Abstract
Lassa virus is a negative-strand RNA virus with only four structural proteins that causes periodic outbreaks in West Africa. The nucleoprotein (NP) encapsidates the viral genome, forming ribonucleoprotein complexes (RNPs) together with the viral RNA and the L protein. RNPs must be continuously restructured during viral genome replication and transcription. The Z protein is important for membrane recruitment of RNPs, viral particle assembly, and budding and has also been shown to interact with the L protein. However, the interaction of NP, viral RNA, and Z is poorly understood. Here, we characterize the interactions between Lassa virus NP, Z, and RNA using structural mass spectrometry. We identify the presence of RNA as the driver for the disassembly of ring-like NP trimers, a storage form, into monomers to subsequently form higher order RNA-bound NP assemblies. We locate the interaction site of Z and NP and demonstrate that while NP binds Z independently of the presence of RNA, this interaction is pH-dependent. These data improve our understanding of RNP assembly, recruitment, and release in Lassa virus.
Introduction
Viruses are a highly diverse group of pathogens that infect all kingdoms of life and cause manifold diseases in humans. A key to developing antiviral strategies is in-depth understanding of viral cell biology. Due to the limited coding capacity in their genomes compared to other pathogens, viruses are greatly dependent on protein–protein and protein–nucleic acid interactions with host factors and between the viral components.1 However, cellular complexity often precludes precise investigations of such interactions, asking for biochemical investigations. Lassa virus (LASV) is the causative agent of Lassa hemorrhagic fever and belongs to the Arenaviridae family within the Bunyavirales order. LASV is rodent-borne and endemic to West Africa, infecting between 100,000 and 300,000 humans each year leading to approximately 5000 deaths.2 Recent predictions estimate up to 186 million people at risk of LASV infections in West Africa by 2030.3 Arenaviruses are bisegmented negative-strand RNA viruses, and each genome segment encodes for two structural proteins using an ambisense coding strategy, meaning two genes are in opposite orientation on the viral genome segment. This implies that some viral genes can only be transcribed from a complementary copy of the genome, called antigenome, which is produced by the viral L protein inside the host cell. The large genome segment encodes for the ∼250 kDa large (L) protein, which contains the RNA-dependent RNA polymerase, and a small RING finger matrix protein Z, which has a molecular weight of ∼11 kDa. The small genome segment encodes for the viral glycoprotein precursor (GPC, ∼75 kDa) and the nucleoprotein (NP, ∼63 kDa).4
Despite the central role of the LASV L protein in viral genome replication and transcription, NP is also essential for these processes.5 It encapsidates the viral RNA genome, forming, together with L, the ribonucleoprotein complex (RNP). Within the RNP, the viral RNA is protected from degradation and recognition by cellular pattern recognition receptors. The RNP is the structural and functional unit of viral genome replication and transcription.6 NP is composed of an N-terminal and a C-terminal domain. The N-terminal domain contains an RNA-binding cavity, and the C-terminal domain has an exonuclease activity.7,8 When not bound to RNA, NP forms homotrimeric rings with a head to tail arrangement of the NP molecules. In this conformation, RNA binding is hindered as the respective pocket is blocked by two helices.9 To allow for RNA binding an RNA gating mechanism was proposed where the C-terminal domain rotates slightly away, allowing one helix to become partly unstructured (α5), whereas the other helix (α6) shifts away from the RNA-binding groove.10 However, the trigger and mechanism of this conformational change are unknown.
The matrix protein Z lines the inner side of the viral envelope and mediates virion assembly and budding by homo-oligomerization and interaction with host factors.11−13 It is anchored into membranes by N-terminal myristoylation14 and has been hypothesized to mediate recruitment of RNPs to the plasma membrane for virion assembly by direct or indirect interaction with NP.11,15,16 Z has also been shown to interact with the L protein, inhibiting its polymerase activity potentially by blocking the formation of an elongation conformation of L.17−21 It remains unclear whether RNP recruitment is driven by the interaction between NP and Z, between L and Z, and/or via a host protein.
In this study, we investigate the mechanism and dynamics of RNP formation and recruitment. In the presence of RNA, even short ones, the ring-like NP trimer undergoes conformational changes leading to its disassembly into RNA-bound NP monomers, which subsequently form higher order NP-RNA assemblies, reminiscent of RNPs. Additionally, we show that the LASV NP can directly interact with the Z protein via the C-terminal domain. We demonstrate that this interaction is strongly reduced at a low pH, which would be in line with the conditions during virus entry via the endosomal pathway to initiate the release of RNPs from the matrix.
Results
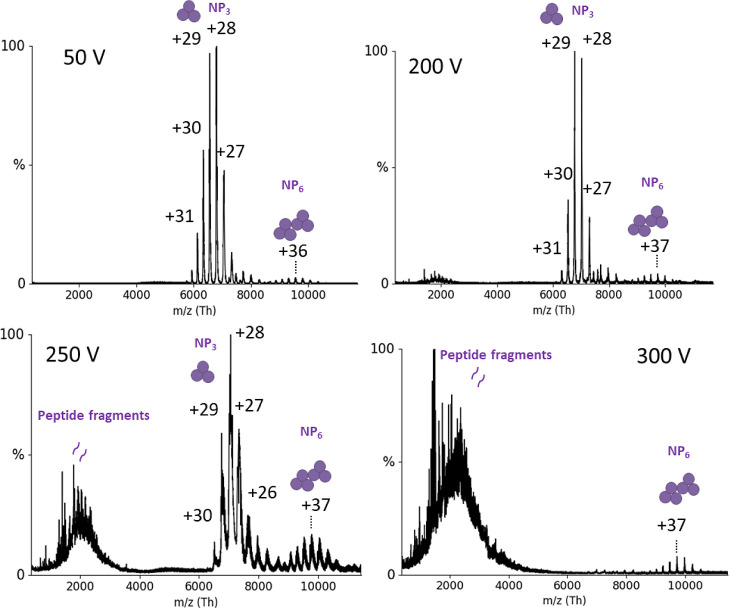
The full-length LASV NP and Z proteins (AV strain) were expressed in Escherichia coli and purified for subsequent interaction studies. As described previously, both NP and Z can form oligomers.22,23 While NP eluted from the size exclusion column (SEC) in one peak corresponding to a trimer, Z eluted in two peaks, indicating a monomeric and oligomeric state. The oligomerization of the proteins was further analyzed by native mass spectrometry (nMS).24 We found that NP was mainly trimeric (189.56 ± 0.03 kDa), which will be referred to as NP3, but traces of NP hexamers could also be detected. The hexameric NP was already found by others with a retained exonuclease function.25 However, it is unclear whether the hexamer is present during viral infection inside cells (or just an artifact from in vitro expression) or what its biological role would be. Further characterizations of NP3 using nMS revealed extraordinarily high gas-phase stability. No collision-induced dissociation (CID) was observed at up to 200 V acceleration into the collision cell (Figure 1). Beyond 200 V, fragmentation into peptides took place, but there was no monomer dissociation, pointing toward high noncovalent complex stability in the gas phase in the absence of disulfide bonds. Although the hexameric NP is not the focus of this study, it is worth mentioning that it also displays a high gas-phase stability. Even at 300 V acceleration voltage, the hexameric NP remained stable, pointing to a specific and strong interaction. Of note, larger molecules are generally more robust to fragmentation due to the lower m/z and hence relatively lower activation. For the Z protein, one of the two fractions from SEC could be identified as monomeric Z (11.35 ± 0.04 kDa), whereas the second fraction contained heterogeneous oligomers of Z (Figure S1) in the range of around 120 kDa. All of the following experiments were performed using the monomeric fraction of Z.
Figure 1.
nMS of NP at different acceleration voltages (V). Six μM NP was measured in 150 mM ammonium acetate buffer surrogate at pH 7.5 with acceleration voltages in the collision cell between 50 and 300 V (indicated in the graphs). The mass of the species between 6000–8000 m/z was determined as 189.56 ± 0.03 kDa which corresponds to an NP trimer. Charge states are indicated. Around 10,000 m/z an NP hexamer is detectable, and above 200 V, peptide fragments appear in the low mass range.
NP Directly Interacts with Monomeric Z Protein
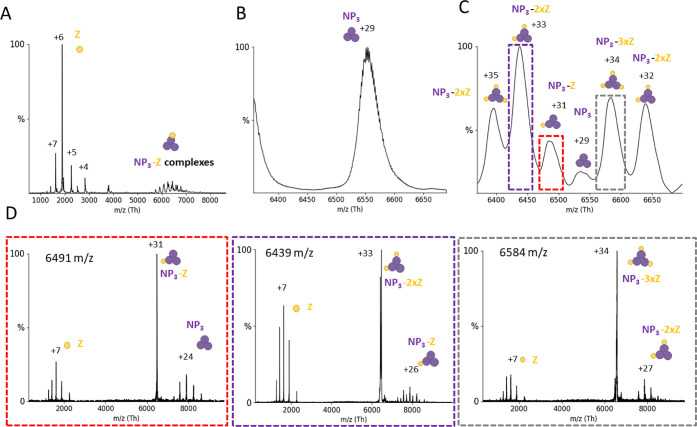
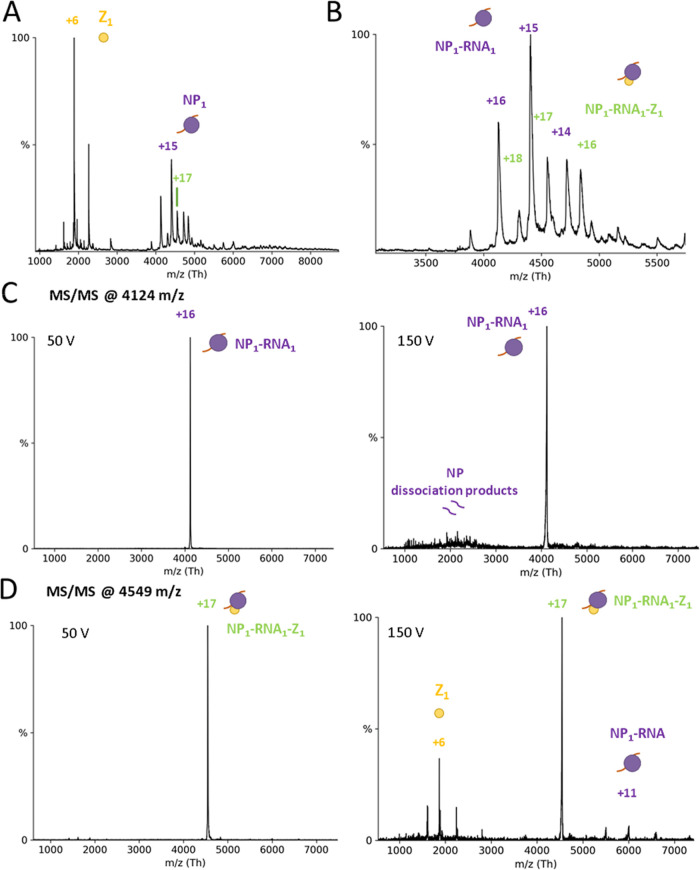
It was previously shown that LASV NP colocalizes with Z in cells.11 We analyzed the ability of NP and Z to form complexes by direct interaction using purified proteins. nMS measurements of the trimeric NP with a 10-fold excess of Z showed new peaks appearing in the mass-to-charge (m/z) range between 5000 and 7000 (Figure 2A–C). To analyze the exact complex composition in this area, we used tandem MS (MS/MS) with CID. CID-MS/MS of peaks corresponding to 1, 2, or 3 Z proteins attached to an NP3 resulted in dissociation products of NP3 (189.56 ± 0.03 kDa), NP3 + Z (201.00 ± 0.09 kDa), and NP3 + 2xZ (212.37 ± 0.13 kDa) (Figure 2D). Overall, these experiments demonstrate a direct interaction between LASV NP3 and Z in an RNA-free context with a stoichiometry of one binding interface per NP monomer. To determine the affinity of Z for NP3, we recorded nMS spectra in triplicate at an NP-to-Z molar ratio of 1:3 and used the deconvoluted spectra to determine the area under the curve (AUC). We determined an apparent KD of 110 (±10) μM for binding of a Z protein to an NP3 (Figure S2).
