/TV), bone surface/volume ratio (BS/BV), bone surface density (BS/TV), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), and trabecular number (Tb.N).
2.5.11. Statistical analysis
The analysis procedure was conducted by using GraphPad Prism 9 (California, USA). All data presented in this project were expressed as the mean and standard deviation. One-way analysis of variance (ANOVA) was used for analyzing single indicator data among three groups of results, while two-way ANOVA was available for the comparison among multiple data to determine the significance of differences. Significance was set at p < 0.05. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3. Results and discussion
3.1. In vitro bone formation within dLhCG through direct MSC osteoblast differentiation and HCC transdifferentiation
Naturally, the long bone is developed by undergoing a series of stages, including chondrogenesis, chondrocyte hypertrophy, and osteogenesis, followed by vascularization and, ultimately, the formation of connective tissue constructing the axial skeleton. The ossification process occurs within a cartilage template. As chondrocytes get hypertrophy, on the paracrine signals generated by HCCs, the initiation of osteogenesis is conducted via activation of the osteogenic transcription factor runt-related transcription Factor 2 (RUNX2), also known as core-binding factor subunit alpha-1 (CBFA1), in the surrounding uncommitted osteogenic progenitor cells (OPCs); at the same time, the chondrogenic transcription factor SOX9 is downregulated [16]. The induction of vascularization by HCCs is executed via the expression of angiogenic cytokines, such as vascular endothelial growth factor (VEGF), that mediate vascular intrusion through interactions with specific HCC receptors, such as neuropilins, and endothelial cells (EC) receptors, such as FLk-1 [17]. Following the invasion of ECs, infiltration of capillaries extends into the perichondrium and hypertrophic regions, from which abundant minerals and osteoclasts (OCs) are introduced [4]. Metallomatrix proteinases-13 (MMP13) are also expressed by HCCs to degrade the residues of the transient cartilage template, further facilitating osteogenic remodeling and vascularization [4,18]. During this osteogenic process, the general ECM composition is dramatically altered, which is marked by upregulation of type X collagen and downregulation of the cartilage-specific proteoglycans and collagen types, namely type II and type IX collagen. Meanwhile, more fibronectin is present to facilitate calcification. Eventually, osteogenesis is achieved by the completion of the differentiation of OPCs into osteoblasts, as marked by the expression of osteocalcin and type I collagen, while HCCs are believed to commit apoptosis [4,5]. However, recent studies suggested that at least a certain type of HCCs could have the capability of direct transdifferentiating into osteoblasts. For investigating the process of HCC transdifferentiation and ECO process in vitro, an artificial cartilage graft that only contains pure cartilaginous extracellular matrices should be constructed as a template. Herein, the dLhCG is designed and fabricated, which is composed of hyaline cartilage ECM and employed as a platform for the study of ECO.
3.1.1. Quantitative polymerase chain reaction (qPCR)
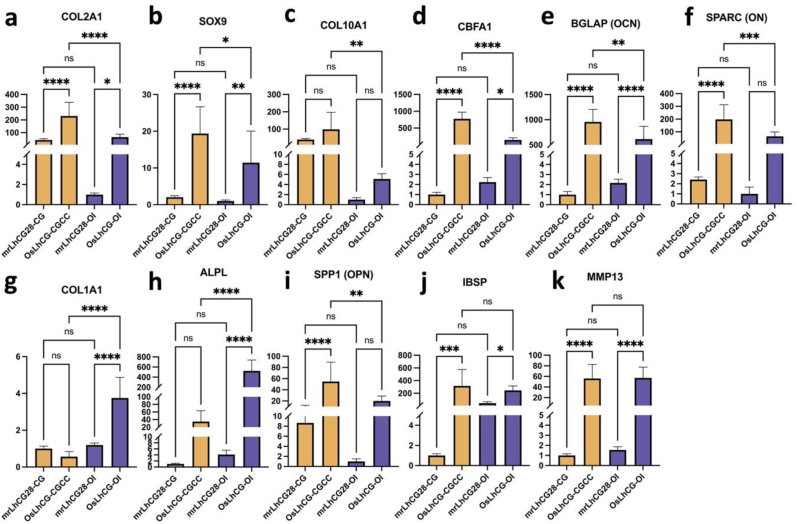
At the gene expression level, the expressions of the genes related to osteogenesis (IBSP, MMP13, BGLAP, SPP1, COL1A1, ALPL and SPARC) (Fig. 2e–k), chondrogenesis (COL2A1, SOX9) (Fig. 2a and b) and chondrocyte hypertrophy (COL10A1, CBFA1) (Fig. 2c and d) were examined by qPCR. COL2A1 is responsible for the formation of type II collagen, which is the main component of cartilage. The significantly increased expression of COL2A1 from mrLhCG28-CG to OsLhCG-CGCC indicates the successful chondrogenic differentiation of MSCs in the CGCC group, while the OI group shows a similar tendency with relatively lower gene expression. Similarly, the expression of SOX9, which is regarded as a regulator of chondrogenic differentiation of MSCs, is also found to be upregulated in both the CGCC group and the OI group [19]. SOX9 is also expressed in early-stage HCCs. Thus, the coexistence of SOX9 and other osteogenic markers indicates the occurrence of osteogenesis through transdifferentiation from HCCs to osteoblasts [20]. However, the actual gene expression level of chondrogenic genes (COL2A1 and SOX9) in the CGCC group is higher than the OI group, indicating that chondrogenesis is a crucial intermediate step during ossification in the CGCC group, while the OI group realize ossification mostly through direct MSCs osteogenic differentiation. Type X collagen is a specific marker of HCCs, and type X collagen provides an environment for further mineralization and bone modeling during the ECO process [21]. The increased expression of COL10A1 demonstrated the formation of HCCs [7]. Fig. 2c shows the COL10A1 gene expression of the CGCC and the OI group, respectively. There is no significance between mrLhCG28-CG and OsLhCG-CGCC, as well as between mrLhCG28-OI and OsLhCG-OI. Similar to COL2A1 and SOX9, the gene expression level shows that the COL10A1 expression of OsLhCG-CGCC is significantly higher than OsLhCG-OI, suggesting that compared with the OI group, the CGCC group is more likely to undergo chondrocytes hypertrophy. A similar difference also happened with CBFA1. CBFA1 (RUNX2), which is encoded by the CBFA1 gene, is a key transcription factor associated with ossification that is essential for the transdifferentiation of HCCs to osteoblasts [22]. In addition to the induction of transdifferentiation, CBFA1 has the capability of inhibiting the apoptosis process of HCCs [23]. Fig. 2d shows the CBFA1 gene expression of the CGCC group and OI group. Although there is significant CBFA1 gene expression upregulation in both the CGCC group and the OI group, the actual gene expression of OsLhCG-OI is significantly lower than OsLhCG-CGCC. Apart from the hypertrophy-related gene, as the ultimate goal is to realize ossification, osteogenesis-related genes should be mentioned to demonstrate whether ossification happened throughout the in vitro culture process. By combining COL10A1 and CBFA1 gene expression with osteogenesis-related gene expression, we concluded that osteogenesis was successfully induced in the CGCC group, and HCCs were intermediate cells, suggesting that the process of the CGCC group was accomplished through ECO. For the OI group, as the expression of chondrogenesis and hypertrophy-related genes are not significant, while osteogenesis-related genes are still highly expressed, a conclusion could be drawn that the ossification process is mostly completed by the direct osteogenic differentiation of MSCs. IBSP and SPP1 are genes encoding bone sialoprotein (BSP) and osteopontin, respectively. Osteopontin is a protein secreted by osteoclasts and osteoblasts [24,25], mediating the formation of mineral crystals and regulating mineralization [26], while BSP is a primary component of the bone ECM, and it acts as a nucleus for the formation of apatite crystals [27,28]. The IBSP gene is normally expressed by osteoblasts, HCCs, and osteoclasts [29]. The expression of IBSP and SPP1 in the CGCC group showed an upward trend and significant differences, indicating the occurrence of the transdifferentiation process, while the expression of IBSP and SPP1 gene in the OI group indicates the happening of MSCs osteogenic differentiation. The SPARC gene is expressed by osteoblasts and HCCs, and its downstream product, osteonectin (ON), can increase the deposition of collagen and promote the maturity of bone tissue [30]. In addition, ON can bind with calcium during the ossification process to initiate mineralization. Osteocalcin (OCN), encoded by the BGLAP gene expressed by osteoblasts, can also bind with calcium as a component of bone tissue. The expression of BGLAP is always regarded as the criterion of the efficacy of bone formation. Compared with the two groups collected on day 28, the gene expression of BGLAP and SPARC of OsLhCG-CGCC and OsLhCG-OI were significantly upregulated. The expression of BGLAP is the strongest evidence of ossification, and OCN is highly correlated with bone mineral density elevation [31]. Matrix metallopeptidase 13 (MMP13) is encoded by the MMP 13 gene, which is highly expressed in terminally differentiated HCCs and osteoblasts [32]. The major function of MMP13 is to realize cleavage and resorption of type II collagen, which is the major component of cartilage [33]. In addition, MMP13 can also function in bone remodeling [34]. The gene expression and significant upregulation of MMP13 suggest the existence of HCCs and/or osteoblasts in both the CGCC and the OI groups, corroborating the happening of the ossification process. Type I collagen, which is encoded by the COL1A1 gene, is the major component of bone tissue. Alkaline phosphatase (ALP) is encoded by the ALPL gene, and it acts as a mediator for the combination with type I collagen and promotes the deposition of calcium phosphate as hydroxyapatite on type I collagen fibers [35]. Fig. 2g&h shows the gene expression of ALPL and COL1A1. For ALPL, comparing with mrLhCG28-CG and mrLhCG-OI, both the CGCC and the OI groups show significant upregulation. However, the COL1A1 gene expression of the CGCC group showed slight downregulation when the construct was further cultured up to day 49, while the OI group still showed upregulation, as predicted. As demonstrated by the expression and elevation of chondrogenesis-, hypertrophy- and osteogenesis-specific genes during the 49 days of cell culturing, samples in the CGCC group underwent chondrocyte proliferation, hypertrophy, and HCC transdifferentiation, while samples in the OI group tend to undergo direct osteoblast differentiation of MSC, proven by the expression of osteogenesis-specific genes and relatively lower expression of chondrogenesis-specific genes. Consequently, from the gene expression at the mRNA level, results of both the CGCC group and the OI group show that the dLhCG, as a pure type II collagen scaffold, has the potential to support the happening of ossification, and the result of CGCC group suggests that dLhCG can be utilized as a cartilage graft for the happening of in vitro ECO.
