Significance
The small GTPase Rac2 is a key regulator of immune cell shape and function. Hyperactivating Rac2 mutations cause rare human immunodeficiencies though the underlying mechanisms are unclear. Based on fundamental cell biology in the fruit fly, we found that hyperactive Rac causes cells to eat other cells alive. We show that activated Rac2 also drives mouse and human macrophages to cannibalize activated T cells and that active Rac enhances engulfment of cancer cell targets by chimeric antigen receptor macrophages (CAR-M). These results suggest that Rac2E62K-stimulated cannibalism may contribute to Rac2+/E62K human immunodeficiency and enhance CAR-M cancer immunotherapy.
Keywords: Rac GTPase, phagocytosis, macrophage, lymphopenia, Drosophila
Abstract
The 21kD GTPase Rac is an evolutionarily ancient regulator of cell shape and behavior. Rac2 is predominantly expressed in hematopoietic cells where it is essential for survival and motility. The hyperactivating mutation Rac2E62K also causes human immunodeficiency, although the mechanism remains unexplained. Here, we report that in Drosophila, hyperactivating Rac stimulates ovarian cells to cannibalize neighboring cells, destroying the tissue. We then show that hyperactive Rac2E62K stimulates human HL60-derived macrophage-like cells to engulf and kill living T cell leukemia cells. Primary mouse Rac2+/E62K bone-marrow-derived macrophages also cannibalize primary Rac2+/E62K T cells due to a combination of macrophage hyperactivity and T cell hypersensitivity to engulfment. Additionally, Rac2+/E62K macrophages non-autonomously stimulate wild-type macrophages to engulf T cells. Rac2E62K also enhances engulfment of target cancer cells by chimeric antigen receptor-expressing macrophages (CAR-M) in a CAR-dependent manner. We propose that Rac-mediated cell cannibalism may contribute to Rac2+/E62K human immunodeficiency and enhance CAR-M cancer immunotherapy.
The Rho family GTPase Rac is a key node in the signaling and cytoskeletal networks that regulate cell shape and movement. The classic study by Ridley et al. (1) demonstrated that constitutively active Rac (Rac1G12V), expressed in serum-starved Swiss 3T3 fibroblasts causes peripheral actin polymerization, membrane ruffling, and macropinocytosis. Rac proteins are deeply conserved in evolution, likely present in the earliest eukaryotes (2). Human and Drosophila Rac proteins are nearly identical, and human Rac can function in Drosophila (3).
Drosophila border cells have long served as an in vivo model for elucidating Rac functions. The requirement for Rac in cell motility was demonstrated in this group of 6 to 10 follicle cells that migrate ~150 μm over ~4 h during Drosophila oogenesis (4). It is now clear that Rac is a nearly universal regulator of cell migration (5) and is required for individually migrating cells such as neutrophils and macrophages as well as collectively migrating cells like border cells and neural crest cells (6–12). Rac is also essential for wound healing, neuronal migration and pathfinding, angiogenesis, and tumor metastasis (13–15).
The human genome encodes three Rac proteins, Rac1, Rac2, and Rac3, which are >95% identical but differ in their expression patterns: Whereas Rac1 is ubiquitously expressed, Rac2 is expressed predominantly in hematopoietic cells, and Rac3 is enriched in neural tissue. Rac2 represents 90% of the Rac protein in immune cells where Rac1 and Rac2 have both redundant and nonredundant functions (16, 17). Rac activity is essential for immune cell motility, which is in turn required for B and T progenitor cells to leave the bone marrow, circulate, and extravasate into the spleen and thymus where B and T cells mature and differentiate, respectively. Cell migration is also essential for cells to move from one compartment to another within the spleen and thymus, to intravasate and re-enter the blood after undergoing positive and negative selection, and then to infiltrate tissues in response to injury or infection. Rac activity is also essential for monocytes, neutrophils, and macrophages to undergo chemotaxis (18, 19). Some of the evidence for these varied roles of Rac in the immune system comes from analysis of the phenotypes of mice with dominant-negative or recessive loss of function mutations in Rac1 and/or Rac2 (20), as well as human patients with mutations in Rac2 (21).
Human patients with dominant-negative or recessive loss-of-function mutations in Rac2 have health problems consistent with the requirement for Rac2 for immune cell migration and survival (17). Paradoxically though, patients with activating mutations in Rac2 also have immune deficiencies. For example, patients with a dominant Rac2+/E62K mutation present with recurrent lung infections and lymphopenia (22). When expressed in cultured cells, Rac2E62K causes ~1.5-fold increase in Rac activity (22). Rac2+/E62K patient-derived neutrophils exhibit increased H2O2 production and macropinocytosis (22), consistent with hyperactivity for known Rac2 functions. Rac2+/E62K mice exhibit similar defects including lymphopenia; however, no defects in T cell trafficking from the bone marrow to the thymus or maturation within the thymus were found. Therefore, the patients’ lymphopenia remains unexplained (22).
Border cells require Rac activity for F-actin-mediated protrusions that drive cell motility (4). Focal activation of Rac in a single cell using photoactivatable Rac causes local F-actin polymerization and can steer the whole cluster (3). However, sustained and uniform expression of constitutively activated Rac impairs border cell motility (23), similar to T cells (24) and neutrophils (22).
Here, we report that high levels of hyperactive Rac in the border cells cause them to engulf living neighbors and destroy the entire egg chamber tissue. We further show that expressing Rac2E62K in human macrophage-like cells is sufficient to cause them to cannibalize Jurkat T cell leukemia cells. Primary macrophages from Rac2+/E62K mice engulf and kill living primary T cells. Interestingly, Rac2E62K expression also renders T cells hypersensitive to engulfment. Rac2+/E62K macrophages also non-autonomously enhance whole T-cell engulfment by Rac2+/+ macrophages. We further show that Rac2E62K expression in CAR (chimeric antigen receptor)-macrophages (CAR-M, aka CAR-P) enhances receptor-dependent engulfment and killing of target cancer cells. We propose cannibalism as a possible mechanism contributing to the unexplained Rac2+/E62K patient T cell lymphopenia. The data also suggest hyperactive Rac may enhance CAR-M cancer immunotherapy.
Results
Expression of Active Rac in a Few Cells Is Sufficient to Cause Tissue Destruction.
Border cells are a group of 4 to 8 migratory cells that originate within an epithelium of ~850 epithelial follicle cells that surround 16 germ cells in a structure called an egg chamber (Fig. 1A). At developmental stage 9, the border cells delaminate from the epithelium and migrate ~150 µm, squeezing in between polyploid nurse cells until they reach the oocyte by stage 10 (Fig. 1B and Movie S1). Inhibiting all three fly Rac proteins prevents migration (4, 23) (Fig. 1C) by blocking both lead cell protrusion (Fig. 1 D and E) and follower cell crawling (25). Constitutively active Rac (RacG12V) also blocks migration (Fig. 1 F and G) (23), presumably by preventing the asymmetry in Rac activity that is required for polarized protrusion.
Fig. 1.
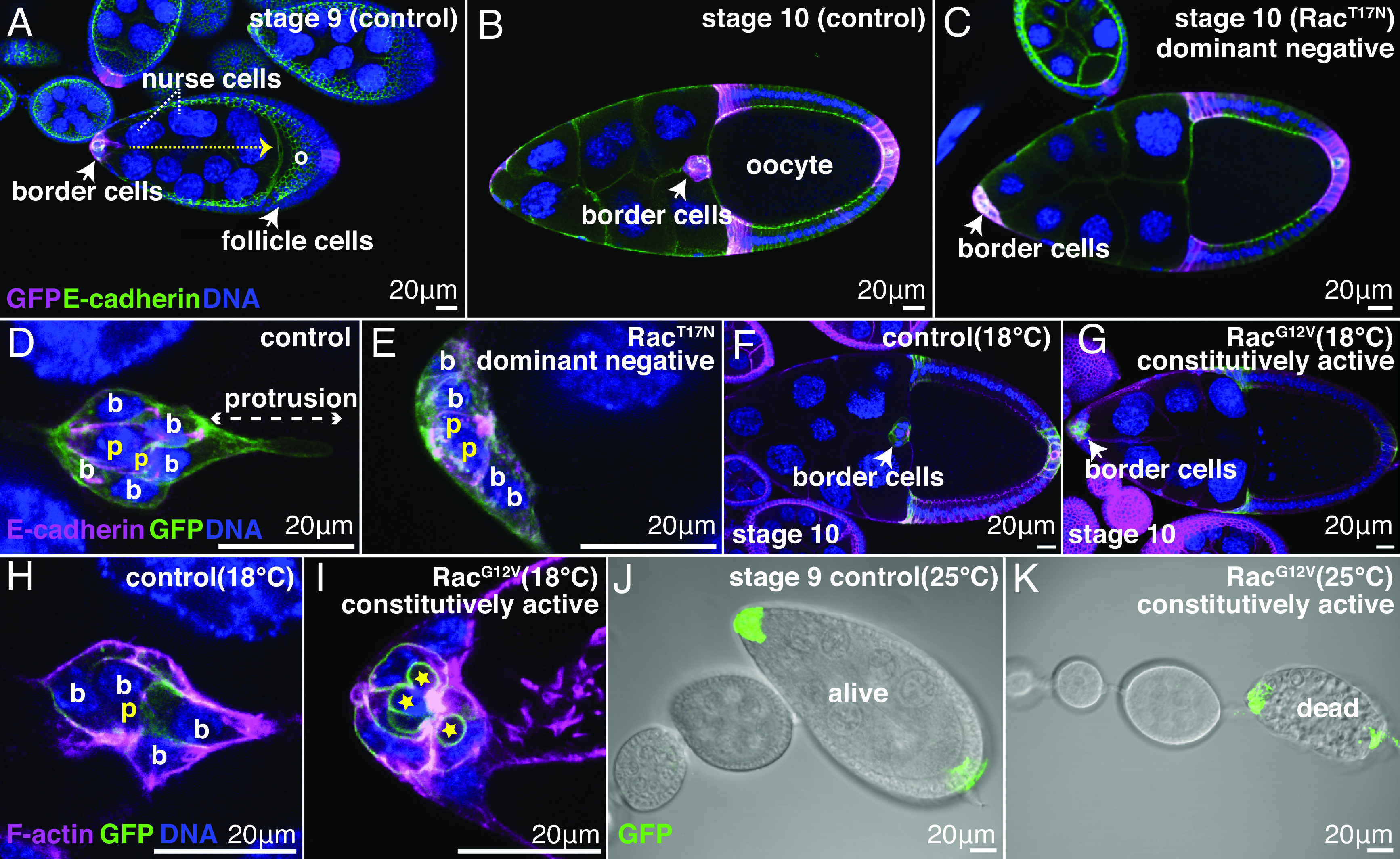
Constitutively active Rac (RacG12V) in a subset of follicle cells causes wholesale tissue destruction. slbo-Gal4; UAS-PLCδ1-PH-GFP egg chambers of the indicated stages and genotypes. (A–I) Confocal micrographs of (A) UAS-lacZ control showing border cell migration path (dotted arrow). Oocyte (o). (B) Stage 10 UAS-lacZ control showing completed migration. (C) Stage 10, UAS-Rac1T17Nshowing failed migration. (D) Control UAS-lacZ cluster. Border cells (b) surround and carry non-motile polar cells (p). The lead border cell protrudes, initiating migration. (E) UAS- Rac1T17N clusters lack protrusions. (F) Completed migration in control, UAS-lacZ. (G) Failed border cell migration in UAS-RacG12V. (H) Normal cluster morphology of UAS-lacZ control, (I) Abnormal cluster morphology in UAS-Rac1G12V. Phalloidin labels F-actin (magenta in H and I). (J and K) DIC imaging of (J) normal egg chamber morphology in a UAS-lacZ control and (K) dead UAS-Rac1G12Vegg chamber. Anterior is on the Left.
