Abstract
The complete nucleotide sequence and gene organization of the three virulence plasmids from Yersinia pestis KIM5 were determined. Plasmid pPCP1 (9,610 bp) has a GC content of 45.3% and encodes two previously known virulence factors, an associated protein, and a single copy of IS100. Plasmid pCD1 (70,504 bp) has a GC content of 44.8%. It is known to encode a number of essential virulence determinants, regulatory functions, and a multiprotein secretory system comprising the low-calcium response stimulation that is shared with the other two Yersinia species pathogenic for humans (Y. pseudotuberculosis and Y. enterocolitica). A new pseudogene, which occurs as an intact gene in the Y. enterocolitica and Y. pseudotuberculosis-derived analogues, was found in pCD1. It corresponds to that encoding the lipoprotein YlpA. Several intact and partial insertion sequences and/or transposons were also found in pCD1, as well as six putative structural genes with high homology to proteins of unknown function in other yersiniae. The sequences of the genes involved in the replication of pCD1 are highly homologous to those of the cognate plasmids in Y. pseudotuberculosis and Y. enterocolitica, but their localization within the plasmid differs markedly from those of the latter. Plasmid pMT1 (100,984 bp) has a GC content of 50.2%. It possesses two copies of IS100, which are located 25 kb apart and in opposite orientations. Adjacent to one of these IS100 inserts is a partial copy of IS285. A single copy of an IS200-like element (recently named IS1541) was also located in pMT1. In addition to 5 previously described genes, such as murine toxin, capsule antigen, capsule anchoring protein, etc., 30 homologues to genes of several bacterial species were found in this plasmid, and another 44 open reading frames without homology to any known or hypothetical protein in the databases were predicted.
Three species of Yersinia, Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis, have been studied extensively because of their ability to cause disease in both humans and animals. These organisms are closely related at the genetic level, as demonstrated by DNA-DNA homology studies against Y. pestis that showed 83 and 23% homology to Y. pseudotuberculosis and Y. enterocolitica, respectively, under conditions of stringent reassociation (30). Nevertheless, the symptoms of disease caused by the three yersiniae are dramatically different, as are their mechanisms of transmission. Enteropathogenic Y. enterocolitica and Y. pseudotuberculosis are mainly food-borne pathogens causing infection of humans that is typically chronic and characterized by diarrhea, fever, and abdominal pain. On the other hand, Y. pestis causes bubonic plague, an acute lethal disease. Following infection of the dermis by flea bite, this organism disseminates to lymph nodes and then to favorite niches within the viscera, eventually promoting marked septicemia; lung involvement in humans may lead to highly infectious pneumonic plague (5, 37).
The observed major distinctions between chronic and acute disease reflect differences in mechanisms of transmission. The enteropathogenic yersiniae must survive in soil and water and then bypass the host gastrointestinal mucosa following ingestion, whereas Y. pestis remains within the closed and protected environment of its flea vector, thereby ensuring transmission by intradermal injection, a route that requires extensive dissemination to achieve favored visceral niches which support the bulk of replication in vivo (5, 20). The most striking genetic difference between Y. pestis and the enteropathogenic species in this regard is the presence in most but not all (14) strains of the former of two unique plasmids: pMT1 (60 to 110 kb) and pPCP1 (9.6 kb) (2, 12, 15, 25). Although some important virulence factors are encoded on pMT (murine toxin and F1 capsular antigen) (60) and pPCP1 (plasminogen activator), a complete catalog of genes present on these two plasmids is not yet available. The three species also share many additional processes that promote disease, as reflected by carriage of a common plasmid in which are clustered a large number of genes encoding virulence factors such as Yop proteins and the Yop protein secretion system, as well as salient regulatory and anti-inflammatory functions. The generic term “low-calcium response,” or Lcr plasmid, has been applied to this plasmid regardless of its origin; it is specifically termed pCD in Y. pestis, pCad or pIB in Y. pseudotuberculosis, and pYV in Y. enterocolitica (10, 22, 37).
Another major difference between Y. pestis and the enteropathogenic yersiniae is the presence in the former of as many as 30 copies of an insertion element termed IS100 both within the chromosome and on all three plasmids (42, 47). The existence of IS100, as well as additional insertion elements (15, 34, 40), accounts for loss of major chromosomal genes either by direct insertion (53) or by reciprocal recombination resulting in their deletion (13). In this publication, we report the entire nucleotide sequence of the three plasmids from Y. pestis KIM5. These sequences define for the first time a large number of genes homologous to those of several unrelated pathogenic bacteria, the presence of numerous insertion elements and transposons, a large number of open reading frames (ORFs) without known homology, and individual origins of replication. Additionally, we show detailed comparative analysis between the genes encoded by the newly sequenced pCD1 plasmid of Y. pestis and those of the analogous Lcr plasmids of Y. pseudotuberculosis and Y. enterocolitica.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains used in this study are listed in Table 1. The three isogenic Y. pestis strains of KIM5 (6, 16) used to isolate the plasmids were obtained from Robert Brubaker (Michigan State University). The cells were grown in brain heart infusion medium at 28°C. Plasmid pMT1 was isolated from strain KIM5-D46 with a plasmid isolation kit (Qiagen, Santa Clarita, Calif.). Elution was achieved by using a heated buffer according to the manufacturer’s recommendations. Plasmid pCD1 was obtained from strain KIM5-D45 and purified by using a 0.7% agarose gel. Plasmid pPCP1 was isolated from strain KIM5-D1 by CsCl gradient ultracentrifugation.
TABLE 1.
Bacterial strains and plasmids
| Strain | Relevant genotype |
|---|---|
| Y. pestis | |
| KIM5-D1 | pPCP1+ pmT1+ pCD1− |
| KIM5-D45 | pPCP1− pMT1+ pCD1+ |
| KIM5-D46 | pPCP1− pMT1+ pCD1− |
| E. coli XL1-Blue | supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lacF′ [proAB+ lacIqlacZΔM15 Tn10 (Tetr)] |
Plasmid subcloning.
Plasmid pPCP1 was subcloned into the BamHI site of pUC18. The resulting recombinant plasmid was subjected to random in vitro transposon bombing with a kit from Perkin-Elmer, Applied Biosystems Division (Foster City, Calif.). A total of 217 clones were sequenced by using dye terminator chemistry and primers SD118 and SD119 (11). M13 libraries were made by shearing purified pMT1 and pCD1 with a nebulizer (3). The instrument used was built at the technical development laboratory of the Center for Genetics in Medicine, Washington University School of Medicine, St. Louis, Mo. The ends of the resultant fragments were repaired with a mixture of T4 DNA polymerase and Klenow fragment as described by Martin-Gallardo et al. (27). Fragments ranging from 1 to 2 kb were ligated into the HincII site of M13mp18. The single-stranded templates for sequencing were isolated by a modified boiling method adapted for a 96-well format (26).
Sequence assembly and gap closure.
A combination of approaches was used to close the gaps between contigs, to obtain sequences from both strands, and to resolve problem regions or compressions. Reads for individual contigs were extended by asymmetrical PCR from end clones, and the PCR products were sequenced directly. Reaction conditions were those described in the AmpliTaq DNA Polymerase Kit from Perkin-Elmer/Roche Biosystems (Branchburg, N.J.). This process quickly joined a number of nearby contigs. A variation of this method involving purification and cloning of the PCR product before sequencing was especially useful for pCD1 because of the scarcity of original plasmid DNA and the high homology found in certain regions shared between pMT1 and pCD1. Regions between contigs were amplified by PCR by using the original plasmid template for pMT1 and M13 clones for pCD1. The PCR products were cloned into pGEM-Easy vector (Promega, Madison, Wisc.). The inserts (0.5 to 1.2 kb) were sequenced by using M13 (−21) and M13 reverse primers or random in vitro transposon bombing as described above. Areas containing compressions and other mobility artifacts were resequenced by using ABI-PRISM dye terminator or ABI BigDye terminator. All sequencing samples were run on ABI PRISM 373 or 377 sequencers.
