Significance
Currently, the majority of nucleic acid–derived therapeutics are restricted to intracellular expression, thereby limiting their therapeutic potential solely to transfected cells. However, there are numerous inflammatory and autoimmune diseases that require systemic circulation of proteins, enzymes, and antibodies for therapeutic benefit. In this study, we identified a broadly applicable mechanism for inducing endogenous production and subsequent secretion of therapeutics into the bloodstream by engineering a host of different signal peptide sequences into mRNA constructs. Through this engineering approach, the body can be utilized as a bioreactor to produce and systemically secrete virtually any encodable protein that would otherwise be confined to the intracellular space of the transfected cell, thus opening up new therapeutic opportunities.
Keywords: lipid nanoparticles, signal peptides, mRNA, nucleic acids
Abstract
Genetic medicines have the potential to treat various diseases; however, certain ailments including inflammatory diseases and cancer would benefit from control over extracellular localization of therapeutic proteins. A critical gap therefore remains the need to develop and incorporate methodologies that allow for posttranslational control over expression dynamics, localization, and stability of nucleic acid–generated protein therapeutics. To address this, we explored how the body’s endogenous machinery controls protein localization through signal peptides (SPs), including how these motifs could be incorporated modularly into therapeutics. SPs serve as a virtual zip code for mRNA transcripts that direct the cell where to send completed proteins within the cell and the body. Utilizing this signaling biology, we incorporated secretory SP sequences upstream of mRNA transcripts coding for reporter, natural, and therapeutic proteins to induce secretion of the proteins into systemic circulation. SP sequences generated secretion of various engineered proteins into the bloodstream following intravenous, intramuscular, and subcutaneous SP mRNA delivery by lipid, polymer, and ionizable phospholipid delivery carriers. SP-engineered etanercept/TNF-α inhibitor proteins demonstrated therapeutic efficacy in an imiquimod-induced psoriasis model by reducing hyperkeratosis and inflammation. An SP-engineered anti-PD-L1 construct mediated mRNA encoded proteins with longer serum half-lives that reduced tumor burden and extended survival in MC38 and B16F10 cancer models. The modular nature of SP platform should enable intracellular and extracellular localization control of various functional proteins for diverse therapeutic applications.
Therapeutic proteins encoded in messenger mRNA (mRNA) hold innumerable potential applications due to the modular nature and ability to provide customizable instructions to create functional proteins (1–3). Clinical studies continue to confirm favorable safety and efficacy profiles, further underscoring the potential of mRNA as a versatile genetic medicine with broad potential to treat various diseases. In parallel with advances in mRNA research (4, 5), there has been substantial progress in the field of delivery using lipid nanoparticles (LNPs), polymer nanoparticles, ionizable phospholipid nanoparticles (iPLNPs), and other approaches to mediate safe and efficacious nucleic acid delivery (3, 6–9). From utility as a vaccine in the COVID-19 pandemic via efficient delivery of mRNA encoding the SARS-CoV-2 spike protein (10) to genome editing in humans with high efficacy and durability for hATTR (11–14), mRNA nanoparticles hold great promise as future medicines for a variety of diseases and cancer. However, most efforts in this area have been directed toward the intracellular generation of proteins that can either be utilized directly within cells (such as protein replacement therapy and antigen production in vaccines) or indirectly to orchestrate the induction of local pathways such as those required for gene editing within cells.
Following a different approach, development of methodologies to control transport and secretion of proteins in a modular fashion would open new therapeutic opportunities. We report the development of a modular signal peptide (SP) platform that can be incorporated into mRNAs encoding reporter, natural, and therapeutic proteins to induce secretion of proteins into systemic circulation, thereby enhancing blood circulation half-lives and enabling therapeutic efficacy in multiple disease models. We focused on posttranslational control of protein localization via exploitation of endogenous protein translocation pathways to produce systemically deliverable protein therapeutics, thus reframing the historical focus from amplifying intracellular expression efficiency via delivery systems to providing control via the nucleic acid cargo. Such temporal control over the expression kinetics of the therapeutic protein production provided by the translational machinery and secretory pathway of the cell can prolong the circulation half-life and help circumnavigate the shortcomings arising from bolus dosing of protein therapeutics.
Within cells, signal peptides function as metaphorical shipping labels to mediate shuttling of certain proteins to organelles such as the nucleus and mitochondria, as well as the export of proteins into the extracellular space via secretion pathways. SP domains consist of a short series of typically ~15 to 30 amino acids located on the N terminus of proteins (15). Individual signal sequences are often specific to their conjugate protein, which coordinates the protein’s emplacement fate via interactions with different pathways. In the cytosol, most proteins are transported to the endoplasmic reticulum (ER) for processing via posttranslational translocation. However, in cotranslational translocation, ribosomal binding and initial translation results in the generation of a signal sequence that is recognized and bound by signal recognition particle (SRP) which halts translation until SRP binds with SRP receptor on the ER membrane (16–19). At this point, translation resumes and an adjacent Sec61 translocon complex—consisting of SEC61A1 or SEC61A2, SEC61B, and SEC61G, wherein the SEC61A subunit contains 10 transmembrane α-helices—forms a conduit through which the SP and newly translated peptides flow into the ER lumen (20, 21). After translation finishes, the ribosome, SRP, and SRP receptor are then released via the hydrolysis of a GTP. This then clears the way for a signal peptidase to cleave the SP from the inchoate protein thus permitting folding and posttranslational modifications in the ER lumen, and finally COPII golgi-mediated vesicular secretion (21, 22). To use these transport systems, the mRNA encoding each protein must contain an SP upstream from the protein sequence.
Inspired by this biology, we hypothesized that engineered signal peptide sequences could be copy and pasted into mRNA sequences coding for proteins that would ordinarily be confined to the intracellular space to instead generate secretion of these proteins into circulation. Through our investigation, we identified several SPs from secreted proteins that when encoded into other mRNA sequences and subsequently complexed with a variety of delivery technologies resulted in the extracellular secretion of these proteins into circulation at sustained, therapeutically relevant concentrations. We incorporated SP sequences into reporter (mCherry), natural (erythropoietin, hEPO), and therapeutic (etanercept/Enbrel and anti-PD-L1) mRNA-encoding proteins. This was accomplished via encoding a secretion SP sequence (FVII, Alb, ApoB, or gLuc) directly upstream from the desired protein sequence in a plasmid DNA (pDNA) transcript. Following this, in vitro transcription (IVT) was performed to generate mRNA containing an optimized 5′ UTR, kozak sequence, secretion signal peptide sequence, protein sequence, 3′ UTR, and an optimized poly-A tail. The resulting mRNA was then encapsulated in multiple delivery systems for in vitro and in vivo delivery. Prior work in this area has focused on proteins that are normally secreted (23–28); here, we demonstrate that the secretory SP approach can be harnessed for proteins not normally secreted. SP mRNAs were effectively delivered wherein the encoded SP sequence efficiently harnessed endogenous protein secretion pathways, thereby allowing tissues including the liver, muscle, and subcutaneous cells to act as independent “protein factories” that subsequently exported the full-length native protein into circulation. Secreted proteins can traffic to the site of action by utilizing the inherent uptake mechanism of the target protein, thereby providing an opportunity to access currently inaccessible organs. The ubiquity of these results indicates that SP-engineered mRNA sequences are an independent and universally applicable nucleic acid technology that is agnostic to delivery carrier and allows for tunable posttranslational governance of protein fate. With further development, the protein factory concept holds promise in enabling intracellular and extracellular localization control of therapeutic proteins for diverse applications.
