Abstract
Extracellular nanocarriers (extracellular vesicles (EVs), lipoproteins, and ribonucleoproteins) of protein and nucleic acids mediate intercellular communication and are clinically adaptable as distinct circulating biomarkers. However, the overlapping size and density of the nanocarriers have so far prevented their efficient physical fractionation, thus impeding independent downstream molecular assays. Here, we report a bias-free high-throughput and high-yield continuous isoelectric fractionation nanocarrier fractionation technique based on their distinct isoelectric points. This nanocarrier fractionation platform is enabled by a robust and tunable linear pH profile provided by water-splitting at a bipolar membrane and stabilized by flow without ampholytes. The linear pH profile that allows easy tuning is a result of rapid equilibration of the water dissociation reaction and stabilization by flow. The platform is automated with a machine learning procedure to allow recalibration for different physiological fluids and nanocarriers. The optimized technique has a resolution of 0.3 ΔpI, sufficient to separate all nanocarriers and even subclasses of nanocarriers. Its performance is then evaluated with several biofluids, including plasma, urine, and saliva samples. Comprehensive, high-purity (plasma: >93%, urine: >95% and saliva: >97%), high-yield (plasma: >78%, urine: >87% and saliva: >96%), and probe-free isolation of ribonucleoproteins in 0.75 mL samples of various biofluids in 30 min is demonstrated, significantly outperforming affinity-based and highly biased gold standards having low yield and day-long protocols. Binary fractionation of EVs and different lipoproteins is also achieved with similar performance.
Keywords: ribonucleoproteins, exRNA nanocarriers, fractionation, isoelectric point, lipoproteins, pH, plasma
Graphical Abstract
Extracellular RNAs (exRNAs) that are secreted in physiological fluids (e.g., blood, urine, lymph fluids) by host cells encode complex cellular communication signatures and thus serve as promising biomarkers for various disease states, even if this has been best investigated in the context of cancers.1–4 ExRNAs are encased and protected by three general classes of nanoscale molecular shuttles: extracellular vesicle (EVs),5 lipoprotein (LLPs),6,7 and ribonucleic protein (RNPs),8,9 many with several subtypes (e.g., small and large EVs or HDL, LDL, VLDL).10,11 It is now understood that these nanocarriers encompass specific functional molecules including metabolites, genetic materials, proteins and act as vehicles by transferring these biomolecules from the donor cells from which they are derived to recipient cells.12,13 Nanocarrier intercellular signaling is hence fundamental to the cellular basis of disease progression.14,15 Studies thus far have suggested that the cargo of different nanocarriers is distinct and functionally diverse, making it difficult to resolve their itineraries upon release.16 For example, it is recently realized that the majority of circulating RNAs in the blood are in RNP complexes with RNA-binding proteins, many of them overlap with those in vesicular EVs and LLPs.17,18 It is likely that these RNPs carry waste RNAs from the cells and should hence be removed if EV or LLP RNAs are of interest.19–22 On the other hand, the >1900 RNPs themselves, particularly RNPs with specific pairings of proteins and RNAs, have become promising biomarkers for cancer and other stress-granule related diseases.23–29 However, the current RNP isolation technologies have extremely low yield. The most common cross-linking and immunoprecipitation (CLIP) and affinity capture technologies often offer very low yield,30,31 thus rendering the quantification of RNAs impractical.
The inherent challenge of physically separating and isolating these nanocarriers from a biofluid stems from their overlapping size and mass distributions, particularly for small nanocarriers like RNP. Currently, the most widely used for nanocarrier separation techniques (LLPs and EVs) are multistage ultracentrifugation (UC)32 and nanoporous membrane-based ultrafiltration (UF).33 However, these conventional technologies are time and labor intensive and can be plagued by low yields, poor isolation purity, and clogging (e.g., UF),34 thus manifesting in inaccurate downstream analysis. Asymmetric-flow field-flow fractionation (AF4) rapidly separates extracellular nanoparticles based on their hydrodynamic size and has a large dynamic range, but similar to UC, heterogeneous nanoparticle populations with overlapping sizes cannot be isolated from each other and require an additional electric field-based separation step in conjunction with AF4 to achieve a better isolation purity.35,36 Other physical fractionation technologies based on deterministic lateral displacement,37 acoustofluidics,38 dielectrophoresis39 and size-exclusion chromatography,40 although promising, have only been successful thus far in isolating larger nanocarriers like EVs and often with low purity.
