Abstract
Red blood cell (RBC)-derived systems offer a potential platform for delivery of biomedical cargos. Although the importance of specific proteins associated with the biodistribution and pharmacokinetics of these particles has been recognized, it remains to be explored whether some of the key transmembrane and cytoskeletal proteins responsible for immune-modulatory effects and mechanical integrity of the particles are retained. Herein, using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and quantitative tandem mass tag mass spectrometry in conjunction with bioinformatics analysis, we have examined the proteomes of micro- and nanosized erythrocyte ghosts doped with indocyanine green and compared them with those of RBCs. We identified a total of 884 proteins in each set of RBCs, micro-, and nanosized particles, of which 8 and 45 proteins were expressed at significantly different relative abundances when comparing micro-sized particles vs RBCs and nanosized particles vs RBCs, respectively. We found greater differences in relative abundances of some mechano-modulatory proteins, such as band 3 and protein 4.2, and immunomodulatory proteins like CD44, CD47, and CD55 in nanosized particles as compared to RBCs. Our findings highlight that the methods utilized in fabricating RBC-based systems can induce substantial effects on their proteomes. Mass spectrometry data are available at ProteomeXchange with the identifier PXD038780.
Keywords: bioinformatics, drug delivery, erythrocyte, tandem mass spectrometry
Graphical Abstract
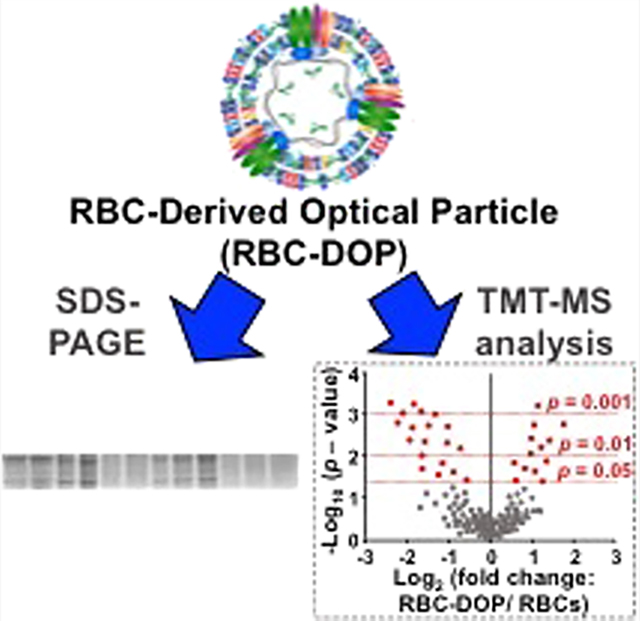
INTRODUCTION
Cell-based platforms have gained increasing attention for the delivery of various therapeutic, diagnostic, and imaging cargos. In particular, red blood cells (RBCs) are promising candidates due to their expected biocompatibility and nontoxicity, especially if derived from autologous blood.1–5 An important issue related to the effectiveness of RBC-derived delivery vehicles in clinical application is their circulation time. To accumulate at sufficient therapeutic or diagnostic quantities within the target site, a sufficiently long circulation time is necessary before the particles are removed by the reticuloendothelial system.
One of the key determinants in the longevity of RBC-derived carriers within circulation is the presence of specific proteins on the surface of their membranes that interact with the immune cells. For example, transmembrane glycoprotein CD47 prevents RBCs from being phagocytosed by macrophages,6 and the decay-accelerating factor (CD55) and CD59 prevent the complement system from activating and forming membrane-attack complexes (MACs) against RBCs.7 The presence of these cell surface proteins (among others) is needed for prolonging the circulation time of RBC-derived vehicles.
Another important factor in determining the circulation time of RBC-derived carriers is their mechanical deformability. Healthy RBCs repeatedly squeeze through narrow capillaries that have diameters similar to those of RBCs (~8 μm) and through the endothelial slits of spleen (~200 to 400 nm),8 the filtration organ involved in sequestering and disassembly of senescent RBCs. In addition to the mechanical properties of the membrane bilayer itself, the deformability of RBCs is attributed to the proteins that link the membrane to the cytoskeleton as well as a network of cytoskeletal proteins.9–11
Toward development of RBC-derived carriers for clinical use, it is important to gain a better understanding of their proteomes, particularly those proteins involved in modulating the immune response and the mechanical characteristics of the carriers as they are related to the circulation time of the particles. While the proteomes of RBCs have been extensively investigated,12,13 this is the first evaluation of the proteome of RBC-derived carriers. In particular, we have used sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and quantitative tandem mass tag (TMT)-based mass spectrometry (MS) in combination with bioinformatics analysis to characterize the proteomes of micro- and nanosized RBC-derived particles. These particles are doped with the FDA-approved near-infrared chromophore, indocyanine green (ICG), which has a long history in clinical applications ranging from ophthalmic angiography14,15 to tumor imaging16–18 and laser therapy.19,20 The nanosized RBC-carriers have relevance to tumor optical imaging and laser-based therapeutics,21–27 while the micro-sized RBC-derived carriers may be useful for laser treatment of cutaneous hypervascular lesions such as port-wine birthmarks.28,29
Herein, we demonstrate that the protein composition of micro-sized erythrocyte ghosts (EGs) doped with ICG (μEGs+ICG) was generally similar to that of RBCs, but there were still 8 proteins with significantly different relative abundance in μEGs+ICG as compared to RBCs. The proteomic profiles of nanosized EGs (nEGs) doped with ICG (nEGs+ICG) showed lower relative abundance levels of some structural proteins (α-/β-Adducin, dematin) and additional differences in their protein compositions, including immunomodulatory surface proteins like CD47, CD44, and complement C4B as compared to those of RBCs and μEGs+ICG. Our findings highlight the importance of methods in engineering RBC-derived carrier systems with appropriate proteomes for clinical translation.
MATERIALS AND METHODS
Fabrication Procedures
RBCs were isolated from human whole blood (Innovative Research Inc., Novi, MI) via centrifugation (1600g for 10 min at 4 °C). The supernatant containing the plasma and buffy coat was removed, and the erythrocyte pellets were washed twice with ~320 mOsm phosphate-buffered saline (PBS) (referred to as 1× PBS, pH = 8) (Fisher Scientific, Hampton, NH). The supernatant was discarded after each wash. The washed erythrocytes were then subjected to hypotonic treatment in 0.25× PBS (~80 mOsm, pH = 8) for 30 min, followed by centrifugation (20 000g for 20 min at 4 °C). The supernatant containing hemoglobin was discarded, and the hypotonic treatment was repeated until a white pellet containing the micro-sized erythrocyte ghosts (μEGs) was obtained. The μEGs were then resuspended in 1× PBS.
To form nEGs, 1 mL of μEGs was first diluted 10-fold in 1× PBS and then extruded sequentially through 800-, 400-, and 200 nm diameter polyester porous membranes (Sterlitech Corp., Kent, WA) using a 10 mL LIPEX extruder (TRANS-FERRA Nanosciences Inc., Burnaby, BC). μEGs were passed through each membrane size at least three times. The pellet containing the nEGs was isolated by centrifugation (100 000g for 1 h at 4 °C) and then resuspended in the original 1 mL of 1× PBS.
To load ICG (MP Biomedicals, Santa Ana, CA) into μEGs and nEGs, particles were incubated in a hypotonic solution made of 1:1:1 volume ratio of particles, Sørenson’s phosphate buffer (Na2HPO4/NaH2PO4, 140 mOsm, pH = 8), and ICG solution for 30 min at 4 °C in the dark. Concentration of ICG in the loading buffer solution was 25 μM. This process results in the formation of μEGs and nEGs doped with ICG, which we refer to as μEGs+ICG and nEGs+ICG, respectively. To obtain the μEGs+ICG and nEGs+ICG pellets, 3 mL of suspension was then centrifuged and washed twice with 1× PBS (20 000g for 20 min at 4 °C and 100 000g for 1 h at 4 °C for μEGs+ICG and nEGs+ICG, respectively). The resulting pellets of μEGs+ICG and nEGs+ICG were then resuspended in their original 1 mL of 1× PBS to restore tonicity. The absorption spectra of the particles were obtained using a spectrophotometer (Jasco-V670, JASCO, Easton, MD).
Size Profiling of Particles
Dynamic light scattering (DLS) (Zetasizer Nanoseries, NanoZS90, Malvern Instruments Ltd., Westborough, MA) was used to estimate the hydrodynamic diameters of the particles suspended in 1× PBS in a polystyrene cuvette with a 1 cm pathlength. Three measurements were collected for each sample. The mean peak diameter ± standard deviation (SD), based on lognormal fits to the DLS-based measurements, were 4,149 ± 200 and 140.87 ± 0.848 nm for μEGs+ICG and nEGs+ICG, respectively.
We also used the Zetasizer to quantify the ζ-potentials of the particles in 1× PBS. The mean ± SD values of ζ-potentials for μEGs+ICG and nEGs+ICG were −13.02 ± 0.85 and −14.4 ± 0.88 mV, respectively.
SDS-PAGE
Samples consisting of RBCs (control), μEGs+ICG or nEGs+ICG were thoroughly mixed in Laemmli buffer (6×; 375 mM Tris-HCl, 6% SDS, 4.8% glycerol, 9% 2-mercaptoethanol, 0.03% bromophenol blue), followed by heating at 95 °C for 5 min to denature the proteins. To quantify the total protein amount in each sample, the Bradford assay (Thermo Fisher Scientific Inc., Rockford), in conjunction with standard curves, was used. The peak absorption of the Bradford reagent (Coomassie blue G-250) changes from 470 to 595 nm when it binds to proteins, which is then measured spectroscopically. After binding, we compared the 595 nm absorbance value to a standard curve that relates the absorbance values to various concentrations of protein, and quantified the amount of the protein in each sample.