Border cells normally surround and carry two non-migratory polar cells (Fig. 1D). In contrast, RacG12V-expressing border cells exhibited complex morphological abnormalities including apparent fragmentation of polar cells (Fig. 1 H and I). Surprisingly, increasing the temperature from 18 to 25 °C, which increases the RacG12V expression level, resulted in non-autonomous destruction of the entire egg chamber (Fig. 1 J and K). Overexpression of wild-type Rac (RacWT) caused a similar phenotype, albeit at lower frequency (SI Appendix, Fig. S1). This effect was perplexing because Rac activity is generally not lethal to cells, and intentionally killing border cells by expressing the pro-apoptotic gene Reaper (23) or by laser ablation (26) does not cause egg chamber death but rather permits development to stage 14.
RacG12V-Stimulated Tissue Destruction Requires the Engulfment Receptor Draper.
In addition to promoting cell migration, Rac is one of hundreds of components of the phagocytic cup (27). Follicle cells normally engulf and kill nurse cells near the end of Drosophila oogenesis (28) via the engulfment receptor Draper (Drpr; Fig. 2A). So could RacG12V-expressing border cells be killing the nurse cells by inappropriately engulfing them?
Fig. 2.
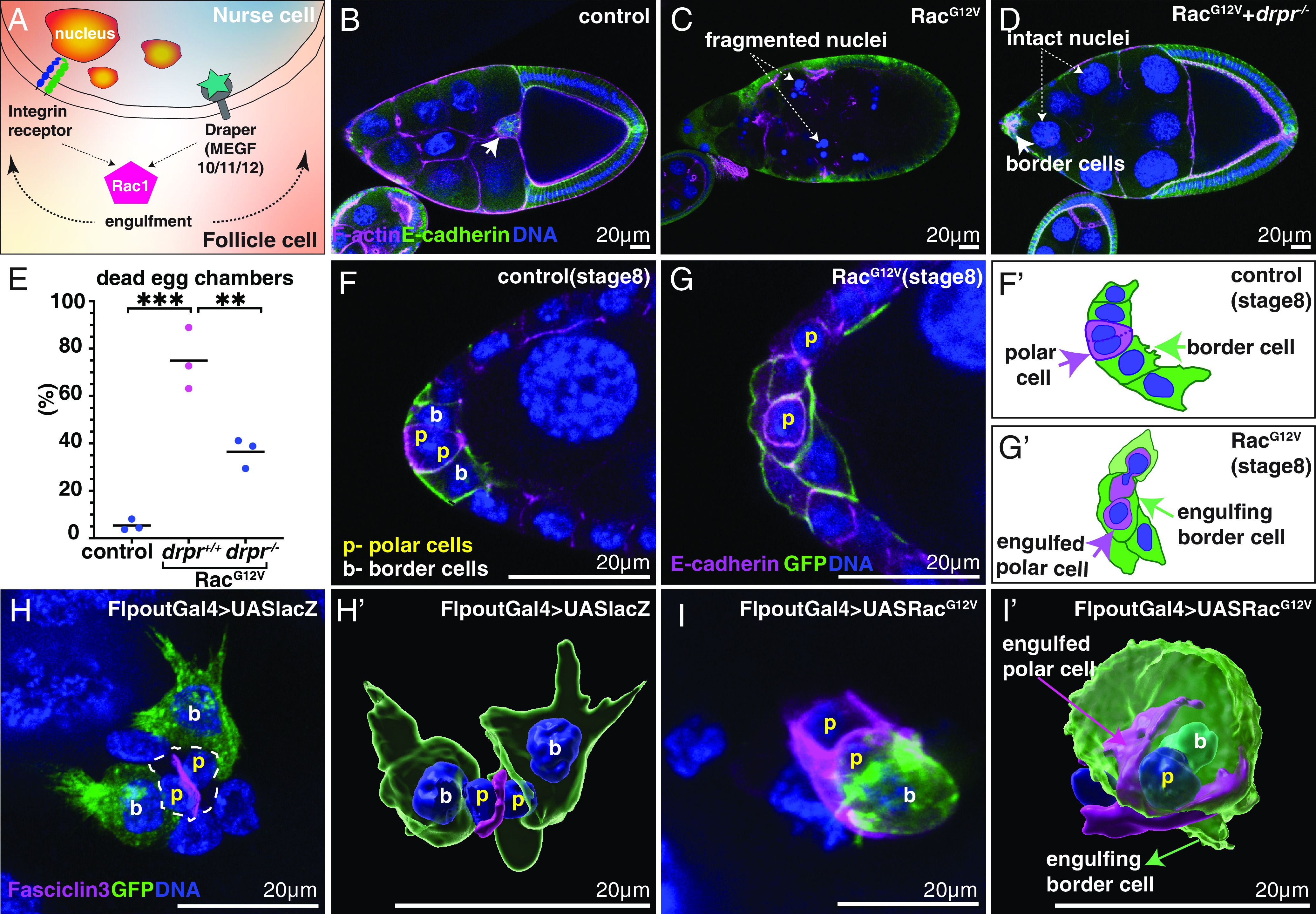
Constitutively active Rac (RacG12V) causes germline (nurse cells and oocyte) death and polar cell engulfment. (A) Schematic showing Draper-receptor-mediated engulfment of nurse cells by follicle cells. (B–G) Confocal micrographs of slbo-Gal4 egg chambers labeled with the indicated markers. Anterior is Left. (B) UAS-lacZ control exhibits normal morphology and completed border cell migration at stage 10. (C) Dying UAS-Rac1G12V egg chamber. (D) The homozygous drpr mutation rescues normal egg chamber morphology but not border cell migration (arrow) in UAS- Rac1G12V. (E) Quantification of dead egg chambers with or without UAS-RacG12V in the wildtype or drpr mutants. All data were analyzed by one-way ANOVA with Tukey’s multiple comparisons test. ** indicates P < 0.005 and ***P < 0.0005. N = 87 (control), 53 (RacG12V) and 66 (RacG12V; drpr−/−) egg chambers from three independent experiments (dots). (F and G) Stage 8 slbo-Gal4; UAS-PLCδ1-PH-GFP egg chambers showing (F) border cells (b) adjacent to polar cells (p) and (G) UAS- Rac1G12V border cells engulf polar cells (p). (F’ and G’) Schematic drawings of F and G. (H) Clonal expression of either UAS-lacZ (control, H and H’) or UAS-Rac1G12V (I and I’) in the GFP-positive subset of border cells (green). Fasciclin 3 (magenta in H–I’) is restricted to the interface between two polar cells in controls (H and H’) whereas border cells expressing RacG12V (I and I’) engulf polar cells, delocalizing Fasciclin 3. H’ and I’ are 3D Imaris surface renderings of H and I.
To test this idea, we expressed RacG12V in border cells in drpr homozygous mutants. In well-fed control flies, egg chamber death during mid oogenesis is rare (Fig. 2B). In contrast, the majority of slboGal4;UAS-RacG12V-expressing egg chambers contained dying nurse cells with condensed and fragmented nuclei and overall abnormal egg chamber morphology (Fig. 2C). However, when homozygous mutant for drpr, the majority of slboGal4;UAS-RacG12V egg chambers exhibited normal morphology (Fig. 2 D and E), except that border cells still failed to migrate (Fig. 2D). We conclude that active Rac kills the germline in a process that depends on Drpr-mediated target recognition.
Control border cells develop within the follicle cell epithelium in response to a cytokine secreted by the polar cells (29) (Fig. 2 F and F’). High magnification imaging revealed that ~75% of slboGal4;UAS-RacG12V border cell clusters engulfed at least one neighboring polar cell (Fig. 2 G and G’), which is never observed in controls (Fig. 2F). Together, these results demonstrate that expression of active Rac in border cells causes them to engulf living neighbors.
To determine how many RacG12V-expressing cells would be sufficient to engulf another living cell, we generated clones of various sizes using the Flpout technique. Control border cell clones expressing GFP and lacZ exhibited normal morphologies, extended outward-directed protrusions, and carried rather than engulfed polar cells (Fig. 2 H and H’ and Movie S2). By contrast, even a single border cell expressing RacG12V and GFP could engulf a polar cell (Fig. 2 I and I’ and Movie S3).
Active Rac Is Sufficient to Cause Human Macrophages to Engulf Living Cells.
