Abstract
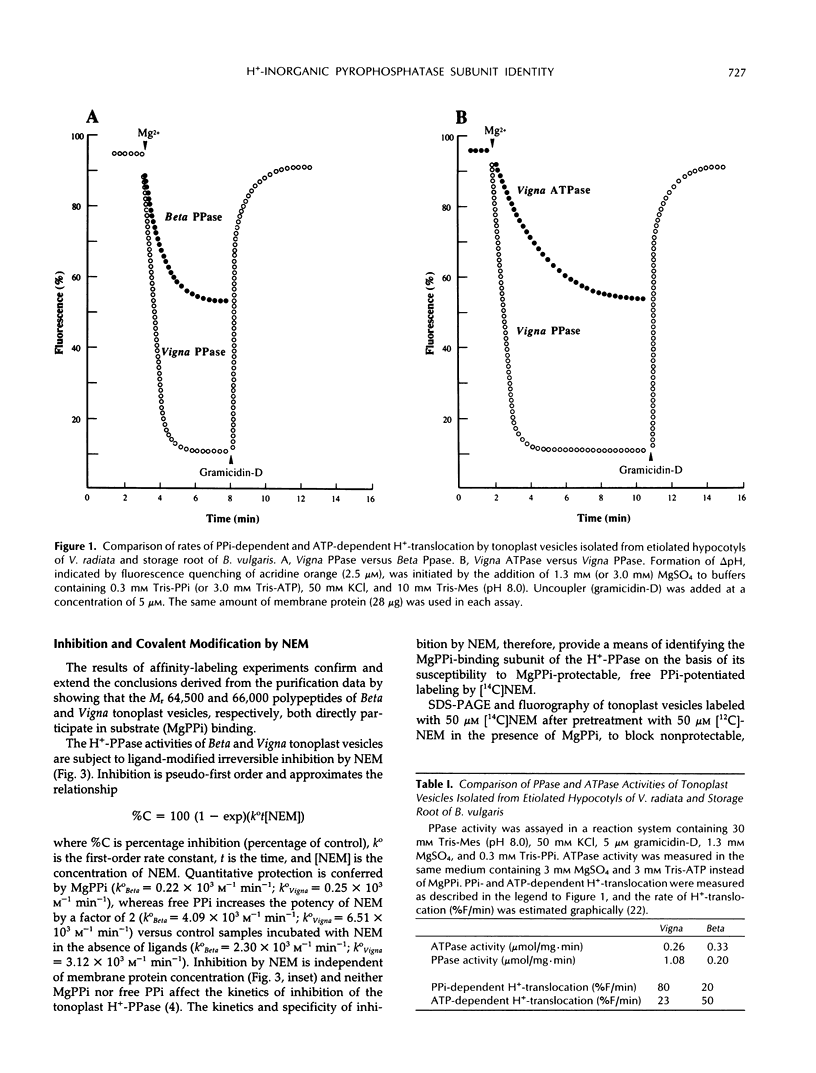
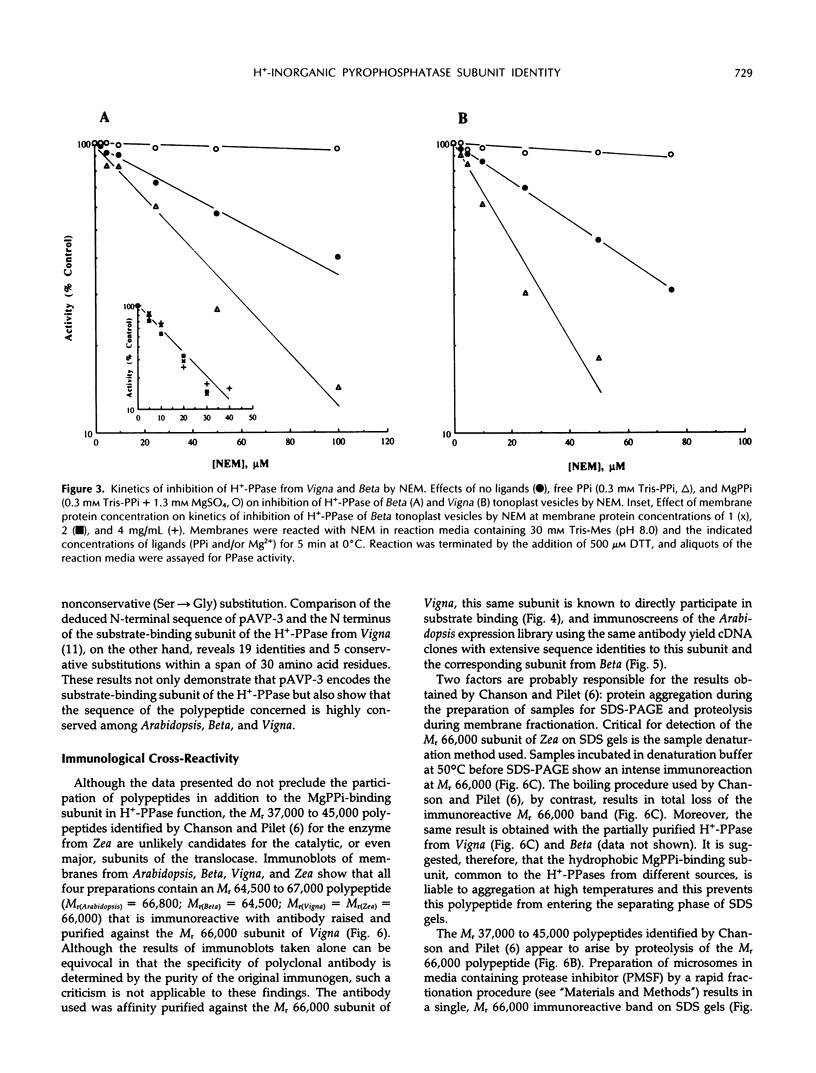
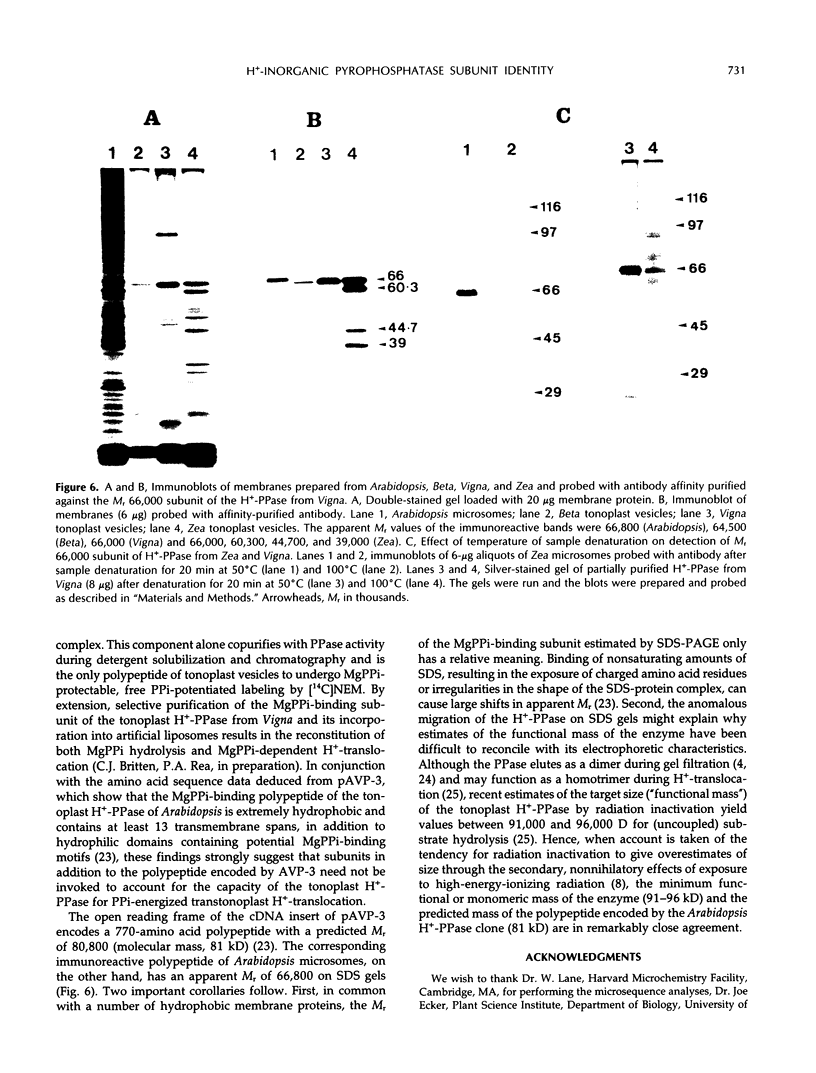
There have been conflicting reports in the literature concerning the polypeptide composition of the vacuolar H+-translocating inorganic pyrophosphatase (tonoplast H+-PPase) of plant cells. The major subunit(s) of the enzyme have been attributed to polypeptides of relative molecular weight (Mr) 64,500 (Beta vulgaris), 67,000 (Beta vulgaris), 73,000 (Vigna radiata), and 37,000 to 45,000 (Zea mays). Here, we reconcile these differences to show, through the combined application of independent purification, affinity-labeling, sequencing, and immunological procedures, that the major polypeptide associated with the H+-PPase from all of these organisms, and Arabidopsis thaliana, corresponds to the same moiety. The principal polypeptide components of the H+-PPase purified from Beta and Vigna by independent procedures have similar apparent subunit masses when subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under identical conditions (Mr(Beta) = 64,500; Mr(Vigna) = 66,000) and exhibit identical kinetics of irreversible inhibition and ligand-modified labeling by [14C]-N-ethylmaleimide. Similarly, the Mr 64,500 and 67,000 polypeptides isolated from Beta by independent methods (cf. C.J. Britten, J.C. Turner, P.A. Rea [1989] FEBS Lett 256: 200-206 versus V. Sarafian and R.J. Poole [1989] Plant Physiol 91: 34-38) are indistinguishable: the two polypeptides comigrate when electrophoresed under the same conditions and yield tryptic fragments with identical overlapping sequences. Because both the N-terminal sequence of the Mr 66,000 subunit of the H+-PPase isolated