We then loaded the samples into a precast, gradient 12% polyacrylamide gel and electrophoretically separated the protein at 100 V for 2 h. The gel was fixed and stained in staining buffer (0.25 g Coomassie blue R-250, 40 mL methanol, 50 mL dH2O, 10 mL acetic acid) overnight to visualize the protein migration. The stained gel was then washed in a destaining buffer (40% methanol, 10% acetic acid in dH2O) overnight to clear the background dye. Visualization of the gel band was obtained using the BioSpectrum Imaging System (UVP system, Anaytik Jena, Upland, CA). To estimate the molecular weight (MW) of unknown proteins, the gel image was then analyzed to determine the relative migration distance ( value) of the protein standard marker and the unknown protein
| (1) |
Finally, a plot of the logarithm of the MW of the protein standards against their values was constructed as the standard curve to determine the MW of various unknown proteins.30,31 The protein identity was predicted by comparing the estimated MWs to those of known proteins in the literature.12,32,33
TMT- MS Analysis
Sample Preparation.
Samples were prepared in triplicate. Following the manufacturer’s instructions (TMT Mass Tag Labeling Kit, Thermo Fisher Scientific), each sample type was lysed to isolate the protein extracts, which were then reduced, alkylated, and digested to obtain the peptides. The TMT reagents were added to label the peptides from each sample. All samples were then mixed together, followed by fractionation prior to MS analysis. Specifically, cell suspensions were first lysed in lysis buffer (0.25% sodium deoxycholate, 1% NP-40, 50 mM triethylammonium bicarbonate (TEAB) (Millipore Sigma), 1 mM phenylmethylsulfonyl fluoride) on ice for 1 h and then adjusted to 500 μL in 50 mM TEAB. To minimize protein degradation and reduce the activity of proteases and the contaminants in each sample, acetone precipitations prior to digestion and labeling process were performed. The proteins derived from lysed cells of each sample were precipitated in equal volumes of trichloroacetic acid and incubated on ice for 1 h. The supernatant was discarded, and the pellet was rinsed twice with 500 μL of cold acetone, which was then removed using a vacuum concentrator (SpeedVac, Thermo Fisher Scientific). These procedures remove the detergents.34 The resulting dried pellet was resuspended in 50 μL of 50 mM TEAB and subsequently reduced by incubating it with 1 μL of 500 mM Tris (2-carboxyethyl) phosphine (Thermo Fisher Scientific, Rockford, IL) at 37 °C for 1 h. Each sample was then added to 3 μL of 500 mM iodoacetamide (Millipore Sigma) and incubated in the dark at room temperature for 1 h.
Next, the digestion of the proteins was performed per the manufacturer’s instructions (Thermo Fisher Scientific). Specifically, proteins of each sample were incubated with 500 μL of 50 mM TEAB and 10 μL (2 μg) of trypsin/lysC mix (Promega, Madison, WI) at 37 °C overnight (16 h). After digestion, the concentrations of peptides from each sample were measured using a colorimetric assay (Thermo Fisher Scientific). The assay was performed following the manufacturer’s instructions using a 96-well microplate which involves protein peptide–copper chelation and detection of the reduced copper. Briefly, the amide backbone of peptides in the sample causes the copper to be reduced under alkaline conditions to form a bright red complex, which can be detected at 480 nm. The peptide concentration of each sample was determined by comparing the absorbance at 480 nm to that of a peptide standard curve.
Lastly, TMT labeling was performed according to the manufacturer’s instructions (10-plex Tandem Mass Tag labeling, Thermo Fisher Scientific). From each sample, 50 μg was mixed with TMT Label Reagent using a 1:1 mass ratio of reagent to peptide and incubated at room temperature for 1 h, followed by quenching with 8 μL of 5% hydroxylamine for 15 min. The labeled aliquots were combined into one sample and then dried in a vacuum concentrator in preparation for high-pH fractions. The Pierce high-pH fraction kit (Thermo Fisher Scientific) was used to separate the peptides by hydrophobicity to increase the number of proteins from the complex sample identified by liquid chromatography–mass spectrometry (LC-MS) analysis.
LC-MS Analysis.
Liquid chromatography was performed on an EASY-nLC1200 liquid chromatography system (Thermo Fisher Scientific) in single-pump trapping mode with a PepMap RSLC C18 EASY-spray column (2 μm, 100 Å, 75 μm × 25 cm) and a Pepmap C18 trap column (3 μm, 100 Å, 75 μm × 20 mm). Samples were separated at 300 nL/min with a 260 min linear gradient (3–85% solvent containing 80% acetonitrile with 0.1% formic acid), and data were acquired on an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) in data-dependent mode. A full scan was conducted using 60k resolution in the Orbitrap in positive mode. Precursors for mass spectrometry 2 (MS2) were filtered by monoisotopic peak determination for peptides. Intensity threshold was set to 5.0 × 103, charge states 2–7 were selected, and dynamic exclusion was set to 60 s after one analysis with a mass tolerance of 10 ppm. Collisionally induced dissociation spectra were collected in ion trap MS2 at 35% energy and isolation window 1.6 m/z. Synchronous precursor selection MS3 was utilized for TMT ratio determination at higher-energy C-trap dissociation (HCD) energy. The sequence of steps for TMT analysis are shown in Figure 1.
Figure 1.
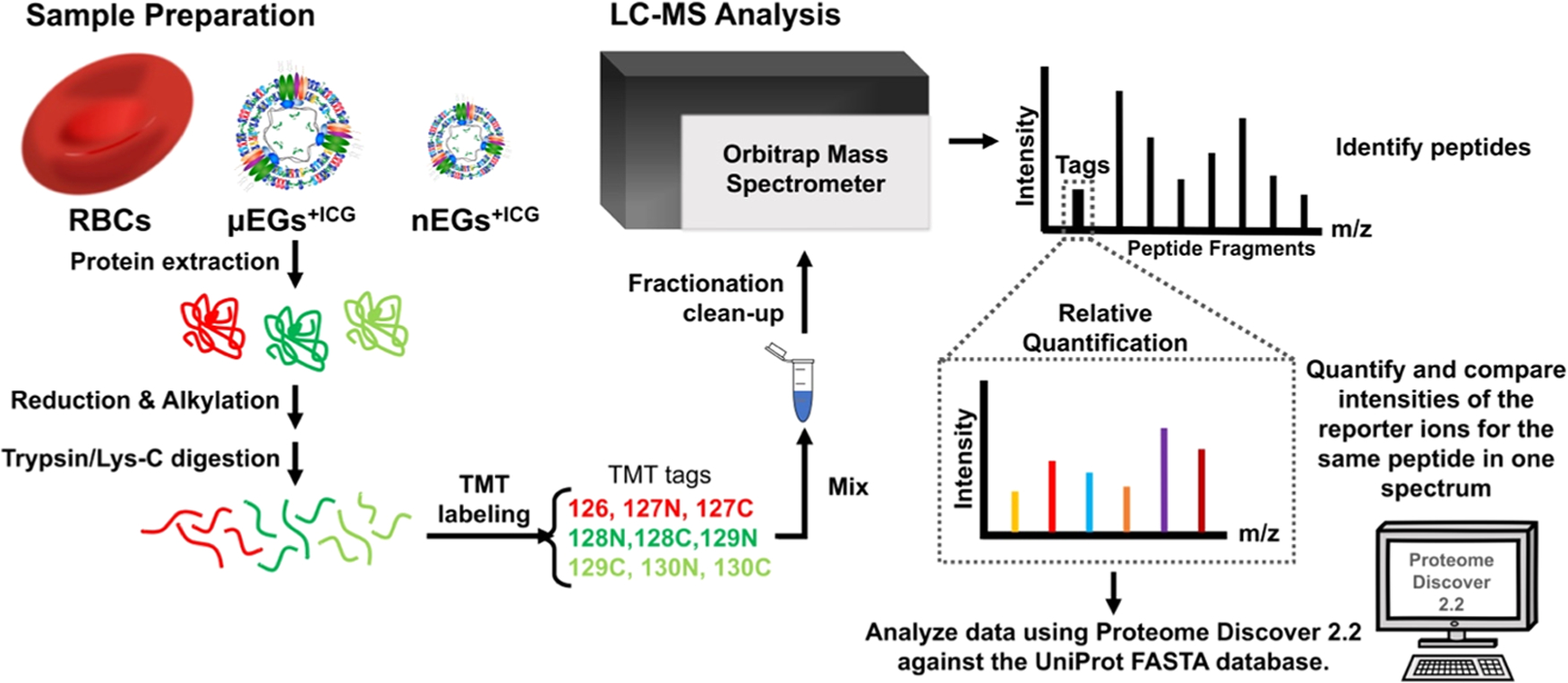
Sequence of steps for TMT experiments. Proteins from each sample type were extracted, followed by reduction, alkylation, and digestion of the proteins prior to specific TMT isobaric tag labeling. Next, all of the samples were combined and fractionated prior to LC-MS analysis. Data analysis and quantification were achieved by Proteome Discover 2.2 against the UniProt FASTA database.
Bioinformatics and Data Analysis.
All obtained data were searched individually in Proteome Discover 2.2 (Thermo Fisher Scientific) against the universal protein resource (UniProt) FASTA database for Homo sapiens. The precursor mass tolerance was set to 10 ppm, and fragment mass tolerance was set to 0.6 Da. Fixed modifications were carbamidomethyl (Cys +57.021 Da) and TMT 6plex (Lys, N-terminus +229.163 Da), and dynamic modifications included methionine oxidation (+15.995 Da) N-terminal acetylation (+42.011 Da). Results were filtered to a strict 1% false discovery rate.
Volcano plots35,36 were generated in Excel 2011 to illustrate the differentially expressed proteins between any two of the three sample types. Specifically, we calculated the log2 fold change of each identified protein from any two sample types and its corresponding −log10 p-value. We used Protein Analysis Through Evolutionary Relationships (PANTHER)37,38 to categorize the proteins that had significantly different relative abundances between any of the two samples according to molecular function, biological process, and protein class.
