Abstract
Microphysiological systems (MPSs), also known as organ-on-chip or disease-on-chip, have recently emerged to reconstitute the in vivo cellular microenvironment of various organs and diseases on in vitro platforms. These microfluidics-based platforms are developed to provide reliable drug discovery and regulatory evaluation testbeds. Despite recent emergences and advances of various MPS platforms, their adoption of drug discovery and evaluation processes still lags. This delay is mainly due to a lack of rigorous standards with reproducibility and reliability, and practical difficulties to be adopted in pharmaceutical research and industry settings. This review discusses the current and potential use of MPS platforms in drug discovery processes while considering the context of several key steps during drug discovery processes, including target identification and validation, preclinical evaluation, and clinical trials. Opportunities and challenges are also discussed for the broader dissemination and adoption of MPSs in various drug discovery and regulatory evaluation steps. Addressing these challenges will transform long and expensive drug discovery and evaluation processes into more efficient discovery, screening, and approval of innovative drugs.
INTRODUCTION
Microphysiological systems (MPSs), also known as organs-on-chips or disease-on-chips, are microfluidics cell culture platforms that aim to replicate physiology and pathology to study diseases and discover new drugs. These systems combine microsystems engineering, microfluidics, and cell biology to create three-dimensional, multi-cellular models with fluid flow, mechanical cues, and tissue–tissue interfaces. MPS platforms have gained attention for their potential to improve drug development by providing more accurate representations of human organ functions compared to traditional cell culture models or animal testing. In drug discovery, clinical drug development has a high failure rate of up to 90%, likely due to overlooked aspects in target validation and drug optimization.1,2 Conventional models used in drug development often fail to accurately predict human-specific physiological features and toxicities, leading to significant challenges in translational success.3,4 With reasonable controllability in developing the system for drug discovery/drug evaluation, MPSs can be a useful alternative to conventional cell culture or animal models.5 MPS platforms to recapitulate the complex features in tissue microenvironment aim to eventually replace animal testing with more human-physiologically relevant models, contributing to drug discovery.
Recent studies have made significant advancements in the development of various MPS models, demonstrating their ability to replicate the complexity of microphysiology. These innovative models combine microsystems engineering, microfluidics, and cell biology, going beyond providing a simple 3D cell culture environment. MPS technology has opened avenues for increased efficiency, sensitivity, and throughput in developing advanced in vitro 3D culture models.6,7 Furthermore, MPSs now allow the faithful reproduction of tissue heterogeneity, encompassing crucial cell-to-cell interactions, such as intratumoral heterogeneity,8 tumor-stroma interactions,9 and interactions between tumor cells and immune cells or endothelial cells.10,11 MPS platforms have also successfully recapitulated the complex functional features of specific organs. For instance, liver-on-chip models have been devised to simulate liver functions, incorporating oxygen tension variations in the liver sinusoids in drug metabolism.12,13 Similarly, kidney-on-chip models have been engineered to replicate the intricate filtration and reabsorption functions of the kidneys,14,15 These advancements have paved the way for more physiologically relevant in vitro models that hold great promise for advancing drug discovery and understanding disease biology.
Despite the recent emergence and advancement of MPS platforms, their integration into drug discovery and evaluation processes still remains significantly limited. The primary reasons for the hesitation arise from the lack of well-defined standards for reproducibility and reliability, as well as the difficulties in adapting the platforms to pharmaceutical research and industrial applications. Drug discovery is a multifaceted and multi-stage process with the primary goal of identifying and validating potential drug targets for treating diseases and medical conditions. The drug discovery pipeline typically comprises several key stages, including target identification and validation, hit generation, hit-to-lead optimization, preclinical testing, and clinical trials, requiring interdisciplinary collaboration among scientists, pharmacologists, chemists, and clinicians, as well as significant financial investment and adherence to stringent regulatory guidelines.1,5
The recent decision by the U.S. Food and Drug Administration (FDA) to eliminate the requirement for animal data in preclinical drug evaluation has paved the way for exciting opportunities in the field of drug discovery and evaluation using microfluidics-based technology.16 The key to realizing these possibilities lies in the widespread adoption of this approach. This review aims to discuss the importance and application of MPSs in drug discovery, focusing on key stages of drug discovery including target identification and validation, preclinical evaluation, and clinical trials. Specifically, we provide perspectives on the context of use for various MPSs in the field of drug discovery, articulating the drug evaluations with MPSs in model description, available outcomes, and the purpose of the tool. The discussion is further extended to the opportunities that MPSs present as well as the challenges that need to be addressed to promote the widespread adoption of these systems in drug development and regulatory evaluation. By incorporating MPSs into these stages, drug discovery can benefit from enhanced predictive capabilities and a more accurate representation of human physiology, potentially leading to more efficient screening and the successful translation of the preclinical study to clinical study. The wider adoption of MPSs in the pharmaceutical industry and regulatory agencies requires rigorous validation and acceptance. Demonstrating the predictive accuracy and reproducibility of MPS data in comparison to traditional models and human clinical outcomes is essential for gaining regulatory approval and industry acceptance.
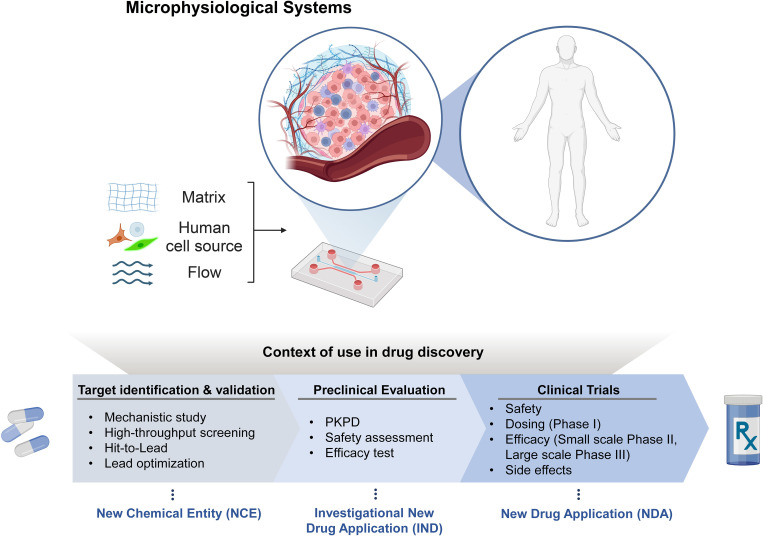
Context of use of MPS in drug discovery
The design paradigm of many MPS platforms has been the recapitulation of the in vivo microenvironment and pathophysiology of diseases. However, this design paradigm needs to be changed considering the context of essential steps in drug discovery pipeline. These contexts include “Target identification and validation,” “Preclinical evaluation,” and “Clinical trials.” During the first stage, known as target discovery, in vitro research is performed to identify targets involved in specific diseases. MPSs provide a better replication of the human environment than the conventional in vitro platforms, allowing to investigate and evaluate the identified targets with mechanistic study, high-throughput screening, hit-to-lead, and lead optimization (Fig. 1). Typically, a target constitutes a molecule essential for gene regulation or intracellular signaling, encompassing entities like nucleic acid sequences or proteins. In order to decide on which target to focus research efforts, one needs to ensure that the molecule is druggable—that its activity can be modulated by an exogenous compound.17 The identified targets, then, should be validated in lead compound identification and lead optimization. The lead compound identification is the process of identifying or creating a compound that can interact with the target selected. This can be repurposing existing compounds or synthesizing new ones. Lead optimization is the refinement of compounds for efficacy, safety and minimum off-target binding. Also, optimal dosage and delivery routes should be tested. The MPS can be facilitated to prepare new chemical entity (NCE) contributing the target identification and validation for FDA drug approval application. The identified targets then are evaluated in various aspects.
FIG. 1.
Application of MPS platforms in drug discovery steps. Created with BioRender.com.
The utilization of MPS platforms enables the recognition of pharmacokinetics and pharmacodynamics (PKPD), safety assessment, and the efficacy of drugs in preclinical assessments.18,19 These evaluations aim to determine the optimal dosage, delivery routes, and drug actions following administration. PKPD studies play a crucial role in addressing optimal dosage and delivery routes. Additionally, the assessment of drug-induced toxic effects on metabolic organs, such as the liver and kidneys, which are essential for drug metabolism and elimination, aids in screening potentially toxic drugs and identifying potential side effects. The outcomes obtained from preclinical studies can be considered during the Investigational New Drug (IND) application process.
In addition to preclinical studies, MPSs can significantly contribute to enhancing and supporting clinical trials by assisting in safety assessments, dose determination (phase I), evaluating efficacy, and identifying potential side effects.20 Ultimately, the successful completion of phase III, coupled with efficient decision-making processes aided by MPSs, can lead to the preparation of a New Drug Application (NDA).
MPS as disease models for target identification and drug efficacy evaluation
Disease-on-a-chip is a type of MPS system being developed as a tool to model and study complex and heterogeneous features of diseases such as cancer. Disease MPS platforms can help to create a controlled pathological environment that mimics the tissue microenvironment of the disease, contributing to the discovery of therapeutic targets and disease-specific markers, which is a crucial drawback of using traditional 2D cell culture models.21–23 In technological routes, advances in microfluidics and micro-scale engineering have significantly propelled the evolution of MPS technology. MPS models have expanded beyond their initial focus on showcasing 3D culture through microscale fluidic physics to include intricate biological elements at an organ level. This progress has been facilitated by a range of technical advancements, encompassing the development of multi-channel systems, integration of porous membranes, and innovations in fabrication techniques such as soft lithography, additive manufacturing, and thermal press methods.24–26 More recently, MPSs have embraced sophisticated molecular and cellular biology techniques integrating intricate biological structures and diverse interactions among multiple cells and their environments.27 In intricating the systems, enhancing MPS technology through modular systems, open microfluidics, and wall-less platforms, signifying a pursuit of improved capabilities and functionalities within MPS technology.26
MPS systems have exhibited a broad spectrum of diseases, including cancer, inflammation, fibrosis, type II diabetes, and many mores.13,28–30 The disease MPS systems have been engineered to replicate specific disease conditions faithfully. For instance, a gut inflammation on-a-chip model was developed simulating intestinal barrier dysfunction,28 while also organ-specific fibrotic transitions were demonstrated in MPSs for fibrosis diseases.31,32 Demonstrating functional crosstalk between organs has been a pivotal aspect in diseases such as type II diabetes. To emulate this, researchers from TissUse and AstraZeneca integrated human pancreatic islets with liver spheroids in two-organ chips, effectively modeling type II diabetes. The study showcased sustained organ crosstalk for 15 days, mimicking the gold standard intravenous glucose tolerance test utilized to evaluate insulin secretion and glucose elimination in humans. Furthermore, it has the potential to develop personalized medicine for various disease stages, as it enables the incorporation of patient-specific cell sources and environments into the system. This integration also leads to improved prediction of drug efficacy during preclinical studies.
