Abstract
Background/Aim
Recently, inactivating somatic mutations of SWI/SNF chromatin-remodeling genes in cancers have been reported. However, few studies have been performed regarding the immunological analysis of the tumor microenvironment (TME) in chromatin remodeling complex gene-mutated tumors. In the present study, we identified cancer patients harboring various mammalian SWI/SNF complex mutations and investigated the immunological features in those mutated cancers.
Patients and Methods
Cancer patients harboring any type of chromatin remodeling complex gene mutation were selected and clinicopathological features were compared between chromatin remodeling complex gene expression-low and expression-high groups. Specifically, expression levels of immune response-associated genes and cancer-associated genes were compared between the SMARCA4 expression-low and expression-high groups using volcano plot analysis.
Results
Among cancers harboring PBRM1, SAMRACA4 and ARID2 gene mutations, T-cell marker and mature B-cell marker genes were up-regulated in the tumor. Specifically, T-cell effector genes (CD8B, CD40LG), central memory marker genes (CD27, CCR7) and mature B-cell marker genes (CD20, CD38, CD79 and IRF4) were up-regulated, and cancer-associated genes including MYB, MYC and AURKB genes were down-regulated in the SMARCA4 expression-low group. Remarkably, heatmap of gene expression and immunohistochemistry (IHC) data demonstrated that the tertiary lymphoid structure (TLS) gene signature of mature B cells was up-regulated in SMACA4 gene-mutated stomach cancers.
Conclusion
These results suggest that immune tumor microenvironment status, such as mature B cell recruitment featuring the TLS gene signature and immune activation mediated by cancer signal down-regulation, might contribute to the classification of SMARCA4 gene-mutated tumors as immune checkpoint blockade therapy-sensitive target tumors.
Keywords: Immunological tumor microenvironment, iTME, mammalian SWI/SNF complex, chromatin remodeling gene, SMARCA4 mutation, tumor-infiltrating lymphocytes, TILs
With the remarkable advance in genetic sequencing technologies, inactivating somatic mutations of mammalian switch/sucrose-nonfermenting (SWI/SNF) chromatin-remodeling genes in cancers, such as BRG1/SMARCA4, PBRM1/BAF180, ARID1A/BAF250A, and ARID2/BAF200, have been reported using clinical genome-wide sequencing and given much attention (1-4). SWI/SNF chromatin-remodeling complex genes have been demonstrated to play a role in gene transcription including epigenetic interaction and DNA double-strand repair (1,2), and such mutations leading to loss of function are likely to be involved in cancer development and progression through a senescence or senescence-associated secretory phenotype (SASP) state (5,6).
Mutations in SWI/SNF chromatin-remodeling complex genes are reported in many solid cancers at a rate of approximately 20% (7), such as ovarian and uterine cancer (ARID1A) (8,9), ovarian and lung cancer (SMARCA4) (10-12), renal cell cancer (PBRM1) (13) and rhabdomyosarcoma (SMARCB1) (14). In particular, almost all rare ovarian cancers, small-cell carcinomas of the ovary, hypercalcemic type (SCCOHTs) have SMARCA4 mutations. In addition, many non-small cell lung cancers (10~20%) harboring SMARCA4 mutations with reduced or absent SMARCA4 expression have recently been reported to show a refractory phenotype to standard regimens, with worse prognosis compared to wild type other non-small cell lung cancer patients (15).
Recently, synthetic lethality therapy development has been studied in SWI/SNF chromatin-remodeling complex gene-deficient cancer patients based on the past achievements of PARP inhibitors against BRCA1/BRCA2-deficient tumors (16-18). On the other hand, few studies have been performed regarding immunological analysis of the TME in chromatin remodeling complex-deficient tumors. Recently, using genome-wide genetic screening with CRISPR, the SWI/SNF chromatin-remodeling complex gene ARID1A, was identified as a novel immune checkpoint target, indicating that down-regulation of the ARID1A gene might induce immune activation through T-cell attraction (19,20).
The HOPE genome project at Shizuoka Cancer Center is currently ongoing and has been successful since 2014 in obtaining substantial genome data leading to suitable drug selection and efficient database development. This effort enables medical researchers to search for necessary information from the genomic database derived from approximately 5,000 cancer patients enrolled in the HOPE project (21).
In the present study, we identified cancer patients harboring various mammalian SWI/SNF complex mutations and down-regulated expression of chromatin remodeling genes through the HOPE project, and investigated the immunological features of those mutant cancers.
Patients and Methods
Patient characteristics. The Shizuoka Cancer Center launched Project HOPE in 2014 using multiomic analyses including whole exome sequencing (WES) and gene expression profiling (GEP). Ethical approval for the HOPE study was obtained from the Institutional Review Board of Shizuoka Cancer Center (Authorization Number: 25–33) (21). All experiments using clinical samples were carried out in accordance with the Helsinki Declaration and the Ethical Guidelines for Human Genome and Genetic Analysis Research. The HOPE cohort comprised 5,143 patients treated at the Shizuoka Cancer Center Hospital from January 2014 to March 2019. The cancer patient cohort harboring SWI/SNF chromatin-remodeling gene mutations, such as in SMARCA4, PBRM1, ARID1A, ARID2 and SMARCB1, was selected and divided into lower expression (below median) and higher expression groups.
DNA microarray-based GEP and WES using next-generation sequencing. The method used to perform GEP and WES analyses has been described previously (20). Mutations that were identified in tumor samples and not observed in matched normal samples were identified as somatic mutations. The methods for determining tumor mutation burden (TMB) and copy number variation (CNV) number have been described previously (21).
Immunohistochemistry (IHC). For analysis of tumor-infiltrating immune cells (TILs), antibodies against SMARCA4 (Abcam, cat. ab110641, Cambridge, UK), CD8 (Leica Microsystems GmbH, cat. NCL-CD8-4B11, Wetzlar, Germany), PD-1 (Abcam, cat. Ab52587) and CD20 (Leica Microsystems, cat. NCL-CD20-L26) were purchased. Three representative marker-positive or -negative cases from the SMARCA4 expression-high or -low group were selected and their formalin-fixed, paraffin-embedded (FFPE) specimens were used for immunohistochemistry.
Immune response-associated genes and SCC820 panel gene expression profiling. The lists of genes in the 204 immune response-associated gene panel and the SCC820 cancer-associated panel have been shown previously (22). Briefly, expression levels of 204 immune response-associated genes and the SCC820 cancer-associated genes of SMARCA4-mutant cancers were compared between the SMARCA4 expression-high (above median) group and SMARCA4 expression-low (below median) group with volcano plot analysis. Genes with altered expression (more than 2-fold) were identified.