Results
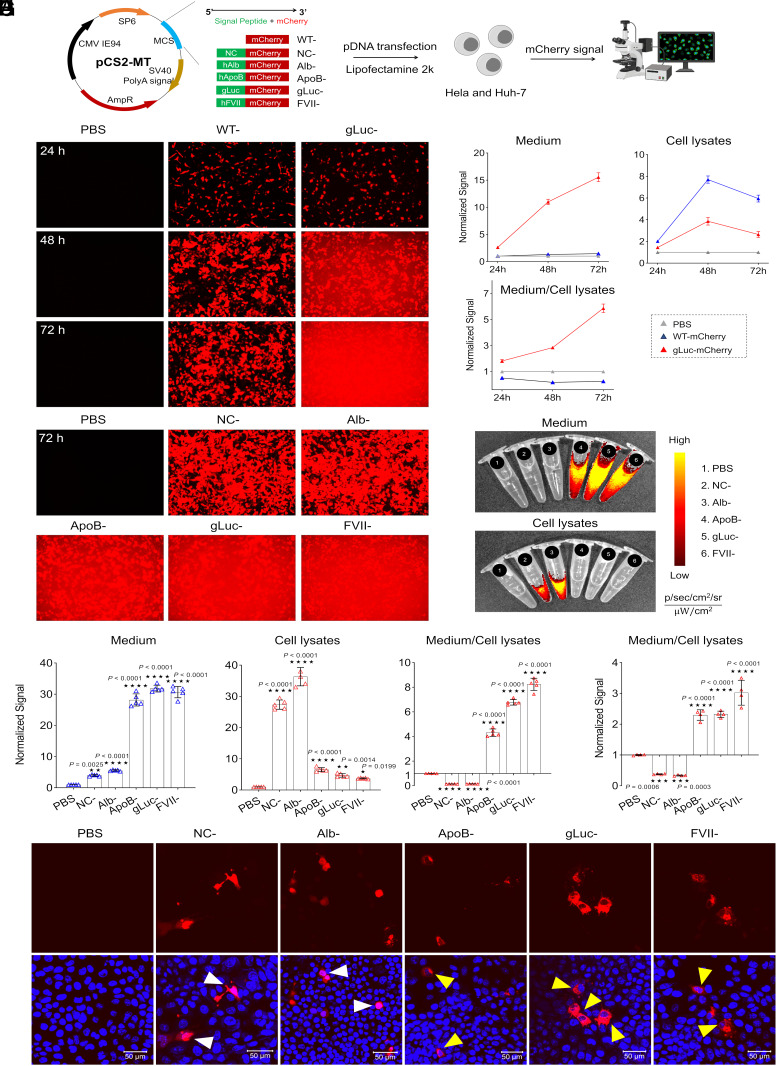
We initially investigated SP sequences and their subsequent effects on protein secretion by integrating several different SPs from known endogenously secreted proteins (albumin, Alb; apolipoprotein B, ApoB; and Factor VII, FVII) and one known synthetic secreted protein (Gaussia luciferase; gLuc) along with a negative control (NC). SP leader sequences were incorporated into a pCS2-MT plasmid backbone directly upstream from a reporter mCherry mRNA sequence. The construct was preceded by the SP6 promoter and an optimized 5′ UTR, followed by an optimized 3′ UTR and polyA tail (Fig. 1A). HeLa cells were transfected with wild-type mCherry pDNA containing no SP (WT) and gLuc-mCherry pDNA via Lipofectamine2000. Intracellular and extracellular fluorescence were quantified via fluorescence microscopy at 24, 48, and 72 h posttransfection. gLuc SP mRNA induced high levels of mCherry secretion into media while mCherry signal in WT was restricted within the HeLa cells (Fig. 1B). Next, mCherry protein content present in cell medium and cell lysate was quantified with a fluorescence plate reader over 72 h and revealed an increase in mCherry secretion into medium over time along with an enhanced medium to cell lysate ratio of mCherry fluorescence in the gLuc SP group (Fig. 1C). The set of SPs was then expanded to include a negative control (scramble sequence), Alb, ApoB, and FVII, in addition to gLuc. HeLa cells were again transfected with the pDNA constructs using Lipofectamine 2000. Images taken 72 h posttransfection via fluorescence microscopy and IVIS demonstrated that the SPs ApoB, gLuc, and FVII all secreted high levels of mCherry protein, while the NC and Alb constructs effectively mediated only intracellular mCherry expression without significant extracellular secretion (Fig. 1 D–F). Projecting to eventual in vivo liver delivery, the same set of SPs were evaluated in a liver cancer cell line, Huh7, wherein the observed trends of mCherry secretion in HeLa cells were the same. Transfected Huh7 cells analyzed by confocal microscopy revealed morphological differences in mCherry signal between SPs driving extracellular secretion of mCherry and those that promoted only intracellular mCherry expression (Fig. 1 G and H). Overall, it was determined that the FVII SP was able to cultivate the highest levels of protein secretion across both cell lines.
Fig. 1.
Optimal signal peptides (SPs) drive protein secretion effectively following plasmid DNA (pDNA) delivery. (A) Engineering secreted mCherry by including a SP upstream of the coding sequence and screening of optimal SP by pDNA transfection in vitro. (B) Time-dependent mCherry secretion by HeLa cells (exposure time, 1/30 s); mCherry signal was observed clearly in the medium 2 d after treatment with gLuc-mCherry. (C) Quantification of mCherry fluorescence in cell lysates and medium at different time points. (D) SP screening in HeLa cells at 72 h (exposure time, 1/70 s). (E) Cell lysates and medium were imaged and (F) quantified at 72 h. Data is presented as mean ± SEM (n = 5 biologically independent samples). HeLa cells were treated with Lipofectamine 2000 containing pDNA in a 96-well plate (50 ng per well). At noted times points, cells were imaged via confocal microscopy; mCherry signal was quantified by plate reader or captured by IVIS. (G) The functionality of SP in Huh7 cells was confirmed by quantifying mCherry signal in cell lysates and medium at 72 h. Data is presented as mean ± SEM (n = 4 biologically independent samples). (H) A “circle-like” signal (indicated by yellow arrows) was observed in Huh7 cells imaged by confocal microscopy indicative of high efficacy secretion mediated by SP. (Scale bar, 50 µm.) WT, wild type; NC, negative control; Alb, human albumin; ApoB, human apolipoprotein B; gLuc, Gaussia luciferase; FVII, human Factor VII. A two-tailed unpaired t test was used to determine the significance of the comparisons of data indicated in F and G (*P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001).
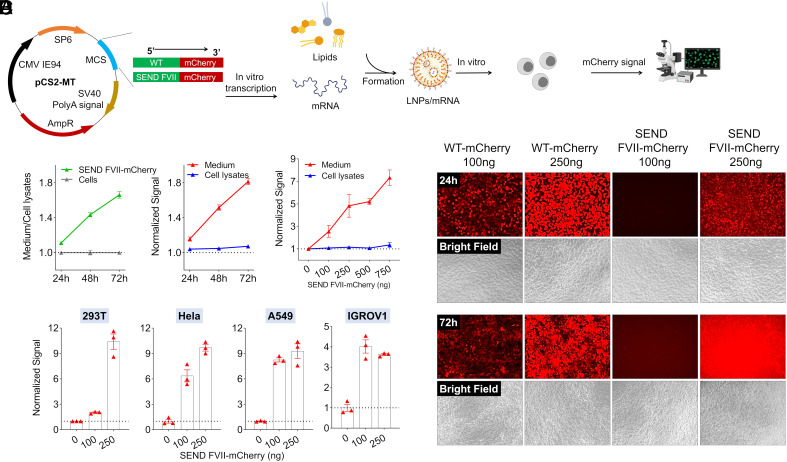
Following these results, we next examined whether mRNA containing an integrated SP sequence would yield similar observations to what was exhibited with pDNA. We generated FVII-mCherry mRNA from the FVII-mCherry-pCS2-MT plasmid via in vitro transcription (IVT) (Fig. 2A). Our lab has reported various libraries of lipids and polymers capable of delivering a variety of cargos including mRNA (29–37). To examine the SP platform, we initially utilized a liver-targeting 4-component LNP that had been previously optimized for mRNA delivery to hepatocytes (33). Transfection of multiple cell lines with LNPs containing FVII-mCherry mRNA indeed demonstrated that protein export into medium positively correlates with time posttransfection as well as the dose, with greater fluorescence signal intensity observed at longer time intervals and higher doses across cell lines (Fig. 2 B–D). The produced mRNA was termed Signal peptide Engineered Nucleic acid Design (SEND) to capture the modular concept of inserting SPs ahead of genes of interest.