Although various disease MPS platforms have been proposed and developed, MPS systems has made most significant advances in modeling tumors which needs reliable but innovative models to recapitulate intricate, heterogenous, and desmoplastic tumor microenvironment (TME) (Fig. 2). They enable systematic investigation of the complex architecture in the tumor microenvironment (TME) and provide insights into the mechanisms underlying drug resistance and metastasis in cancer.8,33,34 With their improved representation of human tumor tissue, MPS systems can serve as a comprehensive testbed for drug efficacy evaluation.35,36 Indeed, cancer is a complex and heterogeneous disease giving high attrition in drug development. The difficulty in defeating invasive cancers, desmoplastic tumor microenvironment, and tumor heterogeneity pose challenges in cancer research. There are various subtypes of cancers that can be identified based on their molecular and genetic characteristics. For instance, breast cancers have been categorized as four subtypes: luminal A, luminal B, HER2-enriched, and triple negative breast cancers (TNBCs).37–39 Unfortunately, TNBC is a heterogeneous disease itself, consisting of multiple entities with distinct histopathological, transcriptomic, and genomic features,40 leading to challenges in finding appropriate drugs or drug combinations for individuals with TNBC.41–43 In addition to the disease-level complexity with the highly heterogeneous cancer cell populations, the desmoplastic stroma components in tumor microenvironment (TME) are multifaceted and can influence the growth, invasion, and drug sensitivity of tumor cells. The stromal tissue, which is a complex environment that stores numerous growth factors and cytokines, can greatly impact tumor growth and drug response. Various stromal cells, including cancer-associated fibroblasts (CAF), as well as diverse immune and inflammatory cell types, are present in the associated stroma along with rich extracellular matrix (ECM) components like type I collagen, hyaluronan acid, fibronectin, and fibrin.44,45 For certain types of cancer, such as pancreatic and prostate cancer, this stroma can constitute up to 90% of the overall tumor volume.45–49 The densely packed stromal tissue can greatly impact tumor growth and drug response, occurring hypoxia and abnormal tumor vasculature. Thus, tumor MPS capable of recapitulating the complex features of TME is crucial to better model the human system, understand physiological processes, facilitate the targeting of drug discovery, and provide a faithful replication of physiological condition for drug transport.
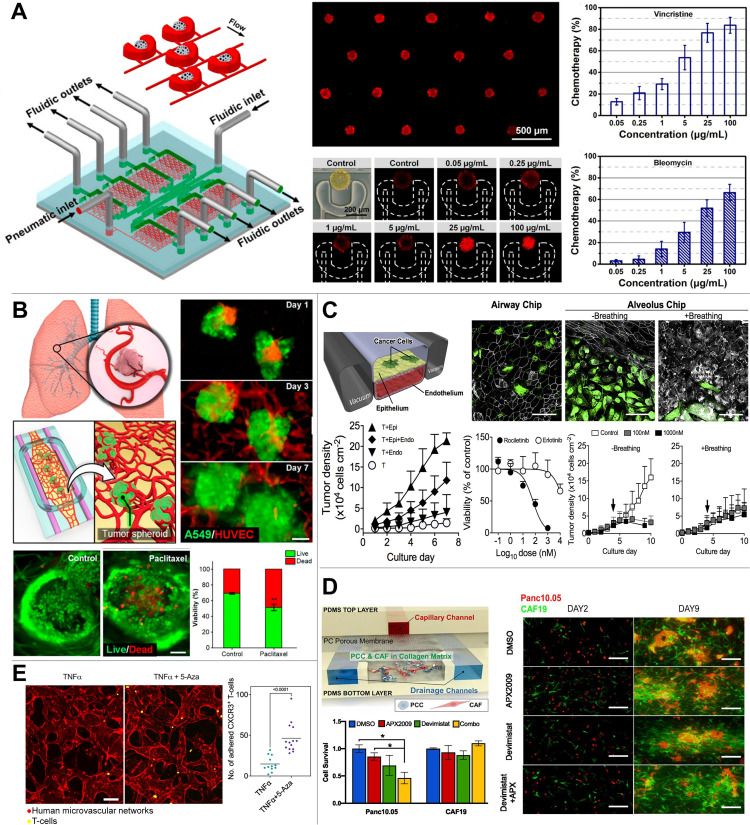
FIG. 2.
Disease MPS models developed to evaluate the drug efficacy. (a) Upscaled multi-well arrays of integrated spheroid culture and microfluidic systems have enhanced throughput, while preserving uniform spheroid formation, cytotoxicity assessment, and molecular analysis. Adapted with permission from Liu et al., Anal. Chem. 87(19), 9752–9760 (2015). Copyright 2015 American Chemical Society. (b) MPS models of 3D cell culture and self-assembled microvascular networks to replicate organ-specific tissue architectures and enable drug efficacy testing. Adapted with permission from Paek et al., ACS Nano 13(7), 7627–7643 (2019). Copyright 2019 American Chemical Society. (c) In vitro orthotopic lung MPS chip recapitulating organ microenvironment-specific tumor responses. Reproduced with permission from Hassell et al., Cell Rep. 21(2), 508–516 (2017). Copyright 2017 Elsevier. (d) A tumor-stroma MPS model recapitulating tumor–CAF interaction within the pancreatic tumor microenvironment was facilitated for the assessment of anti-cancer drug efficacy. Adapted with permission from Gampala et al., J. Exp. Clin. Cancer Res. 40(1), 251 (2021). Copyright 2021 Author(s), licensed under a Creative Commons Attribution 4.0 International License. (e) A human microvasculature network within an MPS model was engineered to study the impact of anti-tumor T-cell activity in response to interactions with tumor angiogenesis. Adapted with permission from Kim et al., Nat. Commun. 14(1), 2122 (2023). Copyright 2023 Author(s), licensed under a Creative Commons Attribution 4.0 International License.
The focus of tumor MPS has primarily been on replicating the complex culture conditions which has impeded the successful prediction of developing anticancer drugs. Instead of using a simple 2D monolayer cell culture, the adoption of 3D culture models has become necessary, as it is now recognized that cellular behaviors vary significantly based on the culture conditions. In this regard, the multicellular spheroid models have provided better 3D modeling compared to 2D monolayer assays, showing advantages in detecting drug resistance for better prediction in drug efficacy.23,50,51 Beyond the spheroid, microfluidic techniques have also been employed by enabling spheroid production with increased efficiency, sensitivity, and throughput.6,7,52,53 By scaling up the multi-well arrays of the spheroid culture integrated with microfluidic systems, the throughput was improved while maintaining consistent spheroid formation, cytotoxicity assay, and signaling pathway analysis [Fig. 2(a)].
While the spheroid model offers improved representation of culture conditions in 3D, it falls short in presenting the complex and heterogeneous microenvironment that significantly impacts tumor progression and metastasis. Intricate interactions between cells and their microenvironment, including the extracellular matrix, various cell types, and fluidic conditions, have been recognized as crucial factors in inducing drug resistance and disease progression. To enhance comprehension of the complex interactions between cells and the microenvironment, MPSs has been developed in increasing the complexity of the 3D culture model.54 This advancement allows for a better representation of the interactions between cells and their microenvironment, enabling the identification of effective drug targets, and more accurate evaluation of drug efficacy using the disease models.36,55–57
Recently, there has been a considerable focus on the role of heterogeneous cell interactions in the tumor microenvironment, which not only influence drug responses but also play crucial roles in pathological processes.58–60 For example, the role of intratumoral heterogeneity in inducing drug resistance has been acknowledged across different cancer types including pancreatic cancer.40,58–62 Building on this perspective, the dynamics of tumor-tumor interactions were effectively illustrated within a tumor MPS model, which integrated cell lines sourced from genetically engineered mouse models (GEMMs) of pancreatic cancer.8 In this research, meticulous selection of the GEMM-driven cells was employed to regulate genotypic and phenotypic heterogeneity. This approach unveiled the impact of cellular interactions between various cell types on gemcitabine resistance, providing valuable insights into the mechanisms underlying drug resistance.
In addition to intratumoral heterogeneity, tumors actively engage with diverse cell types within their microenvironment, including endothelial cells. To enhance the replication of tumor vasculature and facilitate drug delivery studies, the adoption of tumor angiogenesis-on-chip models has been widely adopted.63–67 For instance, the work by Paek et al.65 developed an engineered vasculature beds incorporating with tumor spheroids to use it for drug screening. This concept was successfully validated using Paclitaxel in the context of lung cancer [Fig. 2(b)]. Moreover, the potential of RNAi-based nanomedicine has been assessed using the MPS platform, highlighting interactions between tumor spheroids and endothelial cells.66 The research conducted by the Ingber group67 has revealed intricate interactions between endothelium, epithelium, and tumors for lung tumor growth. The platform was employed to replicate physiological conditions for assessing drug effectiveness and identifying drug resistance [Fig. 2(c)].
