Abstract
In this study, we integrated machine learning (ML), structure-tissue selectivity-activity-relationship (STAR), and wet lab synthesis/testing to design a gastrointestinal (GI) locally activating JAK inhibitor for ulcerative colitis treatment. The JAK inhibitor achieves site-specific efficacy through high local GI tissue selectivity while minimizing the requirement for JAK isoform specificity to reduce systemic toxicity. We used the ML model (CoGT) to classify whether the designed compounds were inhibitors or noninhibitors. Then we used the regression ML model (MTATFP) to predict their IC50 against related JAK isoforms of predicted JAK inhibitors. The ML model predicted MMT3-72, which was retained in the GI tract, to be a weak JAK1 inhibitor, while MMT3-72-M2, which accumulated in only GI tissues, was predicted to be an inhibitor of JAK1/2 and TYK2. ML docking methods were applied to simulate their docking poses in JAK isoforms. Application of these ML models enabled us to limit our synthetic efforts to MMT3-72 and MMT3-72-M2 for subsequent wet lab testing. The kinase assay confirmed MMT3-72 weakly inhibited JAK1, and MMT3-72-M2 inhibited JAK1/2 and TYK2. We found that MMT3-72 accumulated in the GI lumen, but not in GI tissue or plasma, but released MMT3-72-M2 accumulated in colon tissue with minimal exposure in the plasma. MMT3-72 achieved superior efficacy and reduced p-STAT3 in DSS-induced colitis. Overall, the integration of ML, the structure-tissue selectivity-activity-relationship system, and wet lab synthesis/testing could minimize the effort in the optimization of a JAK inhibitor to treat colitis. This site-specific inhibitor reduces systemic toxicity by minimizing the need for JAK isoform specificity.
Keywords: Janus kinase (JAK) inhibitors, GI locally activating, ulcerative colitis, machine learning (ML), structure-tissue selectivity-activity-relationship (STAR)
Ulcerative colitis (UC) affects more than 1 to 2 million patients in the US (1, 2, 3). UC usually starts from the rectum and extends to the colon (4), where the inflammation is restricted to the innermost layer of the intestine (mucosa), resulting in ulceration and bloody diarrhea (2, 5, 6). UC needs life-long treatment (2), and thus the treatment options require not only adequate efficacy but also minimal long-term systemic toxicity.
Recently, inhibition of Janus kinases (JAK1, JAK2, JAK3, and tyrosine kinase 2 [TYK2]) has emerged as a new therapeutic approach for the treatment of UC (7). The activation of JAKs enables the recruitment and phosphorylation of signal transducers and activators of transcription (STAT) family. Phosphorylated STATs translocate to the nucleus to induce the expression of chemokines and cytokines in the UC (8). Several orally bioavailable JAK inhibitors (tofacitinib and upadacitinib) have been developed and approved for the treatment of UC. For instance, tofacitinib, a pan-JAK inhibitor, showed excellent efficacy in the treatment of moderate to severe UC (9, 10). However, tofacitinib and several JAK inhibitors have black box warnings for serious side effects, (https://www.fda.gov/drugs/drug-safety-andavailability/fda-requires-warnings-aboutincreased-risk-serious-heart-related-eventscancer-blood-clots-and-death) which include 1) a high rate of major adverse cardiovascular events (MACEs, such as cardiovascular death, myocardial infarction, and stroke), 2) arterial and venous thrombosis and pulmonary embolism, 3) malignancies of lymphomas and lung cancers, and 4) increased risk of serious infections leading to death.
Different approaches have been explored to reduce the systemic toxicity of JAK inhibitors in the treatment of UC. The first approach is to use a modified release formulation of JAK inhibitors to increase gastrointestinal (GI) local concentration and reduce oral absorption. Although the modified release formulation of tofacitinib delays drug release, increases drug concentration in the GI tract, and reduces drug exposure in the systemic circulation, it is still unable to alleviate the above serious side effects (11). The second approach is to develop JAK isoform-specific inhibitors, such as upadacitinib and filgotinib targeting only one or two isoforms, to prevent their toxicity (12, 13). Unfortunately, the safety review by the food and drug administration has found that all JAK inhibitors, regardless of pan-JAK inhibitors or isoform-specific inhibitors, have similar severe adverse effects, and thus these black box warnings were added to the labels of multiple JAK inhibitors in 2021 (https://www.fda.gov/drugs/drug-safety-andavailability/fda-requires-warnings-aboutincreased-risk-serious-heart-related-eventscancer-blood-clots-and-death). A third approach is to develop a TYK2-specific inhibitor for the treatment of UC. Deucravacitinib was developed to bind to the pseudokinase domain (JH2) of TYK2 for reduced toxicity. Although clinical testing of the TYK2-specific inhibitor showed positive efficacy in treating psoriasis (14, 15), this drug failed in clinical trial in treating UC (https://news.bms.com/news/details/2021/Bristol-Myers-Squibb-Provides-Update-on-Phase-2-Study-of-Deucravacitinib-in-Patients-With-Moderate-to-Severe-Ulcerative-Colitis/default.aspx). Therefore, there is a critical need to develop JAK inhibitors with superior efficacy but minimal toxicity for the long-term treatment of UC. In this study, to overcome the above limitations, we integrated machine learning (ML), structure-tissue selectivity-activity relationship (STAR), and wet lab synthesis/testing to design GI locally activating JAK inhibitor to treat UC. The GI locally activating JAK inhibitor achieves site-specific efficacy through high local GI tissue selectivity while minimizing the requirement for JAK isoform specificity to reduce systemic toxicity.
We utilized a STAR system to design JAK inhibitors for the treatment of UC, by concurrently taking into consideration two factors: the drug’s tissue selectivity and target potency/specificity based on the drug’s STAR (16, 17, 18, 19). The STAR system categorizes drug candidates into four classes, where class I/III candidates (high tissue selectivity, high/medium potency/specificity) may be able to balance clinical dose/efficacy/toxicity for better clinical success. In contrast, class II/IV candidates (low tissue selectivity and high/low potency/specificity) may cause imbalanced clinical dose/efficacy/toxicity, which leads to dose-limiting toxicity or lack of efficacy. Specifically, we intended to design STAR class III JAK inhibitors to achieve high local exposure/selectivity in the GI tissues while minimizing the requirement for JAK isoform specificity since none of the isoform-specific JAK inhibitors could eliminate their systemic toxicity.
We first designed MMT3-72 with poor absorption potential, as modeled to have a low drug-likeness score (low QED score), to ensure the drug will be retained in the GI tract (20). MMT3-72 was then studied to release the potential active compounds of JAK inhibitors in local GI tissues for potential efficacy in treating UC but reduce their exposure in systemic circulation to minimize potential toxicity. Two ML models were used to classify inhibitors/noninhibitors and estimate IC50 values of the predicted inhibitors. Based on the prediction from two ML models, we only needed to synthesize MMT3-72 and MMT3-72-M2 for in vitro and in vivo testing on drug’s potency/specificity, tissue selectivity, and efficacy in colitis models. The integration of ML, the STAR system, and wet lab synthesis/testing could minimize the effort in the optimization of a JAK inhibitor to treat colitis. This site-specific inhibitor reduces systemic toxicity by minimizing the need for JAK isoform specificity. The overall workflow is summarized in Figure 1.
Figure 1.
Overview for the integration of machine learning, structure-tissue selectivity-activity relationship (STAR), and wet lab synthesis/testing of the GI locally activating JAK inhibitor to treat ulcerative colitis. GI, gastrointestinal.
Results
Design rationale for the scaffold of MMT3-72
We chose to design MMT3-72 based on the following three considerations: (A) The scaffold has intrinsic properties to be secreted into the GI tract. We used the scaffold of fedratinib because it could be preferentially secreted to the GI tract intact. For instance, a significant proportion of fedratinib-derived radioactivity (∼77%) was excreted in feces (23% unchanged) after a single oral dose of radiolabeled fedratinib (21). This observation inspired us to further modify fedratinib to create a colon-selective compound for the treatment of colitis. (B) The designed compound needs to be retained in the GI tract without effective absorption to reduce systemic exposure for decreasing its toxicity. This is achieved by the estimation of a low drug-likeness score (low QED score), which was modeled based on eight parameters, including molecular weight, octanol-water partition coefficient (ALOGP), number of hydrogen bond donors, number of hydrogen bond acceptors, molecular polar surface area, number of rotatable bonds (ROTB), number of aromatic rings (AROM), and number of structural alerts (ALERTS) (20). (C) The designed compound could be activated by bacteria in the colon to release its active metabolite for achieving its site-specific efficacy. Based on the X-ray structure of JAK2-bound fedratinib (22), we replaced the solvent-exposed pyrrolidine moiety of fedratinib with 5-aminosalicylic acid linked by an azo bond to N-4-(aminobenzoyl)-beta-alanine (Fig. 2, A and B). The modification is intended to increase the molecular weight and polarity of MMT3-72, which reduces its absorption potential from the GI tract and results in more accumulation in the colon tissues.
