Summary
Most microorganisms found in environmental samples have never been cultured and can often only be explored through molecular or microscopic approaches. Here, we adapt the use of in situ diffusion-based devices to culture “yet-to-be-cultured” microorganisms associated with coral mucus and compare this with a traditional culturing method. The culturability of microorganisms associated with mucus of the coral Pocillopora damicornis increased by 420% and 570% with diffusion growth chambers and microwell chip devices, respectively, compared with the traditional method tested. The obtained cultures represent up to 64.4% of the total diversity of amplicon sequence variants (ASVs) found in the mucus of the coral P. damicornis. In addition, some previously uncultured microorganisms, such as members of the family Nitrosopumilaceae and halophilic/halotolerant bacteria were cultured. Our results validate alternative microbial culturing strategies to culture coral-associated microorganisms, while significantly increasing the culturability of previous microbial dark matter.
Subject areas: Marine organism, Zoology, Microbiology, Marine biotechnology
Graphical abstract
Highlights
-
•
In situ devices boost coral microbe culturability up to 570%
-
•
Bacterial cultures represented up to 64.4% of the total diversity found in coral mucus
-
•
Cultured previously uncultured microbes
-
•
Strategies adaptable to marine organisms and ecosystems
Marine organism; Zoology; Microbiology; Marine biotechnology
Introduction
The large discrepancy between microbial cell counts under the microscope and those from plate counts was coined the “Great Plate Count Anomaly” almost 40 years ago,1 although it is still relevant. Sequencing surveys of the 16S rRNA gene have confirmed that the vast majority of microorganisms on Earth are still uncultured and thus remain largely uncharacterized.2,3 This uncultured fraction is frequently called “microbial dark matter,”2,4,5 and metagenomic analyses have helped predict its metabolic potential. However, these predictions still lack functional validation, which often requires cultured isolates. To address this, and to fully understand the biology, ecological roles, and functions, new approaches to “culture the uncultured” have been proposed and implemented, including the promising strategy of in situ cultivation.5,6,7
Corals are holobionts, as they establish complex symbiotic relationships with a diverse set of microorganisms.8,9 These symbionts include algae, bacteria, fungi, archaea, and viruses, which are capable of shaping the host’s health and performance.10,11,12,13,14 Recent research has shown that microorganisms could be a key component to enhance the growth of corals, as well as their resilience and tolerance to environmental impacts.14,15,16,17,18 Unfortunately, most microbial taxa found in corals through molecular methods belong to the “microbial dark matter,”19 and the genomes of very few have been sequenced.20 In fact, out of 21,100 16S rRNA gene sequences available in the Coral Microbiome Database (CMP—https://vamps2.mbl.edu/portals/CMP), only 6.5% were generated from cultured isolates.19,20 Furthermore, the majority of these cultured species belong to the phylum Pseudomonadota (syn. Proteobacteria, mostly Gamma- and Alphaproteobacteria), Bacillota (syn. Firmicutes), and Actinomycetota.19 In contrast, 16S rRNA gene amplicon sequencing data have revealed that 39 different bacterial phyla plus two archaeal phyla are associated with corals, of which only 14 have cultured representatives.19 Around 87.4% of sequences were identified as uncultured based on PCR-based methods or clone libraries, whereas for the remaining sequences (6.1%) no information on culturing status was available in the CMP.
Culturing the uncultured diversity of coral-associated microbiomes is key for understanding specific mechanisms underlying their ecological roles and symbiotic relationships with corals, as well as for biotechnological and experimental applications, including the development of coral probiotics.11,21 In order to plan an effective cultivation strategy, it is essential to consider that coral microbiomes are highly versatile and are affected by various factors such as environmental conditions,22,23,24,25 geographical location, host-specific variations, the coral’s state of health, and anatomical features.26 The surface mucus layer of corals is an excellent target for the search for novel microorganisms with biotechnological and experimental potential, as it serves as the primary barrier of coral protection27 and harbors a wide diversity of microorganisms able to produce bioactive metabolites with key ecological roles.28 By enhancing the culturability of coral-mucus-associated microorganisms through new cultivation strategies and technologies (i.e., development of new devices or approaches), we aim here to extend the repertoire of cultured microorganisms for the coral microbiome. Specifically, we test in situ culturing strategies to assess the still uncultured coral mucus microbiome. To the best of our knowledge, our study represents the first attempt to expand the cultivability of the coral holobiont using in situ cultivation approaches.
Results
To assess the capability of cell migration from the environment toward the device’s (chips and chambers) inner walls, we performed a leakage assay by semi-quantifying the cell growth inside the devices incubated with and without mucus inoculum, using colony-forming unit (CFU) count in solid culture media. CFU counts were significantly lower in the chips incubated without mucus (4.199, Log10 CFU/mL, ±0.223) in comparison to those incubated with mucus (4.758 Log10 CFU/mL, ±0.237; p value = 0.0008, ANOVA, Figure S1), indicating the possibility of slight contamination through the 0.03-μm pore-size membrane. There were no significant differences between CFU counts for chambers with (6.494 Log10 CFU/mL, ±0.207) and without inoculum (6.494 Log10 CFU/mL, ±0.167; p value = 1.000, ANOVA, Figure S1). This could be explained by the 0.2-μm pore-size membranes used for the chambers allowing a greater range of microorganisms to enter. To compensate for this issue, we only considered the ASVs that were observed in the original coral mucus sample for further analysis.
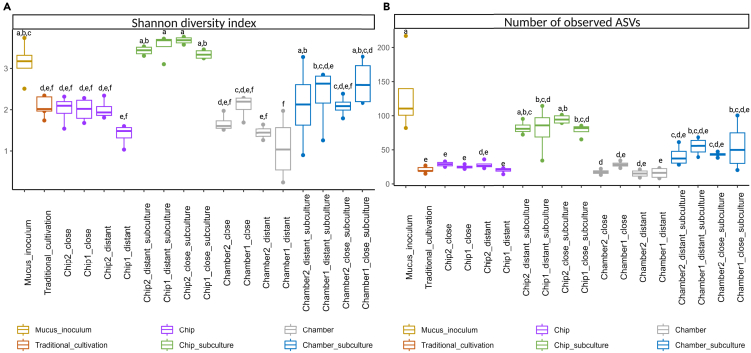
Microbial diversity (Shannon index, Figure 1A) and richness (observed number of ASVs, Figure 1B) increased in the sub-cultivation process (plate wash) for both devices compared with the device contents and traditional cultivation (Table S1). No significant differences were observed in diversity and richness when comparing traditional cultivation and contents of both devices without sub-cultivation (chip and chamber contents, Table S1). The diversity of ASVs obtained from chip sub-cultivation was significantly higher than the diversity obtained using traditional cultivation (p < 0.001) and similar to the diversity obtained from direct sequencing of the mucus inoculum (Figure 1A and Table S1). The incubation distance of the chamber and chip devices from the corals and the different dilutions loaded into the devices did not affect the diversity (Figure 1A) or richness (Figure 1B) of ASVs in the in situ cultivation step (device contents) or in the sub-cultivation processes.
Figure 1.
Alpha diversity indexes
(A) Shannon diversity index and (B) the number of observed amplicon sequence variants (ASVs) of samples from tested cultivation methods with direct amplicon sequencing of the mucus inoculum; different lowercase letters indicate significant differences between treatments (p < 0.05).
