Abstract
Background/Aim
Metastatic lymph node 64 (MLN64) is often co-amplified with ERBB2 (HER2) and plays a role in the progression of breast and prostate cancer. The present study explored the expression of MLN64 in clinical gastric cancer in association with the ERBB family and its impact on drug resistance in patients.
Materials and Methods
Two independent gastric cancer cohorts (n=324; n=87) were used to explore the expression profile of MLN64 in conjunction with ERBB family members in clinical gastric cancer and its association with neoadjuvant chemotherapy responses. Gastric cancer AGS and HCG27 cells with MLN64 knockdown were generated to determine the function of MLN64 in cell behavioural changes.
Results
Gastric tumor tissues expressed significantly higher levels of MLN64 compared with normal tissues (p<0.01); however, MLN64 alone was a weak prognostic indicator. An integrated co-expression of MLN64, ERBB4, and NRG4 was a significant factor in assessing overall survival in both cohorts. MLN64 was a profound indicator of patient response to neoadjuvant chemotherapy. In vitro studies indicated a significant contribution of MLN64 to the response of gastric cancer cells to chemodrugs and Her-2 inhibitors. MLN64 knockdown also contributed to the adhesion and migration and suggested a possible mechanism mediated by the interaction between MLN64 and ERBBs.
Conclusion
MLN64 is an indicator of patient response to neoadjuvant chemotherapy in gastric cancer. Together with the expression pattern of ERBB4, MLN64 is a poor prognostic factor for patients with gastric cancer.
Keywords: MLN64, STARD3, gastric cancer, ERBB4, prognosis, NRG, signalling, drug response
Human metastatic lymph node 64 (MLN64), also known as star related lipid transfer domain containing-3 (STARD3) is an integral membrane protein and its encoding gene is mapped to the q12-q21 region of chromosome 17. The coding region of MLN64 is located in proximity to the amplicons of other important genes associated with cancer, particularly breast cancer, such as BRCA1 and the c-ERBB-2 oncogene (1). MLN64 was found to be identical to the steroidogenic acute regulatory protein (StAR) (2). Among the 15 START domain protein family members, MLN64 is a cholesterol-binding candidate that mediates intracellular non-vesicular cholesterol trafficking and distribution (3).
Cancer cells reprogram cholesterol metabolism to achieve sufficient energy metabolism for uncontrolled cell growth and proliferation. Accumulated mitochondrial cholesterol facilitates cell proliferation, whereas decreased membranous cholesterol levels are associated with anoikis-like apoptosis (4). MLN64 has also been demonstrated to inhibit the maturation of late endosomes to lysosomes, compromising the degradation activity of cancer cells and leading to the over-expression of certain functional proteins (5).
MLN64 is highly expressed in various tissues including the pancreas, heart, placenta, liver, and muscle. Owing to the initially reported role of MLN64 in cancer progression, there have been investigations regarding MLN64 in different cancer types. In breast cancer where MLN64 was initially reported, MLN64 was found to be highly expressed in invasive breast cancers and its expression was associated with the expression of ERBB2 (2,6-8). An elevated expression level of MLN64 was observed in HER2+ (ERBB2) breast cancer, while an opposite trend was observed in triple-negative breast cancer (9). MLN64 over-expression is an indicator of poor patient prognosis (10). In prostate cancer, another endocrine-related cancer, a linear correlation has been found between MLN64 expression and cholesterol levels, and MLN64 was also a prognostic indicator for prostate cancer patients (11). In addition, MLN64 expression was highly correlated with cytochrome P450 Family 17 (CYP17), an enzyme linked to drug metabolism and cholesterol synthesis (12). Co-expression of MLN64 and CYP17 was associated with shorter relapse-free survival (12). In addition to the metabolic link between MLN64 and cancer cells, MLN64 was also shown to be a key regulator of cell-matrix adhesion (10).
Gastric cancer is a highly aggressive cancer type with high incidence in the Far East (13). Over the past decade, improved diet, early detection, refined surgical procedures, and increasingly available drug options have resulted in a steady decrease in the incidence and an improvement in the clinical outcomes of patients with gastric cancer. However, its mortality and therapeutic options remain challenging. In addition to conventional chemotherapies, anti-HER2 therapy has a role in the treatment of gastric cancer, and over-expression of ERBB2 has a significant impact on the prognosis and survival of the patients. Dual silencing of EGFR and HER2 has been shown to increase the potency of gefitinib in gastric cancer cells (14). MLN64, however, has not yet been thoroughly explored in gastric cancer. In a study using a microarray of gastric cancers, MLN64 expression was found to be associated with increased expression of ERBB2 (15). ERBB2, a tyrosine kinase receptor, belongs to the ERBB family that includes ERBB1 (EGFR), ERBB2, ERBB3 and ERBB4. The present study investigated the expression of MLN64 in association with the four ERBBs, along with the expression of the ERBB receptor ligands neuregulin (NRG) 1-4, and its role in the response of patients with gastric cancer to neoadjuvant chemotherapies.
Materials and Methods
Gastric tissue cohorts. Two independent fresh frozen gastric cohorts containing both cancerous and adjacent background tissues were available in the host lab as reported in a previous study (16). Cohort A was a larger cohort for screening, and Cohort B had information on the patient’s response to neoadjuvant chemotherapy. Patient demographic information and clinico-pathological characteristics were also recorded. The expression profile of MLN64 in association with that of the ERBB family members in the tissues was evaluated. The cohorts were collected after obtaining patient consent and approval from the local research Ethics Committee (ethics number: 2006021).
Cell lines and cell culture. The human gastric cancer cell lines, AGS and HGC-27, purchased from ECACC (European Collection of Animal Cell Culture, Salisbury, UK), were maintained in Dulbecco's Modified Eagle’s medium (DMEM) supplemented with 10% foetal calf serum (FCS) and antibiotics (penicillin at 100 unit/ml and streptomycin at 100 μg/ml).
Gastric cancer MLN64kd cell models were established by transfecting the cells with either siRNA (sc-44439) or shRNA plasmid (sc-44439-SH) (Santa Cruz Biotechnologies Inc., Santa Cruz, CA, USA). Transfection with siRNA was carried out using a transfection kit including the transfection medium (SC36868) and reagent (SC-29528) (Santa Cruz Biotechnologies), and shRNA lentiviral transduction of gastric cancer cells was performed using polybrene. After shRNA lentiviral transduction, puromycin was used at 1 μg/ml to select stable knockdown cells and at 0.2 μg/ml to maintain the stability of the transduced cells.