Figure 2.
Design rational of the scaffold of MMT3-72.A, cocrystal structure of JAK2 with fedratinib (PDB code: 6VNE). B, rationale for MMT3-72. C, quantitative estimates of drug-likeness of MMT3-72 and its metabolites.
To evaluate its drug-like property (especially absorption potential), we estimated its drug-likeness (QED, 0–1) according to the published method (20). The QED of MMT3-72 was predicted to be 0.0382, suggesting it has low or no absorption potential (the maximal value of QED is 1 suggesting best absorption potential) (Fig. 2C). As a comparison, the QED of balsalazide is predicted to be 0.5598 (Table S1). When MMT3-72 reaches the colon region, the azo bond would be cleaved by bacterial azoreductases to release the active metabolite(s), which is then absorbed and accumulated in the colon tissues with minimal exposure to the systemic circulation.
Chemistry: synthesis of MMT3-72
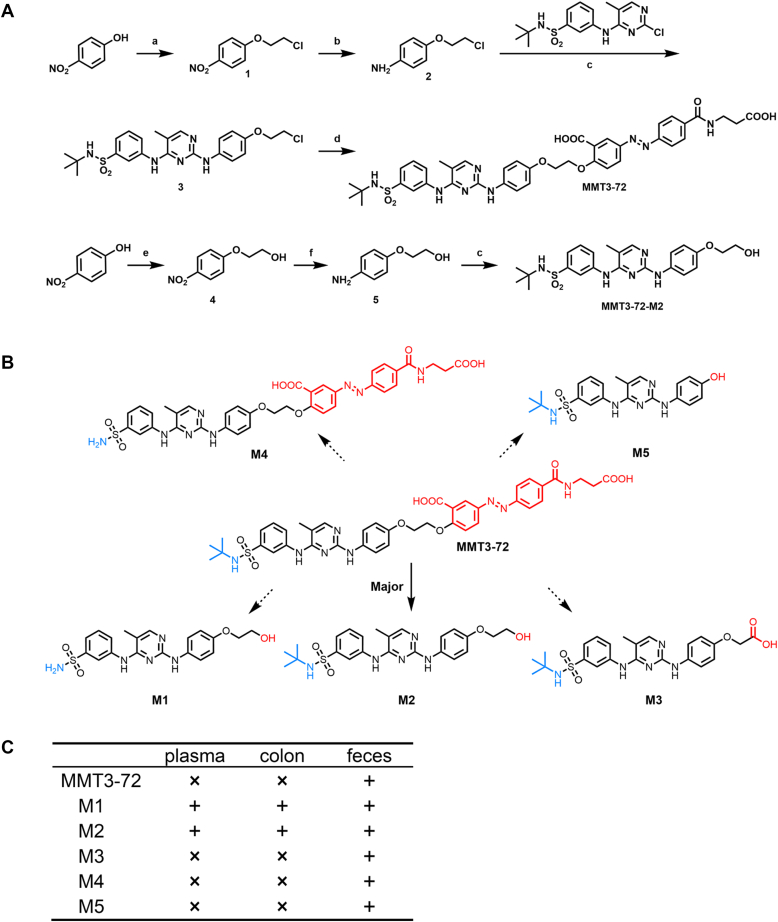
The compound MMT3-72 was synthesized according to the synthetic route as shown in Figure 3A. First, 4-nitrophenol was condensed with dichloroethane to produce intermediates 1. A subsequent nitro reduction of intermediates 1 with Tin chloride afforded the intermediates 2. Then, intermediate 2 was coupled with N-tert-butyl-3-[(2-chloro-5methylpyrimidin-4-yl)amino]benzenesulfonamide in the presence of a few drops of HCl in isopropanol to generate compound 3. Finally, compound 3 was subject to a nucleophilic substitution reaction with balsalazide disodium salt dihydrate to yield the desired compound MMT3-72.
Figure 3.
Synthesis and metabolite identification of MMT3-72. A, synthesis of MMT3-72 and MMT3-72-M2. Reaction conditions: (a) DCE, K2CO3, DMF, 100 °C, 6 h, and 92%; (b) SnCl2 · 2H2O, EtOH, rt, overnight, and 63%; (c) HCl conc, iPrOH, 80 °C, and 79 to 84%; (d) balsalazide disodium, K2CO3, DMF, 100 °C, 6 h, and 68%; (e) 2-chloride ethanol, NaOH, H2O, 8 h, 80 °C, and 79%. (f) H2, Pd/C, MeOH, 50 °C, overnight, and 76%. B, GI locally activating of MMT3-72 and metabolite identification in vivo. Structures of MMT3-72 and its five metabolites are shown as MMT3-72-M1, MMT3-72-M2, MMT3-72-M3, MMT3-72-M4, and MMT3-72-M5. C, concentrations of MMT3-72 and its metabolites in vivo. Mice were orally dosed with 10 mg/kg MMT3-72 and sacrificed at 6 h. ×, not detected; +, detected. GI, gastrointestinal.
GI local activation of MMT3-72 and metabolites identification in GI contents, GI tissues, and plasma
To confirm the activation of our newly synthesized compound MMT3-72 in the colon, mice were dosed orally with 10 mg/kg MMT3-72 and sacrificed at 6 h to collect plasma, colon tissues, and colon contents (feces). Five metabolites (M1 to M5) were identified using LC-MS in the collected samples and their structures are shown in Figure 3B. Interestingly, MMT3-72 was only detected in the feces with no detection in the plasma and the colon tissues. The major metabolite MMT3-72-M2 was only detected in the colon tissues and the feces with limited detection in the plasma. The minor metabolite M1 was detected in all three sampled tissues, with a majority in the feces. The other three minor metabolites M3, M4, and M5 were only identified in the feces with no detection in either the colon tissues or the plasma. Details are summarized in Figure 3C.
Two ML models to classify inhibitors/noninhibitors and predict IC50 of MMT3-72 and its five metabolites against JAK1, 2, 3, and TYK2
In order to know how these five metabolites may inhibit JAK1, 2, 3, and TYK2, we used two ML models to first classify inhibitors/noninhibitors and then predict their IC50 values against these isoforms of predicted inhibitors. This will reduce the efforts to only synthesize the desired compounds for further testing. We first utilized CoGT model to classify whether the compound is an inhibitor or noninhibitor, then used MTATFP model to predict their IC50 values if the compound is predicted as an inhibitor by CoGT. Results are shown in Figure 4, A and B. Further details for CoGT and MTATFP are in Experimental procedures. Both ML methods predicted that MMT3-72 is a very weak JAK1 inhibitor but did not inhibit the other three isoforms. The main metabolite MMT3-72-M2 is predicted as a JAK1 (IC50 19 nM), JAK2 (IC50 34 nM), and TYK2 (IC50 65 nM) inhibitor. This result affirmed our confidence to proceed with the synthesis and evaluation of MMT3-72-M2 for its activity. In addition, MMT3-72-M2 was also confirmed to have a high concentration in the colon tissue. The reason for not proceeding with the evaluation on M1 or M3-M5 is that they have low concentrations in the colon and did not show as good predicted inhibitory activities against different JAK isoforms.
Figure 4.
Two machine learning models to classify inhibitors/noninhibitors and predict IC50values against four JAK isoforms. Prediction results on activities against JAKs of MMT3-72 and its metabolites (A) by CoGT and (B) by MTATFP. Docking simulation was performed using DiffDock. MMT3-72-M2 was docked in the binding sites in JAK1 (C), JAK2 (D), and TYK2 (E) as it is predicted to be inhibitors for these JAK isoforms. The key residues that formed hydrogen bonding contacts with the ligands are colored orange. The dashed lines represent the formed hydrogen bonding contacts between the amino acid residues and MMT3-72-M2 (dark gray) or ground-truth ligands (yellow). Magenta: MMT3-72-M2; cyan: ground-truth ligand (cocrystalized with the proteins). PDB code: JAK1: 6BBU; JAK2: 6VNE; TYK2: 6DBK. The ligand docking results were all visualized in PyMol (http://www.pymol.org/pymol).
To further understand how MMT3-72 and MMT3-72-M2 bind to different JAK isoforms, we also used ML-based docking models to predict molecular pose in JAK proteins. First, we used DiffDock, a diffusion generative model, to predict the binding pose of these two molecules during protein–ligand interaction (23). DiffDock refines the ligand pose space as an (m + 6)-dimensional submanifold M ∈ R3n, in which m and n are the rotatable bond number and the atom number of a ligand, respectively. Results of DiffDock prediction are shown in Figure 4, C–E. The docking simulations consistently revealed highly similar binding poses of MMT3-72-M2 (highlighted in magenta) compared to the cocrystalized ligand data (highlighted in cyan). Detailed analysis of the molecular interactions within the binding sites further confirmed the presence of similar hydrogen bonding contacts. Specifically, MMT3-72-M2 was able to form hydrogen bonding with the residue Leu959 in the binding site of JAK1 similar to its cocrystal structure with its ligand (compound 25 (24)) (Fig. 4C). In addition, MMT3-72-M2 exhibited a similar binding pose to that of fedratinib, and both compounds were capable of forming hydrogen bonding with the Leu932 residue (Fig. 4D). Further, MMT3-72-M2 was predicted to form hydrogen bonding with the residue Val981 as seen in the cocrystal structure with its ligand (Compound 8 (25)) in the binding site of TYK2. In addition, MMT3-72-M2 also forms additional interaction with the residue Arg1027 (Fig. 4E). These findings indicate the potential of MMT3-72-M2 as an effective JAK inhibitor.