In this work, we report the highest throughput (12 mL/h, about 1000 times higher than previous reports), continuous isoelectric fractionation (CIF) platform for bias-free isolation of EVs, lipoproteins (HDL, LDL), and RNPs from biofluids based on their distinct isoelectric points (pIs). Unlike the previous microfluidics free flow isoelectric focusing efforts, there is no external feed of acidic/basic solutions and internal distributors or ampholytes to sustain the pH gradient in our design.41,42 Instead, an on-chip stable linear pH gradient is produced and maintained by a pair of bipolar ion-exchange membranes (IEMs), whose high fields lead to water splitting by the Wien effect and the production of high concentrations of H3O+ and OH− that are spatially segregated by the transverse field. The low and high pH gradients are then extracted and spatially extended in a trapezoidal separation chip to produce a higher resolution gradient. This robust scalability effectively allows us to fractionate multiple nanocarriers with a large dynamic range of pIs (minimum ΔpI of 0.3), and the modular design enables parallel and sequential (high dimension) separations. No prior designs have the resolution to separate nanocarriers, even at their thousand-fold lower throughputs. Furthermore, a machine learning-based approach allows rapid selection of the optimal pH gradients for different physiological fluids and different nanocarriers, in the presence of contamination and equipment noise. We optimize the CIF technology by fractionating various combinations of binary mixtures of exRNA nanocarriers spiked in buffer (yield >80% and purity >90%) and demonstrate its performance with 20× diluted human plasma, 5× diluted urine and 5× diluted saliva. In particular, we demonstrate the utility of the platform for small nanocarriers by isolating RNPs from EVs and LLPs with high-purity (plasma: >93%, urine: >95% and saliva: >97%) and high-yield (plasma: >78%, urine: >87% and saliva: >96%) using only 0.75 mL of several biofluids in 30 min. These performances represent significant improvements over the current gold standard (<1% yield with a day-long protocol).
RESULTS AND DISCUSSION
Design of Integrated CIF Microfluidic Platform.
The CIF device consists of two microfluidic chips: (i) an upstream pH gradient chip for the generation of a coarse pH gradient (pH 3 to 11) from water (see inset of Figure 1a). A portion of the effluent from the pH gradient chip is then injected downstream into (ii) a separation chip to produce a high-resolution pH gradient to enable the isoelectric focusing of different nanocarriers (Figure 1a,c). The only calibration needed is to determine the portion of the effluent of the first chip that should be extracted to separate specific nanocarriers in the downstream separation chip.
Figure 1.
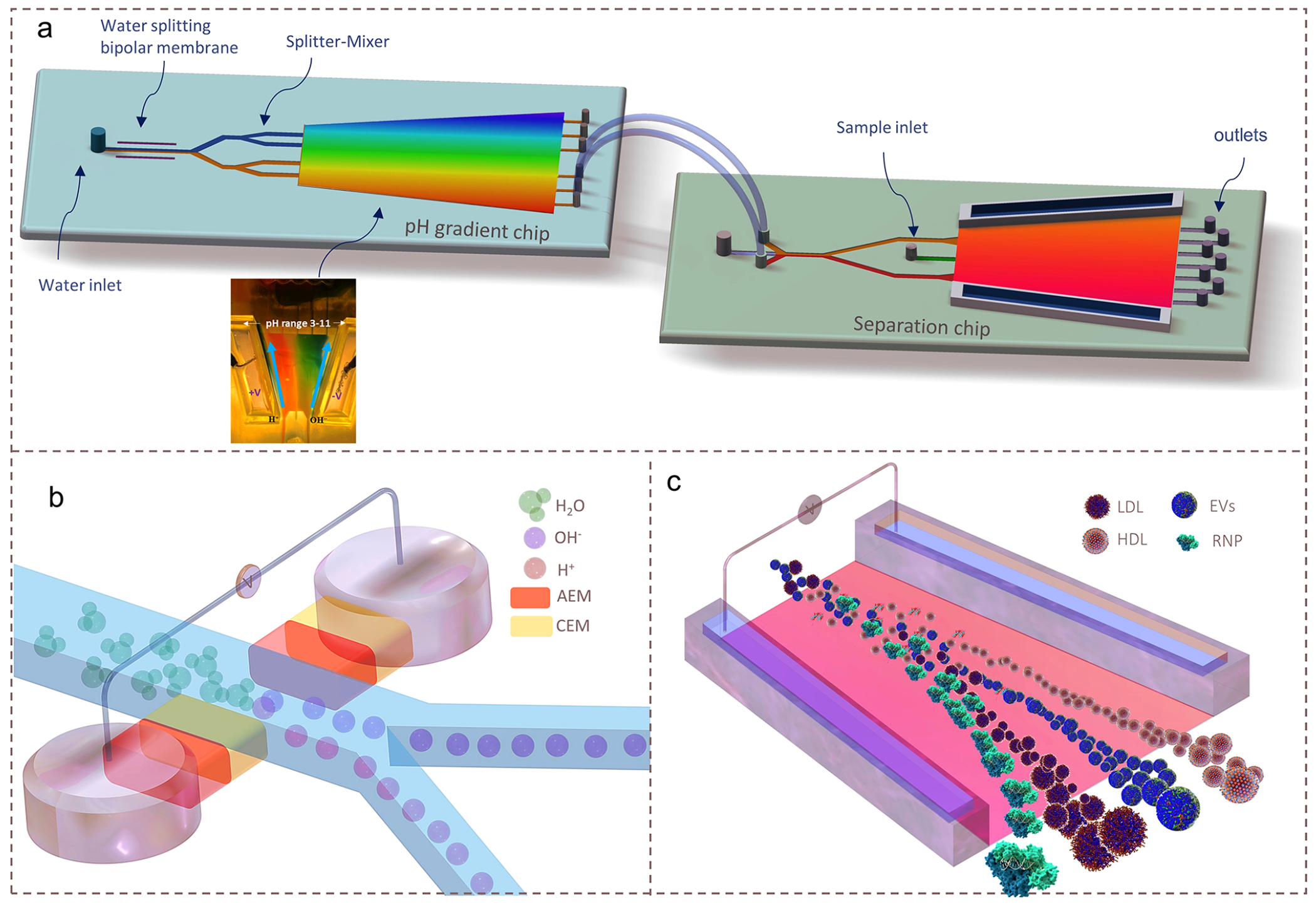
Design and operation of the CIF microfluidic device. (a) Schematics of the integrated CIF microfluidic platform consisting of a pH gradient chip and a separation chip connected by transfer tubes. Inset shows a photograph of an experimentally generated pH profile in the pH gradient chip. (b) Illustration of the water-splitting module of the pH gradient chip which utilizes two bipolar membranes (AEM-CEM sandwiched together; AEM: anion-exchange membrane and CEM: cation-exchange membrane) to dissociate water into H3O+ and OH− ions and subsequently transport them through splitter-mixer microchannels to obtain a stable and high-resolution pH gradient. (c) Illustration of the fractionation of RNP, HDL, LDL, and EVs based on their distinct pIs in the high-resolution pH gradient in the separation chip.
