Abstract
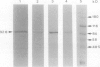
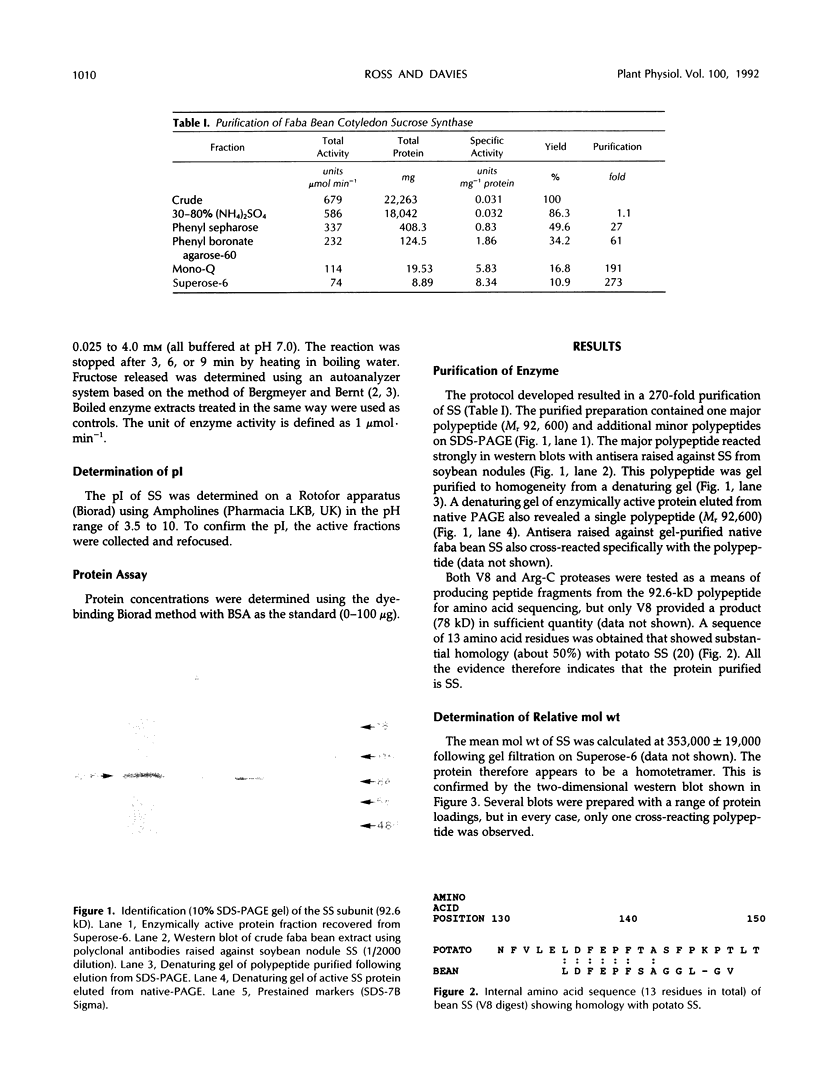
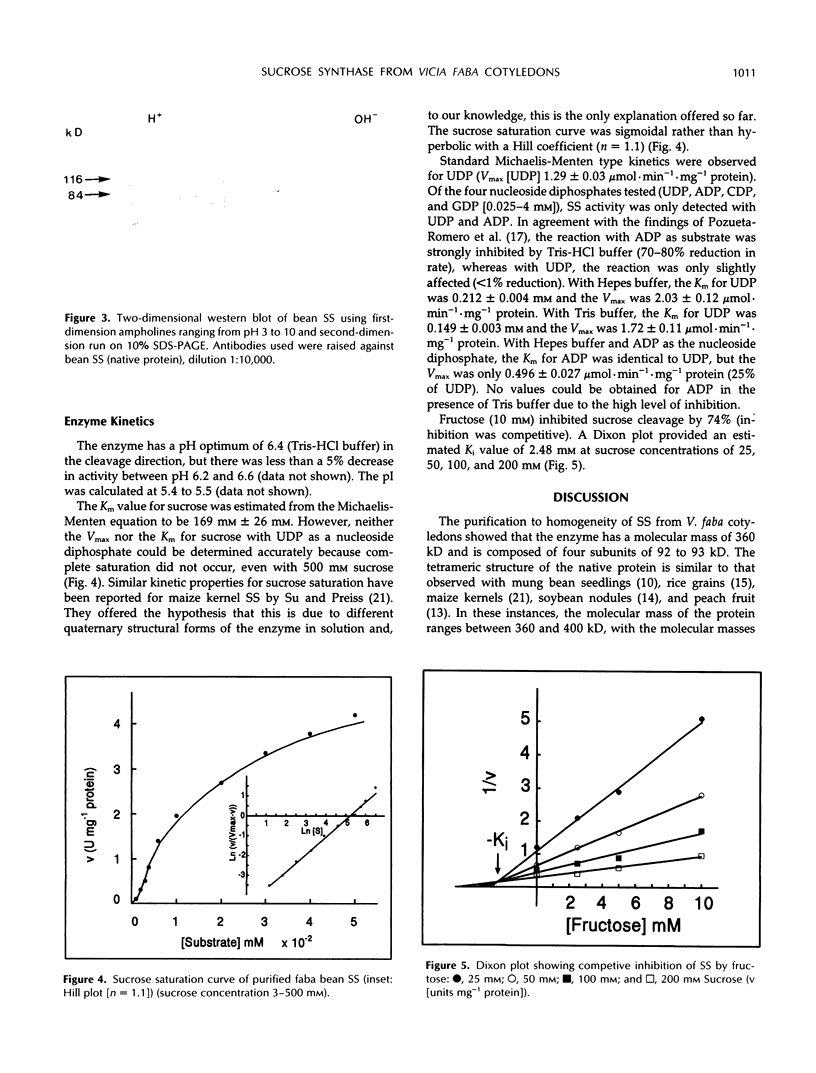
Partial purification (approximately 270-fold) of sucrose synthase (EC 2.4.1.13) from developing cotyledons of Vicia faba L. cv Maris Bead was achieved by ammonium sulfate fractionation and hydrophobic, affinity, anion-exchange, and gel filtration chromatography. Further purification to homogeneity resulted from gel elution of single bands from native and sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The enzyme was identified as a homotetramer with a total molecular mass of 360 kD and subunits of 92 to 93 kD. Antibodies were raised to both native and denatured protein. The identity of the polypeptide was confirmed in western blots using antibodies raised against soybean nodule sucrose synthase. The enzyme has a pH optimum of 6.4 (cleavage direction) and an isoelectric point of 5.5. The affinity of the enzyme for sucrose (Km) was estimated at 169 mm, and for UDP at 0.2 mm. With uridine diphosphate as the nucleoside diphosphate, the Vmax is 4-fold higher than with adenosine diphosphate. Fructose acts as a competitive inhibitor with an inhibitor constant (Ki) of 2.48 mm.