As a comparison, we also predict the binding of MMT3-72-M2 to JAKs using EquiBind (26). Since EquiBind is a regression-based method to predict the ligand position during the process of minimizing the expected square error, the performance is inferior than that of DiffDock. Docking prediction results are shown in Fig. S1.
MMT3-72 was inactive but MMT3-72-M2 was more potent against JAK1, 2, and TYK2 by in vitro kinase assays
To confirm the correctness of the ML prediction of JAK inhibition, biological activities of MMT3-72 and its active metabolite MMT3-72-M2 were evaluated against JAK1, JAK2, JAK3, and TYK2 using kinase assays (Fig. 5). MMT3-72 showed weak activities against JAK1 (367.7 nM), and respectively poor activities against JAK2, JAK3, and TYK2 (630 nM, 5237 nM, and 4697 nM, respectively). On the other hand, the active metabolite MMT3-72-M2 showed potent inhibition of JAK1 (10.8 nM), JAK2 (26.3 nM), and TYK2 (91.6 nM) but weak inhibition of JAK3 (328.7 nM). These experimental results are in agreement with ML prediction.
Figure 5.
Inhibition of different isoforms of JAK by MMT3-72 and active metabolite MMT3-72-M2. Inhibition of JAK activity by MMT3-72 and MMT3-72-M2 (0.01–10,000 nM) was measured using Kinase-Glo Max assay against purified enzymes JAK1 (A), JAK2 (B), JAK3 (C), and TYK2 (D). Assays were run in the presence of 0.1 mM ATP. Data shown as mean ± SD (n = 3 independent biological replicates). E, in vitro inhibitory activities IC50 of MMT3-72 and active metabolite MMT3-72-M2 against different isoforms of JAKs. The IC50 values of compounds to inhibit different JAK isoforms were calculated using GraphPad Prism 9. ∗The IC50 values of tofacitinib were cited from Ref. (42).
MMT3-72 was readily converted to MMT3-72-M2 in the colon contents
To quantify the activation process of the colon-activating MMT3-72, we investigated its release of MMT3-72-M2 in colon content suspensions. Figure 6A shows that fresh colon contents metabolized more than 90% of MMT3-72 within 48 h of incubation. In contrast, most MMT3-72 remained intact in the aqueous environment when the colon contents had been heat-deactivated. Importantly, MMT3-72-M2 was readily converted from MMT3-72 in fresh colon contents (Fig. 6B). Given that the IC50 of MMT3-72-M2 was measured to be 10.8 nM (∼5.0 ng/ml) for JAK1, 26.3 nM (∼12.1 ng/ml) for JAK2, and 91.6 nM (∼42.1 ng/ml) for TYK2 (Fig. 5E), it is reasonable to expect the efficient local conversion to MMT3-72-M2, allowing for a durable treatment after MMT3-72 reaches the colon region to inhibit all three JAK isoforms locally.
Figure 6.
In vitro and in vivo activation of MMT3-72 to release MMT3-72-M2 in the GI tract.A, concentrations of MMT3-72 at the indicated time points (0, 1, 2, 4, 6, 24, and 48 h) were measured after incubation with fresh and boiled colon contents. B, concentrations of MMT3-72-M2 were measured from the same collected samples in (A). C, MMT3-72 concentrations in colon content at 0.5, 2, 4, 12, and 24 h. Concentrations of MMT3-72 in plasma and colon tissue were below the limit of detection of 2 ng/ml or 2 ng/g (not detectable, n.d.) in our analysis. D, MMT3-72-M2 concentrations in plasma, colon tissue, and colon content at 0.5, 2, 4, 12, and 24 h. The inset figure shows the MMT3-72-M2 concentrations in the plasma. The dashed line indicates the limit of detection (2 ng/ml) in our analysis. Data shown as mean ± SD (n = 3 independent biological replicates). GI, gastrointestinal.
MMT3-72 was locally activated in the GI tract to release active metabolite MMT3-72-M2 which achieved high exposure in the GI tissues and minimized exposure in the plasma
To investigate the GI local activation and pharmacokinetics of MMT3-72 and its active metabolite MMT3-72-M2 in vivo, mice were dosed orally with 10 mg/kg MMT3-72 and sacrificed to collect tissues at different time points from 0 to 24 h. As shown in Figure 6C, high concentration (Cmax > 50,000 ng/g) of the MMT3-72 was observed in the colon content. However, MMT3-72 was not detected in either colon tissue or systemic circulation. In contrast, high levels of the active metabolite MMT3-72-M2 were detected in the colon tissue (Cmax > 1500 ng/g) (In Fig. 6D). The effective concentrations lasted for 12 h. At 12 h, the concentration of MMT3-72-M2 was 131.3 ng/g (∼285 nM) in the colon tissues, which exceeded the IC50 values for inhibiting JAK1, JAK2, and TYK2, the relevant targets for treating UC, but was below the IC50 for inhibiting JAK3 (Fig. 5E). Further, the concentration of MMT3-72-M2 in plasma was minimal (Cmax = 8 ng/ml) and was undetectable after 4 h. These findings show that (A) MMT3-72 was not absorbed into the systemic circulation, was retained and activated in the colon region to release active metabolite MMT3-72-M2. (B) The active metabolite MMT3-72-M2 accumulated highly in colon tissues, which may inhibit JAK1, JAK2, and TYK2 for its therapeutic effects. (C) None of MMT3-72 and only low level of the active metabolite MMT3-72-M2 were detected in the systemic circulation, which has the potential to avoid systemic toxicity of JAK inhibitions.
It is worth noting that our design of MMT3-72 is distinctly different from the design of izencitinib (TD-1473), which reduced the absorption potential to limit systemic exposure but without a local-activation mechanism (27). The design of drugs (such as TD-1473) with only reduced absorption potential but without activation mechanism would reduce drug penetration in the colon tissue limiting its efficacy in human trials (28). In contrast, MMT3-72 was designed to not only reduce the GI absorption potential but also have local activation properties to release the active form of MMT3-72-M2 that can easily penetrate colon tissue to reach a therapeutic concentration in the colon tissues while minimizing drug exposure in the systemic circulation.
MMT3-72 exhibited superior efficacy in treating UC in mice
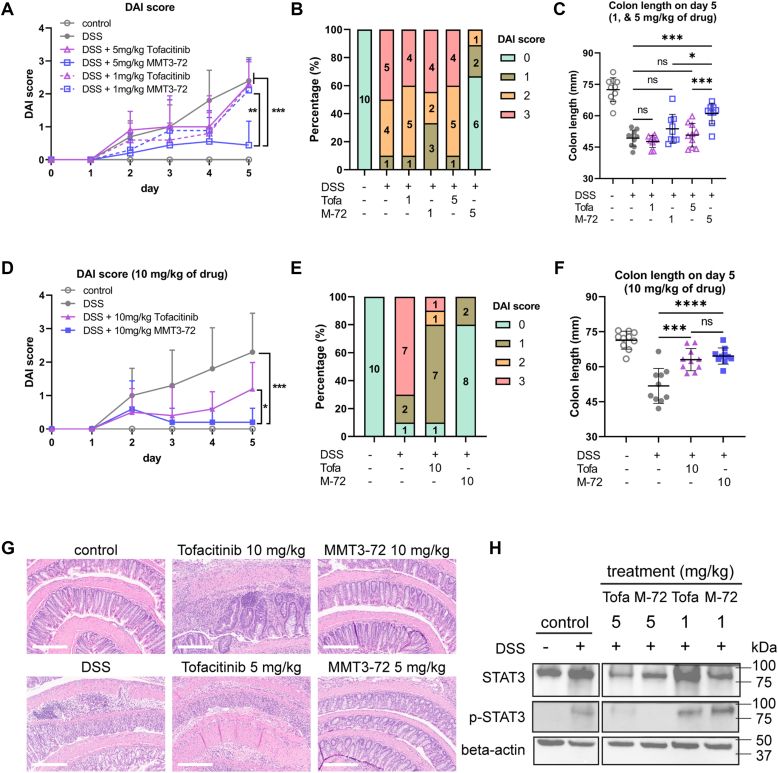
To evaluate the efficacy of MMT3-72 in the treatment of UC in vivo, we established a colitis model in mice using dextran sodium sulfate (DSS). DSS in drinking water could trigger colitis in mice. The DSS-induced colitis model is widely used because of its relatively easy administration and high similarity with human UC (29). In this study, mice treated with 3% DSS water developed symptoms of colitis such as bloody stools and diarrhea on day 5. Disease activity index (DAI) was monitored for the severeness of disease in mice: Normal stool consistency with negative hemoccult: score 0; soft stools with positive hemoccult: score 1; very soft stools with traces of blood: score 2; watery stools with visible rectal bleeding: score 3. To evaluate the efficacy of MMT3-72 in comparison with food and drug administration–approved JAK inhibitor (tofacitinib) for UC treatment, mice were treated orally with 1 mg/kg and 5 mg/kg of both drugs (Fig. 7, A–C). MMT3-72 (5 mg/kg) improved DAI score by 5-fold in comparison with DSS-induced colitis, while tofacitinib (5 mg/kg) did not show any improvement in DAI score (Fig. 7A). In MMT3-72 (5 mg/kg) treatment group, no mice developed severe colitis and only 10% mice (n = 10) developed moderate colitis (Fig. 7B). In contrast, in the tofacitinib treatment group (5 mg/kg), 40% mice (n = 10) developed severe colitis and 40% developed moderate colitis (Fig. 7B). Low dose (1 mg/kg) of both MMT3-72 (1 mg/kg) and tofacitinib (1 mg/kg) did not improve DAI score or disease severity in DSS-induced colitis (Fig. 7, A and B). The inefficacy of tofacitinib was likely due to its lower dosage in our study (1 or 5 mg/kg every other day) than the approved dose of 10 mg twice daily, which is approximately equivalent to ∼4 mg/kg per day in mice. This further underscores the superior efficacy of MMT3-72 at the same dosage.