Extensive research has been conducted on tumor–stroma interactions, wherein the stroma transforms into a desmoplastic environment comprising cancer-associated fibroblasts (CAFs) and immune cells that support tumor progression and metastasis.68–70 Among the stromal cells, CAFs have been recognized for playing either supportive or suppressive roles in tumor progression. Recent advances in single-cell analysis have enabled the identification of distinct CAF subtypes, aiding in the discernment of phenotype distributions within the CAF population. Notably, certain CAF subtypes employ substantial influence over tumor progression and drug resistance through their interactions with tumor cells.9,71,72 The tumor MPS models considering the tumor-CAF interactions have been developed in understanding the pathological processes and drug resistance.9,73,74 Moreover, the tumor–stroma interaction is also integrated in the MPS for drug screening to test developing drugs on pancreatic cancer [Fig. 2(d)]. Gampala et al. investigated the Redox factor-1 (Ref-1) as a novel target of pancreatic tumors, which is a redox signaling protein regulating redox metabolic activity of cancer in hypoxia, known to induce drug resistance and metastasis.75 In testing the drug efficacy of the Ref-1 inhibitor, MPS model integrating human tumor and CAF cells was used following by the conventional screening platforms of spheroids and animal models to test the efficacy of the drugs itself and combination treatment with anti-mitochondrial drugs (Devimistat).
Meanwhile, the recent advances in tumor MPS models have demonstrated the potential of MPSs in the development of immunotherapy, as reviewed by the Tu group.76 Kim et al.77 have achieved significant progress with an innovative tumor-vasculature MPS model that includes cytotoxic CD8+ T-cells [Fig. 2(e)]. These cells were introduced into an in vitro microvascular network to explore the complex interactions between immune and endothelial cells. The study showed that the immunoregulatory effect of a vascular-selective cytokine in the tumor microenvironment affected immune responses and tumor growth. Similarly, Mollica et al. showcased the compact composition of the pancreatic tumor microenvironment within an MPS system, where pancreatic tumor cells, endothelial cells, and pancreatic stellate cells were integrated to study T-cell mobility.78 These investigations underscore the utility of MPSs in exploring the potential improvements in T-cell-focused immunotherapy, aiming to enhance T-cell trafficking and bolster anti-tumor immunity. Moreover, the potency of immune responses is vividly highlighted in MPS tumor models, where the cytotoxic effect of natural killer (NK) cells was effectively demonstrated.79,80 These models furnish critical insights into the intricate and high-fidelity nature of MPS platforms, as they provide a dynamic and representative environment to observe the behavior of immune cells within a tumor context.
Rare disease models and personalized medicine
The integration of human physiological cell sources and environments in MPSs has significant advantages for studying rare diseases and personalized medicine.81,82 By incorporating patient-specific cell sources and considering the unique characteristics of specific diseases, these models can effectively demonstrate disease-specific phenotypes, facilitating target identification and evaluation. This approach enables researchers to study and understand the intricacies of rare diseases at a cellular level, paving the way for tailored therapeutic interventions and personalized treatment strategies. Patient-derived cells were used to model the Shwachman–Diamond syndrome in a bone marrow-on-a-chip, demonstrating the potential of using MPSs for personalized therapy optimization and clinical trial design, particularly for rare diseases that are challenging to test preclinical drugs systematically.83 In addition, intratumoral heterogeneity with pancreatic ductal adenocarcinoma was investigated with genetically engineered mouse model-driven cells.8 This study recapitulated the PDAC's oncogenic heterogeneity which showed a potential of MPS in mimicking patients' specific genetic architecture with available cell sources.8 Advances in stem-cell technology with induced pluripotent stem cells (iPSCs) would accelerate the application of the MPS in personalized medicine.84,85 For instance, the blood–brain barrier (BBB)-on-a-chip, demonstrating the selective barrier that separates the bloodstream from the brain, has been developed by combining it with iPSC-derived brain cells from patients to develop a patient-specific in vitro model for personalized medicine.86,87 This concept was demonstrated by Vatine et al.,87 wherein patient-specific in vitro models of two neurological disorders, Huntington's disease, and MCT8-specific deficiency, were created.
Although many disease models have been integrated into drug efficacy evaluation platforms in preclinical studies, the limited ability to achieve reasonable throughput has hindered the effective screening of developing drugs. This poses a significant barrier to the utilization of these platforms in the pharmaceutical industry.
MPS models for pharmacokinetics and pharmacodynamics (PKPD) evaluation
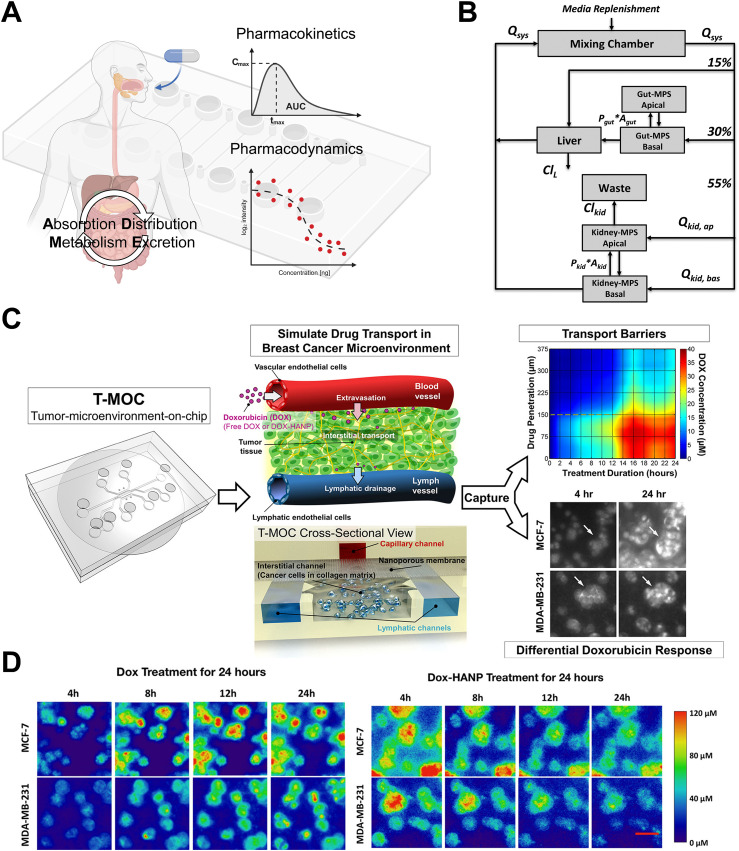
The context of use for MPSs has also expanded greatly in the evaluation of pharmacokinetics (PK) and pharmacodynamics (PD) in drug development. The PK and PD are essential in drug development stages as they provide valuable estimates of drug effectiveness, safety, and dosage regimens for optimal therapeutic outcomes [Fig. 3(a)]. PK focuses on the drug's pathway through the body experiencing drug absorption, distribution, metabolism, and elimination (ADME), governing the consequent concentration in bloodstream. PD refers to the relationship between drug dosage and its pharmacological effects involving its interaction with the targets, enzymes, or other molecules. The application context of PK-PD evaluation using MPSs encompasses several key aspects. First, it involves the replication of functional features of each organ within the system, allowing for the demonstration of drug ADME. Additionally, it entails determining parameters for mathematically driven PK models, which systematically addressing how drugs behave in the body. Furthermore, evaluation platforms are utilized to investigate the PK–PD relationship by simulating the dosage varying drug concentrations mimicking in vivo PK profiles to perfuse them toward the intended drug targets.
FIG. 3.
Employment of MPS models for assessing PKPD. (a) A schematic illustrates drug PKPD based on absorption, distribution, metabolism, and excretion (ADME). Created with BioRender. (b) Multi-functional scaling for muti-organ MPS design was established to optimize the integrated platforms mimicking in vivo drug exposures. Adapted with permission from Maass et al., Integr. Biol. 9(4), 290–302 (2017). Copyright 2017 Oxford University Press. (c) Utilizing the tumor-microenvironment on-chip (T-MOC) platform to emulate drug transport within the breast cancer microenvironment, enabling the study of drug penetration and accumulation. Adapted with permission from Ozcelikkale et al., J. Controlled Release 266, 129–139 (2017). Copyright 2017 Elsevier B.V. (d) Comparison of the distinct drug accumulation patterns of doxorubicin (Dox) and Dox-loaded hyaluronic acid nanoparticles (Dox-HANPs) within the breast cancer T-MOC platform. Reprinted with permission from Shin et al., Mol. Pharm. 13(7), 2214–2223 (2016). Copyright 2016 American Chemical Society.
Accurate prediction of the PK profile is essential for understanding the dosage response of drugs. Over time, mathematical modeling of drug PK has advanced from simplistic compartmental models to more complex physiologically based PK models (PBPK), which represent the entire body as a network of interconnected organs as it was comprehensively reviewed for oral drug PBPK models.88 Construction of PBPK models involves estimating various anatomical and physiological parameters, such as organ volumes, blood or lymphatic perfusion, alongside kinetic constants governing drug ADME behaviors. However, it poses a significant challenge in accurately predicting the parameters for quantitatively determining appropriate drug dosages during clinical trials due to limitations and uncertainties in conventional measurement methods, including in vivo and in vitro studies to extrapolate from preclinical animal data.21,89,90
By integrating MPSs into PBPK modeling, more reliable and relevant parameter values can be obtained with more accurate representation of human physiology, leading to improved predictions of drug behavior and efficacy in humans.18,19,90–93 Several pioneering studies unlocked the potential for demonstrating PKPD in MPSs.18,19 Recently, Korner et al.92 provided the novel pharmacokinetic compartment model using microfluidic droplets and biomimetic artificial cells that significantly improves the prediction of molecular absorption in the jejunum. By mimicking the composition and size of human intestinal cells, the developed microfluidic platform allows for precise quantification of pharmacokinetic parameters, such as half-life, flux, and the apparent permeability coefficient, which closely aligns with absorption of the drugs in actual intestinal tissue. The kidney MPSs have developed and applied to evaluate the renal drug secretion.94 Furthermore, the replication of interconnections and interactions between multiple organs has gained significant attention in the development of MPSs known as multi-organ-on-chip platforms, aiming to achieve more accurate replication of human PKs. These systems allow for the integration of diverse organ-specific models, enabling the simulation of complex physiological processes, including drug ADME. By incorporating multiple organs within a unified system, multi-organ-on-chip platforms provide a more comprehensive and precise representation of human PK, offering promising advancements in drug development and reducing the reliance on animal models.90,91,95–97 By implementing integrated multi-MPS platforms that mimic gut, liver, kidney, and bone-marrow models, researchers successfully applied a multi-functional scaling approach to PBPK modeling. Through these innovative modifications, they were able to quantitatively replicate the in vivo PK profiles observed in human clinical data for nicotine and cisplatin across various doses and administration routes.90 The multi-functional scaling using multi-organ MPS contributed to developing better prediction PBPK model for various drugs91 [Fig. 3(b)]. However, the multi-organ MPS models have not fully replicated the physiologically relevant tissue–tissue barriers between vascular endothelium and parenchymal cells within each organ, which significantly influence ADME responses and are critical for clinically relevant PK modeling.
