Abstract
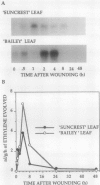
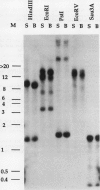
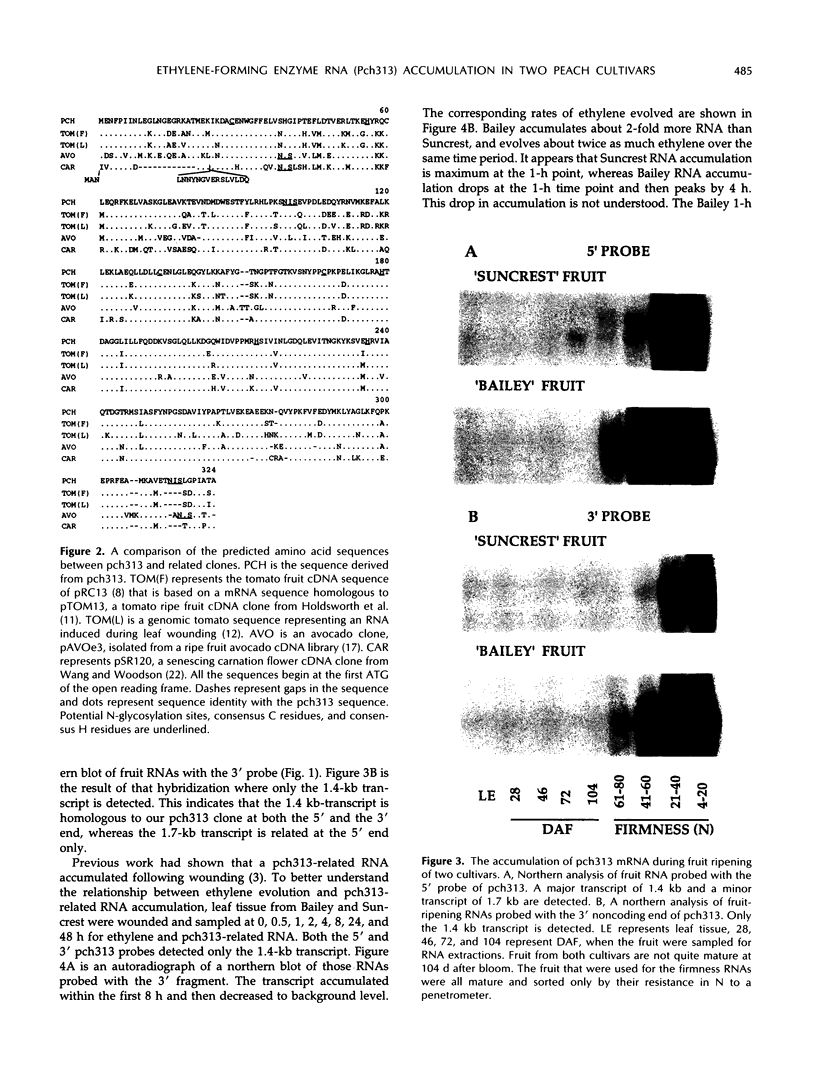
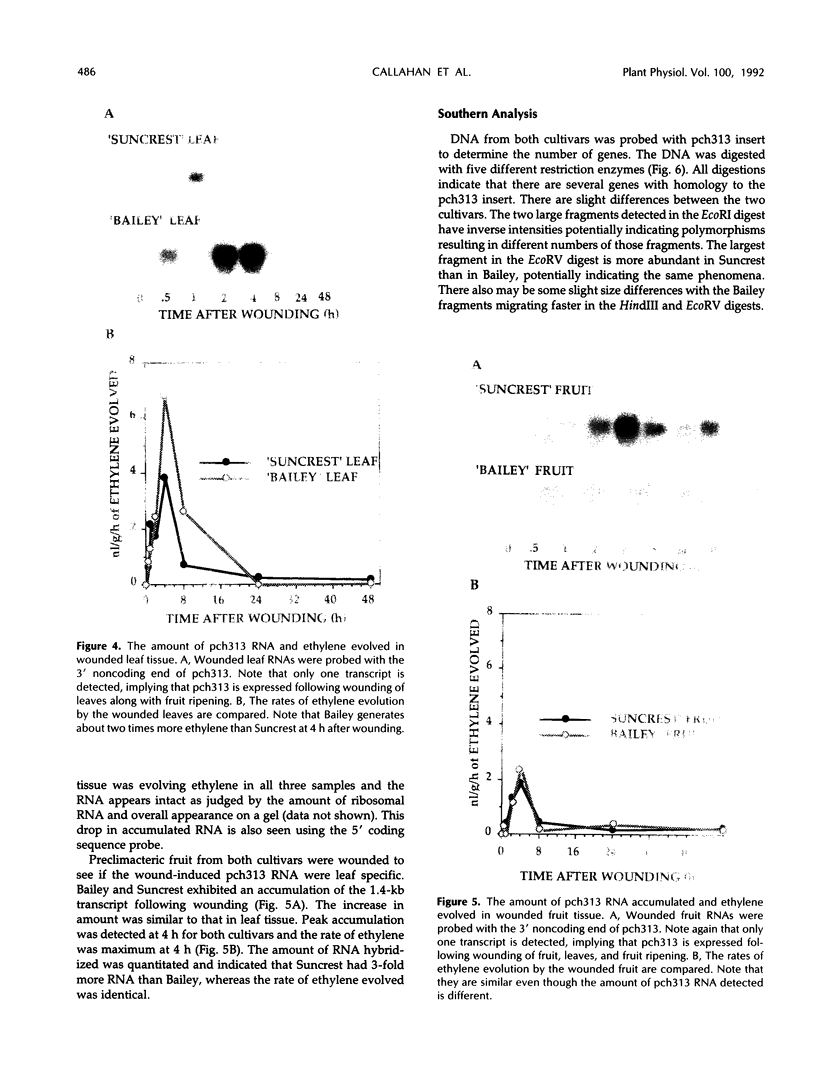
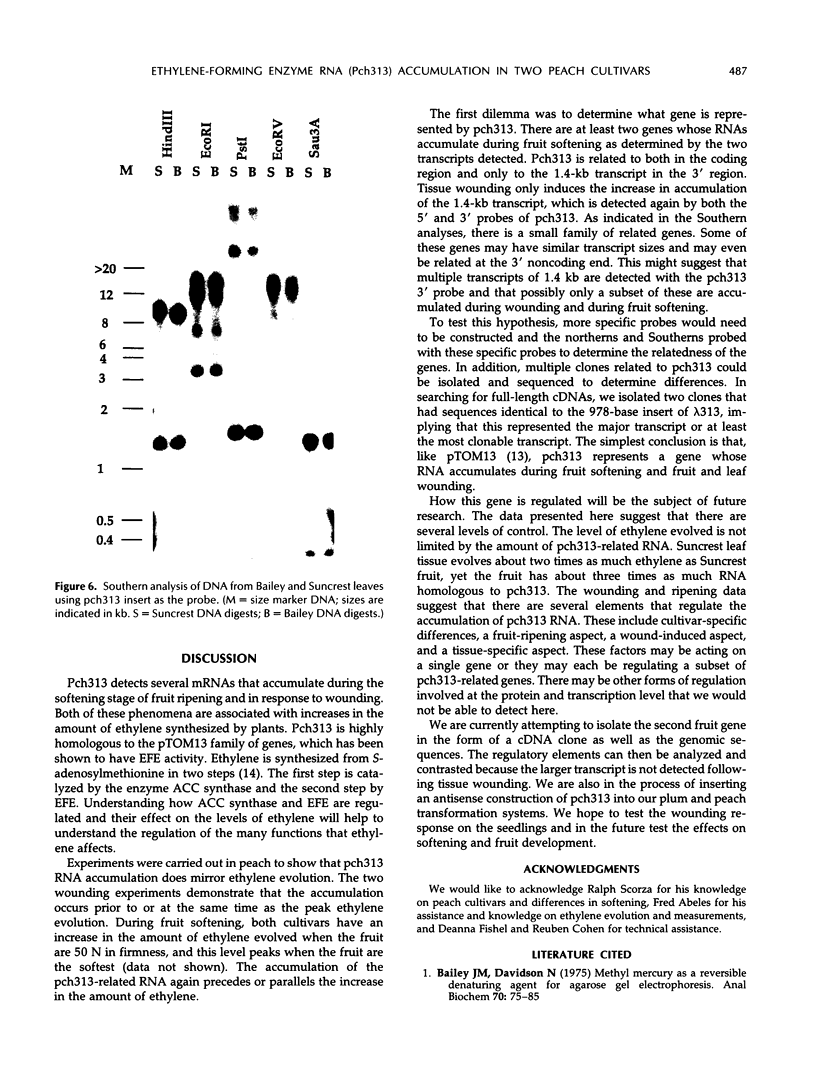
Pch313 was isolated as a cDNA whose RNA accumulated during the softening period of peach (Prunus persica L. Batsch) fruit development. To better understand the role of the gene, we compared the amount of pch313-related RNA detected during fruit softening and tissue wounding between cultivars with different softening characteristics. The cultivar that softened faster, “Bailey,” had a significantly higher amount of pch313-related RNA accumulate during softening than the slower-softening cultivar, “Suncrest.” Pch313 was sequenced and found to be related to a tomato fruit cDNA clone, pTOM13, which has been shown to encode the ethylene-forming enzyme. The derived amino acid sequence of pch313 is 74 to 83% identical to the pTOM13-related sequences. A pch313-3′ noncoding region probe was used to demonstrate that pch313 is related to both a wound-induced RNA transcript and the major fruit-softening transcript. The relationship of pch313 RNA accumulation and ethylene evolution was examined upon wounding and appeared to be both tissue and cultivar specific. When leaves were wounded, more pch313-related RNA was detected in Bailey and the rate