Figure 2.
NP3 binds up to 3 Z monomers. NP and Z were incubated in a 1:10 molar ratio and subjected to nMS in a 150 mM ammonium acetate solution at pH 7.5. (A) Overview spectrum recorded with a 50 V acceleration. Two peak series are observable between 1000 and 4000 m/z (corresponding to the Z protein) and between 5500 and 7000 m/z (NP3 complexes). Zoom to the NP3 29+ charge state (B) without and (C) with Z protein. Several new species corresponding to NP-Z complexes are recorded. (D) CID-MS/MS at 100 V acceleration for NP3-Z (31+ charge state), NP3–2xZ (33+ charge state), and NP3-3xZ (34+ charge state). CID products correspond to masses for Z (11.35 ± 0.04 kDa), NP3 (189.56 ± 0.03 kDa), NP3-Z (201.00 ± 0.09 kDa), and NP3–2xZ (212.37 ± 0.13 kDa). The charge states of the most abundant peaks are indicated.
Next, we tested whether the NP-Z interaction depends on the oligomerization state of NP using an NP trimerization-deficient mutant (R52A). Elution profiles of the SEC for NP-R52A showed two peaks corresponding to a trimeric and a monomeric state. We analyzed both fractions by nMS and found additional traces of dimers in the monomeric fraction (Figures 3A and S3). The monomeric fraction was used for all of the following experiments. Interestingly, we observed heterogeneous mass species with a constant mass difference of around 300 Da (Figure S3). This indicates different numbers of nucleotide monophosphates (NMPs) from the expression host being bound by the NP-R52A. These NMPs can be displaced when adding RNA to NP-R52A. At a 1:3 ratio between NP-R52A and Z, native mass spectra clearly showed the 75.5 ± 0.3 kDa monomeric NP-R52A-Z complex (Figure 3B) with a KD of 33 (±2) μM. CID-MS/MS of the NP-R52A-Z complex further confirmed this interaction (Figure S3). These data show that the monomeric NP-Z interaction is stronger compared to that of trimeric NP, influenced either by the monomeric conformation or by the arginine mutation.
Figure 3.
NP-Z interaction is independent of the oligomerization state of the NP in nMS. (A) NP trimerization mutant R52A was measured with nMS in a 150 mM ammonium acetate solution at pH 7.5 and a concentration of 5.2 μM. Acceleration voltage in the collision cell was at 50 V for the depicted spectrum. R52A appears primarily in the monomeric state between 3500 and 4500 with charge states between +17 and +14 and a corresponding mass of 63.90 ± 0.08 kDa. A small fraction of dimeric NP is visible in the m/z range between 5000 and 6000 with the corresponding charge state of +20, +19, and +18. The most abundant species was normalized to 1. (B) R52A and Z were incubated in a 1:3 ratio and measured with the same instrument settings as described in A. Spectra show new peak series appearing for unbound Z protein in the m/z range between 1500 and 3000. New peaks also appear in the range between 4000 and 5000 m/z with +18, +17, and +16 charge states corresponding to a complexation of R52A and Z (75.5 ± 0.3 kDa).
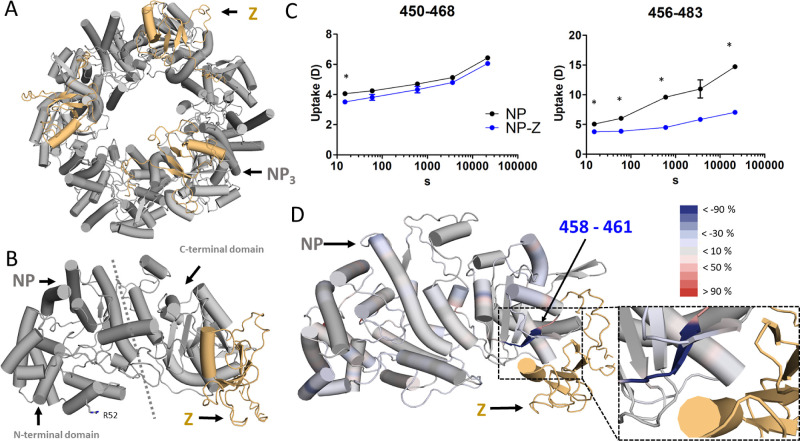
We then used an integrative structural approach to map the interaction site between NP and Z. First, we predicted the structure of the NP-Z complex by AlphaPulldown,26 a python package built upon AlphaFold27 and AlphaFold Multimer28 (Figure 4A,B). The structural model obtained high local and global quality scores as returned by AlphaFold and AlphaPulldown (Figure S4, Table S1) and predicted that Z interacts with the C-terminal domain of NP (Figure 4B). To map changes in the protein dynamics caused by protein–protein interactions, we used hydrogen–deuterium exchange MS (HDX-MS).29 This method is complementary to nMS and can be carried out in standard buffers. We used this method to experimentally validate the predicted interaction site. Labeling experiments were performed with NP in both the presence and absence of Z protein. Differences in deuterium uptake between both states were mapped to the NP sequence (Figures S5 and S6). We found significant differences in deuterium uptake for the peptides 450–483 and 456–483, which align with the predicted NP-Z interface (Figure 4C). The differences based on the deuterium uptake at the 6 h time point were plotted against the predicted NP-Z complex (Figures 4D and S6).
Figure 4.
Identification of the NP-Z interface. The trimeric model of NP-Z (A) was generated by superposing the model of the NP-Z dimer predicted by AlphaFold (Version 2.1.0) (B) onto the crystal structure of the NP trimer (PDB ID 3R3L). NP is colored gray, and the Z protein is shown in gold. (C) HDX differences in NP peptides in the presence of Z. Uptake plots comparing the conditions of NP alone and NP-Z mixture with significant differences (*delta HDX > X D – 95% CI, see Materials and Methods, Table S2) around the predicted NP-Z interaction site (D). Differences in deuterium uptake between both states were mapped to the NP sequence and can be seen in Figure S6. The differences are based on the atomic range, which considers overlapping peptides with no differences. The differences at the 6 h time point were plotted onto the predicted NP-Z complex. Regions that are protected and exposed in HDX in the presence of Z are colored blue and red, respectively. Regions in gray show no change in deuterium uptake in the presence of Z. The Z protein is highlighted in gold.
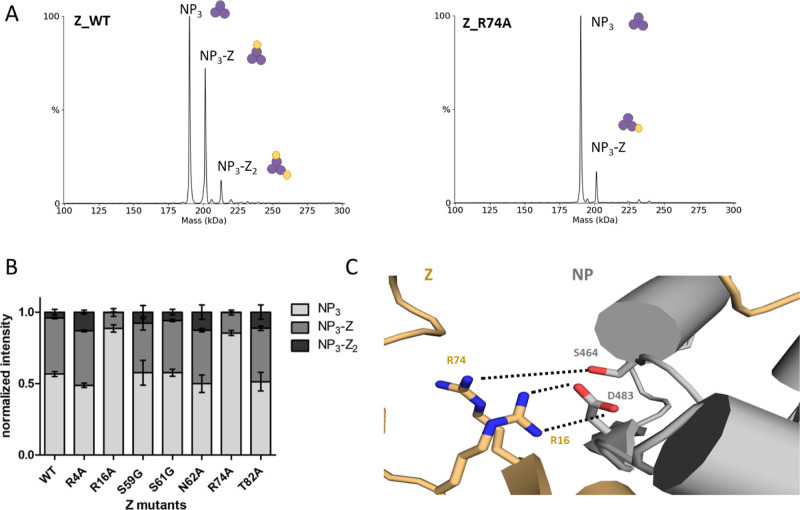
Next, we introduced point mutations into the Z protein based on the NP-Z structural model to further validate the proposed interface. We exchanged polar residues with alanine or glycine at or in close proximity to the predicted interface. These were R4, R16, S59, S61, N62, R74, and T82. We measured all Z mutants together with NP in a 1:3 molar ratio by nMS and compared NP-Z complex formation by using the deconvoluted spectra of at least 3 independent measurements (Figure 5A). Z mutants R16A and R74A showed a strong reduction of NP-Z complex formation compared to wild-type Z, whereas the other mutations had no effect on NP-Z interaction (Figure 5B). This leads to the conclusion that NP-Z interaction is strongly dependent on arginines 16 and 74 of Z, which supports the predicted model (Figure 5C). The other point mutations of polar residues seem to be less critical and were not able to influence NP-Z interaction.
Figure 5.
Z mutant screening identifies important residues for the NP-Z interaction. The different Z mutants were recombinantly expressed and purified. (A) nMS screening of the Z mutants together with NP (6 μM) in a 1:3 (NP/Z) molar ratio in a 150 mM ammonium acetate solution at pH 7.5. (B) Sum of the intensities of every mass species of one measurement was normalized to 1. The normalized intensities for every measurement were plotted. Error bars represent standard deviation of at least three independent measurements. Measurements with the mutants R16A and R74A show reduced abundance of NP-Z complex species compared to wildtype (WT). (C) Detailed closeup to the Z residues R74 and R16 in the AlphaFold Multimer model. Z residues R74 and R16 are in close proximity to NP-S464 and NP-D483, respectively.
Taken together, our structural integrative approach, including AI complex prediction, HDX-MS, and mutational analysis in combination with nMS leads to detailed information about the direct NP-Z interaction. Z seems to bind at the C terminus of NP, where arginines R16 and R74 of Z are likely important for the NP-Z interaction.