RESULTS AND DISCUSSION
SDS-PAGE
SDS-PAGE analysis revealed that the membrane protein band 3 (90 kDa) and other cytoskeletal proteins consisting of α-spectrin (240 kDa), β-spectrin (220 kDa), protein 4.1R (80 kDa), and protein 4.2 (72 kDa)39–41 were retained in μEGs and μEGs+ICG (Figure 2). Based on their MWs, as determined by the comparison of their values with the ladder as well as the known MW of each protein reported in the published literature,33 we expect that the remaining label bands in Figure 2 correspond to dematin (~48 kDa), tropomodulin (~43 kDa), tropomyosin (~27 kDa), and protein 8 (~23 kDa).33,42 These proteins were also retained in μEGs and μEGs+ICG (Figure 2).
Figure 2.
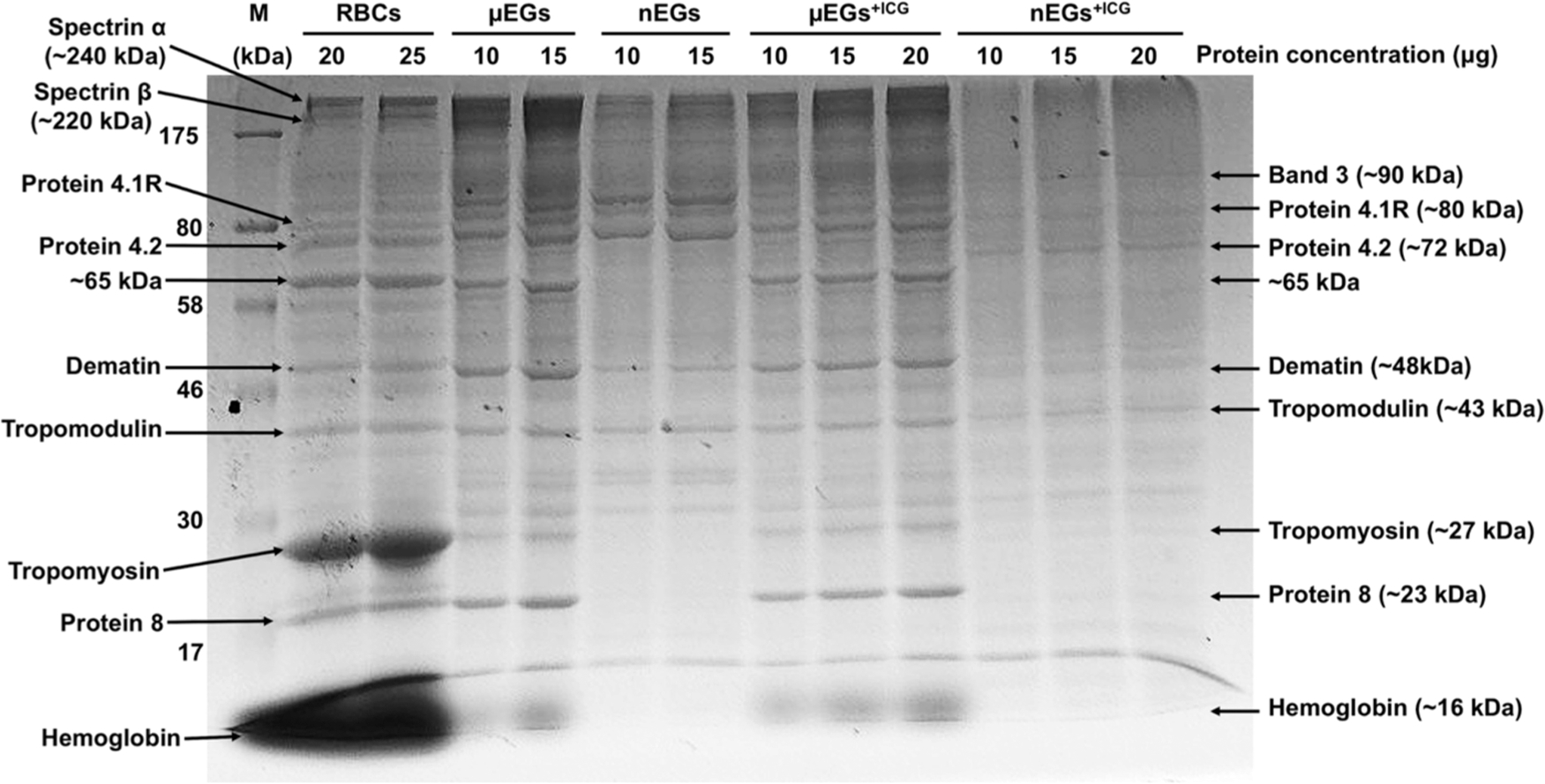
SDS-PAGE analysis of protein expressions in RBCs, μEGs, nEGs, μEGs+ICG, and nEGs+ICG. Each sample was loaded into the gel at different protein concentrations ranging between 10 and 25 μg for electrophoretic running. The nEGs+ICG were formed by sequential mechanical extrusion of μEGs through 800, 400, and 200 nm polyester porous membranes three times. M: protein ladder.
The RBC membrane is anchored to the underlying network of cytoskeletal proteins via ankyrin- and protein 4.1R (actin junctional complex)-based protein complexes.43,44 These complexes have vital roles in maintaining the structural integrity of normal RBCs by serving as a system of vertical linkages that connect the membrane bilayer to the cytoskeleton. Band 3 is one of the main transmembrane proteins that links the bilayer to ankyrin. The principal protein constituents of the RBC cytoskeleton are α- and β-spectrin, actin protofilament, dematin, tropomyosin, and tropomodulin.44,45 These proteins are part of the actin junctional complex and are involved in maintaining the membrane mechanical stability of RBCs.46–49 Other studies have indicated that dematin maintains the integrity of the membrane skeleton by reinforcing the lateral interaction between actin filament and spectrin tetramers,50 and its depletion destabilizes the association of the cytoskeleton to the plasma membrane.45
Our findings suggest that hypotonic treatment and ICG loading had little to no marked effect on the presence of these proteins in μEGs and μEGs+ICG (Figure 2). Nevertheless, it should be recognized that the linkages between the membrane and cytoskeleton may be disrupted, resulting in changes to the membrane mechanical properties and deformability of these RBC-like particles.51 Weakened vertical linkages between the membrane and cytoskeleton can lead to dissociation of the spectrin tetramers into dimers.29,52 Furthermore, hypotonic treatment is associated with clustering and rearrangement of band 3.53,54
Hypotonic treatment did not result in complete depletion of hemoglobin in μEGs and μEGs+ICG. These results are consistent with the absorption spectrum (Figure 3) showing spectral peaks at 412, 542, and 575 nm, respectively, corresponding to the Soret, fundamental (Q0), and its vibronic tone (Qv) associated with oxyhemoglobin,55 as well as quantitative optical phase images, demonstrating the presence of residual oxyhemoglobin in μEGs and μEGs+ICG.51
Figure 3.
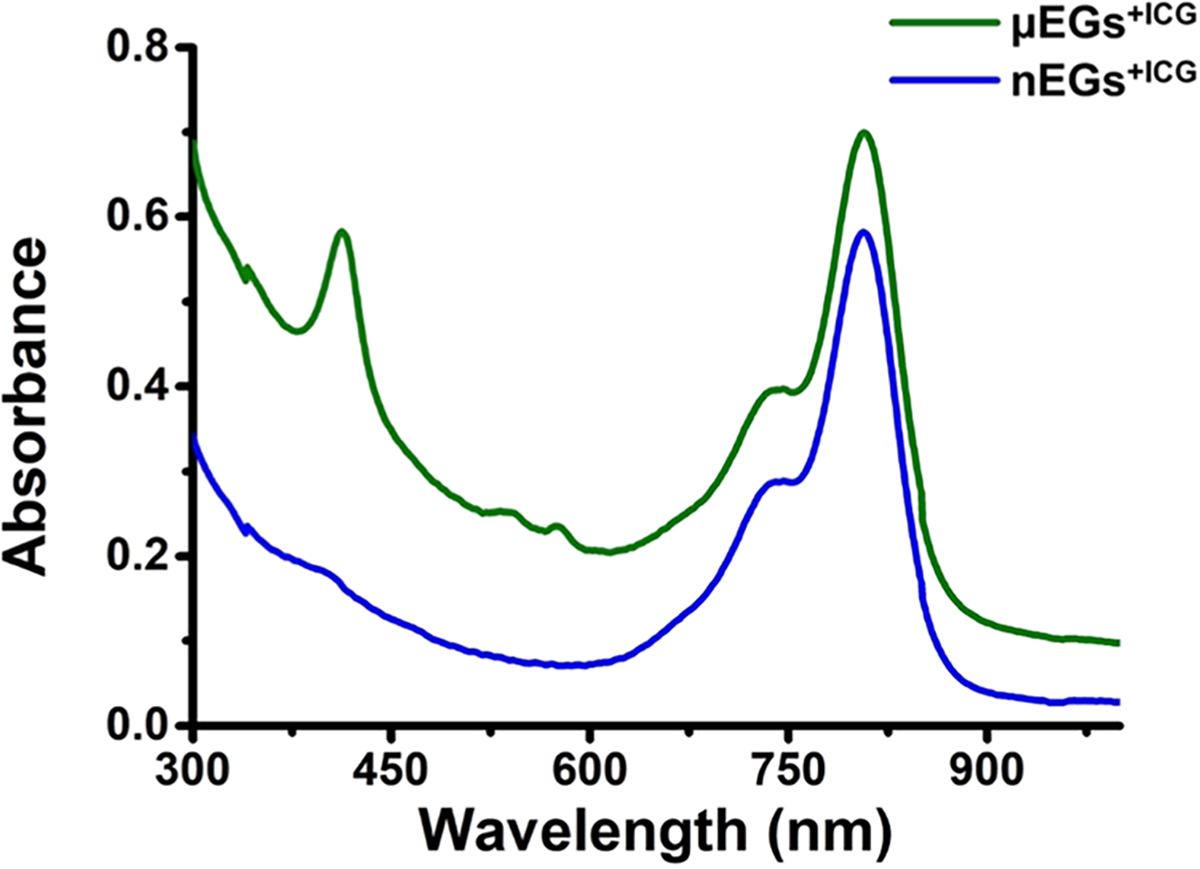
Absorption spectra of μEGs+ICG and nEGs+ICG. Constructs were suspended in 1× PBS for the recordings. Each displayed absorption spectrum is an averaged measurement of triplicate samples.