from Vigna and the direct sequence data from Beta align precisely with the deduced amino acid sequence of cDNAs encoding the H+-PPase of Arabidopsis, all three enzymes are inferred to be highly conserved structurally. Accordingly, immunoblots of membranes prepared from Arabidopsis, Beta, Vigna, and Zea, probed with antibody affinity purified against the magnesium inorganic pyrophosphate-binding, Mr 66,000 polypeptide of Vigna, reveal a single immunoreactive band at Mr 64,500 to 67,000 in all four preparations. The Mr 66,000 polypeptide of Zea membranes is, however, prone to proteolysis during membrane fractionation and selective aggregation during sample denaturation for SDS-PAGE. The anomalous Mr 37,000 to 45,000 subunit pattern previously ascribed to the H+-PPase from Zea (A. Chanson and P.E. Pilet [1989] Plant Physiol 90: 934-938) is attributed to loss of the Mr 66,000 subunit and the appearance of polypeptide fragments of Mr 44,700 and 39,000 through the combined effects of sample aggregation before SDS-PAGE and proteolysis, respectively. It is, therefore, concluded that the substrate-binding subunit of the tonoplast H+-PPase has a common identity in all four organisms.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebersold R. H., Leavitt J., Saavedra R. A., Hood L. E., Kent S. B. Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci U S A. 1987 Oct;84(20):6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanson A., Fichmann J., Spear D., Taiz L. Pyrophosphate-driven proton transport by microsomal membranes of corn coleoptiles. Plant Physiol. 1985 Sep;79(1):159–164. doi: 10.1104/pp.79.