RNA extraction and quantitative PCR. TRI Reagent (Merck Chemicals Ltd., Dorset, England, UK) was used for RNA extraction. All RNA samples were diluted to 500 ng/μl before reverse transcription into cDNA according to the manufactures’ guidelines (Promega, Southampton, UK). To determine MLN64 transcript levels, real time quantitative PCR (qPCR) was carried out using the Amplifuor molecular Beacon system (Fisher Scientific UK, Leicestershire, UK). Primers for qPCRfor MLN64 were 5’gcacctttgtctggattctt’3 and 5’ actgaacctgaccgtacatgaaaggcaaattcaaacat’3 and for GAPDH 5’aagg tcatccatgacaactt’3 and 5’actgaacctgaccgtacagccatccacagtcttctg'3 (actgaacctgaccgtaca was the Z sequence to complement the FAM-tagged Uniprimer™ probe).
Drug response assays. The targeted drugs gefitinib and neratinib, and the commonly used chemodrugs gemcitabine and 5-FU were chosen to determine the toxicity of drugs on gastric cancer cells before and after MLN64kd. Gefitinib is an ERBB1 inhibitor, while neratinib is an irreversible inhibitor of ERBB1, 2 and 4. The assays were conducted over a range of drug concentrations and the IC50 value was calculated from the cell viability 72 h after incubation. On day 3, the cells were fixed in 4% formalin, stained with 0.5% crystal violet and the absorbance was read at 592 nm. Cell viability was calculated as follows: [(absorbance of control cells- absorbance of treated cells)/control ×100].
Real-time monitoring of cell behavior changes using electric cell-substrate sensing (ECIS). The behavioral changes of wild type (WT) and MLN64kd gastric cancer cells were tested using ECIS. Cell adhesion was measured for up to 2 h and migration ability for up to 5 h. Cells were additionally incubated with ERBB activator and inhibitors during the application of NRG1 (ligand of ERBB2, 3, and 4), neratinib and AG825, which is a specific ERBB2 inhibitor.
Cell-matrix adhesion assay. A cell-matrix adhesion assay was conducted in a 96-well plate precoated with Matrigel, a basement membrane matrix extract, at 0.5 μg/well. The cells were resuspended in DMEM and diluted to 50,000 cells/ml. The cells were then seeded into the wells and incubated at 37˚C for 40 min before being fixed with 4% formalin and stained with 0.5% crystal violet staining solution. Cell counting was then performed under a microscope at 20× magnification.
Statistical analysis. Normality testing was performed to determine the distribution pattern of the two gastric cohorts, along with Student’s t-test and ANOVA for gene expression comparisons (version 27; SPSS, Chicago, IL, USA). Kaplan–Meier survival curves classified by receiver operating characteristic curve (ROC) and multivariate regression models were generated.
Results
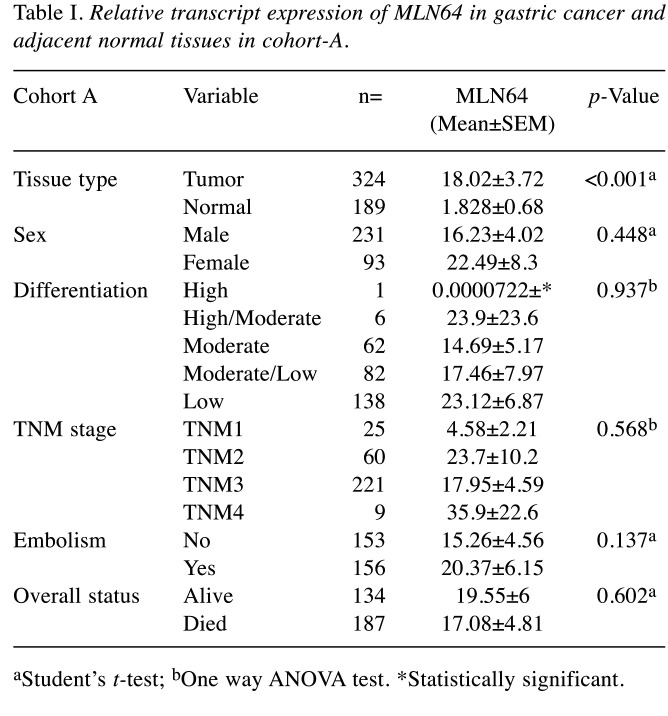
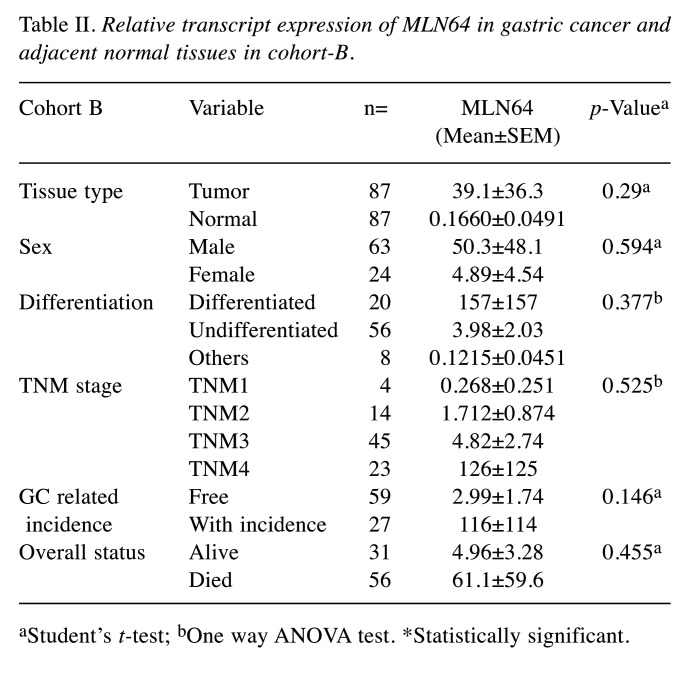
Expression of MLN64 gene transcripts in human normal and tumor gastric tissues. A cohort composed of 324 gastric cancer and 189 matched control gastric tissues (cohort A) was collected at the time of surgery and the expression of MLN64 was analyzed (Table I). MLN64 expression was significantly elevated in gastric tumors compared with normal tissues (p<0.001), and it showed an increasing trend with the increase in TNM stage and nodal positivity in both cohorts. An additional smaller cohort (n=87) (cohort B) specifically containing paired cDNA samples of patients who received neoadjuvant chemotherapy (NAC) was utilized to assess the contribution of MLN64 in gastric cancer progression. The clinicopathological information of patients and the transcript levels of MLN64 were analyzed in both cohorts (Table II). Interestingly, no significant differences in MLN64 expression were observed in cohort B, although a similar trend was observed in cohort A. In both cohorts, the expression levels of MLN64 did not influence the overall clinical outcome of patients.