Fig. 2.
The FVII SP drives effective protein secretion in vitro as validated by mRNA delivery. (A) FVII-mCherry mRNA was synthesized by IVT and encapsulated in LNPs for delivery. (B) Time-dependent (at 100 ng mRNA per well) and dose-dependent (at 72 h) mCherry secretion in Huh7 cells. (C) Dose-dependent (at 72 h) mCherry secretion in different cell lines after treatment with mRNA LNPs in a 96-well plate. At the noted time points, mCherry signal was quantified by plate reader. (D) Obvious mCherry signal was observed in the medium at day 3 after FVII-mCherry mRNA formulation treatment. HEK293T cells in a 96-well plate were treated by mRNA LNPs with different doses and imaged by confocal microscopy (exposure time, 1/6 s for 24 h and 1/15 s for 72 h).
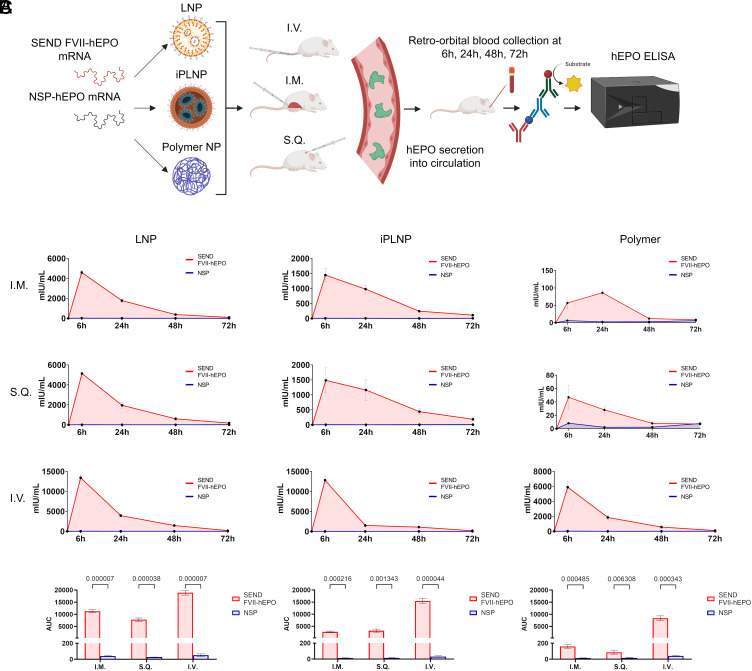
To assess the compatibility of SEND mRNA with different delivery systems and routes of administration (ROA), we designed and compared two mRNA constructs: 1) SEND FVII-hEPO mRNA where the endogenous SP in hEPO was removed and replaced with the FVII SP, and 2) control NSP (no signal peptide)-hEPO mRNA where the endogenous N-terminal SP in hEPO was removed leaving behind only the functional hEPO protein coding sequence. In this design, the FVII SP should enable hEPO secretion, whereas the NSP control should not. We compared three distinctly different delivery vehicles: 4-component LNPs with 5A2-SC8 as the ionizable amino lipid, ionizable phospholipid (iPhos) LNPs (iPLNPs) utilizing the iPhos 9A1-P9, and a polymer nanoparticle comprising a degradable functional polyester PE5.3K-A17-0.33C8 (30, 33, 36, 38). We also compared three different ROAs: intravenous (I.V.) injection, intramuscular (I.M.) injection, and subcutaneous (S.Q.) injection (Fig. 3A). Blood was drawn retro-orbitally from each mouse at 6, 24, 48, and 72 h postinjection and serum was assessed for hEPO concentration via an ELISA assay. As expected, hEPO was practically undetectable in mouse serum following delivery of control NSP-hEPO mRNA across all carriers and all ROAs. In contrast, SEND FVII-hEPO mRNA constructs were able to effectively mediate strong and sustained secretion of hEPO protein into systemic circulation, independent of carrier and ROA. These results establish SEND mRNA as a modular nucleic acid platform compatible across multiple delivery systems and ROAs (Fig. 3B). The most favorable pharmacokinetic outcome was obtained via IV injection of LNPs containing 5A2-SC8 as the ionizable amino lipid, which was therefore chosen for further therapeutic studies.
Fig. 3.
SEND mRNA construct induces secretion of hEPO protein into systemic circulation across multiple delivery systems and routes of administration. (A) hEPO mRNA was synthesized via IVT containing either no endogenous signal peptide sequence (NSP-hEPO) or FVII signal peptide sequence in lieu of hEPO’s endogenous signal peptide (FVII-hEPO). Each mRNA was encapsulated into one of three different nanoparticles (LNPs, iPLNPs, and polymer NPs). Each nanoparticle-mRNA formulation was then injected into mice via one of three different ROAs (I.M., I.V., S.Q.). Blood was drawn from mice at 6 h, 24 h, 48 h, and 72 h postinjection, and hEPO protein concentration was quantified via ELISA. (B) In all cases, FVII-hEPO mRNA was able to effectively induce secretion of hEPO protein into blood using multiple delivery systems and ROAs. (C) Area under the curve (AUC) was calculated for every carrier and ROA. Data are presented as mean ± SEM (n = 4 biologically independent animals).