Rac mediates phagocytosis in organisms ranging from single-celled amoebae to human macrophages (30–32). So we wondered whether activated Rac would be sufficient to cause a human macrophage to eat a living cell. To test this idea, we transduced HL60 cells with a lentivirus expressing either a membrane-associated GFP (lck-GFP) alone as a negative control, or lck-GFP together with Rac2E62K. We differentiated the cells into macrophage-like cells using an established protocol (33, 34) (SI Appendix, Fig. S2) and co-cultured them with human Jurkat T cell leukemia cells (Fig. 3A). We then counted the fraction of macrophages that engulfed at least one Jurkat T cell. Engulfment events were relatively rare in control lck-GFP-expressing cells (Fig. 3B), whereas expression of Rac2E62K increased the fraction of macrophages that contained Jurkat cell fragments (Fig. 3C) from ~10 to ~70% (Fig. 3D). We measured a 2.5-fold increase in RacGTP in Rac2E62K-expressing cells compared to controls (Fig. 3E), consistent with Hsu et al. (22). Overexpressing RacWT or RacG12R, which is another activating mutation, also stimulated Jurkat cell engulfment (Fig. 3 F–I), although Rac2E62K had the strongest effect. We conclude that, as in Drosophila, hyperactive Rac also promotes cannibalistic behavior in human cells.
Fig. 3.
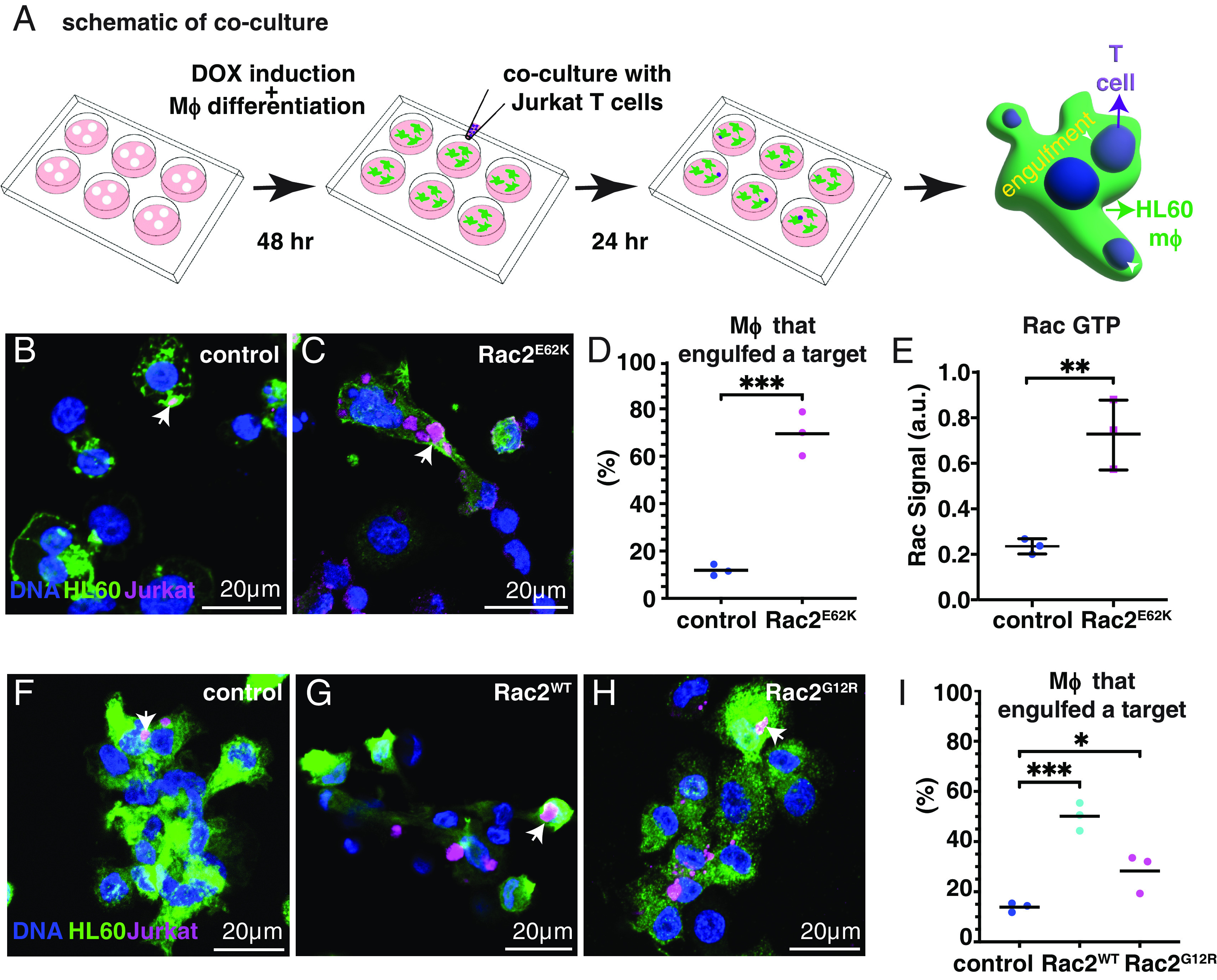
Constitutively active Rac2E62K in HL60-derived macrophage-like cells enhances engulfment and killing of leukemic Jurkat T cells. (A) Schematic of experimental design. (B–I) HL60-derived macrophage-like cells expressing Lck-GFP (green) and the indicated Rac2 variant co-cultured with mCherry-labeled (magenta) Jurkat T cells. DNA (labeled with Hoechst, blue) (B) control (Lck-GFP without Rac2 variant) (C) Lck-GFP with Rac2E62K (D) Quantification of N = 614 (control) and 220 (RacE62K) cells from three independent experiments (dots). (E) GTP-bound (active) Rac measured from differentiated HL60 macrophage lysates using the G-LISA Rac 1,2,3 Activation Assay Kit (Cytoskeleton, Inc.). Each dot is an average of two technical replicates. Statistics: Unpaired t test. **P < 0.005 and ***P < 0.0005. (F) Control (Lck-GFP alone), (G) Rac2WT, (H) Rac2G12R, (I) Quantification of engulfment. N = 1,140 (control), 223 (RacWT), and 653 (RacG12R) samples from three independent experiments (dots). Engulfment data (F–H) were analyzed by one-way ANOVA with Tukey’s multiple comparisons test. * indicates P < 0.05 and ***P < 0.0005.
Rac2E62K Alters the Macrophage Transcriptional State.
Together with Cdc42 and Rac1, Rac2 localizes to the phagocytic cup and promotes phagocytosis by stimulating local F-actin polymerization (30). So Rac2E62K likely stimulates hyper-phagocytosis directly. In addition, Rac2E62K might increase phagocytic activity by changing the cell state. To test whether Rac2E62K causes substantial changes in the macrophage transcriptome, we compared RNAseq profiles for control and Rac2E62K expressing HL60-derived macrophage-like cells. >1,000 genes were differentially expressed between these two cell populations (Fig. 4A). 1,551 genes were upregulated and 790 were downregulated ≥1.5X. The most significantly upregulated pathways were inflammatory, including TNF-α signaling via NF-κB (Fig. 4B). Gene ontology (GO) analysis showed significant enrichment of some genes associated with the cellular process of phagocytosis including receptors that mediate migration and phagocytosis, such as CD302, AXL, and TYRO3, as well as signaling and cytoskeletal regulators such as MYO1G and SH3BP1 (Fig. 4C). Antibody staining confirmed upregulation of CD302 protein (Fig. 4 D and E). Enriched molecular functions included lipid metabolism and ion transporters (SI Appendix, Fig. S3). We conclude that Rac2E62K expression alters the macrophage transcriptional state.
Fig. 4.