Table I. Relative transcript expression of MLN64 in gastric cancer and adjacent normal tissues in cohort-A.
aStudent’s t-test; bOne way ANOVA test. *Statistically significant.
Table II. Relative transcript expression of MLN64 in gastric cancer and adjacent normal tissues in cohort-B.
aStudent’s t-test; bOne way ANOVA test. *Statistically significant.
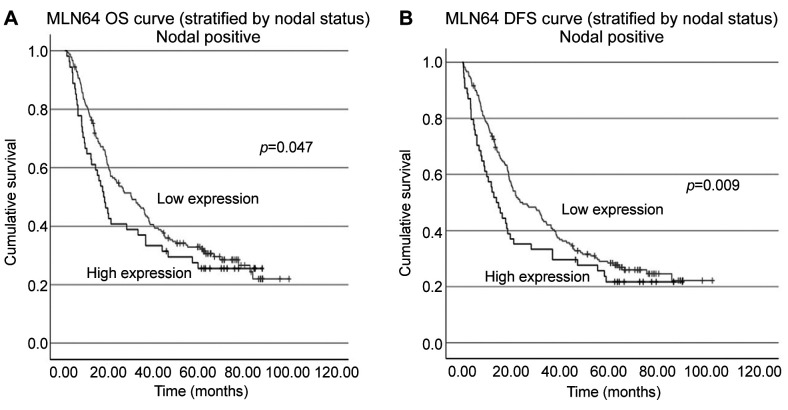
Relationship between MLN64 transcript levels and survival in patients with gastric cancer. The patients were divided into high and low MLN64 expression groups using a cut-off value obtained from the ROC analysis. The MLN64 gene transcript alone had a weak value in predicting overall survival (OS) and disease-free survival (DFS) of patients with gastric cancer. However, among the 247 out of 324 patients with nodal positive status in cohort A, higher MLN64 expression was associated with significantly shorter OS and DFS (Figure 1).
Figure 1. Overall survival (OS) and disease-free survival (DFS) curves of patients with gastric cancer with nodal positive status (A, B).
Correlation between MLN64 expression and the expression of ERBBs and ligand NRGs. In addition to the previously reported co-amplification patterns of MLN64 and ERBB2 in breast cancer (7,8,17), we examined the correlation between MLN64 and all the ERBB family members in cohort A. A statistically positive correlation was observed between MLN64 and ERBB2 in normal gastric tissue (p=0.024), whereas ERBB4 expression was positively associated with MLN64 expression in the cancer tissue (p=0.005). Further analysis was conducted to examine the correlation between MLN64 and NRGs. There was a close association between NRG4 and all four ERBB family members in normal gastric tissues, whereas a relationship was consistently observed with ERBB4 in tumor tissues. No significant correlations were observed between MLN64 and NRG4 levels in both normal and tumor tissues.
The combinational power of ERBB4 (HER4) and MLN64 for patients’ survival. The correlation of the expression led us to evaluate the clinical significance of MLN64 when considered together with ERBBs. In cohort A, none of the ERBBs was a significant prognostic indicator. However, patients with combined high expression of MLN64 and ERBB4 exhibited significantly prolonged OS time (p=0.016, HR=1.322, 95%CI=1.053-1.659). Among patients who expressed lower ERBB4 levels, MLN64 was a significant prognostic factor for both OS (p=0.048) and DFS (p=0.027). A similar non-significant trend was observed in the high ERBB4 group. The combined prognostic effect of MLN64 and other ERBBs (ERBB1, ERBB2, and ERBB3) were not observed in this clinical cohort.
Relationship between NRGs, ERBB4 and MLN64 and their prognostic value. Next, we assessed the impact of the four NRG members alone and in combination with ERBBs and MLN64 on patient’s survival. NRG4 acted as an independent poor prognostic factor for OS in patients with gastric cancer (Figure 2A), but it had limited effect on DFS. Additionally, we found that patients with gastric cancer-induced embolism expressed significantly higher levels of NRG4 than patients without embolism (p=0.0352).
Figure 2. The implications of NRG4 alone and in combination with ERBB4 in the overall survival (OS) of patients with gastric cancer are illustrated in A and B. Patients with integrated expression of MLN64/NRG4/ERBB4 had a significantly shortened OS (C).