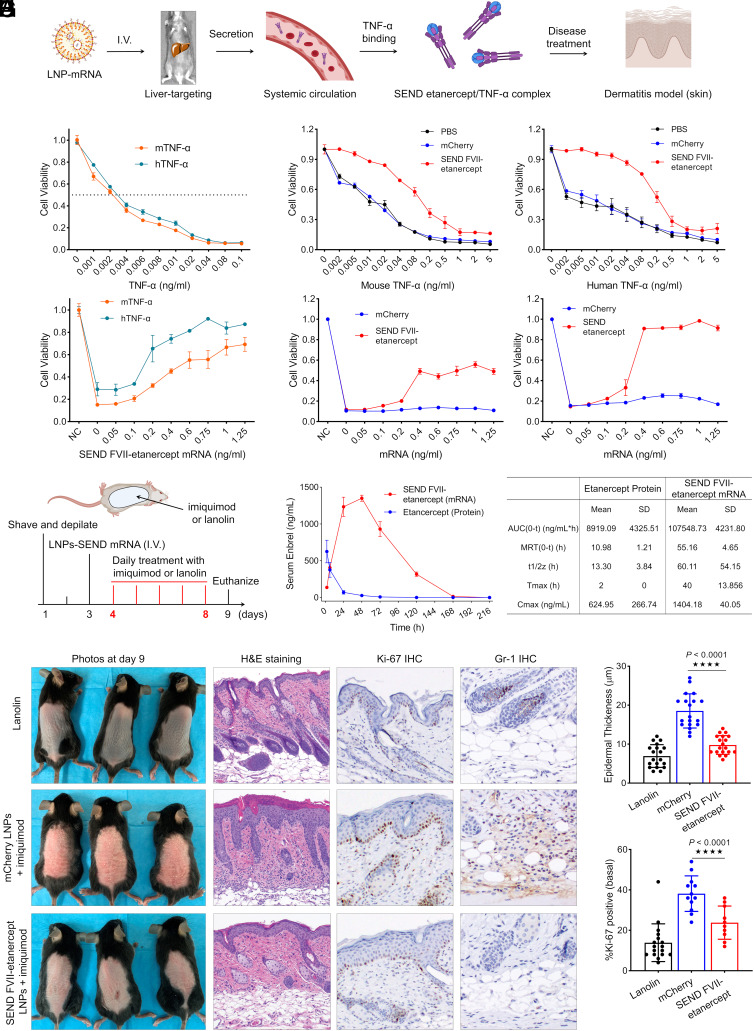
We next examined whether these secretion instructions could be incorporated into mRNAs encoding proteins with therapeutic potential. In that regard, we created an mRNA construct containing FVII SP and coding sequence for etanercept (Enbrel), a synthetic dimeric fusion protein that inhibits TNF-α. This SEND mRNA construct was encapsulated in the LNPs for evaluation in L929 cells and an imiquimod-induced psoriasis in vivo model (Fig. 4A) (39, 40). L929 cells were first treated with mouse and human sourced TNF-α at doses of 0.001 to 0.1 ng/mL. At a low concentration of 0.02 ng/mL, less than 20% of cells remain viable (Fig. 4B). Next, cells were pretreated with 80 ng of FVII-etanercept mRNA loaded LNPs and 48 h later were subsequently challenged with either mouse or human TNF-α at doses ranging from 0.002 ng/mL to 5 ng/mL. In both treatment groups, pretreatment with FVII-etanercept mRNA LNPs resulted in cell viability remaining significantly higher across TNF-α dose ranges when compared with PBS and mCherry mRNA controls (Fig. 4C). Additionally, L929 cells were pretreated with FVII-etanercept mRNA LNPs for 48 h at doses ranging from 0.05 ng/mL to 1.25 ng/mL and then challenged with 0.1 ng/mL of either mouse or human TNF-α. As expected, there was a dose-dependent rise in cell viability in response to increasing pretreatment concentrations of FVII-etanercept mRNA (Fig. 4D). Finally, L929 cells that received 0.1 ng/mL of either mouse or human TNF-α were rescued in a dose-dependent fashion after treatment with medium from cells pretreated with FVII-etanercept mRNA LNPs at doses of 0.05 ng/mL to 1.25 ng/mL, with viability restoration reaching nearly 100% in the human TNF-α group at an mRNA dose of just 0.4 ng/mL (Fig. 4E).
Fig. 4.
SEND FVII-etanercept mRNA LNPs prevent TNF-α induced cell death and achieved a therapeutic benefit in an imiquimod-induced psoriasis model in vivo. (A) Scheme of etanercept (Enbrel) protein production, TNF-α binding, and disease treatment on dermatitis model. (B) Dose-dependent cytotoxicity of human TNF-α (hTNF-α) and mouse TNF-α (mTNF-α) in L929 cells. Cells were incubated 24 h with actinomycin at 1 μg/mL and various concentrations of TNF-α before determining cytotoxicity. (C) L929 cells pretreated by FVII-etanercept mRNA LNPs were resistant to both mouse and human TNF-α. Cells were primed with 80 ng FVII-etanercept mRNA LNPs treatment per well and, 48 h later, were challenged with TNF-α ranging from 0 to 5 ng/mL and a fixed concentration of actinomycin at 1 μg/mL. Twenty-four hours after incubation, cell viability was assessed. (D) Dose-dependent rescue of cell viability following pretreatment by LNPs. Cells were pretreated by FVII-etanercept mRNA LNPs with mRNA doses of 0 to 1.25 ng/mL, after 48 h, cells were challenged by TNF-α with dose of 0.1 ng/mL and fixed actinomycin dose of 1 μg/mL. Twenty-four hours postchallenge, cell viability was measured. (E) Dose-dependent rescue of cell viability after treatment with medium from FVII-etanercept mRNA LNP-treated cells. L929 cells were treated with FVII-etanercept mRNA LNPs with mRNA doses of 0 to 1.25 ng/mL. After 2 d, medium from these cells containing secreted etanercept was collected and transferred into fresh L929 cells, and the cells were challenged by mouse or human TNF-α at a dose of 0.1 ng/mL and actinomycin at 1 μg/mL. Twenty-four hours after media transfer and challenge, cell viability was evaluated. Data are presented as mean ± SEM (n = 5 biologically independent samples) (F) Mice were shaved and depilated on day 1 and subsequently injected with LNPs on day 3. On days 4 to 8, mice were treated with either lanolin or imiquimod to induce dermatitis, and mice were euthanized on day 9. (G) Serum concentration curves for etanercept protein and mRNA following a single I.V. injection at a dose of 0.5 mg/kg native protein or mRNA-LNP formulation. Pharmacokinetic comparison between etanercept protein and mRNA after single dosing. Serum was collected at different time points, and etanercept was detected by ELISA. (H) Images of dorsal skin of lanolin-treated mice (control) and imiquimod-treated mice, which were injected with either mCherry mRNA or FVII-etanercept mRNA formulations. Images of H&E-stained sections, Ki-67 staining, and Gr-1 staining of the dorsal skin of lanolin control and imiquimod-treated mice. (I) Measurement of epidermal thickness in three groups and the percentage of Ki-67+ cells in epidermal basal cells (per 50 cells) quantified from (H). (****P <0.0001).
Having confirmed the effectiveness of both FVII-mCherry mRNA LNP and FVII-etanercept mRNA LNP mediated protein secretion in vitro, we then examined whether this concept would replicate effectively in vivo to facilitate secretion of proteins into systemic circulation. With prior knowledge that the liver is known to generate and secrete plasma proteins, we utilized liver-targeted LNPs with the expectation that the liver could be employed as an endogenous protein factory capable of manufacturing the protein of interest from a specific mRNA and subsequently secreting it via an appropriate set of instructions integrated into the SEND mRNA (23–26). Motivated by the positive outcomes in the in vitro therapeutic models, we designed an in vivo imiquimod-induced psoriasis model to assess the therapeutic potential of LNP encapsulated with FVII-etanercept mRNA. Mice were initially shaved, depilated, and separated into three groups: a negative control group receiving no LNPs and a Lanolin vehicle cream on days 4 to 8; and two experimental groups wherein one group was administered FVII-etanercept mRNA LNPs I.V., and the other group was given mCherry LNPs I.V., both 3 d after shaving and depilation. In the two experimental groups, the dorsal back skin of the mice was treated on days 4 to 8 to induce a psoriasis-like dermatitis. Serum pharmacokinetics of etanercept were first assessed via an etanercept ELISA following I.V. injection with 0.5 mg/kg FVII-etanercept LNPs or etanercept protein. In mice injected with etanercept protein, peak serum concentration occurred 2 h postinjection and quickly diminished thereafter. However, in the group injected with FVII-etanercept mRNA LNPs, serum concentration continued to rise until 48 h postinjection and remained detectible until 168 h postinjection. FVII-etanercept mRNA LNPs demonstrated >10-fold increase in AUC and >20-fold increase in Tmax (h) when compared with etanercept protein (10,7548.73, +/− 4,321.8 vs. 8,919.09 +/− 4,325.51; and 2 h vs. 40 h, respectively) (Fig. 4 D and E). Compared to the lanolin control, mCherry mRNA LNP + imiquimod–treated mice showed obvious scaling and erythema, which was notably decreased in the FVII-etanercept + imiquimod group. H&E staining of sections from the treated skin area confirmed that epidermal thickness was increased in mCherry mRNA LNP + imiquimod–treated mice, which was significantly rescued by FVII-etanercept mRNA. Immunohistochemical staining for Ki-67 revealed significantly decreased proliferation after FVII-etanercept. Staining for Gr-1, a neutrophil marker, also showed decreased inflammation in the skin of FVII-etanercept + imiquimod mice (Fig. 4 D–H). We conclude that FVII-etanercept mRNA LNPs can provide significant therapeutic benefit in an in vivo mouse model of psoriasiform dermatitis.