RNA Triggers Disassembly of Trimeric NP
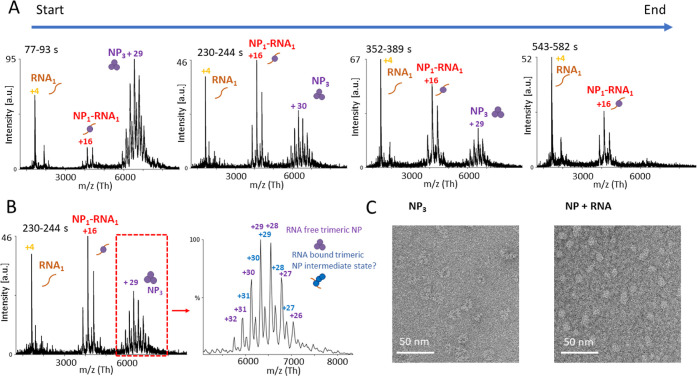
To investigate the effect of RNA binding on the NP quaternary structure, we incubated NP3 with short RNAs. As no sequence specificity of NP for RNA has been described so far, we used RNAs of different sequences and lengths. First, we mixed a 9 nt RNA (Table S2) in an equimolar ratio with NP and monitored the changes in the quaternary structure over at least 500 s in the electrospray ionization (ESI) capillary (Figure 6A). The acquisition started approximately 30 s after mixing the components and mounting the capillary. During the measurement, the fractions belonging to NP3 decreased, whereas peaks assigned to a mass of 66.08 ± 0.02 kDa, corresponding to NP monomer bound to one RNA molecule (NP1-RNA1), increased. Importantly, we also observed NP3 interacting with one RNA molecule (193.00 ± 0.09 kDa) at intermediate time points (Figure 6B). As the RNA-binding groove of NP is inaccessible in the trimeric ring conformation, the observed complex of one NP trimer and one RNA molecule (NP3-RNA1) presumably constitutes an open conformation of NP3, allowing the RNA to bind. After 500 s, the NP3 was mostly disassembled. To further support our results, we used negative-stain electron microscopy to confirm the disassembly of NP3 in the presence of RNA. We observed NP mainly as a ring-shaped trimer in the absence of RNA (Figures 6C and S7A). In the presence of RNA, however, the morphology changes to smaller rounded particles and more heterogeneous species as NP3 is not detectable anymore (Figures 5C and S7B). Furthermore, we validated our results by mass photometry. We observed NP monomer formation in the presence of RNA (Figure S8). We conclude that RNA is sufficient to trigger conformational changes, leading to the disassembly of the trimeric ring structure of NP into RNA-bound NP monomers.
Figure 6.
RNA triggers the dissociation of NP3 into monomers. NP3 and a 9 nt RNA were mixed together in a 150 mM ammonium acetate solution at pH 7.5 on ice in a 1:1 (NP1/RNA) molar ratio. The mixture was placed into an ESI capillary, and the measurement was started approximately 30 s after mixing RNA and NP together. Spectra were recorded over 550 s. (A) Each diagram represents combined spectra recorded at 77–93, 230–244, 355–389, and 543–582 s with the intensity in arbitrary units (a.u.). Main charge states are labeled. Shown is one representative measurement of at least three independent measurements. (B) Recorded spectrum at time point 230–244 s. Zoom into the m/z range between 5000 and 8000 shows an RNA-bound NP3 (193.00 ± 0.08 kDa) species. (C) NP at 160 nM was mixed with a single-stranded 25 nt RNA at a 1:2 molar ratio (NP/RNA) (Table S3) and incubated at room temperature for approximately 15 min. NP was applied to glow-discharged carbon-coated copper grids and stained with uranyl acetate immediately before imaging. Images were collected with a transmission electron microscope.
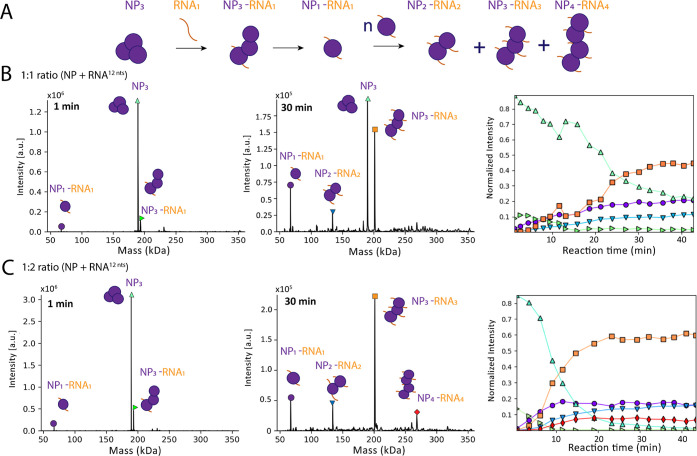
Next, we used a slightly longer RNA of 12 nts (Table S3) to investigate a potential assembly of RNA-bound NP monomers, corresponding to the beginning of higher order NP-RNA assemblies. We again mixed the components and determined the complex species over a duration of 45 min in an ESI capillary. The measurement was started approximately 30 s after mixing RNA and NP together. We first tested an equimolar ratio of NP to the RNA of 12 nts (Figure 7). After the start of the measurement, three species were visible: (i) NP3, (ii) a trimeric NP bound to one RNA molecule (NP3-RNA1, 193.63 ± 0.09 kDa), and (iii) unbound RNA (3.9 kDa) (Figure 7B). One min later, a signal corresponding to an NP monomer bound to one RNA molecule (67.03 ± 0.05 kDa) became visible. Between 5 and 30 min after the start of the measurement, NP-RNA complexes assembled corresponding to NP2-RNA2 (134.03 ± 0.05 kDa) and NP3-RNA3 (201.17 ± 0.13 kDa) species. As in the previous experiment, the signal for ring-structured NP3 decreased over time. The NP3-RNA3 complexes were most abundant after around 30 min of measurement. Notably, NP/RNA ratios of the complexes were always stoichiometric after the initial disassembly of the ring-structured NP3 via NP3-RNA1. We increased the RNA concentration to a molar ratio of 1:2 (NP/RNA) to further evaluate the formation of higher order RNA-NP complexes, which was significantly faster compared with using an equimolar ratio (Figure 7C). Although the NP3 and the NP3-RNA1 complex (193.3 ± 0.09 kDa), likely an intermediate, were observable at early time points of the measurement, the decay rate of the trimeric ring structure was higher than observed in the previous experiment with an equimolar NP/RNA ratio, and NP3 had completely disappeared after 40 min. In this experiment, we observed the formation of NP1RNA1 (67.03 ± 0.05 kDa), NP2-RNA2 (134.03 ± 0.05 kDa), NP3-RNA3 (201.17 ± 0.13 kDa), and NP4-RNA4 (268.25 ± 0.05 kDa) complexes. Small fractions of NP5-RNA5 (335.78 kDa, fwhm: 0.90 kDa) and NP6-RNA6 (393.18 kDa, fwhm: 1.12 kDa) were also detectable (Figure S9A). In addition to the 12 nt RNA-NP complexes, we detected NP monomers bound to truncated RNAs (RNAtr). As we did not observe any RNAtr as free species, the NP1-RNAtr complexes were most likely a result of an onset of CID, clipping off nucleotides not covered by the RNA-binding groove of NP (Figure S9B). These NP1-RNAtr complexes corresponding to 8–9 bases bound to NP were observed with different RNA substrates of varying lengths. This is in line with the previously reported crystal structures of NP bound to RNA, where 6 nts were fit into the RNA-binding groove.10 Furthermore, we can rule out that the assembly between NP molecules happened via base pairing of overhanging RNA, as with our acceleration voltage settings only single RNAs were observed (Figure S10).
Figure 7.
NP3 dissociation and stepwise assembly of nucleocapsids. (A) Schematic summary of NP3 dissociation and NP/RNA complex assembly over time as detected in the experiment. (B) Representative measurement of NP (9 μM) together with a 12 nt RNA in a 1:1 (NP/RNA) molar ratio recorded for 45 min in a 150 mM ammonium acetate solution at pH 7.5. Spectra of all time points were deconvoluted with Unidec.30 Shown are spectra at 1 min after starting the measurement (left) and at 30 min after starting the measurement (middle) with the intensity in arbitrary units (a.u.). The cartoons represent the complexes of NP and NP-RNA. Masses correspond to NP3 (189.56 ± 0.02 kDa), NP3-RNA1 (193.63 ± 0.09 kDa), NP1-RNA1 (67.03 ± 0.05 kDa), NP2-RNA2 (134.03 ± 0.05 kDa), and NP3-RNA3 (201.17 ± 0.13 kDa). The normalized intensity for every mass species from the deconvoluted spectra were plotted according to the different time points. The sum of all mass species of one time point was normalized to 1 (right). (C) Representative measurement of NP together with a 12 nt RNA in a 1:2 (NP/RNA) molar ratio. Spectra were analyzed as in (B). Masses corresponding to NP3 (189.56 ± 0.02 kDa), NP3-RNA1 (193.63 ± 0.09 kDa), NP1-RNA1 (67.03 ± 0.05 kDa), NP2-RNA2 (134.03 ± 0.05 kDa), NP3-RNA3 (201.17 ± 0.13 kDa), and NP4-RNA4 (268.25 ± 0.05 kDa).
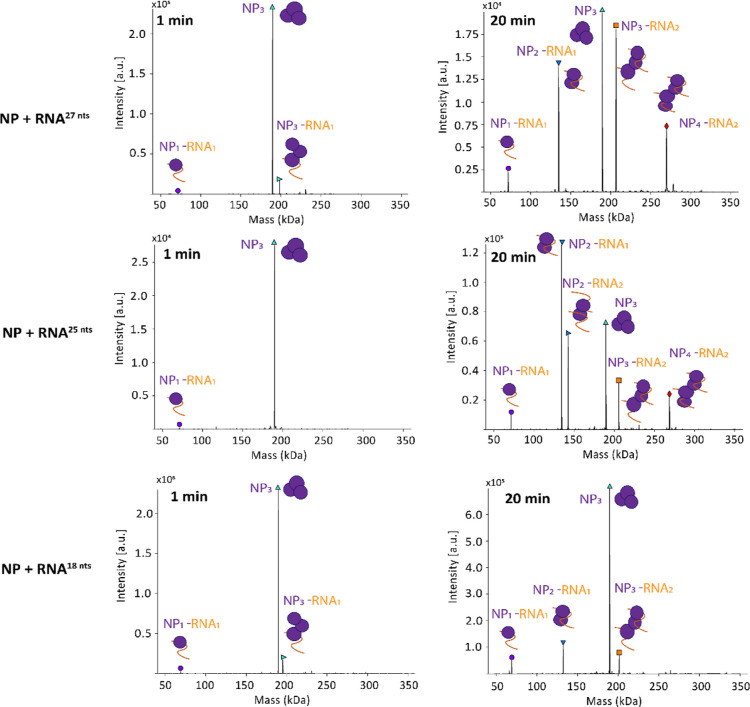
We noticed that the NP-RNA complexes consistently had a 1:1 ratio. We included longer RNAs in our analysis to see whether those would be able to bind to two NPs. For this, we used three different RNAs with lengths of 18, 25, and 27 nucleotides (Figure 8). Like with the previously used RNAs at early time points, the most abundant species was NP3. We also detected masses corresponding to NP3-RNA1 and NP1-RNA1 at the 1 min time point. Also here, NP was assembled with RNA in higher order NP-RNA assemblies. However, we observed NP-RNA complexes in which RNA is bound substoichiometrically. At the 20 min time point, signals corresponding to NP2-RNA1, NP3-RNA2, assemblies were detectable with all tested RNAs and NP4-RNA2 with 25 and 27 nt RNAs. The signal for NP3 and NP3-RNA1 decreased. We concluded that a certain RNA length is needed to allow multiple NPs to bind to the same RNA molecule. The critical RNA length lies between 13 and 18 nucleotides. Notably, the NP-RNA assembly kinetics seemed to be dependent on the RNA length. We detected more assembly products with longer RNA (27 nt) compared to the shorter RNA (18 nt), suggesting a more efficient process.
Figure 8.