Fig. 2.
qPCR results of the CGCC group and the OI for genes that are involved in ECO. a) and b) are gene expressions of chondrogenesis-related genes. c) and d) are HCC gene expressions. e)-k) are gene expressions of osteogenesis-related genes.
3.1.2. mRNA sequencing (mRNA-seq)
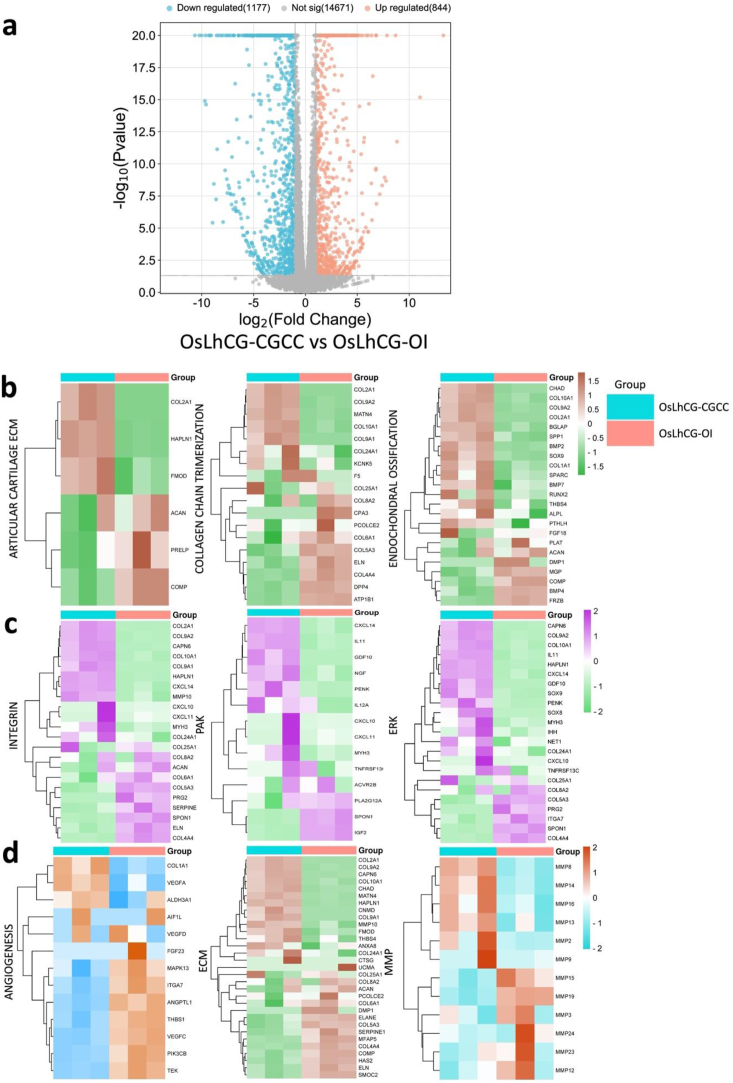
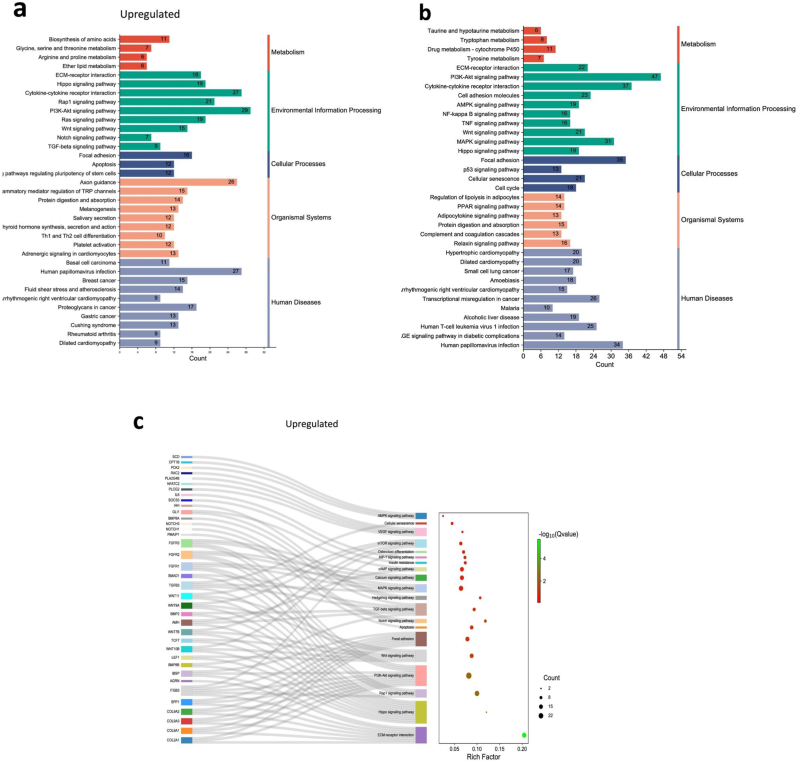
To investigate the overall gene expression of the CGCC group and the OI group in the ossification process, mRNA-seq was performed on OsLhCG-CGCC and OsLhCG-OI using three replicates per group. A total of 16692 genes were identified throughout the mRNA-seq. Among the 16692 genes, 2021 genes were differentially expressed (DEG) based on the selection criteria (p value < 0.05, |log2(fold change)| >1), in which the gene number of upregulation and downregulation are 844 and 1177, respectively (Fig. 3a). This result indicates that two kinds of treatment can induce MSC differentiation into osteoblasts through different pathways.
Fig. 3.