Base calling and assembly of sequences were performed with the PHRED/PHRAP combination of software (developed by Phil Green, University of Washington). All plasmid sequences were subjected to quality control standards by using a program called Swedish, developed in-house, that automatically calculates error rates and ensures a cumulative error rate of less than 1 in 10,000 bases.
Annotation and analysis of sequences.
Sequences were searched against current protein and nucleotide databases (including those from recently sequenced microbial genomes) by using BLAST (1). Only homology scores of less than 10−12 were considered in assigning homologue status during searches of the protein databases. The plasmid sequences were also analyzed by the GeneMark gene prediction program (4). The ORFs predicted by GeneMark were analyzed by MotifFinder (35) and Block (39), which looked for potential motifs and domains. A total of 169, 1,619, and 2,270 fragments were used to assemble pPCP1, pCD1, and pMT1, respectively. This represents an average redundancy of 7-, 9.2-, and 9-fold, respectively. The sequence was determined on both strands for >95% of each plasmid. Since all three plasmids contained at least one copy of IS100, we defined the start of IS100 as position 1 for each of the three plasmids. Only ORFs encoding peptides of more than 50 amino acids were analyzed.
Nucleotide sequence accession numbers.
The sequence of each plasmid was submitted to the GenBank database under accession no. AF053945 for pPCP1, AF053946 for pCD1, and AF053947 for pMT1.
RESULTS
Analysis of pPCP1.
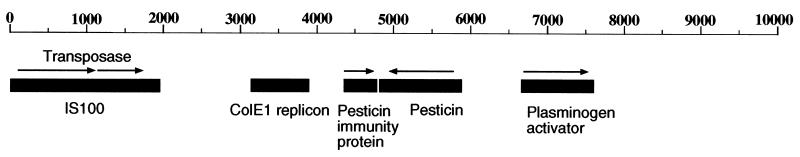
The total length of plasmid pPCP1 is 9,610 bp. Its GC content is 45.3%. As previously described (47), a single copy of insertion element IS100 was found in this plasmid. Three known genes, the pesticin, pesticin immunity protein, and plasminogen activator genes, were located on the plasmid by BLAST searches (Fig. 1). No additional genes were found or predicted. A region between bp 3,119 and 3,899 was found to have high homology to the origin of replication and the immunity region of the ColE1 plasmid of Escherichia coli. It thus defined the origin of replication on pPCP1.
FIG. 1.
Structural organization of the 9,610-bp plasmid pPCP1 derived from Y. pestis KIM5. BLAST searches using the entire nucleotide sequence obtained in this work were performed to precisely localize potential new ORFs, insertion sequence elements, and the three previously described genes present in pPCP. The directions of transcription of these genes are indicated by the arrows. The single IS100 element was used to define position 1 of this plasmid. The characteristics of the genes and proteins involved are described in the text. The numbering above the line is the molecular size in base pairs.
Analysis of pCD1.
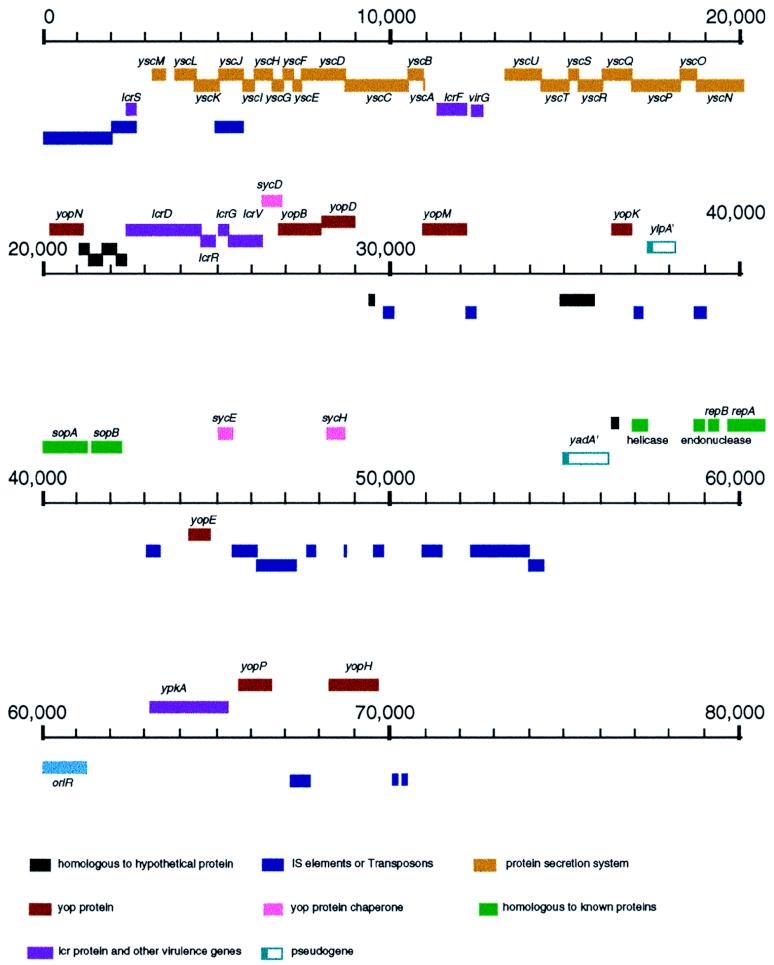
The total length of pCD1 is 70,504 bp, and its GC content is 44.8%. BLAST searches revealed numerous homologues to known virulence genes. Many intact or partial insertion sequences or transposons were found scattered throughout the plasmid, including IS100 and IS285. This phenomenon suggests an earlier transfer or “gathering” of virulence genes among the yersiniae and even among more distantly related organisms, mediated by transposition. Homologues to a large number of proteins previously described in plasmids derived from Y. pseudotuberculosis and Y. enterocolitica were identified by BLAST searches. Among these were Yop proteins, Yop translocation proteins, Yop protein chaperones, V antigen, and other proteins essential for virulence (Fig. 2 and Table 2). Two genes containing premature termination codons (pseudogenes) were found. One of these corresponded to the gene encoding the adhesin YadA, a virulence determinant of the enteropathogenic yersiniae, and the other corresponded to that encoding the lipoprotein YlpA. Both pseudogenes are characterized by frameshifts at the N termini of their putative products (a single deletion of a nucleotide at amino acid 80 in yadA and insertion of a single nucleotide at amino acid 32 in ylpA). Both frameshifts occur at a run of deoxyadenosine nucleotides in the sequence encoding two lysine residues. The ylpA gene is known to be carried by the pYV plasmid of Y. enterocolitica, where it encodes a typical lipoprotein signal peptide (8). The ylpA gene hybridizes with the pYV plasmid of Y. pseudotuberculosis, so that it appears to be conserved among the Yersinia species. ylpA also has significant homology at the protein level with the TraT protein encoded by plasmids pED208, R100, and F (>80% identity for all three) and 77% identity with the protein encoded by the virulence plasmid of Salmonella typhimurium. The yadA gene is known to be nonfunctional in Y. pestis (33, 48, 54); our sequencing results simply confirm this finding.
FIG. 2.
Physical map and genetic organization of pCD1. ORFs, insertion sequences, and other genetic elements were located in the map by using BLAST searches and GeneMark. ORFs and genes in the map are color coded according to function or unique characteristic, and their designations are placed either above or below the colored bars. The scale indicates the number of nucleotides measured from the start of the single IS100 found in this plasmid. Genes in the figures are located precisely in the map and drawn to scale directly from sequence annotation by using an in-house, UNIX-based annotation-rendering program. Genes positioned on top of each line are transcribed from left to right, whereas those placed below the line are encoded by the complementary strand. The two pseudogenes (ylpA and yadA) are represented by partially colored bars. The position of the origin of replication is marked as oriR. The characteristics of the genes, proteins, and sequences depicted are described in the text and in Table 2.