Statistical analysis. Comparison of the proportion of categorical variables between each gene expression-high group and expression-low group was performed with the Mann-Whitney U-test. p<0.05 was considered significant. Data analysis using the volcano plot was performed with GeneSpring GX software version 14.9.1 (Agilent Technologies, Santa Clara, CA, USA). Overall survival was calculated using the Kaplan-Meier method and statistical significance between each gene expression-high group and expression-low group was evaluated by the log-rank test.
Results
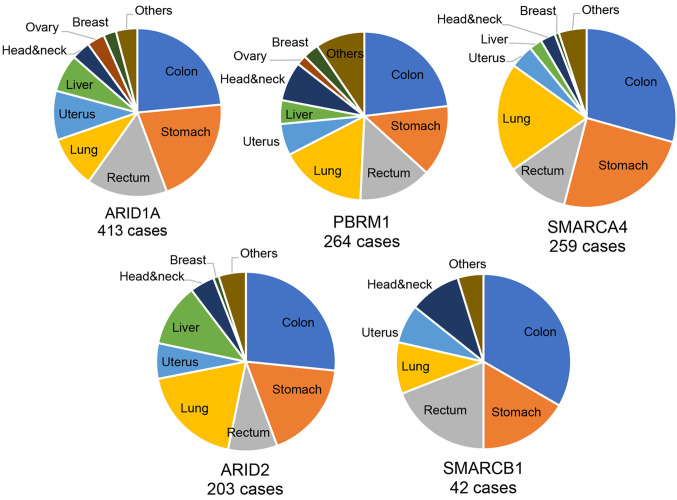
Clinical and genetic characteristics of chromatin remodeling complex gene-mutated cancers. The number of chromatin remodeling complex gene-mutated cancer patients was 413 for ARID1A mutation, 264 for PBRM1 mutation, 259 for SMARCA4 mutation, 203 for ARID2 mutation and 42 for SMARCB1 mutation. The number of cancer patients harboring any type of chromatin remodeling complex gene mutation was 1,181 and comprised 23.0% of all patients. The histological types of those gene-mutated cancers are summarized in Figure 1. There was no significant difference in the frequency of cancer types among each gene-mutated cancer (Figure 1).
Figure 1. Histological frequencies of various chromatin remodeling gene-mutant tumors. The number of chromatin remodeling complex genemutated cancer patients was 413 for ARID1A mutation, 264 for PBRM1 mutation, 259 for SMARCA4 mutation, 203 for ARID2 mutation and 42 for SMARCB1 mutation.
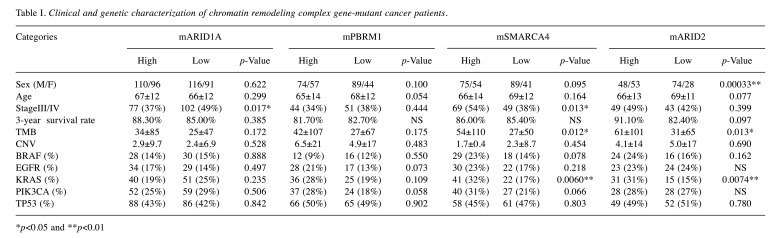
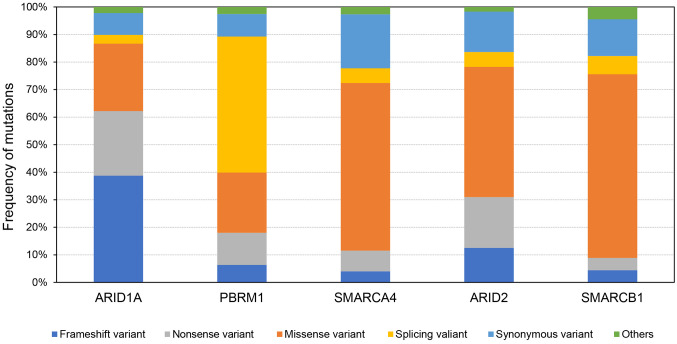
Regarding clinicopathological factors, such as sex, age, pathological staging and performance status, there was no significant difference between the chromatin remodeling gene-mutated cancer groups (Table I). However, in genomic analysis among chromatin remodeling gene-mutated tumors, high TMB and more KRAS-mutated cases were identified in gene expression-high groups for SMARCA4-mutated and ARID2-mutated tumors. Moreover, profiling of mutation patterns in chromatin remodeling gene-mutated tumors indicated that frameshift variants and splicing variants were most frequent in ARID1A- and PBRM1-mutated tumors, respectively, and a higher frequency of missense variants was identified in SMARCA4-, ARID2- and SMARCB1-mutated tumors (Figure 2).
Table I. Clinical and genetic characterization of chromatin remodeling complex gene-mutant cancer patients.
*p<0.05 and **p<0.01
Figure 2. Profiling of various mutations in chromatin remodeling gene-mutant tumors. Frequencies of various patterns of mutations in each chromatin remodeling gene-mutant tumor are shown. The mutation patterns are as follows: frameshift variant, nonsense variant, missense variant, splicing variant, synonymous variant and others.
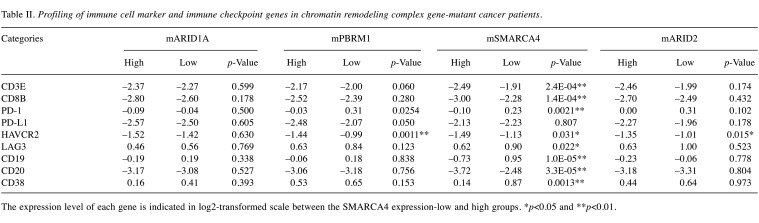
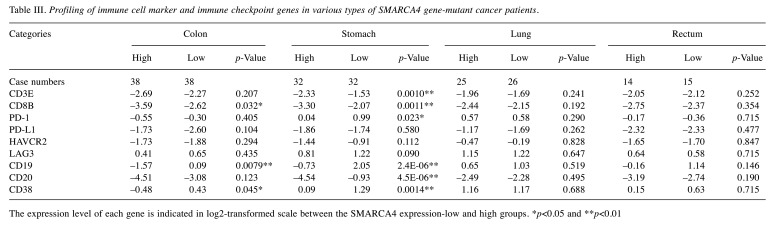
Immune cell-associated marker gene expression of chromatin remodeling complex gene-mutated cancers. Among cancers harboring PBRM1, SAMRACA4 and ARID2 gene mutations, T-cell marker and mature B-cell marker genes were up-regulated in each gene-expression-low group compared with the expression-high group, particularly in SMARCA4-mutated cancers (Table II). Marker genes of exhausted T-cell, such as HAVCR2 and LAG3, were also up-regulated. Regarding the histological type of tumors, PD-1+ T-cell marker and CD38+ mature B-cell marker genes were up-regulated in colon cancers and stomach cancers with SMARCA4 gene-low expression, but not in non-small cell lung and rectal cancers (Table III).