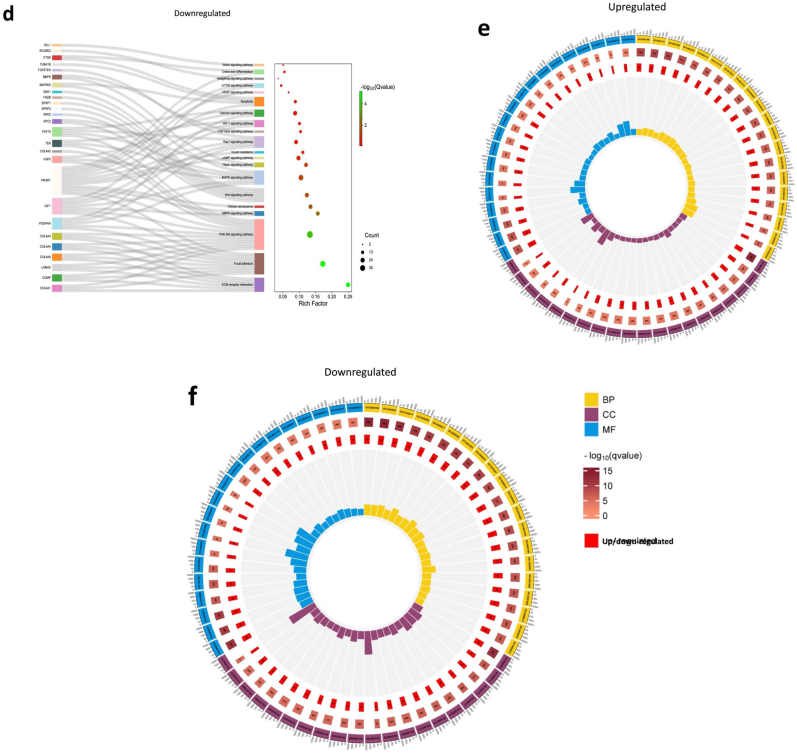
Statistics of the differentially expressed genes and heatmaps of the CGCC group and the OI group. Differentially expressed core genes related to each pathway, component, or biological process are listed. a) Volcano plot of differentially expressed genes. The threshold was defined as a P value < 0.05 and |log2FC|>1. The volcano plot shows that 844 genes were upregulated, 1177 genes of OsLhCG-CGCC were downregulated, and 14671 genes showed no significant difference. b) Heatmaps of genes that are involved in skeletal pathways, including ECO, collagen chain trimerization, and articular cartilage ECM. c) Heatmaps of genes that are involved in some general pathways, including integrin, PAK, and ERK. d) Heatmaps of genes that are involved in angiogenesis, ECM, and MMP.
Fig. 3 illustrates the heatmap of DEGs and categorizes them based on the pathways associated with their functional attributes. The classification was performed using the online tool GeneAnalytics. The expression heatmaps of the genes related to the skeletal pathway are presented in Fig. 3b. The results of gene expression of articular cartilage ECM (COL2A1, SOX9) and endochondral ossification (COL10A1, RUNX1) demonstrate that the gene of chondrogenesis and hypertrophy genes are more likely to be expressed in the CGCC group rather than the OI group. Fig. 3c shows differential gene expression heatmaps of several canonical pathways involved in the ossification process to obtain a straightforward up-/down-regulation of related core genes. Core genes that relate to the ossification process, such as COL2A1 and COL10A1, are involved in the integrin signaling pathway. Integrin can target BM-MSCs, promote differentiation, regulate the expression of downstream genes, and further help promote ossification [36]. ITGA1 is a gene for the encoding of integrin alpha-1, and it shows a downregulation tendency between the CGCC and the OI groups. Integrin expression can lead to ERK activation [37], which is essential during skeletal development and homeostasis [38]. Previous research suggested that the process of the ERK-MAPK pathway promoting bone formation, including osteoblast differentiation, is by activating and phosphorylating CBFA1 and realizing the function of FGF2 [39,40]. The expression heatmap of the genes associated with the pathways presented in Fig. 3c shows that genes related to chondrogenesis (COL2A1, SOX9) and hypertrophy (COL10A1) exhibit upregulation in the CGCC group compared with the OI group. The differentially expressed genes related to phospholipase, which are normally expressed in the late stage of ossification, show downregulation in the CGCC group when compared with the OI group.
A disintegrin and metalloproteinase (ADAM) are a family of metalloendopeptidases produced by both osteoclasts and osteoblasts, such as ADAM12, ADAM9, and ADAM10 [41]. Some ADAM genes, such as ADAM17, can affect osteoblast differentiation, bone formation, and ECM remodeling by influencing RUNX2 and ALPL gene expression [42,43]. Additionally, ADAM12 and ADAM15 were found to be expressed in cartilage since chondrocytes are important sources of metalloproteinase enzymes [44]. Apart from ADAM, matrix metalloproteinase (MMP) is another metalloproteinase that is critical for the degradation of the ECM. During ECO, as chondrocytes transition to HCCs, MMP2, MMP9, and MMP13 are synthesized to promote matrix remodeling and bone formation. Thus, the expression of ADAM and MMP genes is presented with heatmaps (Supplemental Fig. 1b, Fig. 3c). The expression of ADAM and MMP illustrates the existing process of ossification. For example, the CGCC group had a relatively high gene expression level of chondrocyte-related markers, such as ADAM12 and ADAM15, suggesting that the ossification process is achieved through ECO, while ADAM17, which affects osteoblast differentiation, expressed more in the OI group. This phenomenon can also be shown by the gene expression heatmap of ECM, including COL2A1 and COL10A1 (Fig. 3d). During human long bone development, the avascular cartilage is finally replaced with vascularized bone tissue through ECO [45]. The vascularization process is crucial for bone formation, which is accompanied by the production of vascular endothelial growth factor (VEGF). VEGF is a pro-angiogenic factor that can stimulate angiogenesis and osteogenesis [46]. Fig. 3d also shows the expression of the angiogenesis gene, and VEGFA, as a core gene expressed in both the CGCC and the OI, indicating the launch and activation of angiogenesis within the OsLhCG template. Nevertheless, interestingly, OsLhCG-CGCC showed a higher expression level than OsLhCG-OI.
Similar to the results of qPCR, both the two groups have successfully induced the ossification of MSCs in dLhCG while probably undergoing different pathways. The CGCC group realizes osteogenesis through a mechanism similar to ECO, while the OI group, as a control group, realizes osteogenesis by direct MSC osteoblast differentiation. To determine the possible signaling pathways involved in bone formation, KEGG pathway enrichment and GO enrichment analyses based on DEG sets were carried out. DEGs were separated into the “upregulation group” and “downregulation group.”
The top 35 KEGG pathways in which DEGs were selected are listed in Fig. 4a&4b. On the basis of previous research, some pathways related to ossification were presented with the Sankey-dot pathway enrichment plot, and core DEGs involved in the major pathway were listed (Fig. 4c and d). RAP1 has the capability of regulating integrin-mediated cell adhesion through the RAP1 signaling pathway [47] and regulating osteoblastic differentiation through the MAPK signaling pathway. Previous research has illustrated that expression of the RAP1A gene can significantly activate the ERK/p38 signaling pathway and regulate osteoblast differentiation [48,49]. In the CGCC group and the OI group, MAPK1 and RAP1A were highly expressed but showed no difference. Fig. 4a shows that the DEG involved in the RAP1 signaling pathway is mainly enriched in the upregulation group, indicating that the RAP1 signaling pathway was activated in the CGCC group. As shown in the Sankey diagram, FGFR1, FGFR2, and FGFR3 had significantly higher gene expression levels in OsLhCG-CGCC. FGFR1 gene could promote the differentiation of MSCs towards preosteoblasts but inhibit further maturation of the mineralization [50], while FGFR2 can promote the osteogenic differentiation of MSCs, osteoblast maturation, and mineralization [51]. FGFR3, expressed in MSCs and osteoblasts, can support osteogenesis [52]. These genes were also enriched in the MAPK signaling pathway and showed an upregulation tendency in the OsLhCG-CGCC. The platelet-derived growth factor receptor (PDGFRA) gene had a relatively higher expression level in the OI group (Supplemental Fig. 1c). PDGFRA signaling was reported to inhibit osteoblast differentiation in vitro [53].Lower expression of PDGFRA can free BMPRI, allow the formation of the BMPRI-BMPRII complex, and further activate the BMP-Smad1/5/8 signaling pathway to realize osteogenesis [54].Additionally, thrombospondin 1 (THBS1), which also exhibits a higher gene expression level in the OI group, can inhibit osteoblast differentiation and mineralization [55]. The transforming growth factor-beta (TGF-β) signaling pathway is also a key signaling pathway involved in bone formation. To elaborate, Smad-dependent TGF-β first binds with type I receptors and type II receptors, which are serine or threonine kinase receptors, followed by the transduces to Smads. The activated Smads then form a complex with Smad4. The complex then translocates into the nucleus, interacting with other transcription factors, thereby regulating the ossification process. Smad7 can disrupt complex formation and suppress the bone formation process [56]. According to the DEG expression diagram, the smad4, smad2/3, and smad7 did not have significant gene expression differences, while the genes related to this pathway, namely BMP2, and SMAD1, showed an upregulation tendency in the CGCC group. BMP2 is crucial for postnatal chondrogenesis and osteogenesis [[57], [58], [59]], and SMAD1 plays a role in the transduction of BMP signaling by phosphorylation via the BMP type I receptor. Phosphorylated Smad1 is then combined with Smad4, generates a complex, translocates to the cell nucleus, and continues to promote further osteogenesis by combining with other factors, such as RUNX2. RUNX2 is also involved in the canonical WNT signaling pathway, which is a crucial pathway for mediating ECO through the transdifferentiation of HCCs [60]. RUNX2 is a direct target of the WNT signaling pathway. The WNT signaling pathway can activate β-catenin and form a complex by binding with T-cell factor (TCF), and the complex translocates to the cell nucleus and promotes the expression of RUNX2 [61]. TCF7 and LEF1 are genes that are responsible for the transcription of downstream genes, and T-cell factor and lymphoid enhancer factor can recruit β-catenin into the nucleus and bind with β-catenin. For the CGCC group, there is a higher expression of TCF7 and LEF1 genes compared with the OI group, which aligns with the abovementioned results. The overexpression of CCN4 can enhance the capability of BMP-2 to induce MSC osteoblast differentiation [62]. The CCN4 gene is more highly expressed in OsLhCG-OI, suggesting a different osteoblast differentiation pathway.