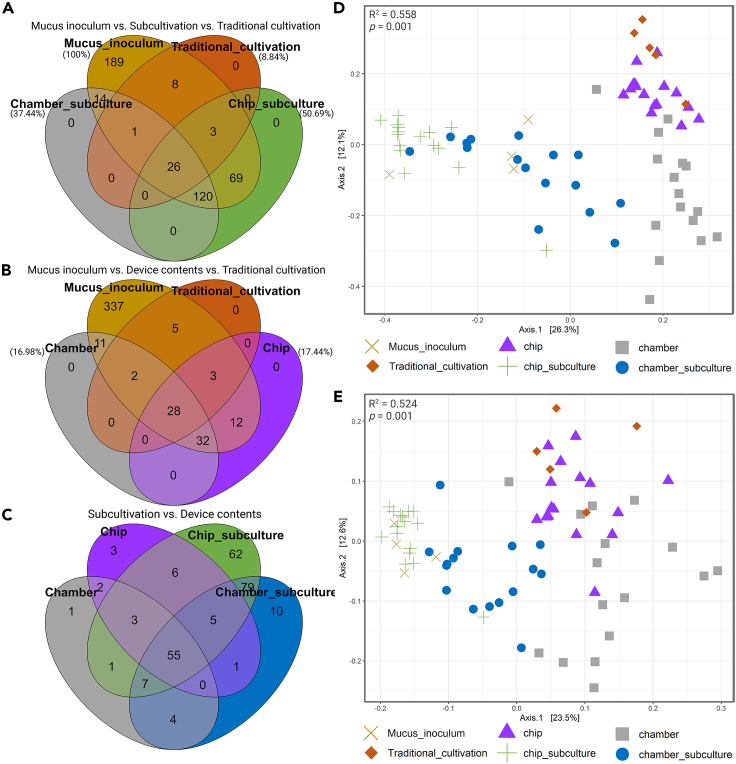
Remarkably, up to 50.69% of the ASVs detected in the mucus inoculum (480 ASVs) were also found in the chips’ sub-cultures (218 ASVs; Figures 2A and 2B), which is a 5.7-fold increase in comparison with traditional cultivation (38 ASVs shared with the mucus inoculum). Interestingly, there was no difference between the microbial community structure (weighted UniFrac distance) at the ASV level of the subculture of chip-1 distant (mucus inoculum at 10−1 dilution, incubated distant from corals) and the mucus inoculum (p value = 0.167; Figure 2D and Table S2), indicating a substantial overlap in the diversity of these samples. In addition, no significant differences, using Sørensen dissimilarity (presence/absence), were found between chip subcultures incubated with a 1/100 dilution and the mucus inoculum (Figure 2E and Table S2). Similar results were observed between chamber subcultures and mucus inoculum (Figure 2E and Table S2). In addition, we were able to retrieve up to 37.44% of the original ASVs via chamber subcultures (161 ASVs, Figure 2A), which is an increase of 4.2-fold compared with traditional cultivation for which only 8.84% (38 ASVs) of the ASVs in the mucus inoculum were found. Combining the sub-cultivation (chamber and chips) with traditional cultivation and the cultured content from the devices, 277 (64.42%) of the total mucus-associated ASVs were recovered and 233 (54.19%) of these had grown in the subcultures from both device types (Figures 2A–2C).
Figure 2.
Venn diagrams and Beta diversity measurements comparing the samples
The numbers of amplicon sequence variants (ASVs) shared between cultivation samples: (A) direct amplicon sequencing of the mucus inoculum versus sub-cultivation versus traditional cultivation, (B) mucus inoculum versus device contents versus traditional cultivation, and (C) sub-cultivation versus device contents. The values in brackets shown in (A) and (B) represent the percentage of ASVs grown from mucus inoculum (100%) in the different culture methods. (D) Comparison of community structure (weighted UniFrac distances) and (E) composition (Sørensen dissimilarity) using Principal Coordinates Analysis (PCoA).
The ASV structure of chip subcultures from closely incubated devices and the chip-2 distant (mucus inoculum at 10−2 dilution, incubated distant from corals) subcultures showed significant differences to those found in the mucus inoculum (Table S2). However, no significant differences were found between the subcultures from chips incubated close to the corals and the subcultures from distant chips (p value [PERMANOVA] = 0.866 and 0.054 for 10−1 and 10−2 dilutions, respectively). In addition, no differences were found between the different dilutions loaded into the chips (p value [PERMANOVA] = 0.452 and 0.301 for distant and close samples, respectively). The same was observed for the chamber subcultures (Table S2), indicating that neither the amount of cells loaded into the in situ cultivation devices nor the distance from the coral affects the diversity of microorganisms cultured.
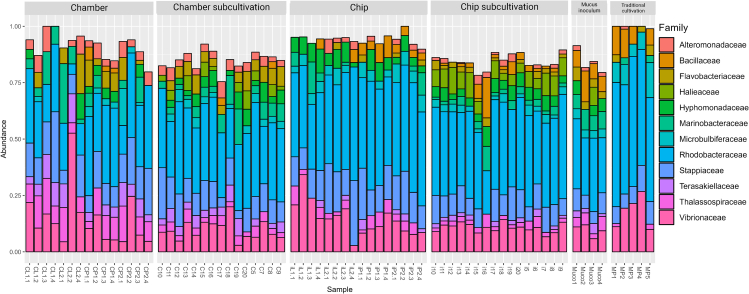
Enrichment of certain microbial taxa was observed with the different tested cultivation techniques (Figure 3). For instance, both the sub-cultivation of chip and chamber contents allowed for the growth of members of the Halomonadaceae, Haliaceae, and other families, whereas traditional cultivation was more selective toward members of the Bacillaceae, which were rarely, or not, recovered by the other techniques (Figure 3).
Figure 3.
The microbial composition of different cultivation methods and the original mucus inoculum
The y axis represents the relative abundance of microbial groups at the family level, whereas the x axis indicates the sample name.
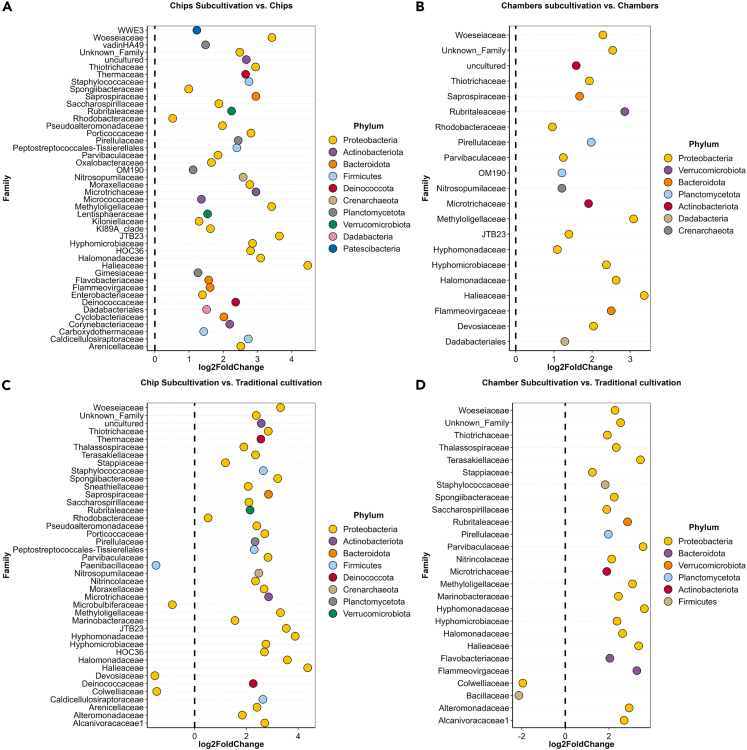
As mentioned above, the sub-cultivation of the device contents allowed for the growth of several microbial taxa found in P. damicornis mucus. Examination of the ASVs enriched in DeSeq2 analysis revealed a number of families were enriched in the chip subcultures in comparison to chip contents, including 44 families from 10 different phyla (Figure 4A). Among those enriched groups, eight ASVs were unclassified (assigned as “uncultured” or “NA”) (see Data S1) at the genus level by the SILVA database. Several families with many uncultured representatives were enriched, including from the placeholder taxa WWE3 (phylum Patescibacteria),29,30 vadinHA49 (phylum Planctomycetota),31,32 OM190 (phylum Planctomycetota),33 JTB2334,35 and HOC3636 (phylum Pseudomonadota), plus one unknown family with incertae sedis taxonomic status (Figure 4A and Data S1). One ASV assigned to the Crenarchaeota was also detected in the chip subcultures.
Figure 4.
Differential abundance analysis
Differential abundance of families between sub-cultivations and contents for (A) chips and (B) chambers, respectively; (C) differential abundance of chip sub-cultivation versus traditional cultivation; and (D) chamber sub-cultivation and traditional cultivation. Differential abundance was determined by using DESeq2 to identify ASVs showing significant differences in abundance between cultivation strategies. Their relative abundance was then aggregated on the family level with fold-change estimates used to quantify the magnitude of these differences.