The pH gradient chip incorporates a pair of bipolar membranes (BiM), which split water into H3O+ and OH− ions under optimized reverse bias voltage43 (Supplementary Note 1). These ions are electrophoretically driven through their respective membranes into the main channel where they are then partitioned (Figure 1b) via a splitter-mixer microfluidics design44 to a trapezoidal channel. The H3O+ and OH− ions at different lateral positions of the pH gradient chip are further transported to a trapezoidal separation chip (or multiple separation chips), where a high-resolution pH gradient is formed (e.g., pH = 3–6 for HDL and LDL, pH = 2–8 for HDL and RNP, pH = 4–5.5 for EVs) (Figure 1c). The geometric design of the separation trapezoidal channel together with optimized flow rates facilitate a high-resolution pH gradient with a high separation efficiency.
Stretchable Linear pH Profile by a Transverse Field.
The robustness of the scalable linear pH profile in both the trapezoidal chambers of the pH gradient generation and separation chips is a key to our design. It removes the need for an immobilized ampholyte to maintain a stable pH gradient in a gel or matrix that does not permit high throughput. We offer a simple mechanistic explanation based on the relative distributions of the H3+ and OH− ions as well as the anions and cations in the buffer solution, which leads to this surprising but desirable feature (Supplementary Note 2). As their generation modules are on opposite sides of the channel, the first set of reactive ions are segregated at two ends of the flow channel upon entry. This stipulates that their charge-compensating ions are inert buffer ions and not each other. The buffer ions will redistribute from the middle of the channel, in the presence of the transverse field, to ensure electroneutrality. Once established, the zero-flux Boltzmann distributions (with equal but opposite electromigration and diffusive flux) of the inert buffer ions are dominant, with H3O+ and OH− equilibrating rapidly via the rapid and reversible water dissociation reaction whose water dissociation kinetics has been favored by the deficiency of either H3O+ or OH−. By introducing two streams of different pH into the separation channel, the field at the separation chip maintains the universal buffer ion Boltzmann distributions and segregation of the reactive H3O+ and OH− ions, but with a different average pH and buffer concentration. This universal feature allows extraction of any linear pH profile and stretching its spatial resolution by an arbitrary factor downstream.
We benchmarked this design concept against finite element method (FEM) simulations.45 Figure 2a shows the concentration profiles of the two dominant ions (labeled A− and B+, respectively) in Boltzmann equilibrium, while the hydroxide and hydronium ions are in negative Boltzmann equilibrium as predicted by the theory. Also, Figure 2b shows that pH profile becomes linear as we move downstream in x confirming the theoretical predictions. Scale invariant pH stretching suggests that instead of the commonly used rectangular microchannels, a trapezoidal geometry can reduce the pH gradient and provide a higher resolution pH profile (Supplementary Note 3). We confirm this experimentally, where the pH profile calculated numerically agrees well with the experimental measurements when normalized by the angle θ and length l of the trapezoidal arm (Supplementary Note 3), as shown in Figure 2e. Furthermore, Figure 2c shows that in a trapezoidal geometry, after an inlet transition region, the pH profile fans out radially parallel to the trapezoidal boundary. Figure 2d shows a section of the pH profile (pH: 3–5) from Figure 2c when fed to a trapezoidal chamber merely stretches out downstream with same pH range and is experimentally confirmed further in Figure 2f. The experimentally measured pH profiles for both the pH gradient and separation chips are in good agreement with the numerically computed ones shown in Figure 2c,d, where a negative Boltzmann distribution offsets the natural Boltzmann distribution of the hydronium ions. Note that because standard pH dye makes it difficult to visually discern narrow pH gradient (Figure 2f inset) later in this article, we developed an automated process using machine learning to accelerate and optimize the repeatable transfer of narrow pH gradients from generation to the separation chip. One other important point we want to highlight is the relative insensitivity of the pH profile on the flow rate if the ratio of the fluid to electrophoretic velocity is small and has been discussed in detail in Supplementary Note 4 and Supplementary Figure 24.