Representative measurement of NP together with a 27, 25, and 18 nt RNA in a 1:1 (NP/RNA) molar ratio. Spectra were analyzed as in Figure 7. Time points at 1 and 20 min are depicted. For the 27 nt RNA, masses corresponding to NP3 (189.56 ± 0.02 kDa), NP3-RNA1 (198.19 ± 0.05 kDa), and NP1-RNA1 (71.79 ± 0.03 kDa) were detectable at the 1 min time point. At the 20 min time point, additional masses corresponding to NP2-RNA1 (134.95 ± 0.04 kDa), NP3-RNA2 (206.69 ± 0.04 kDa), and NP4-RNA2 (269.92 ± 0.05 kDa) were visible. For the 25 nt RNA, masses corresponding to NP3 (189.56 ± 0.02 kDa) and NP1-RNA1 (71.16 ± 0.01 kDa) were detectable at the 1 min time point. At the 20 min time point, additional masses corresponding to NP2-RNA1 (134.29 ± 0.01 kDa), NP3-RNA2 (205.44 ± 0.02 kDa), and NP4-RNA2 (268.62 ± 0.02 kDa) were visible. For the 18 nt RNA, masses corresponding to NP3 (189.56 ± 0.02 kDa), NP3-RNA1 (195.27 ± 0.01 kDa), and NP1-RNA1 (68.97 ± 0.03 kDa) were detectable at the 1 min time point. At the 20 min time point, additional masses corresponding to NP2-RNA1 (132.12 ± 0.03 kDa) and NP3-RNA2 (201.09 ± 0.03 kDa) were visible.
Altogether, these results show the higher-order assembly of NP and RNA, possibly resembling RNP formation during viral replication. Based on our data, we hypothesize that, as a first step, one molecule of NP within the RNA-free NP trimer binds to RNA, leading to an opening of the ring-like conformation, which is less stable than the closed ring-like trimer. After this trigger, the open trimer further disassembles into the RNA-bound NP monomers. Starting from that, higher-order NP-RNA complexes are then formed (Figure 7B). We could demonstrate that this process is dependent on the RNA concentration, as an excess of RNA relative to NP led to a faster trimer disassembly and higher-order RNP-like NP-RNA complex formation (Figure 7C). For multiple NP binding to the same RNA, a certain RNA length is needed, suggesting a critical distance between two NPs inside RNPs. With an extended RNA length, the assembly process appears to be more efficient (Figure 8).
NP Can Bind Z and RNA Simultaneously
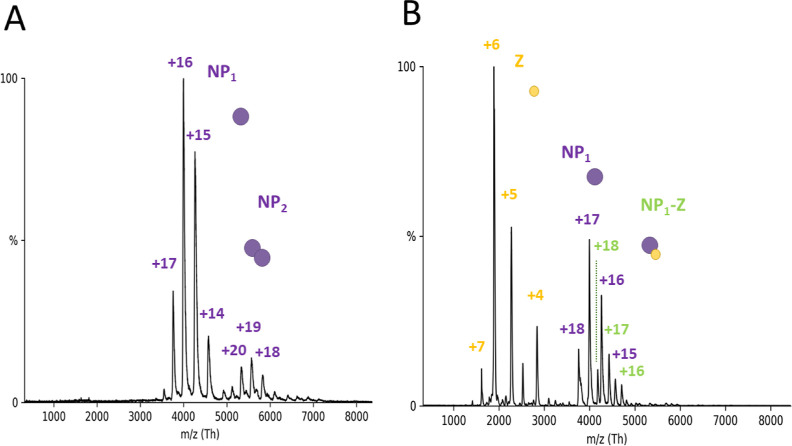
As described above, Z binds to the C-terminal domain of NP, whereas the RNA-binding pocket of NP is located in the N-terminal domain. Co-immunoprecipitation data has previously shown for other arenaviruses that NP and Z, either directly or mediated by other factors, interact with each other.7,15,31 It has been hypothesized that the NP–Z interaction mediates virion assembly by recruiting viral RNPs to the plasma membrane. We therefore investigated whether LASV NP could indeed bind to Z and RNA simultaneously. Mixing a 9 nt RNA, NP, and Z in a 1:1:2 ratio for subsequent nMS measurements, the NP3 ring conformation again dissociated into RNA-bound monomers (Figure 9A) (66.08 ± 0.03 kDa), some of which were associated with Z (77.38 ± 0.06 kDa; Figure 9B). CID-MS/MS confirmed these results (Figure 9C,D). In both cases, the RNA remained bound to NP at high collision voltage, whereas Z dissociated. This indicates a strong ionic interaction between RNA and NP compared to a weaker interaction between NP and Z. This observation fits with our proposed model of NP–Z interaction, which shows a rather small protein–protein interface between the C-terminal domain of NP and Z (Figure 4B). The affinity between wild-type NP and Z for the NP1-Z1-RNA1 complex appeared to be in the same range when using the NP-R52A trimerization mutant with a KD around 30 μM. Taken together, these results show that the NP1–RNA1 complex can indeed interact with Z, supporting the idea that Z mediates the recruitment of RNPs to the plasma membrane by direct interaction of Z with RNA-bound NP.
Figure 9.
NP can bind Z and RNA simultaneously. (A) Full nMS spectrum of NP with Z and 9 nt RNA in a 150 mM ammonium acetate solution at pH 7.5. NP3 completely dissociates into RNA-bound monomers. (B) Closeup of the m/z range 3000–6000 for determination of masses corresponding to NP-Z and RNA-NP-Z complexes. MS/MS of peaks corresponding to NP-Z isolated at 4124 m/z (C) and NP1-Z1-RNA1 isolated at 4549 m/z (D) at low and high acceleration voltage in the collision cell. CID products for the NP1-Z1-RNA1 complexes led to masses corresponding to Z monomers and NP1-RNA1 complexes, whereas no CID products for the NP–RNA complex were observed at a higher acceleration voltage.
NP–Z Interaction is pH-Dependent
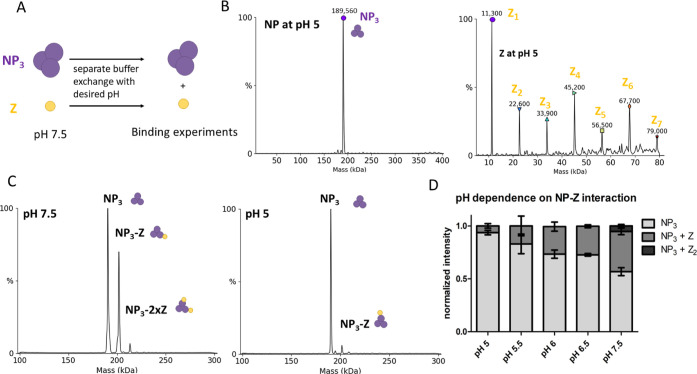
If the NP–Z interaction is responsible for the recruitment of RNPs into budding virions, we wondered what triggers the release of the RNPs upon virus entry and membrane fusion. A potentially important factor could be the pH, as LASV is taken up via the endosomal pathway. It was recently shown that acidification of Lassa virions occurs before membrane fusion via reorganization of the glycoprotein complex.32,33 A low pH could lead to the dissociation of RNPs from the Z protein lining the inner side of the viral membrane. Therefore, we evaluated the pH-dependence of the NP–Z interaction by incubating the proteins at varying pH values between 5 and 7.5 and subsequently measured NP–Z complex formation by nMS (Figure 10A). The first step was to evaluate the protein homogeneity after changing the buffer to a lower pH. NP showed a similar oligomerization behavior at pH 5 and pH 7.5, with the masses still corresponding to a trimer (Figure 10B). In contrast, Z started to homooligomerize after changing the pH to 5, forming up to homohexameric complexes (Figures 10B and S11). This is in line with an observation on influenza matrix protein that also undergoes oligomerization at low pH.34 However, we observed very similar protein concentrations at both pH 5 and pH 7.5 after buffer exchange and high-speed centrifugation, indicating that no unspecific protein aggregation happens during buffer exchange. To test the pH-dependence of the NP–Z interaction, we recorded 3 independent spectra of a mixture of NP and Z at a 1:3 (NP/Z) molar ratio at pH 5.0, 5.5, 6.0, 6.5, and 7.5. We found that almost no NP–Z interaction was observed at pH 5 while complex formation at pH 7.5 was as observed in the previous experiments (Figure 10C). At pH values between 5.5 and 6.5, NP–Z interaction was also less abundant compared to pH 7.5 (Figure 10D). These experiments show that the interaction between NP and Z is highly pH-dependent. This supports our hypothesis that the endosomal acidic pH reduces the NP–Z interaction and, as a result, RNPs are released from the viral matrix (Figure 11). We further investigated the effect of lower pH on NP–RNA binding to support our model. We observed NP–RNA interaction with 9 nt and 12 nt RNA down to pH 5.0. In both cases, we observed trimer dissociation and formation of NP–RNA complexes analogous to pH 7.5. There is no indication of altered affinity. Notably, dissociation kinetics of the trimer in the presence of RNA were accelerated (Figure S12).
Figure 10.
NP–Z interaction is less efficient at lower pH. (A) Scheme of the pH-dependent interaction assay. Proteins were separately exchanged to a 150 mM ammonium acetate surrogate with adjusted pH. NP (6 μM) and Z at the same pH were incubated in a 1:3 (NP/Z) molar ratio and measured by nMS. (B) Deconvoluted spectra of NP and Z after the buffer was changed to pH 5.0. NP oligomerization status was unaltered, whereas Z starts to oligomerize at pH 5.0 and 5.5. (C) Spectra of NP and Z in a 1:3 molar ratio at different pH. (D) Normalized intensity for every mass species from the deconvoluted spectra was plotted according to the different pH conditions. The sum of all mass species for one measurement was set to 1. Error bars represent the standard deviation of at least 3 independent measurements.
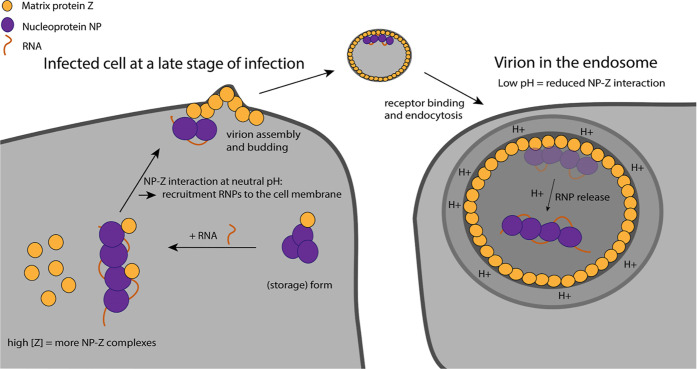
Figure 11.
Proposed model of the NP–Z interaction during the viral life cycle. At a late stage of infection, the cellular concentration of Z is high favoring for NP–Z interaction. This leads to recruitment of RNPs to the cell membrane by myristoylated Z for virion assembly (left). During virus entry into the host cell via the endosomal pathway, the lower pH leads to the dissociation of NP from Z, which coats the inner virion membrane. This leads to the release of RNPs into the cytosol.