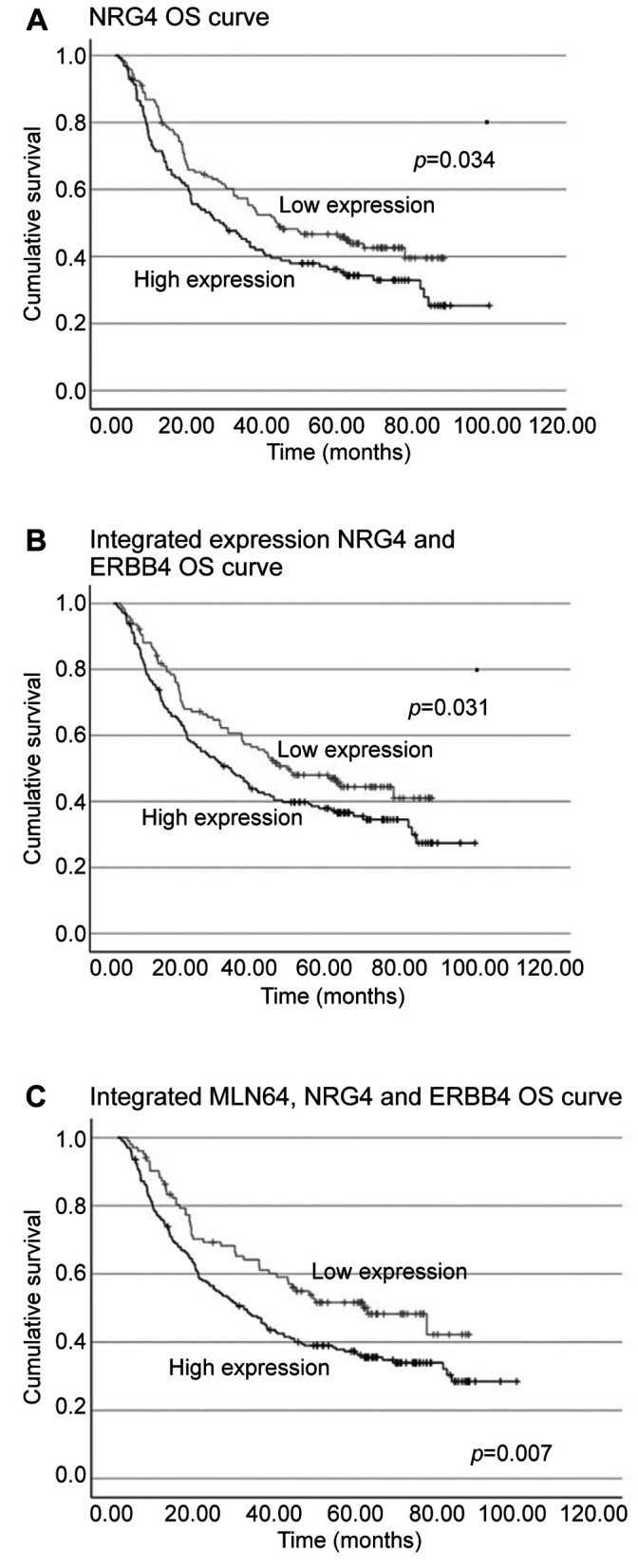
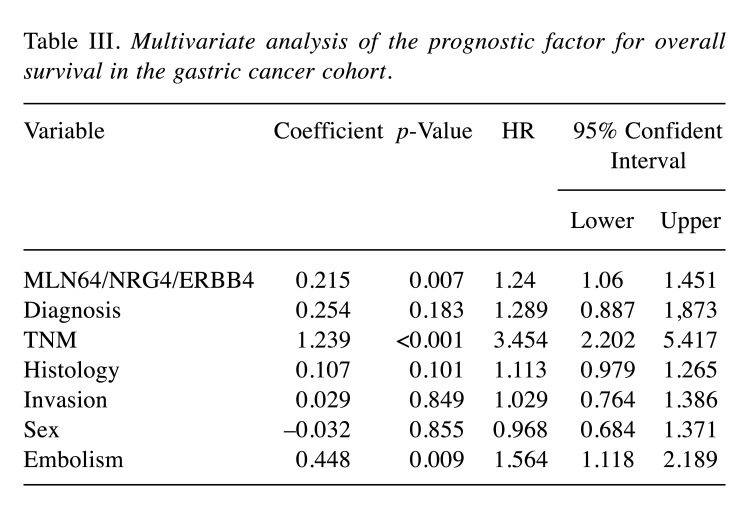
In terms of survival, patients who exhibited dual higher levels of NRG4 and ERBB4 transcripts had significantly longer OS (Figure 2B), with regression analysis confirming the independency of the predictive value of NRG4/ERBB4 (p=0.042, HR=1.168, 95%CI=1.005-1.356). Interestingly, the combined effect of MLN64/ERBB4/NRG4 was more profound (p=0.007) in predicting OS of patients than NRG4/ERBB4 or any of the two molecules alone (Figure 2C). Multivariate analysis identified MLN64/ERBB4/NRG4, TNM staging and embolism status as independent prognostic indicators for patients’ OS (Table III).
Table III. Multivariate analysis of the prognostic factor for overall survival in the gastric cancer cohort.
High levels of MLN64 in gastric tumors tend to confer drug resistance. In cohort B, patients received NAC and were subdivided into resistant and non-resistant groups according to the ROC curve (AUC=0.64; p=0.028). Our data showed a positive contribution of MLN64 to drug resistance; it predicted patients’ drug response independent from the expression of ERBB family members (p=0.007, HR=8.056, 95%CI=1.782-36.414). However, by stratification of patients into high and low ERBB expression groups, we identified a subgroup of patients with high expression of ERBB1 and MLN64 that showed significant drug resistance (p=0.04, HR=2.963, 95%CI=1.05-8.363).
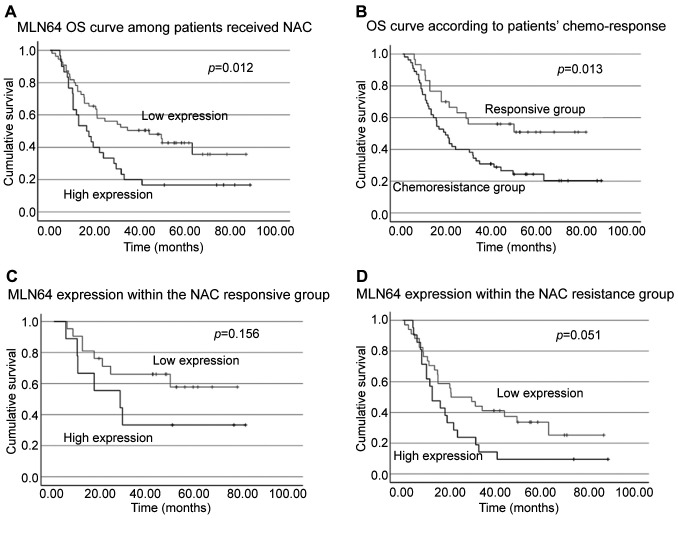
Interestingly, regarding patient survival, in contrast to our finding in cohort A that MLN64 alone was a weak prognostic indicator; following NAC, patients with high MLN64 transcript expression showed significantly reduced OS (Figure 3A). Further exploration of how this might be affected by the expression of ERBBs showed that patients who exhibited high expression of both ERBB2 and MLN64 survived significantly shorter than patients with other ERBB2 and MLN64 expression patterns (p=0.031). A similar trend was also observed within the high ERBB3 and low ERBB1 groups where MLN64 transcripts were associated with shorter survival times (p=0.003 and p=0.033, respectively). Additionally, while the association between MLN64 and ERBB4 with patients’ drug response was not clearly established (p=0.145), the integrated pattern remained a prognostic indicator for patients’ OS (p=0.036).