TABLE 2.
Localization and description of ORFs and noncoding elements in pCD1
| ORF or noncoding element | Position no. | Size (amino acid residues) | Strand or direction | Description (homologue by BLAST) |
|---|---|---|---|---|
| ORFs | ||||
| 1 | 87–1109 | 340 | Direct | Transposase |
| 2 | 1109–1888 | 259 | Direct | Transposase |
| 3 | 1939–2343 | 134 | Complement | Transposase |
| 4 | 2379–2645 | 88 | Complement | lcrS (Y. pseudotuberculosis) |
| 5 | 3193–3540 | 115 | Complement | lcrQ (Y. pseudotuberculosis) and yscM (Y. enterocolitica) |
| 6 | 3765–4430 | 221 | Complement | yscL (Y. enterocolitica) |
| 7 | 4376–5005 | 209 | Complement | yscK (Y. enterocolitica) |
| 8 | 5005–5739 | 244 | Complement | yscJ (Y. enterocolitica) |
| 9 | 5746–6093 | 115 | Complement | ycsI (Y. enterocolitica) and lcrO (Y. pseudotuberculosis) |
| 10 | 6094–6591 | 165 | Complement | yscH (Y. enterocolitica) and lcrP (Y. pseudotuberculosis) |
| 11 | 6588–6935 | 115 | Complement | yscG (Y. enterocolitica) |
| 12 | 6937–7200 | 87 | Complement | yscF (Y. enterocolitica) |
| 13 | 7201–7401 | 66 | Complement | yscE (Y. enterocolitica) |
| 14 | 7398–8657 | 419 | Complement | yscD (Y. enterocolitica) |
| 15 | 8654–10477 | 607 | Complement | yscC (Y. enterocolitica) |
| 16a | 10483–10896 | 137 | Complement | yscB (Y. enterocolitica) |
| 16b | 11121–11220 | 32 | Complement | yscA (Y. enterocolitica) |
| 17 | 11299–12114 | 271 | Complement | lcrF (virF) transcription factor |
| 18 | 12238–12633 | 131 | Complement | virG (Y. enterocolitica) |
| 19 | 13209–14273 | 354 | Complement | yscU (Y. enterocolitica) |
| 20 | 14273–15058 | 261 | Complement | yscT (Y. pseudotuberculosis) |
| 21 | 15055–15321 | 87 | Complement | yscS (Y. pseudotuberculosis) |
| 22 | 15323–15976 | 217 | Complement | yscR (Y. pestis) |
| 23 | 15973–16896 | 307 | Complement | yscQ (Y. pestis) |
| 24 | 16893–18260 | 455 | Complement | yscP (Y. pestis) |
| 25 | 18260–18724 | 154 | Complement | yscO (Y. pestis) |
| 26 | 18721–20040 | 439 | Complement | yscN (Yop secretion ATPase) |
| 27 | 20238–21119 | 293 | Direct | yopN (Y. pseudotuberculosis) |
| 28 | 21100–21378 | 92 | Direct | Y. pseudotuberculosis hypothetical protein |
| 29 | 21365–21736 | 123 | Direct | Y. pseudotuberculosis and Y. enterocolitica hypothetical protein |
| 30 | 21733–22101 | 122 | Direct | Y. enterocolitica hypothetical protein |
| 31 | 22098–22442 | 114 | Direct | Y. enterocolitica hypothetical protein |
| 32 | 22429–24543 | 704 | Direct | lcrD (Y. pseudotuberculosis and Y. enterocolitica) |
| 33 | 24540–24980 | 144 | Direct | lcrR (Y. pestis) |
| 34 | 25022–25309 | 95 | Direct | lcrG (Y. pestis) |
| 35 | 25311–26291 | 326 | Direct | lcrV (V antigen) |
| 36 | 26304–26810 | 168 | Direct | lcrH (sycD) (YopB and YopD chaperones) |
| 37 | 26788–27993 | 401 | Direct | yopB (Y. pseudotuberculosis and Y. enterocolitica) |
| 38 | 28012–28932 | 306 | Direct | yopD (Y. pseudotuberculosis and Y. enterocolitica) |
| 39 | 29345–29512 | 55 | Complement | Y. pestis hypothetical protein |
| 40 | 29778–30038 | 87 | Complement | Y. pseudotuberculosis transposase |
| 41 | 30873–32102 | 409 | Direct | yopM (Y. pestis) |
| 42 | 32145–32444 | 99 | Complement | Transposase |
| 43 | 34860–35828 | 322 | Complement | Y. enterocolitica hypothetical protein |
| 44 | 36328–36876 | 182 | Direct | yopK (Y. pseudotuberculosis) and yopQ (Y. enterocolitica) |
| 45/46 | 37360–38110 | 36 | Direct | ylpA pseudogene |
| 47 | 38624–39016 | 130 | Direct | Transposase |
| 48 | 40080–41288 | 402 | Direct | sopA (E. coli) |
| 49 | 41417–42250 | 277 | Direct | sopB (E. coli) |
| 50 | 44186–44845 | 219 | Complement | yopE (Y. pestis) |
| 51 | 45039–45431 | 130 | Direct | sycE (YopE chaperone) |
| 52 | 45494–46123 | 309 | Complement | Transposase |
| 53 | 46241–47413 | 390 | Direct | Transposase |
| 54 | 47413–47844 | 143 | Direct | Transposase |
| 55 | 48188–48613 | 141 | Direct | sycH (YopH chaperone) (Y. pseudotuberculosis and Y. enterocolitica) |
| 56 | 49594–49860 | 88 | Complement | Transposase |
| 57 | 50911–51462 | 183 | Complement | Transposon gamma-delta resolvase and tnpR (Y. enterocolitica) |
| 58 | 51626–53941 | 771 | Direct | Transposase and tnpA (Y. enterocolitica) |
| 59 | 53938–54318 | 206 | Complement | Transposase |
| 60/61 | 54924–56227 | 50 | Direct | yadA pseudogene |
| 62 | 56488–56297 | 63 | Direct | Hypothetical protein (Y. enterocolitica) |
| 63 | 56928–57344 | 138 | Direct | DNA helicase I (E. coli plasmid F) |
| 64 | 58681–58929 | 82 | Direct | Endonuclease (Y. enterocolitica) |
| 65 | 59067–59321 | 84 | Direct | repB (replication protein) |
| 66 | 59618–60496 | 292 | Direct | repA (replication protein) |
| 67 | 63100–65298 | 732 | Direct | ypkA (protein kinase) (Y. pseudotuberculosis and Y. enterocolitica) |
| 68 | 65694–66557 | 287 | Direct | yopJ (Y. enterocolitica) |
| 69 | 67146–67649 | 167 | Complement | Transposase |
| 70 | 68243–69649 | 468 | Direct | yopH (protein-tyrosine-phosphatase) (Y. pseudotuberculosis and Y. enterocolitica) |
| 71 | 70502–70161 | 113 | Complement | Transposase (partial) |
| Noncoding elements | ||||
| 1–1954 | Forward | IS100 | ||
| 2655–1961 | Reverse | Desulfovibrio vulgaris insertion sequence ISD1 | ||
| 5716–4973 | Reverse | IS285 | ||
| 29501–30801 | Forward | Y. pestis yopM gene, repeat region R1 | ||
| 32118–34817 | Forward | Y. pestis yopM gene, repeat regions R2 and R3 | ||
| 36952–37179 | Forward | Salmonella enteritidis insertion element IS1351 and Yersinia insertion element IS200 | ||
| 38717–39225 | Forward | Shigella dysenteriae insertion sequence IS911 | ||
| 43032–43399 | Forward | Erwinia herbicola IS1327 | ||
| 46137–47226 | Forward | Shigella sonnei insertion sequence IS640 | ||
| 46137–45463 | Reverse | Enterobacter agglomerans nifJ gene and insertion sequence IS2222 | ||
| 49755–49521 | Reverse | Rhizobium insertion element ISR1 | ||
| 48647–48728 | Forward | IS285 (partial) | ||
| 47600–47786 | Forward | DNA IS100 (partial) | ||
| 53976–52230 | Reverse | Y. enterocolitica DNA, partial Tn3 homologue | ||
| 60005–61324 | Forward | E. coli resistance plasmid R100; replication incompatibility, and copy number regions | ||
| 70001–70145 | Reverse | IS285 (partial) | ||
| 70504–70322 | Reverse | S. enteritidis insertion element IS1351 and Yersinia insertion element IS200 |
Other homologues with diverse functions were found in this plasmid, including the genes encoding DNA helicase, DNA resolvase, and DNA replication proteins A and B (sopA and sopB). SopA and SopB function as plasmid partition proteins that ensure the stable and faithful inheritance of the F plasmid (24, 32). Finally, six putative proteins of unknown function that are homologous to those encoded by Lcr plasmids of enteropathogenic yersiniae were found (Table 2).