Discussion
NP is known to form trimers in the absence of RNA and during recombinant purification with its atomic structure solved.9 The trimeric NP can be isolated from RNP-containing cells alongside full RNP complexes, demonstrating the existence of this complex in infected cells.23,35 It was speculated that the trimer serves as a storage form for the NP in cells. The ability of NP to trimerize was found to be essential for viral genome replication and transcription but was dispensable for interferon antagonism and NP recruitment into budding virions.23 Many viruses use specific complexes (e.g., NP–phosphoprotein complex in nonsegmented RNA viruses) to prevent unspecific RNA binding to the NP. It was therefore suggested that the trimeric storage complex of LASV NP acts in a similar way.10,36 Structurally, the trimeric ring conformation of arenavirus NP is unable to bind RNA. To enable this, the C-terminal domain must first rotate away from the RNA-binding pocket, allowing one helix to become partly unstructured (α5) and a second helix (α6) to shift away from the RNA-binding pocket.10,37 To induce this conformational change, it was speculated that specific (host-)factors are needed.10 In our experiments, we demonstrated that short RNAs are sufficient to induce this conformational change and trigger NP trimer dissociation. This was demonstrated by nMS and validated by negative stain electron microscopy and mass photometry, which were performed in physiologically more relevant buffers. Yet, how does NP primarily associate with viral RNA instead of host RNA? We did not observe any sequence specificity as NP bound to all random RNAs used in our experiments, which is in line with other groups reporting randomly bound RNAs to LASV NP.10,25 However, it cannot be ruled out that the LASV RNA genome contains some specific structural and sequence motifs (packaging signals) or additional modifications that could lead to a lower threshold for RNA binding. These signals would have to be different in the genomic RNA compared with viral mRNA, which is usually not encapsidated. It also remains unclear whether and how specific recruitment of all genome segments is controlled in bunyaviruses, as specific RNA secondary structures, as present in influenza viruses,38 seem to be missing. Overall, it is likely that another (host-)factor is needed to block unspecific RNA binding to NP, potentially by further stabilizing the trimeric NP. For the Rift Valley fever virus, nonselective recruitment of viral genome segments into budding virions was found to be the most likely scenario.39−41 Another factor favoring viral RNA binding could be the local RNA concentration. In our experiments, we used relatively high RNA concentrations (1:1 or 1:2 molar ratio) and observed the dissociation and assembly processes in real time, pointing to rather slow RNA-binding kinetics. Several scenarios are conceivable in support of the idea of local viral RNA concentration as a key determinant for the preferred and efficient packaging of viral genomic RNA:
-
(i)
Direct interaction of NP with the L protein for close proximity to nascent viral RNA. NP is a critical component of the arenavirus replication machinery. A direct interaction of NP and L was suggested based on coimmunoprecipitation studies, although an interaction mediated by RNA or host factors could not be ruled out in this case.42,43
-
(ii)
Indirect interaction of NP with L during viral genome replication via a host protein. This has been observed for the influenza virus polymerase complex, where the host acidic nuclear protein (ANP32) enables L–L interaction and also binds to NP to support encapsidation of nascent viral RNA and re-encapsidation of the template.44,45
-
(iii)
Formation of local microenvironments for viral genome replication and transcription within the cell. Bunyavirus NPs have been detected in nonmembranous compartments such as processing bodies and stress granules in cells.46−49 Other studies reported NP-containing cytosolic puncta forming upon arenavirus infection associated with cellular membranes. These puncta were concluded to contain full-length genomic and antigenomic RNAs along with proteins involved in cellular mRNA metabolism reminiscent of classical replication–transcription complexes.50 Interestingly, phosphorylation of LCMV NP at position T206 was found to be critical for the formation of these cytosolic puncta.51 However, there is no evidence that the L protein is indeed present in these puncta, and the presence of these local puncta would not provide an explanation on why mRNAs are not encapsidated. It therefore remains unclear what exactly determines viral RNA packaging by NP in addition to the presence of the RNA molecule itself.
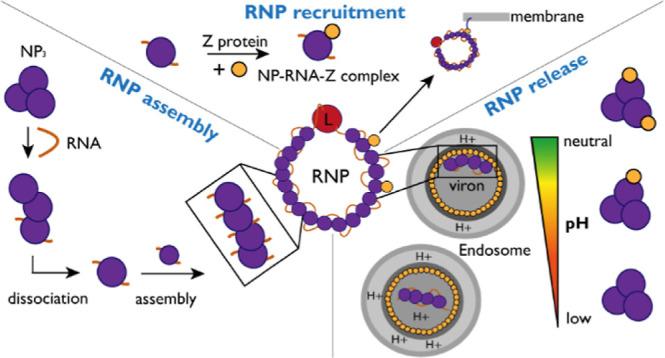
In our experiments, we could observe NP trimer dissociation into RNA-bound NP monomers, which then assembled into higher-order NP–RNA structures. NP–RNA higher-order complex formation seemed to require a certain RNA length (12 nt), indicating an additional stabilizing effect of the RNA or even a secondary RNA binding site in NP as observed for other segmented negative-strand RNA viruses.52 Moreover, we could only identify two NPs per RNA with RNA oligos of 18, 25, and 27 nt length. This demonstrates that a specific RNA length is needed and possibly a secondary binding site for multiple NPs binding to one RNA. We did not observe NP3-RNA1 complexes with the 27 nt RNA after initial disassembly, which would be in theory enough to cover three binding sites as we observe NP2RNA1 complexes with the 18 nt RNA. This further supports a possible secondary RNA binding site required for assembly. Interestingly, during the NP trimer disassembly process we observed a complex of one RNA molecule bound to the NP trimer (NP3-RNA1) although according to our interpretation of the available structural data, RNA binding is not possible in the trimeric ring conformation.9 We propose that the observed NP3-RNA1 state is therefore likely a disassembly intermediate or RNA is bound via a potential secondary binding site. Mechanistically, the process can be explained by two hypotheses. The trimer needs to disassemble, allowing the canonical RNA binding sites to be open. This can either be achieved by (i) RNA binding to a secondary binding site, destabilizing the trimer or (ii) an “open” trimer conformation, which allows one RNA molecule to be bound by the canonical binding site. The open conformation is less stable, and therefore the trimer quickly disassembles into monomers which immediately bind RNA. This indicates overall a cooperative dissociation mechanism.
It has long been known that arenavirus NP and Z are acting together based on cellular experiments, such as VLP assays, coimmunoprecipitation, and colocalization studies.11,13,15,53 Here we showed that NP and Z directly and independent of any host factor interact with each other. Importantly, monomeric NP in the presence or absence of RNA has a substantially higher affinity for Z compared to the NP3 storage form, which is not recruited to the membrane. We mapped the NP–Z interaction site with an integrative structural approach of nMS and Alphafold Multimer-powered complex prediction. We further validated the model with HDX-MS which was performed in a physiologically more related buffer. Our data point toward Z binding to the C-terminal domain of NP, likely involving the side chains of R16 and R74 of Z. Previous coimmunoprecipitation studies already suggested the C-terminal domain of NP to be important for the interaction with Z protein, either directly or mediated by cellular factors.7 Several conserved Z residues, including R16 and T73, were also demonstrated to be important for inhibition of viral RNA synthesis and virus-like particle (VLP) infectivity, although R16 and T73 are not involved in the binding of Z to the L protein.54−56 This points toward the importance of R16 and T73 conservation for Z localization and/or proper folding. However, we could not detect a difference in the protein folding when comparing Z R16A to the wild-type Z in vitro.
It was hypothesized that Z residues R16, T73, and others were involved in RNP recruitment via binding to NP and/or viral RNA.54 Our experimental data and the proposed model support this hypothesis, as we find R16 and T73 to be located in the interface between NP and Z, while we did not detect an interaction between Z and RNA in vitro as observed for the matrix protein of other negative-strand RNA viruses.56 Upon RNA packaging, RNPs are recruited into nascent virions, and Z has been shown to be critical for budding of most arenaviruses.57 While Z alone is sufficient to drive the formation of virus-like particles (VLPs), the process is significantly enhanced when NP is present.15 For the Tacaribe virus, the so-called late-domain motifs ASAP and YxxL of Z have an influence on the NP incorporation, with the Y56 in the YxxL motif being critical. These data suggest a role of the late-domain motif YxxL in the NP–Z interaction.58 Interestingly, although the YxxL motif is also present in LASV Z, the Y48 of the YxxL motif is not in the proposed NP–Z interface in our model but rather on the opposite side of the Z molecule. However, the proposed location of this motif in our model is in agreement with another study reporting the binding of Z to a host protein required for budding via the YxxL motif.59
The data shown here support the concept that the NP–Z interaction is important for the recruitment of RNPs to the plasma membrane and incorporation into nascent virions. We demonstrate that RNA-bound NP is able to bind Z, as would be expected for RNP recruitment.
Arenavirus cell entry is mediated by the glycoprotein complex. After receptor binding, virions enter the host cell via the endosomal pathway.60,61 Our data show that the interaction between NP and Z is highly pH-dependent, with a decreased NP–Z interaction at pH values lower than pH 7.5. Therefore, it is conceivable that RNP-Z dissociation triggered by a lower pH in the endosome leads to the release of RNPs into the cytosol after viral membrane fusion (see model Figure 11). Unlike influenza viruses, arenaviruses do not have proton channels in their lipid envelope that actively acidify the virion upon endocytosis. However, it has recently been shown for LASV that protons passively cross the viral membrane, which leads to acidification of the virus interior prior to membrane fusion.32,33 Similar observations have been reported for Ebola virus, where acidification of virions leads to viral matrix reorganization.62 Besides the pH-dependent NP–Z interaction, we observed that Z starts to oligomerize between pH 5 and 5.5. We hypothesize that stronger Z–Z interaction at low pH, as indicated by oligomerization, further reduces availability of Z for the low-affinity Z–NP interaction. This is in line with several studies that report pH as a critical factor for matrix protein oligomerization and protein conformation.34,63−66 Matching our observations, a lower pH at around 5.4 also induced weakening of influenza virus matrix protein M1-vRNP interactions.67
Our findings support a model in which viral RNA is necessary and sufficient to transform trimeric NP into monomeric RNA-bound, RNP-like NP assemblies. Z then binds to the RNP at a neutral pH in the cytosol or a cellular microenvironment via direct interaction with NP. This interaction facilitates targeted transport of the RNP to the plasma membrane at a high cellular concentration of Z protein during late stages of the viral “life” cycle and subsequent virion budding. Upon entry of a virus into a host cell, the RNPs are released into the cytosol from the virion matrix triggered by the low pH in the endosome and following membrane fusion. Subsequently, viral genome replication and transcription take place.
Materials and Methods
Expression and Purification
Lassa Z (AV strain) was expressed with a pOPIN-J vector with an N-terminal GST tag and purified as described in ref (17). Mutations (if indicated) were introduced by mutagenic PCR before cloning into the pOPIN-J vector and expressed and purified according to the WT.