Table II. Profiling of immune cell marker and immune checkpoint genes in chromatin remodeling complex gene-mutant cancer patients.
The expression level of each gene is indicated in log2-transformed scale between the SMARCA4 expression-low and high groups. *p<0.05 and **p<0.01.
Table III. Profiling of immune cell marker and immune checkpoint genes in various types of SMARCA4 gene-mutant cancer patients.
The expression level of each gene is indicated in log2-transformed scale between the SMARCA4 expression-low and high groups. *p<0.05 and **p<0.01.
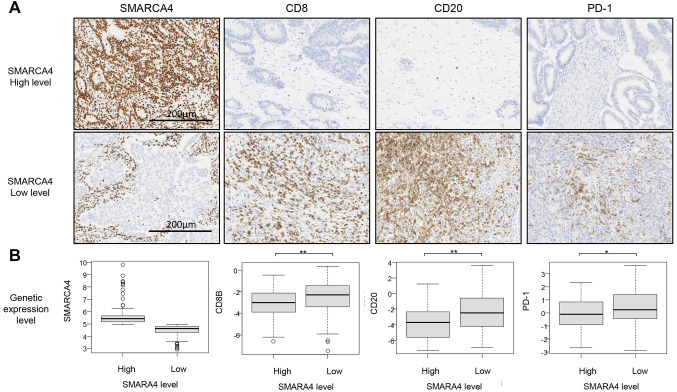
IHC analysis and TMB levels in SMARCA4-mutated cancers. Among SMARCA4-mutated cancers, CD8+PD1+ T cells and CD20+ B cells were increased in SMARCA4 expression-low specimens (Figure 3).
Figure 3. Representative immunohistochemical images of SMARCA4-mutant solid cancer. (A) Antibodies against SMARCA4, CD8, CD20 and PD-1 were used for immunohistochemical (IHC) staining. Images of SMARCA4 and each immune marker staining are shown between SMARCA4 expression-low and -high tumor groups in the upper panel. Magnification: ´200. (B) SMARCA4 and immune marker gene expression levels are shown. The vertical axis shows the expression levels indicated on a log2-transformed scale between the SMARCA4 expression-low and -high groups at the bottom. *p<0.01 and *p<0.05 using the Mann-Whitney U-test.
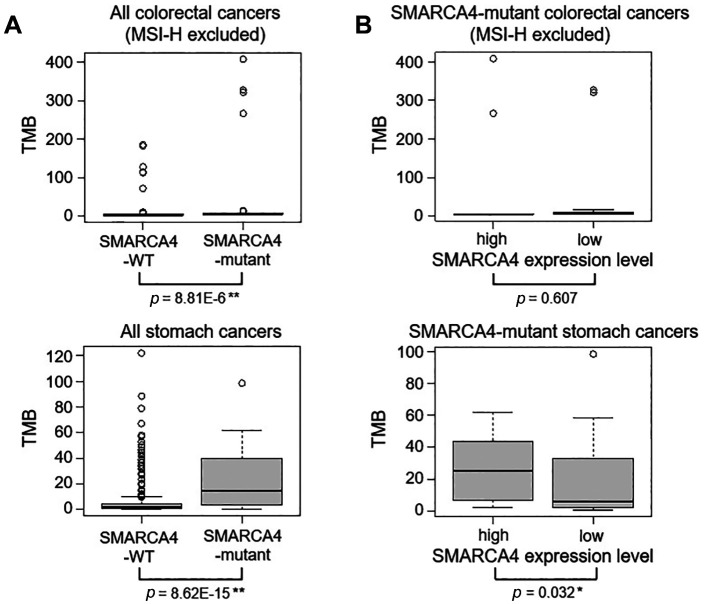
TMB levels were significantly higher in SMARCA4-mutated colon cancers and stomach cancers than in wild-type colon cancers (Figure 4A). Microsatellite instability-high (MSI-H) colon cancers were excluded from all colorectal cancer patients. TMB levels were also higher in the SMARCA4 expression-high group of SMARCA4-mutated stomach cancers; however, TMB levels were not different between the SMARCA4 expression-high and -low groups of SMARCA4-mutated colorectal cancers (Figure 4B).
Figure 4. Tumor mutational burden (TMB) levels in SMARCA4-mutant colon cancers and stomach cancers. Microsatellite instability-high (MSI-H) colon cancers were excluded from all colon cancer patients. (A) Comparison of TMB levels was performed between the SMARCA4-WT and -mutant colon cancer groups and between the SMARCA4-WT and -mutant stomach cancer groups. (B) Comparison of TMB levels was performed between the SMARCA4-high and SMARCA4-low groups of SMARCA4-mutant colon cancers, and between the SMARCA4-high and SMARCA4-low groups of SMARCA4-mutant stomach cancers. *p<0.05 and **p<0.01 using the Mann-Whitney U-test.