of ethylene evolved was also higher in Bailey. When fruits were wounded, the levels of ethylene evolved were nearly identical but Suncrest accumulated more pch313-related RNA. Southern analysis of the DNA indicated a small number of related genes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Deikman J., Fischer R. L. Interaction of a DNA binding factor with the 5'-flanking region of an ethylene-responsive fruit ripening gene from tomato. EMBO J. 1988 Nov;7(11):3315–3320. doi: 10.1002/j.1460-2075.1988.tb03202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A. J., Bouzayen M., Grierson D. Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7434–7437. doi: 10.1073/pnas.88.16.7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrin D. L., Schmidt G. W. Rapid, reversible staining of northern blots prior to hybridization. Biotechniques. 1988 Mar;6(3):196-7, 199-200. [PubMed] [Google Scholar]
- Holdsworth M. J., Bird C. R., Ray J., Schuch W., Grierson D. Structure and expression of an ethylene-related mRNA from tomato. Nucleic Acids Res. 1987 Jan 26;15(2):731–739. doi: 10.1093/nar/15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth M. J., Schuch W., Grierson D. Nucleotide sequence of an ethylene-related gene from tomato. Nucleic Acids Res. 1987 Dec 23;15(24):10600–10600. doi: 10.1093/nar/15.24.10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. Enzymes of ethylene biosynthesis. Plant Physiol. 