MPSs have shown the ability to establish the relationship between PK and PD. Conventional drug screening methods using static perfusion may lead to an overestimation of drug effectiveness. However, by incorporating drug perfusion in MPSs, we can enhance drug evaluation by better mimicking the dynamics of the human system and comprehensively assessing drug efficacy.98–100 Indeed, the MPS platform was utilized to simulate drug transport within the tumor microenvironment.8,101,102 Ozcelikkale et al.101 conducted a study where they perfused doxocuticin (dox) and Dox-encapsulated hyaluronic acid nanoparticles by capturing drug penetration and accumulation in a breast tumor microenvironment-on-chip (T-MOC). They considered PK profile of rapid and slow clearances through the MPS system. In results, drug transport and accumulation are greatly influenced by the PK, revealing the significant relationship between PK and PD in the context of transport101,102 [Figs. 3(c) and 3(d)]. Also, the researchers have approached to develop programmable drug perfusion system integrated with microfluidics mimicking PK and showed drug efficacy for the PK–PD relationship.98–100
Moreover, leveraging PKPD approaches through MPSs offers substantial advantages in deciphering distinct physiological processes such as the BBB and cardiovascular system, as given their pivotal roles in fluidic transportation within the body. The BBB is a guardian of the central nervous system and offers selective permeability to solutes across the barrier. Owing to this high selectivity, the BBB does not allow the passage of medication for the treatment of neurological diseases. The BBB also undergoes physiological alteration under various diseased states, which affects the transport across the barrier. MPS offers the opportunity to mimic the BBB microenvironment under various diseased states, such as Alzheimer's,103,104 Parkinson's,105,106 stroke,107 epilepsy,108 fungal infection,109 glioblastoma multiforme (GBM),110–113 amyotrophic lateral sclerosis (ALS),114,115 and Huntington's disease87 by mimicking three-dimensional intercellular interactions, capillary blood flow, and biochemical gradients. Several studies to evaluate drug response in BBB-on-a-chip have been performed. For instance, a study conducted by Park et al.116 successfully mimicked the in vivo transport of a clinically approved anti-cancer therapeutic antibody, cetuximab, under the reversible osmotic opening of the BBB in vitro. Furthermore, they demonstrated receptor-mediated transcytosis of nanoparticles [20 nm Q-dot coated with Angiopep-2 (AP2) through low-density lipoprotein receptor-related protein 1 (LRP1) and anti-transferrin antibodies (MEM75 and 13E4)] through transferrin receptor.
Safety assessment using organ-on-chips
In drug discovery, safety assessment of the drug evaluation is of utmost important procedures in both preclinical and clinical stages encompassing patient safety and regulatory compliance. Animal testing has been facilitated in drug toxicity evaluation, providing an in vivo condition for studying the effects of drugs on crucial organs and the immune system. However, almost 40% of the safety assessment with animal model has failed in the clinical translation, indicating that the human system is highly desired.1,35 With the advent of organs-on-a-chip devices (OOC), which mimic the human microphysiological environment on an in vitro platform, drug toxicity testing of OOC has become promising in safety assessment for developing drugs.5,82,117 These platforms have been developed for recapitulation of drug metabolism and adverse effects in major organs, including liver (hepatic lobule), lung (acini), kidney (nephron), pancreas (acini), blood vessel and the list go on, which are key factors in preclinical drug development, in particular, to improving decision-making in drug discovery.117
Liver is an organ to metabolize foreign substances that enter the body such as drugs and xenobiotics, converting these substances into forms that can be easily eliminated. Also, the liver is the primary site for drug adverse effects, which can result in liver injury or failure. To ensure the safety of the drug and minimize the risk of liver damage in patients, it is required to screen the hepatotoxic effect of the drug and drug-induced liver injury (DILI) in drug development stages. The liver MPS provides beneficial to recreate the properties of three-dimensional, fluid flows, cell type consideration, and heterogeneous cell to cell interactions to recapitulate the liver function in drug metabolism and hepatotoxicity for predicting clinical drug effects.12,118–121 These features address the limitations of conventional 2D hepatocyte cell culture systems, which often overestimate the hepatotoxicity of drug candidates.118,119 Lee-Montiel et al.12 recapitulated a liver acinus establishing various zonation classified by the oxygen tension. The modeled zonation-dependent hepatocyte functions for liver metabolism were facilitated to test zone-dependent efficacy of acetaminophen.12 Liver-on-chip developed by Bevli et al. integrated with the dynamic environmental manipulation and electrochemical sensor systems to demonstrate the real-time monitoring of metabolic adaptation to mitochondrial dysfunction in response to rotenone and troglitazone, which is the antidiabetic and anti-inflammatory drug withdrawn from the market due to significant drug-induced liver injury.121 The liver sinusoid-on-a-chip device by Deng et al. containing four transformed cell lines arranged in a physiologically relevant distribution and simulating liver functions with engineered blood and bile flow, demonstrated effective drug transport and preserved enzymatic activities, allowing investigation of drug–drug interactions and hepatotoxicity.122 The model was validated with three clinically relevant acute hepatotoxic drugs, acetaminophen (APAP), rifampin (antimycobacterial, RIF), and amiodarone, and also APAP in combination with the other drugs.
The kidney is also a crucial organ to consider in the drug safety assessment, which plays a crucial role in drug metabolism and elimination, serving as a primary filter for blood, expelling surplus fluids and waste materials from the system and in balancing the body's electrolytes, including significant elements like sodium, potassium, and calcium. To effectively use the kidney MPS in drug development, the physiological function of the kidney has been emulated by demonstrating the intricate interactions between various cell types, including podocytes, proximal tubule cells, and endothelial cells, which are key players in the kidney's filtration and reabsorption functions.14 The filtration procedures allow the removal of waste products and excess fluids from the blood while retaining important substances such as glucose, electrolytes, and amino acids, which are transported to the urine. Engineering fluid dynamics and structural architectures of kidney through MPSs allows to mimic the osmotic pressure gradients, structural organization of renal tubular segments, and cellular metabolic and endocrine functions.14,15 The physiological functions of glomerular, proximal tubular, and distal tubular systems have been simulated through the application of MPS as it is well-reviewed in Ashammakhi et al.14 In the context of use in drug discovery, kidney-on-a-chip has demonstrated drug-induced nephrotoxicity which causes when kidney functions are rapidly deteriorated as a result of the toxic effects of drugs.15,123,124
In addition, the skin, as the body's largest organ, has an intricate immune system that holds significant importance in assessing adverse reactions to drugs. These reactions can manifest in various ways, ranging from mild rashes to severe allergic responses, highlighting the crucial role of the skin immune system in identifying drug's adverse effects.125 Thus, reliable skin models capable of reconstituting the skin immune system will be an early indicator to evaluate drug safety and potential adverse side effects during drug development and usage. Recent advancements have seen significant progress in developing skin-on-a-chip systems, as discussed in prior review papers.126,127 These systems aim to replicate the complexities of human skin in vitro, incorporating multiple layers to simulate various aspects, evaluating skin reactions, drug diffusion, and allergies. For example, the use of these skin models in assessing the safety of drug compounds emphasized the drug toxicity testing for the skin by recapitulating the barriers between skin-vasculature,128 and the focus on the immune reactions within the skin. Skin-on-a-Chip by Ramadan et al.129 showed that keratinocytes reduced monocyte cytokine production in condition stimulating with lipopolysaccharides (LPSs) and nickel sulfate, highlighting a strong barrier function of the keratinocyte layer compared to devices without keratinocytes or immune cells.
The adoption of MPS platforms in drug discovery has acquired significant attention in recent discussions with the involvement of multiple institutes, regulatory reagents, and the industry, there is a growing focus on realizing the practical implementation with qualified validation of the platforms.5,130,131 To validate the organ-on-chip platform, some studies have tested drugs that did not advance to clinical development.2,124 For instance, the pro-thrombotic effects of a monoclonal antibody against CD40L were replicated using a blood vessel-on-a-chip system.2 Also, Rubiano et al. assessed the reproducibility and reliability of the liver MPS for the drug evaluation process.132 The study aims to ensure consistent results across multiple test sites and MPS batches, addressing challenges related to inter-batch and inter-site variability. Despite advancements in organ-on-chip technology, there remains a need for a systematic integration of multiple organ systems that can mimic physiological connections within a body-on-chip system.130 Currently, the development of a comprehensive platform that accurately represents the complex interactions and functions of various organs in the human body is still a challenge. Efforts are underway to overcome this limitation and create a more universal body-on-chip system that can provide a realistic and predictive model for studying drug responses, disease mechanisms, and personalized medicine.
Challenges and opportunities
Although numerous innovative and promising MPS have been developed, their application in drug discovery and evaluation remains relatively unexplored. The transition of MPSs from a research tool to an established platform for drug development is crucial. To facilitate this transition, various groups from academic institutions, industry, and regulatory agencies have initiated discussions on the potential utilization of MPSs and the necessary enhancements to replace animal testing and improve the representation of the human system.5,82 In order to successfully recapitulate the human system and replace animal testing, it is crucial to address several challenges. These include the need to improve the similarity between MPSs and the human system, establish reliable and standardized procedures for developing and validating MPS, and overcome throughput challenges. By addressing these limitations, we can enhance the effectiveness of MPSs in mimicking human physiology and enable their widespread adoption as an alternative to animal testing.