Figure 3. Survival curves of patients in subgroups of cohort B. A) Overall survival (OS) curve of patients who received neoadjuvant chemotherapy (NAC). Patients were subclassified into MLN64 high and low groups, B) based on patients’ chemo-response, C) MLN64 expression within the NAC responsive group, D) integrated expression of MLN64/NRG4/ERBB4.
Relationship between chemosensitivity and patients’ survival. According to our results, patients who exhibited chemoresistance had a significantly reduced OS (Figure 3B). Considering the predictive value of MLN64 in patients’ drug response, we then evaluated whether patient chemosensitivity and MLN64 expression were confounding variables in predicting patient survival in this second cohort. As illustrated in Figure 3C and D, low expression of MLN64 in the drug-responsive group and high expression of MLN64 in the drug-resistant group showed favorable and unfavorable outcomes, respectively; however, p-values were not significant, indicating that MLN64 transcript expression was not a significant factor in chemo-response-mediated changes in survival. Additionally, patient response to chemotherapy did not seem to affect the predictive value of MLN64 in accordance with our binary logistic regression results (data not shown).
Consistent with the cohort A, the prognostic value of MLN64/ERBB4/NRG4 in OS was additionally confirmed in the cohort B (p=0.036), with both univariant (p=0.014, HR=1.952, 95%CI=1.148=3.321) and multivariant analysis confirming its independency (p=0.018, HR=2.603, 95%CI=1.781-5.754).
In vitro investigation of the role of MLN64 in drug response using MLN64 knockdown cell models. To test the role of MLN64 in gastric cancer cells’ drug response, we generated MLN64 knockdown (MLN64kd) cell models using siRNA and shRNA. We successfully knocked down the MLN64 transcript in both AGS and HGC27 cells, with over 50% reduction compared to control cells (p<0.01).
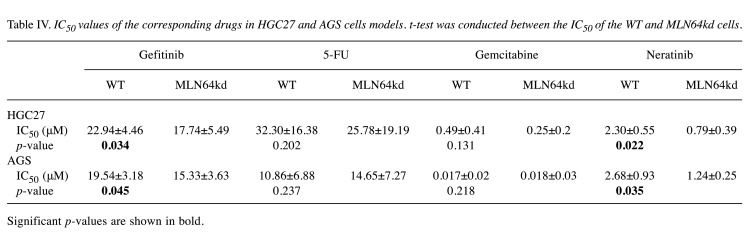
The cell models were treated with four different drugs commonly used in cancer therapy. As reflected by the reduced IC50 values (Table IV), HGC27 cells became more vulnerable to drug treatment after MLN64kd, especially after treatment with gefitinib and neratinib where we observed a significant difference between the WT and the KD cells. Similar trends and statistical significance were also observed in AGS cells after exposure to gefitinib and neratinib. However, in contrast to the HGC27 cell line, AGS cells with decreased MLN64 expression were more resistant to 5-FU treatment compared with WT cells, though the difference was not statistically significant. MLN64kd did not alter cellular response to gemcitabine in both gastric cancer cell lines.
Table IV. IC50 values of the corresponding drugs in HGC27 and AGS cells models. t-test was conducted between the IC50 of the WT and MLN64kd cells.
Significant p-values are shown in bold.
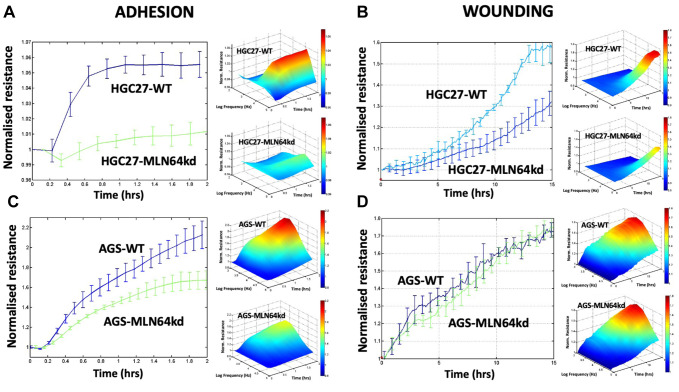
MLN64kd gastric cells showed decreased cell adhesion. Cell behavior changes namely adhesion and migration of cells with and without MLN64 were then assessed using the ECIS assay. As shown in Figure 4A and C, MLN64kd significantly suppressed the adhesion of both gastric cancer cell lines, which was further validated by the Matrigel adhesion assay; fewer MLN64kd cells adhered to the matrix gel compared to the control cells (AGS, p<0.01; HGC27, p<0.001). The changes in the migratory ability after MLN64kd did not follow the same pattern. As shown in Figure 4B, a significant difference between HGC27 WT and MLN64kd cells was only observed 7 h after initiation of the wounding process, whereas MLN64kd did not significantly change AGS cell migration within 15 h of monitoring (Figure 4D).
Figure 4. ECIS-based assessment of cell adhesion and migration at 4,000 Hz for both HGC27 (A, B) and AGS (C, D) cell models.