Figure 2.
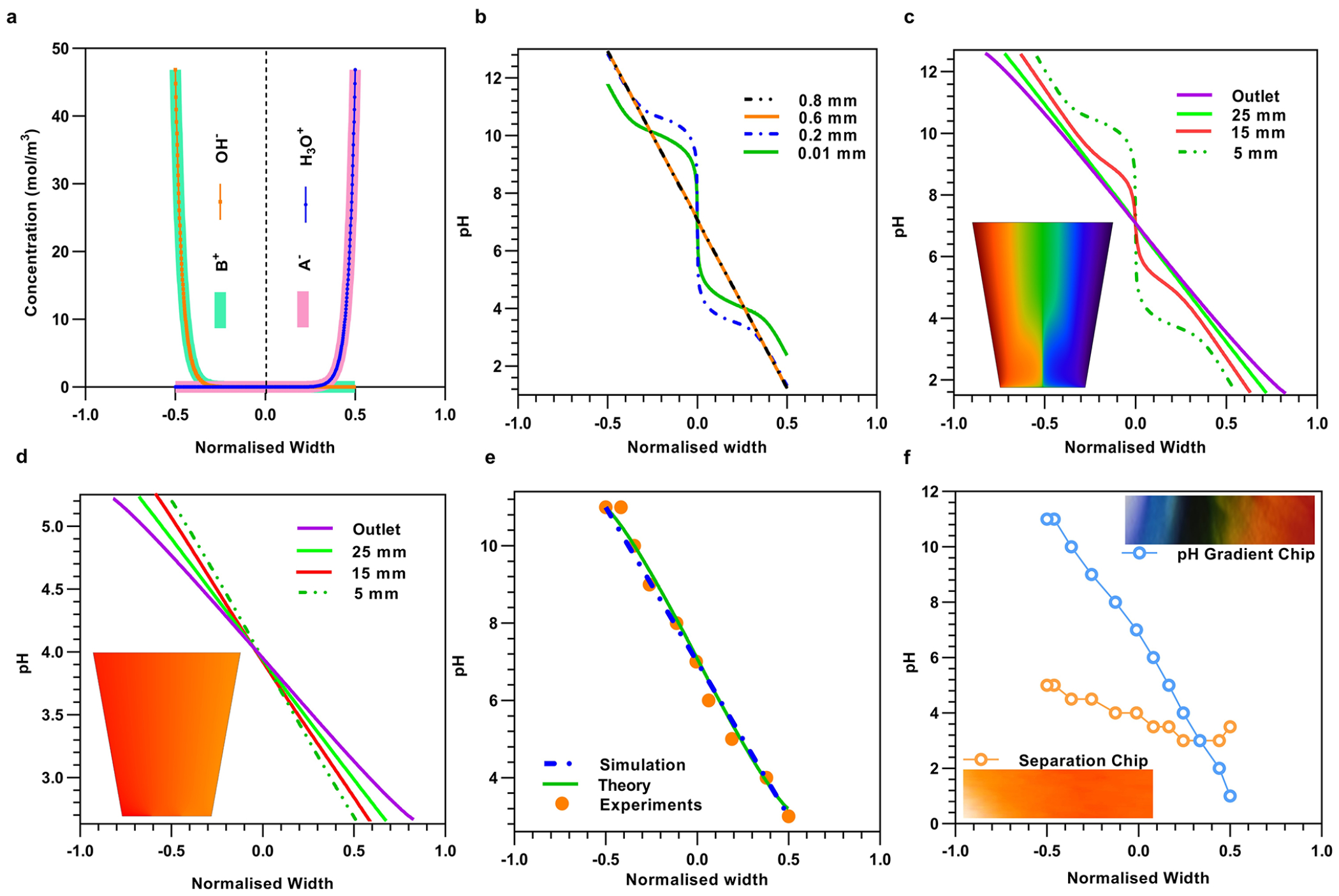
Theoretical, FEM simulations and experimental results of pH gradient generation and separation chip. (a) Simulated concentration profiles of monovalent buffer cation (A−) and anion (B+) in Boltzmann equilibrium and H3O+ and OH− in negative Boltzmann equilibrium along the normalized width of a straight channel. (b) Plot of the simulated pH profile at different downstream locations until stabilizing into a linear pH profile (Supplementary Figure 2c). (c) Inset: pH gradient surface plot obtained by finite element simulation (FEM) of the pH gradient chip. Line plots of the pH profiles at various downstream locations in the trapezoidal geometry, depicting stretching of linear pH profile by 2l × tan(θ)/w as compared to the straight channel (Supplementary Note 3, Supplementary Figure 2a,b). (d) Inset: pH gradient surface plot of the separation chip obtained by FEM with the same geometry parameters as the experimental device. Line plots of the pH profiles at various downstream locations of the trapezoidal geometry of the separation chip depict a high-resolution pH gradient. (e) Collapsed data of the pH profile at the normalized outlet location were obtained experimentally (Supplementary Figure 2d), theoretically, and by FEM numerical simulations of a trapezoidal geometry. (f) Top right inset: An experimental image snapshot of the trapezoidal chamber in the pH gradient chip whose colors are produced by pH indicator dye. The extracted pH profile (blue color) with respect to the normalized width of the trapezoidal chamber. Bottom left inset: Experimental image snapshot of the high-resolution pH gradient obtained in the separation chip by transferring a pH range of 3–5 from the pH gradient generation chip. The extracted pH profile in the separation chip (orange color) is shown with the normalized width of the trapezoidal chamber.