Figure 7.
In vivo efficacy of MMT3-72 in comparison with tofacitinib for UC treatment.A, improvement of UC DAI score after treatment of MMT3-72 and tofacitinib (1, 5 mg/kg). B, percentage of mice within each DAI group on day 5 after treatment of MMT3-72 and tofacitinib (1, 5 mg/kg). C, recovery of colon length after treatment of MMT3-72 and tofacitinib (1, 5 mg/kg). D, improvement of UC DAI score after treatment of MMT3-72 and tofacitinib (10 mg/kg). E, percentage of mice within each DAI group on day 5 after treatment of MMT3-72 and tofacitinib (10 mg/kg). F, recovery of colon length after treatment of MMT3-72 and tofacitinib (10 mg/kg). Data shown as mean ± SD (A and D; n = 10 mice). Data shown as individual data points with mean ± SD (C and F; n = 10 mice). Significance was accepted at p < 0.05. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p ≤ 0.001; ns, not significant. G, H&E staining of colon tissues. Control was H&E staining of healthy mice colon tissue. DSS-induced colitis showed disrupted epithelium and infiltration of immune cells in colon tissues. Treatment of MMT3-72 (5, 10 mg) reduced epithelium disruption and infiltration of immune cells in colon tissues in comparison with tofacitinib (5, 10 mg/kg) in the DSS-induced colitis model. The scale bar, indicated in the lower right corner, represents 300 μm (micrometers). H, the expression level of STAT3 in colon tissues. Abbreviations of treatments in the figure: Tofa: tofacitinib, M-72: MMT3-72. DAI, disease activity index; DSS, dextran sodium sulfate; STAT, signal transducers and activators of transcription; UC, ulcerative colitis.
Encouraged by these data, we tested high dose (10 mg/kg) of both MMT3-72 and tofacitinib for treatment of DSS-induced UC (see Fig. 7, D–F). MMT3-72 (10 mg/kg) improved DAI score by 10-fold in the DSS-induced colitis model, and no mice (n = 10) developed moderate or severe colitis. In comparison, tofacitinib (10 mg/kg) also showed improvement in DAI score, and only 10% of mice developed severe disease with gross bleeding, and only 20% of mice developed moderate colitis. High doses (10 mg/kg) of both MMT3-72 and tofacitinib recovered the colon length from DSS-induced colitis (see Fig. 7F). These data suggest that MMT3-72 has advantages in the treatment of UC.
To further evaluate the efficacy of MMT3-72 in reducing colon inflammation and tissue injury, H&E staining of colon tissues from the above in vivo studies were performed as shown in Figure 7G. The DSS-induced colitis showed severe and diffuse destruction of the epithelial layer with extensive immune cell infiltration in the epithelium. MMT3-72 (5, 10 mg/kg) reduced epithelial loss and decreased infiltration of immune cells in DSS-induced colitis model. In contrast, tofacitinib (5 mg/kg) did not show improvement in epithelial cell loss and infiltration of immune cells in the DSS-induced colitis model while tofacitinib (10 mg/kg) showed moderate improvement. To investigate if the drug could inhibit JAKs and their downstream signaling, we also used Western blot to detect the total STAT3 and phosphorylated STAT3. Consistent with the histology observation, treatment of MMT3-72 could effectively inhibit the phosphorylation of STAT3 (Figs. 7H and S4) in the inflammation tissue, thereby alleviating the production of inflammatory mediators in the colon region.
Discussion
The JAK inhibitors have shown superior efficacy in the treatment of UC. Since UC needs life-long treatment, the therapeutic options require not only adequate efficacy but also minimal toxicity. However, several current JAK inhibitors, regardless of isoform specificity, have black box warnings for serious side effects including a high rate of major adverse cardiovascular events, thrombosis and pulmonary embolism, malignancies, and increased risk of serious infections.
In this study, we integrated ML, STAR, and wet lab synthesis/testing to design a STAR class III JAK inhibitors (MMT3-72, MMT3-72-M2) for localized JAK inhibition, which could achieve high local exposure/selectivity in the GI tissues while minimizing the requirement for JAK isoform specificity since none of the isoform-specific JAK inhibitors could eliminate their systemic toxicity. The molecular modeling showed that MMT3-72 has poor absorption potential with a low QED score and is expected to be retained in the GI tract. Two ML models predicted MMT3-72 is a weak JAK1 inhibitor and MMT3-72-M2 is an inhibitor of JAK1 (IC50 19 nM), JAK2 (IC50 34 nM), TYK2 (IC50 65 nM).
Based on the ML prediction, we only needed to synthesize two compounds (MMT3-72 and MMT372-M2) for in vitro and in vivo testing on drug’s potency/specificity, tissue selectivity, and efficacy in colitis models. In vitro kinase assays confirmed that MMT3-72 weakly inhibited JAK1 (IC50 368 nM) but was less active against JAK2, JAK3, and TYK2. MMT3-72-M2 inhibited JAK1 (IC50 10.8 nM), JAK2 (IC50 26.3 nM), TYK2 (IC50 91.6 nM) and inhibited weakly on JAK3 (IC50 328.7 nM). The pharmacokinetic study showed that a high level of MMT3-72 was accumulated in the lumen of the GI tract (>50,000 ng/g) but was not detected in either GI tissues or plasma. However, MMT3-72 was locally activated primarily in the colon region to release the active form MMT3-72-M2, which showed a high accumulation in colon contents (Cmax > 50,000 ng/g), colon tissues (Cmax > 1500 ng/ml), but a minimal concentration (Cmax < 8 ng/ml) in the plasma. Oral administration of MMT3-72 (5, 10 mg/kg, respectively) achieved superior efficacy, in comparison with tofacitinib, in DSS-induced colitis as measured by DAI score, bleeding, colon length, and pathological H&E staining of colon tissues. The oral dose of MMT3-72 also inhibited the phosphorylation of STAT3 in the colitis tissue. In summary, the integration of ML, the STAR system, and wet lab synthesis/testing could minimize the effort in the optimization of a JAK inhibitor to treat colitis. This site-specific inhibitor reduces systemic toxicity by minimizing the need for JAK isoform specificity.
Experimental procedures
General
All commercially available products and solvents were purchased from Sigma-Aldrich, AK Scientific, and Thermo Fisher Scientific. Solvents were used as received or dried over molecular sieves (4 Å). All water or air-sensitive reactions were performed under an argon atmosphere with dry solvents and anhydrous conditions. All reactions were monitored by TLC that was performed on aluminum-backed silica plates (0.2 mm, 60 F254). Purification by flash chromatography was performed on Merck silica gel 60 (230–400 mesh). Yields refer to chromatographically and spectroscopically (1H NMR) homogeneous materials unless otherwise stated.
NMR spectra were recorded on a Bruker instrument (500 or 300 MHz) and calibrated using a solvent peak as an internal reference. Spectra were processed using MestReNova software (https://mestrelab.com/software/mnova/). Chemical shifts δ are given in ppm and coupling constants (J) in Hz. Peak multiplicities are described as follows: s, singlet; t, triplet; and m, multiplet. High-resolution mass spectra were obtained on an AB Sciex X500R QTOF spectrometer or an AB Sciex 6600+ Triple TOF mass spectrometer. The purity of all compounds subjected to biological tests was determined by analytical HPLC and was found to be ≥95%.