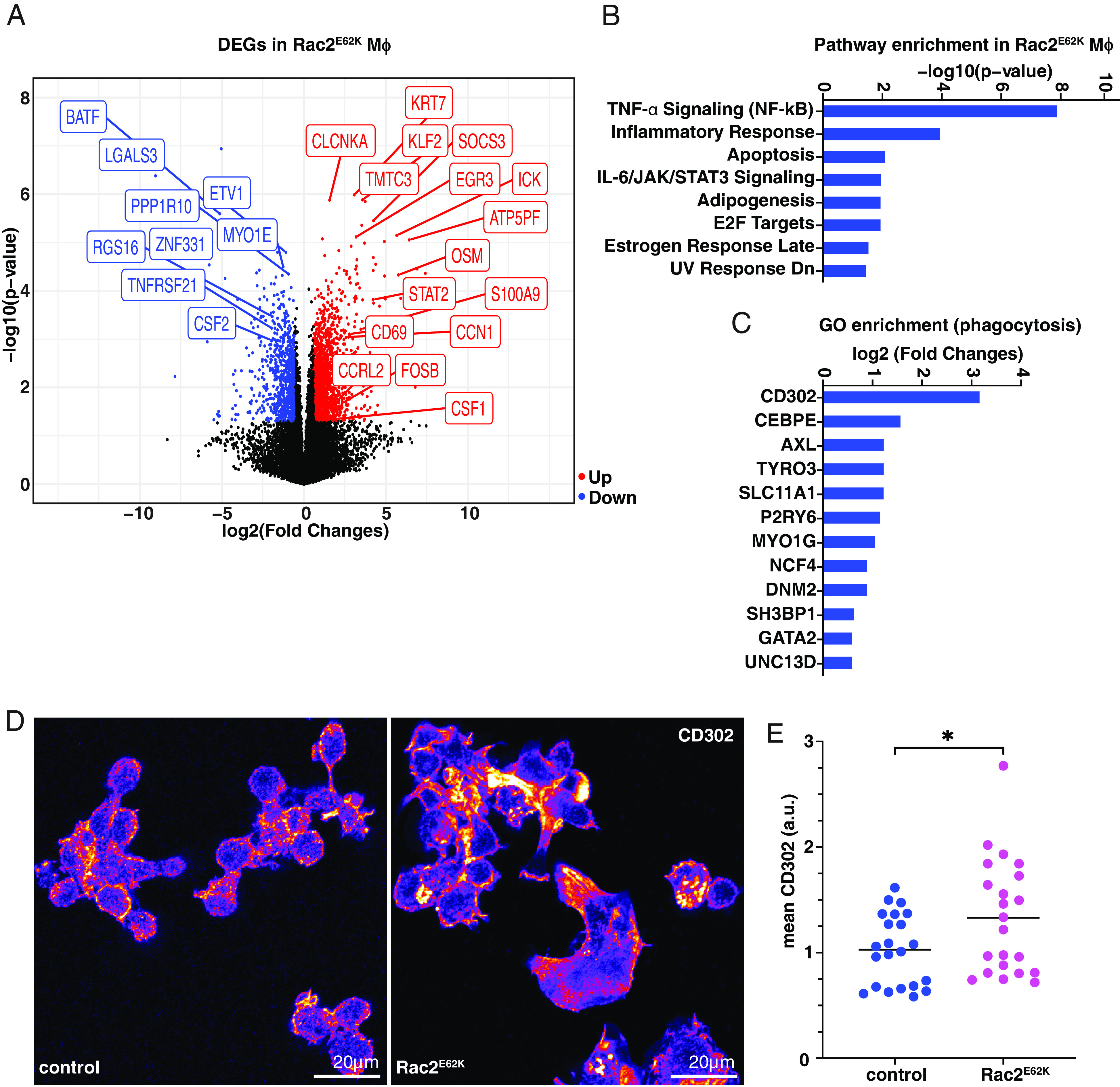
Effect of Rac2E62K on gene expression in HL60-derived macrophage-like cells. (A) Volcano plot representation of DEGs. Red dots (upregulated ≥1.5X), blue dots (downregulated ≥1.5X), and black dots (mRNAs changed <1.5X and insignificant p-value) in Rac2E62K-expressing cells. Data were mapped, analyzed, and extracted from the Subio Platform [v1.24 (Subio, Inc., Japan)] and plotted in R. Select DEGs are labeled. (B) Pathway enrichment analysis was performed in Enrichr (Ma’ayan laboratory) on upregulated genes with a fold change (FC) >3 and the Molecular Signatures Database (MSigDB) Hallmark 2020 output data were plotted. (C) GO term “phagocytosis” in DEGs and upregulated genes with FC > 1.5 and P-value < 0.05. (D) CD302 expression in (control) HL60-LckGFP or Rac2E62K-Lck-GFP macrophages. (E) Mean CD302 fluorescence in control and Rac2E62K-expressing macrophages. Statistics: Unpaired t test, * indicates P < 0.05.
Rac2+/E62K Bone Marrow–Derived Macrophages Engulf Living Rac2+/E62K T Cells.
Hsu et al. established a Rac2+/E62K mouse model (22), which recapitulates the human syndrome, including reduced circulating T cells and hyperactivated neutrophils. Neither T cell trafficking from the bone marrow to the thymus nor T cell maturation within the thymus is impaired in Rac2+/E62K mice, and therefore, the peripheral lymphopenia remains unexplained. We also counted viable and dead CD3+ T cells from the thymus and found no measurable difference between Rac2+/+ and Rac2+/E62K mice (SI Appendix, Fig. S4).
Our results in Drosophila egg chambers and human macrophage-like cells hinted at hyperengulfment of lymphocytes by macrophages as a possible explanation. To test this idea, we obtained the Rac2+/E62K mice, isolated bone marrow–derived monocytes, and differentiated them into macrophages. We isolated splenic T cells from another animal and labeled them with both CellTrace (Far Red) and pHrodo (Red), which is a dye that fluoresces only within the acidic environment of the lysosome. We stimulated the macrophages with interferon‐gamma (IFN‐γ), co-cultured them with T cells (Fig. 5A), and compared engulfment in Rac2+/+ and Rac2+/E62K cells by fixed (Fig. 5 B and C) and live imaging (Fig. 5 D and E and Movies S4 and S5). Rac2+/E62K macrophages engulfed more Rac2+/E62K T cells than wild-type controls. This effect was significant whether we measured pHrodo intensity (Fig. 5 D–F), the depletion of T cell targets after 10 h of co-culture (Fig. 5G), or the percentage of macrophages that engulfed a target (Fig. 5H).
Fig. 5.
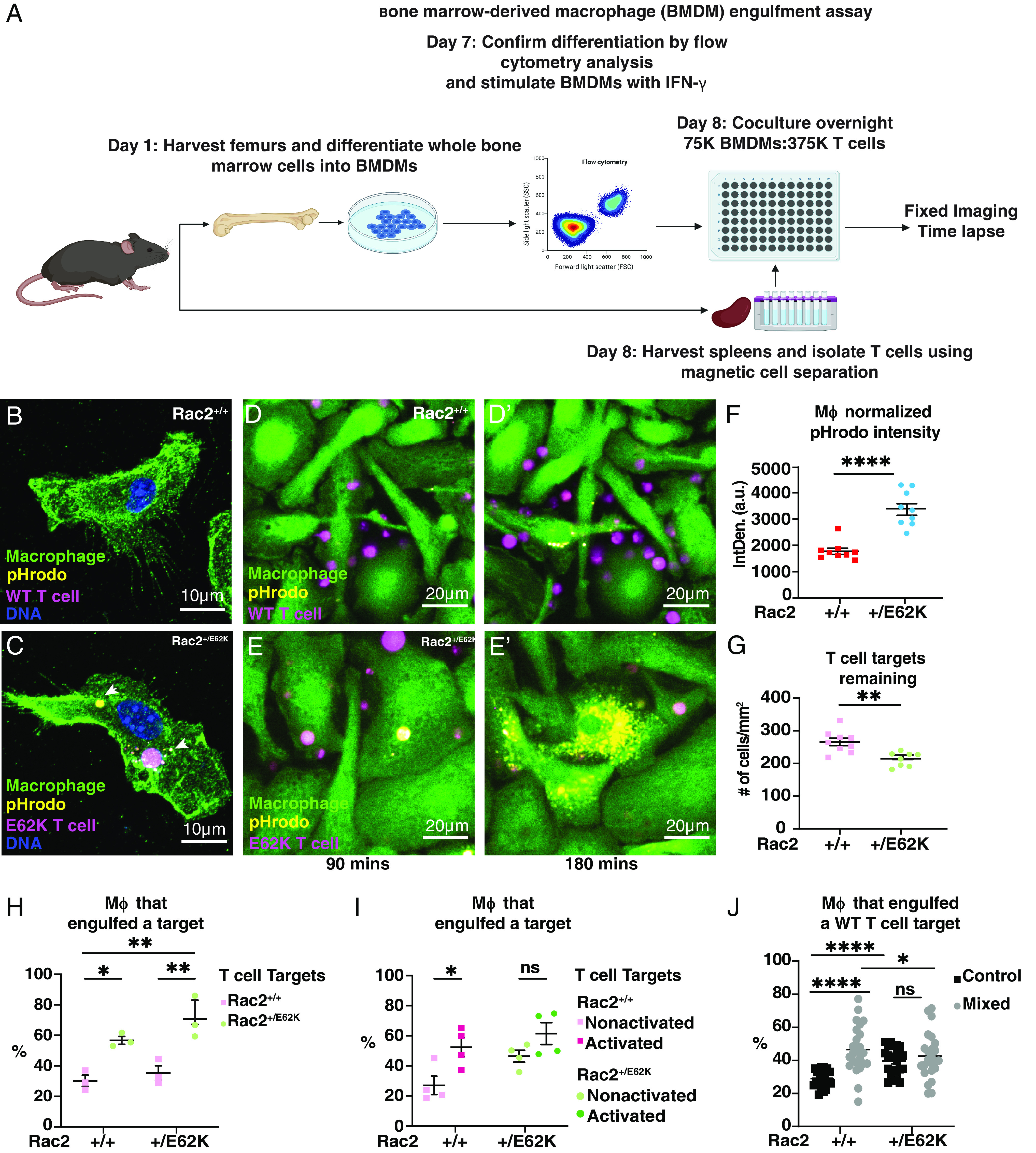
Rac2E62K enhances phagocytosis of T cells by macrophages and sensitizes T cells to engulfment. (A) Schematic of the phagocytosis assay. Rac2+/+ (WT) and Rac2+/E62K BMDMs were incubated overnight with T cells labeled with CellTrace Far Red and pHrodo Red (B and C). Representative confocal images of BMDMs. (D–E’) 10-h time-lapse imaging of macrophage (green) and T cell (magenta) coculture. pHrodo (yellow). Images correspond to Movies S4 and S5. (F) pHrodo intensity as measured by integrated density (a.u.) and normalized to the number of macrophages in the field of view. Each point represents a measurement from one field of view. (G) Total number of T cell targets remaining at the end of a 10-h engulfment time-lapse experiment. Each point represents a measurement from one field of view. (H) Percentage of macrophages that engulfed at least one T cell target. Each point represents the average from two wells. (I) Percentage of macrophages that engulfed at least one T cell target (activated or non-activated). Each point represents the average from two wells. (J) Percentage of macrophages that engulfed at least one WT T cell in control or mixed macrophage conditions. Each point represents a technical replicate. n = 3 biological replicates for experiments (H–J). *, **, and **** indicate P < 0.05, 0.005, and 0.00005 respectively by either an unpaired t test or a two-way ANOVA followed by Sidak’s multiple comparison test. Error bars indicate mean ± SEM.
We then tested whether the phenotype was autonomous to the macrophages, to the T cells, or both. Intriguingly, we found that the enhanced engulfment was due to a combination of increased phagocytosis by Rac2+/E62K macrophages and increased susceptibility to engulfment by Rac2+/E62K T cells, with the greatest engulfment observed when both cell types were Rac2+/E62K (Fig. 5I).