Lassa NP (AV strain) was expressed with a pOPIN-M vector and an N-terminal maltose-binding protein (MBP) fusion protein. Mutations (if indicated) were introduced by mutagenic PCR before cloning into pOPIN-M. Expression was performed in BL21 in E. coli strain BL21 Gold (DE3) (Novagen) at 37 °C and grown in 1 L cultures in terrific growth media to an optical density at 600 nm of 1.0. Cells were induced with 250 μM isopropyl-β-d-thiogalactopyranoside and incubated overnight at 17 °C. Cells were harvested by centrifugation and lysed by sonification in 50 mM NaH2PO4, pH 8.0, 500 mM NaCl, 5% (w/w) glycerol, 1 mM phenylmethylsulfonyl fluoride, 0.05% (v/v) TritonX-100, 0.025% (w/v) lysozyme, and 2 mM dithiothreitol. The lysate was clarified by centrifugation at 30,000g for 30 min at 4 °C and incubated for 45 min with amylose beads (GE Healthcare). The loaded protein was cleaved on the column by GST-tagged 3C protease at 4 °C overnight. The NP without MBP-containing buffer was diluted to reach a 150 mM NaCl concentration by adding 20 mM NaH2PO4 pH 8 and loaded on a heparin column (GE Healthcare) to remove bound bacterial RNA. NP was eluted by an NaCl gradient and concentrated for further purification by size-exclusion chromatography on an SD200 column (GE Healthcare) in 50 mM Tris (HCl) pH 7.5, 150 mM NaCl, 5% (w/w) glycerol, and 2 mM dithiothreitol. Proteins were concentrated using filter units (Amicon Ultra, 30,000 Da cutoff), flash frozen in liquid nitrogen, and stored at −80 °C.
Native MS Sample Preparation
Proteins for native MS analysis were buffer-exchanged into 150 mM ammonium acetate (99.99% purity) at pH 7.5 (if not indicated otherwise) by at least three cycles of centrifugation at 13,000 g and 4 °C in centrifugal filter units (Amicon 3000 or 30,000 molecular cutoff and 500 μL volume). Proteins were centrifuged at 20,000g for 15 min at 4 °C to clear the sample from possible protein aggregates occurring during the buffer exchange. Protein concentrations after buffer exchange ranged from 5 to 100 μM. Proteins were mixed with or without ligands on ice and subsequently introduced to a mass spectrometer.
Samples were introduced to the mass spectrometer by gold-coated capillaries made from borosilicate glass tubes (inner diameter: 0.68 mm; outer diameter: 1.2 mm; World Precision Instruments). Capillaries were pulled in a two-step program using a micropipette puller (P-1000, Sutter Instruments) with a squared box filament (2.5 mm by 2.5 mm, Sutter Instruments). Capillaries were afterward gold-coated using a sputter coater (5.0 × 10–2 mbar, 30.0 mA, 100 s, three runs to vacuum limit 3.0 × 10–2 mbar argon, CCU-010, Safematic).
Native MS Data Acquisition
Data acquisition was performed on a nanoESI Q-ToF II mass spectrometer (Waters/Micromass) modified for high mass experiments in positive ion mode.68 Samples were ionized with voltages of 1450 V at the capillary and 150 V at the cone. The pressure of the in-source region was constantly at 10 mbar. Pressure in the collision cell was at 1.5 × 10–2 mbar argon. Quaternary structure of protein complexes was determined in MS/MS mode with acceleration voltage ranging between 50 and 400 V across the collision cell. The quadrupole profile was 1000 to 10,000 m/z throughout all experiments. Cesium iodide (25 mg/mL) in aqueous solution was used to calibrate the instrument and the data.
The NP-RNA assembly experiment (Figures 7 and 8) was performed on a Q Exactive ultrahigh mass range (UHMR) Orbitrap mass spectrometer (Thermo Fisher Scientific) in positive ion mode with a mass range from 350 to 20,000 m/z and a nano ESI source. The following parameters were used. Capillary voltage was kept at 1.2 kV, source temperature at 50 °C, noise level parameter at 4.64, S-lens radio frequency level 200, in source CID at 5 eV, resolution at 6250 setting value, and ion injection time of 100 ms. Nitrogen was used in the HCD cell with a trapping gas pressure of 5 setting value with ultrahigh vacuum readout at around 2.5 × 10–10 mbar. HCD energy setting was at 100. Instrument detector parameters were set to low m/z. Ten microscans were summed into 1 scan.
Data Analysis for Native MS
Raw data was analyzed with MassLynx V4.1 (Waters) for QToF2 data sets or Xcalibur for UHMR data sets by combining spectra and exporting them into a txt file format. Following that, evaluation regarding full width at half-maximum (fwhm) was done with mMass (Martin Strohalm69). Experimental mass, AUC, and visualization was evaluated with Unidec (Michael T. Marty30). KD determination was calculated as per ref (70). The values for all recorded masses and fwhm were obtained from at least three independent measurements and are provided in Table S4.
HDX-MS
NP protein (50 pmol) was mixed with the Z protein in a 1:5 molar ratio (NP/Z). The deuterium exchange reaction was started by a 1:9 dilution into 99% deuterated buffer (40 mM Tris(HCl) pH 7.5, 150 mM NaCl) at 25 °C. The exchange reaction was quenched after the following time points: 15 s, 1 min, 10 min, 1 h, and 6 h. The quenching was initiated by a 1:1 addition of ice-cold quench buffer (1 M glycin, pH 2.3). After that, the samples were flash-frozen in liquid nitrogen. Additionally, a fully deuterated (FD) control was prepared where NP was diluted 1:9 in 99% deuterated 40 mM Tris(HCl) pH 7.5, 150 mM NaCl, and 6 M urea buffer. The reaction was quenched after 24 h of incubation at room temperature, as described above. All time points were analyzed in three technical replicates.
The samples were shortly centrifuged after thawing and injected onto a cooled (2 °C) HPLC System (Agilent Infinity 1260, Agilent Technologies, Santa Clara, CA, USA) which includes a home-packed pepsin column (IDEX guard column with 60 μL of Porozyme immobilized pepsin beads, Thermo Scientific, Waltham, MA, USA) in a column oven 5 °C), a peptide trap column (OPTI-TRAP for peptides, Optimize Technologies, Oregon City, OR, USA), and a reversed-phase analytical column (PLRP-S for Biomolecules, Agilent Technologies, Santa Clara, CA, USA). Peptide digestion was carried out online at a flow rate of 75 μL/min (0.4% formic acid in water) and washed after every run with 2 M urea, 2% acetonitrile, and 0.4% formic acid. Peptide separation prior to MS analysis was performed on an analytical column at a flow rate of 150 μL/min and a gradient of 8–40% buffer B in 7 min (buffer A: 0.4% formic acid in water; buffer B: 0.4% formic acid in acetonitrile). The analytical column was washed with 100% buffer B after every sample. The HPLC was connected to an Orbitrap Fusion Tribrid in positive ESI MS only mode (Orbitrap resolution of 120,000, 4 microscans, Thermo Scientific, Waltham, MA, USA).
Peptide identification was evaluated with 100 pmol nondeuterated samples. A gradient from 8 to 40% buffer B in 27 min was used for the analytical column with a flow rate of 150 μL/min. MS analysis was performed in data-dependent MS/MS acquisition mode (Orbitrap resolution 120,000, 1 microscan, HCD 30 with dynamic exclusion). Raw data are available in the pride database entry.
HDX Data Analysis
Peptides were searched against the protein sequence in MaxQuant (version 2.1.2.0) with the Andromeda search engine.71 The following MaxQuant settings were used: digestion mode was set to unspecific and peptides between 5 and 30 amino acids length were accepted. The minimum score for the identification of peptides was 20. The default mass tolerance for precursor (4.5 ppm) and fragment (20 ppm) ions was used according to the Thermo Orbitrap instrument. The retention time for peptides of the HDX runs was adjusted to the shorter gradient of the analytical column compared to the longer gradient of the peptide identification runs.
Data sets were analyzed according to the deuterium uptake of peptides via automated centroid analysis by HDExaminer Version 3.3 (Sierra Analytics). All peptides were checked manually based on the presence of overlapping peptide envelopes, correct retention time, m/z range, and charge state. All peptides at different states and time points are plotted as a wood plot (Figure S5) and difference heatmap according to the atomic range which considers partly overlapping peptides with no difference (Figure S6). A summary of all experimental conditions and statistics can be found in Table S2.
GraphPAD Prism (GarphPAD Software, Inc.) and PyMOL (Schrödinger) software were used for visualization. All evaluated data points are available in the database entry.
Negative-Stain Electron Microscopy
Purified NP at 0.01 mg/mL was applied to glow-discharged carbon-coated copper grids (Electron Microscopy Sciences) and stained with uranyl acetate immediately before imaging. Transmission electron microscopy images were collected using a Talos L120c microscope (120 kV) with a LaB6 thermionic source at 92,000× magnification and a defocus of between −0.2 and 0.5 μm. For the NP + RNA condition, NP at 0.01 mg/mL was mixed with a single-stranded 25mer (Table S3) at a 2 molar-excess to the NP monomer. The NP + RNA was incubated at room temperature for approximately 15 min and then imaged as for the NP without RNA condition.
Structural Modeling of LASV NP–Z Complex
To model the NP–Z complex, we used the “custom” mode of AlphaPulldown,26 a python package built upon AlphaFold27 and AlphaFold Multimer.28 Both the initial release of AlphaFold Multimer (version 2.1.0) and the latest version (version 2.3.0) were used. All parameters were set to default except for the max_recycles, which was increased from 3 to 12 for better model quality and to avoid steric clashes. The local quality of the model was assessed by predicted local distance difference test (pLDDT) scores, as returned by AlphaFold. The confidence in the relative arrangement between NP and Z proteins was evaluated by predicted aligned errors (PAE), also as returned by AlphaFold. Other evaluations of the model quality and properties, including Predicted DockQ score (pDockQ),72 protein-interface score (PI-score),73 and biophysical properties of the interaction interface (using PI-score program), were calculated by AlphaPulldown.
Mass Photometry
Mass photometry was performed on a Refeyn OneMP mass photometer using the programs Acquire MP v2.5.0 and Discover MP v2.5.0 (Refeyn Ltd.) and PhotoMol. The workflow was published previously.74 NP was measured in 50 mM Tris (HCl), 150 mM NaCl, 5% glycerol, and a concentration of 25 nM. For the NP + RNA condition, NP at 500 nM was mixed with a single-stranded 9mer (Table S3) at a 4 molar-excess to the NP monomer. NP + RNA was incubated at room temperature for approximately 5 min. The sample was diluted with 50 mM Tris (HCl), 150 mM NaCl, and 5% glycerol to a concentration of 25 nM and subsequently measured.
Experimental Design and Statistical Analysis
The determined masses, fwhm, and AUC from nMS analysis were taken from at least three independent measurements and are shown together with the resulting standard deviation. Values for quantification are shown as the mean of three independent measurements. The error bars represent the standard deviation. The shown standard deviation for KD values are calculated according to the rules of Gaussian error propagation. HDX-MS experimental design and data analysis was evaluated according to HDX-MS community recommendations.75 The quench conditions were optimized for maximum sequence coverage. The back exchange was tested with a fully deuterated control. The labeling time points cover 3–4 orders of magnitude. The deuterium uptake differences between two states were statistically evaluated by using a two paired t-test with the p-value <0.05. Peptides showing differences were only considered to be significant if they passed the t-test and additionally had a difference higher than 0.561 deuteron, which is the variance across all replicates of all peptides.