1989 Sep;91(1):1–4. doi: 10.1104/pp.91.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köck M., Hamilton A., Grierson D. eth1, a gene involved in ethylene synthesis in tomato. Plant Mol Biol. 1991 Jul;17(1):141–142. doi: 10.1007/BF00036816. [DOI] [PubMed] [Google Scholar]
- McGarvey D. J., Yu H., Christoffersen R. E. Nucleotide sequence of a ripening-related cDNA from avocado fruit. Plant Mol Biol. 1990 Jul;15(1):165–167. doi: 10.1007/BF00017736. [DOI] [PubMed] [Google Scholar]
- Sheehy R. E., Kramer M., Hiatt W. R. Reduction of polygalacturonase activity in tomato fruit by antisense RNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8805–8809. doi: 10.1073/pnas.85.23.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu P., Reinhardt D., Boller T. Analysis and cloning of the ethylene-forming enzyme from tomato by functional expression of its mRNA in Xenopus laevis oocytes. EMBO J. 1991 Aug;10(8):2007–2013. doi: 10.1002/j.1460-2075.1991.tb07730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Woodson W. R. A Flower Senescence-Related mRNA from Carnation Shares Sequence Similarity with Fruit Ripening-Related mRNAs Involved in Ethylene Biosynthesis. Plant Physiol. 1991 Jul;96(3):1000–1001. doi: 10.1104/pp.96.3.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]