Abstract
New onset refractory status epilepticus (NORSE), including its subtype with a preceding febrile illness known as febrile infection-related epilepsy syndrome (FIRES), is one of the most severe forms of status epilepticus. The exact causes of NORSE are currently unknown, and there is so far no disease-specific therapy. Identifying the underlying pathophysiology and discovering specific biomarkers, whether immunologic, infectious, genetic, or other, may help physicians in the management of patients with NORSE. A broad spectrum of biomarkers has been proposed for status epilepticus patients, some of which were evaluated for patients with NORSE. Nonetheless, none has been validated, due to significant variabilities in study cohorts, collected biospecimens, applied analytical methods, and defined outcome endpoints, and to small sample sizes. The NORSE Institute established an open NORSE/FIRES biorepository for health-related data and biological samples allowing the collection of biospecimens worldwide, promoting multicenter research and sharing of data and specimens. Here, we suggest standard operating procedures for biospecimen collection and biobanking in this rare condition. We also propose criteria for the appropriate use of previously collected biospecimens. We predict that the widespread use of standardized procedures will reduce heterogeneity, facilitate the future identification of validated biomarkers for NORSE, and provide a better understanding of the pathophysiology and best clinical management for these patients.
Keywords: biomarkers, biospecimens, new onset refractory status epilepticus, standard operating procedures
1 |. INTRODUCTION
Status epilepticus (SE) is a life-threatening prolonged epileptic seizure that affects approximately 40 per 100 000 people yearly, making it the second most common neurological emergency after stroke.1,2 Approximately 25% of SE cases are refractory (RSE) to appropriate antiseizure medications. Patients with new onset refractory status epilepticus (NORSE) often have the most refractory seizures and worst outcomes. NORSE occurs in adults or children without active epilepsy and without a clear acute or active structural, toxic, or metabolic cause.3 Febrile infection-related epilepsy syndrome (FIRES) is a subset of NORSE that involves a prior febrile infection that started between 2 weeks and 24 h before the onset of SE.3 The mortality of NORSE is approximately 12% in children and even higher in adults (16%–27%).4–6 The prognosis is often guarded among survivors, as most patients have long-term neurocognitive sequelae and/or epilepsy.6–8 Although some cases of NORSE and FIRES might be caused by autoimmune or infectious encephalitis or rare genetic disorders, the majority remain unexplained (NORSE of unknown etiology, or cryptogenic NORSE). The pathophysiology of cryptogenic NORSE is largely unknown, and there is so far no disease-specific therapy. Understanding the pathophysiological mechanisms underlying cryptogenic NORSE is crucial to improve patient management and to prevent secondary neuronal injury. Identifying biomarkers including direct causal agents such as as-yet undiagnosed infections or autoantibodies may help elucidate the pathophysiology, promptly identify patients, monitor the disease evolution, guide specific therapy, and predict outcomes.
Although biomarkers are becoming increasingly important in the era of precision medicine, there are concerns over the high attrition of promising biomarkers in neurologic disorders. Although dozens of cerebrospinal fluid (CSF) and blood biomarkers have been proposed in SE, independent studies have not validated them due to significant variabilities in the fundamental aspects of biomarker studies.9 First, there is considerable variability in the populations studied with respect to etiologies, clinical subtypes, concomitant disorders, ages, treatments, and controls used for comparison. Second, specimens are frequently collected at different time points after disease onset, although biomarkers are known to change over time during the course of SE.10 Third, specimens are often retrieved from various anatomical sources including CSF, serum, plasma, whole blood, human peripheral blood mononuclear cells (PBMC), urine, stool, and brain tissue. In addition to these limitations, various preanalytical processing and storage methods are usually applied, and these are known to significantly impact biomarker measurement results.11–13
On a more conceptual level, the identification and validation of NORSE biomarkers are furthermore limited by the lack of sufficient samples that can be obtained at a single center or even by a small consortium and by the prior heterogeneity in criteria used for NORSE diagnosis. To enable improved communication and allow sufficiently powered multicenter research, standardized definitions of NORSE and FIRES were published a few years ago, and an international collection of health-related data and biological samples from patients with NORSE, the NORSE/ FIRES biorepository, was established.3 Recognizing that the field of SE and NORSE can present particularities due to clinical heterogeneity and disease evolution over time, it is important to establish standardization for collecting and analyzing relevant biospecimens.
In the present review, we summarize the proposed NORSE biomarkers and provide standard operating procedures for biospecimen collection and storage. We also propose criteria for the appropriate use of previously collected biospecimens based on the proposed analyses and the prior method of specimen handling and storage. This is a consensus protocol established by an international multidisciplinary team of neurologists, neuropathologists, researchers, and pharmacists involved with the NORSE Institute. The consensus protocol was achieved by intensive discussion during the NORSE Institute Medical and Scientific Advisory Board and NORSE/FIRES Biorepository Steering Committee meetings, followed by email, video conference, and phone discussions. We hope these consensus criteria will promote the collection of high-quality samples and permit multicenter international research to improve the understanding and treatment of NORSE. Standardizing the clinical features, diagnosis, management, and outcome measures of patients with RSE (including NORSE) has been undertaken by a separate (but overlapping) group of investigators.14,15
2 |. CRITERIA FOR GOOD BIOMARKERS
Biomarkers could be qualitative or quantitative and of different types (e.g., biochemical, genetic, neuropathological, imaging, electroencephalographic, or behavioral).16 For patients with NORSE, biomarkers may be used to identify the etiology of NORSE, identify specific pathogenic drivers such as infections or specific immune and inflammatory pathways that may help guide treatment strategy, monitor disease evolution, assess the risk of seizure recurrence during anesthetic weaning, predict both short-and long-term outcomes, and predict the development of post-NORSE epilepsy (Figure S1A).
The ideal NORSE biomarker would be present in patients with NORSE and absent in controls (including in patients with other forms of RSE), that is, it would have good clinical validity. The performance of the biomarkers should be assessed by the measure of their sensitivity and specificity (Figure S1B).17 For a broad-based, low-risk therapeutic strategy to prevent brain damage after NORSE, a biomarker capable of identifying all patients at risk (i.e., a biomarker with good sensitivity) would be required (Figure S1C). On the other hand, for a high-risk therapeutic strategy with benefit only for a limited group of patients, a biomarker identifying specifically this targeted group of patients (i.e., a biomarker with good specificity) is needed (Figure S1D). The ideal biomarker should also have good analytical validity, with a test result reproducible both within the same laboratory between two analyses and between different institutions.
Another important consideration in biomarker research is pathophysiological reliability. A good NORSE biomarker should reflect pathophysiological mechanisms involved either in SE onset (e.g., antineuronal antibodies, proinflammatory cytokines, genetic risk factors) or in its consequences (e.g., neuronal injury biomarkers, neuroinflammation biomarkers, acidosis biomarkers). Although there is currently no published animal model to specifically study pathogenic mechanisms involved in NORSE/FIRES onset, NORSE consequences could be explored by standard animal models of chemically or electrically induced SE.18–20
3 |. BRIEF OVERVIEW OF BIOMARKERS PROPOSED FOR PATIENTS WITH NORSE AND FIRES
We searched the PubMed database for English language articles using the terms “New-O nset Refractory Status Epilepticus”, “Febrile-Infection Related Epilepsy Syndrome” or “Refractory Status Epilepticus”. Articles were selected based on the following criteria: (1) NORSE and FIRES syndromes were well defined3,21; and (2) articles that contained the description of cellular or molecular biomarkers from biological fluids such as serum, plasma, or CSF or results from genetic analyses were included. We expanded the review search to include articles that reported data from patients with an unknown-etiology new onset refractory SE not defined as NORSE/FIRES. Abstracts, review articles, case reports, or retrospective case series with fewer than five patients were excluded.