Pseudomonadota was the phylum with the largest number of differentially abundant families (n = 21) enriched in the chip sub-cultivations compared with the chip contents (Figure 4A). Seven phyla from the mucus inoculum were enriched in the comparison between chamber sub-cultivation and chamber contents, including 21 different microbial families and six unclassified ASVs (assigned as “uncultured” or “NA”) at the genus level (Data S1). The families with the placeholder names Subgroup 9 (phylum Acidobacteriota), OM190 (phylum Planctomycetota), and JTB23 (phylum Pseudomonadota) were also enriched in this comparison. ASVs belonging to the Pseudomonadota were mostly enriched in the chamber sub-cultivation compared with the chamber content, followed by Verrucomibriota and Bacteroidota. Members of the ammonia-oxidizing archaea (AOA) family Nitrosopumilaceae were also enriched in the chamber subculture compared with the chamber content (Figure 4B).
Following the previous observations, we compared the differential abundance between traditional cultivation and subcultures. For chip subcultures (Figure 4C), 41 families from eight phyla were enriched in comparison to traditional cultivation, including six ASVs unclassified (assigned as “uncultured” or “NA”) at the genus level (Data S1), groups JTB23 and HOC36, and a family of AOA (Nitrosopumilaceae), among several others. For chamber subcultures (Figure 4D), 26 families from six phyla were differentially abundant. Pseudomonadota and Bacillota were the only two phyla with differentially abundant families associated with traditional cultivation, compared with subcultures from both devices.
In situ cultivation improved the isolation of abundant taxa from the mucus layer
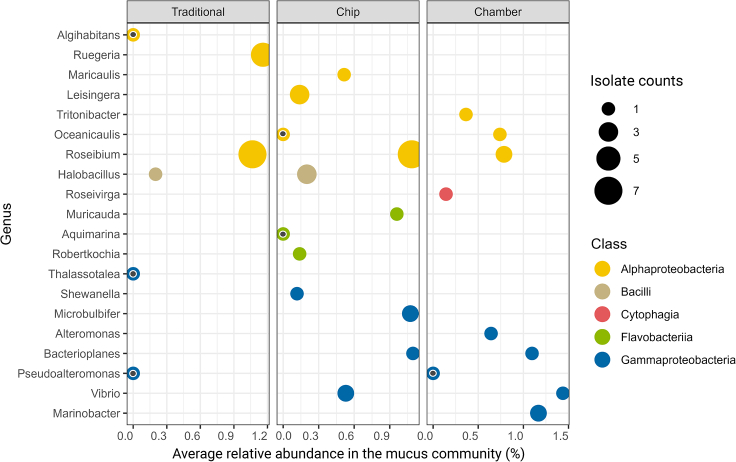
Pure isolates were obtained by plate re-streaking, including 24 isolates from chip subcultures, 11 from chamber subcultures, and 16 from traditional cultivation. Full details of the isolates are available in Table S3, including their NCBI accession numbers. The 16S rRNA gene sequences of several isolates matched, with 100% identity, to ASVs with high relative read abundances in the mucus inoculum (>0.5%); these isolates were mainly recovered from the device sub-cultivations. The genus Roseibium (Alphaproteobacteria) was the only taxon isolated by all culturing approaches, showing >0.8% of average relative read abundance in the original mucus community. Halobacillus isolates were obtained from both chip subcultures and traditional cultivation. Isolates of the genus Pseudoalteromonas were retrieved by traditional cultivation and from chamber subcultures, and four genera were exclusive to the chamber and seven to chip subcultures. Additionally, three genera were isolated only from traditional cultivation (Figure 5). The majority of pure cultures from traditional cultivation belonged to the class Alphaproteobacteria, while we were able to isolate microorganisms from five different classes from chip and four from chamber subcultures (Figure 5). Using 98.6% gene sequence similarity of the full-length 16S rRNA as a threshold for differentiating species level,37 33.3% (n = 8) of the isolates from chips were potentially new species, in comparison to 18.2% (n = 2) from chambers and 6.25% (n = 1) from traditional cultivation (Table S3).
Figure 5.
Genus and class level assignment of the microbial isolates retrieved from different cultivation methods
The average relative abundance of each genus in the mucus microbial community is shown on the x axis. A black inner dot indicates that the average relative abundance was 0%.
Discussion
Studies over the last few decades have recognized the diversity and importance of microorganisms living in association with corals.8,10 More recent studies have indicated that microbial therapies can be used to improve the growth, stress tolerance, and resilience of corals.11,14,15,16,17,18,38 However, the majority of these therapies are biased toward the use of a few microbial groups that can be readily cultured and maintained under laboratory conditions, including bacterial representatives from the phyla Pseudomonadota, Bacillota, Bacteroidota, and Actinomycetota. This study represents the first attempt to expand the culturability of coral-associated microorganisms using in situ cultivation approaches, focusing on the surface mucus layer. We decided to use the mucus layer for two main reasons: (1) it is a non-invasive in situ culturing approach that avoids damage to the coral tissues, which could lead to dysbiosis and consequently affect the retrieval rates and/or improve the growth of opportunistic microorganisms; (2) the mucus layer is considered to be a coral’s first barrier of protection,27,28 harboring a diversity of microorganisms with potential beneficial traits for the host, besides the biotechnological capacities.27
The use of in situ cultivation (chip) allowed us to retrieve up to 50.69% of the microbial community from the mucus layer, an enhancement of up to 5.7-fold compared with traditional cultivation (i.e., direct cultivation on marine agar only allowed the recovery of 8.84% of the ASVs found in the mucus inoculum). The combined use of chips, chambers, and traditional culturing allowed the growth of up to 64.42% of the mucus ASVs. One of the advantages of using in situ approaches is that growth factors are provided in real time and under natural concentrations for a prolonged period.7 Diffusion chambers have also been used to increase the culturability of sponge-associated microbes by up to 339%, including previously uncultured bacteria.6
A sub-cultivation step was found, in this study, to be essential to allow the enrichment of the microorganisms that were retained inside the in situ cultivation devices, increasing the recovery of microbial growth by 420% for chamber and 570% for chip subcultures, compared with traditional cultivation. The capacity of many microorganisms to grow under regular culture media after a prolonged in situ incubation period only has been referred to as “domestication.”7 The mechanisms underlying this domestication are still unknown, but it was postulated that the microorganisms are first “acclimatized” to the culture medium provided in the device’s inner compartment, allowing the cells to better grow under in vitro conditions using the same culture medium. Some experiments have also shown that resubmitting the grown contents from the in situ cultivation devices to subsequent runs of in situ incubation can enhance final culturability.6,7
Current diffusion-based devices and in situ cultivation strategies include the following descriptions: (1) platforms that permit the complete isolation of the microbial cells inside the device using membranes, allowing just growth factors to cross the membranes to the devices’ interior, such as isolation chips7 (or iChips), FACS-iChip,39 and the Microbe Domestication Pod40 (MD Pod); (2) devices based on membranes that allow the diffusion of growth factors and partial cell migration toward the devices’ interior, for example, cultivation chambers,6 diffusion bioreactors,41 and “trap-chambers” for actinobacteria41,42; and lastly, (3) platforms that allow the microbial cell to cross freely through the environment to the device interior and vice versa, such as I-Tips43 and the μMicrobial Domestication Pod44 (μMD Pod).