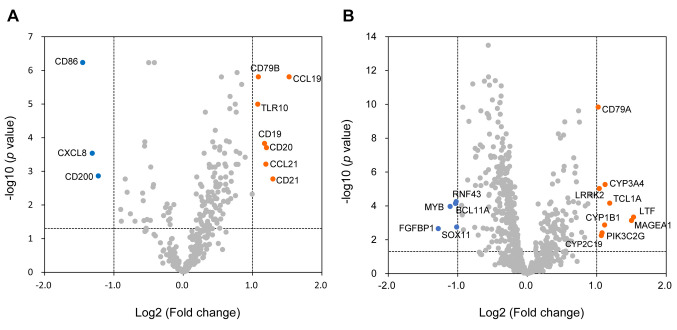
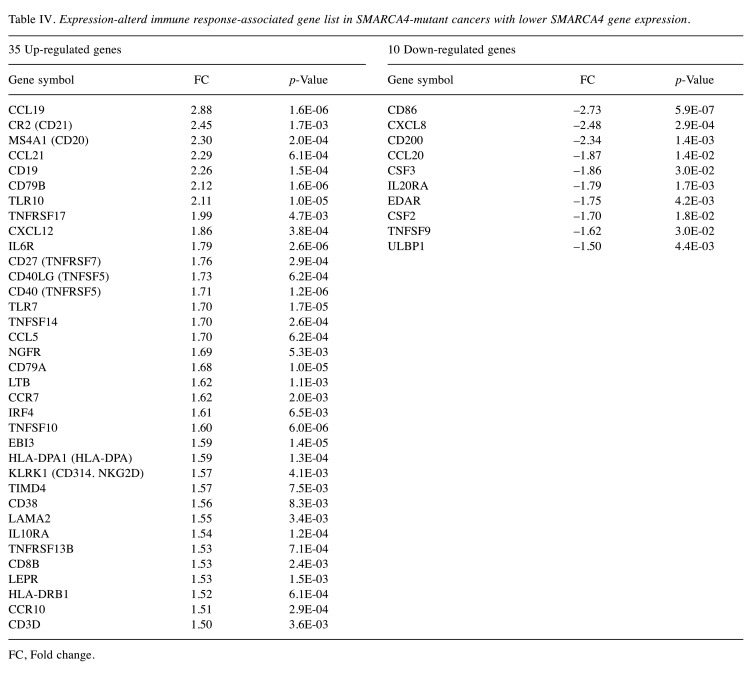
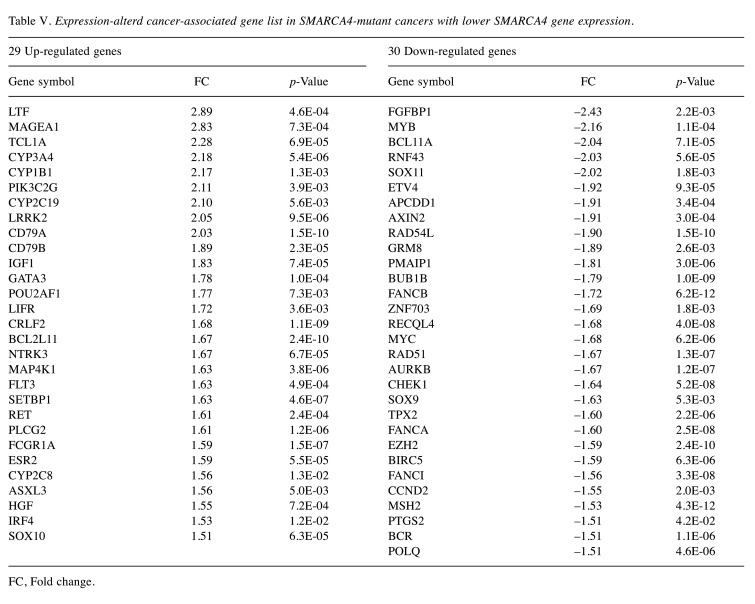
Expression profiling of immune response-associated genes and SCC820 panel genes in SMARCA4-mutated cancers. Volcano plot analysis showed 35 up-regulated and 10 down-regulated genes among immune response-associated genes (Figure 5A, Table IV), and 29 up-regulated and 30 down-regulated genes among the SCC820 cancer-associated genes (Figure 5B, Table V) in the SMARCA4 expression-low group.
Figure 5. Comparison of gene expression between the SMARCA4 expression-low and -high groups of SMARCA4 gene-mutant solid cancers. (A) Immune response-associated genes and (B) cancer-associated SCC820 genes. Up-regulated or down-regulated genes with changes greater than 1.5-fold were identified using volcano plots with Benjamini-Hochberg correction. The horizontal gray line represents a p-value of 0.05. The vertical lines show 2-fold changes in gene expression. Filled circles in orange and blue represent up-regulated and down-regulated genes, respectively.
Table IV. Expression-alterd immune response-associated gene list in SMARCA4-mutant cancers with lower SMARCA4 gene expression.
FC, Fold change.
Table V. Expression-alterd cancer-associated gene list in SMARCA4-mutant cancers with lower SMARCA4 gene expression.
FC, Fold change.
Briefly, T-cell effector genes (CD8B, CD40LG), central memory marker genes (CD27, CCR7) and mature B-cell marker genes (CD20, CD38, CD79, IRF4) were up-regulated. In addition, the CCL19 and CCL21 chemokine genes, which attract functional T- and B-cells inside tumors, increased in expression in the SMARCA4 expression-low group. In contrast, the IL20RA, CD200 and CXCL8 genes, which induce an immunosuppressive TME, were down-regulated.
Cancer-associated genes, including MYB, MYC and AURKB, which correlate with oncogenic signaling and immunosuppression, were down-regulated in the SMARCA4 expression-low group.
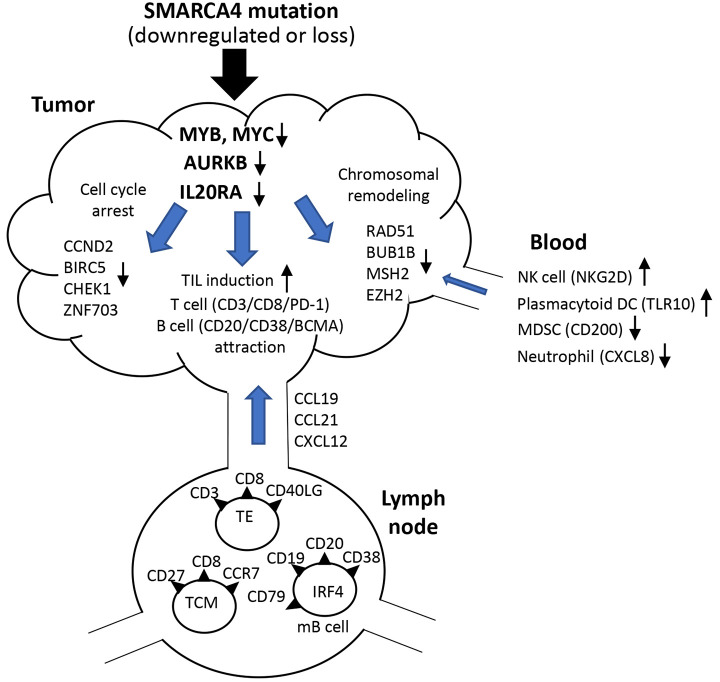
Scheme for the immune-activating state induced by SMARCA4 down-regulation in SMARCA4-mutated cancers. The hypothesis that SMARCA4 down-regulation induces immune-activation events in an immune-suppressive TME is shown in Figure 6. Down-regulation of 3 main genes in the TME, MYC, AURKB and IL20R, might trigger TIL (effector T-cell and mature B-cell) induction, cell cycle arrest and chromatin remodeling insufficiency, which may contribute to immune activation.
Figure 6. Scheme for the immune-activating state induced by SMARCA4 down-regulation in SMARCA4-mutant cancers. Briefly, down-regulations of 3 genes in the TME, namely MYC, AURKB and IL20R, might trigger TIL (effector T-cell and mature B-cell) induction, cell cycle arrest and chromatin remodeling insufficiency, which can contribute to immune activation. In addition, activation of natural killer (NK) cells and dendritic cells (DCs) and inhibition of myeloid derived suppressor cells (MDSCs) and neutrophils in blood may be induced.