The nanosized particles (nEGs and nEGs+ICG), fabricated by mechanical extrusion of the micro-sized particles, had several faint and missing protein bands as compared to nonextruded micro-sized particles (μEGs and μEGs+ICG) and RBCs (Figure 2). During the extrusion process, the cells are passed through cylindrical pores with nanosized diameters, resulting in cell lysis and reassembly of membrane lipids to form nanosized vesicles (e.g., nEGs).56 We attribute the loss or reductions in expression levels of specific proteins such as dematin and tropomyosin to the extrusion process.
To further observe the effects of the extrusion process, we performed the SDS-PAGE analysis after extruding the particles through progressively smaller pores (800, 400, and 200 nm), with each extrusion through each pore size repeated three times. Specifically, we centrifuged the nanosized particles formed following the extrusion through each of the pore sizes and collected both the pellet and the supernatant for SDS-PAGE analysis to identify some of the proteins that were removed from the particles and released into the supernatant as a result of each extrusion. We quantified the relative expression of each band to the RBC bands using ImageJ.
Two bands associated with the pellets and corresponding to dematin and tropomodulin became progressively fainter after each extrusion, whereas the bands corresponding to tropomyosin and protein 8, which have lower molecular weights, became faint immediately after the first extrusion of the μEGs through the 800 nm pore diameter (Figure 4A). The bands associated with these four proteins were detected in the supernatants (Figure 4B), suggesting that they were removed from the RBC ghosts during the extrusion process. In addition, we observed two prominent bands in the supernatants. Based on our proteomic data, we suggest that they may correspond to entries 151–152 (~74 kDa) and entries 183–188 (~65 kDa) in Table S1. These are cytoskeletal proteins such as keratin and part of the actin junctional complex, which mediates binding of protein 4.1R to spectrin.43 Since the aforementioned proteins are involved in maintaining the mechanical stability of normal RBCs, we anticipate that their removal will compromise the mechanical characteristics of the nanosized particles formed from RBCs. Changes in mechanical properties and deformability are thought to modulate the cellular uptake and biodistribution of nanoparticles.57–59
Figure 4.
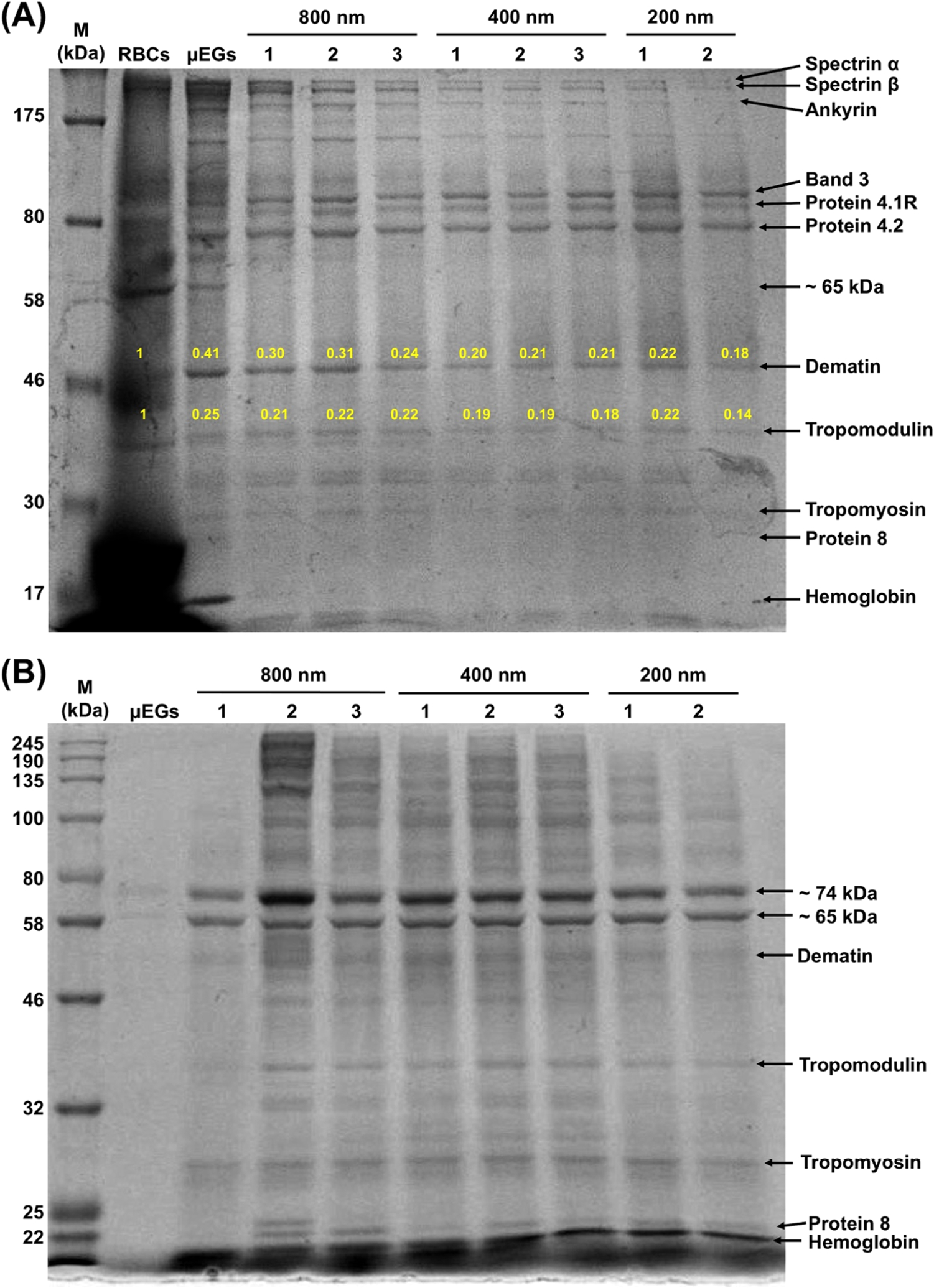
Representative SDS-PAGE analysis of protein expressions following sequential extrusion of μEGs through 800, 400, and 200 nm pore diameters. Particles and supernatants were collected after each extrusion step for analysis. (A) Pellets and (B) supernatants. Numbers 1, 2, and 3, respectively, refer to the first, second, and third extrusion through each of the indicated pore diameters. The yellow numbers in panel (A) correspond to the relative expressions of the bands to that of RBCs as determined from the intensity analysis of the bands using ImageJ. M: protein ladder. Three independent SDS-PAGE experiments were performed.
The band associated with hemoglobin disappeared following the first extrusion through the 800 nm pore diameter (Figure 4A), concomitant with the emergence of a hemoglobin band in the supernatant (Figure 4B). Consistent with these results, the Q0 and Qv peaks associated with oxyhemoglobin absorption were absent in the absorption spectrum of nEGs+ICG, and there was a minimal presence of the Soret band (Figure 3). These results indicate that while the hypotonic treatment of RBCs did not result in the complete depletion of hemoglobin from human RBCs (Figure 3), extruding the RBC ghosts through an 800 nm diameter pore was sufficient to eliminate the residual hemoglobin.
Quantitative Analysis of RBC-Derived Optical Particles by TMT-MS
We performed TMT-MS to identify the proteins and quantify their relative abundances in RBCs, μEGs+ICG, and nEGs+ICG. In total, 884 proteins were detected in each sample. Of these, 710 proteins were quantifiable in RBCs, μEGs+ICG, and nEGs+ICG in all three replicates (Table S1). To determine if the abundance of each identified protein among RBCs, μEGs+ICG, and nEGs+ICG showed statistical significance, we conducted t-test analysis on the remaining 710 proteins. Of these, 363 proteins had sufficient data (abundance values above the detectable threshold from all three TMT channels in the triplicate samples) to perform t-test analysis (see Table S2 for proteins without sufficient data). Using these criteria, we present volcano plots to show the statistical significance of differences in the relative protein expressions for μEGs+ICG vs RBCs (Figure 5A), nEGs+ICG vs RBCs (Figure 5B), and μEGs+ICG vs nEGs+ICG (Figure 5C). In these plots, the −log10 of p-value is plotted against the log2 ratio of the relative protein abundance (fold changes).60 The more spread out the volcano plot, the more differences there are in the protein abundances between two samples.
Figure 5.