Based on our results comparing inoculated with uninoculated devices, the chips used here belong to the second group, whereas chambers should be classified as part of the third group. Specifically, Alkayyali and collaborators40 tested the efficacy of cell migration into or out of devices using different MD Pod platforms and 0.03-μm pore-size membranes, which is the same pore size of chips used here. The success rate of achieving complete isolation of cells varied between 11% and 78%, depending on the complexity and design of the MD pods.40 Interestingly, the study found that MD pod devices with lower complexity (i.e., fewer body parts and less time-consuming assembly) tended to yield better results in terms of isolation effectiveness than more complex designs.40 These results show that even with strategies aimed at complete isolation, there may be some degree of partial leakage due to the design features of the device. Wheatley et al.44 addressed the same issue by suggesting the use of devices manufactured from a single piece and without a membrane. However, the majority of studies employing diffusion-based devices or in situ cultivation do not account for potential migration/leakage,6,39,41 although our results indicate the need for excluding contaminants from the environment when assessing and comparing culturability. Additional experiments, such as those described by Nichols et al.45 with adaptations, could be explored in future research to offer a more comprehensive understanding of microbial migration within these in situ devices. Briefly, these authors assembled and autoclaved their devices in a liquid culture medium,45 and subsequently, the culture medium was inoculated with microbial cells to observe whether the microorganisms could cross the membrane barrier or not. Nevertheless, our data also corroborate the significant improvement in culturability through in situ devices.
The development of cutting-edge in situ devices tailored to the unique needs of microbial cultivation and ecosystem management can provide controlled environments that mimic natural conditions, fostering the growth of diverse microorganisms.7 Moreover, the synergistic combination of different fields, such as microbiology and material science,46 can lead to the use or creation of advanced materials to further enhance microbial colonization and promote the establishment of stable microbial communities. In this study, we successfully used Teflon as the raw material to build the chips that can be instrumental in selecting probiotics for coral reef restoration. This approach opens up opportunities for exploring novel biomaterials and biocompatible coatings that can be used to design innovative devices for various applications beyond coral ecosystems. For instance, the knowledge gained from this research could be extended to developing in situ devices for studying and enhancing microbial communities recovered from other environments, such as wastewater treatment systems,47,48 oil bioremediation,49,50,51 agricultural systems,52,53,54 or human health settings.55,56 Thus, this study contributes to the understanding of coral-microbe interactions and paves the way for exciting advancements at the intersection of microbiology and material science, with far-reaching implications for science, technology, and sustainability.
The growth of moderate/slow growers is one of the benefits of using in situ devices. For example, microorganisms from the ammonia-oxidizing family Nitrosopumilaceae, with specific nutritional demands,57 were obtained from the coral mucus. Halophilic/halotolerant taxa were also cultured and isolated, such as members of the genera Halobacillus, Robertkochia, Maricaulis, and Roseivirga,58,59,60,61 in addition to members of the family Halomonadaceae,62 which are commonly cultured in the regular medium.20 Moreover, our study has resulted in the isolation of several potential new bacterial species, the majority of which were obtained from chip cultivations.
Putative beneficial microorganisms for corals11,12,63 were also cultured in this study. For instance, members of the genera Tritonibacter and Leisingera were isolated, which have the known capacity to degrade dimethylsulfoniopropionate (DMSP),64,65 a trait that has been recently validated as beneficial for the coral holobiont.14 Taxa with the potential for hydrocarbon degradation were also cultured, such as members of the families Thalassospiraceae66 and Haliaceae,67,68 and this might benefit hydrocarbon-contaminated reefs. Moreover, the isolates of the genera Muricauda and Aquimarina (class Flavobacteriia) have been reported as scavengers of reactive oxygen species (ROS).69,70,71 ROS mitigation was suggested as an important trait for beneficial microorganisms for corals,11 as ROS is associated with the coral bleaching process.72 In addition, several previously unclassified (“uncultured” or “NA”) taxa were enriched in the sub-cultivations, including members of the phyla Planctomycetota, Actinobacteriota, Acidobacteriota, and Pseudomonadota, with no description of the function for the host attributed to these, hitherto, uncultured groups.
In summary, the wide diversity of microorganisms cultured in this study includes many taxa that are still uncharacterized and could harbor unknown functions that can be beneficial to the coral host. This study is the start of a broader exploration of the diversity and ecology of host-associated microorganisms and their relationships with their host, including the discovery of new beneficial traits21 to be used as part of the customized medicine for corals platforms.73
Limitations of the study
This study represents the first attempt to develop in situ microbial culturing strategies for coral-associated microorganisms, thereby increasing the culturability of previously inaccessible microbial dark matter. Our analysis, along with previous research, suggest that there can still be partial leakage due to the design features of the devices for in situ cultivation, even when devices are designed for complete isolation. This highlights the need to account for potential migration or leakage when assessing and comparing culturability, a factor often overlooked in previous studies using diffusion-based devices. To address this limitation and gain a more comprehensive understanding of microbial migration within these devices, future research could explore experimental adaptations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Bacterial isolates (n = 51) | P. damicornis mucus | Available at Table S3 |
| Biological samples | ||
| Pocillopora damicornis | Ocyan Reef coral farm (Itú, São Paulo, Brazil) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Marine Agar 2216 | BD DIFCO™ | Cat#212185 |
| Marine Broth 2216 | BD DIFCO™ | Cat#279110 |
| Agar | BD DIFCO™ | Cat#281210 |
| Critical commercial assays | ||
| DNeasy PowerSoil Pro Kit | QIAGEN | Cat#47016 |
| Genomic DNA Purification Kit | Wizard® | Cat#A1120 |
| Deposited data | ||
| Raw amplicon sequencing data | This study/NCBI’s SRA | PRJNA934364 |
| 16S rRNA sequences from all 51 isolates | This study/NCBI’s GenBank | Available at Table S3 |
| Oligonucleotides | ||
| 515F | Apprill et al.74 | 5′-GTGYCAGCMGCCGCGGTAA-3′ |
| 806R | Apprill et al.74 | 5′-GGACTACNVGGGTWTCTAAT-3′ |
| 27F | Weisburg et al.75 | 5′-AGAGTTTGATCCTGGCTCAG -3′ |
| 1492R | Weisburg et al.75 | 5′-CTACGGCTACCTTGTTACGA -3′ |
| Software and algorithms | ||
| QIIME2 (v.2022.2) | Bolyen et al.76 | https://forum.qiime2.org/ |
| DADA2 | Callahan et al.77 | https://benjjneb.github.io/dada2/ |
| Phyloseq | McMurdie and Holmes78 | https://github.com/joey711/phyloseq |
| DESeq2 | Love et al.79 | doi:10.18129/B9.bioc.DESeq2 |
| Vegan (v.2.6–4) | https://rdrr.io/cran/vegan/ | https://github.com/vegandevs/vegan |
| ggplot2 (v.3.4.0) | https://cran.r-project.org/web/packages/ggplot2/index.html | https://github.com/tidyverse/ggplot2 |
| BioEdit 7.2 | Hall80 | https://bioedit.software.informer.com/7.2/ |
| Other | ||
| Centrifugal Microfilters | Whatman™ Centrex™ | Cat#32-10467015 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Raquel Peixoto (raquel.peixoto@kaust.edu.sa).
Materials availability
Bacterial strains isolated in this study have been conserved at the Molecular Microbial Ecology Lab’s biobank at the Federal University of Rio de Janeiro - Rio de Janeiro, Brazil (zip code 21941-902), under the same unique IDs as available in Table S3.
Materials reported in this paper may be provided by Fluvio Modolon (modolonfluvio@gmail.com) or Raquel Peixoto upon request.
Data and code availability
-
•
Raw amplicon sequencing data are available on NCBI’s Read Archive under BioProject: PRJNA934364. The 16S rRNA sequences from isolates are available on NCBI’s GenBank under the IDs listed in Table S3. All datasets are publicly available as of the date of publication.