Twenty-four original investigations reported CSF abnormalities, antineuronal antibodies, or increased levels of inflammatory markers in patients with NORSE, including FIRES. Nonetheless, most studies were conducted on small patient cohorts (median n = 16, interquartile range = 7–32), and often lacked control patients, precluding the validation of those biomarkers and their utilization in clinical settings. Six case–control studies were performed after the publication of NORSE and FIRES consensus definitions.3 Those studies aimed to compare data from patients with NORSE to healthy donors or patients with febrile SE (defined by the emergence of fever within 24 h before SE onset, but not present earlier).22–27 Two other studies were conducted in 2015 and 2016 to compare pediatric patients with FIRES to healthy controls or patients with noninflammatory neurological disorders.28,29 Both of those studies used the original definition of FIRES proposed in 2010: a non-encephalitic encephalopathy defined by the occurrence of recurrent seizures or SE, which was not necessarily refractory, and was preceded by febrile infection in previously healthy children.21
3.1 |. CSF abnormalities
CSF analyses were reported in 13 original articles or case series (study sample size from 5 to 130 patients). Abnormal CSF, with mild, predominantly lymphocytic pleocytosis and/or elevated protein levels, was commonly found in patients with NORSE on admission.4,6,26,30–35 Patients with FIRES more frequently had CSF abnormalities than patients with febrile SE.23,26 Protein levels were not different between cryptogenic SE and cases with an identified etiology.6 CSF pleocytosis was less prevalent in cryptogenic cases.6,36 There was no significant difference in clinical outcomes based on CSF white cell count or CSF protein in four studies, each involving between 20 and 77 patients with NORSE.5,31,35,37
3.2 |. Inflammatory markers
Six studies reported CSF and/or serum cytokine levels in patients with NORSE including FIRES (study sample size from 5 to 14 patients).22,25–28,38 The levels of C-X -C motif chemokine ligand 10 (CXCL10), CXCL9, interferon γ, and tumor necrosis factor α (TNFα) were higher in the CSF of patients with NORSE compared to those in patients with chronic epilepsy or noninflammatory neurological disorders.22,25,26,28,38 Those cytokines, classically defined as Th1-associated cytokines but also produced by neurons and glia, were also found to be elevated in patients with febrile SE but to a lesser extent compared to children with FIRES.26,39 Proinflammatory cytokines interleukin (IL)-6, CXCL8/IL-8, and IL-1ß levels were found to be increased in the CSF for the majority of patients with NORSE, even if the samples were drawn after immunotherapy.22,25–28,38 Data were conflicting for changes in serum cytokine levels. In one study, the serum IL-1 ß levels were reported to be higher in pediatric patients with FIRES compared to healthy donors.27 However, in another, an increase of serum IL-1ß levels was noted in only 1 of 9 patients with FIRES.38 The increased levels of proinflammatory cytokines were associated with the production of anti-inflammatory cytokines IL-1RA, IL-4, and IL-10 in both CSF and serum.25,27
The correlation between cytokine levels and patient outcomes has not been evaluated in these studies. However, the CSF cytokine levels were significantly decreased after intrathecal dexamethasone therapy, corresponding with clinical improvement in one study involving six patients with FIRES.22 Furthermore, febrile SE patients with T2 magnetic resonance imaging (MRI) hippocampal hyperintensity had higher plasma IL-6 and IL-8 levels as well as lower IL-1RA/IL-6 and IL-1RA/IL-8 ratios than patients with normal brain imaging, highlighting the possible relevance of those cytokines (used individually or combined) for predicting MRI abnormalities and the development of hippocampal sclerosis and its associated effects, including post-SE epilepsy and cognitive sequelae.40
In summary, although various abnormal inflammatory markers have been reported, most of the findings have not been confirmed in independent studies, and the current state of the literature does not suggest a single, consistent abnormality in NORSE. Additionally, current data do not allow us to conclude the superiority of CSF compared to serum for looking at inflammation biomarkers. Moreover, although gut dysbiosis was found able to modulate cytokine responses in healthy subjects,41 no study has been conducted to date to evaluate the benefit of examination of stool in identifying biomarkers for patients with NORSE. We expect that more systematic biosampling through the standard operating procedures suggested in this review may help alleviate some of the discrepancies, enabling reproducible research.
3.3 |. Antineuronal antibodies
Nonparaneoplastic and paraneoplastic autoimmune encephalitides represent the most commonly identified etiologies of NORSE, at least in adults, whereas infectious encephalitis is less common.6 A retrospective review of 130 adult patients admitted to the hospital for NORSE reported that nonparaneoplastic autoimmune encephalitis and paraneoplastic autoimmune encephalitis occurred in 19% and 15% of patients, respectively.6 Staining was most frequently positive for anti-N-methyl-D-aspartate receptor antibodies and anti-voltage-gated potassium channel complex antibodies.5,6,37,38,42,43 There were no significant differences in outcomes based on the presence or absence of these autoantibodies.37,42
3.4 |. Genetic markers
Children with NORSE (including FIRES) share clinical features with individuals presenting with disease-causing variants in SCN1A, PCDH19, or POLG.44 In a single-center retrospective study, three patients with NORSE of 40 were found to have genetic mutations, in DNM1L, KCNT1, POLG, and CTSD.35 However, no mutation was found for those genes in another cohort of 15 children with FIRES.39 Subsequent whole-exome studies and human leukocyte antigen (HLA) sequencing in a cohort of 50 individuals with FIRES identified several candidate genes (NYP2R, MYO1D, UNC50, NAV1, LRIF1); however, none of these genes was considered disease causing and, ultimately, no single causative genetic etiology was found.45 This lack of evidence for genetic etiologies is striking compared to other epilepsies.45 In a recent systematic review, NORSE/FIRES stands out as having the lowest genetic diagnostic yield of all the epilepsies, suggesting fundamental differences between NORSE/FIRES and other severe, nonlesional epilepsies.46,47 Nowadays, the absence of the HLA–FIRES association might also be explained by the small size of the cohort studied. Given the low frequency of NORSE/FIRES, identifying autoantibodies might be more effective for assessing autoimmune components in NORSE/FIRES rather than HLA association studies.
4 |. STANDARD OPERATING PROCEDURES FOR PROSPECTIVE BIOSPECIMEN COLLECTION
For harmonization of clinical data registries and biospecimen collection for NORSE research, we established a list of criteria for the following points: (1) criteria of eligibility to participate in a NORSE/FIRES biorepository; (2) time of biospecimen collection; (3) types of biospecimens to be collected; and (4) procedures for biospecimen processing, storage, and transportation.