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanson A., Pilet P. E. Target Molecular Size and Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis Analysis of the ATP-and Pyrophosphate-Dependent Proton Pumps from Maize Root Tonoplast. Plant Physiol. 1989 Jul;90(3):934–938. doi: 10.1104/pp.90.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempner E. S., Miller J. H. Radiation inactivation of glutamate dehydrogenase hexamer: lack of energy transfer between subunits. Science. 1983 Nov 11;222(4624):586–589. doi: 10.1126/science.6635656. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai S. P., Randall S. K., Sze H. Peripheral and integral subunits of the tonoplast H+-ATPase from oat roots. J Biol Chem. 1988 Nov 15;263(32):16731–16737. [PubMed] [Google Scholar]
- Maeshima M., Yoshida S. Purification and properties of vacuolar membrane proton-translocating inorganic pyrophosphatase from mung bean. J Biol Chem. 1989 Nov 25;264(33):20068–20073. [PubMed] [Google Scholar]
- Nelson N., Taiz L. The evolution of H+-ATPases. Trends Biochem Sci. 1989 Mar;14(3):113–116. doi: 10.1016/0968-0004(89)90134-5. [DOI] [PubMed] [Google Scholar]
- Parry R. V., Turner J. C., Rea P. A. High purity preparations of higher plant vacuolar H+-ATPase reveal additional subunits. Revised subunit composition. J Biol Chem. 1989 Nov 25;264(33):20025–20032. [PubMed] [Google Scholar]
- Peterson G. L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977 Dec;83(2):346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Rea P. A., Griffith C. J., Sanders D. Purification of the N,N'-dicyclohexylcarbodiimide-binding proteolipid of a higher plant tonoplast H+-ATPase. J Biol Chem. 1987 Oct 25;262(30):14745–14752. [PubMed] [Google Scholar]