-
•
The complete bioinformatic pipeline including scripts for figure reproduction is publicly available through the GitHub repository at <https://github.com/modolon/insitu_cultivation>.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Devices for in situ cultivation
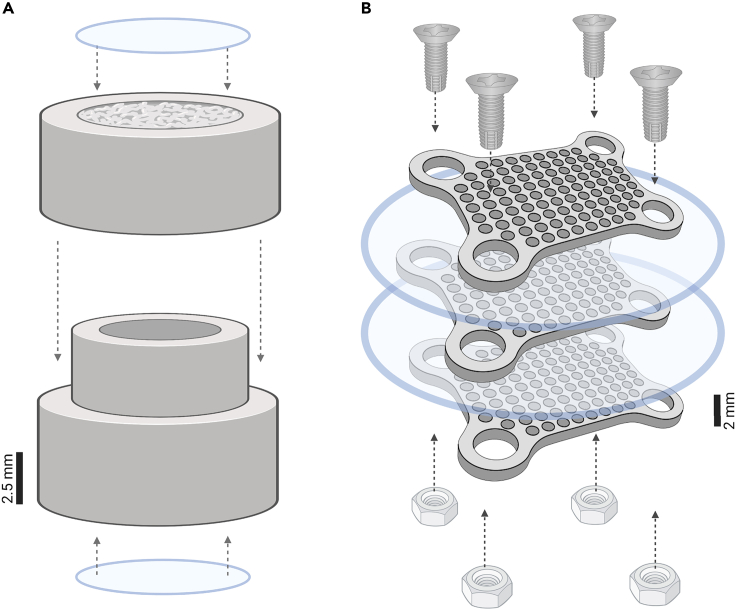
The devices used here for in situ cultivation are based on the assumption that the diffusion of growth factors from the environment or the coral through membranes toward the interior of the devices will facilitate the growth of microorganisms. Two different devices were adapted for this purpose: (i) diffusion growth chambers (or ‘chambers’) and (ii) microwell chips (or ‘chips’). The chambers were adapted from Steinert et al.,6 in which they were originally developed for the cultivation of sponge-associated microorganisms. The chambers consist of two sterile centrifuge tubes with membranes (Whatman Centrex, pore size 0.2 μm), cut using a scalpel and attached to each other (see below figure [A]) to form a single chamber, and sealed with high-purity silicone sealant for aquariums (Maxx Tekbond, Brazil). The microwell chips consist of three identical layers made from Teflon with 63 microchambers (1 mm3) and two membranes (0.03 μm pore size), sealed with four screws and nuts (see below figure [B]). The chip components were sterilized by autoclaving (20 min, 121°C) before their assembly. Technical details on the chip design are shown in Figure S2.
Design of devices for in situ cultivation
Schematic representation of (A) chamber and (B) chip devices. The arrows indicate the direction of closing and sealing the devices. Panel A was created with Biorender.com.
Aquarium setup
The experimental set-up encompassed a 10 L aquarium, placed in a water bath to maintain the temperature around 26°C. The aquarium had an individual 26 L sump to form a circulating loop using a water pump (Mini A, Sarlo Better, São Caetano do Sul, Brazil), with no water exchange between the aquarium and water bath. The aquarium received artificial light, with day/night cycles to simulate natural conditions. The temperature of the water bath was controlled with Full Gauge controls MT-518ri (Canoas, Brazil) connected to heaters and pumps that fed the cooled water from a master tank to the water baths. Partial water changes (30%) were performed three times a week. Pocillopora damicornis clonal colonies were obtained from Ocyan Reef coral farm (Itú, São Paulo, Brazil) and placed into the aquarium for acclimation for 10 days, after which the in situ cultivation was started.
In situ incubation and microbial cultivation
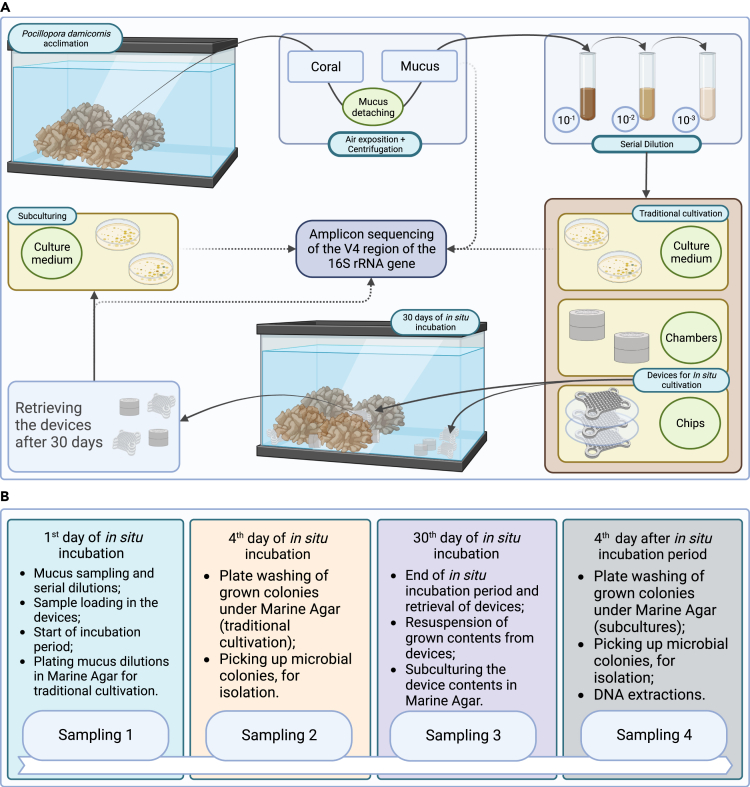
The schematic representation of the experiment, as well as the timeline for sampling, are shown in below figure. On the first day, four replicate coral fragments were sampled. Since the aim of the experiment was to validate the microbial culturing methods, devices and culturing processes were replicated, and only one coral fragment was used for inoculation to minimize the impact of biological variation on subsequent cultivation.
Experimental workflow and sampling timeline in in situ incubation
Schematic figure showing (A) the experiment workflow and (B) the timeline of sampling points starting on the first day of in situ incubation. This figure was created with Biorender.com.
The surface mucus layer material of the selected coral was sampled. First, the coral was rinsed with sterile artificial seawater and exposed to the air for 5 min to induce mucus exudation, and immediately subjected to centrifugation at room temperature (8000 g, 2 min). The mucus layer collected was diluted in sterile saline solution (0.85%) up to 10-3. Aliquots of all dilutions were plated onto Marine Agar (MA) in five replicates and incubated at 26°C for up to four days, representing a traditional cultivation approach.6,20 Once grown, all colonies on the plates were retrieved by plate-wash in 4 mL of sterile saline solution (0.85%) (adapted from Keller-Costa et al.81). Some colonies were also randomly picked from non-washed plates and successively streaked twice on MA for further purification.
The same mucus sample used in the previous dilutions for plating was also used for in situ incubation using the devices. Aliquots for in situ incubation were diluted (10-1 and 10-2) in Marine Broth +0.25% agar (MA 0.25) to make a viscous medium to adhere to the wells of the chips or the chambers. Each chip well was filled with 1 μL of the diluted sample. Chambers were filled with 300 μL of dilution. To assess the migration of microbial cells into or out of the cultivation devices, a leakage assay was carried out, where replicates of both devices were incubated with sterile MA 0.25 plus sterile saline solution (0.85%) before they were placed into the aquarium. We tested whether the distance from the coral influenced the cultivation by placing replicate devices (n = 4) close (2–3 cm) or distant (13–15 cm) from the corals (Figure S3). After 30 days of incubation, the devices were recovered in order to proceed with the next steps.
Microorganisms growing inside the chips (chip content) and chambers (chamber content) were retrieved by aspiration and DNA was extracted. For the subculturing, the device contents were diluted in four replicates (up to 10-3) and plated onto MA and incubated at 26°C for four days. Grown colonies were randomly picked and streaked in MA for further isolation, and the full content of the plates was retrieved by plate wash as described above.
Method details
Sampling, DNA extraction, and sequencing
Aliquots of 0.5 mL of mucus and plate-washed contents as well as the remaining aspirated contents from the devices were used for total DNA extraction with a DNeasy PowerSoil Pro Kit (QIAGEN). The V4 region of the 16S rRNA gene was amplified by PCR using the primers 515F 5′-GTGYCAGCMGCCGCGGTAA-3′ and 806R 5′-GGACTACNVGGGTWTCTAAT-3’ (appended with the universal Illumina tags) and thermocycling of 3 min initial denaturation at 94 °C followed by 32 cycles of 45 s at 94 °C, 1 min at 50 °C and 90 s at 72 °C, with a final extension step of 10 min at 72 °C. The resulting amplicons were barcoded, pooled, and sequenced with the Illumina Miseq platform according to the manufacturer’s instructions at the Ramaciotti Center for Genomics (UNSW). In total, 81 samples were amplicon sequenced (Table S4).