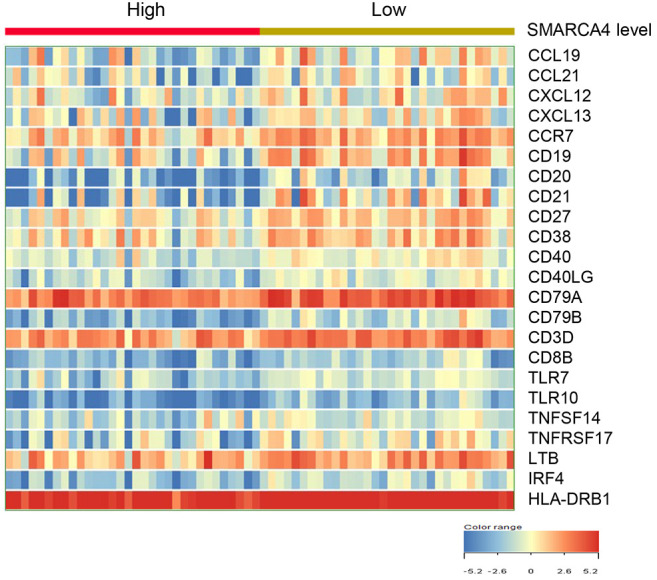
Heatmap of 23 tertiary lymphoid structure (TLS)-associated gene expression between SMARCA4 expression-high and -low groups in SMARCA4-mutated stomach cancers. Among 23 TLS-associated genes, mature B cell marker genes (CD19, CD20, CD27, CD38, CD40), follicular DC marker genes (CD21, CCR7) and chemokine genes (CCL19, CCL21, CXCL13) were up-regulated in the SMARCA4-low group compared to the SMARCA4-high group (Figure 7).
Figure 7. Comparison of the expression levels of TLS-associated 23 genes between SMARCA4-high group and -low groups in SMARCA4-mutated stomach cancers. The data are presented in matrix format, where each row represents an individual case, and each column represents a gene. The red and green colors reflect the gene expression levels, as indicated in the color scale (log2-transformed scale) in the bottom right corner.
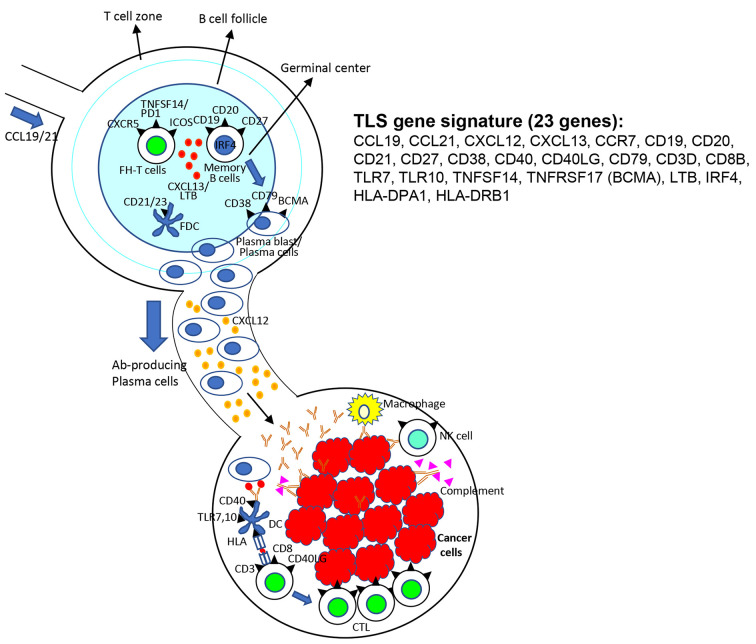
Scheme for TLS signature contributing lymphoid structure development and TLS-associated antitumor effect. Lymphoid stromal fibroblasts and follicular dendritic cells producing lymphotoxins and CXCL13 could be developing the budding of lymphoid follicles. CXCL19 and CXCL21 produced by stromal fibroblasts can attract T-cells like follicular helper (FH) T-cells and memory B-cells to form mature TLS with germinal centers. Memory B-cells can proliferate and become mature plasma cells producing specific antibodies, which can migrate, mediated by CXCL12, to tumor tissues leading to antibody-based antitumor effects (Figure 8).
Figure 8. Scheme for tertiary lymphoid structure (TLS) signature gene contributing lymphoid structure development and TLS-associated antitumor effect. Lymphoid stromal fibroblasts and follicular dendritic cells producing lymphotoxins and CXCL13 are likely to develop the budding of lymphoid follicles. Chemokines produced by stromal fibroblasts, such as CCL19 and CCL21, can attract follicular helper (FH) T-cells and memory B-cells to form mature TLS. Memory Bcells become mature plasma cells producing specific antibodies, which can migrate, mediated by CXCL12, to tumor tissues, leading to antibodybased antitumor effect by antibody-dependent cellular cytotoxicity, complement activation, and CTL inductions.
Discussion
The SWI/SNF complex is a conserved ATP-dependent chromatin remodeling complex that is closely involved in gene transcription and DNA damage repair mechanisms (23). Each complex comprises approximately 15 subunits and is classified into 3 categories: the BRG1/BRM-associated factor (BAF) complex, polybromo-associated BAF (PBAF) complex and noncanonical BAF (ncBAF) complex. The component genes comprise SMARCA4, ARID1A, ARID2, PBRM1 and SMARCB1. Based on the observation that the SWI/SNF complex shows tumor suppressor function, it has been reported that approximately 20% of human cancers harbor any type of mutations in the SWI/SNF complex (24,25). Briefly, more than 95% of malignant rhabdoid tumors and epithelioid sarcomas harbor SMARCB1 mutations leading to loss of SMARCB1 protein expression (14). Recently, an inactivating SMARCA4 mutation has been demonstrated in almost all small-cell carcinomas of the ovary, hypercalcemic type (SCCOHTs) (8).
Furthermore, very recently, rare round-cell thoracic sarcomas harboring SMARCA4-inactivated mutations have been defined as having the following features: rapid course and worse prognosis, heavy smoking exposure, frequent presentation at a younger age, SOX2 up-regulation and claudin-4 loss (26,27). However, thoracic sarcoma tumors are distinguishable from other SMARCA4-mutated (down-regulated) lung adenocarcinomas showing characteristics, such as SMARCA4-deficient adenocarcinoma, CK-positivity and TTF-1 negativity, absence of EGFR driver mutations, and concurrent SMARCA4 and TP53 mutations (28).
Next, synthetic lethality-based therapies should be evaluated because SWI/SNF chromatin remodeling complex-deficient tumors are considered to be good targets for those approaches, particularly in ARID1A-, SMARCA2- and SMARCA4-mutant tumors, which have resulted in the development of PARP and EZH2 inhibitors (17,18). From a cancer metabolic point of view, OXPHOS inhibitors have been demonstrated to be effective for SMARCA4-mutated tumors (29).