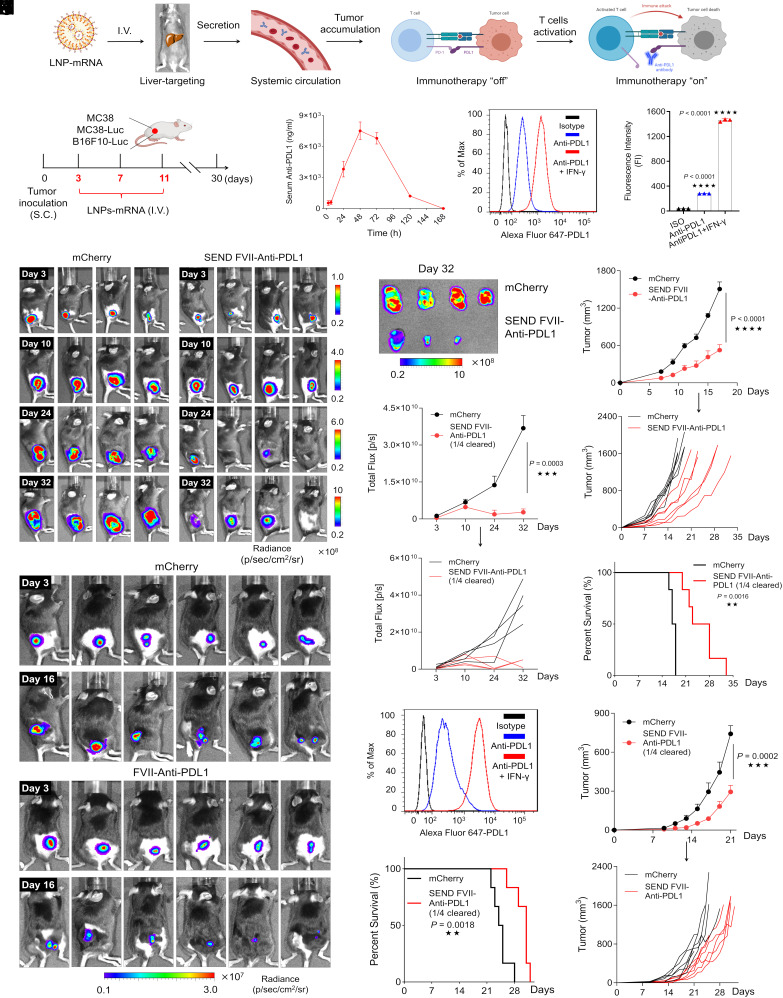
In addition to autoimmune disorders, we evaluated the utility of the SEND platform as an anticancer therapeutic in three syngeneic xenograft tumor mouse models. MC38 (native) and MC38-Luc cells (containing a luciferase reporter construct for tracking)—an aggressive murine adenocarcinoma cell line, and B16F10-Luc cells—a murine melanoma cell line (containing a luciferase reporter construct for tracking), were injected subcutaneously into the right hind leg of C57BL/6 mice and tumors were allowed to grow. To gauge the anticancer therapeutic potential of SEND mRNA LNPs, mice were first inoculated with tumor cells and then injected intravenously on days 3, 7, and 11 postinoculation with LNPs containing either control mCherry mRNA or LNPs encapsulating mRNA that encoded anti-PD-L1 antibody with an upstream FVII SP (Fig. 5 A and B). A pharmacokinetic distribution of serum anti-PDL1 levels was established with peak serum concentration occurring 48 h post-IV injection—essentially mirroring the curve seen previously with FVII-etanercept mRNA LNPs. PD-L1 expression on the surface of MC38 cells was determined using flow cytometry (Fig. 5 C and D). Tumor growth was evaluated in the MC38-Luc model on days 3, 10, 24, and 32 via IVIS imaging for luminescence. In the group receiving mCherry mRNA LNPs, tumor development was rapid and saw a corresponding increase in luminescence and tumor volume at the time points tested. The tumor mass occupied the entire hind leg of all mice by day 32. However, in the group treated with FVII-anti-PDL1 mRNA LNPs, tumor growth noticeably declined by day 24, suggesting that treatment with FVII-anti-PDL1 mRNA LNPs was effective at inhibiting tumor progression. As expected, this reduction was paralleled by a decrease in luminescence, significantly slower tumor development, and smaller tumor volume following resection on day 32 (Fig. 5 E–H). To examine potential survival benefit, we employed native MC38 cells (no Luc reporter) as they grow faster than MC38-Luc cells in vivo. Notably, survival of the FVII-anti-PDL1 mRNA LNP injected mice was almost twofold longer than that of the control group (Fig. 5I). Similarly, mice inoculated with the B16F10-Luc tumors were injected with LNPs containing mCherry mRNA or FVII-anti-PDL1 mRNA via IV injection. IVIS imaging at days 3 and 16 revealed significant declines in tumor mass and luminescence, slower growth rates, and extension of overall survival in the FVII-anti-PDL1 mRNA group when compared to the mCherry mRNA group (Fig. 5 J–M).
Fig. 5.
SEND FVII-anti-PDL1 LNPs reduce tumor burden and extend survival in vivo. (A) Scheme of anti-PDL1 antibody production and tumor immunotherapy. (B) Scheme of experimental design utilizing MC38, MC38-Luc, and B16F10-Luc xenograft mice models. (C) Pharmacokinetic profile of serum anti-PDL1 antibody secretion after single dosing with SEND mRNA LNPs. Mice were I.V. injected with a dose of 0.5 mg/kg mRNA. Serum was collected at different time points, and anti-PDL1 was detected by ELISA. (D) Representative PD-L1 expression on the membrane surface of MC38 cells determined by flow cytometry. (E) Luminescence images of MC38-Luc tumors at different time points captured by IVIS. (F) Luminescence images of isolated MC38-Luc tumors at day 32. (G) Quantified luminescence signal in MC38-Luc tumor area at different time points. (H) Tumor growth of the native MC38 model and (I) mice survival during treatments by mRNA formulations. (J) Luminescence images of B16F10-Luc tumors on day 3 and day 16 treated with different formulations. (K) Representative PDL1 expression on the membrane surface of B16F10-Luc cells determined by flow cytometry. (L) Tumor growth of B16F10-Luc model and (M) mice survival during treatments by mRNA formulations. (*P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001).
Discussion
To date, nonviral delivery technologies for nucleic acid therapeutics have been relatively limited with respect to tissue specificity, with the majority of LNP formulations being restricted to hepatic delivery following intravenous injection. Breakthrough technologies such as Selective Organ Targeting (SORT) have helped to overcome this delivery hurdle through the expansion of targeted delivery to extrahepatic tissues such as the lung and spleen (34). However, once the nucleic acids are successfully delivered to the target organ and the respective proteins are translated, they are essentially trapped inside the transfected cell and therefore restricted to exert therapeutic benefit intracellularly. This is an effective therapeutic modality for medicaments with localized mechanisms of action. However, many diseases with generalized areas of affliction such as autoimmune and inflammatory disorders would benefit from therapeutics with more extensive biodistribution throughout systemic circulation.