Analysis of pMT1.
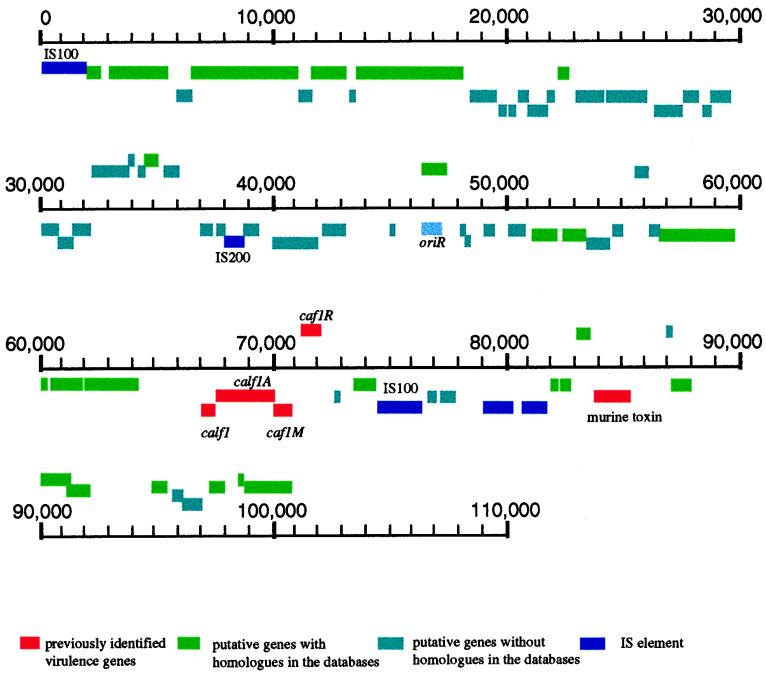
The total length of pMT1 is 100,984 bp, and its GC content is 50.2%. This plasmid contains two copies of IS100 in opposite orientations. The start of one of the IS100 inserts was defined as position 1 of pMT1. The second IS100 copy was found to be located between positions 74592 and 76545. To rule out the possibility that sequences are incorrectly assembled due to the presence of these two identical IS100 elements, an extensive restriction digestion analysis was carried out on this plasmid. One copy of an IS200-like element (also known as IS1541) (34) was found (Table 3 and Fig. 3). Its orientation is the same as that of the second copy of IS100. Two putative transposases were found not far from the latter (positions 79222 to 80430 and 80899 to 81922), indicating the presence of an insertion element or a transposon at this location. BLAST searches showed that IS285 had the closest similarity to these sequences.
TABLE 3.
Localization and description of ORFs and noncoding elements in pMT1
| ORF or noncoding element | Position no. | Size (amino acid residues) | Strand or direction | Description (homologue by BLAST) |
|---|---|---|---|---|
| ORFs | ||||
| 1 | 87–1109 | 340 | Direct | Transposase |
| 2 | 1109–1888 | 259 | Direct | Transposase |
| 3 | 2022–2618 | 198 | Complement | Similar to hypothetical E. coli protein |
| 4 | 2932–5514 | 860 | Complement | Similar to tail fiber protein gp37 |
| 5 | 5901–6518 | 205 | Complement | Unknown |
| 6 | 6571–11049 | 1,492 | Complement | Putative protein, similar to lambda host-specific protein J and Northern European squid neurofilament-like protein |
| 7 | 11188–11739 | 183 | Complement | Unknown |
| 8 | 11763–12521 | 252 | Complement | Similar to hypothetical Coxiella burnetii protein (motif: RGD cell attachment site) |
| 9 | 12553–13251 | 232 | Complement | Similar to phage lambda minor tail protein L |
| 10 | 13341–13676 | 111 | Complement | Unknown |
| 11 | 13718–18295 | 1,525 | Complement | Putative protein, similar to hypothetical Haemophilus influenzae protein HI1514 |
| 12 | 18653–18970 | 105 | Complement | Unknown |
| 13 | 19030–19776 | 248 | Complement | Unknown |
| 14 | 19851–20207 | 118 | Complement | Unknown (Block hit: complement C1Q domain signature) |
| 15 | 20236–20616 | 126 | Complement | Unknown (Block hit: NSF attachment protein) |
| 16 | 20700–21044 | 114 | Complement | Unknown (Block hit: geminivirus AR1 coat protein) |
| 17 | 21142–21975 | 277 | Complement | Unknown (Block hit: histone H5 signature) |
| 18 | 21975–22274 | 99 | Complement | Unknown |
| 19 | 22453–22884 | 143 | Complement | Similar to glucan endo-1,3-β-d-glucosidase |
| 20 | 23190–24065 | 291 | Complement | Unknown |
| 21 | 24092–24409 | 105 | Complement | Unknown |
| 22 | 24537–24977 | 146 | Complement | Unknown |
| 23 | 25015–26253 | 412 | Complement | Unknown |
| 24 | 26623–27831 | 402 | Complement | Unknown (motif: ATP/GTP binding motif A [P loop]) |
| 25 | 27882–28523 | 213 | Complement | Unknown |
| 26 | 28719–28985 | 88 | Complement | Unknown (motif: RGD cell attachment site) |
| 27 | 28995–29885 | 296 | Complement | Unknown |
| 28 | 30138–30776 | 212 | Complement | Similar to ABC transporter (motif: ATP/GTP binding motif A [P loop]) |
| 29 | 30773–31441 | 222 | Complement | Unknown (motif: aminoacyl-RNA synthetase class II, signature 2) |
| 30 | 31441–32121 | 226 | Complement | Unknown |
| 31 | 32285–33763 | 492 | Direct | Unknown |
| 32 | 33766–34044 | 92 | Direct | Unknown |
| 33 | 34269–34526 | 85 | Direct | Unknown |
| 34 | 34531–35058 | 175 | Direct | Similar to S. typhimurium repressor of phase 1 flagellin gene |
| 35 | 35382–36032 | 216 | Direct | Unknown |
| 36 | 36982–37464 | 160 | Complement | Unknown |
| 37 | 37669–37950 | 93 | Complement | Unknown |
| 38 | 38797–39384 | 195 | Complement | Unknown |
| 39 | 40132–42015 | 627 | Complement | Unknown (motifs: ABC transporter, aminoacyl-tRNA synthetase class II signature 2, ATP/GTP binding motif A [P loop]) |
| 40 | 42278–43231 | 317 | Complement | Unknown |
| 41 | 45161–45385 | 74 | Complement | Unknown |
| 42 | 46550–47614 | 354 | Direct | Similar to E. coli replication protein A (repA) |
| 43 | 48184–48384 | 66 | Complement | Unknown |
| 44 | 48396–48656 | 86 | Complement | Unknown |
| 45 | 49291–49587 | 98 | Complement | Unknown |
| 46 | 50211–50996 | 261 | Complement | Unknown |
| 47 | 51336–52412 | 358 | Complement | Similar to recA (motif: ATP/GTP binding motif A [P loop]) |
| 48 | 52681–53550 | 289 | Complement | Similar to Streptococcus pneumoniae DNA Pol Ia |
| 49 | 53686–54714 | 342 | Complement | Unknown (motif: RGD cell attachment site) (Block hit: T. cruzi P2 protein signature) |
| 50 | 54834–55265 | 143 | Complement | Unknown |
| 51 | 55810–56373 | 187 | Direct | Unknown |
| 52 | 56403–56846 | 147 | Complement | Unknown |
| 53 | 56843–60250 | 1135 | Complement | Similar to DNA Pol III alpha subunit |
| 54 | 60548–61783 | 411 | Complement | Similar to CobS protein |
| 55 | 61881–64235 | 784 | Complement | Similar to CobT protein |
| 56 | 67074–67588 | 175 | Complement | F1 capsule antigen |
| 57 | 67669–70170 | 833 | Complement | F1 capsule anchoring protein |
| 58 | 70195–70971 | 258 | Complement | caf1M |
| 59 | 71299–72204 | 301 | Direct | caf1R |
| 60 | 72719–73024 | 101 | Complement | Unknown |
| 61 | 73545–74582 | 347 | Complement | Similar to DNA ligase |
| 62 | 74658–75437 | 259 | Complement | Transposase |
| 63 | 75437–76459 | 340 | Complement | Transposase |
| 64 | 76861–77118 | 85 | Complement | Unknown |
| 65 | 77419–78048 | 209 | Complement | Unknown |
| 66 | 79222–80430 | 402 | Direct | Transposase |
| 67 | 80899–81921 | 340 | Direct | Transposase |
| 68 | 82120–82437 | 105 | Complement | Similar to E. coli yhgA |
| 69 | 82617–82988 | 123 | Complement | Similar to hypothetical protein |
| 70 | 83297–83752 | 151 | Direct | Similar to E. coli hypothetical protein and reverse transcriptase-like protein |
| 71 | 83997–85595 | 532 | Complement | Murine toxin |
| 72 | 87149–87340 | 63 | Direct | Unknown |
| 73 | 87363–88220 | 285 | Complement | Putative protein, similar to E. coli yhgA |
| 74 | 90045–91250 | 401 | Direct | Similar to phage P7 parA |
| 75 | 91247–92218 | 323 | Direct | Similar to phage P7 parB |