4.1 |. Identifying a potential participant and collecting clinical and paraclinical data
Eligibility for studies on NORSE is determined by the patient’s clinical presentation with NORSE, as per the published consensus definition.3 Subjects will be critically ill pediatric or adult patients with RSE undergoing acute treatment with antiseizure medications. For our studies, we have excluded patients with major ongoing acute or subacute medical conditions such as organ failure, malignancy, or sepsis at the time of onset of SE. If these issues (organ failure, sepsis, etc.) developed after the onset of RSE as complications during the management of NORSE, patients should not be excluded (and these should be reported as complications of NORSE or its treatment).
Clinical information extracted from the records should include details of the patient’s history (demographic data, medical history), the clinical presentation (fever prior to admission, including its timing, and other prodromes), treatments used (antiseizure medications, anesthetics, immunotherapy), and paraclinical data including brain imaging, CSF analysis, and autoimmune testing. Outcomes should be obtained at 3–6, 12, and 24 months after SE onset, including the assessment of seizures, antiseizure medications, functional scales such as the Glasgow Outcome Scale–E xtended, and quality of life scales such as the Neuro-Quality of Life (neuro-QOL), or pediatric versions of those scales (e.g., neuro-QOL pediatric form and Pediatric Cerebral Performance Category Scale).
4.2 |. Biospecimen collection at various phases of the disease
4.2.1 |. Acute phase
We recommend collecting biospecimens as soon as possible after NORSE onset. When a legally authorized representative (LAR) is not immediately available, consider the use of emergency research consent if possible to enable appropriate storage of early samples while awaiting LAR consent. Certain conditions such as sepsis or renal/hepatic failure may develop in the course of SE, increase in parallel with the duration of stay in the intensive care unit (ICU), and alter biomarker composition.48 Similarly, some treatments are known to alter biomarkers studies and as a consequence single-cell transcriptomic studies should either not be performed if the patient was previously treated by immunotherapy (including isolated courses of corticosteroids) or be interpreted with caution if performed. However, we do not recommend delaying immunotherapy only to allow specimen collection; it is recommended to start immunotherapy quickly in patients with NORSE.14 A decrease of cytokine storm was observed after plasma exchange in patients with COVID-19.49 Therefore, one should be cautious when measuring cytokine levels after plasma exchange, especially if the cytokine levels were not also measured before immunotherapy. All appropriate treatments used before sample collection should be reported. The remaining samples previously collected for clinical purposes and that had been processed appropriately could also be useful for analysis.
4.2.2 |. Collection at different time points during the acute phase
Biomarkers are known to change over time during the course of SE.10 Those changes might reflect variations in the daily ictal seizure burden, comorbidities (e.g., infections), or treatments used for SE management.22 We suggest collecting the first samples, when possible, before the introduction of immunotherapy and reassessing the levels of inflammatory biomarkers (specifically cytokines/chemokines) during the SE course, especially before and after the introduction of immunotherapy. When CSF presents with abnormalities or when positive staining was found for an antineuronal antibody in serum or CSF, it is also relevant to collect new biospecimens (CSF, serum) after the immunotherapy course to look at the impact of treatment on abnormalities/antibody persistence. We cannot provide a specific time window after which the disturbances should no longer be present. As previously performed for SE patients,10 daily measurements of neuronal injury biomarkers (in serum or plasma) might be useful to investigate the relationships between treatments, daily ictal burden, and biomarker levels. Therefore, whereas there is no argument for a longitudinal collection of whole blood and saliva for DNA, serial collection of serum, plasma, and CSF (in the case of iterative clinical procedures) might be useful for biomarker studies.
4.2.3 |. Chronic phase (during follow-up evaluation after resolution of the initial RSE ended)
Most NORSE survivors have neurological sequelae, including functional disability, and many have unprovoked seizures (epilepsy) after hospital discharge. The emergence of new seizures, or even recurrent SE, may be due to the consequences of the initial SE, but a chronic underlying inflammatory process may also persist in some cases. Polymorphisms in cytokine-related genes were observed more commonly in children with FIRES than in healthy controls.27,29 Therefore, some NORSE cases could be related to a genetic predisposition, likely associated with a response to infection or other aspects of inflammation. We advise collecting whole blood for DNA extraction and genetic studies for patients lacking such analysis during the acute phase of the disease. Examining the levels of inflammatory biomarkers during the chronic phase might be useful for both patients with pharmacoresistant post-NORSE epilepsy and patients who did not present any seizure recurrence and for whom biomarkers were evaluated during the acute phase. However, further research is needed to delineate the exact time when the acute neuroinflammatory phase ends.
4.3 |. Biospecimen collection: site and method of sample acquisition
Biological samples that should be collected include (but are not limited to) whole blood, serum, CSF, urine, stool, nasopharyngeal or nasal swabs, saliva, and brain tissue if obtained via biopsy or postmortem examination (Figure 1). The time of sample collection should be recorded.
FIGURE 1.
Flowchart for new onset refractory status epilepticus/febrile infection-related epilepsy syndrome (NORSE/FIRES) biorepository. mn, minutes; PBMC, peripheral blood mononuclear cells; PBS, phosphate-buffered saline.
K2 EDTA tubes are preferred for DNA extraction and genetic studies. We recommend using tubes without anticoagulant or preservative, typically red-top tubes in Europe and the United States, to isolate serum for other studies. It is important to report which tubes were used to collect blood and whether the supernatant obtained after spinning corresponds to plasma or serum. Although plasma and serum can generate similar results in biological studies, different collecting procedures and corresponding coagulation cascades might influence concentrations of proteins and metabolites.50
When obtaining CSF, a volume of 10 mL or more is preferred. In accordance with previous guidelines, we recommend using polypropylene tubes with no additive for collecting CSF.51,52 It is important to collect blood and CSF simultaneously to provide additional information on biomarker distribution across different anatomical compartments and highlight the origin of the biomarker.
For saliva, we recommend placing a flocked nylon fiber tip swab under the tongue for 1 or 2 min and then inserting the swab into a storage tube containing liquid Amies medium (Appendix S1).