Current state-of-the-art protein therapeutics have shown great promise in addressing the clinical challenges surrounding their specificity and selectivity compared to their small molecule counterparts (41). However, recombinant protein-based therapies suffer from manufacturing and purification limitations where the host utilized to generate the protein is often unable to mimic the posttranslational modifications (PTMs) on the target protein, thereby diminishing their therapeutic performance. Additionally, administration of bolus dose of the therapeutic protein can cause rapid clearance from the system thereby resulting in sub-optimal pharmacokinetics and elicit an inflammatory immunological cascade in the form of cytokine release syndrome (CRS) (42, 43). With these design challenges in mind, we developed a platform that allows in situ generation of therapeutic proteins engineered for efficient extracellular secretion through incorporation of SPs into pDNA and mRNA constructs. By introducing SPs directly preceding the gene of interest, we have appropriated the body’s endogenous mechanism of protein shuttling and localization to orchestrate the generation and subsequent secretion of newly translated therapeutic proteins. In principle, in situ production of therapeutic proteins should minimize potential immunogenicity arising from differences in PTM patterns. Additionally, the SEND platform can be incorporated into any carrier for use in any tissue, opening new deployment options for nucleic acid nanoparticle medicines.
Through a comparison of natural and unnatural SP sequences, we identified the FVII SP as particularly amenable to the goal of enabling ubiquitous secretion of engineered proteins. This SP-modified mRNA construct was packaged in LNPs, iPLNPs, or polymer NPs and evaluated following different ROAs. The liver, muscle, and subcutaneous cells can all be utilized as protein factories, efficiently mediating secretion of the protein product into circulation. Beyond simple augmentation of control over protein localization, enhanced AUC and Cmax were observed, indicating improved performance of the SP platform over direct administration of native therapeutic protein. We hypothesize that the in situ production of biologics may further improve biocompatibility and reduce the likelihood of CRS due to posttranslational modifications such as glycosylation taking place within one’s own body, as well as enhanced control over protein production kinetics utilizing a signal peptide engineered therapeutic approach (42, 44–46). These potential effects will be examined in future studies. We evaluated the ability of this platform to provide therapeutic advantages in a series of animal disease models that would benefit from widespread expression of a specific therapeutic protein. As hypothesized, the incorporation of FVII SP into anti-hTNF-α (etanercept/Enbrel) mRNA or anti-PDL1 mRNA resulted in the genesis and secretion of intact, functional proteins into circulation, thereby providing therapeutic benefit in an imiquimod-induced psoriasis model and MC38/MC38-Luc/B16F10-Luc cancer models, respectively.
Given these results, we believe that the SEND platform could impact nucleic acid-based therapeutics for a variety of disorders ranging from rheumatoid indications and cancer, to clotting disorders and potentially diabetes. Moving forward, as with all emerging technologies, preclinical evaluation, manufacturing, safety, and translatability into humans will need to be evaluated. Additionally, work remains to fully elucidate the mechanics behind SP design and effects on corresponding therapeutic durability, systemic serum concentration, and efficacy. We also anticipate that computational design of signal peptides could further increase secretion capacity. The present results provide a method for introducing a previously nonexistent level of posttranslational spatiotemporal control for protein-based therapeutics, a strategy for extending the circulation time of current antibody-based therapeutics, and a modular platform that could reduce patient burden and dosing frequency in a variety of therapeutic applications.
Materials and Methods
SP-mCherry Plasmid (pDNA) Construction.
SP-modified mCherry plasmids were constructed to determine the optimal signal peptide (SP) for secretion. Briefly, the SP-mCherry coding region (SI Appendix, Table S1) was obtained directly by PCR with well-designed primers. Several different SPs were selected for the constructs including Alb (human albumin), ApoB (human Apolipoprotein B), gLuc (Gaussia luciferase), and FVII (human Factor VII). BamH1 and EcoR1 restriction enzyme-digested SP-mCherry products were cloned into a pCS2-MT vector backbone via ligation at 16 °C overnight and subsequent transformation. After validation by sequencing, SP-mCherry plasmids were ready for in vitro screening.
In Vitro SP Evaluation by pDNA Transfection.
To perform SP screening, pDNA transfection was performed using Lipofectamine 2000 in HeLa and Huh7 cells. Each cell line was seeded into 96-well plates with 1 × 104 cells per well in 100 μL of respective cell culture media and placed at 37 °C. After incubating for 24 h, cells were treated by Lipofectamine 2000–pDNA formulations with 50 ng pDNA per well. Directly after treatment, 100 μL of additional media was added to each well. At 24, 48, or 72 h after treatment, cells were imaged immediately using a Keyence microscope for fluorescence. Following imaging, 100 μL of cell media was collected and placed into a new white bottom 96-well plate. The remaining media were aspirated from each well, and the cells were lysed in 1X cell lysis buffer. Cell lysate was then collected and added to the 96-well white bottom plate. After collection of lysate and media, mCherry signal was quantified using a Tecan plate reader with excitation/emission spectra of 563/610, respectively. To observe mCherry signal clearly, cell lysates and medium were further transferred into 1.5-mL Eppendorf tubes and imaged by an IVIS Lumina system using mCherry filter settings. Finally, subcellular mCherry signal distribution was determined via confocal microscopy. Huh7 cells were treated as described above. After 3 days, cells were washed three timed by 1×PBS, stained by Hoechst 33342, and imaged by confocal microscopy.
mRNA Synthesis.
All mRNAs this work used were produced by in vitro transcription (IVT) as previously described (26). Briefly, linear pDNA with optimized 5′(3′)-untranslated regions (UTR) and poly A sequences were obtained first by enzyme digestion, and IVT reactions were subsequently prepared using standard protocols with N1-methylpseudouridine-5′-triphosphate modification. Finally, mRNA was capped (Cap-1 structure) by Vaccinia Capping Enzyme and 2′-O-methyltransferase (NEB).
mRNA LNP Nanoparticle Formation.
mRNA-loaded LNP formulations were formed using the ethanol dilution method as described previously (31). A liver-targeted mRNA LNP formulation reported in our previous paper was utilized. Briefly, all lipids with specified molar ratios—LNP (15:15:30:3; 5A2-SC8:DOPE:Cholesterol:PEG2000DMG) were dissolved in ethanol, and RNA was dissolved in 10 mM citrate buffer (pH 4.0). Then, the two solutions were rapidly mixed at an aqueous to ethanol ratio of 3:1 by volume (3:1, aq.:ethanol, vol:vol) to satisfy a final total lipid weight ratio of 40:1 (total lipids:mRNA). After incubation for 10 min at room temperature (RT), the mRNA LNP formulations were diluted using 1× PBS and added directly to cells or dialyzed against 1× PBS for 2 h for in vivo experiments.
mRNA iPLNP Nanoparticle Formation.
For mRNA-encapsulating iPLNPs, all particles were created using the ethanol dilution method as previously described (28). Lipids of the following molar ratios (25:30:30:1, 9A1-P9:DLin-MC3-DMA:Cholesterol:PEG2000DMG) were dissolved in ethanol, and mRNA was dissolved in 10 mM citrate buffer (pH 4.0). Both solutions were subsequently mixed at a ratio of 3:1 by volume (3:1, aq.:ethanol, vol:vol), with a 9A1-P9:mRNA weight ratio of 18:1. Particles were incubated at RT for 10 min before being dialyzed against 1× PBS for 2 h for in vivo experiments.
mRNA Polymer Nanoparticle Formation.
For polymer NPs, PE5.3 K-A17-0.33C8 and 5 wt% of F127 in DMSO and ethanol solution were mixed with mRNA-containing citrate buffer (pH = 4, 10 mM) at a PE5.3 K-A17-0.33C8/mRNA ratio of 30:1 (wt/wt) through vortex mixing method (23, 30). The final mRNA-polyplex NPs with the final mRNA concentration of 50 ng/µL were obtained by dialysis (cutoff MW 3500 Da) against PBS for 2 h prior to injection to mice.
In Vitro mCherry Secretion Driven by Optimal FVII SP Verified with mRNA Formulation.