Importantly, the SWI/SNF chromatin remodeling complex-deficient state leads to chromatin instability, which activates or triggers the cGAS-STING pathway to sensitize the immune system to broken DNA or RNA released from collapsed nuclei (30-32). Our preliminary genomic analysis data revealed that MMR-deficient (MSI-high) colorectal cancers show up-regulation of cGAS-STING mRNAs, resulting in functional type-I interferon production (unpublished data).
Remarkably, a few studies have demonstrated that immune checkpoint blockade (ICB) therapy leads to positive antitumor responses in PBRM1-deficient clear cell renal cell cancer (33) and ARID1A-deficient ovarian cancers (19). Furthermore, Alessi et al. reported the response to ICB therapy against SMARCA4-mutated non-small cell lung cancer and demonstrated no difference in the objective clinical response rate between SMARCA4-wild type and SMARCA4-mutated non-small cell lung cancer (NSCLC) groups; however, the concurrent SMARCA4 and KRAS-mutated NSCLC group showed a significantly lower overall response rate (ORR) and shorter median overall survival than the SMARCA4-mutant alone group (34). Other clinical researchers have reported similar observations except one case report, which showed a responder to the combination of chemotherapy and ICB (35-37).
Despite previous reports regarding the clinical response of ICB therapy in SMARCA4-mutated cancers, few studies have investigated the immune TME derived from SWI/SNF chromatin remodeling complex gene-mutated tumors (38,39). Ganzer et al. characterized nine cases of thoracic SMARCA4-deficient undifferentiated tumors and found that all specimens had an immune-desert TME phenotype; four patients were given ICB therapy, but only one responded (38).
In the current study, T-cell (CD3, CD8 and PD-1) and mature B-cell marker (CD19, CD20, and CD38) genes were up-regulated inside the tumor in SMARCA4-mutant solid tumors with low SMARCA4 expression (below median). To the best of our knowledge, this is the first report of SMARCA4-mutant tumors. With regard to the association of TMB with TIL accumulation, considering that TMB levels were higher in the SMARCA4 expression-high group of SMARCA4-mutated stomach cancers, TMB is unlikely to be involved in TIL accumulation.
Our hypothesis scheme shown in Figure 6 suggests that immune-activating events inducing T- and B-cell attraction derived from MYC, AURKB and IL20RA gene down-regulation (40,41) involved in SMARCA4-inactivating mutation might be triggered. These results may suggest that the immune TME signature, such as TIL recruitment and immune activation, could contribute to classification of SWI/SNF chromatin remodeling complex gene-mutated tumors as immune checkpoint blockade therapy-sensitive target tumors.
Conclusion
The tertiary lymphoid structure (TLS)-associated gene signature has been proposed in the present study; 23 genes, such as mature B cell marker genes (CD19, CD20, CD27, CD38, CD40, TNFRSF17, IRF4), follicular DC marker genes (CD21, CCR7) and chemokine genes (CCL19, CCL21, CXCL13), were up-regulated in SMARCA4-mutated stomach cancers. Previous research has demonstrated that TLS signature genes are closely involved in budding of lymphoid tissue, development, and maturation of lymphoid follicles with germinal center inside the tumor (42,43).
At the moment specific reasons or mechanisms that induced TLS development and mature B cell accumulation in SMARCA4-mutated stomach cancers have not been elucidated. In the future, TLS gene signature could become a possible biomarker for immune checkpoint-based immunotherapy promoting functional antibody production against cancers. Thus, TLS might be a novel source for cancer neoantigen-specific antibodies and a promising target for single-cell RNAseq-based analysis.
Conflicts of Interest
The Authors declare that they have no conflicts of interest.
Authors’ Contributions
CH and YA participated in the design of the study and drafting of the manuscript and were responsible for supervising the study. TN, KO, YS, and KU performed the genetic analysis using NGS and gene microarray. AI, HM, CM, and TA mainly performed the immunological in vitro experiments. TI performed the statistical analysis. KM and TS contributed to the preparation and staining of pathological specimens. AS, YO, EB, KF, Teiichi Sugiura, TM, SN, YH, Koichi Mitsuya, SY, YT, HK, and MN were involved in collecting the clinical samples and clinical data. Hirotsugu Kenmotsu and KY reviewed the manuscript. All the authors have read and approved the final draft.
Acknowledgements
We thank the staff of the Shizuoka Cancer Center Hospital for assistance in sample preparation and the members of the Shizuoka Cancer Center Research Institute for discussions. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28(14):1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 2.Biegel JA, Busse TM, Weissman BE. SWI/SNF chromatin remodeling complexes and cancer. Am J Med Genet C Semin Med Genet. 2014;166C(3):350–366. doi: 10.1002/ajmg.c.31410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oike T, Ogiwara H, Nakano T, Yokota J, Kohno T. Inactivating mutations in SWI/SNF chromatin remodeling genes in human cancer. Jpn J Clin Oncol. 2013;43(9):849–855. doi: 10.1093/jjco/hyt101. [DOI] [PubMed] [Google Scholar]