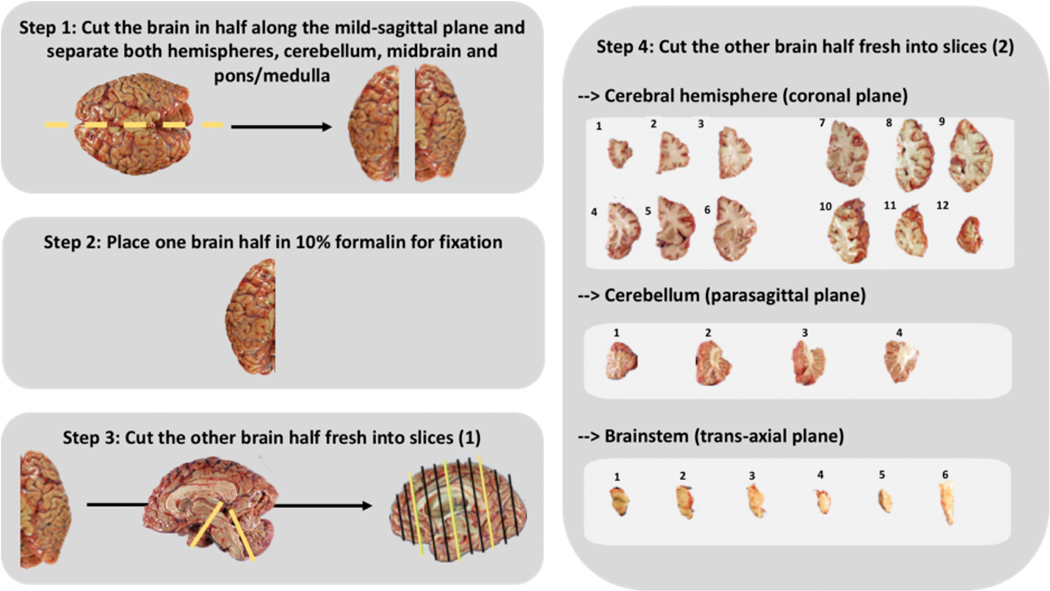
The first neuropathology report in patients with SE highlighted the hippocampus as the most affected brain area but also revealed the vulnerability of the cerebellum, thalamus, and neocortices.53 Not to miss neuropathology abnormalities, we suggest examining the entire brain for autopsy tissue. We suggest four options for the collection and preparation of brain samples. The detailed procedure is provided in Figure 1, and a graphical representation is represented in Figure 2. For brain postmortem tissue, we recommend collecting samples promptly after death to have high-quality brain tissue.54
FIGURE 2.
Standard operating procedures for brain tissue preparation.
The location of the biopsy should be guided by the clinical findings as well as electroencephalographic and imaging data (e.g., areas with maximum enhancement, fluid-attenuated inversion recovery abnormalities, or suspected brain lesions). By following these criteria, some studies revealed astrogliosis, prominent microglial reactivity, and lymphocytic infiltration in brain biopsies performed at the acute phase of NORSE.4,5,55–57 Those neuropathology findings informed a significant modification of the patient’s management for some cases.56,57 For brain biopsy tissue, we recommend snap freezing the samples immediately and reporting the brain region biopsied.
Protein levels, quantity and quality of brain autopsy tissue RNA, and DNA yield and integrity were found unchanged after long-term storage at −80°C.58–60 Therefore, it remains unresolved as to whether sample storage at −150°C, which is less accessible and with an added cost, provides significant advantages relative to long-term storage at −80°C.61 We, therefore, suggest freezing and storing both autopsy postmortem tissues and biopsy tissues at −80°C. Samples can later be sectioned frozen with a cryostat to determine the distribution of mRNA within tissue (i.e., for in situ hybridization studies).62 Nowadays, such studies can still be conducted on paraffin-embedded tissues.62
A separate (but overlapping) group of investigators is working on providing further, more detailed guidelines for neuropathologists related to the collection and evaluation of brain specimens from patients with NORSE and FIRES.
Figure 1 provides additional standard operating procedure guidelines for collecting urine, stool, nasal and nasopharyngeal swabs, and PBMC.
Other biological samples might be of interest for patients with NORSE, for diagnosis or research purposes. Mitochondrial disorders were reported as a cause of NORSE for a few patients.63,64 Whole genome sequencing may allow the identification of the concerned variants, but muscle biopsy and skin fibroblasts could provide additional information by looking at the function of the respiratory chain complexes.63,65 Previous recommendations suggest cutting the muscle tissue into several pieces for formalin fixation, glutaraldehyde fixation, and rapid freezing in isopentane.65 We cannot yet provide specific recommendations for rare patients with NORSE due to mitochondrial disorders.66,67
4.4 |. Biospecimen processing, storage, and shipment
Preanalytical methods can significantly alter the molecular composition of biospecimens and influence experimental results.11–13 We provide recommendations to harmonize protocols and allow multicenter research.
4.4.1 |. Collection times
Some biomarkers can be influenced by circadian rhythms.68 Because it would be too difficult to standardize the collection time in all hospitals, we recommend recording the time of sample collection and obtaining the specimens in the morning when practical.
4.4.2 |. Sample processing and storage conditions
Whole blood should preferably be stored at 4°C for up to 3 days, at −20°C for up to 1 month, or at −80°C for long-term DNA extraction. Although DNA can still be extracted up to 15 days from the time of collection if the sample is kept as whole blood at 4°C, the DNA quantity will decrease dramatically.69 Therefore, we recommend storing whole blood at −80°C and isolating DNA in a batch to maintain good quality and reduce the batch effect.
For PBMC analyses, a specific protocol should be followed for the cell preparation tube with a first spin-down at 800 × g for 20 min at room temperature. After spin-down, the PBMC layer should be transferred to a conical tube filled with phosphate-buffered saline (PBS). After a new spin-down at 400 × g for 8 min, PBMC should be resuspended in PBS, and the concentration of PBMC should be measured. After the last spin-down at 400 × g for 5 min, PBMC should be resuspended with serum-free cell long-term conservation medium (e.g., Bambanker) at 10 × 106 cells per milliliter and stored in cryovials at a preferred volume of 500 μL to allow good cell survival. The PBMC layer can also be isolated from EDTA whole blood by adding a density gradient medium (e.g., Lymphoprep or Ficoll-P aque) to the blood or by using prefilled tubes with a separation medium (e.g., Leucosep tubes; Appendix S1).70
The effect of storing blood or CSF at different temperatures before spinning has not yet been studied extensively.71 Whereas CSF neurofilament and progranulin levels seem not to be affected by temperature changes and delays prior to spinning,11,72 serum levels of some cytokines have declined if centrifugation was delayed by 3 h,73 whereas the levels of other cytokines (IL-1ß, TNFα) increased significantly with cell deterioration over time (personal observation). To match with daily clinical practice, we recommend keeping the blood at room temperature and the CSF on ice before spinning and centrifuge tubes within 2 h after sample collection. The time of centrifugation should be recorded. We propose to spin the CSF and the serum collection tube at 1500 × g for 10 min at room temperature. The supernatant should be divided into single-u se microtubes made from polypropylene (e.g., Eppendorf tubes), with a preferred volume of 250 μL per tube. Splitting the supernatant into multiple small aliquots is optimal to avoid freezing/thawing cycles. Aliquots should be stored immediately at −80°C. If this is not done, the time of storage should at least be reported.
For saliva samples, we recommend centrifuging the collection tube at 1500 × g for 15 min at room temperature to extract the saliva. The swab can then be removed and saliva should be divided into single-u se microtubes made from polypropylene (e.g., Eppendorf tubes), with a preferred volume of 500 μL per tube. Aliquots should be stored immediately at −80°C. If this is not done, the time of storage should at least be reported.
All aliquots should be labeled with the patient study number, the date of sample collection, and the sample type (serum, CSF, PBMC, saliva, etc.), and stored immediately at −80°C. Labels should be water-resistant and stable at −80°C.