The Wizard Genomic DNA Purification Kit was used for the DNA extraction of isolates following the manufacturer’s protocol. Sanger sequencing was carried out to the full length of 16S rRNA gene of the isolates, using the universal primers 27F 5′- AGAGTTTGATCCTGGCTCAG -3′ and 1492R 5′- CTACGGCTACCTTGTTACGA -3’.
Data analysis
The 16S rRNA gene amplicon sequencing reads were imported into QIIME2 (v.2022.2)76 using the q2-toolsimport command and primers were trimmed using the q2-cutadapt command. Forward and reverse reads were truncated at positions 132 and 130, respectively. DADA2 implemented into QIIME2 was used for denoising and clustering of the amplicon sequence variants (ASVs).77 The command q2-feature-classifier classify-sklearn and the SILVA database v138.1 were used to infer taxonomy. Finally, phylogenetic trees were built using q2-align-to-tree-mafft-fasttree. Qiime2 data (feature table, taxonomy table, and rooted tree), in addition to metadata, were exported as phyloseq objects for further analysis in R (version 4.1.2). Rarefied data were used to generate alpha-diversity indexes (observed number of ASV and Shannon diversity). Raw ASV data were normalized with DESeq279 and used to calculate weighted Unifrac distance or Sørensen dissimilarity, which were visualized via Principal coordinates analysis (PCoA) using the phyloseq package implemented in R. The package DESeq2 was used to evaluate the differential relative read abundances between sample groups. Plots and graphs were generated in the package ggplot2 (version 3.4.0).
The 16S rRNA gene sequences of the isolates were manually trimmed and the contigs were assembled in BioEdit 7.2. Following exploratory analysis, BLASTN analysis was carried out against the NT database of the National Institute of Biotechnology Information (NCBI) to identify isolates.
Quantification and statistical analysis
The statistical differences in the indices (alpha-diversity) were assessed by Analysis of Variance (ANOVA) for normal data and Wilcoxon for non-normal data (R, version 4.1.2, package stats v.3.6.2).
Permutational multivariate analysis of variance (PERMANOVA) was performed on the data matrix82 to compare the structure (weighted Unifrac) or composition (Sørensen dissimilarity) of the microbial communities. Permutational multivariate analysis of dispersion (PERMDISP) was also performed using the betadisper function (implemented in vegan 2.6–4) with 999 permutations.
Acknowledgments
This research paper was carried out in association with the ongoing R&D project registered as ANP 21005-4, “PROBIO-DEEP—survey of potential impacts caused by oil and gas exploration on deep-sea marine holobionts and selection of potential bioindicators and bioremediation processes for these ecosystems” (UFRJ/Shell Brasil/ANP), sponsored by Shell Brasil under the ANP R&D levy as “Compromisso de Investimentos com Pesquisa e Desenvolvimento.” We thank the Aquario Marinho do Rio (AquaRio), the AquaRio team, and the Programa de Pós-Graduação em Ciências (Microbiologia)—PPG-MICRO—Instituto de Microbiologia Paulo de Góes (IMPG) da Universidade Federal do Rio de Janeiro (UFRJ) for the support and resources provided during the experiments. R.S.P. was supported through KAUST grant number BAS/1/1095-01-01 and KAUST Center Competitive Funding (CCF) FCC/1/1973-51-01). F.M. received support from the National Council for Scientific and Technological Development (CNPq).
Author contributions
F.M., J.S., C.L.V., G.D., R.P., and T.T.: study conception and design and drafting of the manuscript. F.M., J.S., and RP: design of devices. F.M., J.S., G.D., and C.L.V.: development of the experiment. F.M. and G.D.: aquarium work. F.M. and T.T.: bioinformatic analyses. All authors: data analysis and interpretation and involved in critical revision.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Published: November 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108374.
Supplemental information
References
- 1.Staley J.T., Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 2.Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.-F., Darling A., Malfatti S., Swan B.K., Gies E.A., et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 3.Zamkovaya T., Foster J.S., de Crécy-Lagard V., Conesa A. A network approach to elucidate and prioritize microbial dark matter in microbial communities. ISME J. 2021;15:228–244. doi: 10.1038/s41396-020-00777-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiao J.-Y., Liu L., Hua Z.-S., Fang B.-Z., Zhou E.-M., Salam N., Hedlund B.P., Li W.-J. Microbial dark matter coming to light: challenges and opportunities. Natl. Sci. Rev. 2021;8:nwaa280. doi: 10.1093/nsr/nwaa280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schultz J., Modolon F., Rosado A.S., Voolstra C.R., Sweet M., Peixoto R.S. Methods and Strategies to Uncover Coral-Associated Microbial Dark Matter. mSystems. 2022;7:e0036722. doi: 10.1128/msystems.00367-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinert G., Whitfield S., Taylor M.W., Thoms C., Schupp P.J. Application of diffusion growth chambers for the cultivation of marine sponge-associated bacteria. Mar. Biotechnol. 2014;16:594–603. doi: 10.1007/s10126-014-9575-y. [DOI] [PubMed] [Google Scholar]
- 7.Berdy B., Spoering A.L., Ling L.L., Epstein S.S. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat. Protoc. 2017;12:2232–2242. doi: 10.1038/nprot.2017.074. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 9.Bourne D.G., Garren M., Work T.M., Rosenberg E., Smith G.W., Harvell C.D. Microbial disease and the coral holobiont. Trends Microbiol. 2009;17:554–562. doi: 10.1016/j.tim.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Reshef L., Koren O., Loya Y., Zilber-Rosenberg I., Rosenberg E. The coral probiotic hypothesis. Environ. Microbiol. 2006;8:2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- 11.Peixoto R.S., Rosado P.M., Leite D.C.d.A., Rosado A.S., Bourne D.G. Beneficial Microorganisms for Corals (BMC): Proposed Mechanisms for Coral Health and Resilience. Front. Microbiol. 2017;8:341. doi: 10.3389/fmicb.2017.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peixoto R.S., Sweet M., Villela H.D.M., Cardoso P., Thomas T., Voolstra C.R., Høj L., Bourne D.G. Coral Probiotics: Premise, Promise, Prospects. Annu. Rev. Anim. Biosci. 2021;9:265–288. doi: 10.1146/annurev-animal-090120-115444. [DOI] [PubMed] [Google Scholar]
- 13.Voolstra C.R., Suggett D.J., Peixoto R.S., Parkinson J.E., Quigley K.M., Silveira C.B., Sweet M., Muller E.M., Barshis D.J., Bourne D.G., Aranda M. Extending the natural adaptive capacity of coral holobionts. Nat. Rev. Earth Environ. 2021;2:747–762. [Google Scholar]