| 76 | 94893–95534 | 213 | Direct | Similar to adenine DNA methyltransferase |
| 77 | 95766–96185 | 139 | Direct | Unknown (Block hit: NSF attachment site) |
| 78 | 96239–97018 | 259 | Direct | Unknown |
| 79 | 97417–97923 | 168 | Direct | Similar to antirestriction protein |
| 80 | 98635–98865 | 76 | Direct | Similar to E. coli hypothetical protein |
| 81 | 98938–100947 | 669 | Direct | Similar to Rhizobium meliloti protein and S. sonnei protein |
| Noncoding elements | ||||
| 1–1954 | Forward | IS100 | ||
| 38040–38757 | Reverse | IS200 | ||
| 46565–47425 | Forward | IncF plasmid RepFIB replicon | ||
| 74592–76545 | Reverse | IS100 |
Pol, polymerase.
FIG. 3.
Physical map and genetic organization of pMT1. ORFs, genes, and other features displayed in the map are depicted as described in Fig. 2. The characteristics of all of the elements described in the map are defined in the text and in Table 3. The caf1A and caf1 genes located at about 70,000 nucleotides are incorrectly labeled calf1A and calf1, respectively.
Five previously sequenced genes were located: F1 capsular antigen, F1 capsule anchoring protein, its chaperone (Caf1M), the regulatory protein (Caf1R), and murine toxin. These genes are clustered in the region between positions 67669 and 85595 (Fig. 3). Twenty-six homologues (excluding transposases in transposons and insertion elements) were found by BLAST searches (Table 3). Three homologues are similar to the E. coli tail fiber protein and the lambda phage host-specific protein. Homology searches also revealed the presence of two possible new operons. One contains homologues to the phage p7 parA and parB genes, which are involved in plasmid partition (19). The other operon contains homologues to the genes cobS and cobT in Pseudomonas denitrificans. cobS and cobT were isolated as an independent cluster of the cobalamin biosynthetic genes, likely to be involved in cobalt insertion-mediating reactions and the transformation of precorrin-3 (7). The function of the Y. pestis homologue is not known.
An additional 44 ORFs were predicted by GeneMark. These ORFs have no homology with any known or hypothetical proteins currently in the databases. MotifFinder found 14 types of motifs (including transposase) in the PROSITE database. Several interesting motifs, including an ATP/GTP binding site (P loop), cell attachment site (RGD), ABC (ATP binding cassette) transporter, and sigma 54 interaction domain, were observed. Extensive experience with these searches suggests that, due to the problems inherent in the search algorithm, predicted phosphorylation, glycosylation, amidation, and myristoylation sites do not tend to have significant biological relevance; thus, such findings are not discussed here. However, a number of motifs identified by Block searches with potentially interesting and relevant biological functions included the Trypanosoma cruzi P2 protein signature, complement C1q domain signature, N-ethylmaleimide-sensitive factor (NSF) attachment site, and Rho family GDP dissociation inhibitor signature.
DISCUSSION
Plasmid pPCP1.
All structural genes identified during the sequencing of plasmid pPCP1 have been described previously (47, 55). The organization of the genes encoded in this plasmid was the same as that previously reported (Fig. 1). At the protein level, pesticin and pesticin immunity protein were found to be identical to those described in the databases. The predicted sequence of the plasminogen activator was identical to that described by McDonough and Falkow (28), even though their sequences were obtained from a different strain of Y. pestis (EV76). Two putative transposases were found within IS100. Their genes are transcribed in the same direction as the pesticin immunity protein and the plasminogen activator genes, while the pesticin gene is transcribed in the opposite direction. We did not find new ORFs larger than 50 amino acids in this plasmid. Replication of the plasmid is controlled by a mechanism highly homologous to the ColE1 replicon of E. coli. This is consistent with the fact that yersiniae and E. coli are taxonomically related.
Plasmid pCD1.
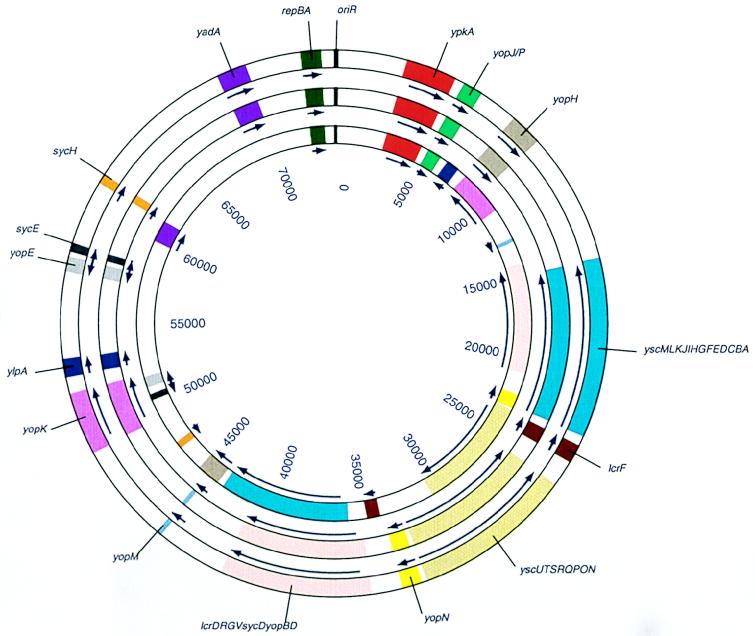
Plasmid pCD1 mediates the low-calcium response. Salient genes include those encoding the Yop proteins and their chaperones, secretory mediators, and regulatory genes. The Lcr plasmids are essential for virulence in all three species of Yersinia pathogenic for humans. The laboratories of Susan C. Straley, Hans Wolf-Watz, and Guy R. Cornelis were instrumental in defining the structures and functions of the Lcr plasmids in Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, respectively (10, 17, 21, 43, 44, 56, 57). As expected, all functional homologues to Yops and their related proteins were found on pCD1. The organization of the operons is highly conserved among the Lcr plasmids of the three pathogenic yersiniae (22, 37). However, a number of global structural differences can readily be discerned from the completed sequence of the Y. pestis Lcr plasmid. For example, in spite of the fact that the origins of replication in pCD1 and the corresponding pYV of Y. enterocolitica lie between the yadA and pkA genes in both plasmids, they map in entirely different positions within the plasmid (with respect, for example, to the yopBD and the yopM genes). YlpA maps near the origin of replication in the Y. enterocolitica plasmid but some 20 kb away in pCD1. ylpA, lcrVGRD, yopM, yopD, and a series of other homologous genes are transcribed in pCD1 in orientations opposite those in the Y. enterocolitica pYV plasmid. It is clear that in spite of the high degree of functional conservation observed among the virulence genes of all three plasmids, a number of rearrangements and internal translocations have taken place as the plasmids have proceeded to diverge and evolve (Fig. 4). It is interesting that more than 10 partial insertion sequences and other sequences homologous to those of diverse eubacteria (Salmonella, Erwinia, Rhizobium, and Desulfovibrio spp.) are scattered throughout the plasmid. This high proportion of insertions and mosaic sequences opens up the possibility that during evolution these virulence-associated genes were gathered from a diverse bacterial assemblage through transposition.