To fully realize the potential of MPSs in accurately representing the human system, several advancements are still required. One significant challenge is the lack of reliable and standardized methods for obtaining physiological materials, including cellular and matrix components.133 While recent progress has been made in utilizing patient-specific cell sources, such as stem cells, the current approaches still struggle to recapitulate the highly heterogeneous nature of cellular compositions. In particular, the inclusion of immune cell populations, which play a crucial role in drug evaluation, has been largely overlooked. Despite recent advancements in incorporating immune cells into MPS systems,76,134,135 there is still significant room for improvement to reconstitute functional immune systems. The opportunities persist in effectively demonstrating immune surveillance in disease states and patient-specific contexts. Notably, in pancreatic tumors, it has been observed that the intricate and heterogeneous immune surveillance could determine the disease progression as well as effectiveness of drugs and immune therapy.136,137 However, the absence of human immune cells within MPS poses a limitation. The tumor microenvironment is heavily influenced by immune cell interactions, making their presence essential for a comprehensive evaluation of drug responses. Reconstituting functional immune systems into MPS would provide a more accurate representation of the complex interplay between tumor cells and the immune system, enabling better assessment of drug efficacy and immunomodulatory effects.
Efforts should also be directed towards improving the physiological relevance of matrix compositions within MPSs. Currently, the selection and composition of matrix materials do not fully capture the diverse characteristics of native tissues. Developing standardized protocols for incorporating a range of matrix components, including extracellular matrix proteins and soluble factors, would enhance the biomimetic properties of MPS and better replicate the intricate cellular interactions and signaling pathways present in vivo.
Furthermore, the field would benefit from the establishment of standardized protocols and quality control measures to ensure consistency and reproducibility across different MPS platforms. Standardization would facilitate comparison and validation of results obtained from various laboratories, enabling better collaboration and advancement of MPS technology. The recent momentum around standardization, highlighted in a perspective report derived from the 2022 Innovation and Quality MPS Affiliate workshop, underscores the urgency of defining a clear context of use for MPSs.131 However, as outlined in the report, creating context-specific standards faces significant technical challenges. The diversity of applications and functions within MPSs, whether for drug development, toxicity testing, disease modeling, or other biomedical uses, presents distinct complexities and demands unique requirements.
Developing standards for MPSs to be applied in PKPD is crucial, especially considering the conventional reliance on animal and human studies in this field. While MPSs offer a closer emulation of human physiology than animal models, they encounter challenges in fully replicating the intricate interactions among multiple organs involved in ADME parameters. The main hurdle lies in accurately mimicking the dynamic relationships governing how the body absorbs, distributes, metabolizes, and eliminates drugs, as traditionally predicted in animal models. To address this, several innovative contexts have been proposed for validating MPS in PKPD.131 These suggestions involve evaluating MPS against in vivo predictive models or leveraging robust mechanistic insights gathered from clinical studies, including enzyme/transporter interplay and multiple interaction scenarios.
Furthermore, insufficient reliable throughput production has also been considered as a critical barrier in the adoption of MPS. The pharmaceutical industry recognizes the challenge of achieving high throughput in evaluating drug toxicity and efficacy within MPS, which involve multiple cells and complex tissue architectures. Thus, there is a demand for the advancement of high-throughput design and engineering techniques in MPS production and maintenance, enabling more efficient and scalable drug screening processes.
By addressing the identified limitations and challenges associated with MPSs, we can significantly enhance their utility in the field of drug discovery. Overcoming the discrepancies between MPSs and the human system, establishing reliable and standardized procedures for MPS development and validation, and addressing the throughput issues will enable MPSs to become a translational research tool in accelerating drug discovery processes and reducing reliance on animal testing.
ACKNOWLEDGMENTS
This study was partially supported by grants from NIH (Nos. U01 HL143403, R01 CA254110, R61 HL159948, and U01 CA274304), NSF (MCB-2134603), and the Purdue University Institute for Cancer Research (No. P30 CA023168), and a Program Grant from Purdue Institute for Drug Discovery.
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
Hye-Ran Moon: Conceptualization (equal); Writing – original draft (lead); Writing – review & editing (equal). Nishanth Surianarayanan: Writing – original draft (supporting). Tarun Singh: Writing – original draft (supporting). Bumsoo Han: Conceptualization (equal); Funding acquisition (lead); Writing – review & editing (lead).
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created in this study.
REFERENCES
- 1.Sun D., Gao W., Hu H., and Zhou S., Acta Pharmaceut. Sin. B 12(7), 3049–3062 (2022). 10.1016/j.apsb.2022.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrile R., van der Meer A. D., Park H., Fraser J. P., Simic D., Teng F., Conegliano D., Nguyen J., Jain A., Zhou M., Karalis K., Ingber D. E., Hamilton G. A., and Otieno M. A., Clin. Pharmacol. Ther. 104(6), 1240–1248 (2018). 10.1002/cpt.1054 [DOI] [PubMed] [Google Scholar]
- 3.Baillie T. A. and Rettie A. E., Drug Metab. Pharmacokinet. 26(1), 15–29 (2011). 10.2133/dmpk.DMPK-10-RV-089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Norman G. A., JACC: Basic Transl. Sci. 5(4), 387–397 (2020). 10.1016/j.jacbts.2020.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baran S. W., Brown P. C., Baudy A. R., Fitzpatrick S. C., Frantz C., Fullerton A., Gan J., Hardwick R. N., Hillgren K. M., Kopec A. K., Liras J. L., Mendrick D. L., Nagao R., Proctor W. R., Ramsden D., Ribeiro A. J. S., Stresser D., Sung K. E., Sura R., Tetsuka K., Tomlinson L., Van Vleet T., Wagoner M. P., Wang Q., Arslan S. Y., Yoder G., and Ekert J. E., ALTEX-Altern. Animal Experiment. 39(2), 297–314 (2022). 10.14573/altex.2112203 [DOI] [PubMed] [Google Scholar]
- 6.Chen Y., Gao D., Liu H., Lin S., and Jiang Y., Anal. Chim. Acta 898, 85–92 (2015). 10.1016/j.aca.2015.10.006 [DOI] [PubMed] [Google Scholar]
- 7.Lee D. W., Choi Y.-S., Seo Y. J., Lee M.-Y., Jeon S. Y., Ku B., Kim S., Yi S. H., and Nam D.-H., Anal. Chem. 86(1), 535–542 (2014). 10.1021/ac402546b [DOI] [PubMed] [Google Scholar]
- 8.Moon H.-R., Ozcelikkale A., Yang Y., Elzey B. D., Konieczny S. F., and Han B., Lab Chip 20(20), 3720–3732 (2020). 10.1039/D0LC00707B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labernadie A., Kato T., Brugués A., Serra-Picamal X., Derzsi S., Arwert E., Weston A., González-Tarragó V., Elosegui-Artola A., Albertazzi L., Alcaraz J., Roca-Cusachs P., Sahai E., and Trepat X., Nat. Cell Biol. 19(3), 224–237 (2017). 10.1038/ncb3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angelidakis E., Chen S., Zhang S., Wan Z., Kamm R. D., and Shelton S. E., Adv. Healthcare Mater. 12, 2202984 (2023). 10.1002/adhm.202202984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H., Park W., Ryu H., and Jeon N. L., Biomicrofluidics 8(5), 054102 (2014). 10.1063/1.4894595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee-Montiel F. T., George S. M., Gough A. H., Sharma A. D., Wu J., DeBiasio R., Vernetti L. A., and Taylor D. L., Exp. Biol. Med. 242(16), 1617–1632 (2017). 10.1177/1535370217703978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer S., Wennberg Huldt C., Kanebratt K. P., Durieux I., Gunne D., Andersson S., Ewart L., Haynes W. G., Maschmeyer I., Winter A., Ämmälä C., Marx U., and Andersson T. B., Sci. Rep. 7(1), 14620 (2017). 10.1038/s41598-017-14815-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashammakhi N., Wesseling-Perry K., Hasan A., Elkhammas E., and Zhang Y. S., Kidney Int. 94(6), 1073–1086 (2018). 10.1016/j.kint.2018.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilmer M. J., Ng C. P., Lanz H. L., Vulto P., Suter-Dick L., and Masereeuw R., Trends Biotechnol. 34(2), 156–170 (2016). 10.1016/j.tibtech.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 16.Wadman M., Science 379(6628), 127–128 (2023). 10.1126/science.adg6276 [DOI] [PubMed] [Google Scholar]