Full text
PDF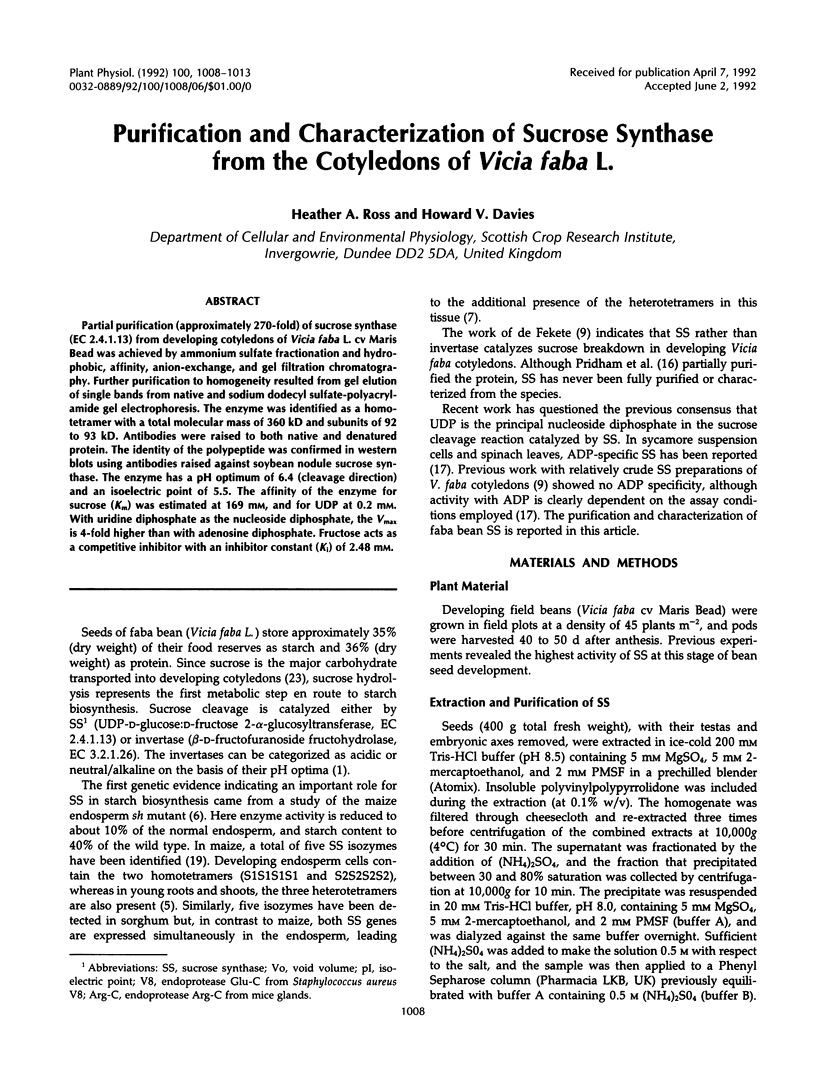



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chourey P. S., Nelson O. E. The enzymatic deficiency conditioned by the shrunken-1 mutations in maize. Biochem Genet. 1976 Dec;14(11-12):1041–1055. doi: 10.1007/BF00485135. [DOI] [PubMed] [Google Scholar]
- Chourey P. S., Taliercio E. W., Kane E. J. Tissue-Specific Expression and Anaerobically Induced Posttranscriptional Modulation of Sucrose Synthase Genes in Sorghum bicolor M. Plant Physiol. 1991 Jun;96(2):485–490. doi: 10.1104/pp.96.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Delmer D. P. The purification and properties of sucrose synthetase from etiolated Phaseolus aureus seedlings. J Biol Chem. 1972 Jun 25;247(12):3822–3828. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Morell M., Copeland L. Sucrose synthase of soybean nodules. Plant Physiol. 1985 May;78(1):149–154. doi: 10.1104/pp.78.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Akazawa T. Enzymic mechanism of starch stynthesis in ripening rice grains. VII. Purification and enzymic properties of sucrose synthetase. Arch Biochem Biophys. 1973 Jun;156(2):644–652. doi: 10.1016/0003-9861(73)90316-0. [DOI] [PubMed] [Google Scholar]