- 14.Santoro E.P., Borges R.M., Espinoza J.L., Freire M., Messias C.S.M.A., Villela H.D.M., Pereira L.M., Vilela C.L.S., Rosado J.G., Cardoso P.M., et al. Coral microbiome manipulation elicits metabolic and genetic restructuring to mitigate heat stress and evade mortality. Sci. Adv. 2021;7:eabg3088. doi: 10.1126/sciadv.abg3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fragoso Ados Santos H., Duarte G.A.S., Rachid C.T.d.C., Chaloub R.M., Calderon E.N., Marangoni L.F.d.B., Bianchini A., Nudi A.H., do Carmo F.L., van Elsas J.D., et al. Impact of oil spills on coral reefs can be reduced by bioremediation using probiotic microbiota. Sci. Rep. 2015;5:18268. doi: 10.1038/srep18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosado P.M., Leite D.C.A., Duarte G.A.S., Chaloub R.M., Jospin G., Nunes da Rocha U., P Saraiva J., Dini-Andreote F., Eisen J.A., Bourne D.G., Peixoto R.S. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. 2019;13:921–936. doi: 10.1038/s41396-018-0323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva D.P., Villela H.D.M., Santos H.F., Duarte G.A.S., Ribeiro J.R., Ghizelini A.M., Vilela C.L.S., Rosado P.M., Fazolato C.S., Santoro E.P., et al. Multi-domain probiotic consortium as an alternative to chemical remediation of oil spills at coral reefs and adjacent sites. Microbiome. 2021;9:118. doi: 10.1186/s40168-021-01041-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Yang Q., Ling J., Long L., Huang H., Yin J., Wu M., Tang X., Lin X., Zhang Y., Dong J. Shifting the microbiome of a coral holobiont and improving host physiology by inoculation with a potentially beneficial bacterial consortium. BMC Microbiol. 2021;21:130. doi: 10.1186/s12866-021-02167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huggett M.J., Apprill A. Coral microbiome database: Integration of sequences reveals high diversity and relatedness of coral-associated microbes. Environ. Microbiol. Rep. 2019;11:372–385. doi: 10.1111/1758-2229.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sweet M., Villela H., Keller-Costa T., Costa R., Romano S., Bourne D.G., Cárdenas A., Huggett M.J., Kerwin A.H., Kuek F., et al. Insights into the Cultured Bacterial Fraction of Corals. mSystems. 2021;6:e0124920. doi: 10.1128/mSystems.01249-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peixoto R.S., Voolstra C.R., Sweet M., Duarte C.M., Carvalho S., Villela H., Lunshof J.E., Gram L., Woodhams D.C., Walter J., et al. Harnessing the microbiome to prevent global biodiversity loss. Nat. Microbiol. 2022;7:1726–1735. doi: 10.1038/s41564-022-01173-1. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez-Agreda A., Leggat W., Bongaerts P., Herrera C., Ainsworth T.D. Rethinking the Coral Microbiome: Simplicity Exists within a Diverse Microbial Biosphere. mBio. 2018;9:e00812–e00818. doi: 10.1128/mBio.00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leite D.C.A., Salles J.F., Calderon E.N., Castro C.B., Bianchini A., Marques J.A., van Elsas J.D., Peixoto R.S. Coral Bacterial-Core Abundance and Network Complexity as Proxies for Anthropogenic Pollution. Front. Microbiol. 2018;9:833. doi: 10.3389/fmicb.2018.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Oppen M.J.H., Blackall L.L. Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 2019;17:557–567. doi: 10.1038/s41579-019-0223-4. [DOI] [PubMed] [Google Scholar]
- 25.Maher R.L., Rice M.M., McMinds R., Burkepile D.E., Vega Thurber R. Multiple stressors interact primarily through antagonism to drive changes in the coral microbiome. Sci. Rep. 2019;9:6834. doi: 10.1038/s41598-019-43274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollock F.J., McMinds R., Smith S., Bourne D.G., Willis B.L., Medina M., Thurber R.V., Zaneveld J.R. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 2018;9:4921. doi: 10.1038/s41467-018-07275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shnit-Orland M., Kushmaro A. Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol. Ecol. 2009;67:371–380. doi: 10.1111/j.1574-6941.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 28.Modolon F., Barno A.R., Villela H.D.M., Peixoto R.S. Ecological and biotechnological importance of secondary metabolites produced by coral-associated bacteria. J. Appl. Microbiol. 2020;129:1441–1457. doi: 10.1111/jam.14766. [DOI] [PubMed] [Google Scholar]
- 29.Guermazi S., Daegelen P., Dauga C., Rivière D., Bouchez T., Godon J.J., Gyapay G., Sghir A., Pelletier E., Weissenbach J., Le Paslier D. Discovery and characterization of a new bacterial candidate division by an anaerobic sludge digester metagenomic approach. Environ. Microbiol. 2008;10:2111–2123. doi: 10.1111/j.1462-2920.2008.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luef B., Frischkorn K.R., Wrighton K.C., Holman H.-Y.N., Birarda G., Thomas B.C., Singh A., Williams K.H., Siegerist C.E., Tringe S.G., et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 2015;6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 31.Hervé V., Liu P., Dietrich C., Sillam-Dussès D., Stiblik P., Šobotník J., Brune A. Phylogenomic analysis of 589 metagenome-assembled genomes encompassing all major prokaryotic lineages from the gut of higher termites. PeerJ. 2020;8:e8614. doi: 10.7717/peerj.8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaboré O.D., Godreuil S., Drancourt M. Planctomycetes as Host-Associated Bacteria: A Perspective That Holds Promise for Their Future Isolations, by Mimicking Their Native Environmental Niches in Clinical Microbiology Laboratories. Front. Cell. Infect. Microbiol. 2020;10:519301. doi: 10.3389/fcimb.2020.519301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pushpakumara B.L.D.U., Tandon K., Willis A., Verbruggen H. Unravelling microalgal-bacterial interactions in aquatic ecosystems through 16S rRNA gene-based co-occurrence networks. Sci. Rep. 2023;13:2743. doi: 10.1038/s41598-023-27816-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen S., Fortunato S.A.V., Hoffmann F., Hoem S., Rapp H.T., Øvreås L., Torsvik V.L. The Relative Abundance and Transcriptional Activity of Marine Sponge-Associated Microorganisms Emphasizing Groups Involved in Sulfur Cycle. Microb. Ecol. 2017;73:668–676. doi: 10.1007/s00248-016-0836-3. [DOI] [PubMed] [Google Scholar]
- 35.Schellenberg J., Reichert J., Hardt M., Klingelhöfer I., Morlock G., Schubert P., Bižić M., Grossart H.-P., Kämpfer P., Wilke T., Glaeser S.P. The Bacterial Microbiome of the Long-Term Aquarium Cultured High-Microbial Abundance Sponge Haliclona cnidata – Sustained Bioactivity Despite Community Shifts Under Detrimental Conditions. Front. Mar. Sci. 2020;7 [Google Scholar]
- 36.Vavourakis C.D., Andrei A.-S., Mehrshad M., Ghai R., Sorokin D.Y., Muyzer G. A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments. Microbiome. 2018;6:168–218. doi: 10.1186/s40168-018-0548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim M., Oh H.-S., Park S.-C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 38.Doering T., Wall M., Putchim L., Rattanawongwan T., Schroeder R., Hentschel U., Roik A. Towards enhancing coral heat tolerance: a “microbiome transplantation” treatment using inoculations of homogenized coral tissues. Microbiome. 2021;9:102. doi: 10.1186/s40168-021-01053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H., Xue R., Wang Y., Stirling E., Ye S., Xu J., Ma B. FACS-iChip: a high-efficiency iChip system for microbial “dark matter” mining. Mar. Life Sci. Technol. 2021;3:162–168. doi: 10.1007/s42995-020-00067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alkayyali T., Pope E., Wheatley S.K., Cartmell C., Haltli B., Kerr R.G., Ahmadi A. Development of a microbe domestication pod (MD Pod) for in situ cultivation of micro-encapsulated marine bacteria. Biotechnol. Bioeng. 2021;118:1166–1176. doi: 10.1002/bit.27633. [DOI] [PubMed] [Google Scholar]
- 41.Chaudhary D.K., Khulan A., Kim J. Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 2019;9:6666. doi: 10.1038/s41598-019-43182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gavrish E., Bollmann A., Epstein S., Lewis K. A trap for in situ cultivation of filamentous actinobacteria. J. Microbiol. Methods. 2008;72:257–262. doi: 10.1016/j.mimet.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung D., Seo E.-Y., Epstein S.S., Joung Y., Han J., Parfenova V.V., Belykh O.I., Gladkikh A.S., Ahn T.S. Application of a new cultivation technology, I-tip, for studying microbial diversity in freshwater sponges of Lake Baikal, Russia. FEMS Microbiol. Ecol. 2014;90:417–423. doi: 10.1111/1574-6941.12399. [DOI] [PubMed] [Google Scholar]