Acknowledgments
We thank Prof. H. Schlüter (UKE, University of Hamburg, Germany) for access to the high-resolution mass spectrometer for the HDX experiments. We would like to thank Janine-Denise Kopicki for helpful advice on the KD calculation. We thank Kay Grünewald for support and all current and past members of the Uetrecht and Rosenthal group for discussions and data interpretation. Part of this work was performed at the Multi-User CryoEM Facility at the Centre for Structural Systems Biology, Hamburg, supported by the Universität Hamburg and DFG grant numbers (INST 152/772-1|152/774-1|152/775-1|152/776-1|152/777-1 FUGG). We acknowledge the technical support by the SPC facility at EMBL Hamburg.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.3c07325.
nMS spectra of different oligomerization states of Z; overall area under the curve for the NP-Z mass species for NP WT and NP_R52A KD determination; characterization of the NP trimerization mutant R52A; estimated quality of structural models for LASV NP and Z protein complex; wood plot of HDX-MS data of NP together with Z: HDX-MS analysis of NP and NP together with Z; negative-stain electron microscopy of NP; AlphaFold model of the N–Z dimer (https://www.modelarchive.org/); and mass spectrometry raw data and the analyzed HDX data set (PRIDE database, Identifier PXD040186) (PDF)
Leibniz Center Infection Graduate School and Leibniz Interact (to M.R., C.U.), The Leibniz Institute of Virology was supported by the Free and Hanseatic City of Hamburg and the Federal Ministry of Health (to L.S., C.U.), German Federal Ministry of Education and Research 01KI2019 (to MR), DFG RO 5954/2–1 (to M.R.), DFG KO 5979/2–1 (to J.K.), Leibniz Association’s Leibniz competition program K72/2017 (to M.R.) and SAW-2014-HPI-4 grant (to C.U.), Horizon 2020 ERC StG-2017 759661 (to C.U.), BWFBG of the Free and Hanseatic City of Hamburg for an equipment grant (C.U.), Wilhelm and Maria Kirmser-Stiftung (to M.R.)
The authors declare no competing financial interest.
Supplementary Material
References
- Brito A. F.; Pinney J. W. Protein-Protein Interactions in Virus-Host Systems. Front. Microbiol. 2017, 8, 1557. 10.3389/fmicb.2017.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam H. J.; Hallam S.; Rodriguez S. E.; Barrett A. D.; Beasley D. W.; Chua A.; Ksiazek T. G.; Milligan G. N.; Sathiyamoorthy V.; Reece L. M. Baseline mapping of Lassa fever virology, epidemiology and vaccine research and development. npj Vaccines 2018, 3 (1), 11. 10.1038/s41541-018-0049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitting R.; Kafetzopoulou L. E.; Thiery W.; Dudas G.; Gryseels S.; Kotamarthi A.; Vrancken B.; Gangavarapu K.; Momoh M.; Sandi J. D.; et al. Predicting the evolution of the Lassa virus endemic area and population at risk over the next decades. Nat. Commun. 2022, 13 (1), 5596. 10.1038/s41467-022-33112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auperin D. D.; Romanowski V.; Galinski M.; Bishop D. H. Sequencing studies of pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J. Virol. 1984, 52 (3), 897–904. 10.1128/jvi.52.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J.; Novella I. S.; Teng M. N.; Oldstone M. B.; de La Torre J. C. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 2000, 74 (8), 3470–3477. 10.1128/JVI.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinschewer D. D.; Perez M.; de la Torre J. C. Role of the Virus Nucleoprotein in the Regulation of Lymphocytic Choriomeningitis Virus Transcription and RNA Replication. J. Virol. 2003, 77 (6), 3882–3887. 10.1128/JVI.77.6.3882-3887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levingston Macleod J. M.; D’Antuono A.; Loureiro M. E.; Casabona J. C.; Gomez G. A.; Lopez N. Identification of two functional domains within the arenavirus nucleoprotein. J. Virol. 2011, 85 (5), 2012–2023. 10.1128/JVI.01875-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K. M.; Kimberlin C. R.; Zandonatti M. A.; MacRae I. J.; Saphire E. O. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc. Natl. Acad. Sci. U.S.A. 2011, 108 (6), 2396–2401. 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunotte L.; Kerber R.; Shang W.; Hauer F.; Hass M.; Gabriel M.; Lelke M.; Busch C.; Stark H.; Svergun D. I.; et al. Structure of the Lassa Virus Nucleoprotein Revealed by X-ray Crystallography, Small-angle X-ray Scattering, and Electron Microscopy *. J. Biol. Chem. 2011, 286 (44), 38748–38756. 10.1074/jbc.M111.278838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K. M.; Liu T.; Li S.; King L. B.; Ngo N.; Zandonatti M. A.; Woods V. L.; de la Torre J. C.; Saphire E. O. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc. Natl. Acad. Sci. U.S.A. 2011, 108 (48), 19365–19370. 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler R.; Strecker T.; Kolesnikova L.; ter Meulen J.; Weissenhorn W.; Becker S.; Klenk H. D.; Garten W.; Lenz O. Characterization of the Lassa virus matrix protein Z: electron microscopic study of virus-like particles and interaction with the nucleoprotein (NP). Virus Res. 2004, 100 (2), 249–255. 10.1016/j.virusres.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Salvato M. S.; Schweighofer K. J.; Burns J.; Shimomaye E. M. Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res. 1992, 22 (3), 185–198. 10.1016/0168-1702(92)90050-J. [DOI] [PubMed] [Google Scholar]
- Perez M.; Craven R. C.; de la Torre J. C. The small RING finger protein Z drives arenavirus budding: Implications for antiviral strategies. Proc. Natl. Acad. Sci. U.S.A. 2003, 100 (22), 12978–12983. 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M.; Greenwald D. L.; de La Torre J. C. Myristoylation of the RING Finger Z Protein Is Essential for Arenavirus Budding. J. Virol. 2004, 78 (20), 11443–11448. 10.1128/JVI.78.20.11443-11448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabona J. C.; Levingston Macleod J. M.; Loureiro M. E.; Gomez G. A.; Lopez N. The RING Domain and the L79 Residue of Z Protein Are Involved in both the Rescue of Nucleocapsids and the Incorporation of Glycoproteins into Infectious Chimeric Arenavirus-Like Particles. J. Virol. 2009, 83 (14), 7029–7039. 10.1128/JVI.00329-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro M. E.; D’Antuono A.; Levingston Macleod J. M.; López N. Uncovering Viral Protein-Protein Interactions and their Role in Arenavirus Life Cycle. Viruses 2012, 4 (9), 1651–1667. 10.3390/v4091651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel D.; Rosenthal M.; Gogrefe N.; Reindl S.; Günther S. Biochemical characterization of the Lassa virus L protein. J. Biol. Chem. 2019, 294 (20), 8088–8100. 10.1074/jbc.RA118.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouba T.; Vogel D.; Thorkelsson S. R.; Quemin E. R. J.; Williams H. M.; Milewski M.; Busch C.; Günther S.; Grünewald K.; Rosenthal M.; et al. Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity. Nat. Commun. 2021, 12 (1), 7018. 10.1038/s41467-021-27305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch P. J.; Whelan S. P. J. Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc. Natl. Acad. Sci. U.S.A. 2011, 108 (49), 19743–19748. 10.1073/pnas.1112742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.; Cong J.; Wang C.; Ji W.; Xin Y.; Qian Y.; Li X.; Chen Y.; Rao Z. Structural basis for recognition and regulation of arenavirus polymerase L by Z protein. Nat. Commun. 2021, 12 (1), 4134. 10.1038/s41467-021-24458-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Zhang S.; Zhang X. Structure of Machupo virus polymerase in complex with matrix protein Z. Nat. Commun. 2021, 12 (1), 6163. 10.1038/s41467-021-26432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K. M.; Zandonatti M.; Liu T.; Li S.; Woods V. L.; Saphire E. O.; Sundquist W. I. Crystal Structure of the Oligomeric Form of Lassa Virus Matrix Protein Z. J. Virol. 2016, 90 (9), 4556–4562. 10.1128/JVI.02896-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartz F.; Hoenen T.; Lehmann M.; Groseth A.; Garten W. The role of oligomerization for the biological functions of the arenavirus nucleoprotein. Arch. Virol. 2013, 158 (9), 1895–1905. 10.1007/s00705-013-1684-9. [DOI] [PubMed] [Google Scholar]
- Dülfer J.; Kadek A.; Kopicki J.-D.; Krichel B.; Uetrecht C. Structural mass spectrometry goes viral. Adv. Virus Res. 2019, 105, 189–238. 10.1016/bs.aivir.2019.07.003. [DOI] [PubMed] [Google Scholar]
- Qi X.; Lan S.; Wang W.; Schelde L. M.; Dong H.; Wallat G. D.; Ly H.; Liang Y.; Dong C. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 2010, 468 (7325), 779–783. 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D.; Chojnowski G.; Rosenthal M.; Kosinski J. AlphaPulldown≡a python package for protein-protein interaction screens using AlphaFold-Multimer. Bioinformatics 2023, 39 (1), 749. 10.1093/bioinformatics/btac749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J.; Evans R.; Pritzel A.; Green T.; Figurnov M.; Ronneberger O.; Tunyasuvunakool K.; Bates R.; Žídek A.; Potapenko A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596 (7873), 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R.; O’Neill M.; Pritzel A.; Antropova N.; Senior A.; Green T.. Protein complex prediction with AlphaFold-Multimer. bioRxiv [Preprint] 2021.10.04.463034, October 2021. [Google Scholar]
- Dülfer J.; Yan H.; Brodmerkel M. N.; Creutznacher R.; Mallagaray A.; Peters T.; Caleman C.; Marklund E. G.; Uetrecht C. Glycan-induced protein dynamics in human norovirus P dimers depend on virus strain and deamidation status. Molecules 2021, 26 (8), 2125. 10.3390/molecules26082125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty M. T.; Baldwin A. J.; Marklund E. G.; Hochberg G. K. A.; Benesch J. L. P.; Robinson C. V. Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Anal. Chem. 2015, 87 (8), 4370–4376. 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Riaño E.; Cheng B. Y. H.; de la Torre J. C.; Martínez-Sobrido L. The C-Terminal Region of Lymphocytic Choriomeningitis Virus Nucleoprotein Contains Distinct and Segregable Functional Domains Involved in NP-Z Interaction and Counteraction of the Type I Interferon Response. J. Virol. 2011, 85 (24), 13038–13048. 10.1128/JVI.05834-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Carlos de la Torre J.; Melikyan G. B. Human LAMP1 accelerates Lassa virus fusion and potently promotes fusion pore dilation upon forcing viral fusion with non-endosomal membrane. PLoS Pathog. 2022, 18 (8), e1010625 10.1371/journal.ppat.1010625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; York J.; Brindley M. A.; Nunberg J. H.; Melikyan G. B. Fusogenic structural changes in arenavirus glycoproteins are associated with viroporin activity. PLoS Pathog. 2023, 19 (7), e1011217 10.1371/journal.ppat.1011217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtykova E. V.; Baratova L. A.; Fedorova N. V.; Radyukhin V. A.; Ksenofontov A. L.; Volkov V. V.; Shishkov A. V.; Dolgov A. A.; Shilova L. A.; Batishchev O. V.; et al. Structural Analysis of Influenza A Virus Matrix Protein M1 and Its Self-Assemblies at Low pH. PLoS One 2013, 8 (12), e82431 10.1371/journal.pone.0082431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle J. D.; Whelan S. P. J. Isolation of Reconstructed Functional Ribonucleoprotein Complexes of Machupo Virus. J. Virol. 