FIG. 4.
Diagram comparing the organization of selected genes and elements of the Lcr plasmid in yersiniae. Shown are circular maps of the pCD1 plasmid and of the homologous pYV and pCad plasmids derived from Y. enterocolitica and Y. pseudotuberculosis, respectively. The relative positions of selected loci with respect to the origin of replication of pCD1 are shown. Outer circle, pCD1; middle circle, pIB1 (Y. pseudotuberculosis); inner circle, pYVe O:9 (Y. enterocolitica). The nomenclature and approximate positions of genes in pYV and pIB1 are from Iriarte and Cornelis (22), Persson et al. (38), and Salyers and Whitt (51). The genes and sequence features of pCD1 and the corresponding regions in pYV and pIB1 are depicted in the same color to aid in their visualization (e.g., the repBA, oriR, and ypkA regions are presented in green, black, and red, respectively). Numbering inside the circles indicates the approximate sizes of the plasmids in nucleotides, measured from the start of their origins of replication. Arrows above each color segment representing a gene or gene group point to the direction of transcription.
A notable finding was the identification of a new pseudogene in pCD1 and the confirmation of a frameshift mutation in yadA. The genes yadA and ylpA, which are fully functional in Y. enterocolitica (37), have frameshift mutations that create premature termination of transcription in pCD1. Since yadA encodes an adhesin protein involved in attachment to epithelial cells, inactivation of this gene should represent no essential loss of function for Y. pestis pathogenesis. YlpA, on the other hand, is homologous to the TraT protein, encoded by the virulence plasmid of Salmonella (8). Since TraT is involved in serum resistance in Salmonella, YlpA is also likely to be involved in serum resistance in Y. enterocolitica. In Y. pestis, however, resistance to serum occurs independently of pCD1 as an evident function of lipopolysaccharide structure (41). It is nevertheless possible that a potential truncated protein could be translated from a downstream start codon in ylpA, leading to the production of a YlpA protein lacking the first 45 amino acids. Biochemical evidence, however, is not presently available to ascertain whether YlpA is absent or whether a truncated version of this protein is still expressed in Y. pestis. It is interesting that both of the frameshifts in these genes are caused by either deletion or insertion of a single deoxyadenosine nucleotide within a run of seven to eight deoxyadenosine nucleotides. Such stretches of redundancy are known to be hot spots for mutations and could be responsible for this phenomenon (49).
Although Y. pestis and Y. pseudotuberculosis are generally thought to be the most closely related species, we found that YopJ in pCD1 had higher homology to the Y. enterocolitica homologue (called YopP in this organism) than to its YopJ counterpart in Y. pseudotuberculosis. However, further inspection indicated that the YopJ protein from Y. pestis and its YopJ counterpart in Y. pseudotuberculosis are 99% identical (with a single amino acid difference) in the first 241 residues. After residue 241, the amino acid sequences differ markedly, due to an apparent change in the reading frame in the previously described Y. pseudotuberculosis yopJ gene. Our finding is entirely consistent with that reported by Mills et al. (29) during their studies of the Y. enterocolitica YopP. The apparent frameshift in Y. pseudotuberculosis has recently been shown to be a sequencing mistake (36). Since YopJ was first discovered in Y. pestis KIM by Straley and Bowmer (58), we have retained this terminology in pCD1.
In summary, most regions comprising the genetic material of plasmid pCD1 were identified as homologues of known or hypothetical proteins, or as occupied by insertion elements or transposons. Six putative proteins were found to have homologues in the databases, but their functions are unknown. Plasmid pCD1 contains very few large intergenic regions; its coding ratio is approximately 1 ORF per kb.
Plasmid pMT1.
The third plasmid and the largest, pMT1, is also the least studied of the Y. pestis plasmids. Although it was initially considered a cryptic plasmid, subsequent studies localized five important genes on this plasmid, encoding F1 capsular antigen, F1 capsule anchoring protein, Caf1M, Caf1R, and the plague murine toxin (45, 46). These genes are clustered in a region spanning approximately 18 kb of the entire plasmid. Interestingly, the DNA encoding this cluster of genes has a GC ratio of 45.8%, compared with 51.1% for the remainder of the plasmid. Such regions of atypical base composition have been found in several gram-negative and gram-positive organisms to be associated with what has been termed pathogenicity islands (18). These genetic elements, which cumulatively participate in pathogenicity, are likely acquired by genetic transfer among bacterial pathogens and sometimes contribute to differences in host specificity, tissue tropism, and disease manifestation (9). It is thus intriguing to conjecture if a plasmid such as pMT1, which readily integrates into the bacterial chromosome, may have arisen by a mechanism involving such a genetic mechanism.
Several ORFs (ORFs 34, 9, 4, and 6) were found to encode proteins that resemble E. coli flagellin or phage host-specific proteins (Table 3). Whether these proteins participate or aid in the actual biogenesis of pili or are involved in pathogenesis in Y. pestis is unknown. Interestingly, one putative protein is homologous to both phage p7 ParB and Shigella flexneri VirB protein or Shigella dysenteriae IpaR. VirB was implicated as a transcriptional activator of several invasion genes, and IpaR was found to induce apoptosis of macrophages (61). It is thus tempting to speculate that this gene may act as a virB or ipaA homologue. However, based on the following two observations, we suspect that this protein may instead function as a plasmid partition protein. First, although there is clear homology between this putative protein and VirB, BLAST searches showed a higher degree of homology with ParB. Second, whereas there are no other potential ipa genes in this plasmid, the next ORF upstream from the parB homologue is highly homologous to phage p7 parA. Thus, the region seems to constitute a parA parB operon containing the upstream element important for parA autoregulation and the parS site important for parB function. Thus, it is likely that ParA and ParB are fully functional as plasmid partition proteins. Plasmid pCD1, on the other hand, has at least two different and distinct proteins dedicated to plasmid partition which are homologous to the E. coli F plasmid-associated genes sopA and sopB (31). Thus, whereas in pMT1 the partition apparatus appears to resemble that of the p7 phage more closely, in pCD1 this apparatus is more akin to that of the F family of plasmids. The two plasmids have to use different partition systems in order to maintain the faithful inheritance of low-copy-number plasmids. Otherwise, the segregation of the plasmids would come disastrously close to random distribution.