- 44.Wheatley S.K., Cartmell C., Madadian E., Badr S., Haltli B.A., Kerr R.G., Ahmadi A. Microfabrication of a micron-scale microbial-domestication pod for cultivation of marine bacteria. RSC Adv. 2022;12:28123–28127. doi: 10.1039/d2ra05420e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nichols D., Cahoon N., Trakhtenberg E.M., Pham L., Mehta A., Belanger A., Kanigan T., Lewis K., Epstein S.S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010;76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muralimohan A., Eun Y.-J., Bhattacharyya B., Weibel D.B. Dissecting microbiological systems using materials science. Trends Microbiol. 2009;17:100–108. doi: 10.1016/j.tim.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Bassin J.P., Rachid C.T., Vilela C., Cao S.M., Peixoto R.S., Dezotti M. Revealing the bacterial profile of an anoxic-aerobic moving-bed biofilm reactor system treating a chemical industry wastewater. International. 2017;120:152–160. [Google Scholar]
- 48.Obotey Ezugbe E., Rathilal S. Membrane Technologies in Wastewater Treatment: A Review. Membranes. 2020;10:89. doi: 10.3390/membranes10050089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villela H.D.M., Peixoto R.S., Soriano A.U., Carmo F.L. Microbial bioremediation of oil contaminated seawater: A survey of patent deposits and the characterization of the top genera applied. Sci. Total Environ. 2019;666:743–758. doi: 10.1016/j.scitotenv.2019.02.153. [DOI] [PubMed] [Google Scholar]
- 50.de Jesus H.E., Peixoto R.S., Rosado A.S. Bioremediation in Antarctic soils. J. Petrol Environ. Biotechnol. 2015;06 [Google Scholar]
- 51.do Carmo F.L., dos Santos H.F., Martins E.F., van Elsas J.D., Rosado A.S., Peixoto R.S. Bacterial structure and characterization of plant growth promoting and oil degrading bacteria from the rhizospheres of mangrove plants. J. Microbiol. 2011;49:535–543. doi: 10.1007/s12275-011-0528-0. [DOI] [PubMed] [Google Scholar]
- 52.Rachid C.T.C.C., Piccolo M.C., Leite D.C.A., Balieiro F.C., Coutinho H.L.C., van Elsas J.D., Peixoto R.S., Rosado A.S. Physical-chemical and microbiological changes in Cerrado Soil under differing sugarcane harvest management systems. BMC Microbiol. 2012;12:170. doi: 10.1186/1471-2180-12-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saad M.M., Eida A.A., Hirt H. Tailoring plant-associated microbial inoculants in agriculture: a roadmap for successful application. J. Exp. Bot. 2020;71:3878–3901. doi: 10.1093/jxb/eraa111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schloter M., Nannipieri P., Sørensen S.J., van Elsas J.D. Microbial indicators for soil quality. Biol. Fertil. Soils. 2018;54:1–10. [Google Scholar]
- 55.Wilkins T., Sequoia J. Probiotics for Gastrointestinal Conditions: A Summary of the Evidence. Am. Fam. Physician. 2017;96:170–178. [PubMed] [Google Scholar]
- 56.Kumar R., Sood U., Gupta V., Singh M., Scaria J., Lal R. Recent Advancements in the Development of Modern Probiotics for Restoring Human Gut Microbiome Dysbiosis. Indian J. Microbiol. 2020;60:12–25. doi: 10.1007/s12088-019-00808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Könneke M., Bernhard A.E., de la Torre J.R., Walker C.B., Waterbury J.B., Stahl D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 58.Poindexter J.S. 2015. Maricaulis. Bergey's Manual of Systematics of Archaea and Bacteria; pp. 1–12. [Google Scholar]
- 59.Selvaratnam C., Thevarajoo S., Goh K.M., Chan K.-G., Chong C.S. Genomic analyses of five Roseivirga species: Insights into marine adaptation. Mar. Genomics. 2018;38:97–101. doi: 10.1016/j.margen.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 60.Spring S. 2015. Halobacillus. Bergey's Manual of Systematics of Archaea and Bacteria; pp. 1–10. [Google Scholar]
- 61.Lam M.Q., Vodovnik M., Zorec M., Chen S.J., Goh K.M., Yahya A., Md Salleh M., Ibrahim Z., Tokiman L., McQueen-Mason S.J., et al. sp. nov., isolated from mangrove soil, and emended description of the genus. Int. J. Syst. Evol. Microbiol. 2020;70:1769–1776. doi: 10.1099/ijsem.0.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vreeland R.H. 2015. Halomonas. Bergey's Manual of Systematics of Archaea and Bacteria; pp. 1–19. [Google Scholar]
- 63.Rosado P.M., Cardoso P.M., Rosado J.G., Schultz J., Nunes da Rocha U., Keller-Costa T., Peixoto R.S. Exploring the Potential Molecular Mechanisms of Interactions between a Probiotic Consortium and Its Coral Host. mSystems. 2023;8:e0092122. doi: 10.1128/msystems.00921-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li N., He X.-Y., Liu N.-H., Gu T.-J., Li J., Geng Y.-H., Zhang S., Wang P., Fu H.-H., Shi M., et al. Tritonibacter aquimaris sp. nov. and Tritonibacter litoralis sp. nov., two novel members of the Roseobacter group isolated from coastal seawater. Antonie Leeuwenhoek. 2021;114:787–798. doi: 10.1007/s10482-021-01558-y. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y., Zheng L., Wang S., Zhao Y., Xu X., Han B., Hu T. Quorum Sensing Bacteria in the Phycosphere of HAB Microalgae and Their Ecological Functions Related to Cross-Kingdom Interactions. Int. J. Environ. Res. Publ. Health. 2021;19:163. doi: 10.3390/ijerph19010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kayama G., Kanaly R.A., Mori J.F. Comprehensive Genomic Characterization of Marine Bacteria spp. Provides Insights into Their Ecological Roles in Aromatic Hydrocarbon-Exposed Environments. Microbiol. Spectr. 2022;10:e0314922. doi: 10.1128/spectrum.03149-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peeb A., Dang N.P., Truu M., Nõlvak H., Petrich C., Truu J. Assessment of Hydrocarbon Degradation Potential in Microbial Communities in Arctic Sea Ice. Microorganisms. 2022;10:328. doi: 10.3390/microorganisms10020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y.-N., Zhang T.-S., Chen X.-Y., Gong Y., Du Z.-J. Pseudohalioglobus sediminis sp. nov., isolated from coastal sediment. Arch. Microbiol. 2022;204:207. doi: 10.1007/s00203-022-02816-x. [DOI] [PubMed] [Google Scholar]
- 69.Dungan A.M., Bulach D., Lin H., van Oppen M.J.H., Blackall L.L. Development of a free radical scavenging bacterial consortium to mitigate oxidative stress in cnidarians. Microb. Biotechnol. 2021;14:2025–2040. doi: 10.1111/1751-7915.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varasteh T., Tschoeke D., Garcia G., Lima A.S., Moreira A.P.B., Thompson C., Thompson F. Insights into the genomic repertoire of Aquimarina litoralis CCMR20, a symbiont of coral Mussismilia braziliensis. Arch. Microbiol. 2021;203:2743–2746. doi: 10.1007/s00203-021-02194-w. [DOI] [PubMed] [Google Scholar]
- 71.Xu T., Yu M., Lin H., Zhang Z., Liu J., Zhang X.-H. Genomic insight into Aquimarina longa SW024 T: its ultra-oligotrophic adapting mechanisms and biogeochemical functions. BMC Genom. 2015;16:772. doi: 10.1186/s12864-015-2005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weis V.M. Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 2008;211:3059–3066. doi: 10.1242/jeb.009597. [DOI] [PubMed] [Google Scholar]
- 73.Peixoto R.S., Sweet M., Bourne D.G. Customized Medicine for Corals. Front. Mar. Sci. 2019;6 [Google Scholar]
- 74.Apprill A., McNally S., Parsons R., Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015;75:129–137. [Google Scholar]
- 75.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McMurdie P.J., Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 81.Keller-Costa T., Eriksson D., Gonçalves J.M.S., Gomes N.C., Lago-Lestón A., Costa R. The gorgonian coral Eunicella labiata hosts a distinct prokaryotic consortium amenable to cultivation. FEMS Microbiol. Ecol. 2017;93 doi: 10.1093/femsec/fix143. [DOI] [PubMed] [Google Scholar]
- 82.Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw amplicon sequencing data are available on NCBI’s Read Archive under BioProject: PRJNA934364. The 16S rRNA sequences from isolates are available on NCBI’s GenBank under the IDs listed in Table S3. All datasets are publicly available as of the date of publication.
-
•
The complete bioinformatic pipeline including scripts for figure reproduction is publicly available through the GitHub repository at <https://github.com/modolon/insitu_cultivation>.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.