4.4.3 |. Bloody CSF/hemolyzed samples
In general, hemolyzed serum and plasma samples should not be used for biomarker studies. For example, neuron-specific enolase, a biomarker proposed for the assessment of brain injury, is highly sensitive to hemolysis.74 Blood contamination of CSF in the case of traumatic tap can induce a bias for biomarker studies, especially if the biomarker is present at a higher concentration in blood compared to CSF. To reduce the impact of blood on CSF, it is particularly important to spin down the CSF and collect only the supernatant before initial freezing. It is recommended not to use samples with an erythrocyte count of >500/μL for biomarker studies.51
4.4.4 |. Storage and shipping
With the exception of formalin-fixed paraffin-embedded tissues, which are stable at room temperature, all other samples (serum, CSF, PBMC, whole blood, urine, stool, nasal/nasopharyngeal swab, saliva swab, unfixed brain tissues) should be stored at −80°C in alarm-controlled freezers until shipping. Samples should be shipped on dry ice to avoid the thawing of samples. We recommend using temperature-logging devices for international shipping of frozen samples. Those disposals are not necessarily required for national shipments or shipments between neighboring countries without customs (e.g., between European countries).
5 |. USE OF PREVIOUSLY COLLECTED SAMPLES
Although it is crucial to gather biological specimens prospectively, it is also important to establish criteria for the effective use of previously collected samples.
Here, we propose specific criteria depending on the type of specimen (serum, plasma, whole blood, DNA, CSF, PBMC, tissue), the time of sample collection (acute or chronic phase of NORSE/FIRES), the processing time, delay, and method, storage conditions, and the type of planned analyses (Tables 1 and Table 2).
TABLE 1.
Criteria for the accurate use of previously collected blood and CSF samples.
| Sample type | Processing methods: Spinning | Storage | Appropriate for use in the biorepository? (Examples of molecules to be measured) |
|---|---|---|---|
| Blood tube without anticoagulant, typically red top (for serum) AND Polypropylene tube (for CSF) | ≤2 h = serum/CSF supernatant | −80°C | Yes (Cytokines, antibody panel, neurofilament, NSE, S100beta, etc.) For the chronic phase, only if samples were also collected in the acute phasea |
| Other | No | ||
| 2–24 h = serum/CSF supernatant | −80°C | Yes (Only for stable molecules such as antibody panel) For the chronic phase, only if samples were also collected in the acute phasea |
|
| Other | No | ||
| >24 h = serum/CSF supernatant | - | No | |
| K2 EDTA tube (for plasma or whole blood) | No spinning = whole blood | −80°C | Yes (Genetic studies) For acute and chronic phasesa |
| ≤2 h = plasma | −80°C | Yes (Neurofilament, progranulin) For the chronic phase, only if samples were also collected in the acute phasea |
|
| Other | No | ||
| >2 h = plasma | - | No | |
| K2 EDTA tube or cell preparation tube (for PBMC) | ≤3 h = PBMC | −80°C in serum-free cell medium (e.g., Bambanker) | Yes (Single cell RNA sequencing) Only if no previous immunotherapy used |
| >3 h = PBMC | - | No |
Abbreviations: CSF, cerebrospinal fluid; NSE, neuron specific enolase; PBMC, peripheral blood mononuclear cells; SE, status epilepticus.
The acute phase refers to specimens collected as soon as possible after SE onset and for patients with ongoing SE. The chronic phase refers to specimens collected during the follow-up evaluations after the initial refractory SE ends.
TABLE 2.
Criteria for the accurate use of previously collected brain tissues.
| Sample type | Clinical phase at time of sample collection | Processing methods: Preparation | Storage | Appropriate for use in the biorepository? (Examples of molecules to be measured) |
|---|---|---|---|---|
| Brain biopsy samples | Acute | Snap frozen immediately | −80°C | Yes (DNA, RNA, and protein analyses) |
| Fixation/paraffin embedding | Room temperature | Yes (Histology, immunohistochemistry, in situ hybridization) |
||
| Chronic | Snap frozen immediately | −80°C | Yes (DNA, RNA, and protein analyses) |
|
| Fixation/paraffin embedding | Room temperature | Yes (Histology, immunohistochemistry, in situ hybridization) |
||
| Brain postmortem samples | Acute | Snap frozen immediately | −80°C | Yes (DNA, RNA, and protein analyses) |
| Fixation/paraffin embedding | Room temperature | Yes (Histology, immunohistochemistry, in situ hybridization) |
||
| Chronic | Snap frozen immediately | −80°C | Yes (DNA, RNA, and protein analyses) |
|
| Fixation/paraffin embedding | Room temperature | Yes (Histology, immunohistochemistry, in situ hybridization) |
The following information should be provided for all already collected samples: demographic data (age, sex), clinical information (date of SE onset, date of SE end, preceding prodromes including fever, etiology if found, the functional state before SE, the functional state at ICU discharge, post-NORSE epilepsy), and biological information (date of sample collection, treatments used before sample collection, spinning delay, storage temperature, etc.).
6 |. SAMPLES SHARING AND SHIPMENT
Given the rarity of NORSE/FIRES, it is crucial to conduct international collaborations and share data and specimens as much as possible.
Due to the data protection and privacy regulations, specifically in the European Economic Area, sharing data and specimens outside Europe might be a complex issue. To allow this sharing, we suggest requiring a signature on a notification letter from patients residing in Europe for the international use of their study data. This form needs to provide the following statements: (1) the study is protected by General Data Protection Regulation (GDPR), (2) the research team will collect and use the following types of study data for this research (provide a detailed list of data collected such as demographic, clinical, and paraclinical), (3) genetic analysis will be performed on the specimen collected, and (4) the research protocol requires the research team to enter data into a computer. We also suggest reminding patients of their rights related to the study data given by the GDPR, such as for (1) accessing, correcting, or withdrawing the study data; (2) restricting the types of activities the research team can do with the study data (for example, excluding genetic analyses); and (3) withdrawing the consent to the study. Detailed contact information for the study principal investigator should be provided.
7 |. CONCLUSIONS AND FUTURE DIRECTIONS
Careful investigations in as many patients as possible, including various approaches and biomarkers, can help physicians to identify NORSE etiology, monitor patient responses to treatment, optimize individual treatments, and predict NORSE outcomes. Although some biomarkers may reflect mechanisms involved in seizure onset, the majority of previously proposed biomarkers aimed to assess NORSE consequences. To date, no biomarker has been validated in NORSE using extensive prospective studies. The marked heterogeneity in NORSE etiology and clinical subtypes as well as treatments used before sample collection, irregular time points for biospecimen collection, and lack of standardization for biospecimen processing and storage preclude the validation of findings from previous investigations and limit their utility for clinical practice. An international collection of health-related data and biological samples from patients with NORSE has been established to permit multicenter research. We now provide standard operating procedures for biospecimen collection, storage, shipping, and banking for future patients, as well as criteria for the appropriate use of previously collected biospecimens. The success of future research studies on NORSE will likely depend on high-quality, multicenter, international collaborations using standardized procedures and sharing data and ideas as much as possible.
Supplementary Material
Key Points.
Identifying the pathophysiology and discovering specific biomarkers may help physicians in the management of patients with NORSE
Prior research, including attempts to find biomarkers, has been limited by significant variability in the collection, storage, and analysis of biospecimens
Widespread use of standardized procedures will reduce heterogeneity and facilitate research regarding NORSE, including the identification of validated biomarkers
This report suggests standard operating procedures for biospecimen collection and biobanking
Criteria for the appropriate use of previously collected biospecimens are proposed as well
ACKNOWLEDGMENTS
The NORSE/FIRES biorepository is supported by the NORSE/FIRES Research Fund at Yale and the NORSE Institute. A.V. received seeds grants from AES in conjunction with the NORSE Institute. C.L.H. received support from the National Organization of Rare Diseases (#18008) with funding from the NORSE Institute.