- 17.Pei H., Guo W., Peng Y., Xiong H., and Chen Y., Med. Res. Rev. 42(4), 1607–1660 (2022). 10.1002/med.21886 [DOI] [PubMed] [Google Scholar]
- 18.Sung J. H., Esch M. B., and Shuler M. L., Expert Opin. Drug Metab. Toxicol. 6(9), 1063–1081 (2010). 10.1517/17425255.2010.496251 [DOI] [PubMed] [Google Scholar]
- 19.Sung J. H., Kam C., and Shuler M. L., Lab Chip 10(4), 446–455 (2010). 10.1039/b917763a [DOI] [PubMed] [Google Scholar]
- 20.Cirit M. and Stokes C. L., Lab Chip 18(13), 1831–1837 (2018). 10.1039/C8LC00039E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H., Brown P. C., Chow E. C. Y., Ewart L., Ferguson S. S., Fitzpatrick S., Freedman B. S., Guo G. L., Hedrich W., Heyward S., Hickman J., Isoherranen N., Li A. P., Liu Q., Mumenthaler S. M., Polli J., Proctor W. R., Ribeiro A., Wang J.-Y., Wange R. L., and Huang S.-M., Clin. Transl. Sci. 14(5), 1659–1680 (2021). 10.1111/cts.13066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osuna de la Peña D., Trabulo S. M. D., Collin E., Liu Y., Sharma S., Tatari M., Behrens D., Erkan M., Lawlor R. T., Scarpa A., Heeschen C., Mata A., and Loessner D., Nat. Commun. 12(1), 5623 (2021). 10.1038/s41467-021-25921-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stock K., Estrada M. F., Vidic S., Gjerde K., Rudisch A., Santo V. E., Barbier M., Blom S., Arundkar S. C., Selvam I., Osswald A., Stein Y., Gruenewald S., Brito C., van Weerden W., Rotter V., Boghaert E., Oren M., Sommergruber W., Chong Y., de Hoogt R., and Graeser R., Sci. Rep. 6, 28951 (2016). 10.1038/srep28951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., Man K., Liu J., Liu Y., Chen Q., Zhou Y., and Yang Y., ACS Biomater. Sci. Eng. 6(6), 3231–3257 (2020). 10.1021/acsbiomaterials.9b01667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell S. B., Wu Q., Yazbeck J., Liu C., Okhovatian S., and Radisic M., ACS Biomater. Sci. Eng. 7(7), 2880–2899 (2021). 10.1021/acsbiomaterials.0c00640 [DOI] [PubMed] [Google Scholar]
- 26.Virumbrales-Muñoz M. and Ayuso J. M., Org. Chip 4, 100015 (2022). 10.1016/j.ooc.2022.100015 [DOI] [Google Scholar]
- 27.Sohn L. L., Schwille P., Hierlemann A., Tay S., Samitier J., Fu J., and Loskill P., Cell Syst. 11(3), 209–211 (2020). 10.1016/j.cels.2020.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin W. and Kim H. J., Proc. Natl. Acad. Sci. U.S.A. 115(45), E10539–E10547 (2018). 10.1073/pnas.1810819115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mastikhina O., Moon B.-U., Williams K., Hatkar R., Gustafson D., Mourad O., Sun X., Koo M., Lam A. Y. L., Sun Y., Fish J. E., Young E. W. K., and Nunes S. S., Biomaterials 233, 119741 (2020). 10.1016/j.biomaterials.2019.119741 [DOI] [PubMed] [Google Scholar]
- 30.Plebani R., Potla R., Soong M., Bai H., Izadifar Z., Jiang A., Travis R. N., Belgur C., Dinis A., Cartwright M. J., Prantil-Baun R., Jolly P., Gilpin S. E., Romano M., and Ingber D. E., J. Cyst. Fibros. 21(4), 606–615 (2022). 10.1016/j.jcf.2021.10.004 [DOI] [PubMed] [Google Scholar]
- 31.Hayward K. L., Kouthouridis S., and Zhang B., ACS Biomater. Sci. Eng. 7(7), 2900–2925 (2021). 10.1021/acsbiomaterials.0c01089 [DOI] [PubMed] [Google Scholar]
- 32.Asmani M., Velumani S., Li Y., Wawrzyniak N., Hsia I., Chen Z., Hinz B., and Zhao R., Nat. Commun. 9(1), 2066 (2018). 10.1038/s41467-018-04336-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baka Z., Stiefel M., Figarol A., Godier C., Mallick A., Joubert O., Ashammakhi N., Gaffet E., and Alem H., Prog. Biomed. Eng. 4(3), 32011 (2022). [Google Scholar]
- 34.Moon H. R., Ospina-Munoz N., Noe-Kim V., Yang Y., Elzey B. D., Konieczny S. F., and Han B., PLoS One 15(6), e0234012 (2020). 10.1371/journal.pone.0234012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagle D. A., Curr. Opin. Pharmacol. 48, 146–154 (2019). 10.1016/j.coph.2019.09.007 [DOI] [PubMed] [Google Scholar]
- 36.Moon H.-R. and Han B., Biomaterials for Cancer Therapeutics (Elsevier, 2020), pp. 423–443. [Google Scholar]
- 37.Chavez K. J., Garimella S. V., and Lipkowitz S., Breast Dis. 32, 35–48 (2011). 10.3233/BD-2010-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheang M. C. U., Voduc D., Bajdik C., Leung S., McKinney S., Chia S. K., Perou C. M., and Nielsen T. O., Clin. Cancer Res. 14, 1368–1376 (2008). 10.1158/1078-0432.CCR-07-1658 [DOI] [PubMed] [Google Scholar]
- 39.Livasy C. A., Karaca G., and Nanda R., Mod. Pathol. 19, 264–271 (2006). 10.1038/modpathol.3800528 [DOI] [PubMed] [Google Scholar]
- 40.Marra A., Trapani D., Viale G., Criscitiello C., and Curigliano G., NPJ Breast Cancer 6(1), 54 (2020). 10.1038/s41523-020-00197-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andre F. and Zielinski C. C., Ann. Oncol. 23, vi46–vi51 (2012). 10.1093/annonc/mds195 [DOI] [PubMed] [Google Scholar]
- 42.Hudis C. A. and Gianni L., Oncologist 16, 1–11 (2011). 10.1634/theoncologist.2011-S1-01 [DOI] [PubMed] [Google Scholar]
- 43.Barton S. and Turner N. C., in Handbook of Metastatic Breast Cancer, edited by Swanton C. and Johnston S. R. D. (Informa Healthcare, London, 2012), pp. 50–66. [Google Scholar]
- 44.Erkan M., Hausmann S., Michalski C. W., Fingerle A. A., Dobritz M., Kleeff J., and Friess H., Nat. Rev. Gastroenterol. Hepatol. 9, 454–467 (2012). 10.1038/nrgastro.2012.115 [DOI] [PubMed] [Google Scholar]
- 45.Mahadevan D. and Hoff D. D. V., Mol. Cancer Ther. 6(4), 1186–1197 (2007). 10.1158/1535-7163.MCT-06-0686 [DOI] [PubMed] [Google Scholar]
- 46.Drifka C. R., Tod J., Loeffler A. G., Liu Y., Thomas G. J., Eliceiri K. W., and Kao W. J., Mod. Pathol. 28(11), 1470–1480 (2015). 10.1038/modpathol.2015.97 [DOI] [PubMed] [Google Scholar]
- 47.Neesse A., Michl P., Frese K. K., Feig C., Cook N., Jacobetz M. A., Lolkema M. P., Buchholz M., Olive K. P., Gress T. M., and Tuveson D. A., Gut 60(6), 861–868 (2011). 10.1136/gut.2010.226092 [DOI] [PubMed] [Google Scholar]
- 48.Gore J. and Korc M., Cancer Cell 25(6), 711–712 (2014). 10.1016/j.ccr.2014.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie D. and Xie K., Genes Dis. 2(2), 133–143 (2015). 10.1016/j.gendis.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voskoglou-Nomikos T., Pater J. L., and Seymour L., Clin. Cancer Res. 9(11), 4227–4239 (2003). [PubMed] [Google Scholar]
- 51.Esch E. W., Bahinski A., and Huh D., Nat. Rev. Drug Discovery 14(4), 248–260 (2015). 10.1038/nrd4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X., Zhang X., Zhao S., Wang J., Liu G., and Du Y., Lab Chip 14(3), 471–481 (2014). 10.1039/C3LC51103K [DOI] [PubMed] [Google Scholar]
- 53.Liu W., Xu J., Li T., Zhao L., Ma C., Shen S., and Wang J., Anal. Chem. 87(19), 9752–9760 (2015). 10.1021/acs.analchem.5b01915 [DOI] [PubMed] [Google Scholar]
- 54.Irrechukwu O., Yeager R., David R., Ekert J., Saravanakumar A., and Choi C. K., ALTEX–Alternatives to Animal Experimentation 40(3), 485–518 (2023). 10.14573/altex.2204071 [DOI] [PubMed] [Google Scholar]
- 55.Kim H. N., Habbit N. L., Su C.-Y., Choi N., Ahn E. H., Lipke E. A., and Kim D.-H., Adv. Funct. Mater. 29(22), 1807553 (2019). 10.1002/adfm.201807553 [DOI] [Google Scholar]
- 56.Moon H.-R. and Han B., in Biomaterials for Cancer Therapeutics, 2nd ed., edited by Park K. (Woodhead Publishing, 2020), pp. 423–443. [Google Scholar]
- 57.Fong E. L., Harrington D. A., Farach-Carson M. C., and Yu H., Biomaterials 108, 197–213 (2016). 10.1016/j.biomaterials.2016.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Felsenstein M., Hruban R. H., and Wood L. D., Adv. Anatomic Pathol. 25(2), 131–142 (2018). 10.1097/PAP.0000000000000172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burrell R. A., McGranahan N., Bartek J., and Swanton C., Nature 501(7467), 338–345 (2013). 10.1038/nature12625 [DOI] [PubMed] [Google Scholar]
- 60.Fisher R., Pusztai L., and Swanton C., Br. J. Cancer 108(3), 479–485 (2013). 10.1038/bjc.2012.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samuel N. and Hudson T. J., Nat. Rev. Gastroenterol. Hepatol. 9(2), 77–87 (2012). 10.1038/nrgastro.2011.215 [DOI] [PubMed] [Google Scholar]
- 62.Patel A. P., Tirosh I., Trombetta J. J., Shalek A. K., Gillespie S. M., Wakimoto H., Cahill D. P., Nahed B. V., Curry W. T., Martuza R. L., Louis D. N., Rozenblatt-Rosen O., Suvà M. L., Regev A., and Bernstein B. E., Science 344(6190), 1396–1401 (2014). 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang S., Wan Z., and Kamm R. D., Lab Chip 21(3), 473–488 (2021). 10.1039/D0LC01186J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ko J., Ahn J., Kim S., Lee Y., Lee J., Park D., and Jeon N. L., Lab Chip 19(17), 2822–2833 (2019). 10.1039/C9LC00140A [DOI] [PubMed] [Google Scholar]