One putative protein (ORF 19) is homologous to Bacillus circulans glucan endo-1,3-β-d-glucosidase (Table 3). Further work may show that it mediates an interaction between the organism and some polysaccharide moiety on the host cell surface. ORFs 18, 26, and 49 were found to contain an RGD (arginine-glycine-aspartate) cell attachment site. This protein sequence is a characteristic eukaryotic recognition motif which binds to cell surface integrins (50). It also has been found in an array of bacterial virulence factors, such as the Bordetella pertussis adherence factor filamentous hemagglutinin (FHA), pertactin, pertussis toxin, and BrkA. Studies have shown that the RGD sequence of FHA mimics that of the host cell (52). It binds to host cell CD11b/CD18, which mediates the uptake of the bacteria into macrophages without triggering an oxidative burst, thus protecting the bacteria (23). Although not all proteins containing RGD are involved in cell attachment, those containing properly presented RGD sequences have a strong potential for binding to host cell integrin or extracellular matrices. These proteins could thus be important candidates in adherence or in resistance to phagocytosis in Y. pestis. ORF 39 was found by MotifFinder to have an ABC transporter signature. ABC transporter superfamily members are found in both prokaryotes and eukaryotes, where they are involved in drug resistance and in the transport of substrates ranging from ions to large proteins (59). Thus, this ORF could have a role in the transport of substrates. In addition to the potential ORFs discussed above, some 40% of the ORFs predicted by GeneMark on plasmid pMT1 had neither homologous counterparts in the publicly available databases nor any manifestation of motif-associated features. Hence, we are presently unable to predict or speculate on the possible functions of these genes or if, in fact, any of these ORFs are translated into functional proteins. This issue is further complicated by the fact that pMT can integrate into the chromosome (46) and thus may contain copies of chromosomal genes required for normal vegetative functions.
The availability of the entire nucleotide sequence of these three plasmids should enable the global study of the gene complement encoded in them, as well as of the mechanism of expression that underlies their regulation. Such studies should help to elucidate the functions of presently unknown genes and should provide insight into the interplay of those virulence factors which are common to the three human pathogens and those that are unique to Y. pestis.
ACKNOWLEDGMENTS
We thank all the members of the sequencing core facility at Lawrence Livermore National Laboratory for their contribution to this work, Matt Nolan for providing the sequence quality analysis program (Swedish), Aaron Adamson for his assistance during the assembly process, and Vladimir Motin for helpful discussions and review of the manuscript.
This work was performed under the auspices of the U.S. DOE by Lawrence Livermore National Laboratory under contract W-7405-Eng-48.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Gurion R, Shafferman A. Essential virulence determinants of different Yersinia species are carried on a common plasmid. Plasmid. 1981;5:183–187. doi: 10.1016/0147-619x(81)90019-6. [DOI] [PubMed] [Google Scholar]
- 3.Bodenteich A, Chissoe S, Wang Y-F, Roe B A. Shotgun cloning as the strategy of choice to generate templates for high-throughput dideoxynucleotide sequencing. In: Adams M, Fields C, Venter J C, editors. Automated DNA sequencing and analysis. London, England: Academic Press; 1994. pp. 42–49. [Google Scholar]
- 4.Borodovsky M, Koonin E V, Rudd K E. New genes in old sequences: a strategy for finding genes in the bacterial genome. Trends Biochem Sci. 1994;19:309–313. doi: 10.1016/0968-0004(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 5.Brubaker R R. Factors promoting acute and chronic diseases caused by yersiniae. Clin Microbiol Rev. 1991;4:309–324. doi: 10.1128/cmr.4.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brubaker R R. Interconversion of purine mononucleotides in Pasteurella pestis. Infect Immun. 1970;1:446–454. doi: 10.1128/iai.1.5.446-454.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron B, Guilhot C, Blanche F, Cauchois L, Rouyez M-C, Rigault S, Levy-Schil S, Crouzet J. Genetic and sequence analysis of a Pseudomonas dentrificans DNA fragment containing two cob genes. J Bacteriol. 1991;173:6058–6065. doi: 10.1128/jb.173.19.6058-6065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.China B, Michiels T, Cornelis G R. The pYV plasmid of Yersinia encodes a lipoprotein, YlpA, related to TraT. Mol Microbiol. 1990;4:1585–1593. doi: 10.1111/j.1365-2958.1990.tb02070.x. [DOI] [PubMed] [Google Scholar]
- 9.Conner C, Heithoff D M, Julio S M, Sinsheimer R L, Mahan M J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 11.Devine S E, Boeke J D. Efficient integration of artificial transposons into plasmid targets in vivo: a useful tool for DNA mapping, sequencing and genetic analysis. Nucleic Acids Res. 1994;22:3765–3772. doi: 10.1093/nar/22.18.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferber D M, Brubaker R R. Plasmids in Yersinia pestis. Infect Immun. 1981;31:839–841. doi: 10.1128/iai.31.2.839-841.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 14.Filippov A A, Solodovnikov N S, Protsenko O A. Plasmid content in Yersinia pestis strains of different origin. FEMS Microbiol Lett. 1990;67:45–48. doi: 10.1016/0378-1097(90)90165-m. [DOI] [PubMed] [Google Scholar]
- 15.Filippov A A, Oleinikov P N, Motin V L, Protsenko O A, Smirnov G B. Sequencing of two Yersinia pestis IS elements, IS285 and IS100. Contrib Microbiol Immunol. 1995;13:306–309. [PubMed] [Google Scholar]
- 16.Finegold M J, Petery J J, Berendt R F, Adams H V. Studies on the pathogenesis of plague. Am J Pathol. 1961;53:99–114. [PMC free article] [PubMed] [Google Scholar]
- 17.Galyov E E, Håkansson S, Wolf-Watz H. Characterization of the operon encoding the YpkA Ser/Thr protein kinase and the YopJ protein of Yersinia pseudotuberculosis. J Bacteriol. 1994;176:4543–4548. doi: 10.1128/jb.176.15.4543-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 19.Hayes F, Radnedge L, Davis M A, Austin S J. The homologous operons for P1 and P7 plasmid partition are autoregulated from similar operator sites. Mol Microbiol. 1994;11:825–830. doi: 10.1111/j.1365-2958.1994.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 20.Hinnebusch B J. Bubonic plague: a molecular genetic case history of the emergence of an infectious disease. J Mol Med. 1997;75:645–652. doi: 10.1007/s001090050148. [DOI] [PubMed] [Google Scholar]
- 21.Holmstrom A, Petterson J, Rosqvist R, Hakansson S, Tafazoli F, Fallman M, Magnusson K E, Wolf-Watz H, Forsberg A. YopK of Yersinia pseudotuberculosis controls translocation of Yop effectors across the eukaryotic cell membrane. Mol Microbiol. 1997;24:73–91. doi: 10.1046/j.1365-2958.1997.3211681.x. [DOI] [PubMed] [Google Scholar]
- 22.Iriarte M, Cornelis G R. Molecular determinants of Yersinia pathogenesis. Microbiol SEM. 1996;12:267–280. [PubMed] [Google Scholar]
- 23.Ishibashi Y, Claus S, Relman D A. Bordetella pertussis filamentous hemagglutinin interacts with a leukocyte signal transduction complex and stimulates bacterial adherence to monocyte CR3 (CD11b/CD18) J Exp Med. 1994;180:1225–1233. doi: 10.1084/jem.180.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S K, Wang J C. Localization of F plasmid SopB protein to positions near the poles of Escherichia coli cells. Proc Natl Acad Sci USA. 1998;95:1523–1527. doi: 10.1073/pnas.95.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutyrev V V, Popov Y A, Protsenko O A. Pathogenicity plasmids of the plague microbe (Yersinia pestis) Mol Genet Mikrobiol Virusol. 1986;6:3–11. [PubMed] [Google Scholar]