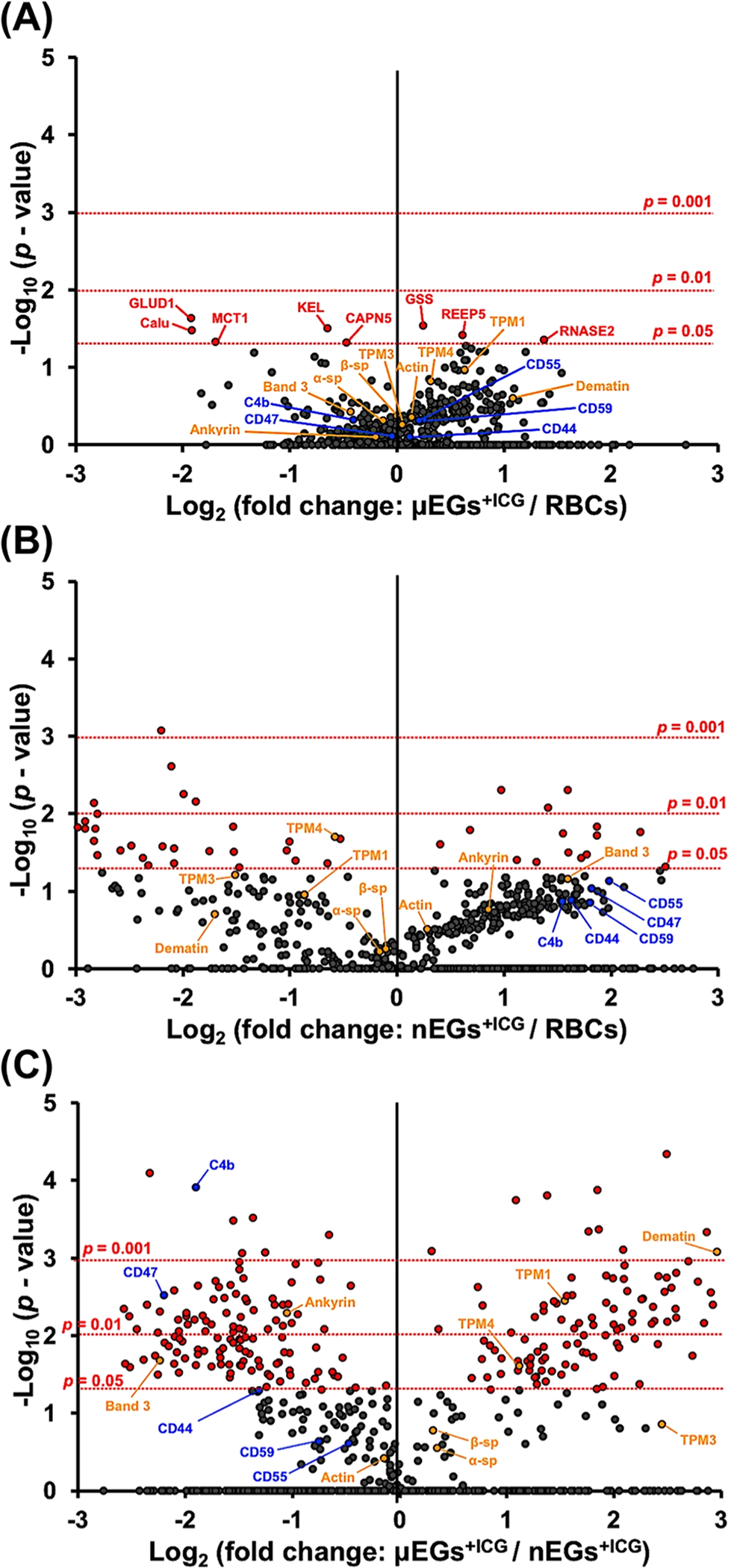
Volcano plots associated with proteomics analysis of (A) μEGs+ICG vs RBCs, (B) nEGs+ICG vs RBCs, and (C) μEGs+ICG vs nEGs+ICG. Horizontal dotted lines correspond to p = 0.05, 0.01, or 0.001. Each dot represents a particular protein. The fold change is calculated by the abundance value of the protein identified in a given sample divided by the abundance value of the same protein quantified in another sample. Three independent fabrications were used to generate triplicate experiments for each sample. C4b: complement component 4b; TPM: tropomyosin; α-sp: α-spectrin; β-sp: β-spectrin; GLUD1: glutamate dehydrogenase 1; GSS: glutathione synthetase; KEL: Kell blood group glycoprotein; Calu: calumenin; REEP5: receptor expression-enhancing protein 5; RNASE2: nonsecretory ribonuclease; MCT1: monocarboxylate transporter 1; CAPN5: calpain-5.
The protein profile of μEGs+ICG is more similar to that of RBCs, while nEGs+ICG present more differences in their proteomic composition as compared to both RBCs and μEGs+ICG. Specifically, our statistical analysis demonstrated that in comparison with RBCs, only 8 proteins (glutamate dehydrogenase 1, glutathione synthetase, kell blood group glycoprotein, calumenin, receptor expression-enhancing protein 5, nonsecretory ribonuclease, monocarboxylate transporter 1, calpain-5) had significantly different relative abundance in μEGs+ICG (Table S3). However, 45 proteins in nEGs+ICG had significantly different abundance as compared to RBCs. Comparing nEGs+ICG to μEGs+ICG, 230 proteins had statistically significant relative abundances (Table S3), accounting for ~63% of the total proteins identified in these two samples, suggesting that the extrusion processes induced substantial effects on the proteome of nEGs and nEGs+ICG.
The biodistribution and circulation time of RBC-based particles are influenced by proteins that provide mechanical stability as well as those that mediate evasion from the immune cells. We provide a partial listing of these mechano- and immunomodulatory proteins in Table 1. Through our proteomics analysis, we have estimated that the relative abundance levels of ankyrin, band 3, and protein 4.2 are, respectively, reduced by about 12.8, 25.8, and 36.5% in μEGs+ICG as compared to RBCs (reduction percentages are calculated from the abundance ratios provided in Table S1). However, other proteins that participate in the actin junctional complex showed increased expression levels in μEGs+ICG as compared to RBCs. These proteins included actin with increased expression of 10%, tropomodulin (36.8%), α-/β-adducin (96.8 and 56.6%, respectively), tropomyosin (76.8%), and dematin (112.4%). Combined with reductions in ankyrin, band 3, and protein 4.2, these changes may be sufficient to alter the membrane mechanical properties of μEGs+ICG and weaken the adhesion between the membrane and the cytoskeleton.51
Table 1.
Quantitative Comparisons of Mechano- and Immunomodulatory Proteins
| accession | protein | MW (kDa) | abundance ratio (μEGs+ICG)/(RBCs) | abundance ratio p-value (μEGs+ICG)/(RBC) | abundance ratio (nEGs+ICG)/(RBC) | abundance ratio p-value (nEGs+ICG)/(RBC) | abundance ratio (μEGs+ICG)/(nEGs+ICG) | abundance ratio p-value (μEGs+ICG)/(nEGs+ICG) |
|---|---|---|---|---|---|---|---|---|
| Mechano-Modulatory Proteins | ||||||||
| P02549 | spectrin α chain | 279.8 | 0.915 | 0.490 | 0.894 | 0.606 | 1.278 | 0.283 |
| P11277 | spectrin β chain | 246.3 | 0.999 | 0.430 | 0.931 | 0.556 | 1.243 | 0.167 |
| P16157 | ankyrin | 206.1 | 0.872 | 0.802 | 1.807 | 0.172 | 0.482 | 0.005 |
| P35611 | α-adducin | 80.9 | 1.968 | 0.306 | 0.552 | 0.742 | 2.317 | 0.029 |
| P35612 | β-adducin | 80.8 | 1.566 | 0.255 | 0.395 | 0.750 | 3.503 | 0.007 |
| P02730 | band 3 anion transport protein | 101.7 | 0.742 | 0.379 | 3.022 | 0.070 | 0.212 | 0.021 |
| A0A2R8Y6D0 | protein 4.1 | 61.4 | 0.851 | 0.658 | 2.045 | 0.110 | 0.451 | 0.006 |
| P16452 | erythrocyte membrane protein band 4.2 | 77 | 0.635 | 0.454 | 2.635 | 0.066 | 0.212 | 0.005 |
| Q08495 | dematin | 45.5 | 2.124 | 0.252 | 0.308 | 0.198 | 7.803 | 0.001 |
| Q00013 | 55 kDa erythrocyte membrane protein (p55) | 52.3 | 0.877 | 0.563 | 1.449 | 0.230 | 0.600 | 0.002 |
| P60709 | actin | 41.7 | 1.1 | 0.444 | 1.217 | 0.312 | 0.904 | 0.386 |
| P28289 | tropomodulin-1 | 40.5 | 1.368 | 0.277 | 0.565 | 0.616 | 3.102 | 0.008 |
| H7BYY1 | tropomyosin 1 (TPM 1) | 28.7 | 1.55 | 0.108 | 0.551 | 0.112 | 2.910 | 0.004 |
| J3KN67 | tropomyosin α-3 chain (TPM 3) | 33.2 | 1.043 | 0.616 | 0.352 | 0.062 | 5.459 | 0.140 |
| A0A2R8Y5V9 | tropomyosin α-4 chain (TPM 4) | 28.6 | 1.244 | 0.151 | 0.669 | 0.020 | 2.166 | 0.025 |
| Immunomodulatory Membrane Proteins | ||||||||
| Q08722 | leukocyte surface antigen CD47 | 35.2 | 0.97 | 0.778 | 3.523 | 0.093 | 0.218 | 0.003 |
| H7BY55 | complement decay-accelerating factor (CD55) | 58.9 | 1.158 | 0.492 | 3.963 | 0.074 | 0.720 | 0.244 |
| P13987 | CD59 glycoprotein | 14.2 | 1.194 | 0.483 | 3.495 | 0.141 | 0.593 | 0.233 |
| P16070 | CD44 antigen | 81.5 | 1.086 | 0.792 | 3.091 | 0.132 | 0.403 | 0.051 |
| A0A140TA29 | complement C4-B (C4B) | 187.5 | 0.755 | 0.482 | 2.926 | 0.136 | 0.268 | 0.000 |
Consistent with our SDS-PAGE analysis (Figure 4), the proteomic profiles obtained from TMT-MS showed relatively lower abundance levels of dematin (69.2%), tropomodulin (43.5%), and tropomyosin (33.1–64.8%) in nEGs+ICG as compared to normal RBCs. In addition, the actin junctional complex-related proteins α- and β-adducin in nEGs+ICG also showed lower relative abundance levels of about 44.8 and 60.5%, respectively, as compared to RBCs.