FVII-mCherry mRNA was transfected into several different cell lines, including Huh7, HEK293T, HeLa, A549, and IGROV1. Cells were seeded into 96-well plates at a density of 1 × 104 cells per well and allowed to incubate at 37 °C for 24 h. mRNA LNP formulations were prepared as described above, and cells were then treated with mRNA at doses ranging from 0 to 750 ng mRNA per well. At 24, 48, or 72 h posttransfection, cells were imaged directly via a Keyence microscope, and mCherry signal in medium and cell lysates were quantified using the Tecan plate reader as described above. To ensure accuracy of the observed signal, wild-type mCherry mRNA was transfected into cells as a control.
Cytotoxicity Rescue of FVII-Etanercept mRNA.
L929 cells were used to evaluate TNF-α mediated cytotoxicity. Both mouse TNF-α (mTNF-α) and human TNF-α (hTNF-α) were selected. Cells were seeded into 96-well plates at a density of 1 × 104 cells per well and allowed to incubate for 24 h at 37 °C. Following this incubation, medium was replaced with 180 µL of fresh medium containing actinomycin and TNF-α with the final concentration of actinomycin being 1 µg/mL and TNF-α concentrations ranging from 0 to 0.1 ng/mL. After the media change, cells were allowed to incubate at 37 °C for another period of 24 h, and cell viability was detected using the CellTiter-Glo kit using the manufacturer’s standard protocol.
To evaluate the cytotoxicity rescue capability of FVII-etanercept mRNA formulations, cells were pretreated with FVII-etanercept mRNA LNPs for 48 h prior to challenging them with TNF-α. In this study, we tested a dose-dependent of rescue for both the mRNA and TNF-α. For the dose-dependent rescue of mRNA formulations, we utilized mRNA doses ranging from 0 ng/mL to 1.25 ng/mL per well, and then challenged with a TNF-α dose of 0.1 ng/mL for 24 h. For the dose-dependent rescue of TNF-α, we fixed the transfected mRNA concentration at 0.4 ng/mL, and then challenged the cells with TNF-α at a concentration of 0 ng/mL to 5 ng/mL.
To further validate that the rescue effects were from etanercept being secreted by the cells in medium, we measured the cytotoxicity rescue after pretreatment with functional medium. We seeded and treated the cells as described above, with mRNA concentrations of 0 ng/mL to 1.25 ng/mL, then collected the medium containing the secreted etanercept and transferred it into a 96-well plate containing attached L929 cells that had been allowed to incubate at 37 °C for 24 h prior to the media transfer. At the same time as the media transfer, the plate containing the attached L929 cells was challenged with TNF-α (0.1 ng/mL) and actinomycin (1 µg/mL), and cell viability was measured 24 h after the media transfer/challenge.
Pharmacokinetics Studies.
Male C57BL/6 mice weighing ~20 g were randomly divided into groups. Either etanercept protein or FVII-etanercept mRNA LNPs formulated as described above were I.V. injected into mice at a dose of 0.5 mg/kg. Starting at 2 h post-I.V. injection, serum was collected at 24, 48, 72, 120, 168, and 216 h, and etanercept serum concentration was quantified via ELISA (MyBioSource) for etanercept.
Imiquimod-Induced Psoriasis Model.
For the imiquimod-induced psoriasiform hyperplasia model (32), 8-wk-old female C57BL/6 mice were shaved and chemically depilated with Nair (indicated on day 1). On day 3, the mice were I.V. injected with LNPs containing FVII-etanercept mRNA at a dose of 0.5 mg/kg. LNPs were prepared as described above. After injection, the shaved dorsal-skin samples were treated topically with 60 mg of Aldara cream (5% imiquimod) (Aldara, 3 M Pharmaceuticals) daily for a total of 5 d. On day 9, photos of Lanolin+PBS, imiquimod + FVII-etanercept LNP, and imiquimod + mCherry LNP were taken. After taking photos, mice were euthanized. The dorsal skin of the mice was harvested, fixed overnight in 4% paraformaldehyde, and sectioning for H&E staining. IHC was performed as previously described (31). The primary antibodies, Ki-67 (1:400, Abcam, ab16667) and Gr-1 (1:20, BD Biosciences 550291), were used manufacturer’s instructions.
Assessment of PDL1 Expression.
MC38, MC38-Luc, and B16F10-Luc cell lines were incorporated to study in vivo tumor immunotherapy. Flow cytometry was utilized to ascertain levels of PDL1 expression in cell membranes. Cells were seeded into 6-well plates at a density of 3 × 105 cells per well and allowed to incubate for 24 h at 37 °C. After the initial incubation for cell adhesion, the cells were subsequently incubated with IFN-γ (100 ng/mL) for an additional 24 h. Following the antibody staining protocols, the cells were tagged with a primary anti-PDL1 antibody and an Alexa Fluor 647–labeled second antibody wherein PDL1 expression was next determined via flow cytometry for Alexa Fluor 647–positive cells. Isotype antibody stained cells were used for gating.
Tumor Immunotherapy.
MC38 or MC38-Luc (stably expressing luciferase) cells were grown in DMEM medium with 10% FBS and collected once sufficiently confluent for the generation of xenograft tumors. At day 0, a total of 1 × 106 cells suspended in 100 μL PBS were subcutaneously injected into the right flank of C57BL6/j mice. In the treatment group, FVII-anti-PDL1 LNPs were formulated as previously described and injected via I.V. into the mice at a dose of 0.5 mg/kg starting at day 3 and then again at day 7 and day 11, comprising three injections in total. The same dosing protocol was followed for mice in the control group which were I.V. injected with LNPs containing mCherry mRNA. For mice in the MC38 xenograft tumor model group, tumors were physically measured using a digital caliper for volume (0.5 × length × width2) and a survival curve was generated. For mice in the MC38-Luc xenograft tumor group, luciferase expression was continuously captured by IVIS at day 3, day 10, day 24, and day 32, and luminescence was quantified via IVIS software. Mice were sacrificed and recorded as deceased (end point reached) when the tumor volume reached 1,500 cm3 or larger.
For the B16F10-Luc model, cells were grown and collected as described above, and a total of 4 × 105 cells were subcutaneously injected into the right flank of C57BL/6 mice. LNPs containing FVII-anti-PDL1 were formulated as described above and subsequently I.V. injected into the mice tails following the same dosing schedule and dosing concentration described in the MC38/MC38-Luc xenograft tumor model groups. Likewise, mCherry LNPs were injected following the same schedule to serve as a control. Luciferase signal, tumor size, and survival were monitored from day 0 to day 32. Tumor mass was again measured with a digital caliper, and the tumor sizes were calculated using the formula: volume = 0.5 × length × width2. Once tumor volume reached 1,500 cm3 or larger, the mice were euthanized and recorded as deceased for the survival curve.
SEND hEPO Experiment.
All hEPO pDNA constructs were created utilizing restriction enzyme digestions followed by PCR and cloning as described above. mRNAs were also created as described above via IVT of pDNA constructs. Both NSP-hEPO mRNA and SEND FVII-hEPO mRNA were loaded into one of three nanoparticles: LNPs, iPLNPs, or polymer NPs. The particles were then administered to C57BL6 mice via one of three injection routes: I.V., I.M., or S.Q., and blood was collected retro-orbitally using hematocrit tubes. Blood was then allowed to clot at RT for 30 min before being centrifuged at 2,000 G for 15 min at 4 °C to separate serum from whole blood. Serum was then collected and kept at −80 °C until the ELISA was performed. To quantify the protein yield of secreted Human Erythropoietin (hEPO), an ELISA was conducted according to the manufacturer’s protocol (DY286-05 and DuoSet Ancillary Reagent kit, R&D systems). Briefly, microplates were coated overnight with 100 μL of capture antibody (5 μg/mL). Microplates were washed three times with 1× wash buffer and blocked with reagent diluent for at least an hour. Following another wash step, 100 μL of appropriately diluted sample was added and incubated for 1 h at RT under shaking conditions. After incubation, sample wells were washed, and 100 μL of detection antibody (50 ng/mL) was applied to the samples at RT for 1 h under shaking conditions. After repeating the wash step, Streptavidin–HRP was added and incubated at RT for 20 min. Microplates were washed, and 100 μL of TMB substrate was added. Samples were incubated at RT for 20 min under shaking conditions, and reaction was stopped with 2 N sulfuric acid. Optical density was measured at 450 nm and wavelength correction at 540 nm. Results were plotted as hEPO concentration (mIU/mL) vs. time (h).