Fig. 4.
mRNA sequencing results, including KEGG pathway annotation of differentially expressed genes and differentially expressed gene GO annotation classification. Upregulation is defined as the gene expression of OsLhCG-CGCC being higher than OsLhCG-OI, while downregulation is defined as the gene expression of OsLhCG being higher. a) and b) are KEGG pathway classifications, and the top 35 KEGG pathways of the “upregulation” and “downregulation” groups are listed, respectively. c) and d) are Sankey-dot pathway enrichment plots of the “upregulation” and “downregulation” groups, where differentially expressed core genes involved in the major pathway are listed. The curve in the figure indicates the affiliation of genes to KEGG pathways, and the bubble plot reveals the significance, rich factor, and count number of each KEGG pathway. e) and f) are GO pathway enrichment circle diagrams. Each rectangle in the outermost layer of the circular plot represents a GO term, the middle layer represents the number of genes included in each GO term, and the inner layer represents the number of overlapping genes between the input genes and the genes included in each GO term. The bar chart in the center represents the rich factor of input genes in each GO term.
Gene Ontology (GO) analysis was conducted to highlight the biological process (BP), cellular component (CC), and molecular function (MF) of DEGs between the CGCC group and OI group. The GO pathway enrichment circle diagram for the “upregulated” and “downregulated” is presented in Fig. 4e and f, respectively. The enrichment circle diagrams list the top 20 in GO categories of BP, CC, and MF, respectively. Correspondences between GO-ID and GO description are presented in tabular form (Supplemental Table 1). Furthermore, the GO terms closely related to the ossification process are ossification (GO:0001503), biomineral tissue development (GO:0031214), biomineralization (GO:0110148), cartilage development (GO:0051216), mesenchymal cell differentiation (GO:0048762), and collagen-containing extracellular matrix (GO:0062023). Apart from the genes mentioned above, additional genes involved in osteogenesis were found in GO analysis. For example, phosphoethanolamine is an enzyme that is encoded by the PHOSPHO1 gene, and it is normally localized to sites of mineralization in cartilage and bone. The PHOSPHO1 gene is expressed in osteoblast-like cells, and this gene is involved in the GO terms of ossification, biomineral tissue development, and biomineralization [63]. For the downregulated genes, the GO pathways these genes enriched in are not directly related to the osteogenic process, while the upregulated genes are. Some relevant GO pathways are extracellular matrix organization (GO:0030198), extracellular structure organization (GO:0043062), collagen-containing extracellular matrix (GO:0062023), glycosaminoglycan binding (GO:0005539), extracellular matrix binding (GO:0050840), collagen binding (GO:0005518), and genes related to osteoblast differentiation have been mentioned above.
Gene expression of some core genes involved in the abovementioned KEGG pathways and GO pathways are presented in the violin diagram (Supplemental Fig. 1c).
3.1.3. General quantification analysis, including calcium quantification, DNA assay, GAG assay, and ELISA
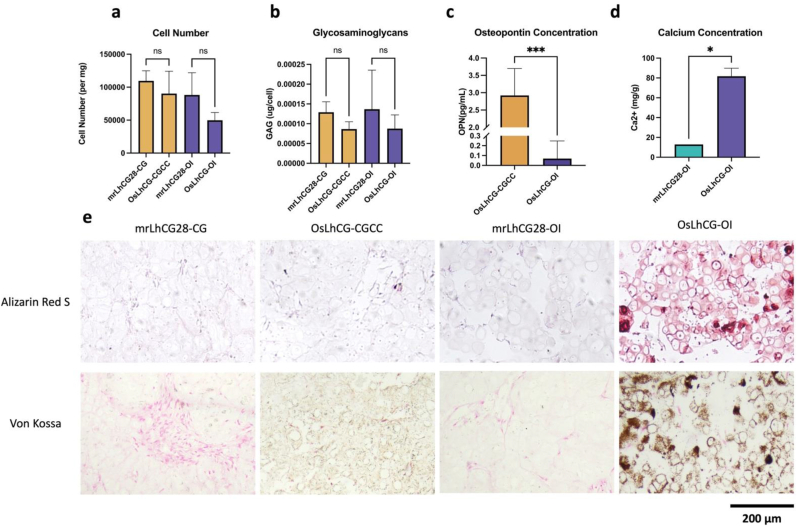
Calcium quantification was conducted to evaluate the bone formation process. During the bone formation process, osteoblasts absorb the acid formed by mineral deposition through the alkalization of calcium and phosphate salts, thereby enabling calcium deposition. Therefore, calcium deposition is a relevant function of osteoblasts and an important step in the process of bone formation [64]. Fig. 5d shows the calcium content of the OI group. The calcium content showed an upward trend, indicating the deposition of calcium ions within the dLhCG scaffold. Calcium deposition of the OI group can also be observed straightforwardly by Alizarin Red S staining and Von Kossa staining (Fig. 5e). Alizarin Red S can bind with calcium and form an orange-colored lake pigment, which can be observed visually. Von Kossa is a staining method based on the metal displacement reaction, in which silver ions displace calcium to form silver salt, which can be reduced to metallic silver under UV light. However, both calcium quantification and staining results show no calcium deposition in the mrLhCG28-CG, while the OsLhCG-CGCC has slight calcium deposition, which can be shown by Von Kossa staining. The increased calcium deposition further demonstrates the osteogenesis of MSCs in the dLhCG scaffold after 49 days of in vitro culturing.
Fig. 5.
General analysis of samples collected at each time point. Figures a, b, c, and d demonstrate the cell number (per mg sample dry weight), glycosaminoglycans (μg/cell number), osteopontin concentration (pg/mL), and calcium concentration (mg/g), respectively. e) Alizarin Red S and Von Kossa staining. Positive expression of calcium deposition is represented in the figure by the sample being stained dark red in Alizarin Red S staining. In Von Kossa, calcium deposition is presented in brown, and cells are presented in pink. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
DNA contents were quantified with Hoechst dye. By quantifying the DNA concentration, the total cell number could be calculated. Fig. 5a shows the total cell number per milligram of samples in different groups. The cell number was normalized to sample dry weight; the actual cell counts per construct ranged from 250,000 to 350,000, which was significantly increased compared with the initial 25,000 MSCs seeded in the dLhCG scaffold. These results indicate that dLhCG has the capability of supporting cell proliferation and differentiation of MSCs within the scaffold, providing a prerequisite for completing the in vitro process of chondrogenesis and osteogenesis. Compared with the mrLhCG28 group, OsLhCG-CGCC, and OsLhCG-OI showed slight downregulation without a significant difference. The concentration of GAG was determined by DMMB solution, and serially diluted chondroitin sulfate was used to set up the standard curve. For both the CGCC and the OI groups, a slight downregulation of GAG concentration existed without a significant difference, probably because of the transformation from cartilage graft to bone graft (Fig. 5b).
Osteopontin, as a protein that is encoded by the SPP1 gene, is responsible for bone remodeling and biomineralization. ELISA was conducted to determine the OPN concentration in each group. For mrLhCG28-CG and mrLhCG28-OI, OPN could not be detected, while for OsLhCG-CGCC, the concentration of OPN was significantly higher than that in the OsLhCG-OI group (Fig. 5c).