Other studies have indicated that the rigidity of the nanoparticles plays a part in their effectiveness as drug delivery vehicles where shorter circulation times are associated with nanoparticles that have higher Young’s moduli.48,49,61 These findings may be attributed to faster uptake rates of more rigid nanoparticles by the immune cells.58,62 Therefore, changes in the expression levels of the aforementioned structural proteins in nEGs+ICG may influence the mechanical characteristics of these nanoparticles and, ultimately their circulation pharmacokinetics.
Another contributing factor to the circulation time of RBC-based carriers relates to specific membrane proteins involved in regulating the interaction with the immune system. Specifically, the decay-accelerating factor (CD55) and MAC-inhibitory protein (CD59) provide protection against complement activation by binding to C3b and C4b proteins to prevent the formation of MACs.63 CD47 serves as a “self-marker” on RBCs that impedes phagocytosis through interaction with the inhibitory phagocytic receptor, SIRPα expressed on macrophages.6,64,65 Our proteomic analysis indicates that the relative abundance of the aforementioned immune markers on both μEGs+ICG and nEGs+ICG were not significantly different from RBCs (Figure 5A,B and Table 1). These results are consistent with our previous study, where using fluorescence immunostaining, we showed that CD47 is retained on μEGs+ICG and nEGs+ICG.66 Nevertheless, we point out that the extrusion process may induce conformational changes to CD47, possibly switching its role to activate phagocytosis.67 Such a switch would be consistent with our in vivo fluorescence imaging of C3H mice microvasculature, where the half-life in the bloodstream was reduced from ~49 min for μEGs+ICG to ~15 min for nEGs+ICG.29 Since CD47 resides within the ankyrin-based complex, it is possible that conformational changes to CD47 in μEGs+ICG may occur as a result of disruptions in the vertical linkages in the complex.68
Additionally, CD44, a multifunctional transmembrane glycoprotein expressed on RBCs and leukocytes, is involved in the regulation of hyaluronic acid, mediating the adhesion of leukocytes, and triggering phagocytosis.69–71 Increased relative abundance of CD44 on nEGs+ICG (Figure 5B and Table 1) may also play a role in activating their phagocytic removal and reducing the circulation time of these nanoparticles.
We evaluated the reproducibility of each sample type by calculating the coefficient of variation (CV) of protein abundances in RBCs, μEGs+ICG, and nEGs+ICG. The mean CV values were ~52.53, 25.81, and 17.39% for RBCs, μEGs+ICG, and nEGs+ICG, respectively. The greater CV value for RBCs may be attributable to the mixture of the young and senescent RBCs in the sample. For example, aging of RBCs is associated with changes in band 3 and the protein composition of the membrane.72–74 The lower values of CV for μEGs+ICG and nEGs+ICG as compared to RBCs is suggestive of the lower dispersion in protein abundances for these samples and their fabrication reproducibility. Since increased binding of modified hemoglobin to band 3 is thought to be associated with the aging of RBCs,72 the reduced hemoglobin content in μEGs+ICG and nEGs+ICG may be a contributing factor to the lower CV values in these constructs.
Categorization of Identified Proteins with Significantly Different Abundances
To further understand the characteristics of the identified proteins with significantly different abundances, we categorized those proteins according to gene ontology (molecular functions, biological processes, and protein class) based on the PANTHER classification system.37,75 (Figure 6). A detailed list of all proteins with significantly different relative abundances is provided in Table S3.
Figure 6.
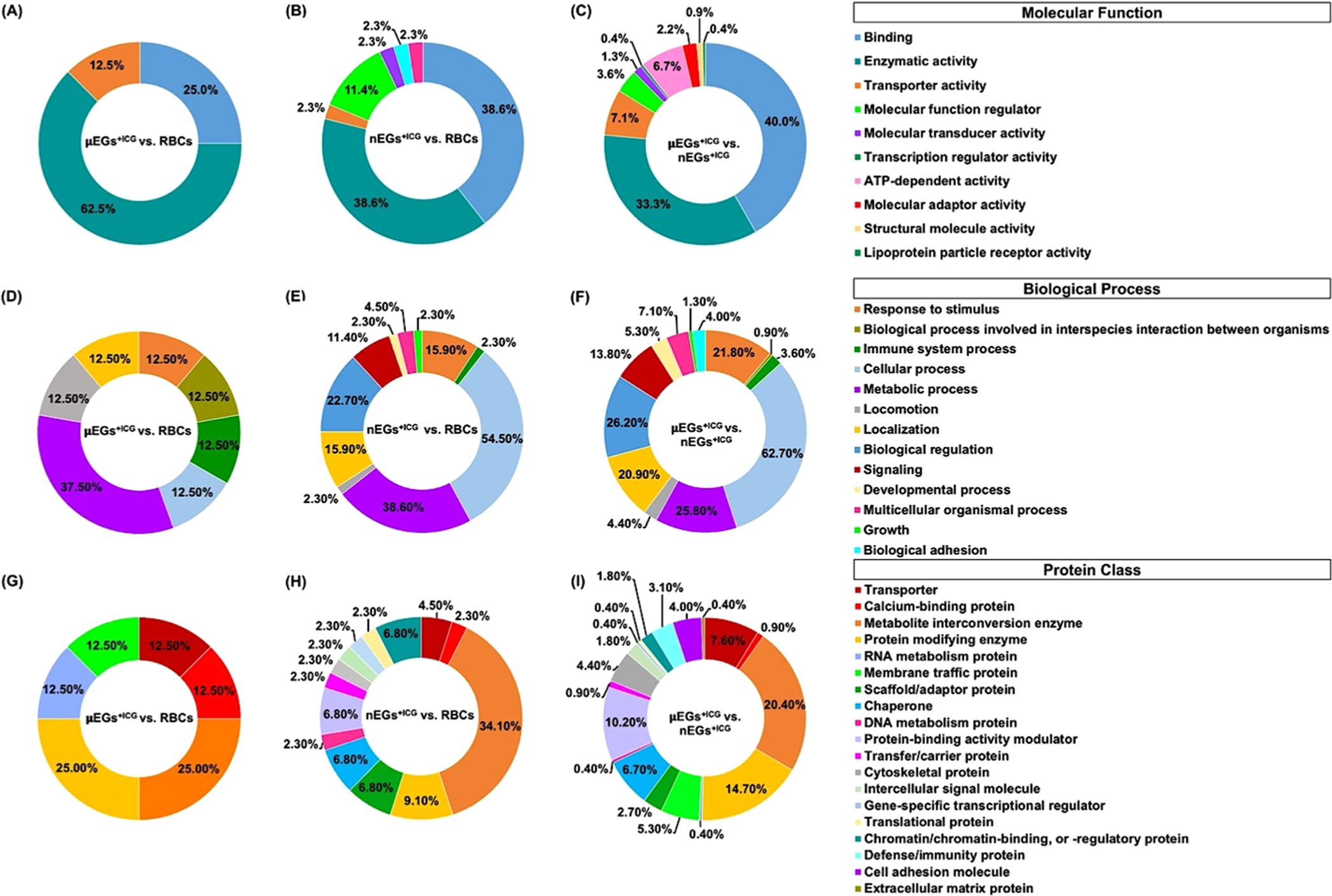
Categorization of identified proteins with significantly different relative abundances among RBCs, μEGs+ICG, and nEGs+ICG according to gene ontology by PANTHER classification system. Categorization by (A–C) molecular function, (D–F) biological process, and (G–I) protein class.
When categorized by molecular function, comparing μEGs+ICG with RBCs, the largest fraction of proteins with significantly different relative abundances were those related to the enzymatic activity (62.5% of the proteins, corresponding to 5 proteins) (Figure 6A). In this category, when comparing nEGs+ICG vs RBCs, and μEGs+ICG vs nEGs+ICG, the largest fractions were those associated with binding with approximate values of 38.6% (17 proteins) and 40% (90 proteins), respectively (Figure 6B,C).
When comparing μEGs+ICG vs RBCs by biological function, the greatest fraction of proteins (37.5% of the proteins, corresponding to 3 proteins) were those involved in metabolic processes, including catabolism and macromolecular processes, such as protein synthesis and degradation (Figure 6D). When comparing nEGs+ICG vs RBCs, and nEGs+ICG vs μEGs+ICG, the majority of proteins with significantly different abundances were related with cellular processes, defined as any process that occurs at the cellular level such as cellular development, protein folding, and transmembrane transport. Specifically, 54.5% of the proteins (corresponding to 24 proteins) when comparing nEGs+ICG vs μEGs+ICG (Figure 6E), and 62.7% of the proteins (corresponding to 141 proteins) when comparing nEGs+ICG vs RBCs (Figure 6F) were involved in such processes.
When comparing μEGs+ICG vs RBCs by protein class, metabolite interconversion enzymes such as transferases, ligases, and lyases, and protein modifying enzymes such as proteases and protein phosphatases each accounted for 25% of the proteins (corresponding to a total of 4 proteins) (Figure 6G). Metabolite conversion enzymes also comprised the largest fraction when comparing nEGs+ICG vs RBCs (9.1%, corresponding to 4 proteins) (Figure 6H), and μEGs+ICG vs RBCs (14.7%, corresponding to 33 proteins) (Figure 6I). When comparing μEGs+ICG vs RBCs, six classes of proteins accounted for the proteomic differences (Figure 6G); however, comparing nEGs+ICG vs RBCs (Figure 6H) and nEGs+ICG vs μEGs+ICG (Figure 6I), the number of protein classes increased to 14 and 19, respectively.
Finally, we point out that while the present study indicates that the physical and biochemical treatments of RBCs are the primary determinants of the proteomes of the particles, it has been reported that the protein composition of the membrane of RBCs and RBC ghosts changes with storage time.72,73 For example, changes to the band 3 complex, including its association with flotillin-2, have been reported.74 Therefore, the effects of storage time on the proteomes of RBC-based carrier systems should be considered in future studies.