Macrophages engulf activated T cells to limit inflammatory responses (35) and promote tolerance (36). Rac is activated downstream of the T cell receptor (37), so the Rac2E62K mutation might mimic chronic activation. To test whether activating T cells renders them more susceptible to engulfment, we treated wild-type and Rac2+/E62K T cells with anti-CD3 and anti-CD28 antibodies (38) for 3 d, collected the cells and washed them to remove any remaining antibody, and then co-cultured them with macrophages. We found that activated Rac2+/+ T cells were engulfed to about the same extent as Rac2+/E62K T cells (Fig. 5I). Furthermore, anti-CD3 and anti-CD28 pretreatment only slightly increased engulfment of Rac2+/E62K T cells (Fig. 5I), consistent with an interpretation that they are already activated. We conclude that activation sensitizes T cells to engulfment to a similar degree as the Rac2+/E62K mutation.
We next tested whether mixing Rac2+/E62K BMDMs with wild-type BMDMs would alter their behavior. Co-culture with Rac2+/E62K BMDMs increased T cell engulfment by wild-type BMDMs (Fig. 5J).
Rac2+/E62K Macrophages Are in a Unique State.
Macrophages have classically been described as M1 (inflammatory) and M2 (anti-inflammatory) though there are substates, possibly intermediate states, and the states may be dynamic (39–41). To characterize the Rac2+/E62K BMDMs, we first measured RacGTP and found ~two-fold increase relative to Rac2+/+ cells (Fig. 6A), similar to HL60-derived macrophage-like cells (Fig. 3E) and Rac2E62K-transfected COS cells (22). We then isolated BMDMs from Rac2+/+ mice and either did not stimulate them or induced them to a classic M1 state using IFN‐γ and LPS or to an M2 state with IL-4. We also isolated primary BMDMs from Rac2+/E62K mice. We then carried out RNAseq and identified differentially expressed genes (DEGs) characteristic of M1, M2, and/or Rac2+/E62K. Remarkably, a ~two-fold increase in Rac2 activity was sufficient to cause differential expression of 2,259 genes (930 up and 1,329 down) (Fig. 6B), indicating a significant change in cell state. GO enrichment analysis with search terms “phagocytosis” and “positive regulation of phagocytosis” returned Ticam2, Rap1gap, Mfge8, Tlr4, and Pik3ca as top hits (Fig. 6C). Similar to the HL60 cells, TNF-α signaling and inflammatory responses were top enriched pathways (Fig. 6D). The twofold increase in Rac2 activity also caused gene expression changes that significantly overlapped with cytokine stimulation. A total of 1087 of the Rac2+/E62K DEGs overlapped with those differentially expressed in the M1 state, while 75 overlapped with the M2 state specifically, and 485 DEGs were common to all three states compared to unstimulated control BMDMs (Fig. 6E). Common upregulated GO enrichment terms (molecular function) between M1 and Rac2+/E62K included genes associated with membrane transporter activity (SI Appendix, Fig. S5). We conclude that Rac hyperactivation causes macrophages to adopt a uniquely stimulated state.
Fig. 6.
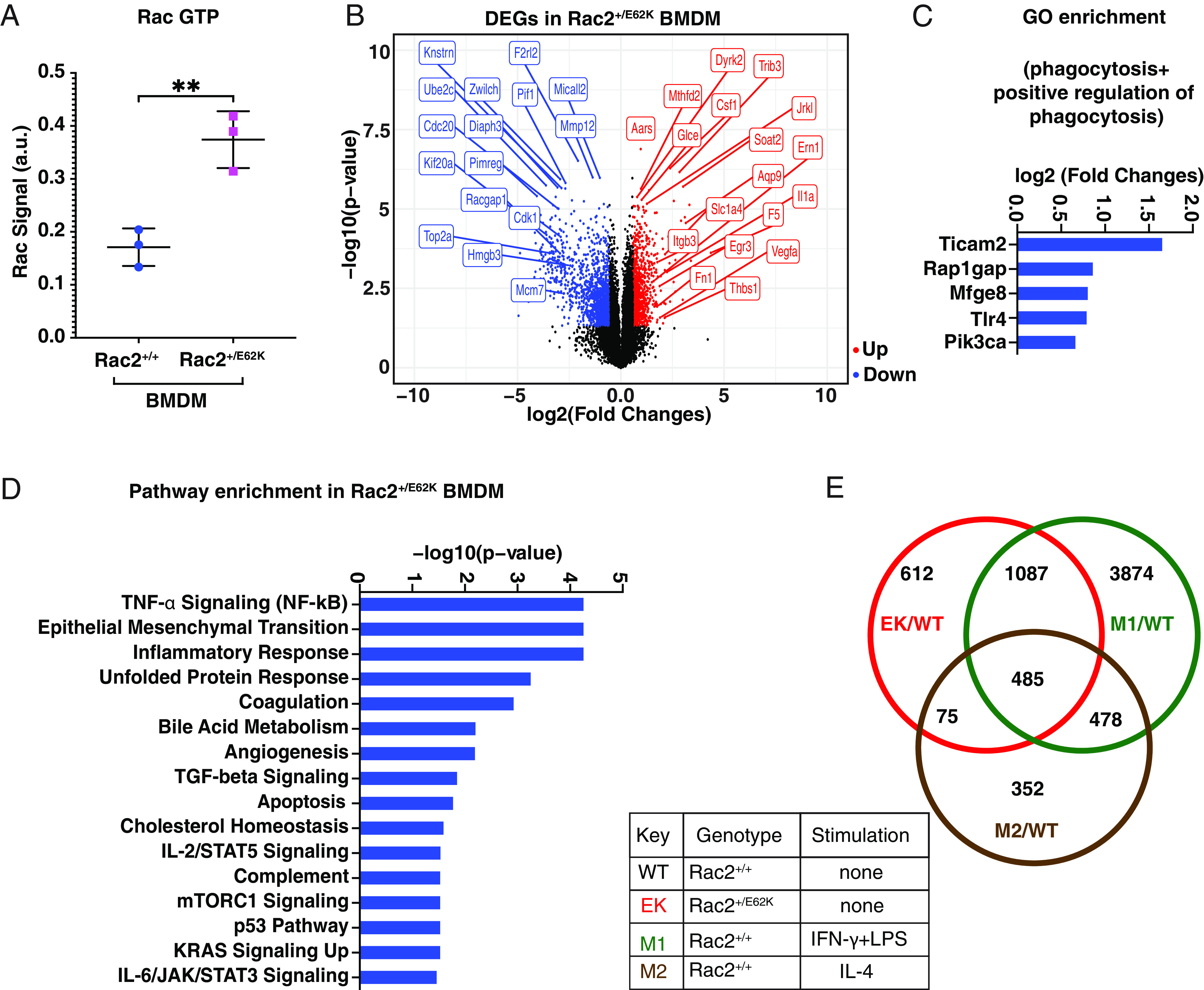
Effect of Rac2+/E62K on Rac activity and gene expression in primary BMDMs. (A) GTP-bound (active) Rac was measured from Rac2+/+ and Rac2+/E62K BMDM lysates using the G-LISA Rac 1,2,3 Activation Assay Kit (Cytoskeleton, Inc.). Data were analyzed by unpaired t test. ** indicates P < 0.005. (B) Volcano plot representation of differential expression analysis of genes in Rac2+/+ and Rac2+/E62K macrophages. Data was mapped, analyzed, and extracted from the Subio Platform (v1.24) and plotted in R. Red dots (upregulated genes), blue dots (downregulated genes) and black dots (insignificant genes). Select DEGs are labeled. (C) GO term phagocytosis and positive regulation of phagocytosis in DEGs and upregulated genes with (FC > 1.5) and P-value < 0.05. (D) Pathway enrichment analysis on DEGs was performed in Enrichr (Ma’ayan laboratory) on upregulated genes with (FC > 3) and the Molecular Signatures Database (MSigDB) Hallmark 2020 output data were plotted. (E) DEGs in groups EK/WT, M1/WT, and M2/WT were co-analyzed to assess the inflammatory profile of Rac2+/E62K BMDMs.
Rac2E62K Enhances CAR-M Phagocytosis of Target Cancer Cells.
CAR therapies are promising cancer immunotherapies (42). In CAR-T therapy, T cells are removed from a cancer patient, engineered ex vivo to express a CAR that recognizes a tumor antigen, and then re-infused into the patient. While there have been exciting successes, there are limitations, including physical exclusion of T cells from solid tumors, antigen escape, and cytokine storms (42, 43). A newer cousin is CAR-M in which patient monocytes and/or macrophages are engineered to express a CAR (44–46). Potential advantages include the unrivaled ability of macrophages to infiltrate the tumor microenvironment, removal of tumor cells by engulfment, and the possibility of initiating a broader adaptive immune response via antigen presentation. A limitation of CAR-M however is that CAR macrophages tend to nibble on cells rather than engulfing and killing them, in a process known as trogocytosis (44). Therefore, enhancing whole target cell engulfment and killing is an important goal.
To test whether expressing Rac2E62K in CAR-M cells could enhance their ability to engulf and kill target cells, we isolated BMDMs from Rac2+/E62K mice and from littermate controls, and used lentiviral infection to express an anti-CD19 CAR or a GFP-CAAX control (Fig. 7A). We then co-cultured the cells with CD19+ Raji B cell lymphoma cells and carried out live imaging. Little to no engulfment was observed in GFP-CAAX expressing macrophages in the absence of a CAR, regardless of whether they were Rac2+/+ (Fig. 7B and Movie S6) or Rac2+/E62K (Fig. 7C and Movie S7). Expression of the CAR significantly enhanced whole cell engulfment (Fig. 7D and Movie S8), as previously described (44, 45). Expression of the CAR in Rac2+/E62K macrophages further stimulated whole cell engulfment (Fig. 7 E and F and Movie S9). We conclude that Rac activity specifically enhances CAR-dependent engulfment of target cells.
Fig. 7.