- Rea P. A., Poole R. J. Chromatographic resolution of h-translocating pyrophosphatase from h-translocating ATPase of higher plant tonoplast. Plant Physiol. 1986 May;81(1):126–129. doi: 10.1104/pp.81.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea P. A., Poole R. J. Proton-Translocating Inorganic Pyrophosphatase in Red Beet (Beta vulgaris L.) Tonoplast Vesicles. Plant Physiol. 1985 Jan;77(1):46–52. doi: 10.1104/pp.77.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafian V., Kim Y., Poole R. J., Rea P. A. Molecular cloning and sequence of cDNA encoding the pyrophosphate-energized vacuolar membrane proton pump of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1775–1779. doi: 10.1073/pnas.89.5.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafian V., Poole R. J. Purification of an h-translocating inorganic pyrophosphatase from vacuole membranes of red beet. Plant Physiol. 1989 Sep;91(1):34–38. doi: 10.1104/pp.91.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafian V., Potier M., Poole R. J. Radiation-inactivation analysis of vacuolar H(+)-ATPase and H(+)-pyrophosphatase from Beta vulgaris L. Functional sizes for substrate hydrolysis and for H+ transport. Biochem J. 1992 Apr 15;283(Pt 2):493–497. doi: 10.1042/bj2830493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K., Tazawa M. Determination of the inorganic pyrophosphate level and its subcellular localization in Chara corallina. J Biol Chem. 1989 Feb 25;264(6):3262–3266. [PubMed] [Google Scholar]
- White P. J., Marshall J., Smith J. A. Substrate kinetics of the tonoplast h-translocating inorganic pyrophosphatase and its activation by free mg. Plant Physiol. 1990 Jul;93(3):1063–1070. doi: 10.1104/pp.93.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]