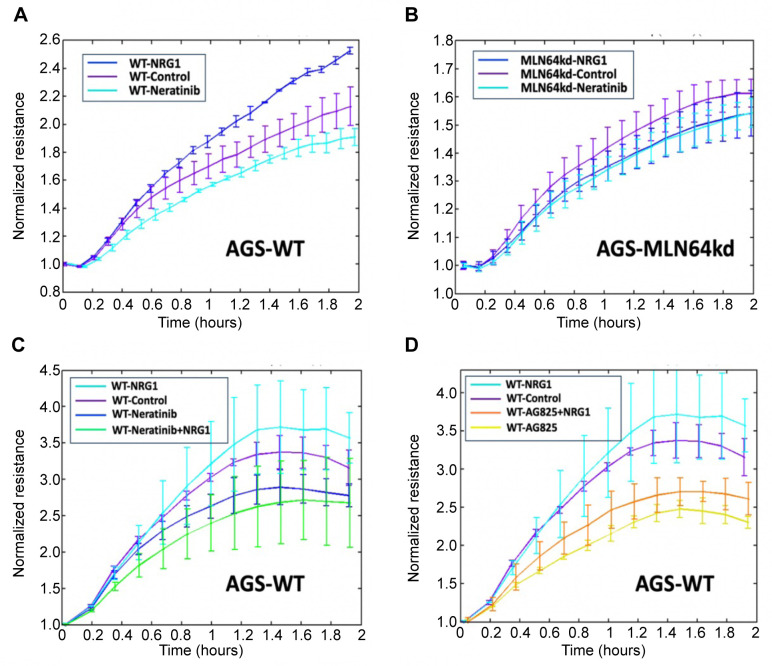
MLN64kd hindered the action of NRG1, neratinib and AG825. The effect of NRG1 and neratinib on AGS cell models was examined using the ECIS assay. The results showed that NRG1 significantly facilitated the adhesion of WT cells (Figure 5A), whereas this effect was diminished in the MLN64kd cells (Figure 5B). Similarly, neratinib exhibited a reduced inhibitory effect in cells with decreased expression of MLN64 (Figure 5B). When WT cells were treated with both NRG1 and neratinib, the resistance was even lower than that following treatment with neratinib alone; however, the effect was not significant (Figure 5C). In contrast, coadministration of NRG1 and the ERBB2 specific inhibitor AG825 showed a trend of partially alleviating the repressive effect of AG825 on AGS WT cell adhesion. However, it is noteworthy that the observed resistance remained significantly lower than that of the WT control cells and the cells treated with NRG1. This suggested the potential involvement of ERBBs other than ERBB2 in MLN64 signalling to regulate cell adhesion (Figure 5D).
Figure 5. ECIS-based assessment of cell adhesion at 4,000 Hz for AGS (C, D) cell models under different treatments.
Discussion
Amplification of MLN64 along with other genes mapped at 17q12-21 has been reported in different types of solid cancers and advanced disease, especially for breast carcinomas where MLN64 was found to be co-amplified with ERBB2 (2,6,12,15). In rare cases of gastrointestinal carcinomas, up-regulation of MLN64 was also observed (15,18). By analyzing an available public dataset of patients with gastric cancer, MLN64 was found to be a potential gastric cancer-promoting gene that was co-amplified with ERBB2 (19); however, this co-amplification pattern was not observed. This may be explained by patient heterogeneity since ERBB2 up-regulation in gastric cancer has been reported to be less than 20% (20). Instead, we identified a significant positive correlation between MLN64 and ERBB4, a gene that promotes the oncogenic PI3K/Akt signalling pathway in gastric cancer (21). In addition to ERBBs, we examined the contribution of NRGs, a family of ligands that induce ERBB activation and downstream oncogenic signalling events (22,23). Our results showed that MLN64 transcript level alone was insufficient to predict gastric patients’ survival, but the transcript levels of both MLN64 and ERBB4, or ERBB4/NRG4 was a highly powerful prognostic indicator.
Study limitations. First, clinical information regarding NAC use was not available for the large cohort. Although the results from the second cohort indicated that NAC response did not affect the predictive value of MLN64, this cohort had a limited sample size, especially after stratification of patients according to their responsiveness to chemotherapy. The results of a meta-analysis showed an improvement in the survival of patients with gastric cancer receiving NAC (24). More importantly, NAC can result in tumor downstaging, eventually increasing the curative rate in patients gastric cancer and improving survival (25,26). Although MLN64 alone was a weak predictor, we indeed highlighted its involvement in drug response from both clinical and in vitro perspectives.
Owing to different drug selections and synergism, the significant association between MLN64 and chemotherapeutic drug resistance was not obvious in our in vitro study. Instead, the differences in IC50 values between MLN64kd and WT cells were only observed when the cells were treated with ERBB inhibitors. The current study proposes a possible signalling pathway between MLN64 and ERBBs, potentially involving ERBB1, 2 and 4, in regulating drug response. ERBB3 is known to dimerize with ERBB1 and ERBB2, which may in turn lead to MLN64 signalling. This indirect communication may also explain the significant prognostic value of MLN64 after stratification by ERBB3.
Knockdown of MLN64 resulted in reduced cell adhesion, which is a critical component of cancer progression and development of drug resistance (27). ECIS results within this study further validated the potential interaction between MLN64 and ERBB1 and 4. Following addition of neratinib, an inhibitor which can irreversibly bind to ERBB1, 2 and 4 (28), the cells showed significantly reduced adhesion compared with the MLN64kd cells. However, NRG1 significantly facilitated the adhesion of WT cells that was not observed in MLN64 cells. As shown in Figure 5C, NRG1 did not mitigate the effect brought by inhibition of ERBB1, 2 and 4. In contrast, Figure 5D shows that adhesion could still be stimulated through activation of ERBBs other than ERBB2. NRG1 may directly interact with ERBB3 and ERBB4 (29). This further suggested that cell adhesion may be achieved via the NRG1/ERBB4/MLN64 axis.
The contribution of ERBB2 to MLN64kd-mediated changes in cancer cell behavior was not negligible. Although ERBB2 does not have an extracellular domain to interact with ligands, NRG1 and 2 have been reported to be functionally related to ERBB2. Indeed, the activation of the NRG1/ERBB2/ERBB3 axis has been shown to induce cell growth in triple-negative breast cancer and proliferation of cancer stem cells (30,31). It has also been proposed that ERBB2 and ERBB4 form a dimer that contributes to tumor advancement, as ERBB2 is required for ERBB4-dependent breast cancer progression (32).
Interestingly, MLN64 expression had different roles in cell migration in the two gastric cancer cell lines. While no difference was observed in AGS cells, a significant difference in resistance was observed between HGC27 cell models 7 h after initiation of the wounding process. Wound healing is a multi-dimensional process that includes polarization, adhesion to the underlying matrix, contraction, and detachment from the original site. The resistance generated by the cell layer and assessed using ECIS not only reflected the overall cell coverage of the wound, but also the junction formation between cells and between the cells and the surface. Our results indicated a slower migration and reduced junction formation in cells with reduced MLN64 expression. The established stable cell-cell adhesions in epithelial cells also influence cell migration (33). An altered junction formation caused by MLN64kd might therefore be another factor affecting cell migration. Further studies should be conducted to evaluate the possible contribution of MLN64 in cell junction properties.