Synthesis
The synthesis and characterization of compounds 1 and 4 have been previously described (30, 31).
1-(2-chloroethoxy)-4-nitrobenzene
To a mixture of 4-nitrophenol (4 g, 28.754 mmol) and 1,2-dichloromethane (20 ml, 5 vol) in DMF (25 ml) was added K2CO3 (6 g, 43.131 mmol, 1.5 equiv), and the resulting mixture was stirred at 100 °C for 6 h and monitored by TLC. Upon completion, the reaction mixture was quenched with water and the product was extracted three times with CH2Cl2. The combined organic phase was washed with water, brine, dried over Na2SO4, and concentrated under vacuum to give compound 1 (5.35 g; yield, 92%). This intermediate was taken forward to the next step without further purification.
4-(2-chloroethoxy)aniline
To a mixture of compound 1 (1 g, 4.96 mmol) in EtOH (30 ml) was added SnCl2 ·2H2O (4.5 g, 19.84 mmol, 4 equiv) and the reaction mixture was stirred at 90 °C for overnight. Upon completion of the reaction, the solvent was evaporated under reduced pressure and the residue was taken into 5% aqueous NaOH and extracted three times with CH2Cl2. The combined organic phase was washed with 5% aqueous NaOH, water, brine, and dried over Na2SO4. The solvent was concentrated under vacuum and the residue was purified by silica gel column chromatography to provide the compound 2 (532.4 mg, 63% yield). 1H NMR (500 MHz, CDCl3) δ 6.82 to 6.69 (m, 2H), 6.69 to 6.58 (m, 2H), 4.16 (t, J = 5.9 Hz, 2H), 3.77 (t, J = 5.9 Hz, 2H). HRMS (ESI): mass calcd. for C8H10ClNO, 171.05; m/z found, 172.0425 [M + H]+.
N-(tert-butyl)-3-((2-((4-(2-chloroethoxy)phenyl)amino)-5methylpyrimidin-4-yl)amino)benzenesulfonamide
To a mixture of compound 2 (400 mg, 2.330 mmol, 2 equiv) and N-tert-butyl-3-[(2-chloro-5-methylpyrimidin-4-yl)amino]benzenesulfonamide (413.53 mg, 1.165 mmol) in isopropanol (8 ml) was added three drops of concentrated HCl 37% and the reaction mixture was stirred at 80 °C for overnight. Upon completion of the reaction, the solvent was evaporated under reduced pressure and the residue was taken into aqueous NaHCO3 and extracted three times with CH2Cl2. The combined organic phase was washed with water, brine and dried over Na2SO4. The solvent was concentrated under vacuum and the obtained solid was washed three times with EtOAc to provide the compound 3 (482 mg, 84% yield). 1H NMR (500 MHz, DMSO-d6) δ 8.82 (s, 1H), 8.55 (s, 1H), 8.12 (d, J = 5.6 Hz, 2H), 7.91 (d, J = 1.0 Hz, 1H), 7.56 (d, J = 4.3 Hz, 2H), 7.51 to 7.45 (m, 2H), 6.87 to 6.80 (m, 2H), 4.18 (d, J = 6.0 Hz, 2H), 3.92 (d, J = 5.9 Hz, 2H), 2.16 to 2.06 (m, 3H), 1.12 (s, 9H). HRMS (ESI): mass calcd. for C23H28ClN5O3S, 489.16; m/z found, 490.1506 [M + H]+.
2-(4-nitrophenoxy)ethan-1-ol
To a mixture of 4-nitrophenol (3 g, 21.56 mmol) and 2-chloride ethanol (2.89 ml, 43.16 mmol, 2 equiv) in H2O (10 ml) was added NaOH (1.73 g, 43.16 mmol, 2 equiv) and the reaction mixture was stirred at 80 °C for overnight. Upon completion of the reaction, the reaction mixture was cooled down to room temperature, diluted with H2O, and extracted three times with EtOAc. The combined organic phase was washed with water, brine and dried over Na2SO4. The solvent was concentrated under vacuum to give compound 4 (3.1 g, 79% yield), which was taken forward to the next step without further purification.
2-(4-aminophenoxy)ethan-1-ol
To a mixture of compound 4 (1 g, 5.460 mmol) in MeOH (20 ml) was added Pd/C (0.1 g, 10% equiv) and the reaction mixture was stirred at 50 °C for overnight under H2 atmosphere. Upon completion of the reaction, the Pd/C was filtered off on Celite and the solvent was evaporated under reduced pressure. The residue was purified by silica gel column chromatography to provide compound 5 (635.2 mg, 76% yield). 1H NMR (300 MHz, CDCl3) δ 6.82 to 6.72 (m, 2H), 6.70 to 6.59 (m, 2H), 4.01 (dd, J = 5.1, 3.5 Hz, 2H), 3.92 (dd, J = 5.1, 3.5 Hz, 2H). HRMS (ESI): mass calcd. for C8H11NO2, 153.18; m/z found, 154.0770 [M + H]+.
(E)-2-(2-(4-((4-((3-(N-(tertbutyl)sulfamoyl)phenyl)amino)-5-methylpyrimidin-2-yl)amino)phenoxy)ethoxy)-5-((4-((2-carboxyethyl)carbamoyl)phenyl)diazenyl)benzoic acid (MMT3-72)
To a mixture of compound 3 (45 mg, 0.09 mmol, 1.5 equiv) and balsalazide disodium salt dehydrate (27.29 mg, 0.068 mmol, 1 equiv) in DMF (2 ml) was added K2CO3 (37.6 mg, 0.272 mmol, 4 equiv), and the resulting mixture was stirred at 100 °C for overnight and monitored by TLC. Upon completion, the solvent was evaporated under reduced pressure. The residue was taken into H2O and the solution was acidified with H3PO4 until pH 2 to 3. The precipitate was filtered and recrystallization in CH2Cl2 provided the desired compound MMT3-72 (50.4 mg, 68%). 1H NMR (500 MHz, DMSO-d6) δ 8.68 (s, 1H), 8.29 (s, 1H), 8.08 (s, 2H), 7.96 (d, J = 8.3 Hz, 2H), 7.88 (d, J = 10.5 Hz, 2H), 7.82 (d, J = 8.3 Hz, 2H), 7.57 (s, 1H), 7.49 (t, J = 14.4 Hz, 4H), 6.86 (d, J = 9.7 Hz, 1H), 6.80 (d, J = 8.5 Hz, 2H), 4.36 (t, J = 4.5 Hz, 2H), 4.13 (t, J = 4.8 Hz, 2H), 3.61 to 3.48 (m, 2H), 2.66 (t, J = 6.9 Hz, 2H), 2.11 (s, 3H), 1.11 (s, 9H). HRMS (ESI): mass calcd. for C40H42N8O9S, 810.2; m/z found, 811.2758 [M + H]+.
N-(tert-butyl)-3-((2-((4-(2-hydroxyethoxy)phenyl)amino)-5methylpyrimidin-4-yl)amino)benzenesulfonamide (MMT3-72-M2)
To a mixture of compound 5 (130 mg, 0.846 mmol, 3 equiv) and N-tert-butyl-3-[(2-chloro-5-methylpyrimidin-4-yl)amino]benzenesulfonamide (100 mg, 0.282 mmol) in isopropanol (2 ml) was added three drops of concentrated HCl 37% and the reaction mixture was stirred at 80 °C for overnight. Upon completion of the reaction, the solvent was evaporated under reduced pressure and the residue was taken into aqueous NaHCO3 and extracted three times with CH2Cl2. The combined organic phase was washed with water, brine, and dried over Na2SO4. The solvent was concentrated under vacuum and the obtained solid was washed three times with EtOAc to provide the compound MMT3-72-M2 (105 mg, 79% yield). 1H NMR (300 MHz, DMSO-d6) δ 8.79 (s, 1H), 8.55 (s, 1H), 8.12 (d, J = 3.4 Hz, 2H), 7.90 (s, 1H), 7.62 to 7.39 (m, 4H), 6.80 (d, J = 8.9 Hz, 2H), 3.92 (t, J = 5.1 Hz, 2H), 3.69 (t, J = 5.0 Hz, 2H), 2.12 (s, 3H), 1.12 (s, 9H). HRMS (ESI): mass calcd. for C23H29N5O4S, 471.58; m/z found, 472.1847 [M + H]+.