Funding information
AES -NORSE Institute seeds grants; National Organization of Rare Diseases -NORSE Institute, Grant/Award Number: 18008; NORSE Institute, Grant/Award Number: NORSE/FIRES Research Fund at Yale and The Daniel Raymond Neurology Research Fund; Paratonnerre Association; Philippe Foundation; Servier laboratory
CONFLICT OF INTEREST STATEMENT
Disclosures related to the article: A.Ha. received postdoctoral grants from the Paratonnerre Association, Servier Laboratory, and Philippe Foundation for NORSE-related research. L.J.H. received support for investigator-initiated studies from the Daniel Raymond Wong Neurology Research Fund and from the NORSE/FIRES Research Fund at Yale. C.L.H., A.V., O.T., E.A., A.Hu., S.K., M.R.W., N.G., and L.J.H. are members of the Scientific Board of the NORSE Institute. Disclosures not related to the article: E.A. has received speaker honoraria from Novartis, Nutricia, and UCB; served as an investigator for UCB and Nutricia; and served on scientific advisory boards for Novartis and UCB. S.K. has served on scientific advisory boards for Zogenix and Neurelis. V.N. has received unrelated grants from the Fondation AP-H P pour la Recherche (EPIRES, Marie Laure PLV Merchandising); consulting fees from UCB, Eisai, Jazz, and Angellini; and honoraria from UCB and Eisai. O.T. has received salary and research support from the NIH P20GM120447 Cognitive NeuroScience and Development of Aging Award and Nebraska Stem Cell Grant. M.R.W. has received unrelated grants from Roche/Genentech and Novartis; has received speaking honoraria from Novartis, Takeda, WebMD, and Genentech; and is an advisor for Delve Bio. N.G. has received unrelated grants from Innoviris and Fonds Erasme pour la Recherche Médicale, and honoraria from UCB and Ligue Francophone Belge contre l’Epilepsie. L.J.H. has received consultation fees for advising from Ceribell, Eisai, Marinus, Neurelis, Neuropace, Rafa Laboratories, and UCB; royalties from Wolters-Kluwer for authoring chapters for UpToDate–Neurology and from Wiley for coauthoring the book Atlas of EEG in Critical Care, first and second editions; and honoraria for speaking from Neuropace, Natus, and UCB. The other authors have no disclosures to report.
Footnotes
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Leitinger M, Trinka E, Giovannini G, Zimmermann G, Florea C, Rohracher A, et al. Epidemiology of status epilepticus in adults: a population-based study on incidence, causes, and outcomes. Epilepsia. 2019;60(1):53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trinka E, Cock H, Hesdorffer D, Rossetti AO, Scheffer IE, Shinnar S, et al. A definition and classification of status epilepticus –report of the ILAE task force on classification of status epilepticus. Epilepsia. 2015;56(10):1515–23. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch LJ, Gaspard N, van Baalen A, Nabbout R, Demeret S, Loddenkemper T, et al. Proposed consensus definitions for new-onset refractory status epilepticus (NORSE), febrile infection-related epilepsy syndrome (FIRES), and related conditions. Epilepsia. 2018;59(4):739–44. [DOI] [PubMed] [Google Scholar]
- 4.Costello DJ, Kilbride RD, Cole AJ. Cryptogenic new onset refractory status epilepticus (NORSE) in adults-infectious or not? J Neurol Sci. 2009;277(1–2):26–31. [DOI] [PubMed] [Google Scholar]
- 5.Kramer U, Chi C-S, Lin K-L, Specchio N, Sahin M, Olson H, et al. Febrile infection-related epilepsy syndrome (FIRES): pathogenesis, treatment, and outcome: a multicenter study on 77 children. Epilepsia. 2011;52(11):1956–65. [DOI] [PubMed] [Google Scholar]
- 6.Gaspard N, Foreman BP, Alvarez V, Cabrera Kang C, Probasco JC, Jongeling AC, et al. New-onset refractory status epilepticus: etiology, clinical features, and outcome. Neurology. 2015;85(18):1604–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howell KB, Katanyuwong K, Mackay MT, Bailey CA, Scheffer IE, Freeman JL, et al. Long-term follow-up of febrile infection-related epilepsy syndrome. Epilepsia. 2012;53(1):101–10. [DOI] [PubMed] [Google Scholar]
- 8.Tan TH-L, Perucca P, O’Brien TJ, Kwan P, Monif M. Inflammation, ictogenesis, and epileptogenesis: an exploration through human disease. Epilepsia. 2021;62(2):303–2 4. [DOI] [PubMed] [Google Scholar]
- 9.Hanin A, Lambrecq V, Denis JA, Imbert-Bismut F, Rucheton B, Lamari F, et al. Cerebrospinal fluid and blood biomarkers of status epilepticus. Epilepsia. 2020;61(1):6–18. [DOI] [PubMed] [Google Scholar]
- 10.Hanin A, Demeret S, Denis JA, Nguyen-Michel V-H, Rohaut B, Marois C, et al. Serum neuron-specific enolase: a new tool for seizure risk monitoring after status epilepticus. Eur J Neurol. 2021;29(3):883–9. [DOI] [PubMed] [Google Scholar]
- 11.Fourier A, Portelius E, Zetterberg H, Blennow K, Quadrio I, Perret-Liaudet A. Pre-analytical and analytical factors influencing Alzheimer’s disease cerebrospinal fluid biomarker variability. Clin Chim Acta. 2015;449:9–15. [DOI] [PubMed] [Google Scholar]
- 12.Kang F, Li W, Xia X, Shan Z. Three years’ experience of quality monitoring program on pre-analytical errors in China. J Clin lab Anal. 2021;35(3):e23699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lima-Oliveira G, Volanski W, Lippi G, Picheth G, Guidi GC. Pre-analytical phase management: a review of the procedures from patient preparation to laboratory analysis. Scand J Clin lab Invest. 2017;77(3):153–63. [DOI] [PubMed] [Google Scholar]
- 14.Wickstrom R, Taraschenko O, Dilena R, Payne ET, Specchio N, Nabbout R, et al. International consensus recommendations for management of new onset refractory status epilepticus (NORSE) including febrile infection-related epilepsy syndrome (FIRES): summary and clinical tools. Epilepsia. 2022;63(11):2827–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickstrom R, Taraschenko O, Dilena R, Payne ET, Specchio N, Nabbout R, et al. International consensus recommendations for management of new onset refractory status epilepticus (NORSE) incl. Febrile infection-related epilepsy syndrome (FIRES): statements and supporting evidence. Epilepsia. 2022;63(11):2827–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource [Internet]. Silver Spring (MD): Food and Drug Administration (US). 2016. [cited 2021]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK326791/ [PubMed] [Google Scholar]
- 17.Fluss R, Faraggi D, Reiser B. Estimation of the Youden index and its associated cutoff point. Biom J. 2005;47(4):458–72. [DOI] [PubMed] [Google Scholar]