Conclusion
The present study demonstrated potential crosstalk between MLN64 and ERBBs in gastric cancer progression, suggesting a pivotal role of interaction between MLN64 and ERBB4 in disease progression of this cancer type. It also showed a significant contribution of MLN64 in predicting patients’ NAC response. Further studies are required to uncover and validate the underlying mechanisms and downstream pathways involved in this signalling.
Conflicts of Interest
The Authors declare no conflicts of interest in relation to this study.
Authors’ Contributions
Conceptualization, W. G. J; Formal analysis, A.X.L, J.Z, X.Z F.R and W.G.J; Investigation, A.X.L, J.Z, F.R and W.G.J; Methodology, F.R, K.J, S.J, A.X.L, X.Z and W.G.J; Resources, W.G.J, S.J; Supervision, W.G.J, T.A.M; Validation, W.G.J, J.Z and A.X.L; Writing – original draft, A.X.L, W.G.J, Q.P.Q, E.K, J.Z; Writing – review & editing, W.G.J, Q.P.D, T.A.M.
Acknowledgements
The Authors wish to acknowledge the Cardiff University China Medical Scholarship program. Andrew J Sanders is a RealCan Fellow.
Funding
The study was supported by the Cardiff University China Medical Scholarship program.
References
- 1.Muss HB, Thor AD, Berry DA, Kute T, Liu ET, Koerner F, Cirrincione CT, Budman DR, Wood WC, Barcos M, Henderson IC. c-erbB-2 expression and response to adjuvant therapy in women with node-positive early breast cancer. N Engl J Med. 1994;330(18):1260–1266. doi: 10.1056/NEJM199405053301802. [DOI] [PubMed] [Google Scholar]
- 2.Moog-Lutz C, Tomasetto C, Regnier CH, Wendling C, Lutz Y, Muller D, Chenard MP, Basset P, Rio MC. MLN64 exhibits homology with the steroidogenic acute regulatory protein (STAR) and is over-expressed in human breast carcinomas. Int J Cancer. 1997;71(2):183–191. doi: 10.1002/(sici)1097-0215(19970410)71:2<183::aid-ijc10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 3.Wilhelm LP, Wendling C, Védie B, Kobayashi T, Chenard MP, Tomasetto C, Drin G, Alpy F. STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J. 2017;36(10):1412–1433. doi: 10.15252/embj.201695917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park EK, Park MJ, Lee SH, Li YC, Kim J, Lee JS, Lee JW, Ye SK, Park JW, Kim CW, Park BK, Kim YN. Cholesterol depletion induces anoikis-like apoptosis via FAK down-regulation and caveolae internalization. J Pathol. 2009;218(3):337–349. doi: 10.1002/path.2531. [DOI] [PubMed] [Google Scholar]
- 5.Asif K, Memeo L, Palazzolo S, Frión-Herrera Y, Parisi S, Caligiuri I, Canzonieri V, Granchi C, Tuccinardi T, Rizzolio F. STARD3: a prospective target for cancer therapy. Cancers (Basel) 2021;13(18):4693. doi: 10.3390/cancers13184693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alpy F, Boulay A, Moog-lutz C, Andarawewa KL, Degot S, Stoll I, Rio MC, Tomasetto C. Metastatic lymph node 64 (MLN64), a gene overexpressed in breast cancers, is regulated by Sp/KLF transcription factors. Oncogene. 2003;22(24):3770–3780. doi: 10.1038/sj.onc.1206500. [DOI] [PubMed] [Google Scholar]
- 7.Dressman MA, Baras A, Malinowski R, Alvis LB, Kwon I, Walz TM, Polymeropoulos M. Gene expression profiling detects gene amplification and differentiates tumor types in breast cancer. Cancer Res. 2003;63(9):2194–2199. [PubMed] [Google Scholar]
- 8.Vinatzer U, Dampier B, Streubel B, Pacher M, Seewald MJ, Stratowa C, Kaserer K, Schreiber M. Expression of HER2 and the coamplified genes GRB7 and MLN64 in human breast cancer: Quantitative real-time reverse transcription-PCR as a diagnostic alternative to immunohistochemistry and fluorescence in situ hybridization. Clin Cancer Res. 2005;11(23):8348–8357. doi: 10.1158/1078-0432.CCR-05-0841. [DOI] [PubMed] [Google Scholar]
- 9.Fararjeh AFS, Al Khader A, Kaddumi E, Obeidat M, Al-Fawares O. Differential expression and prognostic significance of STARD3 gene in breast carcinoma. Int J Mol Cell Med. 2021;10(1):34–41. doi: 10.22088/IJMCM.BUMS.10.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai W, Ye L, Sun J, Mansel RE, Jiang WG. Expression of MLN64 influences cellular matrix adhesion of breast cancer cells, the role for focal adhesion kinase. Int J Mol Med. 2010;25(4):573–80. [PubMed] [Google Scholar]
- 11.Dong JT. Prevalent mutations in prostate cancer. J Cell Biochem. 2006;97(3):433–447. doi: 10.1002/jcb.20696. [DOI] [PubMed] [Google Scholar]
- 12.Stigliano A, Gandini O, Cerquetti L, Gazzaniga P, Misiti S, Monti S, Gradilone A, Falasca P, Poggi M, Brunetti E, Aglianò AM, Toscano V. Increased metastatic lymph node 64 and CYP17 expression are associated with high stage prostate cancer. J Endocrinol. 2007;194(1):55–61. doi: 10.1677/JOE-07-0131. [DOI] [PubMed] [Google Scholar]
- 13.Shin WS, Xie F, Chen B, Yu P, Yu J, To KF, Kang W. Updated epidemiology of gastric cancer in Asia: Decreased incidence but still a big challenge. Cancers (Basel) 2023;15(9):2639. doi: 10.3390/cancers15092639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Zhang H, Zheng J, Wei X, Du J, Lu H, Sun Q, Zhou W, Zhang R, Han Y. Dual silencing of EGFR and HER2 enhances the sensitivity of gastric cancer cells to gefitinib. Mol Carcinog. 2018;57(8):1008–1016. doi: 10.1002/mc.22821. [DOI] [PubMed] [Google Scholar]
- 15.Maqani N, Belkhiri A, Moskaluk C, Knuutila S, Dar AA, El-Rifai W. Molecular dissection of 17q12 amplicon in upper gastrointestinal adenocarcinomas. Mol Cancer Res. 2006;4(7):449–455. doi: 10.1158/1541-7786.MCR-06-0058. [DOI] [PubMed] [Google Scholar]