3.1.4. MSC viability within the mrLhCG constructs
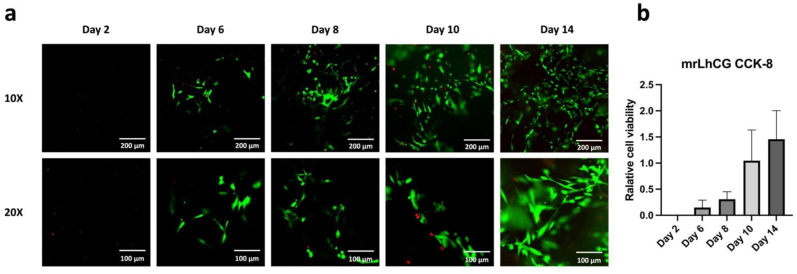
Fig. 6 illustrates the cell viability of mrLhCG within 14 days after seeding MSCs. Fig. 6a presents the distribution of live and dead cells in the constructs on Day 2, 6, 8, 10, and 14, while Fig. 6b displays the changes in relative cell viability measured by the CCK-8 assay at different time points. From the graph, it can be observed that on the second day after cell seeding, the number of cells in the construct was relatively low, below the detection limit of the CCK-8 assay, and no distinct cell distribution could be observed under confocal microscopy. As the cultivation period increased, both the number of live cells and dead cells showed a noticeable increase. In the microscopic images, the green region represents live cells, while the red region represents dead cells. By the sixth day, live cells within the construct became clearly visible. By the tenth day, the proportion of the green region increased compared to previous time points, and by the fourteenth day, the number of live cells significantly increased compared to earlier time points. Additionally, the relative cell viability also exhibited a clear increase over time, indicating successful expansion of MSCs within dLhCG with good cell viability.
Fig. 6.
Cell viability testing. a) The confocal microscope images of mrLhCG stained with live/dead dyes on Day 2, 6, 8, 10, and 14 after MSC implantation. The green regions in the images represent live cells, while the red regions represent dead cells. b) The results of the CCK-8 assay for mrLhCG on Day 2, 6, 8, 10, and 14 after MSC seeding. The vertical axis of the bar chart represents the relative cell viability, which was obtained by normalizing the absorbance values of mrLhCG at different time points with the absorbance value of a certain number of MSCs. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.1.5. Surface morphology evaluation based on SEM images
SEM was employed to characterize the surface morphology of mrLhCG, mrLhCG28-CG, mrLhCG28-OI, OsLhCG-CGCC, and OsLhCG-OI. As depicted in the figures (Supplemental Fig. 2), the surface of mrLhCG exhibited a relatively porous state. In comparison to mrLhCG, the experimental groups subjected to differentiation, including osteogenic induction and chondrogenic induction followed by chondrocyte hypertrophy and HCC transdifferentiation, displayed relatively denser surface morphology, accompanied by the presence of pores of varying sizes.
3.1.6. In vitro histological staining
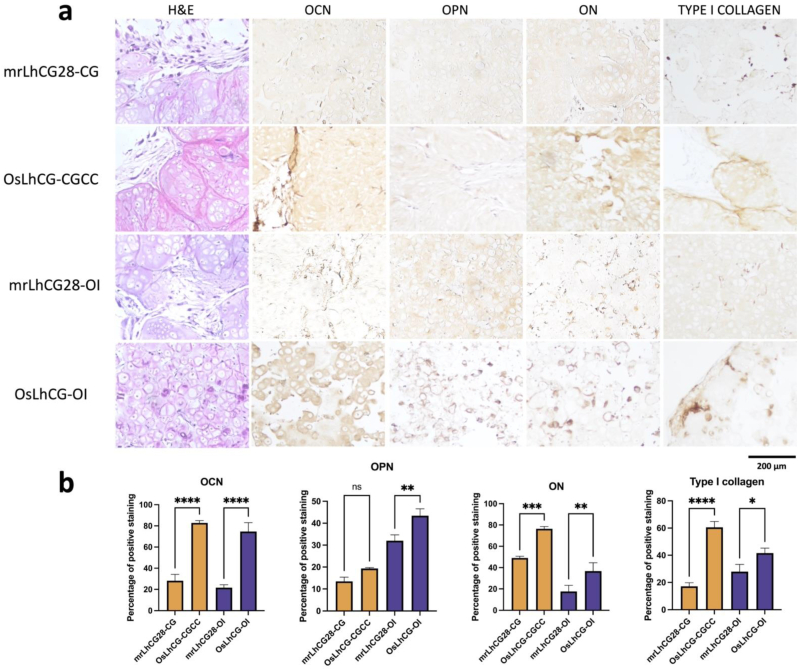
mrLhCG and OsLhCG samples were collected at predetermined time points and fixed with PFA solution before paraffin embedding. The paraffin-embedded samples were sectioned at a thickness of 7 μm and stained with H&E, IHC (OCN, OPN, ON, and Type I collagen), Alizarin red S, and von Kossa. Positive staining with Alizarin red S and von Kossa (Fig. 5e) indicates calcium deposition in OsLhCG-CGCC and OsLhCG-OI. Fig. 7a shows the H&E staining of the CGCC group and the OI group and gives a general overview of the distribution of cells and ECM. To investigate the extent of ossification at the protein expression level, IHC staining was conducted. The sections were incubated with different antibodies prior to DAB staining. As shown in Fig. 7a, both mrLhCG28-CG and mrLhCG28-OI showed relatively lower depositions of osteocalcin, osteopontin, osteonectin and type I collagen than OsLhCG-CGCC and OsLhCG-OI, indicating that the existing cultivation conditions and differentiation induction process have a positive effect on the ossification process and contribute to the deposition of bone formation related proteins. The result can be proved by the percentage of positive staining shown in Fig. 7b.
Fig. 7.
In vitro samples staining and analyzing. a) In vitro samples characterization of H&E and immunohistochemistry staining (OCN, OPN, ON, type I collagen). In H&E staining, ECM is presented in light purple/red, and dark purple dots represent cells. In IHC staining, the brown area represents the positive expression of the related protein. b) The percentage of positive staining. (OCN, OPN, ON, Type I collagen). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. In vivo evaluation of OsLhCG for vascularization capability and bone repair efficacy
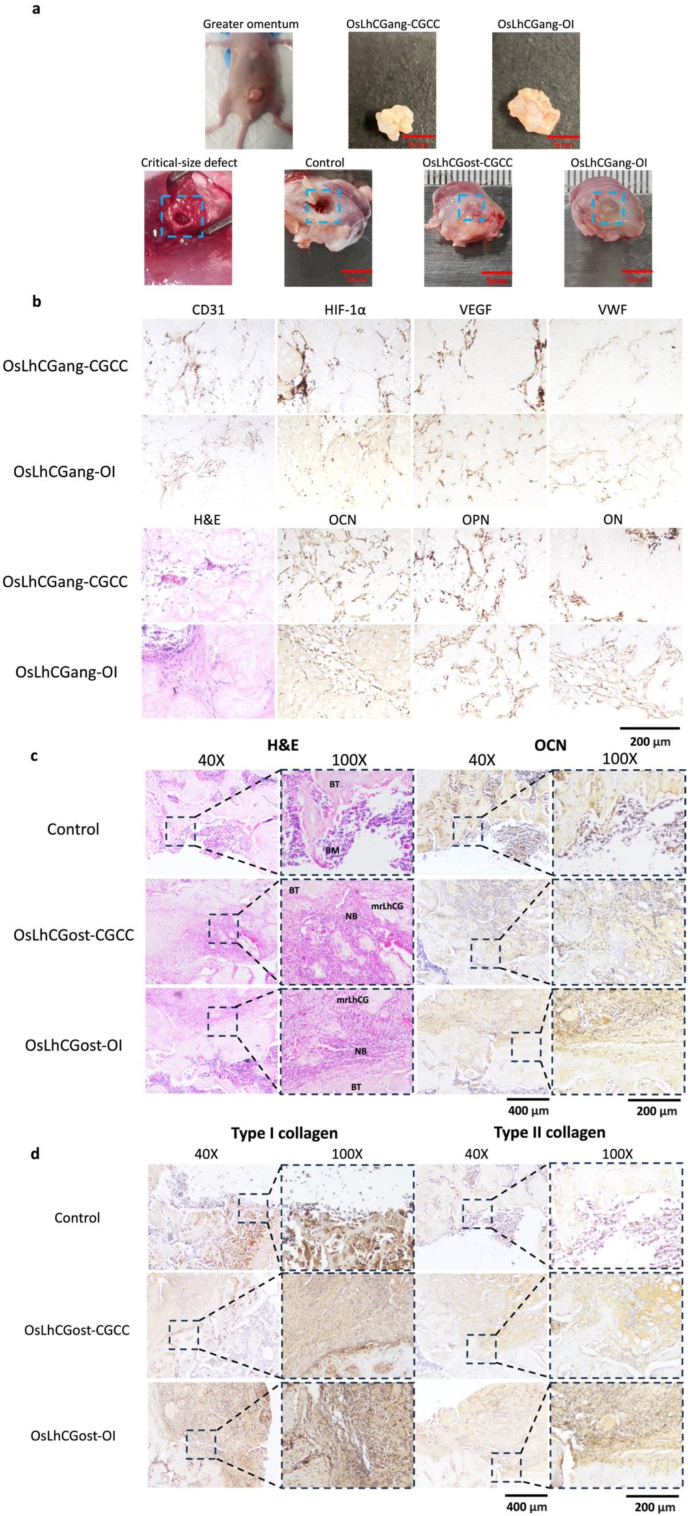
Vascularization plays a crucial role in bone development, bone regeneration, and bone remodeling. Blood vessels supply oxygen and nutrients to the skeletal system while removing metabolic waste products from the bone tissue [[65], [66], [67], [68]]. In tissue engineering, bone grafts require increased oxygen and nutrient supply for the repair of bone defects [69]. Insufficient oxygen and nutrient delivery can lead to incomplete bone integration and impaired bone defect healing [70,71]. The greater omentum is a part of the peritoneum, which is a broad, thin, and transparent peritoneal fold. The greater omentum is a highly vascularized tissue and is commonly utilized to assess the vascularization potential of a biomaterial. Given that the transplanted material possesses angiogenic-inducing capabilities, the greater omentum can facilitate the formation of blood vessels within the material. This, in turn, can provide an adequate supply of oxygen and nutrients to the cells within the graft [69], supporting their proliferation and enabling them to fulfill their functional roles [72,73]. OsLhCG samples that contain living cells were implanted in the greater omentum of nude mice. The desired angiogenic intrusion proceeds as stretches of the host's vasculature system into the osteogenic domain, and the resulting neo-capillaries inside the domain connect back to the host's vasculature. Following the establishment of such a vascularized model, the introduction of abundant blood vessels into the cell-laden OsLhCG provides sufficient oxygen and nutrient supply, creating favorable conditions for further cell proliferation and functional activities within the OsLhCG. Fig. 8b shows the IHC