CONCLUSIONS
We have studied the proteomic profiles of μEGs+ICG, nEGs+ICG, and RBCs by SDS-PAGE analysis and quantitative TMT-MS. While the protein composition was generally retained in μEGs+ICG, nEGs+ICG, fabricated by mechanical extrusion of μEGs, showed greater differences in their protein composition as compared to RBCs and μEGs+ICG. Specifically, only 8 proteins had significantly different relative abundance in μEGs+ICG, while there were 45 proteins with significant differences in abundance in nEGs+ICG as compared to RBCs. Moreover, greater differences in the relative abundances of some of the mechano-modulatory proteins, such as band 3 and dematin, and immunomodulatory proteins such as CD47, CD55, and CD44 were found in nEGs+ICG as compared to RBCs. The differences in the proteomes of nEGs+ICG vs RBCs may provide a mechanism for altered biodistribution dynamics and circulation times of nEGs+ICG as compared to μEGs+ICG and RBCs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (2R01-AR068067). N.G.H. was supported by a grant from the National Institutes of Health (NIH-S10-OD010669). The SDS-PAGE analysis experiments were graciously assisted by Vipul Madahar and George Way from Dr. Jiayu Liao’ laboratory in the Department of Bioengineering at UCR. The TMT mass spectrometry and bioinformatics were performed at the Proteomics core at University of California, Riverside. The authors thank Dr. Quanqing Zhang for assistance in depositing the mass spectrometry data in ProteomeXchange.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.2c00695.
Complete set of the identified proteins data from TMT mass spectrometry analysis (Table S1), list of all proteins with unknown identification or under detectable threshold (Table S2), and proteins with significantly different relative abundance between any of the two particle types (Table S3) (XLSX)
Contributor Information
Chi-Hua Lee, Department of Biochemistry, University of California, Riverside, Riverside, California 92521, United States.
Jack C. Tang, Department of Bioengineering, University of California, Riverside, Riverside, California 92521, United States
Nathan G. Hendricks, Institute for Integrative Genome Biology, Proteomics Core, University of California, Riverside, Riverside, California 92521, United States
Bahman Anvari, Department of Biochemistry, University of California, Riverside, Riverside, California 92521, United States; Department of Bioengineering, University of California, Riverside, Riverside, California 92521, United States.
REFERENCES
- (1).Pierigè F; Serafini S; Rossi L; Magnani M Cell-based drug delivery. Adv. Drug Delivery Rev. 2008, 60, 286–295. [DOI] [PubMed] [Google Scholar]
- (2).Millán CG; Gandarillas CIC; Marinero MLS; Lanao JM Cell-based drug-delivery platforms. Ther. Delivery 2012, 3, 25–41. [DOI] [PubMed] [Google Scholar]
- (3).Timin AS; Litvak MM; Gorin DA; Atochina-Vasserman EN; Atochin DN; Sukhorukov GB Cell-based drug delivery and use of nano-and microcarriers for cell functionalization. Adv. Healthcare Mater. 2018, 7, No. 1700818. [DOI] [PubMed] [Google Scholar]
- (4).Yang L; Zang GC; Li JW; Li XY; Li YZ; Zhao YP Cell-derived biomimetic nanoparticles as a novel drug delivery system for atherosclerosis: predecessors and perspectives. Regen. Biomater. 2020, 7, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Bush LM; Healy CP; Javdan SB; Emmons JC; Deans TL Biological cells as therapeutic delivery vehicles. Trends Pharmacol. Sci. 2021, 42, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Oldenborg PA; Zheleznyak A; Fang YF; Lagenaur CF; Gresham HD; Lindberg FP Role of CD47 as a marker of self on red blood cells. Science 2000, 288, 2051–2054. [DOI] [PubMed] [Google Scholar]
- (7).Makrides SC Therapeutic inhibition of the complement system. Pharmacol. Rev. 1998, 50, 59–87. [PubMed] [Google Scholar]
- (8).Chien S Red cell deformability and its relevance to blood flow. Annu. Rev. Physiol. 1987, 49, 177–192. [DOI] [PubMed] [Google Scholar]
- (9).Huisjes R; Bogdanova A; van Solinge WW; Schiffelers RM; Kaestner L; van Wijk R Squeezing for life - properties of red blood cell deformability. Front. Physiol. 2018, 9, 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Pivkin IV; Peng Z; Karniadakis GE; Buffet PA; Dao M; Suresh S Biomechanics of red blood cells in human spleen and consequences for physiology and disease. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 7804–7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Safeukui I; Buffet PA; Deplaine G; Perrot S; Brousse V; Sauvanet A; Aussilhou B; Dokmak S; Couvelard A; Cazals-Hatem D; et al. Sensing of red blood cells with decreased membrane deformability by the human spleen. Blood Adv. 2018, 2, 2581–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kakhniashvili DG; Bulla LA Jr.; Goodman SR The human erythrocyte proteome: analysis by ion trap mass spectrometry. Mol. Cell. Proteomics 2004, 3, 501–509. [DOI] [PubMed] [Google Scholar]
- (13).Bryk AH; Wisniewski JR Quantitative analysis of human red blood cell proteome. J. Proteome Res. 2017, 16, 2752–2761. [DOI] [PubMed] [Google Scholar]
- (14).Yannuzzi LA Indocyanine green angiography: a perspective on use in the clinical setting. Am. J. Ophthalmol. 2011, 151, 745–751. [DOI] [PubMed] [Google Scholar]
- (15).Klufas MA; Yannuzzi NA; Pang CE; Srinivas S; Sadda SR; Freund KB; Kiss S Feasibility and clinical utility of ultra-widefield indocyanine green angiography. Retina 2015, 35, 508–520. [DOI] [PubMed] [Google Scholar]
- (16).Kitai T; Inomoto T; Miwa M; Shikayama T Fluorescence navigation with indocyanine green for detecting sentinel lymph nodes in breast cancer. Breast Cancer 2005, 12, 211–215. [DOI] [PubMed] [Google Scholar]
- (17).Onda N; Kimura M; Yoshida T; Shibutani M Preferential tumor cellular uptake and retention of indocyanine green for in vivo tumor imaging. Int. J. Cancer 2016, 139, 673–682. [DOI] [PubMed] [Google Scholar]
- (18).Teng CW; Huang V; Arguelles GR; Zhou C; Cho SS; Harmsen S; Lee JYK Applications of indocyanine green in brain tumor surgery: review of clinical evidence and emerging technologies. Neurosurg. Focus 2021, 50, No. E4. [DOI] [PubMed] [Google Scholar]
- (19).An F; Yang Z; Zheng M; Mei T; Deng G; Guo P; Li Y; Sheng R Rationally assembled albumin/indocyanine green nanocomplex for enhanced tumor imaging to guide photothermal therapy. J. Nanobiotechnol. 2020, 18, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Klein A; Szeimies RM; Baumler W; Zeman F; Schreml S; Hohenleutner U; Landthaler M; Koller M; Babilas P Indocyanine green-augmented diode laser treatment of port-wine stains: clinical and histological evidence for a new treatment option from a randomized controlled trial. Br. J. Dermatol. 2012, 167, 333–342. [DOI] [PubMed] [Google Scholar]
- (21).Ren X; Zheng R; Fang X; Wang X; Zhang X; Yang W; Sha X Red blood cell membrane camouflaged magnetic nanoclusters for imaging-guided photothermal therapy. Biomaterials 2016, 92, 13–24. [DOI] [PubMed] [Google Scholar]
- (22).Liu W; Ruan M; Wang Y; Song R; Ji X; Xu J; Dai J; Xue W Light-triggered biomimetic nanoerythrocyte for tumor-targeted lung metastatic combination therapy of malignant melanoma. Small 2018, 14, No. 1801754. [DOI] [PubMed] [Google Scholar]
- (23).Burns JM; Vankayala R; Mac JT; Anvari B Erythrocyte-derived theranostic nanoplatforms for near infrared fluorescence imaging and photodestruction of tumors. ACS Appl. Mater. Interfaces 2018, 10, 27621–27630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ye S; Wang F; Fan Z; Zhu Q; Tian H; Zhang Y; Jiang B; Hou Z; Li Y; Su G Light/pH-triggered biomimetic red blood cell membranes camouflaged small molecular drug assemblies for imaging-guided combinational chemo-photothermal therapy. ACS Appl. Mater. Interfaces 2019, 11, 15262–15275. [DOI] [PubMed] [Google Scholar]
- (25).Wang P; Jiang F; Chen B; Tang H; Zeng X; Cai D; Zhu M; Long R; Yang D; Kankala RK; et al. Bioinspired red blood cell membrane-encapsulated biomimetic nanoconstructs for synergistic and efficacious chemo-photothermal therapy. Colloids Surf., B 2020, 189, No. 110842. [DOI] [PubMed] [Google Scholar]
- (26).Pan Y; He Y; Zhao X; Pan Y; Meng X; Lv Z; Hu Z; Mou X; Cai Y Engineered red blood cell membrane-coating salidroside/indocyanine green nanovesicles for high-Eefficiency hypoxic targeting phototherapy of triple-negative breast cancer. Adv. Healthcare Mater. 2022, 11, No. 2200962. [DOI] [PubMed] [Google Scholar]
- (27).Mac JT; Vankayala R; Lee CH; Anvari B Erythrocyte-derived nanoparticles with folate functionalization for near infrared pulsed laser-mediated photo-chemotherapy of tumors. Int. J. Mol. Sci. 2022, 23, 10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Burns JM; Jia W; Nelson JS; Majaron B; Anvari B Photothermal treatment of port-wine stains using erythrocyte-derived particles doped with indocyanine green: a theoretical study. J. Biomed. Opt. 2018, 23, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Jia W; Burns JM; Villantay B; Tang JC; Vankayala R; Lertsakdadet B; Choi B; Nelson JS; Anvari B Intravital vascular phototheranostics and real-time circulation dynamics of micro- and nanosized erythrocyte-derived carriers. ACS Appl. Mater. Interfaces 2020, 12, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Garfin DE One-dimensional gel electrophoresis. Methods Enzymol. 1990, 182, 425–441. [DOI] [PubMed] [Google Scholar]
