Abstract
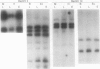
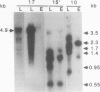
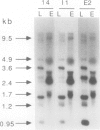
Amyloplasts in storage organs such as maize (Zea mays L.) endosperm are plastid-derived, nonphotosynthetic, starch-accumulating organelles. This study was initiated to characterize the plastid genome in maize endosperm cells containing differentiated amyloplasts and to determine whether plastid genes are transcribed during the period of amyloplast biogenesis in endosperm development. Four cosmid clones representing the total sequence diversity of the maize plastid genome were hybridized to restriction digests of total cellular DNA from isolated 16-day-old endosperms. The hybridization patterns indicated that the plastid DNA present in endosperm tissue was indistinguishable from that in leaf total DNA. Methylation of maize endosperm amyloplast DNA or leaf chloroplast DNA was not detected with the methylation-sensitive enzymes HpaII and EcoRII. Transcripts homologous to the 17 specific plastid DNA BamHI fragments tested were detectable in total RNA prepared from 16-day-old endosperm tissue. Compared with leaf transcripts, the abundance of endosperm transcripts was substantially lower for transcripts detected by 12 different BamHI fragments and was similar or relatively higher for some transcripts homologous to five BamHi fragments. Transcripts homologous to genes for plastid ribosomal small subunit proteins 7 and 12 on fragments 10 and 23 and to an open reading frame on fragment 14 accumulated primarily as unprocessed or partially processed species in endosperm RNA. The demonstration that maize endosperm cells contain an intact, transcriptionally active plastid genome indicates that plastid genes could contribute to amyloplast biogenesis, although no transcripts unique to endosperm were identified.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Deng X. W., Gruissem W. Constitutive transcription and regulation of gene expression in non-photosynthetic plastids of higher plants. EMBO J. 1988 Nov;7(11):3301–3308. doi: 10.1002/j.1460-2075.1988.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Fish L. E., Kück U., Bogorad L. Two partially homologous adjacent light-inducible maize chloroplast genes encoding polypeptides of the P700 chlorophyll a-protein complex of photosystem I. J Biol Chem. 1985 Feb 10;260(3):1413–1421. [PubMed] [Google Scholar]
- Giese K., Subramanian A. R., Larrinua I. M., Bogorad L. Nucleotide sequence, promoter analysis, and linkage mapping of the unusually organized operon encoding ribosomal proteins S7 and S12 in maize chloroplast. J Biol Chem. 1987 Nov 5;262(31):15251–15255. [PubMed] [Google Scholar]
- Haley J., Bogorad L. Alternative promoters are used for genes within maize chloroplast polycistronic transcription units. Plant Cell. 1990 Apr;2(4):323–333. doi: 10.1105/tpc.2.4.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C. R., Meng B. Y. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989 Jun;217(2-3):185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Hunt C. M., Hardison R. C., Boyer C. D. Restriction enzyme analysis of tomato chloroplast and chromoplast DNA. Plant Physiol. 1986 Dec;82(4):1145–1147. doi: 10.1104/pp.82.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangasjärvi J., McCullough A., Gengenbach B. G. Nucleotide sequence and transcription of maize plastid genome Bam HI fragment 14 containing ORF170. Plant Mol Biol. 1991 Sep;17(3):513–515. doi: 10.1007/BF00040647. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Ngernprasirtsiri J., Akazawa T. Transcriptional regulation and DNA methylation in plastids during transitional conversion of chloroplasts to chromoplasts. EMBO J. 1990 Feb;9(2):307–313. doi: 10.1002/j.1460-2075.1990.tb08112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marano M. R., Carrillo N. Chromoplast formation during tomato fruit ripening. No evidence for plastid DNA methylation. Plant Mol Biol. 1991 Jan;16(1):11–19. doi: 10.1007/BF00017913. [DOI] [PubMed] [Google Scholar]
- Ngernprasirtsiri J., Chollet R., Kobayashi H., Sugiyama T., Akazawa T. DNA methylation and the differential expression of C4 photosynthesis genes in mesophyll and bundle sheath cells of greening maize leaves. J Biol Chem. 1989 May 15;264(14):8241–8248. [PubMed] [Google Scholar]
- Ngernprasirtsiri J., Kobayashi H., Akazawa T. DNA Methylation Occurred around Lowly Expressed Genes of Plastid DNA during Tomato Fruit Development. Plant Physiol. 1988 Sep;88(1):16–20. doi: 10.1104/pp.88.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngernprasirtsiri J., Kobayashi H., Akazawa T. DNA methylation as a mechanism of transcriptional regulation in nonphotosynthetic plastids in plant cells. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4750–4754. doi: 10.1073/pnas.85.13.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngernprasirtsiri J., Macherel D., Kobayashi H., Akazawa T. Expression of Amyloplast and Chloroplast DNA in Suspension-Cultured Cells of Sycamore (Acer pseudoplatanus L.). Plant Physiol. 1988 Jan;86(1):137–142. doi: 10.1104/pp.86.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]