staining of vascular markers after 14 days of implantation, including VEGFA, HIF1-α, VWF, and CD31, as well as several ossification markers, including OCN, OPN, and ON, where the OsLhCGang-CGCC and OsLhCGang-OI represent the OsLhCG-CGCC and OsLhCG-OI that obtained after 14 days of the greater omentum implantation, respectively. The IHC staining results demonstrated the positive expression of VEGFA, HIF1-α, VWF, and CD31, indicating the occurrence of vascularization in OsLhCG. This finding also suggests that OsLhCG, as an artificial bone graft intended for transplantation, possesses the capability to support vascular invasion and angiogenesis in vivo. Moreover, despite our primary focus being the investigation of OsLhCG angiogenic capabilities, osteogenic-related protein IHC staining (i.e., OCN, OPN, and ON) was also performed, and the experimental results revealed that the expression levels of osteogenic markers in OsLhCGang samples exhibited an increase compared to the expression of osteogenic markers in the in vitro samples. This finding suggests that osteoblasts within OsLhCG have the ability to continue secreting osteogenic-related proteins following implantation in the nude mouse greater omentum, concomitant with the invasion of blood vessels.
Fig. 8.
In vivo experiment sample demonstration and staining. (H&E, IHC). a) Images of the greater omentum, critical size defects, and samples obtained after implantation. b) H&E and IHC staining of OsLhCGang. Angiogenesis-related markers include CD31, HIF1-α, VEGF, and VWF. Osteogenesis-related genes include OCN, OPN, and ON. c) H&E and IHC staining of OsLhCGost. 40X photos give a general view of bone defect and newly deposited bone, while 100X photos reveal more detail of protein deposition, which is presented in brown color. (BT: bone tissue; NB: newly deposited bone; BM: bone marrow). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Subsequently, critical-size femoral defect models were established to evaluate the capacity of decellularized OsLhCG. The decellularized OsLhCG-CGCC and OsLhCG-OI were implanted into a cylindrical bone defect created on rat femurs for 30 days. The samples were collected for micro-CT scanning and PFA fixing, named OsLhCGost-CGCC and OsLhCGost-OI, respectively. In a macroscopic view, the surface of the defect appears to have been largely restored to smoothness and even for both OsLhCGost-CGCC and OsLhCGost-OI groups, albeit the defect sites can still be distinguished. A significant defect could be observed in the control group, demonstrating the repair capability of decellularized OsLhCG (Fig. 8a). These observations could also be shown by the 3D reconstructed micro-CT image and cross-sectional diagrams in three directions. From a microscopic perspective, bone repair could be observed through H&E staining and IHC staining (Fig. 8c). A significant defect could be observed in the H&E staining of the control group. Only bone marrow (BM, dense purple dots) and a very small amount of matrix (pink substrate beneath the purple dots) could be observed within the defect of the control group. The defects of OsLhCG-CGCC and OsLhCG-OI are relatively difficult to observe compared with the control group. Bone tissue (BT), newly deposited bone (NB), and decellularized OsLhCG samples could be distinguished under a 100X microscope. After 30 days of in vivo implantation, the whole block of decellularized OsLhCG became dispersed, and a major part of decellularized OsLhCG was replaced with newly deposited bone. This result suggests that decellularized OsLhCG can achieve bone repair and promote the regeneration of bone tissue. Furthermore, the newly deposited bone and decellularized OsLhCG were closely adhered to the original bone tissue, making it possible to fuse with the bone and achieve bone functionality. IHC staining shows that both OsLhCGost-CGCC and OsLhCGost-OI can finely fuse to the original bone tissue with significant deposition of osteogenesis markers such as osteocalcin and type I collagen, while these markers only deposit on the matrix beneath the bone marrow in the control group. For type II collagen, both experimental groups showed positive results, basically because the decellularized OsLhCG samples were derived from dLhCG, which is a pure type II collagen scaffold. To summarize, decellularized OsLhCG samples that underwent different osteogenic induction methods have been modified to be an artificial bone graft and could be used for implantation to realize bone reparation and regeneration with an ideal osteogenic effect.
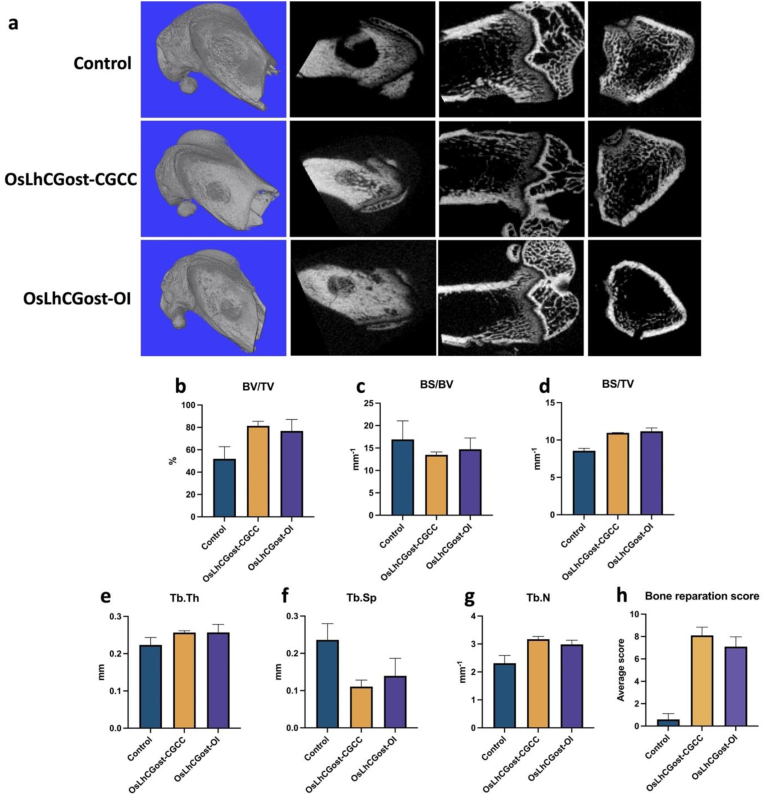
Fig. 9a shows the 3D reconstructed micro-CT image and cross-sectional diagrams in three directions. Moreover, related data reflecting the bone repair efficacy are shown in Fig. 9b. Ten research personnel not involved in this study were asked to rate the bone reparation degree on a scale of 0–10, with 10 indicating full recovery. The average score of each group is presented in Fig. 9h. The result of the average score shows that the control group has not achieved bone repair while both OsLhCG-CGCC and OsLhCG-OI have realized bone repair markedly. The bone repair degree of the OsLhCGost-CGCC group was slightly higher than that of the OsLhCGost-OI group, and this can also be suggested based on the BV/TV result shown in Fig. 9b. Bone volume/tissue volume ratio (BV/TV) suggests the bone volume of different groups [74], and bone surface/volume ratio (BS/BV) suggests the bone surface area per unit bone volume [75]. Bone surface density (BS/TV) can reflect bone mass [76]. The results of BV/TV, BS/BV, and BS/TV clearly indicate that both OsLhCG-CGCC and OsLhCG-OI can repair critical bone defects to a large extent after 30 days of in vivo implantation. Trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), and trabecular number (Tb.N) can reflect the trabecular density of the repaired region [74]. The number and thickness of trabecular bone in the decellularized OsLhCG group are larger than the control group, suggesting the bone reparation efficacy of decellularized OsLhCG.