- 4.Wilson BG, Roberts CWM. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 5.Marin I, Boix O, Garcia-Garijo A, Sirois I, Caballe A, Zarzuela E, Ruano I, Attolini CS, Prats N, López-Domínguez JA, Kovatcheva M, Garralda E, Muñoz J, Caron E, Abad M, Gros A, Pietrocola F, Serrano M. Cellular senescence is immunogenic and promotes antitumor immunity. Cancer Discov. 2023;13(2):410–431. doi: 10.1158/2159-8290.CD-22-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuollo L, Antonangeli F, Santoni A, Soriani A. The senescence-associated secretory phenotype (SASP) in the challenging future of cancer therapy and age-related diseases. Biology (Basel) 2020;9(12):485. doi: 10.3390/biology9120485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45(6):592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda T, Banno K, Okawa R, Yanokura M, Iijima M, Irie-Kunitomi H, Nakamura K, Iida M, Adachi M, Umene K, Nogami Y, Masuda K, Kobayashi Y, Tominaga E, Aoki D. ARID1A gene mutation in ovarian and endometrial cancers (Review) Oncol Rep. 2016;35(2):607–613. doi: 10.3892/or.2015.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toumpeki C, Liberis A, Tsirkas I, Tsirka T, Kalagasidou S, Inagamova L, Anthoulaki X, Tsatsaris G, Kontomanolis EN. The role of ARID1A in endometrial cancer and the molecular pathways associated with pathogenesis and cancer progression. In Vivo. 2019;33(3):659–667. doi: 10.21873/invivo.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jelinic P, Mueller JJ, Olvera N, Dao F, Scott SN, Shah R, Gao J, Schultz N, Gonen M, Soslow RA, Berger MF, Levine DA. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46(5):424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herpel E, Rieker RJ, Dienemann H, Muley T, Meister M, Hartmann A, Warth A, Agaimy A. SMARCA4 and SMARCA2 deficiency in non-small cell lung cancer: immunohistochemical survey of 316 consecutive specimens. Ann Diagn Pathol. 2017;26:47–51. doi: 10.1016/j.anndiagpath.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Reisman DN, Sciarrotta J, Wang W, Funkhauser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63(3):560–566. [PubMed] [Google Scholar]
- 13.Linehan WM, Ricketts CJ. The Cancer Genome Atlas of renal cell carcinoma: findings and clinical implications. Nat Rev Urol. 2019;16(9):539–552. doi: 10.1038/s41585-019-0211-5. [DOI] [PubMed] [Google Scholar]
- 14.Sigauke E, Rakheja D, Maddox DL, Hladik CL, White CL, Timmons CF, Raisanen J. Absence of expression of SMARCB1/INI1 in malignant rhabdoid tumors of the central nervous system, kidneys and soft tissue: an immunohistochemical study with implications for diagnosis. Mod Pathol. 2006;19(5):717–725. doi: 10.1038/modpathol.3800581. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida A, Kobayashi E, Kubo T, Kodaira M, Motoi T, Motoi N, Yonemori K, Ohe Y, Watanabe S, Kawai A, Kohno T, Kishimoto H, Ichikawa H, Hiraoka N. Clinicopathological and molecular characterization of SMARCA4-deficient thoracic sarcomas with comparison to potentially related entities. Mod Pathol. 2017;30(6):797–809. doi: 10.1038/modpathol.2017.11. [DOI] [PubMed] [Google Scholar]
- 16.Pilié PG, Gay CM, Byers LA, O’Connor MJ, Yap TA. PARP inhibitors: Extending benefit beyond BRCA-mutant cancers. Clin Cancer Res. 2019;25(13):3759–3771. doi: 10.1158/1078-0432.CCR-18-0968. [DOI] [PubMed] [Google Scholar]
- 17.Oike T, Ogiwara H, Tominaga Y, Ito K, Ando O, Tsuta K, Mizukami T, Shimada Y, Isomura H, Komachi M, Furuta K, Watanabe S, Nakano T, Yokota J, Kohno T. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013;73(17):5508–5518. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 18.Chan-Penebre E, Armstrong K, Drew A, Grassian AR, Feldman I, Knutson SK, Kuplast-Barr K, Roche M, Campbell J, Ho P, Copeland RA, Chesworth R, Smith JJ, Keilhack H, Ribich SA. Selective killing of SMARCA2- and SMARCA4-deficient small cell carcinoma of the ovary, hypercalcemic type cells by inhibition of EZH2: In vitro and in vivo preclinical models. Mol Cancer Ther. 2017;16(5):850–860. doi: 10.1158/1535-7163.MCT-16-0678. [DOI] [PubMed] [Google Scholar]
- 19.Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, Nagel ZD, Zou J, Wang C, Kapoor P, Ma X, Ma D, Liang J, Song S, Liu J, Samson LD, Ajani JA, Li GM, Liang H, Shen X, Mills GB, Peng G. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24(5):556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belk JA, Yao W, Ly N, Freitas KA, Chen YT, Shi Q, Valencia AM, Shifrut E, Kale N, Yost KE, Duffy CV, Daniel B, Hwee MA, Miao Z, Ashworth A, Mackall CL, Marson A, Carnevale J, Vardhana SA, Satpathy AT. Genome-wide CRISPR screens of T cell exhaustion identify chromatin remodeling factors that limit T cell persistence. Cancer Cell. 2022;40(7):768–786.e7. doi: 10.1016/j.ccell.2022.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagashima T, Yamaguchi K, Urakami K, Shimoda Y, Ohnami S, Ohshima K, Tanabe T, Naruoka A, Kamada F, Serizawa M, Hatakeyama K, Matsumura K, Ohnami S, Maruyama K, Mochizuki T, Kusuhara M, Shiomi A, Ohde Y, Terashima M, Uesaka K, Onitsuka T, Nishimura S, Hirashima Y, Hayashi N, Kiyohara Y, Tsubosa Y, Katagiri H, Niwakawa M, Takahashi K, Kashiwagi H, Nakagawa M, Ishida Y, Sugino T, Takahashi M, Akiyama Y. Japanese version of The Cancer Genome Atlas, JCGA, established using fresh frozen tumors obtained from 5143 cancer patients. Cancer Sci. 2020;111(2):687–699. doi: 10.1111/cas.14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasui K, Kondou R, Miyata H, Iizuka A, Ashizawa T, Nagashima T, Ohshima K, Urakami K, Muramatsu K, Sugino T, Yamaguchi K, Ogawa H, Onoe T, Harada H, Asakura H, Murayama S, Nishimura T, Goto S, Okada S, Mukaigawa T, Hamauchi S, Yokota T, Onozawa Y, Akiyama Y. Immunological and genetic characterization of patients with head and neck cancer who developed recurrence. Anticancer Res. 2022;42(9):4417–4428. doi: 10.21873/anticanres.15942. [DOI] [PubMed] [Google Scholar]
- 23.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1(5):e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hohmann AF, Vakoc CR. A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet. 2014;30(8):356–363. doi: 10.1016/j.tig.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jancewicz I, Siedlecki JA, Sarnowski TJ, Sarnowska E. BRM: the core ATPase subunit of SWI/SNF chromatin-remodelling complex-a tumour suppressor or tumour-promoting factor. Epigenetics Chromatin. 2019;12(1):68. doi: 10.1186/s13072-019-0315-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Loarer F, Watson S, Pierron G, de Montpreville VT, Ballet S, Firmin N, Auguste A, Pissaloux D, Boyault S, Paindavoine S, Dechelotte PJ, Besse B, Vignaud JM, Brevet M, Fadel E, Richer W, Treilleux I, Masliah-Planchon J, Devouassoux-Shisheboran M, Zalcman G, Allory Y, Bourdeaut F, Thivolet-Bejui F, Ranchere-Vince D, Girard N, Lantuejoul S, Galateau-Sallé F, Coindre JM, Leary A, Delattre O, Blay JY, Tirode F. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet. 2015;47(10):1200–1205. doi: 10.1038/ng.3399. [DOI] [PubMed] [Google Scholar]