- (31).Walker JM SDS Polyacrylamide Gel Electrophoresis of Proteins. In Springer Protocols Handbook; Springer, 2009; pp 177–185. [Google Scholar]
- (32).Low TY; Seow TK; Chung MC Separation of human erythrocyte membrane associated proteins with one-dimensional and two-dimensional gel electrophoresis followed by identification with matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2002, 2, 1229–1239. [DOI] [PubMed] [Google Scholar]
- (33).Yawata Y Cell Membrane: The Red Blood Cell as a Model; Wiley-VCH: Weinheim, Germany, 2006; pp 4–9. [Google Scholar]
- (34).Feist P; Hummon AB Proteomic challenges: sample preparation techniques for microgram-quantity protein analysis from biological samples. Int. J. Mol. Sci. 2015, 16, 3537–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Cui X; Churchill GA Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 2003, 4, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Aguilan JT; Kulej K; Sidoli S Guide for protein fold change and p-value calculation for non-experts in proteomics. Mol. Omics 2020, 16, 573–582. [DOI] [PubMed] [Google Scholar]
- (37).Mi H; Muruganujan A; Huang X; Ebert D; Mills C; Guo X; Thomas PD Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Bathla S; Sindhu A; Kumar S; Dubey SK; Pattnaik S; Rawat P; Chopra A; Dang A; Kaushik JK; Mohanty AK Tandem Mass Tag (TMT)-based quantitative proteomics reveals potential targets associated with onset of Sub-clinical Mastitis in cows. Sci. Rep. 2020, 10, No. 9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Fairbanks G; Steck TL; Wallach DF Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry 1971, 10, 2606–2617. [DOI] [PubMed] [Google Scholar]
- (40).Ballas SK; Krasnow SH Structure of erythrocyte membrane and its transport functions. Ann. Clin. Lab. Sci. 1980, 10, 209–219. [PubMed] [Google Scholar]
- (41).Aoki T A comprehensive review of our current understanding of red blood cell (RBC) glycoproteins. Membranes 2017, 7, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Matei H; Frentescu L; Benga G Comparative studies of the protein composition of red blood cell membranes from eight mammalian species. J. Cell. Mol. Med. 2000, 4, 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Lux SE t. Anatomy of the red cell membrane skeleton: unanswered questions. Blood 2016, 127, 187–199. [DOI] [PubMed] [Google Scholar]
- (44).Mohandas N; Gallagher PG Red cell membrane: past, present, and future. Blood 2008, 112, 3939–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Khanna R; Chang SH; Andrabi S; Azam M; Kim A; Rivera A; Brugnara C; Low PS; Liu SC; Chishti AH Headpiece domain of dematin is required for the stability of the erythrocyte membrane. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 6637–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Bennett V The spectrin-actin junction of erythrocyte membrane skeletons. Biochim. Biophys. Acta 1989, 988, 107–121. [DOI] [PubMed] [Google Scholar]
- (47).Bennett V Spectrin-based membrane skeleton - a multipotential adapter between plasma-membrane and cytoplasm. Physiol. Rev. 1990, 70, 1029–1065. [DOI] [PubMed] [Google Scholar]
- (48).Hui Y; Yi X; Hou F; Wibowo D; Zhang F; Zhao D; Gao H; Zhao CX Role of nanoparticle mechanical properties in cancer drug delivery. ACS Nano 2019, 13, 7410–7424. [DOI] [PubMed] [Google Scholar]
- (49).Baranov MV; Kumar M; Sacanna S; Thutupalli S; van den Bogaart G Modulation of immune responses by particle size and shape. Front. Immunol. 2020, 11, No. 607945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Koshino I; Mohandas N; Takakuwa Y Identification of a novel role for dematin in regulating red cell membrane function by modulating spectrin-actin interaction. J. Biol. Chem. 2012, 287, 35244–35250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Lu T; Lee CH; Anvari B Morphological characteristics, hemoglobin content, and membrane mechanical properties of red blood cell delivery systems. ACS Appl. Mater. Interfaces 2022, 14, 18219–18232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).An X; Lecomte MC; Chasis JA; Mohandas N; Gratzer W Shear-response of the spectrin dimer-tetramer equilibrium in the red blood cell membrane. J. Biol. Chem. 2002, 277, 31796–31800. [DOI] [PubMed] [Google Scholar]
- (53).Knowles DW; Tilley L; Mohandas N; Chasis JA Erythrocyte membrane vesiculation: model for the molecular mechanism of protein sorting. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 12969–12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Pajic-Lijakovic I Erythrocytes under osmotic stress - modeling considerations. Prog. Biophys. Mol. Biol. 2015, 117, 113–124. [DOI] [PubMed] [Google Scholar]
- (55).Tang JC; Vankayala R; Mac JT; Anvari B RBC-derived optical nanoparticles remain stable after a freeze-thaw cycle. Langmuir 2020, 36, 10003–10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Hunter DG; Frisken BJ Effect of extrusion pressure and lipid properties on the size and polydispersity of lipid vesicles. Biophys. J. 1998, 74, 2996–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Sun X; Rossin R; Turner JL; Becker ML; Joralemon MJ; Welch MJ; Wooley KL An assessment of the effects of shell cross-linked nanoparticle size, core composition, and surface PEGylation on in vivo biodistribution. Biomacromolecules 2005, 6, 2541–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Anselmo AC; Mitragotri S Impact of particle elasticity on particle-based drug delivery systems. Adv. Drug Delivery Rev. 2017, 108, 51–67. [DOI] [PubMed] [Google Scholar]
- (59).Guo P; Liu D; Subramanyam K; Wang B; Yang J; Huang J; Auguste DT; Moses MA Nanoparticle elasticity directs tumor uptake. Nat. Commun. 2018, 9, No. 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Mirzaei M; Pascovici D; Wu JX; Chick J; Wu Y; Cooke B; Haynes P; Molloy MP TMT one-stop shop: from reliable sample preparation to computational analysis platform. Methods Mol. Biol. 2017, 1549, 45–66. [DOI] [PubMed] [Google Scholar]
- (61).Anselmo AC; Zhang M; Kumar S; Vogus DR; Menegatti S; Helgeson ME; Mitragotri S Elasticity of nanoparticles influences their blood circulation, phagocytosis, endocytosis, and targeting. ACS Nano 2015, 9, 3169–3177. [DOI] [PubMed] [Google Scholar]
- (62).Key J; Palange AL; Gentile F; Aryal S; Stigliano C; Di Mascolo D; De Rosa E; Cho M; Lee Y; Singh J; Decuzzi P Soft discoidal polymeric nanoconstructs resist macrophage uptake and enhance vascular targeting in tumors. ACS Nano 2015, 9, 11628–11641. [DOI] [PubMed] [Google Scholar]
- (63).Ruiz-Argüelles A; Llorente L The role of complement regulatory proteins (CD55 and CD59) in the pathogenesis of autoimmune hemocytopenias. Autoimmun. Rev 2007, 6, 155–161. [DOI] [PubMed] [Google Scholar]
- (64).Lutz HU; Bogdanova A Mechanisms tagging senescent red blood cells for clearance in healthy humans. Front. Physiol. 2013, 4, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Oronsky B; Carter C; Reid T; Brinkhaus F; Knox SJ Just eat it: A review of CD47 and SIRP-alpha antagonism. Semin. Oncol 2020, 47, 117–124. [DOI] [PubMed] [Google Scholar]
- (66).Mac JT; Nunez V; Burns JM; Guerrero YA; Vullev VI; Anvari B Erythrocyte-derived nano-probes functionalized with antibodies for targeted near infrared fluorescence imaging of cancer cells. Biomed. Opt. Express 2016, 7, 1311–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Burger P; Hilarius-Stokman P; de Korte D; van den Berg TK; van Bruggen R CD47 functions as a molecular switch for erythrocyte phagocytosis. Blood 2012, 119, 5512–5521. [DOI] [PubMed] [Google Scholar]
- (68).Mordue KE; Hawley BR; Satchwell TJ; Toye AM CD47 surface stability is sensitive to actin disruption prior to inclusion within the band 3 macrocomplex. Sci. Rep. 2017, 7, No. 2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Aruffo A; Stamenkovic I; Melnick M; Underhill CB; Seed B Cd44 is the principal cell-surface receptor for hyaluronate. Cell 1990, 61, 1303–1313. [DOI] [PubMed] [Google Scholar]
- (70).Vachon E; Martin R; Kwok V; Cherepanov V; Chow CW; Doerschuk CM; Plumb J; Grinstein S; Downey GP CD44-mediated phagocytosis induces inside-out activation of complement receptor-3 in murine macrophages. Blood 2007, 110, 4492–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Vachon E; Martin R; Plumb J; Kwok V; Vandivier RW; Glogauer M; Kapus A; Wang XM; Chow CW; Grinstein S; et al. CD44 is a phagocytic receptor. Blood 2006, 107, 4149–4158. [DOI] [PubMed] [Google Scholar]
- (72).Bosman GJ; Lasonder E; Groenen-Döpp YA; Willekens FL; Werre JM; Novotný VM Comparative proteomics of erythrocyte aging in vivo and in vitro. J. Proteomics 2010, 73, 396–402. [DOI] [PubMed] [Google Scholar]
- (73).Bosman GJ; Lasonder E; Luten M; Roerdinkholder-Stoelwinder B; Novotny VM; Bos H; De Grip WJ The proteome of red cell membranes and vesicles during storage in blood bank conditions. Transfusion 2008, 48, 827–835. [DOI] [PubMed] [Google Scholar]
- (74).Prudent M; Delobel J; Hübner A; Benay C; Lion N; Tissot JD Proteomics of Stored Red Blood Cell Membrane and Storage-Induced Microvesicles Reveals the Association of Flotillin-2 With Band 3 Complexes. Front. Physiol 2018, 9, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Mi H; Ebert D; Muruganujan A; Mills C; Albou LP; Mushayamaha T; Thomas PD PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