Statistical Analysis.
Statistical analysis was performed using a two-tailed unpaired t test for Fig. 1. For Figs. 4 and 5, a two-tailed unpaired t test and ANOVA with Tukey’s post hoc correction were used where appropriate.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
D.J.S. acknowledges financial support from the NIH National Institute of Biomedical Imaging and Bioengineering (NIBIB) (R01 EB025192-01A1). We acknowledge the UTSW Small Animal Imaging Shared Resource, which is supported in part by the NIH NCI Support Grant (P30 CA142543).
Author contributions
Q.C., L.F., R.C.W., and D.J.S. designed research; Q.C., L.F., A.V., E.G., E.E.L., E.K.R., X.W., J.R., S.M.L., T.W., W.E.M., E.A.B., and X.L. performed research; L.F., S.M.L., and R.C.W. contributed new reagents/analytic tools; Q.C., L.F., and D.J.S. analyzed data; R.C.W. and D.J.S. supervised research; and L.F., A.V., R.C.W., and D.J.S. wrote the paper.
Competing interests
D.J.S. is on the scientific advisory boards of ReCode Therapeutics and Tome Biosciences. D.J.S. owns stock or stock options in Signify Bio, ReCode Therapeutics, and Tome Biosciences. L.F. owns stock in Signify Bio. The University of Texas System has filed patent applications related to the described technology. L.F. and D.J.S. are co-founders of Signify Bio, which has licensed intellectual property from UT Southwestern.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Hajj K. A., Whitehead K. A., Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2, 17056 (2017). [Google Scholar]
- 2.Wei T., et al. , Delivery of tissue-targeted scalpels: Opportunities and challenges for in vivo CRISPR/Cas-based genome editing. ACS Nano 14, 9243–9262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Sun C., Wang C., Jankovic K. E., Dong Y., Lipids and lipid derivatives for RNA delivery. Chem. Rev. 121, 12181–12277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kariko K., Buckstein M., Ni H., Weissman D., Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Li B., Luo X., Dong Y., Effects of chemically modified messenger RNA on protein expression. Bioconjugate Chem. 27, 849–853 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Hou X., Zaks T., Langer R., Dong Y., Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A., Andresen J. L., Manan R. S., Langer R., Nucleic acid delivery for therapeutic applications. Adv. Drug Deliv. Rev. 178, 113834 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Han X., et al. , An ionizable lipid toolbox for RNA delivery. Nat. Commun. 12, 7233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eygeris Y., Gupta M., Kim J., Sahay G., Chemistry of lipid nanoparticles for RNA delivery. Acc. Chem. Res. 55, 2–12 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Chaudhary N., Weissman D., Whitehead K. A., mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nat. Rev. Drug Discov. 20, 817–838 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polack F. P., et al. , Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. New. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbett K. S., et al. , SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baden L. R., et al. , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. New. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillmore J. D., et al. , CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. New. Engl. J. Med. 385, 493–502 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Owji H., Nezafat N., Negahdaripour M., Hajiebrahimi A., Ghasemi Y., A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 97, 422–441 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Gilmore R., Blobel G., Transient involvement of signal recognition particle and its receptor in the microsomal membrane prior to protein translocation. Cell 35, 677–685 (1983). [DOI] [PubMed] [Google Scholar]
- 17.Walter P., Ibrahimi I., Blobel G., Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 91, 545–550 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walter P., Blobel G., Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 91, 551–556 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter P., Blobel G., Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 91, 557–561 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Görlich D., Prehn S., Hartmann E., Kalies K.-U., Rapoport T. A., A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 71, 489–503 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Viotti C., “ER to golgi-dependent protein secretion: The conventional pathway” In Unconventional Protein Secretion, Methods in Molecular Biology, A. Pompa, F. De Marchis, F. Eds. (Springer, New York, 2016), 10.1007/978-1-4939-3804-9_1 vol. 1459, pp. 3–29. [DOI] [PubMed] [Google Scholar]
- 22.Sun Z., Brodsky J. L., Protein quality control in the secretory pathway. J. Cell Biol. 218, 3171–3187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knappskog S., et al. , The level of synthesis and secretion of Gaussia princeps luciferase in transfected CHO cells is heavily dependent on the choice of signal peptide. J. Biotechnol. 128, 705–715 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Haryadi R., et al. , Optimization of heavy chain and light chain signal peptides for high level expression of therapeutic antibodies in CHO cells. PLoS One 10, e0116878 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Liu H., Yuan W., Cheng Y., Han W., Efficient production of CYTL1 protein using mouse IgGkappa signal peptide in the CHO cell expression system. Acta Biochim. Biophys. Sin. 48, 391–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybakova Y., et al. , mRNA delivery for therapeutic anti-HER2 antibody expression in vivo. Mol. Ther. 27, 1415–1423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kedmi R., et al. , A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 13, 214–219 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Dammes N., et al. , Conformation-sensitive targeting of lipid nanoparticles for RNA therapeutics. Nat. Nanotechnol. 16, 1030–1038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao J., et al. , Rapid synthesis of a lipocationic polyester library via ring-opening polymerization of functional valerolactones for efficacious siRNA delivery. J. Am. Chem. Soc. 137, 9206–9209 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Zhou K., et al. , Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc. Natl. Acad. Sci. U.S.A. 113, 520–525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan Y., et al. , Functional polyesters enable selective siRNA delivery to lung cancer over matched normal cells. Proc. Natl. Acad. Sci. U.S.A. 113, E5702–E5710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J. B., et al. , Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. 56, 1059–1063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Q., et al. , Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv. Mater. 30, e1805308 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Cheng Q., et al. , Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei T., Cheng Q., Min Y.-L., Olson E. N., Siegwart D. J., Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 11, 3232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S., et al. , Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater. 20, 701–710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D., et al. , Enhancing CRISPR/Cas gene editing through modulating cellular mechanical properties for cancer therapy. Nat. Nanotechnol. 17, 777–787 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Y., Xiong H., Zhang X., Cheng Q., Siegwart D. J., Systemic mRNA delivery to the lungs by functional polyester-based carriers. Biomacromolecules 18, 4307–4315 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., et al. , Differential glucose requirement in skin homeostasis and injury identifies a therapeutic target for psoriasis. Nat. Med. 24, 617–627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Fits L., et al. , Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J. Immunol. 182, 5836–5845 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Leader B., Baca Q. J., Golan D. E., Protein therapeutics: A summary and pharmacological classification. Nat. Rev. Drug Discov. 7, 21–39 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Godel P., Shimabukuro-Vornhagen A., von Bergwelt-Baildon M., Understanding cytokine release syndrome. Intensive Care Med. 44, 371–373 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Shimabukuro-Vornhagen A., et al. , Cytokine release syndrome. J. Immunother. C ancer 6, 56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf B., et al. , Therapeutic antibody glycosylation impacts antigen recognition and immunogenicity. Immunology 166, 380–407 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Schiestl M., et al. , Acceptable changes in quality attributes of glycosylated biopharmaceuticals. Nat. Biotechnol. 29, 310–312 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Dumont J., Euwart D., Mei B., Estes S., Kshirsagar R., Human cell lines for biopharmaceutical manufacturing: History, status, and future perspectives. Crit. Rev. Biotechnol. 36, 1110–1122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.