Fig. 9.
Micro-CT Scanning results of untreated group and critical-size defects that implanted with decellularized OsLhCGost-CGCC and OsLhCGost-OI. a) 3D reconstructed micro-CT image and cross-sectional diagrams in three directions of control, OsLhCGost-CGCC, and OsLhCGost-OI. b)-d) General parameters that reflect bone volume, including bone volume/tissue volume ratio (BV/TV), bone surface/volume ratio (BS/BV), and bone surface density (BS/TV). e)-g) Parameters for trabecular bone microarchitecture, including trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), and trabecular number (Tb.N). h) Bone reparation score.
4. Conclusion
In this study, the dLhCG served as a scaffold to develop a bone graft by mimicking the ECO of MSCs. The MSCs laden mrLhCGs were successively cultured in the MSC expansion medium for 14 days, in the CG medium for 14 days, and in the CC medium for 21 days, and successively induced ECO. The increased expression of COL2A1 and COL10A1 after treatment with CG medium and CC medium indicates the occurrence of chondrogenesis and chondrocyte hypertrophy, respectively, which provides an ideal model to study the underlying mechanism of chondrocyte hypertrophy. Significantly, the elevated deposition of calcium nodules and expression of OCN demonstrate the successful transdifferentiation of the HCCs, establishing the OsLhCG bone graft. The in vivo trials demonstrate that the OsLhCG not only realizes vascularization but also promotes the repair of critical-size femoral defects, showing great potential in clinical applications.
CRediT authorship contribution statement
Cheng Ma: Data curation, Formal analysis, Investigation, Validation, Writing - original draft, Writing - review & editing. Chao Tao: Investigation. Zhen Zhang: Data curation, Formal analysis, Investigation. Huiqun Zhou: Investigation. Changjiang Fan: Funding acquisition, Investigation, Methodology, Supervision, Validation, Writing - review & editing. Dong-an Wang: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The work was financially supported by General Research Fund, Research Grants Council, University Grants Committee, Hong Kong SAR (CityU 11205520 to Dong-An Wang); National Natural Science Foundation of China (51973180 to Dong-An Wang, and 82172108 to Changjiang Fan); Grant from Karolinska Institute Ming Wai Lau Centre of Reparative Medicine (to Dong-An Wang); Grants from City University of Hong Kong (7020028, 7005949, 9231486 to Dong-An Wang); and, Natural Science Foundation of Shandong Province, China (Grant No. ZR2021MB036 to Changjiang Fan).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2023.100893.
Contributor Information
Changjiang Fan, Email: cjfan@qdu.edu.cn.
Dong-an Wang, Email: dwang229@cityu.edu.hk.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Flierl M.A., et al. Outcomes and complication rates of different bone grafting modalities in long bone fracture nonunions: a retrospective cohort study in 182 patients. J. Orthop. Surg. Res. 2013;8(1):1–10. doi: 10.1186/1749-799X-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebraheim N.A., Elgafy H., Xu R. Bone-graft harvesting from iliac and fibular donor sites: techniques and complications. JAAOS-J. Am. Acad. Orthopaedic Surgeons. 2001;9(3):210–218. doi: 10.5435/00124635-200105000-00007. [DOI] [PubMed] [Google Scholar]
- 3.St John T.A., et al. Physical and monetary costs associated with autogenous bone graft harvesting. Am. J. Orthoped. 2003;32(1):18–23. [PubMed] [Google Scholar]
- 4.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 5.Mackie E., et al. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int. J. Biochem. Cell Biol. 2008;40(1):46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X., et al. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10(12) doi: 10.1371/journal.pgen.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L., et al. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. USA. 2014;111(33):12097–12102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang G., et al. Osteogenic fate of hypertrophic chondrocytes. Cell Res. 2014;24(10):1266–1269. doi: 10.1038/cr.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su K., et al. Creating a living hyaline cartilage graft free from non‐cartilaginous constituents: an intermediate role of a biomaterial scaffold. Adv. Funct. Mater. 2012;22(5):972–978. [Google Scholar]
- 10.Peck Y., et al. A preclinical evaluation of an autologous living hyaline-like cartilaginous graft for articular cartilage repair: a pilot study. Sci. Rep. 2015;5(1) doi: 10.1038/srep1622510.1038/srep16225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nie X., et al. Decellularized tissue engineered hyaline cartilage graft for articular cartilage repair. Biomaterials. 2020;235 doi: 10.1016/j.biomaterials.2020.119821. [DOI] [PubMed] [Google Scholar]
- 12.Dang Y., et al. Freeze-thaw decellularization of the trabecular meshwork in an ex vivo eye perfusion model. PeerJ. 2017;5:e3629. doi: 10.7717/peerj.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burk J., et al. Freeze-thaw cycles enhance decellularization of large tendons. Tissue Eng. C Methods. 2014;20(4):276–284. doi: 10.1089/ten.tec.2012.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syedain Z.H., et al. Decellularized tissue-engineered heart valve leaflets with recellularization potential. Tissue Eng. 2013;19(5–6):759–769. doi: 10.1089/ten.tea.2012.0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaretsky J., et al. Ultra-processed food targets bone quality via endochondral ossification. Bone Res. 2021;9(1):1–13. doi: 10.1038/s41413-020-00127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akiyama H., et al. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16(21):2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber H.-P., et al. VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat. Med. 1999;5(6):623–628. doi: 10.1038/9467. [DOI] [PubMed] [Google Scholar]
- 18.Stickens D., et al. 2004. Altered Endochondral Bone Development in Matrix Metalloproteinase 13-deficient Mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bi W., et al. Sox9 is required for cartilage formation. Nat. Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 20.Lefebvre V., Angelozzi M., Haseeb A. SOX9 in cartilage development and disease. Curr. Opin. Cell Biol. 2019;61:39–47. doi: 10.1016/j.ceb.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., et al. Hypertrophic chondrocyte-specific Col10a1 controlling elements in Cre recombinase transgenic studies. Am. J. Tourism Res. 2019;11(10):6672. [PMC free article] [PubMed] [Google Scholar]
- 22.Qin X., et al. Runx2 is essential for the transdifferentiation of chondrocytes into osteoblasts. PLoS Genet. 2020;16(11) doi: 10.1371/journal.pgen.1009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komori T. Whole aspect of Runx2 functions in skeletal development. Int. J. Mol. Sci. 2022;23(10):5776. doi: 10.3390/ijms23105776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamate T., et al. Osteopontin expression by osteoclast and osteoblast progenitors in the murine bone marrow: demonstration of its requirement for osteoclastogenesis and its increase after ovariectomy. Endocrinology. 1997;138(7):3047–3055. doi: 10.1210/endo.138.7.5285. [DOI] [PubMed] [Google Scholar]
- 25.Morinobu M., et al. Osteopontin expression in osteoblasts and osteocytes during bone formation under mechanical stress in the calvarial suture in vivo. J. Bone Miner. Res. 2003;18(9):1706–1715. doi: 10.1359/jbmr.2003.18.9.1706. [DOI] [PubMed] [Google Scholar]
- 26.Sodek J., Ganss B., McKee M. Osteopontin. Crit. Rev. Oral Biol. Med. 2000;11(3):279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- 27.Hunter G.K., Goldberg H.A. Modulation of crystal formation by bone phosphoproteins: role of glutamic acid-rich sequences in the nucleation of hydroxyapatite by bone sialoprotein. Biochem. J. 1994;302(1):175–179. doi: 10.1042/bj3020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher L., et al. Human bone sialoprotein. Deduced protein sequence and chromosomal localization. J. Biol. Chem. 1990;265(4):2347–2351. [PubMed] [Google Scholar]
- 29.Bianco P., et al. Expression of bone sialoprotein (BSP) in developing human tissues. Calcif. Tissue Int. 1991;49:421–426. doi: 10.1007/BF02555854. [DOI] [PubMed] [Google Scholar]