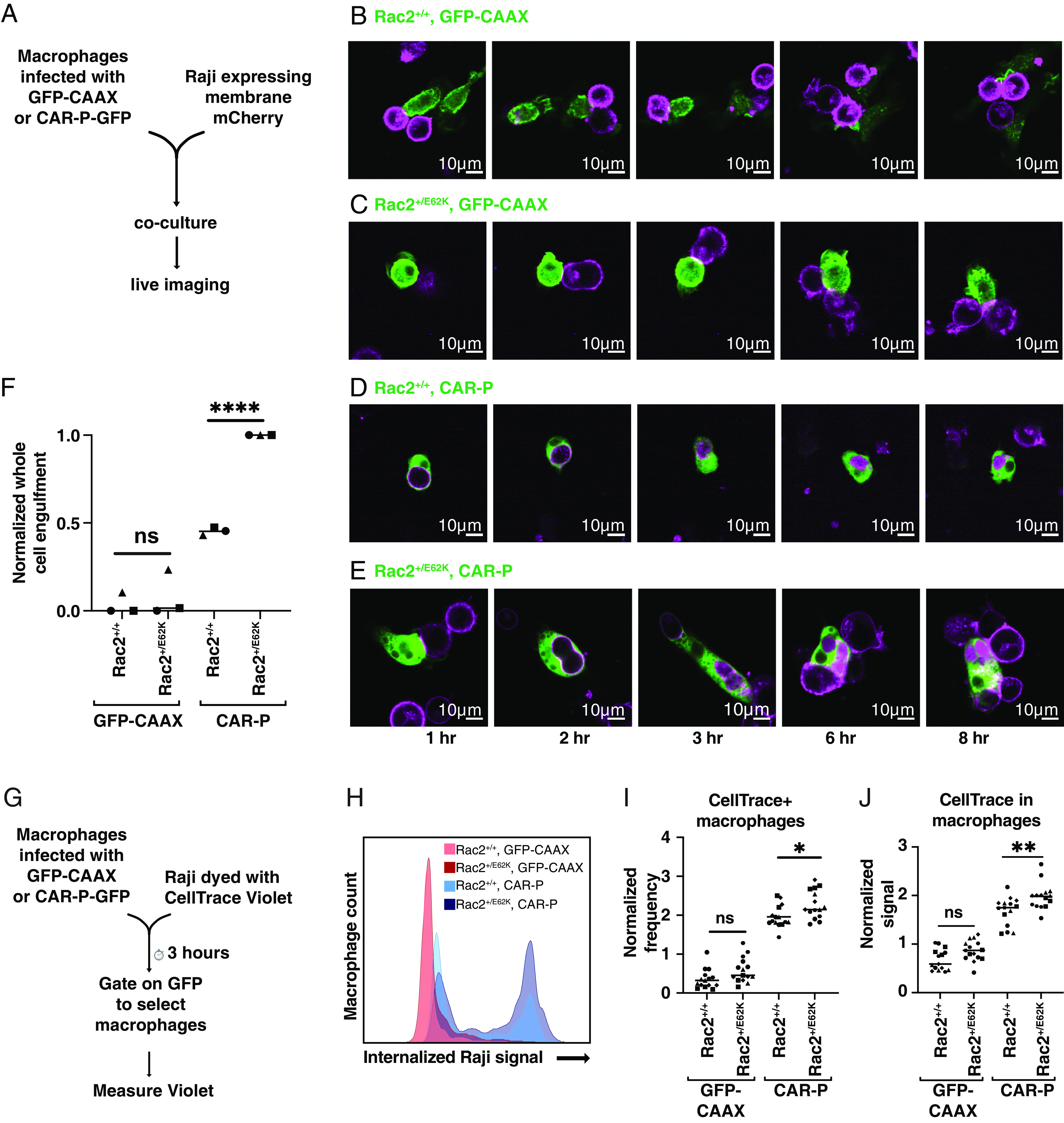
Rac2+/E62K enhances whole cell phagocytosis of cancer cells by CAR macrophages. (A) Schematic of the experiment in (B–F). Rac2+/+ and Rac2+/E62K BMDMs were infected with lentivirus encoding membrane-tethered GFP (GFP-CAAX) or a CAR that recognizes the B cell surface antigen CD19 and signals through the Fc Receptor intracellular signaling domain. (B–E) To distinguish between trogocytosis or whole cell phagocytosis, control GFP-CAAX (B and C) or CAR-GFP (D and E) BMDMs (green) were mixed with mCherry-CAAX-Raji B cell lymphoma cells (magenta) and imaged via spinning disc confocal. Images correspond to Movies S6–S9. (F) Graph indicates the number of whole Raji cells eaten per macrophage in 8 h, normalized to the maximum observed on that day. *,**, and **** indicate P < 0.05, 0.005, and 0.00005 respectively by an ordinary one-way ANOVA followed by Sidak’s multiple comparison test. In graphs, each point represents data from one well and wells collected on the same day share a symbol. The line indicates the mean of each condition. (G) A schematic of the flow cytometry phagocytosis assay in (H and I). (H) Histogram shows representative flow cytometry data of CellTrace Violet signal within macrophage (GFP+) population. (I) Graph depicts the fraction of macrophages that were CellTrace+, normalized to the average of the day. (J) Graph depicts the mean CellTrace Violet intensity of GFP+ macrophages, normalized to the daily average.
We further confirmed this result using flow cytometry (Fig. 7 G and H), which detects both trogocytosis and whole cell engulfment. Rac2+/E62K or Rac2+/+ macrophages were infected with lentivirus expressing the GFP-CAAX control or the CAR-M-GFP. Target Raji cells were labeled with CellTrace Violet and co-cultured with macrophages. Both the frequency of CellTrace-positive macrophages (Fig. 7I) and the intensity of the CellTrace signal in macrophages (Fig. 7J) were highest in macrophages expressing both the CAR and Rac2E62K.
Discussion
The results reported here leverage insights gleaned from studying a perplexing non-cell-autonomous egg-chamber-death phenotype in the Drosophila ovary to offer a possible explanation for a rare human immunodeficiency and then to enhance CAR-M-mediated engulfment and killing of cancer cells. These findings demonstrate the power of basic science in an experimental organism like Drosophila to lead to insights of potential clinical significance.
From Flies to Humans, Activated Rac Enhances Engulfment of Whole, Living Cells.
In migrating cells, Rac proteins generally function downstream of a variety of receptors to promote actin polymerization, protrusion dynamics, and motility (1, 4, 47). Rac has also long been known to stimulate macropinocytosis, which is fluid engulfment (1), as well as phagocytosis (30, 48, 49). Proteomic studies have identified hundreds of different proteins within phagosomes from multiple species but surprisingly little overlap from one cell type to the next (32). Interestingly, the most conserved components are actin and Rac. Yutin et al. (32) even propose that horizontal gene transfer of an early form of Rac from a bacterium to an archeon could have conferred phagocytic ability, enabling engulfment of a bacterial symbiont, generating the first eukaryotic cell. What is clear is that Rac-driven actin polymerization is a fundamental feature of the engulfment machinery. Here, we show that hyperactive Rac drives cannibalistic behavior in diverse metazoan cell types and organisms.
In the context of the fly egg chamber and CAR-M, engulfment of living cells that is stimulated by hyperactive Rac is directed to specific targets by receptor-mediated recognition. For example, in the fly ovary, mutation of the engulfment receptor Drpr significantly reduces germ cell engulfment and egg chamber killing by border cells expressing activated Rac. Drpr normally mediates follicle cell recognition and engulfment of living nurse cells at the end of oogenesis (50). This raises the question as to how constitutively active Rac, which functions downstream of receptors, actually still requires the “upstream” engulfment receptor. There are at least two possible explanations. One is that active Rac amplifies the signal through feedback but still requires an initiating signal, for example, in a feedforward loop with phosphoinositide 3-kinase (7, 51, 52) or through upregulated expression of phagocytic receptors. Another possibility, which is not mutually exclusive, is that Rac functions downstream of a second receptor and that activation of both parallel pathways is essential for engulfment. For example, in both flies and in human macrophages, integrin adhesion receptor activation is required in parallel to engulfment receptor activation. During engulfment, Rac may normally be activated downstream of integrins, as it is in cells migrating on integrin ligands (53).
Acute membrane recruitment of Rac and a phosphatidylserine receptor is sufficient to cause HeLa cells, which are not usually phagocytic, to engulf dead cells (54). Consistent with our results, neither active Rac nor the receptor alone is sufficient in that context either. Active Rac also stimulates engulfment of dead cells in Caenorhabditis elegans (55) and enhances mammalian cell phagocytosis of dead cells, beads (56), and opsonized red blood cells (57). Here, we show that hyperactive Rac stimulates engulfment and killing of unadulterated living cells.
The hyperactivating mutation Rac1P29S is a driver mutation in melanoma (58). Cancer cells in general, and metastatic melanoma in particular, have been reported to cannibalize immune cells as well as other cancer cells (59). Cannibalism is defined as the internalization of one living cell by another (60), and this behavior may fuel cancer growth and suppress immune responses (61). When it has been examined, cell cannibalism and other cell internalization processes require Rac activity in the engulfing cell (62, 63). Intriguingly, melanoma is an aggressive cancer that is highly cannibalistic (59), though a possible connection between the Rac1P29S driver mutation and cannibalism in metastatic melanoma has not been explored.
Rac1 overexpression is implicated in metastatic behavior in numerous additional cancers (64, 65). Furthermore, elevated Rac expression and/or activity correlate with poor prognosis in most cancers (66). The mechanism by which Rac is generally thought to drive metastasis is through its effects on migration, invasion, and survival, whereas its known effects on phagocytosis have been mostly overlooked (15, 64, 66). Based on the results presented here and the existing literature on Rac-mediated engulfment, it seems possible that Rac hyperactivity causes tumor cells to engulf and kill other living cells including anti-tumor T cells. Studies of the functional relationships between Rac overexpression and hyperactivity, cannibalistic behavior, and poor prognosis in cancer are warranted.
Rac2E62K Lymphopenia May Result from Hyperactivation of Both Phagocytes and Lymphocytes.