Activation of MMT3-72 and metabolites identification
In vivo metabolites identification was conducted using mouse plasma, colon, and feces samples that were collected at 6 h after oral administration of MMT3-72 (10 mg/kg). Liquid chromatography-tandem high-resolution mass spectrometry was employed to separate and identify the possible metabolites. The LC–MS method consisted of a Shimadzu LC20AD HPLC system and a high-resolution AB Sciex X500R QTOF mass spectrometer (AB Sciex). Chromatographic separation of MMT3-72 and its metabolites was achieved using a Waters XBridage reverse phase C18 column (15 cm × 2.1 mm I.D., packed with 3.5 μm). For metabolites identification, mass spectrometer was adjusted into the positive-ion information-dependent acquisition mode. The mass range was recorded from m/z 100 to 1000 Da. The collision energy was set to 50 V for TOF MSMS. Data were collected with the software SCIEX OS (https://sciex.com/products/software/sciex-os-software) and then processed with the software MetabolitePilot 2.0 (https://sciex.com/cl/products/software/metabolitepilot-software) (AB Sciex). The metabolites in mouse plasma, colon, and feces were then detected with LC-MS using TOFMS scan with exacted ions 416.1387 ± 0.01 for M1, 472.2013 ± 0.01 for M2, 486.1806 ± 0.01 for M3, 755.2242 ± 0.01 for M4, and 428.1751 ± 0.01 for M5.
ML models for JAK inhibition prediction
CoGT
We applied our previously trained ensemble model CoGT (Conventional ML models + Graph-based models + Transformer-based models) for classification of JAK inhibitors or noninhibitors (32). CoGT is specifically trained for the JAK inhibitor classification task. Briefly, a dataset was collected from ChEMBL (33, 34), BindingDB (35), PubChem (36, 37), and Liu et al. (38), consisting of N molecule-label pairs and was assumed to be i.i.d. (independently and identically distributed random variables). Tanimoto similarity analysis for the data set used is visualized in Fig. S2A. xi is the feature of one molecule, and yi is its true label (1 if the molecule is JAK inhibitor, 0 otherwise, 10 μM is the threshold for IC50). For the feature xi, it could either be MACCS fingerprint, graph, or simplified molecular input line entry systems (SMILES) representation based on different ML models. The aim is to do binary classification given an unknown drug compound x, the model outputs the probability p of the compound being a JAK inhibitor (if p > 0.5, the molecule is predicted by CoGT as a JAK inhibitor with IC50 < 10 μM, else it is predicted to be a noninhibitor). For model architecture, CoGT is an ensemble model including graph-based model (GraphVAE), transformer-based model (chemBERTa), and conventional ML models Support Vector Machine, Random Forest, and XGBoost. By integrating different ML methods with different input representation of compounds using multilayer perceptron, CoGT outperformed individual ML models and leveraged the predicting ability for JAK inhibition prediction.
To train CoGT, the dataset was first randomly split into training set, validation set, and test set with ratio 8:1:1. Training data were used to train the model, validation data was used for fine tuning, and test set was used for model evaluation of the generalization ability. BCELoss was used as loss function for this binary classification task to calculate the difference between the model prediction and the compound labels (0 if noninhibitor; 1 if inhibitor for specific JAK). Training was terminated if loss did not decrease and model with lowest loss was saved as the optimal model.
MTATFP
Wang et al. (39) built a multitask regression model based on the attentive fingerprint framework (MTATFP) to predict compounds’ IC50 for predicted JAK inhibitors. Briefly, data were collected from ChEMBL (33, 34), BindingDB (35), and PubChem (36, 37). Tanimoto similarity analysis for the data set used is visualized in Fig. S2B and the data distribution are summarized in Fig. S3. The model was trained as a regression task, aiming to predict compounds’ pIC50 values based on the molecular graph constructed by atom matrix and bond matrix. The Attentive FP framework was used to learn atomic and molecular properties through the graph-attention network mechanism (40). MTATFP used a multitask learning strategy for four JAKs to enhance the performance by data augmentation through eavesdropping between tasks, allowing simultaneous prediction of compounds' IC50 values for four JAK isoforms. For model architecture, atomic features are extracted by Attentive FP convolutional layers and the molecular embedding was obtained using a readout layer. Transformation of molecular embedding is performed using linear layers with LeakyReLU.
To elaborate the training process, collected data set was randomly split into training set, validation set, and test set with a ratio of 8:1:1. MSELoss was used as loss function to calculate the difference between the model prediction and the true pIC50 of compounds. If JAK pIC50 values were missing, masking strategy was used to ignore the loss computation for the missing data entry. Early stopping approach was applied to avoid overfitting.
Molecular docking
DiffDock
We employed DiffDock for its speed and accuracy to accelerate this process (23). Briefly, instead of considering ligand pose in which n is the atom number of the ligand like EquiBind, DiffDock constrains the ligand pose in an (m + 6)-dimensional submanifold , in which m represents the number of rotatable bonds, and six comes from the rotation and translation of the ligand relative to the given protein position. DiffDock utilizes a generative diffusion model to gradually update the random-initialized molecules’ torsion, rotation, and translation to fit into the fixed protein.
To elaborate on how to define the (m + 6)-dimensional submanifold , for a seed conformation , let be the position of the ith atom, be the mass center of the ligand, we have
translation
| (1) |
rotation
| (2) |
Define and let be the kth rotatable bond torsion update by θk and all updates for m rotatable bonds are θ = (θ1, · · ·, θm). To make sure the infinitesimal effect of torsion is orthogonal to rototranslation, the torsion is defined as
| (3) |
Let the product space , and define the overall pose update as
| (4) |
Thus the space of ligand poses is formally defined.
To apply a diffusion model on the product manifold , it is sufficient enough to sample from the diffusion kernel and compute the score independently in each manifold, and the tangent space is a direct sum, where g = (r, R, θ). For ligand pose x and protein structure y, the authors construct the score model to output SE(3)-equivariant translation vectors, rotation vectors, and SE(3)-invariant torsion angles. An SE(3)-invariant confidence model assigns scores for different ligand poses, and the top-ranked pose is then taken as DiffDock’s top-1 prediction. We set the diffusion step to 20 and evaluated the docking poses of MMT3-72, M2, and fedratinib. The crystal structures of JAKs were retrieved from the Protein Data Bank (https://www.rcsb.org/): JAK1: 6BBU, JAK2: 6VNE, JAK3: 4HVD, and TYK2: 6DBK. The code is available at https://github.com/gcorso/DiffDock.
EquiBind
EquiBind was built to perform direct-shot prediction (26). In detail, both ligands and proteins were represented as spatial k-nearest neighbor (k-NN) graphs . is the atom/residue information for ligand and protein, respectively, consisting of node 3D coordinates X and initial features F for each node. For edges , ligand edges include atom pairs < 4 Å and the protein edges consist of connection of the closest 10 other nodes <30 Å.
It is desired to predict the same binding complex regardless of the initial position and orientation of ligand/protein in 3D space, thus EquiBind incorporated Independent E(3)-Equivariant Graph Matching Network (IEGMN) to make sure the coordinate transformations are E(3)-equivariant while the feature embeddings are E(3)-invariant (41). In other words, the output of the IEGMN is
| (5) |
in which Z, Z′ represent transformed coordinates and H, H′ are feature embeddings of molecules and targets, respectively. If applied a transformation in 3D space for both the ligand and/or target, the initial location of the ligand now is UX + b and the protein location is U′X′ + b′, in which rotation matrices U, U′ ∈ SO (3) and translation vectors b, b′ ∈ 3, IEGMN guarantees that
| (6) |
For updating transformed coordinates during training IEGMN, molecules were assumed to be flexible only with rotatable bonds, while bond lengths, adjacent bond angles, and small rings were treated as rigid, which could be defined as minimizing the following function defined by the authors:
| (7) |
These constraints could be satisfied by incorporating T gradient descent layers and update .
However, implausible conformers may be produced during this process, thus the authors came up with the idea of only changing the torsion angles of the conformer initialized by RDKit. In detail, the output C ∈ 3×n is initialized as X and thus . Then, only the rotatable bonds’ torsion angles are changed to match those of Z to avoid implausible bond angles/lengths. Instead of computing the gradient of a point cloud with respect to torsion angles which is computationally expensive, the authors compute the rotatable bonds’ dihedral angles of C as maximum likelihood estimates of von Mises distributions on those of Z’s without requiring optimization, that is, to minimize the torsion angle difference between Z and C, a closed-form solution exists which could provide all dihedral angle values.
By using an SE(3)-equivariant multi-head attention mechanism, ligand and receptor keypoints Y, Y′ ∈ 3×K were trained to match the ground truth-binding pocket points using the coordinate outputs Z, Z′ computed by IEGMN above. Once trained, performing SE(3) transformation to superimpose Y and Y′ would be equivalent to performing ligand docking. The code is available at https://github.com/HannesStark/EquiBind.