- 65.Paek J., Park S. E., Lu Q., Park K.-T., Cho M., Oh J. M., Kwon K. W., Yi Y.-s., Song J. W., Edelstein H. I., Ishibashi J., Yang W., Myerson J. W., Kiseleva R. Y., Aprelev P., Hood E. D., Stambolian D., Seale P., Muzykantov V. R., and Huh D., ACS Nano 13(7), 7627–7643 (2019). 10.1021/acsnano.9b00686 [DOI] [PubMed] [Google Scholar]
- 66.Lee S., Kim S., Koo D.-J., Yu J., Cho H., Lee H., Song J. M., Kim S.-Y., Min D.-H., and Jeon N. L., ACS Nano 15(1), 338–350 (2021). 10.1021/acsnano.0c05110 [DOI] [PubMed] [Google Scholar]
- 67.Hassell B. A., Goyal G., Lee E., Sontheimer-Phelps A., Levy O., Chen C. S., and Ingber D. E., Cell Rep. 21(2), 508–516 (2017). 10.1016/j.celrep.2017.09.043 [DOI] [PubMed] [Google Scholar]
- 68.González A. and Tapia J. A., J. Physiol. Biochem. 79 175–177 (2023). 10.1007/s13105-022-00942-0 [DOI] [PubMed] [Google Scholar]
- 69.Ni Y., Zhou X., Yang J., Shi H., Li H., Zhao X., and Ma X., Front. Cell Develop.Biol. 9, 637675 (2021). 10.3389/fcell.2021.637675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fiori M. E., Di Franco S., Villanova L., Bianca P., Stassi G., and De Maria R., Mol. Cancer 18(1), 1–16 (2019). 10.1186/s12943-019-0994-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei L., Ye H., Li G., Lu Y., Zhou Q., Zheng S., Lin Q., Liu Y., Li Z., and Chen R., Cell Death Dis. 9(11), 1065 (2018). 10.1038/s41419-018-1104-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McAndrews K. M., Chen Y., Darpolor J. K., Zheng X., Yang S., Carstens J. L., Li B., Wang H., Miyake T., Correa de Sampaio P., Kirtley M. L., Natale M., Wu C.-C., Sugimoto H., LeBleu V. S., and Kalluri R., Cancer Discov. 12(6), 1580–1597 (2022). 10.1158/2159-8290.CD-20-1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee J.-H., Kim S.-K., Khawar I. A., Jeong S.-Y., Chung S., and Kuh H.-J., J. Exp. Clin. Cancer Res. 37, 4 (2018). 10.1186/s13046-017-0654-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ying L., Zhu Z., Xu Z., He T., Li E., Guo Z., Liu F., Jiang C., and Wang Q., PLoS One 10(6), e0129593 (2015). 10.1371/journal.pone.0129593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gampala S., Shah F., Lu X., Moon H.-r., Babb O., Umesh Ganesh N., Sandusky G., Hulsey E., Armstrong L., Mosely A. L., Han B., Ivan M., Yeh J.-R. J., Kelley M. R., Zhang C., and Fishel M. L., J. Exp. Clin. Cancer Res. 40(1), 251 (2021). 10.1186/s13046-021-02046-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ngan Ngo T. K., Kuo C.-H., and Tu T.-Y., Biomicrofluidics 17(1), 011501 (2023). 10.1063/5.0108792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D. J., Anandh S., Null J. L., Przanowski P., Bhatnagar S., Kumar P., Shelton S. E., Grundy E. E., Chiappinelli K. B., Kamm R. D., Barbie D. A., and Dudley A. C., Nat. Commun. 14(1), 2122 (2023). 10.1038/s41467-023-37807-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mollica H., Teo Y. J., Tan A. S. M., Tan D. Z. M., Decuzzi P., Pavesi A., and Adriani G., Biomater. Sci. 9(22), 7420–7431 (2021). 10.1039/D1BM00210D [DOI] [PubMed] [Google Scholar]
- 79.Ayuso J. M., Truttschel R., Gong M. M., Humayun M., Virumbrales-Munoz M., Vitek R., Felder M., Gillies S. D., Sondel P., Wisinski K. B., Patankar M., Beebe D. J., and Skala M. C., Oncoimmunology 8(3), 1553477 (2019). 10.1080/2162402X.2018.1553477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen O. T. P., Misun P. M., Lohasz C., Lee J., Wang W., Schroeder T., and Hierlemann A., Front. Immunol. 12, 781337 (2021). 10.3389/fimmu.2021.781337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Mello C. P. P., Rumsey J., Slaughter V., and Hickman J. J., Drug Discovery Today 24(11), 2139–2151 (2019). 10.1016/j.drudis.2019.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roth A. and Berlin M.-W., Science 373(6561), 1304–1306 (2021). 10.1126/science.abc3734 [DOI] [PubMed] [Google Scholar]
- 83.Chou D. B., Frismantas V., Milton Y., David R., Pop-Damkov P., Ferguson D., MacDonald A., Vargel Bölükbaşı Ö., Joyce C. E., Moreira Teixeira L. S., Rech A., Jiang A., Calamari E., Jalili-Firoozinezhad S., Furlong B. A., O’Sullivan L. R., Ng C. F., Choe Y., Marquez S., Myers K. C., Weinberg O. K., Hasserjian R. P., Novak R., Levy O., Prantil-Baun R., Novina C. D., Shimamura A., Ewart L., and Ingber D. E., Nat. Biomed. Eng. 4(4), 394–406 (2020). 10.1038/s41551-019-0495-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ingber D. E., Nat. Rev. Genet. 23(8), 467–491 (2022). 10.1038/s41576-022-00466-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morad G., Carman C. V., Hagedorn E. J., Perlin J. R., Zon L. I., Mustafaoglu N., Park T.-E., Ingber D. E., Daisy C. C., and Moses M. A., ACS Nano 13(12), 13853–13865 (2019). 10.1021/acsnano.9b04397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Linville R. M., DeStefano J. G., Sklar M. B., Xu Z., Farrell A. M., Bogorad M. I., Chu C., Walczak P., Cheng L., Mahairaki V., Whartenby K. A., Calabresi P. A., and Searson P. C., Biomaterials 190–191, 24–37 (2019). 10.1016/j.biomaterials.2018.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vatine G. D., Barrile R., Workman M. J., Sances S., Barriga B. K., Rahnama M., Barthakur S., Kasendra M., Lucchesi C., Kerns J., Wen N., Spivia W. R., Chen Z., Van Eyk J., and Svendsen C. N., Cell Stem Cell 24(6), 995–1005.e6 (2019). 10.1016/j.stem.2019.05.011 [DOI] [PubMed] [Google Scholar]
- 88.Kostewicz E. S., Aarons L., Bergstrand M., Bolger M. B., Galetin A., Hatley O., Jamei M., Lloyd R., Pepin X., Rostami-Hodjegan A., Sjögren E., Tannergren C., Turner D. B., Wagner C., Weitschies W., and Dressman J., Eur. J. Pharm. Sci. 57, 300–321 (2014). 10.1016/j.ejps.2013.09.008 [DOI] [PubMed] [Google Scholar]
- 89.Isoherranen N., Madabushi R., and Huang S.-M., Clin. Transl. Sci. 12(2), 113–121 (2019). 10.1111/cts.12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Herland A., Maoz B. M., Das D., Somayaji M. R., Prantil-Baun R., Novak R., Cronce M., Huffstater T., Jeanty S. S. F., Ingram M., Chalkiadaki A., Benson Chou D., Marquez S., Delahanty A., Jalili-Firoozinezhad S., Milton Y., Sontheimer-Phelps A., Swenor B., Levy O., Parker K. K., Przekwas A., and Ingber D. E., Nat. Biomed. Eng. 4(4), 421–436 (2020). 10.1038/s41551-019-0498-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maass C., Stokes C. L., Griffith L. G., and Cirit M., Integr. Biol. 9(4), 290–302 (2017). 10.1039/C6IB00243A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Korner J. L., Stephenson E. B., and Elvira K. S., Lab Chip 20(11), 1898–1906 (2020). 10.1039/D0LC00263A [DOI] [PubMed] [Google Scholar]
- 93.Yang Y., Chen Y., Wang L., Xu S., Fang G., Guo X., Chen Z., and Gu Z., Front. Bioeng. Biotechnol. 10, 900481 (2022). 10.3389/fbioe.2022.900481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caetano-Pinto P., Nordell P., Nieskens T., Haughan K., Fenner K. S., and Stahl S. H., ALTEX—Alternatives to Animal Experimentation 40(3), 408–424 (2023). 10.14573/altex.2204011 [DOI] [PubMed] [Google Scholar]
- 95.Picollet-D’hahan N., Zuchowska A., Lemeunier I., and Le Gac S., Trends Biotechnol. 39(8), 788–810 (2021). 10.1016/j.tibtech.2020.11.014 [DOI] [PubMed] [Google Scholar]
- 96.Ronaldson-Bouchard K., Teles D., Yeager K., Tavakol D. N., Zhao Y., Chramiec A., Tagore S., Summers M., Stylianos S., Tamargo M., Lee B. M., Halligan S. P., Abaci E. H., Guo Z., Jacków J., Pappalardo A., Shih J., Soni R. K., Sonar S., German C., Christiano A. M., Califano A., Hirschi K. K., Chen C. S., Przekwas A., and Vunjak-Novakovic G., Nat. Biomed. Eng. 6(4), 351–371 (2022). 10.1038/s41551-022-00882-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shinha K., Nihei W., Ono T., Nakazato R., and Kimura H., Biomicrofluidics 14(4), 044108 (2020). 10.1063/5.0011545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petreus T., Cadogan E., Hughes G., Smith A., Pilla Reddy V., Lau A., O’Connor M. J., Critchlow S., Ashford M., and Oplustil O’Connor L., Commun. Biol. 4(1), 1001 (2021). 10.1038/s42003-021-02526-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Komen J., Westerbeek E. Y., Kolkman R. W., Roesthuis J., Lievens C., Van Den Berg A., and Van Der Meer A. D., Lab Chip 20(17), 3167–3178 (2020). 10.1039/D0LC00419G [DOI] [PubMed] [Google Scholar]
- 100.Guerrero Y. A., Desai D., Sullivan C., Kindt E., Spilker M. E., Maurer T. S., Solomon D. E., and Bartlett D. W., AAPS J. 22, 1–10 (2020). 10.1208/s12248-020-0430-y [DOI] [PubMed] [Google Scholar]