- 16.Gong W, Zeng J, Ji J, Jia Y, Jia S, Sanders AJ, Jiang WG. EPLIN expression in gastric cancer and impact on prognosis and chemoresistance. Biomolecules. 2021;11(4):547. doi: 10.3390/biom11040547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rondón-Lagos M, Verdun Di Cantogno L, Rangel N, Mele T, Ramírez-Clavijo SR, Scagliotti G, Marchiò C, Sapino A. Unraveling the chromosome 17 patterns of FISH in interphase nuclei: an in-depth analysis of the HER2 amplicon and chromosome 17 centromere by karyotyping, FISH and M-FISH in breast cancer cells. BMC Cancer. 2014;14:922. doi: 10.1186/1471-2407-14-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varis A, Wolf M, Monni O, Vakkari ML, Kokkola A, Moskaluk C, Frierson H Jr, Powell SM, Knuutila S, Kallioniemi A, El-Rifai W. Targets of gene amplification and overexpression at 17q in gastric cancer. Cancer Res. 2002;62(9):2625–2659. [PubMed] [Google Scholar]
- 19.Kwon MJ, Kim RN, Song K, Jeon S, Jeong HM, Kim JS, Han J, Hong S, Oh E, Choi JS, An J, Pollack JR, Choi YL, Park CK, Shin YK. Genes co-amplified with ERBB2 or MET as novel potential cancer-promoting genes in gastric cancer. Oncotarget. 2017;8(54):92209–92226. doi: 10.18632/oncotarget.21150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei YY, Huang JY, Zhao QR, Jiang N, Xu HM, Wang ZN, Li HQ, Zhang SB, Sun Z. The clinicopathological parameters and prognostic significance of HER2 expression in gastric cancer patients: a meta-analysis of literature. World J Surg Oncol. 2017;15(1):68. doi: 10.1186/s12957-017-1132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Gong L, Qian Z, Song G, Liu J. ERBB4 promotes the proliferation of gastric cancer cells via the PI3K/Akt signaling pathway. Oncol Rep. 2018;39(6):2892–2898. doi: 10.3892/or.2018.6343. [DOI] [PubMed] [Google Scholar]
- 22.Almaraz Postigo S, Montero JC. Neuregulin modulates hormone receptor levels in breast cancer through concerted action on multiple signaling pathways. Clin Sci (Lond) 2023;137(1):1–15. doi: 10.1042/CS20220472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haskins JW, Nguyen DX, Stern DF. Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci Signal. 2014;7(355):ra116. doi: 10.1126/scisignal.2005770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao ZF, Liu XY, Wang ZN, Zhao TT, Xu YY, Song YX, Huang JY, Xu H, Xu HM. Effect of neoadjuvant chemotherapy in patients with gastric cancer: a PRISMA-compliant systematic review and meta-analysis. BMC Cancer. 2018;18(1):118. doi: 10.1186/s12885-018-4027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu JJ, Shi YR, Liu FR, Ma SQ, Ma FY. A clinical study of paclitaxel combined with FOLFOX4 regimen as neoadjuvant chemotherapy for advanced gastric cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 2010;13(9):664–667. [PubMed] [Google Scholar]
- 26.Al-Batran SE, Hofheinz RD, Pauligk C, Kopp HG, Haag GM, Luley KB, Meiler J, Homann N, Lorenzen S, Schmalenberg H, Probst S, Koenigsmann M, Egger M, Prasnikar N, Caca K, Trojan J, Martens UM, Block A, Fischbach W, Mahlberg R, Clemens M, Illerhaus G, Zirlik K, Behringer DM, Schmiegel W, Pohl M, Heike M, Ronellenfitsch U, Schuler M, Bechstein WO, Königsrainer A, Gaiser T, Schirmacher P, Hozaeel W, Reichart A, Goetze TO, Sievert M, Jäger E, Mönig S, Tannapfel A. Histopathological regression after neoadjuvant docetaxel, oxaliplatin, fluorouracil, and leucovorin versus epirubicin, cisplatin, and fluorouracil or capecitabine in patients with resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4-AIO): results from the phase 2 part of a multicentre, open-label, randomised phase 2/3 trial. Lancet Oncol. 2016;17(12):1697–1708. doi: 10.1016/S1470-2045(16)30531-9. [DOI] [PubMed] [Google Scholar]
- 27.Hartmann TN. Editorial: Metabolism and cell adhesion in cancer. Front Cell Dev Biol. 2022;10:871471. doi: 10.3389/fcell.2022.871471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian J, Katta A, Masood A, Vudem DR, Kancha RK. Emergence of ERBB2 mutation as a biomarker and an actionable target in solid cancers. Oncologist. 2019;24(12):e1303–e1314. doi: 10.1634/theoncologist.2018-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benzel I, Bansal A, Browning BL, Galwey NW, Maycox PR, McGinnis R, Smart D, St Clair D, Yates P, Purvis I. Interactions among genes in the ErbB-Neuregulin signalling network are associated with increased susceptibility to schizophrenia. Behav Brain Funct. 2007;3:31. doi: 10.1186/1744-9081-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miano C, Romaniello D, Mazzeschi M, Morselli A, Da Pra S, Sacchi F, Bongiovanni C, Sgarzi M, Pantano E, Lauriola M, D’Uva G. Neuregulin 4 boosts the efficacy of anti-ERBB2 neutralizing antibodies. Front Oncol. 2022;12:831105. doi: 10.3389/fonc.2022.831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeong H, Kim J, Lee Y, Seo JH, Hong SR, Kim A. Neuregulin-1 induces cancer stem cell characteristics in breast cancer cell lines. Oncol Rep. 2014;32(3):1218–1224. doi: 10.3892/or.2014.3330. [DOI] [PubMed] [Google Scholar]
- 32.Mill CP, Zordan MD, Rothenberg SM, Settleman J, Leary JF, Riese DJ 2nd. ErbB2 is necessary for ErbB4 ligands to stimulate oncogenic activities in models of human breast cancer. Genes Cancer. 2011;2(8):792–804. doi: 10.1177/1947601911431080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theveneau E, Mayor R. Collective cell migration of epithelial and mesenchymal cells. Cell Mol Life Sci. 2013;70(19):3481–3492. doi: 10.1007/s00018-012-1251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]