- 30.Rosset E.M., Bradshaw A.D. SPARC/osteonectin in mineralized tissue. Matrix Biol. 2016;52:78–87. doi: 10.1016/j.matbio.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee N.K., et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130(3):456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponte F., et al. Mmp13 deletion in mesenchymal cells increases bone mass and may attenuate the cortical bone loss caused by estrogen deficiency. Sci. Rep. 2022;12(1):1–14. doi: 10.1038/s41598-022-14470-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Q., Ecker M. Overview of MMP-13 as a promising target for the treatment of osteoarthritis. Int. J. Mol. Sci. 2021;22(4):1742. doi: 10.3390/ijms22041742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behonick D.J., et al. Role of matrix metalloproteinase 13 in both endochondral and intramembranous ossification during skeletal regeneration. PLoS One. 2007;2(11):e1150. doi: 10.1371/journal.pone.0001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Root A.W. In: Pediatric Endocrinology. fourth ed. Sperling M.A., editor. W.B. Saunders; 2014. Chapter 8 - disorders of calcium and phosphorus homeostasis in the newborn and infant; pp. 209–276.e1. [Google Scholar]
- 36.Marie P.J. Targeting integrins to promote bone formation and repair. Nat. Rev. Endocrinol. 2013;9(5):288–295. doi: 10.1038/nrendo.2013.4. [DOI] [PubMed] [Google Scholar]
- 37.Pang X., et al. Targeting integrin pathways: mechanisms and advances in therapy. Signal Transduct. Targeted Ther. 2023;8(1):1. doi: 10.1038/s41392-022-01259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J.-M., et al. The ERK MAPK pathway is essential for skeletal development and homeostasis. Int. J. Mol. Sci. 2019;20(8):1803. doi: 10.3390/ijms20081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao G., et al. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J. Biol. Chem. 2002;277(39):36181–36187. doi: 10.1074/jbc.M206057200. [DOI] [PubMed] [Google Scholar]
- 40.Xiao G., et al. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J. Biol. Chem. 2000;275(6):4453–4459. doi: 10.1074/jbc.275.6.4453. [DOI] [PubMed] [Google Scholar]
- 41.Verrier S., et al. ADAM gene expression and regulation during human osteoclast formation. Bone. 2004;35(1):34–46. doi: 10.1016/j.bone.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 42.Scheller J., et al. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32(8):380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Araya H.F., et al. Expression of the ectodomain‐releasing protease ADAM17 is directly regulated by the osteosarcoma and bone‐related transcription factor RUNX2. J. Cell. Biochem. 2018;119(10):8204–8219. doi: 10.1002/jcb.26832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKie N., et al. Expression of members of a novel membrane linked metalloproteinase family (ADAM) in human articular chondrocytes. Biochem. Biophys. Res. Commun. 1997;230(2):335–339. doi: 10.1006/bbrc.1996.5957. [DOI] [PubMed] [Google Scholar]
- 45.Ruhrberg C., Maes C., Carmeliet G. VEGF in Development; 2008. Vascular and Nonvascular Roles of VEGF in Bone Development; pp. 79–90. [Google Scholar]
- 46.Han Y., et al. Paracrine and endocrine actions of bone—the functions of secretory proteins from osteoblasts, osteocytes, and osteoclasts. Bone Res. 2018;6(1):16. doi: 10.1038/s41413-018-0019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bos J.L., de Rooij J., Reedquist K.A. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2001;2(5):369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 48.Wu Y., et al. Rap1A regulates osteoblastic differentiation via the ERK and p38 mediated signaling. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0143777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H., Jeon T.J. Activated Rap1A induces osteoblastic differentiation and cell adhesion. J. Chosun Nat. Sci. 2016;9(3):171–176. [Google Scholar]
- 50.Su N., Jin M., Chen L. Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models. Bone research. 2014;2(1):1–24. doi: 10.1038/boneres.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miraoui H., et al. Fibroblast growth factor receptor 2 promotes osteogenic differentiation in mesenchymal cells via ERK1/2 and protein kinase C signaling. J. Biol. Chem. 2009;284(8):4897–4904. doi: 10.1074/jbc.M805432200. [DOI] [PubMed] [Google Scholar]
- 52.Fan C., et al. A mussel-inspired double-crosslinked tissue adhesive intended for internal medical use. Acta Biomater. 2016;33:51–63. doi: 10.1016/j.actbio.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Brun J., et al. PDGF receptor signaling in osteoblast lineage cells controls bone resorption through upregulation of Csf1 expression. J. Bone Miner. Res. 2020;35(12):2458–2469. doi: 10.1002/jbmr.4150. [DOI] [PubMed] [Google Scholar]
- 54.Li A., et al. PDGF-AA promotes osteogenic differentiation and migration of mesenchymal stem cell by down-regulating PDGFRα and derepressing BMP-Smad1/5/8 signaling. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0113785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ueno A., et al. Constitutive expression of thrombospondin 1 in MC3T3‐E1 osteoblastic cells inhibits mineralization. J. Cell. Physiol. 2006;209(2):322–332. doi: 10.1002/jcp.20735. [DOI] [PubMed] [Google Scholar]
- 56.Chen G., Deng C., Li Y.-P. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 2012;8(2):272. doi: 10.7150/ijbs.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon B.S., et al. 2006. BMPs Regulate Multiple Aspects of Growth-Plate Chondrogenesis through Opposing Actions on FGF Pathways. [DOI] [PubMed] [Google Scholar]
- 58.Haas A.R., Tuan R.S. Chondrogenic differentiation of murine C3H10T1/2 multipotential mesenchymal cells: II. Stimulation by bone morphogenetic protein-2 requires modulation of N-cadherin expression and function. Differentiation. 1999;64(2):77–89. doi: 10.1046/j.1432-0436.1999.6420077.x. [DOI] [PubMed] [Google Scholar]
- 59.Cao X., Chen D. The BMP signaling and in vivo bone formation. Gene. 2005;357(1):1–8. doi: 10.1016/j.gene.2005.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nadine S., et al. Close-to-native bone repair via tissue engineered endochondral ossification approaches. iScience. 2022 doi: 10.1016/j.isci.2022.105370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaur T., et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J. Biol. Chem. 2005;280(39):33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 62.Ono M., et al. WISP‐1/CCN4 regulates osteogenesis by enhancing BMP‐2 activity. J. Bone Miner. Res. 2011;26(1):193–208. doi: 10.1002/jbmr.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Houston B., Stewart A.J., Farquharson C. PHOSPHO1—a novel phosphatase specifically expressed at sites of mineralisation in bone and cartilage. Bone. 2004;34(4):629–637. doi: 10.1016/j.bone.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 64.Blair H.C., et al. Calcium and bone disease. Biofactors. 2011;37(3):159–167. doi: 10.1002/biof.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Filipowska J., et al. The role of vasculature in bone development, regeneration and proper systemic functioning. Angiogenesis. 2017;20:291–302. doi: 10.1007/s10456-017-9541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dai J., Rabie A. VEGF: an essential mediator of both angiogenesis and endochondral ossification. J. Dent. Res. 2007;86(10):937–950. doi: 10.1177/154405910708601006. [DOI] [PubMed] [Google Scholar]
- 67.Hankenson K.D., et al. Angiogenesis in bone regeneration. Injury. 2011;42(6):556–561. doi: 10.1016/j.injury.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang B., et al. Osteoblasts secrete Cxcl9 to regulate angiogenesis in bone. Nat. Commun. 2016;7(1) doi: 10.1038/ncomms13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei J., et al. Guided self-generation of vascularized neo-bone for autologous reconstruction of large mandibular defects. J. Craniofac. Surg. 2016;27(4):958–962. doi: 10.1097/SCS.0000000000002680. [DOI] [PubMed] [Google Scholar]
- 70.Sathy B.N., et al. Bone tissue engineering with multilayered scaffolds—Part I: an approach for vascularizing engineered constructs in vivo. Tissue Eng. 2015;21(19–20):2480–2494. doi: 10.1089/ten.tea.2015.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mercado-Pagán Á.E., et al. Vascularization in bone tissue engineering constructs. Ann. Biomed. Eng. 2015;43:718–729. doi: 10.1007/s10439-015-1253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pellicciaro M., et al. The greater omentum as a site for pancreatic islet transplantation. CellR4--repair, replacement, regeneration, & reprogramming. 2017;5(3) [PMC free article] [PubMed] [Google Scholar]
- 73.Liu H., et al. Vascularization of engineered organoids. BMEMat. 2023;1(3) [Google Scholar]
- 74.Mohan S., et al. In vivo evidence of IGF-I–estrogen crosstalk in mediating the cortical bone response to mechanical strain. Bone Res. 2014;2(1):1–6. doi: 10.1038/boneres.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adams G., et al. Microarchitecture and morphology of bone tissue over a wide range of BV/TV assessed by micro-computed tomography and three different threshold backgrounds. Med. Eng. Phys. 2022;106 doi: 10.1016/j.medengphy.2022.103828. [DOI] [PubMed] [Google Scholar]
- 76.Kiyomoto K., et al. High bone turnover state under osteoporotic changes induces pain-like behaviors in mild osteoarthritis model mice. J. Bone Miner. Metabol. 2020;38:806–818. doi: 10.1007/s00774-020-01124-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.