Effluent Selection for Separating LDL and HDL Mixture Spiked in Buffer.
HDL (pI = 4.62) and LDL (pI = 5.01) cholesterol levels are important in determining the risk of coronary artery disease.46–48 They are also known to carry functional molecules that are biomarker candidates.49 Yet, a comprehensive understanding of their functional cargo is currently limited, as the separation of these biomarkers from plasma at high throughput is challenging owing to very small differences in size and charge.50–52 Initial studies with individual nanocarrier show both HDL and LDL respond to a pH gradient and moved to their respective isoelectric points (Supplementary Figures S4–S6 and Note 5). Figure 3a shows the measured ζ potential of HDL and LDL to be −16.56 ± 3.73 mV and −12.44 ± 1.85 mV, respectively, suggesting that their isoelectric fractionation will be challenging. Hence, to achieve their fractionation, a high-resolution gradient is created by transferring the pH = 3–6 band from the pH gradient chip to the separation chip.
Figure 3.
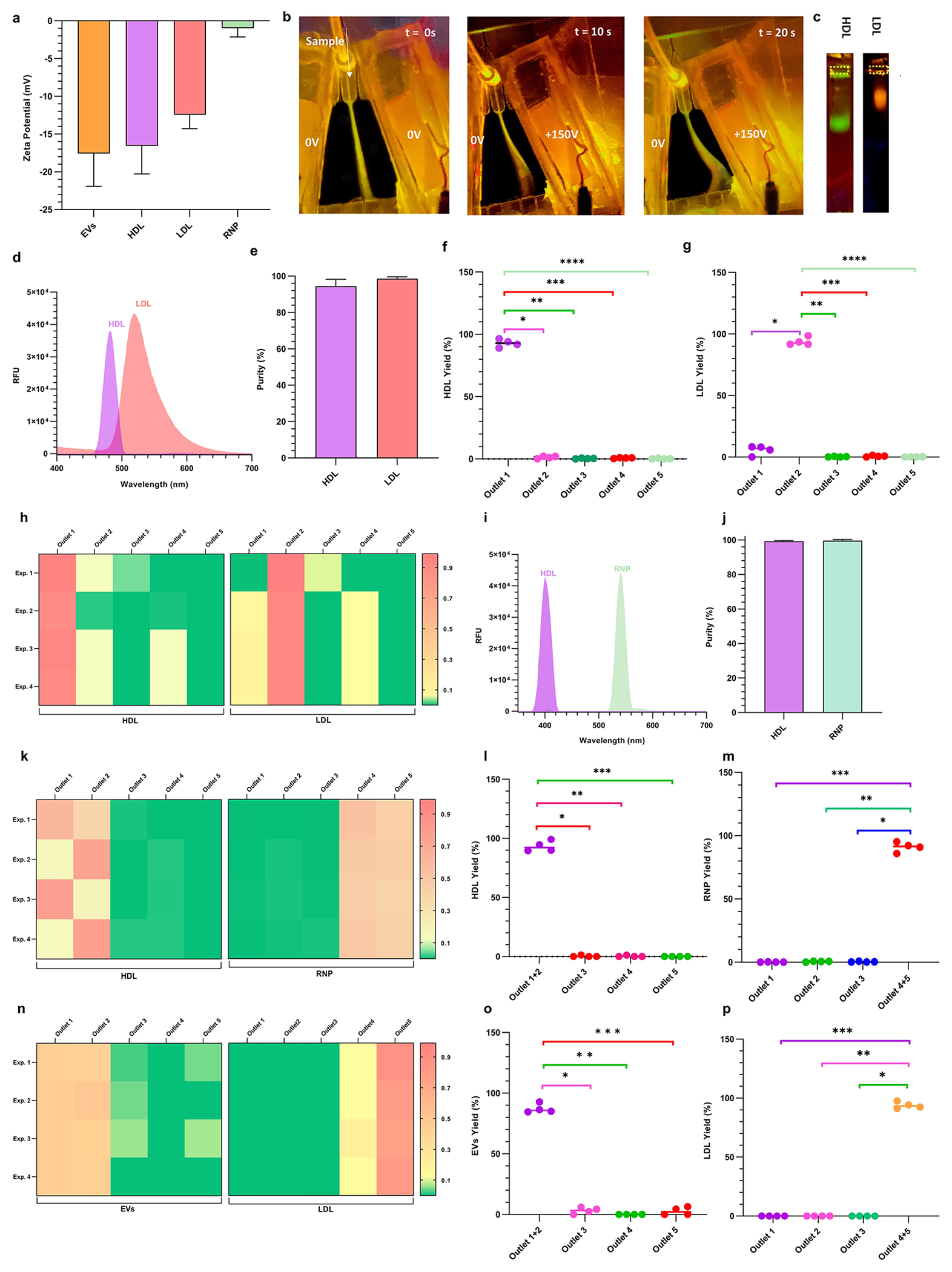
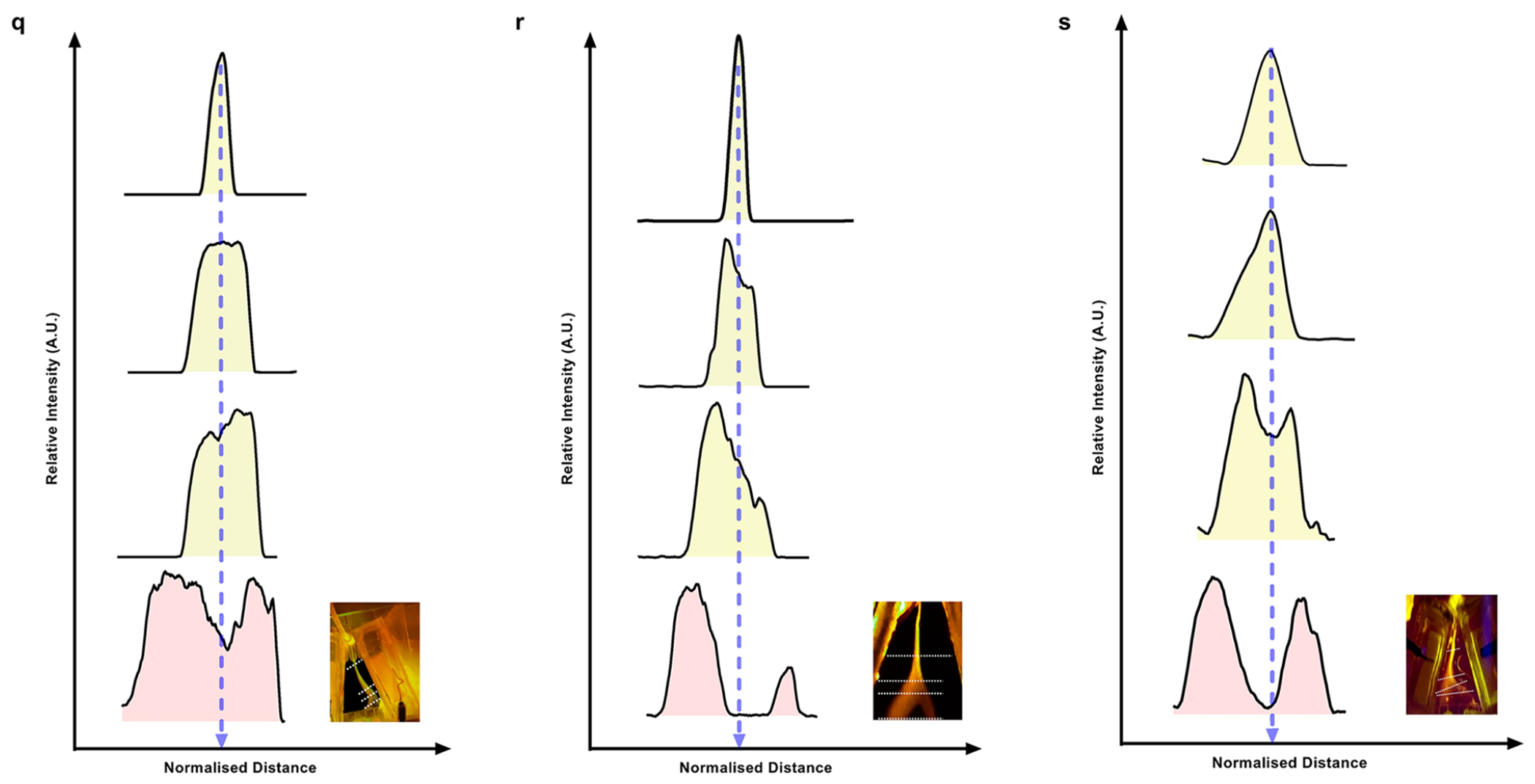
Demonstration of the separation of a binary equimolar mixture in 1× PBS buffer solution of HDL and LDL, HDL and RNP, and LDL and EVs. (a) Bar graphs of the ζ potential of EVs, HDL, LDL, and RNP measured in PBS buffer at pH = 7.4 (n = 7). (b) Sequential images of CFSE dye-tagged HDL (green) and Atto dye-tagged LDL (orange) mixture illustrating isoelectric focusing in the separation chip. When no voltage is applied, the mixture sample follows the injection streamline. At 150 V, the LLPs are deflected toward their respective isoelectric points, and eventually, two distinct streams are observed which are directed to two different outlets. (c) On-chip gel electrophoresis of samples collected from the HDL outlet (outlet 1) and LDL outlet (outlet 2) showing negligible cross-contamination during CIF separation. The outlet numbering starts from right to left. (d) Fluorescence spectrophotometer data of collected from the respective sample outlets 1 and 2 show a high fraction of HDL and LDL, respectively, with very little cross-contamination. (e) Purities of HDL and LDL are 94.43 ± 3.82% and 98.59 ± 1.01% for n = 4. (f) Box plots showing yield (obtained by Apo A1 ELISA) of HDL (n = 4) obtained at different outlets with statistically significant difference between HDL outlet (outlet 1) as compared to all other outlets (* indicates p = 2.51 × 10−6, ** indicates p = 9.37 × 10−6, *** indicates p = 8.94 × 