- 27.Rekhtman N, Montecalvo J, Chang JC, Alex D, Ptashkin RN, Ai N, Sauter JL, Kezlarian B, Jungbluth A, Desmeules P, Beras A, Bishop JA, Plodkowski AJ, Gounder MM, Schoenfeld AJ, Namakydoust A, Li BT, Rudin CM, Riely GJ, Jones DR, Ladanyi M, Travis WD. SMARCA4-deficient thoracic sarcomatoid tumors represent primarily smoking-related undifferentiated carcinomas rather than primary thoracic sarcomas. J Thorac Oncol. 2020;15(2):231–247. doi: 10.1016/j.jtho.2019.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agaimy A, Fuchs F, Moskalev EA, Sirbu H, Hartmann A, Haller F. SMARCA4-deficient pulmonary adenocarcinoma: clinicopathological, immunohistochemical, and molecular characteristics of a novel aggressive neoplasm with a consistent TTF1neg/CK7pos/HepPar-1pos immunophenotype. Virchows Arch. 2017;471(5):599–609. doi: 10.1007/s00428-017-2148-5. [DOI] [PubMed] [Google Scholar]
- 29.Lissanu Deribe Y, Sun Y, Terranova C, Khan F, Martinez-Ledesma J, Gay J, Gao G, Mullinax RA, Khor T, Feng N, Lin YH, Wu CC, Reyes C, Peng Q, Robinson F, Inoue A, Kochat V, Liu CG, Asara JM, Moran C, Muller F, Wang J, Fang B, Papadimitrakopoulou V, Wistuba II, Rai K, Marszalek J, Futreal PA. Mutations in the SWI/SNF complex induce a targetable dependence on oxidative phosphorylation in lung cancer. Nat Med. 2018;24(7):1047–1057. doi: 10.1038/s41591-018-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chabanon RM, Rouanne M, Lord CJ, Soria JC, Pasero P, Postel-Vinay S. Targetable the DNA damage response in immune-oncology: developments and opportunities. Nat Rev Cancer. 2021;21(11):701–717. doi: 10.1038/s41568-021-00386-6. [DOI] [PubMed] [Google Scholar]
- 31.Reisländer T, Groelly FJ, Tarsounas M. DNA damage and cancer immunotherapy: a STING in the tale. Mol Cell. 2020;80(1):21–28. doi: 10.1016/j.molcel.2020.07.026. [DOI] [PubMed] [Google Scholar]
- 32.Zheng J, Mo J, Zhu T, Zhuo W, Yi Y, Hu S, Yin J, Zhang W, Zhou H, Liu Z. Comprehensive elaboration of the cGAS-STING signaling axis in cancer development and immunotherapy. Mol Cancer. 2020;19(1):133. doi: 10.1186/s12943-020-01250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, Norton C, Bossé D, Wankowicz SM, Cullen D, Horak C, Wind-Rotolo M, Tracy A, Giannakis M, Hodi FS, Drake CG, Ball MW, Allaf ME, Snyder A, Hellmann MD, Ho T, Motzer RJ, Signoretti S, Kaelin WG Jr, Choueiri TK, Van Allen EM. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alessi JV, Ricciuti B, Spurr LF, Gupta H, Li YY, Glass C, Nishino M, Cherniack AD, Lindsay J, Sharma B, Felt KD, Rodig SJ, Cheng ML, Sholl LM, Awad MM. SMARCA4 and other SWItch/sucrose nonfermentable family genomic alterations in NSCLC: Clinicopathologic characteristics and outcomes to immune checkpoint inhibition. J Thorac Oncol. 2021;16(7):1176–1187. doi: 10.1016/j.jtho.2021.03.024. [DOI] [PubMed] [Google Scholar]
- 35.Schoenfeld AJ, Bandlamudi C, Lavery JA, Montecalvo J, Namakydoust A, Rizvi H, Egger J, Concepcion CP, Paul S, Arcila ME, Daneshbod Y, Chang J, Sauter JL, Beras A, Ladanyi M, Jacks T, Rudin CM, Taylor BS, Donoghue MTA, Heller G, Hellmann MD, Rekhtman N, Riely GJ. The genomic landscape of SMARCA4 alterations and associations with outcomes in patients with lung cancer. Clin Cancer Res. 2020;26(21):5701–5708. doi: 10.1158/1078-0432.CCR-20-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H, Shen J, Liu J, Fang W, Zhang L. Efficacy of immune checkpoint inhibitors in SMARCA4-mutant NSCLC. J Thorac Oncol. 2020;15(8):e133–e136. doi: 10.1016/j.jtho.2020.03.030. [DOI] [PubMed] [Google Scholar]
- 37.Utsumi T, Taniguchi Y, Noda Y, Fukai M, Kibata K, Murakawa T. SMARCA4-deficient undifferentiated tumor that responded to chemotherapy in combination with immune checkpoint inhibitors: A case report. Thorac Cancer. 2022;13(15):2264–2266. doi: 10.1111/1759-7714.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gantzer J, Davidson G, Vokshi B, Weingertner N, Bougoüin A, Moreira M, Lindner V, Lacroix G, Mascaux C, Chenard MP, Bertucci F, Davidson I, Kurtz JE, Sautès-Fridman C, Fridman WH, Malouf GG. Immune-desert tumor microenvironment in thoracic SMARCA4-deficient undifferentiated tumors with limited efficacy of immune checkpoint inhibitors. Oncologist. 2022;27(6):501–511. doi: 10.1093/oncolo/oyac040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Fan R, Chen D, Hou J, Chen H, Lu M. Pathological characteristics and immune microenvironment of SMARCA4-deficient undifferentiated uterine sarcoma. Diagn Pathol. 2023;18(1):62. doi: 10.1186/s13000-023-01347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang J, Wang J, Yue M, Cai X, Wang T, Wu C, Su H, Wang Y, Han M, Zhang Y, Zhu X, Jiang P, Li P, Sun Y, Xiao W, Feng H, Qing G, Liu H. Direct phosphorylation and stabilization of MYC by Aurora B kinase promote T-cell leukemogenesis. Cancer Cell. 2020;37(2):200–215.e5. doi: 10.1016/j.ccell.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao W, Wen H, Liang L, Dong X, Du R, Zhou W, Zhang X, Zhang C, Xiang R, Li N. IL20RA signaling enhances stemness and promotes the formation of an immunosuppressive microenvironment in breast cancer. Theranostics. 2021;11(6):2564–2580. doi: 10.7150/thno.45280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randall TD, Carragher DM, Rangel-Moreno J. Development of secondary lymphoid organs. Annu Rev Immunol. 2008;26:627–650. doi: 10.1146/annurev.immunol.26.021607.090257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaeuble K, Britschgi MR, Scarpellino L, Favre S, Xu Y, Koroleva E, Lissandrin TK, Link A, Matloubian M, Ware CF, Nedospasov SA, Tumanov AV, Cyster JG, Luther SA. Perivascular fibroblasts of the developing spleen act as LTα1β2-dependent precursors of both T and B zone organizer cells. Cell Rep. 2017;21(9):2500–2514. doi: 10.1016/j.celrep.2017.10.119. [DOI] [PubMed] [Google Scholar]