- Pozueta-Romero J., Yamaguchi J., Akazawa T. ADPG formation by the ADP-specific cleavage of sucrose-reassessment of sucrose synthase. FEBS Lett. 1991 Oct 21;291(2):233–237. doi: 10.1016/0014-5793(91)81292-g. [DOI] [PubMed] [Google Scholar]
- Ross H. A., Davies H. V. Sucrose Metabolism in Tubers of Potato (Solanum tuberosum L.): Effects of Sink Removal and Sucrose Flux on Sucrose-Degrading Enzymes. Plant Physiol. 1992 Jan;98(1):287–293. doi: 10.1104/pp.98.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat M., Belliard G. Molecular cloning and sequencing of sucrose synthase cDNA from potato (Solanum tuberosum L.): preliminary characterization of sucrose synthase mRNA distribution. Gene. 1987;60(1):47–56. doi: 10.1016/0378-1119(87)90212-5. [DOI] [PubMed] [Google Scholar]
- Su J. C., Preiss J. Purification and properties of sucrose synthase from maize kernels. Plant Physiol. 1978 Mar;61(3):389–393. doi: 10.1104/pp.61.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolosiuk R. A., Pontis H. G. Studies on sucrose synthetase. Kinetic mechanism. Arch Biochem Biophys. 1974 Nov;165(1):140–145. doi: 10.1016/0003-9861(74)90151-9. [DOI] [PubMed] [Google Scholar]