In vitro activity to inhibit JAK enzymes
JAK1, JAK2, JAK3, and TYK2 assay kits were obtained from BPS Bioscience. The assays were conducted according to the manufacturer’s protocols in 96-well microplates. Briefly, master mixtures (25 μl per well) were prepared for JAK1 and TYK2 assays (6 μl 5 × kinase assay buffer + 1 μl ATP (500 μM) + 5 μl 10 × IRS1-tide + 13 μl distilled water) or for JAK2 and JAK3 assays (6 μl 5 × kinase assay buffer + 1 μl ATP (500 μM) + 1 μl PTK substrate Poly(Glu:Tyr 4:1) (10 mg/ml) + 17 μl distilled water), respectively. Then 5 μl of fedratinib, MMT3-72, MMT3-72-M2 solutions at different concentrations were added to the above-prepared master mixtures, which were followed by 20 μl of enzymes (JAK1 at 5 ng/μl, JAK2 at 2.5 ng/μl, JAK3 at 0.4 ng/μl, or TYK2 at 0.5 ng/μl), respectively. The reaction mixtures were incubated at 30 °C for 40 min. Finally, 50 μl of Kinase-Glo Max reagent (Promega) were added to each well and the reactions were performed in darkness for 15 min at room temperature. The luminescence of the reaction mixture was read on a Synergy 2 microplate reader (Biotek).
LC-MS analysis of MMT3-72 and MMT3-72-M2 in biological samples
MMT3-72 and MMT3-72-M2 concentrations in plasma (ng/ml) and tissues (ng/g) were determined by the liquid chromatography-tandem mass spectrometry method that was developed and validated for this study. The HPLC method was conducted on a Shimadzu LC-20AD HPLC system, and chromatographic separation was achieved using a Waters XBridage reverse phase C18 column (5 cm × 2.1 mm I.D., packed with 3.5 μm). The flow rate of gradient elution was 0.4 ml/min with mobile phase A (0.1% formic acid in purified deionized water) and mobile phase B (0.1% formic acid in acetonitrile). An AB Sciex QTrap 4500 mass spectrometer (AB Sciex) in the positive-ion multiple reaction monitoring mode was used for detection. Protonated molecular ions and the respective ion products were monitored at the transitions of m/z 811.3 > 737.4 for MMT3-72 and 472.3 > 416.0 for MMT3-72-M2. Data were processed with the software Analyst (version 1.6; https://www.sciex.com/products/software/analyst-software).
In vitro conversion of MMT3-72 to MMT3-72-M2 by active colon contents
C57BL/6 female mice (aged 6–8 weeks) were purchased from Charles River Laboratories. Fresh colon contents were isolated from the mice and suspended in PBS. The resulting 10% (w/v) colon content suspension was used as the source of azoreductases. A stock solution of MMT3-72 was added to the suspension to achieve a final concentration of 0.1 mM. The mixture was thoroughly combined and incubated on a shaker at 1000 rpm and 37 °C. At 0, 1, 2, 4, 6, 24, and 48 h, a sample aliquot was collected, followed by 10-fold dilution and extraction using acetonitrile. Additionally, a parallel experiment was conducted using deactivated colon contents (boiled). The extracted supernatants were stored at −20 °C until quantitative analysis. The amounts of MMT3-72 and MMT3-72-M2 were determined via LC-MS analysis using the method described above.
Animals
Female C57BL/6 mice were obtained from Charles River Laboratories at 4 to 6 weeks of age. Mice were housed in groups of five animals per cage on a 12:12 h light/dark cycle. Water and food were provided ad libitum. Mice aged 6 to 8 weeks were used for experiments. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Michigan.
Pharmacokinetic study in mice
Briefly, C57BL/6 female mice (6–8 weeks old) were orally dosed with 10 mg/kg MMT3-72 dissolved in a pH-adjusted solution comprising an 80% v/v mixture of beta-cyclodextrin and water (at a ratio of 1:4), 15% hyaluronic acid, and 5% phosphate-buffered saline. At 0.5, 2, 4, 12, and 24 h, mice were sacrificed, and blood samples were collected by drawing directly from heart. Intestinal tissue samples were collected and homogenized in PBS to 10% homogenate. The contents of small and large intestines were collected and homogenized in PBS. Afterward, concentrations of MMT3-72, MMT3-72-M2 in the plasma, colon tissues, and colon content were determined by the quantitation method described above.
In vivo efficacy of MMT3-72 in the treatment of DSS-induced colitis in mice
C57BL/6 female mice (6–8 weeks old) were randomly divided into different treatment groups. Acute colitis was induced by administering 3% DSS (MP Biomedicals) in distilled water continuously for 5 days, and the control group received pure water (29). MMT3-72 or tofacitinib was dissolved in a pH-adjusted solution comprising an 80% v/v mixture of beta-cyclodextrin and water (at a ratio of 1:4), 15% hyaluronic acid, and 5% PBS. For the noninhibitor treatment groups, the same formulation solution was given as the vehicle control. Drugs were administered every other day orally by gavage in a volume of 0.1 ml/10g body weight. During the model establishment, body weight, stool consistency, and gross blood in feces were monitored and recorded daily (29). DAI was monitored for the severeness of disease in mice using the fecal occult blood slide test system (HemoCue America) according to the manufacturer’s instruction. After 5 days, mice were sacrificed, and blood was collected. Serum was obtained by centrifugation and stored at −80 °C until further assay. The colon was excised, and the length was measured.
H&E staining of colon tissues
After dissecting and transecting the colon, a feeding needle and 5 ml syringe were used to incubate and flush the colon with ice-cold PBS until the stool was flushed out. Scissors were used to incise longitudinally from the distal to the proximal end of the colon and the colon tissue could then be expanded as a flat sheet. The edge of the distal colon was grasped using a pair of forceps and the colon tissue was rotated into a Swiss roll. The roll was firmly grasped and transected using a 27G 1/2 needle. Then the sample was placed in 4% Paraformaldehyde Fix Solution (Thermo Fisher Scientific) at room temperature for 24 h. The Swiss roll was then paraffin-embedded, sectioned, mounted, and stained with H&E to determine the extent of damage to the colon from the distal (inside end) to the proximal (outside end).
Western blot assay of colon tissues
The dissected colon tissue was rinsed in cold PBS and homogenized in RIPA buffer. The homogenates were subjected to ultrasonication and centrifugation for protein extraction. The resulting supernatants were collected, and the total protein was quantified using the BCA kit (Thermo Fisher Scientific). For each treatment group, 200 μg of total protein was used for SDS-electrophoresis. The separated proteins were then transferred onto polyvinylidene difluoride membranes. These membranes were incubated overnight at 4 °C with the indicated primary antibodies, followed by a one-hour incubation with horseradish peroxidase–labeled secondary antibodies at room temperature. The blots were visualized using the ChemiDoc Imaging System (Bio-Rad). Antibodies against JAK2, STAT3, Phospho-STAT3 (Tyr705), and GAPDH (Cell Signaling Technology) were used for immunoblotting.
Data analysis
Data are presented as mean ± SD. For statistical difference determination between treatment groups, one-way ANOVA or two-way ANOVA was performed with Tukey’s post hoc correction (GraphPad Prism 9). Significance was accepted at p < 0.05. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p ≤ 0.001; ns, not significant. IC50 values were derived from fits to Hill equations.
Data availability
All data will be available upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The University of Michigan filed a patent for these compounds, where some authors are inventors. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the NMR core and Pharmacokinetics core at the College of Pharmacy for their technical support. We thank the University of Michigan for internal funding support.
Author contributions
Y. B., M. D. M. T., and D. S. conceptualization; Y. B., M. D. M. T., H. H., and D. S. methodology; Y. B. software; Y. B. and M. D. M. T. validation; Y. B., L. Z., L. W., Z. L., and M. W. formal analysis; Y. B., M. D. M. T., L. Z., L. W., Z. L., H. H., and C. L. investigation; Y. B., M. D. M. T., L. Z., and D. S. writing–original draft; writing–original draft; Y. B., L. Z., and D. S. writing–review and editing; L. Z. and H. H. visualization; H. H., M. W., and C. L. resources; D. S. supervision; D. S. funding acquisition.