- 101.Ozcelikkale A., Shin K., Noe-Kim V., Elzey B. D., Dong Z., Zhang J.-T., Kim K., Kwon I. C., Park K., and Han B., J. Controlled Release 266, 129–139 (2017). 10.1016/j.jconrel.2017.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shin K., Klosterhoff B. S., and Han B., Mol. Pharm. 13(7), 2214–2223 (2016). 10.1021/acs.molpharmaceut.6b00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shin Y., Choi S. H., Kim E., Bylykbashi E., Kim J. A., Chung S., Kim D. Y., Kamm R. D., and Tanzi R. E., Adv. Sci. 6(20), 1900962 (2019). 10.1002/advs.201900962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park J., Wetzel I., Marriott I., Dréau D., D'Avanzo C., Kim D. Y., Tanzi R. E., and Cho H., Nat. Neurosci. 21(7), 941–951 (2018). 10.1038/s41593-018-0175-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pediaditakis I., Kodella K. R., Manatakis D. V., Le C. Y., Hinojosa C. D., Tien-Street W., Manolakos E. S., Vekrellis K., Hamilton G. A., Ewart L., Rubin L. L., and Karalis K., Nat. Commun. 12(1), 5907 (2021). 10.1038/s41467-021-26066-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bolognin S., Fossépré M., Qing X., Jarazo J., Ščančar J., Moreno E. L., Nickels S. L., Wasner K., Ouzren N., Walter J., Grünewald A., Glaab E., Salamanca L., Fleming R. M. T., Antony P. M. A., and Schwamborn J. C., Adv. Sci. 6(1), 1800927 (2019). 10.1002/advs.201800927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lyu Z., Park J., Kim K.-M., Jin H.-J., Wu H., Rajadas J., Kim D.-H., Steinberg G. K., and Lee W., Nat. Biomed. Eng. 5(8), 847–863 (2021). 10.1038/s41551-021-00744-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu J., Sternberg A. R., Ghiasvand S., and Berdichevsky Y., IEEE Trans. Biomed. Eng. 66(5), 1231–1241 (2019). 10.1109/TBME.2018.2871415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim J., Lee K.-T., Lee J. S., Shin J., Cui B., Yang K., Choi Y. S., Choi N., Lee S. H., Lee J.-H., Bahn Y.-S., and Cho S.-W., Nat. Biomed. Eng. 5(8), 830–846 (2021). 10.1038/s41551-021-00743-8 [DOI] [PubMed] [Google Scholar]
- 110.Straehla J. P., Hajal C., Safford H. C., Offeddu G. S., Boehnke N., Dacoba T. G., Wyckoff J., Kamm R. D., and Hammond P. T., Proc. Natl. Acad. Sci. U.S.A. 119(23), e2118697119 (2022). 10.1073/pnas.2118697119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yi H.-G., Jeong Y. H., Kim Y., Choi Y.-J., Moon H. E., Park S. H., Kang K. S., Bae M., Jang J., Youn H., Paek S. H., and Cho D.-W., Nat. Biomed. Eng. 3(7), 509–519 (2019). 10.1038/s41551-019-0363-x [DOI] [PubMed] [Google Scholar]
- 112.Seo S., Nah S. Y., Lee K., Choi N., and Kim H., Adv. Funct. Mater. 32(10), 2106860 (2022). 10.1002/adfm.202106860 [DOI] [Google Scholar]
- 113.Fan Y., Nguyen D. T., Akay Y., Xu F., and Akay M., Sci. Rep. 6, 25062 (2016). 10.1038/srep25062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Machado C. B., Pluchon P., Harley P., Rigby M., Gonzalez Sabater V., Stevenson D. C., Hynes S., Lowe A., Burrone J., Viasnoff V., and Lieberam I., Adv. Biosyst. 3(7), 1800307 (2019). 10.1002/adbi.201800307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Osaki T., Uzel S. G. M., and Kamm R. D., Sci. Adv. 4(10), eaat5847 (2018). 10.1126/sciadv.aat5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Park T.-E., Mustafaoglu N., Herland A., Hasselkus R., Mannix R., FitzGerald E. A., Prantil-Baun R., Watters A., Henry O., Benz M., Sanchez H., McCrea H. J., Goumnerova L. C., Song H. W., Palecek S. P., Shusta E., and Ingber D. E., Nat. Commun. 10(1), 2621 (2019). 10.1038/s41467-019-10588-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vulto P. and Joore J., Nat. Rev. Drug Discovery 20(12), 961–962 (2021). 10.1038/s41573-021-00323-0 [DOI] [PubMed] [Google Scholar]
- 118.Ribeiro A. J. S., Yang X., Patel V., Madabushi R., and Strauss D. G., Clin. Pharmacol. Therap. 106(1), 139–147 (2019). 10.1002/cpt.1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vernetti L. A., Senutovitch N., Boltz R., DeBiasio R., Ying Shun T., Gough A., and Taylor D. L., Exp. Biol. Med. 241(1), 101–114 (2016). 10.1177/1535370215592121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jang K.-J., Otieno M. A., Ronxhi J., Lim H.-K., Ewart L., Kodella K. R., Petropolis D. B., Kulkarni G., Rubins J. E., Conegliano D., Nawroth J., Simic D., Lam W., Singer M., Barale E., Singh B., Sonee M., Streeter A. J., Manthey C., Jones B., Srivastava A., Andersson L. C., Williams D., Park H., Barrile R., Sliz J., Herland A., Haney S., Karalis K., Ingber D. E., and Hamilton G. A., Sci. Transl. Med. 11(517), eaax5516 (2019). 10.1126/scitranslmed.aax5516 [DOI] [PubMed] [Google Scholar]
- 121.Bavli D., Prill S., Ezra E., Levy G., Cohen M., Vinken M., Vanfleteren J., Jaeger M., and Nahmias Y., Proc. Natl. Acad. Sci. U.S.A. 113(16), E2231–E2240 (2016). 10.1073/pnas.1522556113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deng J., Zhang X., Chen Z., Luo Y., Lu Y., Liu T., Wu Z., Jin Y., Zhao W., and Lin B., Biomicrofluidics 13(2), 024101 (2019). 10.1063/1.5070088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jang K.-J., Mehr A. P., Hamilton G. A., McPartlin L. A., Chung S., Suh K.-Y., and Ingber D. E., Integr. Biol. 5(9), 1119–1129 (2013). 10.1039/c3ib40049b [DOI] [PubMed] [Google Scholar]
- 124.Nieskens T. T. G., Magnusson O., Andersson P., Söderberg M., Persson M., and Sjögren A.-K., Arch. Toxicol. 95(6), 2123–2136 (2021). 10.1007/s00204-021-03062-8 [DOI] [PubMed] [Google Scholar]
- 125.Zhang Q., Sito L., Mao M., He J., Zhang Y. S., and Zhao X., Microphysiol. Syst. 2, 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Risueño I., Valencia L., Jorcano J. L., and Velasco D., APL Bioeng. 5(3), 030901 (2021). 10.1063/5.0046376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Monteduro A. G., Rizzato S., Caragnano G., Trapani A., Giannelli G., and Maruccio G., Biosens. Bioelectron. 231, 115271 (2023). 10.1016/j.bios.2023.115271 [DOI] [PubMed] [Google Scholar]
- 128.Kwak B. S., Jin S.-P., Kim S. J., Kim E. J., Chung J. H., and Sung J. H., Biotechnol. Bioeng. 117(6), 1853–1863 (2020). 10.1002/bit.27320 [DOI] [PubMed] [Google Scholar]
- 129.Ramadan Q. and Ting F. C. W., Lab Chip 16(10), 1899–1908 (2016). 10.1039/C6LC00229C [DOI] [PubMed] [Google Scholar]
- 130.Marx U., Akabane T., Andersson T. B., Baker E., Beilmann M., Beken S., Brendler-Schwaab S., Cirit M., David R., Dehne E.-M., Durieux I., Ewart L., Fitzpatrick S. C., Frey O., Fuchs F., Griffith L. G., Hamilton G. A., Hartung T., Hoeng J., Hogberg H., Hughes D. J., Ingber D. E., Iskandar A., Kanamori T., Kojima H., Kuehnl J., Leist M., Li B., Loskill P., Mendrick D. L., Neumann T., Pallocca G., Rusyn I., Smirnova L., Steger-Hartmann T., Tagle D. A., Tonevitsky A., Tsyb S., Trapecar M., van de Water B., van den Eijnden-van Raaij J., Vulto P., Watanabe K., Wolf A., Zhou X., and Roth A., Altex 37(3), 365 (2020). 10.14573/altex.2001241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tomlinson L., Ramsden D., Leite S. B., Beken S., Bonzo J. A., Brown P., Candarlioglu P. L., Chan T. S., Chen E., Choi C. K., David R., Delrue N., Devine P. J., Ford K., Garcia M. I., Gosset J. R., Hewitt P., Homan K., Irrechukwu O., Kopec A. K., Liras J. L., Mandlekar S., Raczynski A., Sadrieh N., Sakatis M. Z., Siegel J., Sung K., Sunyovszki I., Van Vleet T. R., Ekert J. E., and Hardwick R. N., “Considerations from an International Regulatory and Pharmaceutical Industry (IQ MPS Affiliate) Workshop on the standardization of complex in vitro models in drug development,” Adv. Biol. (published online 2023). 10.1002/adbi.202300131 [DOI] [PubMed] [Google Scholar]
- 132.Rubiano A., Indapurkar A., Yokosawa R., Miedzik A., Rosenzweig B., Arefin A., Moulin C. M., Dame K., Hartman N., Volpe D. A., Matta M. K., Hughes D. J., Strauss D. G., Kostrzewski T., and Ribeiro A. J. S., Clin. Transl. Sci. 14(3), 1049–1061 (2021). 10.1111/cts.12969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Choi S. R., Yang Y., Huang K.-Y., Kong H. J., Flick M. J., and Han B., Mater. Today Adv. 8, 100117 (2020). 10.1016/j.mtadv.2020.100117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Miller C. P., Shin W., Ahn E. H., Kim H. J., and Kim D.-H., Trends Biotechnol. 38(8), 857–872 (2020). 10.1016/j.tibtech.2020.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sasserath T., Rumsey J. W., McAleer C. W., Bridges L. R., Long C. J., Elbrecht D., Schuler F., Roth A., Bertinetti-LaPatki C., and Shuler M. L., Adv. Sci. 7(13), 2000323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang W.-Q., Liu L., Xu H.-X., Wu C.-T., Xiang J.-F., Xu J., Liu C., Long J., Ni Q.-X., and Yu X.-J., Br. J. Surgery 103(9), 1189–1199 (2016). 10.1002/bjs.10187 [DOI] [PubMed] [Google Scholar]
- 137.Karamitopoulou E., Br. J. Cancer 121(1), 5–14 (2019). 10.1038/s41416-019-0479-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created in this study.