Rac GTPases cycle between active, guanosine triphosphate (GTP) bound and inactive, guanosine diphosphate (GDP) bound forms. Guanine nucleotide exchange factors (GEFs) activate Rac by replacing GDP with GTP. GTPase-activating proteins (GAPs) accelerate the intrinsic GTPase activity of the Rac enzymes, thus inactivating the proteins by hydrolyzing GTP to GDP. Mutations that damage the catalytic activity such as RacG12V, RacG12R, and RacQ61L are strongly activating as they lock the protein in the GTP-bound state. By contrast, Rac2E62K disrupts interaction with specific regulatory proteins, including the GEF Tiam1 as well as a GAP. The net effect is a mild 1.5 to 2.5 fold increase in Rac-GTP (this study and ref. 22). RacG12V strongly impairs cell migration (24) and causes T cell apoptosis in the thymus (67), and Rac1Q61L transgene expression in T cells drives their negative selection in the mouse thymus (68). These strongly activating mutations may mimic a highly autoreactive state because Rac functions downstream of the TCR, and strong TCR activation causes negative selection (aka deletion) of autoreactive T cells (69, 70). Patients with Rac2G12R mutations exhibit severe combined immune deficiency and hypocellularity in the thymus (21), which may be due to a failure of innate and adaptive immune cell precursors to migrate out of the bone marrow combined with apoptotic death of T cells that do develop.
By contrast, Hsu et al. (22) reported normal lymphocyte development in the bone marrow and normal cellularity in the thymus in Rac2+/E62K mice despite a profound reduction in the number of circulating T cells. So the T cell lymphopenia, which likely underlies the chronic infections that Rac2+/E62K patients suffer, remained unexplained. We find that Rac2+/E62K T cells and activated T cells are more likely to be engulfed by the wildtype as well as Rac2+/E62K macrophages. These results suggest that mild activation of T cells insufficient to cause apoptosis may nevertheless sensitize them to removal by phagocytosis. Our results also suggest that the combination of hyperactive phagocytosis by Rac2+/E62K macrophages and hypersensitivity to engulfment of Rac2+/E62K T cells may contribute to the dramatic reduction in circulating T cells in Rac2+/E62K mice and in human patients. However, additional experiments will be required to establish the relative contributions of cannibalism and other mechanisms—such as trafficking defects and negative selection—to the in vivo phenotype.
Rac-Enhanced CAR (RaceCAR) Therapy.
Although clearly detrimental as a germline mutation, our results suggest that it may be possible to harness the receptor-dependent hyperactivity that Rac2E62K confers to macrophages to enhance CAR-M therapy. It is encouraging that Rac2E62K enhanced the specific CD19/CAR-dependent engulfment with little to no detectable effect on receptor-independent engulfment. The observation that mixing Rac2+/+ and Rac2+/E62K macrophages increased phagocytosis by the wild-type macrophages is intriguing and raises the possibility that RaceCAR macrophages might be able to inflame tumor-associated macrophages and thereby enhance the anti-tumor immune response both non-autonomously as well as autonomously. Cancer patients to be treated with CAR-M or RaceCAR-M would not have the Rac2E62K mutation in their lymphocytes. So, we would not expect lymphopenia as a side effect of RaceCAR-M therapy. The viability of human patients with germline Rac2+/E62K mutations also implies that engineered RaceCAR macrophages would be unlikely to engulf cell types other than the intended targets. Together, these results suggest that expressing Rac2E62K together with a CAR may enhance the efficacy of CAR-M cancer immunotherapy without compromising safety.
Materials and Methods
Detailed materials and methods are available in SI Appendix. Briefly, Drosophila stocks and crosses were maintained under standard conditions and ovaries dissected, fixed, stained, and imaged as described (25). HL60 cells were infected with lentiviruses to express Rac constructs and Lck-GFP to identify infected cells. Lentivirus constructs were transfected into HEK293T cells and supernatants prepared as described (44). HL60 cells were differentiated into macrophage-like cells as described (33). Mouse BMDMs were prepared as described (44). RNA-seq library preparation and sequencing was performed by Genewiz (Azenta Life Sciences) and data was trimmed, filtered, and aligned to the reference genome to perform differential gene expression analysis on the Subio platform. RaceCAR-M/P BMDMs were generated by lentivirus infection, and phagocytosis was assessed by time-lapse imaging and flow cytometry as described (44).
Supplementary Material
Appendix 01
Confocal time-lapse imaging of border cell migration.
Movie of 3D surface renderings of Figure 2H and H’.
Movie of 3D surface renderings of Figure 2I and I’. Border cell nucleus is cyan and polar cell nuclei are blue.
Rac2+/+ macrophages do not show enhanced engulfment of target cells. Spinning disc confocal microscopy shows macrophages (green) do not engulf many Rac2+/+ T cells (magenta) overtime. Reduced pHrodo signal (yellow) within the macrophages (green) represents fewer phagocytosis events. Frames were acquired every 3 mins for 10 hours. Scale bar denotes 20 μm.
Rac2+/E62K macrophages hyperphagocytose target cells. Spinning disc confocal microscopy shows Rac2+/E62K macrophages (green) more rapidly and frequently phagocytosing Rac2+/E62K T cells (magenta). Robust pHrodo signal (yellow) within the macrophages (green) suggests enhanced phagocytosis. Frames were acquired every 3 mins for 10 hours. Scale bar denotes 20 μm.
Unedited Rac2+/+ macrophages do not phagocytose whole cancer cells. Spinning disc confocal microscopy shows BMDMs expressing a membrane tethered GFP (GFPcaax) fail to phagocytose whole Raji B cells (mCherry-CAAX; magenta). Frames were acquired every 5 min for 10 hours. Scale bar denotes 10 μm.
Unedited Rac2+/E62Kmacrophages do not phagocytose many whole cancer cells. Spinning disc confocal microscopy shows Rac2+/E62K BMDMs expressing GFP-CAAX (green) show little to no engulfment of whole Raji B cells (mCherry-CAAX; magenta). Frames were acquired every 5 min for 10 hours. Scale bar denotes 10 μm.
Rac2+/+ CAR-P macrophages phagocytose whole cancer cells. Spinning disc confocal microscopy shows BMDMs expressing the CAR-P-GFP (green) phagocytose whole Raji B cells (mCherry-CAAX; magenta). Frames were acquired every 5 min for 10 hours. Scale bar denotes 10 μm.
Rac2+/E62KCAR-P macrophages phagocytose many whole cancer cells. Spinning disc confocal microscopy shows a Rac2+/E62K BMDM expressing the CAR-P-GFP (green) phagocytose multiple whole Raji B cells (mCherry-CAAX; magenta). Frames were acquired every 5 min for 10 hours. Scale bar denotes 10 μm.
Acknowledgments
Heartfelt thanks go to Amy Hsu and Steve Holland for sharing the Rac2+/E62K mouse despite the pandemic and to Orion Weiner for HL60 cells. We thank Varuzhan Balasanyan, Anshul Rao, Amelia Tsark, and Wyatt Miller for technical advice and assistance. We thank all members of the Montell lab for feedback on the project. We acknowledge Flybase, Bloomington Drosophila Stock Center, and Developmental Studies Hybridoma Bank. This work was supported by a gift from Inceptor Bio and NIH grant R01 GM046425-30 to D.J.M., R35GM146935 to M.A.M., and NSF MRI grant DBI-1625770 to the Neuroscience Research Institute and Molecular, Cellular and Developmental Biology department Microscopy Facility. The research was supported in part with an unrestricted gift from Inceptor Bio. However they had no input into the design or execution of the experiments nor interpretation of the results and no input into the writing of the manuscript.
Author contributions
A.K.M., M.R., M.A.M., and D.J.M. designed research; A.K.M., M.R., A.Y.T., M.S., A.R., and M.A.M. performed research; A.B. offered technical advice; A.K.M., M.R., and M.A.M. contributed new reagents/analytic tools; A.K.M., M.R., M.S., A.R., D.J.M. and M.A.M. analyzed data; D.J.M. supervision; and A.K.M., M.R., M.A.M., and D.J.M. wrote the paper.
Competing interests
The University of California, Santa Barbara has filed a patent application on the basis of these findings.
Footnotes
Reviewers: A.H., University of Wisconsin-Madison; and O.D.W., University of California, San Francisco.
Data, Materials, and Software Availability
Data including all raw image files in this study will be made available upon request. RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE247106 (71). All reagents generated in this study will be available upon request.