2021, 95 (22), e01054–01021 10.1128/JVI.01054-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyrat C.; Yabukarski F.; Tarbouriech N.; Ribeiro E. A. Jr; Jensen M. R.; Blackledge M.; Ruigrok R. W.; Jamin M. Structure of the vesicular stomatitis virus N0-P complex. PLoS Pathog. 2011, 7 (9), e1002248 10.1371/journal.ppat.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattis J. G.; May E. R. Markov State Model of Lassa Virus Nucleoprotein Reveals Large Structural Changes during the Trimer to Monomer Transition. Structure 2020, 28 (5), 548–554.e3. 10.1016/j.str.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y.; Goto H.; Watanabe T.; Yoshida T.; Kawaoka Y. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. U.S.A. 2003, 100 (4), 2002–2007. 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond D. D.; Piper M. E.; Gerrard S. R.; Smith J. L. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc. Natl. Acad. Sci. U.S.A. 2010, 107 (26), 11769–11774. 10.1073/pnas.1001760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichgers Schreur P. J.; Kortekaas J. Single-Molecule FISH Reveals Non-selective Packaging of Rift Valley Fever Virus Genome Segments. PLoS Pathog. 2016, 12 (8), e1005800 10.1371/journal.ppat.1005800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez-Méndez E.; Katrukha E. A.; Spruit C. M.; Kortekaas J.; Wichgers Schreur P. J. Visualizing the ribonucleoprotein content of single bunyavirus virions reveals more efficient genome packaging in the arthropod host. Commun. Biol. 2021, 4 (1), 345. 10.1038/s42003-021-01821-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle J. D.; Whelan S. P. Isolation of reconstructed functional ribonucleoprotein complexes of Machupo virus. J. Virol. 2021, 95 (22), e01054–01021 10.1128/jvi.01054-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber R.; Rieger T.; Busch C.; Flatz L.; Pinschewer D. D.; Kümmerer B. M.; Günther S. Cross-species analysis of the replication complex of Old World arenaviruses reveals two nucleoprotein sites involved in L protein function. J. Virol. 2011, 85 (23), 12518–12528. 10.1128/JVI.05091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Sheppard C. M.; Mistry B.; Staller E.; Barclay W. S.; Grimes J. M.; Fodor E.; Fan H. The C-terminal LCAR of host ANP32 proteins interacts with the influenza A virus nucleoprotein to promote the replication of the viral RNA genome. Nucleic Acids Res. 2022, 50 (10), 5713–5725. 10.1093/nar/gkac410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrique L.; Fan H.; Walker A. P.; Keown J. R.; Sharps J.; Staller E.; Barclay W. S.; Fodor E.; Grimes J. M. Host ANP32A mediates the assembly of the influenza virus replicase. Nature 2020, 587 (7835), 638–643. 10.1038/s41586-020-2927-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.; Mazur M.; Gulickx N.; Hong H.; Overmars H.; Tao X.; Kormelink R. Bunyaviral N Proteins Localize at RNA Processing Bodies and Stress Granules: The Enigma of Cytoplasmic Sources of Capped RNA for Cap Snatching. Viruses 2022, 14 (8), 1679. 10.3390/v14081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M. A.; Duran W. A.; Hjelle B. L.; Ye C.; Panganiban A. T. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (49), 19294–19299. 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins K. C.; Tartell M. A.; Herrmann C.; Hackett B. A.; Taschuk F.; Panda D.; Menghani S. V.; Sabin L. R.; Cherry S. Virus-induced translational arrest through 4EBP1/2-dependent decay of 5′-TOP mRNAs restricts viral infection. Proc. Natl. Acad. Sci. U.S.A. 2015, 112 (22), E2920–E2929. 10.1073/pnas.1418805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins K. C.; McLane L. M.; Maqbool T.; Panda D.; Gordesky-Gold B.; Cherry S. A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of Dcp2-accessible targets for cap-snatching. Genes Dev. 2013, 27 (13), 1511–1525. 10.1101/gad.215384.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird N. L.; York J.; Nunberg J. H. Arenavirus Infection Induces Discrete Cytosolic Structures for RNA Replication. J. Virol. 2012, 86 (20), 11301–11310. 10.1128/JVI.01635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopp K. A.; Ngo T.; Gershon P. D.; Buchmeier M. J. Single Nucleoprotein Residue Modulates Arenavirus Replication Complex Formation. mBio 2015, 6 (3), e00524 10.1128/mBio.00524-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng A. K.-L.; Zhang H.; Tan K.; Li Z.; Liu J.-h.; Chan P. K.-S.; Li S.-M.; Chan W.-Y.; Au S. W.-N.; Joachimiak A.; et al. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008, 22 (10), 3638–3647. 10.1096/fj.08-112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtanko O.; Imai M.; Goto H.; Lukashevich I. S.; Neumann G.; Watanabe T.; Kawaoka Y. A role for the C terminus of Mopeia virus nucleoprotein in its incorporation into Z protein-induced virus-like particles. J. Virol. 2010, 84 (10), 5415–5422. 10.1128/JVI.02417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capul A. A.; de la Torre J. C.; Buchmeier M. J. Conserved Residues in Lassa Fever Virus Z Protein Modulate Viral Infectivity at the Level of the Ribonucleoprotein. J. Virol. 2011, 85 (7), 3172–3178. 10.1128/JVI.02081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.; Peng R.; Peng Q.; Wang M.; Xu Y.; Liu S.; Tian X.; Deng H.; Tong Y.; Hu X.; et al. Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals. Nat. Microbiol. 2021, 6 (7), 921–931. 10.1038/s41564-021-00916-w. [DOI] [PubMed] [Google Scholar]
- Rodríguez L.; Cuesta I.; Asenjo A.; Villanueva N. Human respiratory syncytial virus matrix protein is an RNA-binding protein: binding properties, location and identity of the RNA contact residues. J. Gen. Virol. 2004, 85 (3), 709–719. 10.1099/vir.0.19707-0. [DOI] [PubMed] [Google Scholar]
- Strecker T.; Eichler R.; Meulen J. t.; Weissenhorn W.; Dieter Klenk H.; Garten W.; Lenz O. Lassa virus Z protein is a matrix protein sufficient for the release of virus-like particles. J. Virol. 2003, 77 (19), 10700–10705. 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A.; Wolff S.; Strecker T.; Hoenen T.; Becker S. Efficient Budding of the Tacaribe Virus Matrix Protein Z Requires the Nucleoprotein. J. Virol. 2010, 84 (7), 3603–3611. 10.1128/JVI.02429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtanko O.; Watanabe S.; Jasenosky L. D.; Watanabe T.; Kawaoka Y. ALIX/AIP1 is required for NP incorporation into Mopeia virus Z-induced virus-like particles. J. Virol. 2011, 85 (7), 3631–3641. 10.1128/JVI.01984-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jae L. T.; Raaben M.; Herbert A. S.; Kuehne A. I.; Wirchnianski A. S.; Soh T. K.; Stubbs S. H.; Janssen H.; Damme M.; Saftig P.; et al. Lassa virus entry requires a trigger-induced receptor switch. Science 2014, 344 (6191), 1506–1510. 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W.; Henry M. D.; Borrow P.; Yamada H.; Elder J. H.; Ravkov E. V.; Nichol S. T.; Compans R. W.; Campbell K. P.; Oldstone M. B. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 1998, 282 (5396), 2079–2081. 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- Winter S. L.; Golani G.; Lolicato F.; Vallbracht M.; Thiyagarajah K.; Ahmed S. S.; Lüchtenborg C.; Fackler O. T.; Brügger B.; Hoenen T.; et al. The Ebola virus VP40 matrix layer undergoes endosomal disassembly essential for membrane fusion. EMBO J. 2023, 42 (11), e113578 10.15252/embj.2023113578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safo M. K.; Musayev F. N.; Mosier P. D.; Zhou Q.; Xie H.; Desai U. R. Crystal Structures of Influenza A Virus Matrix Protein M1: Variations on a Theme. PLoS One 2014, 9 (10), e109510 10.1371/journal.pone.0109510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A.; Forouhar F.; Qiu S.; Sha B.; Luo M. The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology 2001, 289 (1), 34–44. 10.1006/viro.2001.1119. [DOI] [PubMed] [Google Scholar]
- Fontana J.; Steven A. C. At low pH, influenza virus matrix protein M1 undergoes a conformational change prior to dissociating from the membrane. J. Virol. 2013, 87 (10), 5621–5628. 10.1128/JVI.00276-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J.; Cardone G.; Heymann J. B.; Winkler D. C.; Steven A. C. Structural changes in Influenza virus at low pH characterized by cryo-electron tomography. J. Virol. 2012, 86 (6), 2919–2929. 10.1128/JVI.06698-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer S.; Feng Y.; Nebioglu F.; Heilig R.; Picotti P.; Helenius A. Stepwise Priming by Acidic pH and a High K+ Concentration Is Required for Efficient Uncoating of Influenza A Virus Cores after Penetration. J. Virol. 2014, 88 (22), 13029–13046. 10.1128/JVI.01430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Heuvel R. H. H.; Van Duijn E.; Mazon H.; Synowsky S. A.; Lorenzen K.; Versluis C.; Brouns S. J. J.; Langridge D.; Van Der Oost J.; Hoyes J.; et al. Improving the Performance of a Quadrupole Time-of-Flight Instrument for Macromolecular Mass Spectrometry. Anal. Chem. 2006, 78 (21), 7473–7483. 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- Strohalm M.; Hassman M.; Košata B.; Kodíček M. mMass data miner: an open source alternative for mass spectrometric data analysis. Rapid Commun. Mass Spectrom. 2008, 22 (6), 905–908. 10.1002/rcm.3444. [DOI] [PubMed] [Google Scholar]
- Kopicki J.-D.; Saikia A.; Niebling S.; Günther C.; Anjanappa R.; Garcia-Alai M.; Springer S.; Uetrecht C. Opening opportunities for Kd determination and screening of MHC peptide complexes. Commun. Biol. 2022, 5 (1), 488. 10.1038/s42003-022-03366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S.; Temu T.; Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11 (12), 2301–2319. 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Bryant P.; Pozzati G.; Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 2022, 13 (1), 1265–1311. 10.1038/s41467-022-28865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra S.; Joseph A. P.; Thiyagalingam J.; Topf M. Assessment of protein-protein interfaces in cryo-EM derived assemblies. Nat. Commun. 2021, 12 (1), 3399–3412. 10.1038/s41467-021-23692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebling S.; Veith K.; Vollmer B.; Lizarrondo J.; Burastero O.; Schiller J.; Struve García A.; Lewe P.; Seuring C.; Witt S.; et al. Biophysical Screening Pipeline for Cryo-EM Grid Preparation of Membrane Proteins. Front. Mol. Biosci. 2022, 9, 882288. 10.3389/fmolb.2022.882288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson G. R.; Burke J. E.; Ahn N. G.; Anand G. S.; Borchers C.; Brier S.; Bou-Assaf G. M.; Engen J. R.; Englander S. W.; Faber J.; et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 2019, 16 (7), 595–602. 10.1038/s41592-019-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.