Reviewed by members of the JBC Editorial Board. Edited by Qi-Qun Tang
Supporting information
References
- 1.Cosnes J., Gower-Rousseau C., Seksik P., Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 2.Silvio D., Claudio F. Ulcerative colitis new England. J. Med. 2011;365:1713–1725. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan G.G. The global burden of IBD: from 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015;12:720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 4.Ordas I., Eckmann L., Talamini M., Baumgart D.C., Sandborn W.J. Ulcerative colitis. Lancet. 2012;380:1606–1619. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 5.Collins R.H., Jr., Feldman M., Fordtran J.S. Colon cancer, dysplasia, and surveillance in patients with ulcerative colitis. N. Engl. J. Med. 1987;316:1654–1658. doi: 10.1056/NEJM198706253162609. [DOI] [PubMed] [Google Scholar]
- 6.Eisenstein M. Ulcerative colitis: towards remission. Nature. 2018;563:S33. doi: 10.1038/d41586-018-07276-2. [DOI] [PubMed] [Google Scholar]
- 7.Salas A., Hernandez-Rocha C., Duijvestein M., Faubion W., McGovern D., Vermeire S., et al. JAK--STAT pathway targeting for the treatment of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020;17:323–337. doi: 10.1038/s41575-020-0273-0. [DOI] [PubMed] [Google Scholar]
- 8.Neurath M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 9.Lamb C.A., Kennedy N.A., Raine T., Hendy P.A., Smith P.J., Limdi J.K., et al. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut. 2019;68:s1–s106. doi: 10.1136/gutjnl-2019-318484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh S., Facciorusso A., Dulai P.S., Jairath V., Sandborn W.J. Comparative risk of serious infections with biologic and/or immunosuppressive therapy in patients with inflammatory bowel diseases: a systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2020;18:69–81. doi: 10.1016/j.cgh.2019.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamba M., Wang R., Fletcher T., Alvey C., Kushner Iv J., Stock T.C. Extended-release once-daily formulation of tofacitinib: evaluation of pharmacokinetics compared with immediate-release tofacitinib and impact of food. J. Clin. Pharmacol. 2016;56:1362–1371. doi: 10.1002/jcph.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubbert-Roth A., Enejosa J., Pangan A.L., Haraoui B., Rischmueller M., Khan N., et al. Trial of upadacitinib or abatacept in rheumatoid arthritis. New Engl. J. Med. 2020;383:1511–1521. doi: 10.1056/NEJMoa2008250. [DOI] [PubMed] [Google Scholar]
- 13.Traves P.G., Murray B., Campigotto F., Galien R., Meng A., Di Paolo J.A. JAK selectivity and the implications for clinical inhibition of pharmacodynamic cytokine signalling by filgotinib, upadacitinib, tofacitinib and baricitinib. Ann. Rheum. Dis. 2021;80:865–875. doi: 10.1136/annrheumdis-2020-219012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krueger J.G., McInnes I.B., Blauvelt A. Tyrosine kinase 2 and Janus kinase--signal transducer and activator of transcription signaling and inhibition in plaque psoriasis. J. Am. Acad. Dermatol. 2022;86:148–157. doi: 10.1016/j.jaad.2021.06.869. [DOI] [PubMed] [Google Scholar]
- 15.Papp K., Gordon K., Thai D., Morita A., Gooderham M., Foley P., et al. Phase 2 trial of selective tyrosine kinase 2 inhibition in psoriasis. New Engl. J. Med. 2018;379:1313–1321. doi: 10.1056/NEJMoa1806382. [DOI] [PubMed] [Google Scholar]
- 16.Gao W., Hu H., Dai L., He M., Yuan H., Zhang H., et al. Structure--tissue exposure/selectivity relationship (STR) correlates with clinical efficacy/safety. Acta Pharm. Sin. B. 2022;12:2462–2478. doi: 10.1016/j.apsb.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu H., Mady Traore M.D., Li R., Yuan H., He M., Wen B., et al. Optimization of the prodrug moiety of Remdesivir to improve lung exposure/selectivity and enhance anti-SARS-CoV-2 activity. J. Med. Chem. 2022;65:12044–12054. doi: 10.1021/acs.jmedchem.2c00758. [DOI] [PubMed] [Google Scholar]
- 18.Lin A., Giuliano C.J., Palladino A., John K.M., Abramowicz C., Yuan M.L., et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun D., Gao W., Hu H., Zhou S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B. 2022;12:3049–3062. doi: 10.1016/j.apsb.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bickerton G.R., Paolini G.V., Besnard J., Muresan S., Hopkins A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012;4:90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogasawara K., Xu C., Kanamaluru V., Siebers N., Surapaneni S., Ridoux L., et al. Excretion balance and pharmacokinetics following a single oral dose of 14 C -fedratinib in healthy subjects. Cancer Chemother. Pharmacol. 2020;86:307–314. doi: 10.1007/s00280-020-04121-0. [DOI] [PubMed] [Google Scholar]
- 22.Davis R.R., Li B., Yun S.Y., Chan A., Nareddy P., Gunawan S., et al. Structural insights into JAK2 inhibition by ruxolitinib, fedratinib, and derivatives thereof. J. Med. Chem. 2021;64:2228–2241. doi: 10.1021/acs.jmedchem.0c01952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corso G., Strk H., Jing B., Barzilay R., Jaakkola T. Diffdock: diffusion steps, twists, and turns for molecular docking. arXiv. 2022 doi: 10.48550/arXiv.2210.01776. [preprint] [DOI] [Google Scholar]
- 24.Vazquez M.L., Kaila N., Strohbach J.W., Trzupek J.D., Brown M.F., Flanagan M.E., et al. Identification of N-cis-3-methyl(7H-pyrrolo 2, 3-d pyrimidin-4-yl) amino cyclobutylpropane-1-sulfonamide (PF-04965842): a selective JAK1 clinical candidate for the treatment of autoimmune diseases. J. Med. Chem. 2018;61:1130–1152. doi: 10.1021/acs.jmedchem.7b01598. [DOI] [PubMed] [Google Scholar]
- 25.Fensome A., Ambler C.M., Arnold E., Banker M.E., Brown M.F., Chrencik J., et al. Dual inhibition of TYK2 and JAK1 for the treatment of autoimmune diseases: discovery of ((S)-2, 2-difluorocyclopropyl)((1 R, 5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl) amino) pyrimidin-4-yl)-3, 8-diazabicyclo 3.2. 1 octan-8-yl) methanone (PF-06700841) J. Med. Chem. 2018;61:8597–8612. doi: 10.1021/acs.jmedchem.8b00917. [DOI] [PubMed] [Google Scholar]
- 26.Strk H., Ganea O., Pattanaik L., Barzilay R., Jaakkola T. Equibind: geometric deep learning for drug binding structure prediction. arXiv. 2022 doi: 10.48550/arXiv.2202.05146. [preprint] [DOI] [Google Scholar]
- 27.Hardwick R.N., Brassil P., Badagnani I., Perkins K., Obedencio G.P., Kim A.S., et al. Gut-selective design of orally administered izencitinib (TD-1473) limits systemic exposure and effects of Janus kinase inhibition in nonclinical species. Toxicol. Sci. 2022;186:323–337. doi: 10.1093/toxsci/kfac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandborn W.J., Nguyen D.D., Beattie D.T., Brassil P., Krey W., Woo J., et al. Development of gut-selective pan-Janus kinase inhibitor TD-1473 for ulcerative colitis: a translational medicine programme. J. Crohn's Colitis. 2020;14:1202–1213. doi: 10.1093/ecco-jcc/jjaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snider A.J., Bialkowska A.B., Ghaleb A.M., Yang V.W., Obeid L.M., Hannun Y.A. Murine model for Colitis-associated cancer of the colon. Methods Mol. Biol. 2016;1438:245–254. doi: 10.1007/978-1-4939-3661-8_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Šukalović V., Andrić D., Roglić G., Kostić-Rajačić S., Schrattenholz A., Šoškić V. Synthesis, dopamine D2 receptor binding studies and docking analysis of 5- 3-(4-arylpiperazin-1-yl) propyl -1H-benzimidazole, 5- 2-(4-arylpiperazin-1-yl) ethoxy -1H-benzimidazole and their analogs. Eur. J. Med. Chem. 2005;40:481–493. doi: 10.1016/j.ejmech.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Luo G., Chen M., Lyu W., Zhao R., Xu Q., You Q., et al. Design, synthesis, biological evaluation and molecular docking studies of novel 3-aryl-4-anilino-2H-chromen-2-one derivatives targeting ER$∖alpha$ as anti-breast cancer agents. Bioorg. Med. Chem. Lett. 2017;27:2668–2673. doi: 10.1016/j.bmcl.2017.04.029. [DOI] [PubMed] [Google Scholar]
- 32.Bu Y., Gao R., Zhang B., Zhang L., Sun D. CoGT: ensemble machine learning method and its application on JAK inhibitor discovery. ACS Omega. 2023;8:13232–13234. doi: 10.1021/acsomega.3c00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45:D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Bryant S.H. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Wu Y., Shen X., Xie L. COVID-19 multi-targeted drug repurposing using few-shot learning. Front. Bioinform. 2021;1:693177. doi: 10.3389/fbinf.2021.693177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Gu Y., Lou C., Gong Y., Wu Z., Li W., et al. A multitask GNN-based interpretable model for discovery of selective JAK inhibitors. J. Cheminform. 2022;14:16. doi: 10.1186/s13321-022-00593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Z., Wang D., Liu X., Zhong F., Wan X., Li X., et al. Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J. Med. Chem. 2019;63:8749–8760. doi: 10.1021/acs.jmedchem.9b00959. [DOI] [PubMed] [Google Scholar]
- 41.Ganea O.-E., Huang X., Bunne C., Bian Y., Barzilay R., Jaakkola T., et al. Independent se (3)-equivariant models for end-to-end rigid protein docking. arXiv. 2021 doi: 10.48550/arXiv.2111.07786. [preprint] [DOI] [Google Scholar]
- 42.Clark J.D., Flanagan M.E., Telliez J.-B. Discovery and development of Janus Kinase (JAK) inhibitors for inflammatory diseases. J. Med. Chem. 2014;57:5023–5038. doi: 10.1021/jm401490p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data will be available upon request.