10−6, **** indicates p = 9.01 × 10−6). (g) Box plots showing yield (obtained by ApoB ELISA) of LDL (n = 4) obtained at different outlets with statistically significant difference between the primary LDL outlet (outlet 2) as compared to all other outlets (* indicates p = 5.40 × 10−8, ** indicates p = 9.90 × 10−6, *** indicates p = 6.68 × 10−6, **** indicates p = 1.09 × 10−5). (h) Heatmaps of the yield for the separation of HDL and LDL across 4 different experiments for all 5 outlets of the separation chip. (i) Fluorescence spectrophotometer data of the sample collected at HDL outlet (outlet 1 + outlet 2) and RNP outlet (outlet 4 + outlet 5) show a pure separation of a binary mixture of HDL and RNP with very small cross-contamination. (j) Purities of HDL and RNP are 99.28 ± 0.37% and 99.61 ± 0.63%, respectively for n = 4. (k) Heatmaps of the yield for the separation of HDL (obtained by Apo A1 ELISA) and RNP (obtained by cas9 ELISA) across 4 different experiments for all 5 outlets of the separation chip. The HDL primarily exits outlet 1 and outlet 2, whereas RNP is coming out from the outlets 4 and 5. (l) Box plots showing a yield of HDL (n = 4) obtained at different outlets with statistically significant difference between HDL outlet (outlet 1 + outlet 2) as compared to all other outlets (* indicates p = 2.35 × 10−5, ** indicates p = 2.44 × 10−5, *** indicates p = 3.12 × 10−5). (m) Box plots showing the yield of RNP (n = 4) obtained at different outlets with statistically significant difference between RNP outlet (outlet 4 + outlet 5) as compared to all other outlets (* indicates p = 1.85 × 10−5, ** indicates p = 1.87 × 10−5, *** indicates p = 2.03 × 10−5). (n) Heatmaps of yield for the separation of LDL (obtained by ApoB ELISA) and commercial EVs (obtained by CD63 ELISA) across 4 different experiments for all 5 outlets of the separation chip. The LDL primarily exits outlets 4 and outlet 5, whereas EVs exit outlets 1 and 2. (o) Box plots showing the yield of commercial EVs (n = 4) obtained at different outlets with statistically significant differences between EVs outlet (outlet 1 + outlet 2) as compared to all other outlets (* indicates p = 1.63 × 10−7, ** indicates p = 6.33 × 10−8, *** indicates p = 2.30 × 10−5). (p) Box plots showing the yield of commercial EVs (n = 4) obtained at different outlets with statistically significant differences between LDL outlet (outlet 4 + outlet 5) as compared to all other outlets (* indicates p = 7.01 × 10−6, ** indicates p = 7.01 × 10−6, *** indicates p = 7.01 × 10−6). Line intensity plots were obtained from the fluorescence image of the separation chip for the fractionation of binary mixtures of (q) HDL and LDL, (r) HDL and RNP, and (s) LDL and EVs, respectively. All data in bar plots are shown as mean α standard deviation. For statistical significance, a two-tailed Student’s t test was used with Welch’s correction, and a p-value <0.05 is considered significant.
As shown in the sequence of fluorescence images of Figure 3b, when no voltage is applied, the LLPs mixture (HDL is green and LDL is orange) followed its streamline. When 150 V was applied across the trapezoidal channel, the labeled mixture is directed toward the positive electrode and two distinct bands become visible, green for HDL and orange for LDL after nearly 10 s (Figure 3b). Line intensity plots were extracted at the indicated four axial locations along the separation channel of an image taken at a steady state (Figure 3q). A clear separation of peaks is observed as the sample progresses downstream, and the line intensity plots denote the spatial separation of the LLPs mixture as the nanocarriers are gradually focused on their respective pIs. Moreover, the collected effluents from the two outlets were subsequently analyzed using gel electrophoresis, and as shown in Figure 3c, two distinct bands are observed with qualitatively no apparent cross-contamination. For further qualitative assessment, the collected effluents were analyzed using a fluorescence spectrophotometer (Figure 3d), which also confirmed little cross-contamination between the HDL and LDL fractions. The calculated recovery of the HDL and LDL from the mixture was 91.33 ± 3.22% and 88.48 ± 3.62% respectively (Supplementary Figure 11a). Furthermore, we also analyzed the effluents from all five outlets of the separation chip using ApoA1 and ApoB ELISA kits53,54 for quantitative detection of HDL and LDL. Figure 3h shows two heatmaps placed adjacent to each other depicting the yield of HDL and LDL from each outlet. As shown in the heatmaps, the isolated HDL exclusively came from outlet 1, while the isolated LDL from outlet 2, thus illustrating very little cross-contamination of the two targets. Moreover, Figure 3f,g shows that the yields obtained from the LDL outlet (93.87 ± 3.21%) and HDL outlet (92.87 ± 3.14%) are statistically significant across all four (n = 4) experiments. The purity of the LLPs mixture separation at the HDL and LDL outlets was evaluated to be 94.43 ± 3.82% and 98.59 ± 1.01%, respectively (Figure 3e).