- 18.Reddy DS, Kuruba R. Experimental models of status epilepticus and neuronal injury for evaluation of therapeutic interventions. Int J Mol Sci. 2013;14(9):18284–3 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Curtis M, Rossetti AO, Verde DV, van Vliet EA, Ekdahl CT. Brain pathology in focal status epilepticus: evidence from experimental models. Neurosci Biobehav Rev. 2021;131:834–46. [DOI] [PubMed] [Google Scholar]
- 20.Pitkänen A, Buckmaster PS, Galanopoulou AS, Moshé SL. Models of seizures and epilepsy. 2nd ed. Elsevier Inc.; 2017. [Google Scholar]
- 21.van Baalen A, Häusler M, Boor R, Rohr A, Sperner J, Kurlemann G, et al. Febrile infection-related epilepsy syndrome (FIRES): a nonencephalitic encephalopathy in childhood. Epilepsia. 2010;51(7):1323–8. [DOI] [PubMed] [Google Scholar]
- 22.Horino A, Kuki I, Inoue T, Nukui M, Okazaki S, Kawawaki H, et al. Intrathecal dexamethasone therapy for febrile infection-related epilepsy syndrome. Ann Clin Transl Neurol. 2021;8(3):645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sculier C, Barcia Aguilar C, Gaspard N, Gaínza-L ein M, Sánchez Fernández I, Amengual-Gual M, et al. Clinical presentation of new onset refractory status epilepticus in children (the pSERG cohort). Epilepsia. 2021;62(7):1629–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh M-Y, Lin J-J, Hsia S-H, Huang J-L, Yeh K-W, Chang K-W, et al. Diminished toll-l ike receptor response in febrile infection-related epilepsy syndrome (FIRES). Biom J. 2020;43(3):293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jun J-S, Lee S-T, Kim R, Chu K, Lee SK. Tocilizumab treatment for new onset refractory status epilepticus. Ann Neurol. 2018;84(6):940–5. [DOI] [PubMed] [Google Scholar]
- 26.Kothur K, Bandodkar S, Wienholt L, Chu S, Pope A, Gill D, et al. Etiology is the key determinant of neuroinflammation in epilepsy: elevation of cerebrospinal fluid cytokines and chemokines in febrile infection-related epilepsy syndrome and febrile status epilepticus. Epilepsia. 2019;60(8):1678–88. [DOI] [PubMed] [Google Scholar]
- 27.Clarkson BDS, LaFrance-Corey RG, Kahoud RJ, Farias-Moeller R, Payne ET, Howe CL. Functional deficiency in endogenous interleukin-1 receptor antagonist in patients with febrile infection-related epilepsy syndrome. Ann Neurol. 2019;85(4):526–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakuma H, Tanuma N, Kuki I, Takahashi Y, Shiomi M, Hayashi M. Intrathecal overproduction of proinflammatory cytokines and chemokines in febrile infection-related refractory status epilepticus. J Neurol Neurosurg Psychiatry. 2015;86(7):820–2 . [DOI] [PubMed] [Google Scholar]
- 29.Saitoh M, Kobayashi K, Ohmori I, Tanaka Y, Tanaka K, Inoue T, et al. Cytokine-related and sodium channel polymorphism as candidate predisposing factors for childhood encephalopathy FIRES/AERRPS. J Neurol Sci. 2016;368:272–6. [DOI] [PubMed] [Google Scholar]
- 30.Aurangzeb S, Prisco L, Adcock J, Speirs M, Raby S, Westbrook J, et al. New-onset super refractory status epilepticus: a case-series. Seizure. 2020;75:174–84. [DOI] [PubMed] [Google Scholar]
- 31.La S-K, L W-Y, Wen W-C, Fa P-C, Le W-T. The short-term and long-term outcome of febrile infection-related epilepsy syndrome in children. Epilepsy Behav. 2019;95:117–23. [DOI] [PubMed] [Google Scholar]
- 32.Lee H-F, Chi C-S. Febrile infection-related epilepsy syndrome (FIRES): therapeutic complications, long-term neurological and neuroimaging follow-up. Seizure. 2018;56:53–9. [DOI] [PubMed] [Google Scholar]
- 33.Gall CRE, Jumma O, Mohanraj R. Five cases of new onset refractory status epilepticus (NORSE) syndrome: outcomes with early immunotherapy. Seizure. 2013;22(3):217–20. [DOI] [PubMed] [Google Scholar]
- 34.Wilder-Smith EPV, Lim ECH, Teoh HL, Sharma VK, Tan JJH, Chan BPL, et al. The NORSE (new-onset refractory status epilepticus) syndrome: defining a disease entity. Ann Acad Med Singapore. 2005;34(7):417–20. [PubMed] [Google Scholar]
- 35.Husari KS, Labiner K, Huang R, Said RR. New-onset refractory status epilepticus in children: etiologies, treatments, and outcomes. Pediatr Crit Care Med. 2020;21(1):59–66. [DOI] [PubMed] [Google Scholar]
- 36.Yanagida A, Kanazawa N, Kaneko J, Kaneko A, Iwase R, Suga H, et al. Clinically based score predicting cryptogenic NORSE at the early stage of status epilepticus. Neurol Neuroimmunol Neuroinflamm. 2020;7(5):e849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gugger JJ, Husari K, Probasco JC, Cervenka MC. New-onset refractory status epilepticus: a retrospective cohort study. Seizure. 2020;74:41–8. [DOI] [PubMed] [Google Scholar]
- 38.Lai Y-C, Muscal E, Wells E, Shukla N, Eschbach K, Hyeong Lee K, et al. Anakinra usage in febrile infection related epilepsy syndrome: an international cohort. Ann Clin Transl Neurol. 2020;7(12):2467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satarkar D, Patra C. Evolution, expression and functional analysis of CXCR3 in neuronal and cardiovascular diseases: a narrative review. Front Cell Dev Biol. 2022;10:882017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallentine WB, Shinnar S, Hesdorffer DC, Epstein L, Nordli DR, Lewis DV, et al. Plasma cytokines associated with febrile status epilepticus in children: a potential biomarker for acute hippocampal injury. Epilepsia. 2017;58(6):1102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167(4):1125–1136.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D, Pan Y, Huang K, Lin Z, Xie Z, Liu G, et al. Is rat hippocampus section immunostaining an indicator for immunotherapy in cryptogenic adult new-onset refractory status epilepticus (NORSE)? Seizure. 2020;76:131–6. [DOI] [PubMed] [Google Scholar]
- 43.Khawaja AM, de Wolfe JL, Miller DW, Szaflarski JP. New-onset refractory status epilepticus (NORSE) — the potential role for immunotherapy. Epilepsy Behav. 2015;47:17–23. [DOI] [PubMed] [Google Scholar]
- 44.Appenzeller S, Helbig I, Stephani U, Häusler M, Kluger G, Bungeroth M, et al. Febrile infection-related epilepsy syndrome (FIRES) is not caused by SCN1A, POLG, PCDH19 mutations or rare copy number variations. Dev Med Child Neurol. 2012;54(12):1144–8. [DOI] [PubMed] [Google Scholar]
- 45.Helbig I, Barcia G, Pendziwiat M, Ganesan S, Mueller SH, Helbig KL, et al. Whole-exome and HLA sequencing in febrile infection-related epilepsy syndrome. Ann Clin Transl Neurol. 2020;7(8):1429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith L, Malinowski J, Ceulemans S, Peck K, Walton N, Sheidley BR, et al. Genetic testing and counseling for the unexplained epilepsies: an evidence-based practice guideline of the National Society of genetic counselors. J Genet Couns. 2023;32(2):266–80. [DOI] [PubMed] [Google Scholar]