Supporting Information
References
- 1.Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A., The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Boureux A., Vignal E., Faure S., Fort P., Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol. Biol. Evol. 24, 203–216 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X., He L., Wu Y. I., Hahn K. M., Montell D. J., Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12, 591–597 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy A. M., Montell D. J., Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J. Cell Biol. 133, 617–630 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridley A. J., Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 36, 103–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theveneau E., et al. , Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell 19, 39–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiner O. D., Regulation of cell polarity during eukaryotic chemotaxis: The chemotactic compass. Curr. Opin. Cell Biol. 14, 196–202 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dandekar S. N., et al. , Actin dynamics rapidly reset chemoattractant receptor sensitivity following adaptation in neutrophils. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20130008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levskaya A., Weiner O. D., Lim W. A., Voigt C. A., Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedl P., Wolf K., Zegers M. M., Rho-directed forces in collective migration. Nat. Cell Biol. 16, 208–210 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Deng Q., Yoo S. K., Cavnar P. J., Green J. M., Huttenlocher A., Dual roles for Rac2 in neutrophil motility and active retention in zebrafish hematopoietic tissue. Dev. Cell 21, 735–745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambruso D. R., et al. , Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc. Natl. Acad. Sci. U.S.A. 97, 4654–4659 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tscharntke M., et al. , Impaired epidermal wound healing in vivo upon inhibition or deletion of Rac1. J. Cell Sci. 120, 1480–1490 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Steven R., et al. , UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell 92, 785–795 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Maldonado M. D. M., Dharmawardhane S., Targeting rac and cdc42 gtpases in cancer. Cancer Res. 78, 3101–3111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bokoch G. M., Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 15, 163–171 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Pai S.-Y., Kim C., Williams D. A., Rac GTPases in human diseases. Dis. Markers 29, 177–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Artemenko Y., Lampert T. J., Devreotes P. N., Moving towards a paradigm: Common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell. Mol. Life Sci. 71, 3711–3747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somesh B. P., et al. , RacG regulates morphology, phagocytosis, and chemotaxis. Eukaryot. Cell 5, 1648–1663 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo F., Cancelas J. A., Hildeman D., Williams D. A., Zheng Y., Rac GTPase isoforms Rac1 and Rac2 play a redundant and crucial role in T-cell development. Blood 112, 1767–1775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lougaris V., Baronio M., Gazzurelli L., Benvenuto A., Plebani A., RAC2 and primary human immune deficiencies. J. Leukoc. Biol. 108, 687–696 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Hsu A. P., et al. , Dominant activating RAC2 mutation with lymphopenia, immunodeficiency, and cytoskeletal defects. Blood 133, 1977–1988 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisbrecht E. R., Montell D. J., A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell 118, 111–125 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Cernuda-Morollón E., Millán J., Shipman M., Marelli-Berg F. M., Ridley A. J., Rac activation by the T-cell receptor inhibits T cell migration. PLoS One 5, e12393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campanale J. P., Mondo J. A., Montell D. J., A scribble/Cdep/Rac pathway controls follower-cell crawling and cluster cohesion during collective border-cell migration. Dev. Cell 57, 2483–2496.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montell D. J., Keshishian H., Spradling A. C., Laser ablation studies of the role of the Drosophila oocyte nucleus in pattern formation. Science 254, 290–293 (1991). [DOI] [PubMed] [Google Scholar]
- 27.Uribe-Querol E., Rosales C., Phagocytosis: Our current understanding of a universal biological process. Front. Immunol. 11, 1066 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serizier S. B., McCall K., Scrambled eggs: Apoptotic cell clearance by non-professional phagocytes in the Drosophila ovary. Front. Immunol. 8, 1642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silver D. L., Montell D. J., Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell 107, 831–841 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Hoppe A. D., Swanson J. A., Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol. Biol. Cell 15, 3509–3519 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddien P. W., Cameron S., Horvitz H. R., Phagocytosis promotes programmed cell death in C. elegans. Nature 412, 198–202 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Yutin N., Wolf M. Y., Wolf Y. I., Koonin E. V., The origins of phagocytosis and eukaryogenesis. Biol. Direct 4, 9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murao S., Gemmell M. A., Callaham M. F., Anderson N. L., Huberman E., Control of macrophage cell differentiation in human promyelocytic HL-60 leukemia cells by 1,25-dihydroxyvitamin D3 and phorbol-12-myristate-13-acetate. Cancer Res. 43, 4989–4996 (1983). [PubMed] [Google Scholar]
- 34.Rovera G., Santoli D., Damsky C., Human promyelocytic leukemia cells in culture differentiate into macrophage-like cells when treated with a phorbol diester. Proc. Natl. Acad. Sci. U.S.A. 76, 2779–2783 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albacker L. A., et al. , TIM-4, a receptor for phosphatidylserine, controls adaptive immunity by regulating the removal of antigen-specific T cells. J. Immunol. 185, 6839–6849 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albacker L. A., et al. , TIM-4, expressed by medullary macrophages, regulates respiratory tolerance by mediating phagocytosis of antigen-specific T cells. Mucosal Immunol. 6, 580–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H., Leitenberg D., Li B., Flavell R. A., Deficiency of small GTPase Rac2 affects T cell activation. J. Exp. Med. 194, 915–926 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim J. F., Berger H., Su I.-H., Isolation and activation of murine lymphocytes. J. Vis. Exp. 116, e54596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mills C. D., Ley K., M1 and M2 macrophages: The chicken and the egg of immunity. J. Innate Immun. 6, 716–726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosser D. M., Edwards J. P., Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian B.-Z., Pollard J. W., Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim W. A., June C. H., The principles of engineering immune cells to treat cancer. Cell 168, 724–740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou A. J., Chen L. C., Chen Y. Y., Navigating CAR-T cells through the solid-tumour microenvironment. Nat. Rev. Drug Discov. 20, 531–550 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Morrissey M. A., et al. , Chimeric antigen receptors that trigger phagocytosis. Elife 7, e36688 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klichinsky M., et al. , Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 38, 947–953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sloas C., Gill S., Klichinsky M., Engineered CAR-macrophages as adoptive immunotherapies for solid tumors. Front. Immunol. 12, 783305 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones G. E., Allen W. E., Ridley A. J., The Rho GTPases in macrophage motility and chemotaxis. Cell Adhes. Commun. 6, 237–245 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Reddien P. W., Horvitz H. R., CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2, 131–136 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Massol P., Montcourrier P., Guillemot J. C., Chavrier P., Fc receptor-mediated phagocytosis requires CDC42 and Rac1. EMBO J. 17, 6219–6229 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebo D. P. V., McCall K., Murder on the ovarian express: A tale of non-autonomous cell death in the drosophila ovary. Cells 10, 1454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campa C. C., Ciraolo E., Ghigo A., Germena G., Hirsch E., Crossroads of PI3K and Rac pathways. Small GTPases 6, 71–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swaney K. F., Huang C.-H., Devreotes P. N., Eukaryotic chemotaxis: A network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys. 39, 265–289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huttenlocher A., Horwitz A. R., Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 3, a005074 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Onuma H., et al. , Rapidly rendering cells phagocytic through a cell surface display technique and concurrent Rac activation. Sci. Signal. 7, rs4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y. C., et al. , elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev. Cell 1, 491–502 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Tosello-Trampont A.-C., Nakada-Tsukui K., Ravichandran K. S., Engulfment of apoptotic cells is negatively regulated by Rho-mediated signaling. J. Biol. Chem. 278, 49911–49919 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Ai J., et al. , The inositol phosphatase SHIP-2 down-regulates FcgammaR-mediated phagocytosis in murine macrophages independently of SHIP-1. Blood 107, 813–820 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krauthammer M., et al. , Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 44, 1006–1014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lugini L., et al. , Cannibalism of live lymphocytes by human metastatic but not primary melanoma cells. Cancer Res. 66, 3629–3638 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Borensztejn K., et al. , Classification of cell-in-cell structures: Different phenomena with similar appearance. Cells 10, 2569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fais S., Overholtzer M., Cell-in-cell phenomena in cancer. Nat. Rev. Cancer 18, 758–766 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Sun Q., et al. , Competition between human cells by entosis. Cell Res. 24, 1299–1310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishidome T., Yoshida T., Hanayama R., Induction of live cell phagocytosis by a specific combination of inflammatory stimuli. EBioMedicine 22, 89–99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang J., et al. , Rac1, A potential target for tumor therapy. Front. Oncol. 11, 674426 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cardama G. A., et al. , Relevance of small GTPase Rac1 pathway in drug and radio-resistance mechanisms: Opportunities in cancer therapeutics. Crit. Rev. Oncol. Hematol. 124, 29–36 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Cannon A. C., Uribe-Alvarez C., Chernoff J., RAC1 as a therapeutic target in malignant melanoma. Trends Cancer 6, 478–488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lorès P., Morin L., Luna R., Gacon G., Enhanced apoptosis in the thymus of transgenic mice expressing constitutively activated forms of human Rac2GTPase. Oncogene 15, 601–605 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Gomez M., Kioussis D., Cantrell D. A., The GTPase Rac-1 controls cell fate in the thymus by diverting thymocytes from positive to negative selection. Immunity 15, 703–713 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Lancaster J. N., et al. , Live-cell imaging reveals the relative contributions of antigen-presenting cell subsets to thymic central tolerance. Nat. Commun. 10, 2220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gallegos A. M., Bevan M. J., Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. J. Exp. Med. 200, 1039–1049 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.A. K. Mishra et al. , Hyperactive Rac stimulates cannibalism of living target cells and enhances CAR-M-mediated cancer cell killing. GEO DataSets. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE247106. Deposited 3 November 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01
Confocal time-lapse imaging of border cell migration.
Movie of 3D surface renderings of Figure 2H and H’.
Movie of 3D surface renderings of Figure 2I and I’. Border cell nucleus is cyan and polar cell nuclei are blue.
Rac2+/+ macrophages do not show enhanced engulfment of target cells. Spinning disc confocal microscopy shows macrophages (green) do not engulf many Rac2+/+ T cells (magenta) overtime. Reduced pHrodo signal (yellow) within the macrophages (green) represents fewer phagocytosis events. Frames were acquired every 3 mins for 10 hours. Scale bar denotes 20 μm.
Rac2+/E62K macrophages hyperphagocytose target cells. Spinning disc confocal microscopy shows Rac2+/E62K macrophages (green) more rapidly and frequently phagocytosing Rac2+/E62K T cells (magenta). Robust pHrodo signal (yellow) within the macrophages (green) suggests enhanced phagocytosis. Frames were acquired every 3 mins for 10 hours. Scale bar denotes 20 μm.
Unedited Rac2+/+ macrophages do not phagocytose whole cancer cells. Spinning disc confocal microscopy shows BMDMs expressing a membrane tethered GFP (GFPcaax) fail to phagocytose whole Raji B cells (mCherry-CAAX; magenta). Frames were acquired every 5 min for 10 hours. Scale bar denotes 10 μm.
Unedited Rac2+/E62Kmacrophages do not phagocytose many whole cancer cells. Spinning disc confocal microscopy shows Rac2+/E62K BMDMs expressing GFP-CAAX (green) show little to no engulfment of whole Raji B cells (mCherry-CAAX; magenta). Frames were acquired every 5 min for 10 hours. Scale bar denotes 10 μm.
Rac2+/+ CAR-P macrophages phagocytose whole cancer cells. Spinning disc confocal microscopy shows BMDMs expressing the CAR-P-GFP (green) phagocytose whole Raji B cells (mCherry-CAAX; magenta). Frames were acquired every 5 min for 10 hours. Scale bar denotes 10 μm.
Rac2+/E62KCAR-P macrophages phagocytose many whole cancer cells. Spinning disc confocal microscopy shows a Rac2+/E62K BMDM expressing the CAR-P-GFP (green) phagocytose multiple whole Raji B cells (mCherry-CAAX; magenta). Frames were acquired every 5 min for 10 hours. Scale bar denotes 10 μm.
Data Availability Statement
Data including all raw image files in this study will be made available upon request. RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE247106 (71). All reagents generated in this study will be available upon request.