- 26.Mardis E R. High-throughput detergent extraction of M13 subclones for fluorescent DNA sequencing. Nucleic Acids Res. 1994;22:2173–2175. doi: 10.1093/nar/22.11.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Gallardo A, Lamerdin J, Carrano A V. Shot-gun sequencing. In: Adams M, Fields C, Venter J C, editors. Automated DNA sequencing and analysis. London, England: Academic Press; 1994. pp. 37–41. [Google Scholar]
- 28.McDonough K A, Falkow S. A Yersinia pestis-specific DNA fragment encodes temperature-dependent coagulase and fibrinolysin-associated phenotypes. Mol Microbiol. 1989;3:767–775. doi: 10.1111/j.1365-2958.1989.tb00225.x. [DOI] [PubMed] [Google Scholar]
- 29.Mills D S, Boland A, Sory M-P, Van der Smissen P, Kerbourch C, Finlay B B, Cornelis G R. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore R L, Brubaker R R. Hybridization of deoxyribonucleotide sequences of Yersinia enterocolitica and other selected members of Enterobacteriaceae. Int J Syst Bacteriol. 1975;25:336–339. [Google Scholar]
- 31.Mori H, Kondo A, Oshima A, Ogura T, Hiaga S. Structure and function of the F plasmid genes essential for partitioning. J Mol Biol. 1986;192:1–15. doi: 10.1016/0022-2836(86)90459-6. [DOI] [PubMed] [Google Scholar]
- 32.Mori H, Mori Y, Ichinose C, Niki H, Ogura T, Kato A, Hiraga S. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J Biol Chem. 1989;264:15535–15541. [PubMed] [Google Scholar]
- 33.Motin V L, Pokrovskaya M S, Telepnev M V, Kutyrev V V, Vidyaeva N A, Filippov A A, Smirnov G B. The difference in the lcrV sequences between Yersinia pestis and Yersinia pseudotuberculosis and its application for characterization of Y. pseudotuberculosis strains. Microb Pathog. 1992;12:165–175. doi: 10.1016/0882-4010(92)90050-x. [DOI] [PubMed] [Google Scholar]
- 34.Odaert M, Devalckenaere A, Trieu-Cuot P, Simonet M. Molecular characterization of IS1541 insertions in the genome of Yersinia pestis. J Bacteriol. 1998;180:178–181. doi: 10.1128/jb.180.1.178-181.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogiwara A, Uchiyama I, Seto Y, Kanchisa M. Construction of a dictionary of sequence motifs that characterize groups of related proteins. Protein Eng. 1992;5:479–488. doi: 10.1093/protein/5.6.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer L E, Hobbie S, Galan J E, Bliska J B. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-α production and down regulation of the MAP kinase p38 and JNK. Mol Microbiol. 1998;27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- 37.Perry R D, Fetherston J D. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persson C, Nordfelth R, Holmstrom A, Hakansson S, Rosqvist R, Wolf-Watz H. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 39.Pietrokovski S, Henikoff J G, Henikoff S. The Blocks database—a system for protein classification. Nucleic Acids Res. 1996;24:197–200. doi: 10.1093/nar/24.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Podladchikova O N, Dikhanov G G, Rakin A V, Heesemann J. Nucleotide sequence and structural organization of Yersinia pestis insertion sequence IS100. FEMS Microbiol Lett. 1994;121:269–274. doi: 10.1111/j.1574-6968.1994.tb07111.x. [DOI] [PubMed] [Google Scholar]
- 41.Porat R, McCabe W R, Brubaker R R. Lipopolysaccharide-associated resistance to killing of yersiniae by complement. J Endotoxin Res. 1995;2:91–97. [Google Scholar]
- 42.Portnoy D A, Falkow S. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J Bacteriol. 1981;148:877–883. doi: 10.1128/jb.148.3.877-883.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price S B, Cowan C, Perry R D, Straley S C. The Yersinia pestis V antigen is a regulatory protein necessary for Ca2+-dependent growth and maximal expression of low-Ca2+ response virulence genes. J Bacteriol. 1991;173:2649–2657. doi: 10.1128/jb.173.8.2649-2657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price S B, Straley S C. lcrH, a gene necessary for virulence of Yersinia pestis and for the normal response of Y. pestis to ATP and calcium. Infect Immun. 1989;57:1491–1498. doi: 10.1128/iai.57.5.1491-1498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Protsenko O A, Anisimov P I, Mozharov O T, Konnov N P, Popov I. Detection and characterization of Yersinia pestis plasmids determining pesticin I, fraction 1 antigen and mouse toxin synthesis. Genetika. 1983;19:1081–1090. [PubMed] [Google Scholar]
- 46.Protsenko O A, Filippov A A, Kutyrev V V. Integration of the plasmid encoding the synthesis of capsular antigen and murine toxin into Yersinia pestis chromosome. Microb Pathog. 1991;11:123–128. doi: 10.1016/0882-4010(91)90005-u. [DOI] [PubMed] [Google Scholar]
- 47.Rakin A, Boogakona J, Heesemann J. Structural and functional organization of the Yersinia pestis bacteriocin pesticin gene cluster. Microbiology. 1996;142:3415–3424. doi: 10.1099/13500872-142-12-3415. [DOI] [PubMed] [Google Scholar]
- 48.Rosqvist R, Skurnik M, Wolf-Watz H. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature. 1988;334:522–525. doi: 10.1038/334522a0. [DOI] [PubMed] [Google Scholar]
- 49.Roth J R A. Frameshift mutations. Annu Rev Genet. 1974;8:319–346. doi: 10.1146/annurev.ge.08.120174.001535. [DOI] [PubMed] [Google Scholar]
- 50.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 51.Salyers A A, Whitt D D. Bacterial pathogenesis: a molecular approach. Washington D.C: ASM Press; 1994. pp. 217–219. [Google Scholar]
- 52.Sandros J, Tuomanen E. Attachment factors of Bordetella pertussis: mimicry of eukaryotic cell recognition molecules. Trends Microbiol. 1993;1:192–196. doi: 10.1016/0966-842x(93)90090-e. [DOI] [PubMed] [Google Scholar]
- 53.Simonet M, Riot B, Fortineau N, Berche P. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect Immun. 1996;64:375–379. doi: 10.1128/iai.64.1.375-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skurnik M, Wolf-Watz H. Analysis of the yopA gene encoding the Yop1 virulence determinant of Yersinia spp. Mol Microbiol. 1989;3:517–529. doi: 10.1111/j.1365-2958.1989.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 55.Sodeinde O A, Goguen J D. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to ompT of Escherichia coli and gene E of Salmonella typhimurium. Infect Immun. 1989;57:1517–1523. doi: 10.1128/iai.57.5.1517-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Straley S C. The low-Ca2+ response virulence regulon of human-pathogenic Yersiniae. Microb Pathog. 1991;10:87–91. doi: 10.1016/0882-4010(91)90069-m. [DOI] [PubMed] [Google Scholar]
- 57.Straley, S. C. 1988. The plasmid-encoded outer-membrane proteins of Yersinia pestis. Rev. Infect. Dis. 10(Suppl. 2):S323–S326. [DOI] [PubMed]
- 58.Straley S C, Bowmer W S. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect Immun. 1986;51:445–454. doi: 10.1128/iai.51.2.445-454.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Veen H W, Callaghan R, Soceneantu L, Sardini A, Konings W N, Higgins C F. A bacterial antibiotic resistance gene that complements the human multidrug-resistance P-glycoprotein gene. Nature. 1998;391:291–295. doi: 10.1038/34669. [DOI] [PubMed] [Google Scholar]
- 60.Welkos S L, Davis K M, Pitt L M, Worsham P L, Frielander A M. Studies on the contribution of the F1 capsule-associated plasmid pFra to the virulence of Yersinia pestis. Contrib Microbiol Immunol. 1995;13:299–305. [PubMed] [Google Scholar]
- 61.Zychlinsky A M, Prevost M C, Sansonetti P J. Shigella flexneri induces apoptosis in infected macrophages. Nature. 1992;358:167–169. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]