- 47.Sheidley BR, Malinowski J, Bergner AL, Bier L, Gloss DS, Mu W, et al. Genetic testing for the epilepsies: a systematic review. Epilepsia. 2022;63(2):375–87. [DOI] [PubMed] [Google Scholar]
- 48.Oddo M, Crippa IA, Mehta S, Menon D, Payen J-F, Taccone FS, et al. Optimizing sedation in patients with acute brain injury. Crit Care. 2016;20:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beraud M, Hashami SA, Lozano M, Bah A, Keith P. Role of therapeutic plasma exchange in the management of COVID-19-induced cytokine storm syndrome. Transfus Apher Sci. 2022;61(4):103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Z, Kastenmüller G, He Y, Belcredi P, Möller G, Prehn C, et al. Differences between human plasma and serum metabolite profiles. PLoS One. 2011;6(7):e21230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teunissen CE, Petzold A, Bennett JL, Berven FS, Brundin L, Comabella M, et al. A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology. 2009;73(22):1914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore HM, Kelly A, Jewell SD, McShane LM, Clark DP, Greenspan R, et al. Biospecimen reporting for improved study quality (BRISQ). J Proteome Res. 2011;10(8):3429–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corsellis JA, Bruton CJ. Neuropathology of status epilepticus in humans. Adv Neurol. 1983;34:129–39. [PubMed] [Google Scholar]
- 54.Scholefield M, Church SJ, Xu J, Robinson AC, Gardiner NJ, Roncaroli F, et al. Effects of alterations of post-mortem delay and other tissue-collection variables on metabolite levels in human and rat brain. Metabolites. 2020;10(11):438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donnelly JP, Kasatwar N, Hafeez S, Seifi A, Gilbert A, Barthol C, et al. Resolution of cryptogenic new onset refractory status epilepticus with tocilizumab. Epilepsy Behav Rep. 2021;15:100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dilena R, Mauri E, Aronica E, Bernasconi P, Bana C, Cappelletti C, et al. Therapeutic effect of anakinra in the relapsing chronic phase of febrile infection-related epilepsy syndrome. Epilepsia Open. 2019;4(2):344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matar RK, Alshamsan B, Alsaleh S, Alhindi H, Alahmedi KO, Khairy S, et al. New onset refractory status epilepticus due to primary angiitis of the central nervous system. Epilepsy Behav Case Rep. 2017;8:100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ericsson C, Franzén B, Nistér M. Frozen tissue biobanks. Tissue handling, cryopreservation, extraction, and use for proteomic analysis. Acta Oncol. 2006;45(6):643–61. [DOI] [PubMed] [Google Scholar]
- 59.Yasojima K, McGeer EG, McGeer PL. High stability of mRNAs postmortem and protocols for their assessment by RT-PCR. Brain Res Brain Res Protoc. 2001;8(3):212–8. [DOI] [PubMed] [Google Scholar]
- 60.Chu T-Y, Hwang K-S, Yu M-H, Lee H-S, Lai H-C, Liu J-Y. A research-b ased tumor tissue bank of gynecologic oncology: characteristics of nucleic acids extracted from normal and tumor tissues from different sites. Int J Gynecol Cancer. 2002;12(2):171–6. [DOI] [PubMed] [Google Scholar]
- 61.Shabihkhani M, Lucey GM, Wei B, Mareninov S, Lou JJ, Vinters HV, et al. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin Biochem. 2014;47:258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lammers CH, Hara Y, Maral Mouradian M. In Situ Hybridization on Brain Tissue. Methods Mol Med. 2001;62:229–46. [DOI] [PubMed] [Google Scholar]
- 63.Astner-Rohrach r A, Mauri z M, Leiting r M, Rossi i F, Kal s G, Neur y C, et al. l. A case report: new-onset refractory status epilepticus in a patient with FASTKD2-related mitochondrial disease. Front Neurol. 2022;13:1063733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morrison HD, Morgan C, Urankar K, Wylde J, O’Beirne M, Krolikowski K, et al. New-onset refractory status epilepticus (NORSE) in a 23-year-old female: answer. J Clin Neurosci. 2020;82:271–2. [DOI] [PubMed] [Google Scholar]
- 65.Kušíková K, Feichtinger RG, Csillag B, Kalev OK, Weis S, Duba H-C, et al. Case report and review of the literature: a new and a recurrent variant in the VARS2 gene are associated with isolated lethal hypertrophic cardiomyopathy, hyperlactatemia, and pulmonary hypertension in early infancy. Front Pediatr. 2021;9:660076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Araki T, Suzuki J, Taniwaki Y, Ishido K, Kamikaseda K, Turuta Y, et al. A case of MELAS presenting complex partial status epilepticus. Rinsho Shinkeigaku. 2001;41(8):487–9 0. [PubMed] [Google Scholar]
- 67.Steriade C, Andrade DM, Faghfoury H, Tarnopolsky MA, Tai P. Mitochondrial encephalopathy with lactic acidosis and stroke-like episodes (MELAS) may respond to adjunctive ketogenic diet. Pediatr Neurol. 2014;50(5):498–502. [DOI] [PubMed] [Google Scholar]
- 68.Laing EE, Möller-Levet CS, Poh N, Santhi N, Archer SN, Dijk D-J. Blood transcriptome based biomarkers for human circadian phase. eLife. 2017;6:e20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang L-H, Lin P-H, Tsai K-W, Wang L-J, Huang Y-H, Kuo H-C, et al. The effects of storage temperature and duration of blood samples on DNA and RNA qualities. PLoS One. 2017;12(9):e0184692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson RK, Overlee BL, Sagen JA, Howe CL. Peripheral blood mononuclear cell phenotype and function are maintained after overnight shipping of whole blood. Sci Rep. 2022;12(1):19920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hok-A-Hin YS, Willemse EAJ, Teunissen CE, del Campo M. Guidelines for CSF processing and biobanking: impact on the identification and development of optimal CSF protein biomarkers. Methods Mol Biol. 2019;2044:27–50. [DOI] [PubMed] [Google Scholar]
- 72.Koel-Simmelink MJA, Vennegoor A, Killestein J, Blankenstein MA, Norgren N, Korth C, et al. The impact of pre-a nalytical variables on the stability of neurofilament proteins in CSF, determined by a novel validated SinglePlex Luminex assay and ELISA. J Immunol Methods. 2014;402(1–2):43–9. [DOI] [PubMed] [Google Scholar]
- 73.Verberk IM, Nossent EJ, Bontkes HJ, Teunissen CE. Pre-analytical sample handling effects on blood cytokine levels: quality control of a COVID-1 9 biobank. Biomark Med. 2021;15(12):987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramont L, Thoannes H, Volondat A, Chastang F, Millet M-C, Maquart F-X. Effects of hemolysis and storage condition on neuron-specific enolase (NSE) in cerebrospinal fluid and serum: implications in clinical practice. Clin Chem lab Med. 2005;43(11):1215–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.