Effluent Selection for Separating hDl and RNP Mixture Spiked in Buffer.
RNPs, another important exRNA nanocarrier, has been implicated in transcription, regulating gene expression, and protection of free-floating mRNA/miRNA biomarkers.55,56 We used a mixture of RNP and HDL to check the feasibility of RNP separation based on its isoelectric point. As seen in Figure 3a, the measured ζ potential of HDL and RNP is −16.56 ± 3.73 mV and −0.98 ± 1.16 mV, respectively. Given the large difference in pIs of RNP (pI = 7.4–8.1) and HDL (pI = 5.01), we transferred a pH gradient in the range of 2–8 into the separation chip after initial optimization (Supplementary Figure 7 and Note 7). As expected, under the application of no voltage bias, the injected HDL (green)-RNP (orange) mixture in the separation chip remains inseparable. Under 200 V, the mixture sample starts separating with HDL moving toward the positive electrode and RNP moving toward the negative electrode until they reach their isoelectric point (Supplementary Figure 8a) with two distinct bands, showing a clear separation of the mixture which is collected into different outlets. Line intensity plots in four different spatial locations show that as the sample moves downstream, the intensity peaks broaden out which eventually divides into two peaks, thus depicting the separation of HDL and RNP from the mixture (Figure 3r).
Qualitative analysis of the collected samples from these outlets was conducted using gel electrophoresis, which shows a bright band of HDL and RNP in their respective outlets with negligible cross-contamination (Supplementary Figure 8b). The collected samples were further analyzed using a fluorescence spectrophotometer which showed very little cross-contamination in the two outlets containing HDL and RNP respectively (Figure 3i). The calculated recovery of the HDL and RNP from the mixture was 91.26 ± 2.71% and 84.48 ± 2.59%, respectively (Supplementary Figure 11b). Figure 3k shows two adjacently placed heatmaps of HDL and RNP yield from each outlet of the separation chip for different experiments (n = 4). The majority of the HDL is separated into outlets 1 and 2, while RNP exits from outlets 4 and 5. Moreover, quantitative measurements show that the yield obtained from HDL outlets 1 and 2 (93.35 ± 4.52%) and RNP outlets 4 and 5 (91.05 ± 3.82%) are statistically significant across all experiments (Figure 3l,m). The purity of the fractionation at the HDL outlets was evaluated to be 99.28 ± 0.37% whereas at the RNP outlets were 99.61 ± 0.63% (Figure 3j).
Effluent Selection for Separating EVs and LDL with Sequential Separation Chips.
Finally, the device was used to separate EVs and LPPs, which are abundant circulating biomarkers in blood and have a significant overlap in size distribution and density. Cryogenic electron microscopy images and morphological properties of lipoproteins and EVs are identical to a large extent.57 A recent study also found that LDL can mimic the properties of blood plasma-derived EVs, skewing subsequent downstream analysis.58 We performed gel electrophoresis of pure samples of commercially available EVs, LDL, HDL, and RNP to qualitatively examine their electrophoretic mobilities. The band of commercial EVs lies in between and with some overlap with the LDL and HDL band, suggesting a very small difference between their isoelectric points (Supplementary Figure 9). Moreover, the overlapping error bars for ζ potential measurements (Figure 3a) of the commercial EVs (−17.57 ± 4.35 mV), HDL (−16.56 ± 3.73 mV), and LDL (−12.44 ± 1.85 mV) confirm the gel electrophoresis observation. This suggests that the successful separation of EVs and LDL, for example, will require a very high-resolution pH gradient.
Our attempts at CIF separation of LDL and EVs by transferring the pH in the range from 2 to 6 from the pH gradient chip to the separation chip were unsuccessful. To generate an even finer pH gradient, two separation chips were connected in series along with the pH gradient chip. A pH in the range of 2–6 was first transferred downstream into the first separation chip, and subsequently, the pH roughly in the range of 4–5.5 was transferred into the second separation chip; the sample of a mixture of EVs and LDL in 1× PBS buffer solution was then injected into the second separation chip. As shown in Supplementary Figure 10a, the series of images of the second separation chip at different time points show that the LDL particles are deflected toward the negative electrode and EVs are deflected toward the positive electrode, illustrating successful isoelectric focusing. Image analysis of the fluorescence image with line intensity plots in four different spatial locations shows that as the sample moves downstream, the intensity peaks broaden out which eventually divides into two peaks, thus depicting the separation of LDL and EVs from the mixture (Figure 3s). The fractionated effluents were collected from two separate outlets, and qualitatively analyzed using gel electrophoresis (Supplementary Figure 10b) showed insignificant cross-contamination between different outlets.
Figure 3n shows two adjacently placed heatmaps of LDL and EVs yield from each outlet of the separation chip for four different experiments. As shown in heatmaps, the majority of EVs exit outlets 1 and 2, while the LDL exit outlets 4 and 5, with very little cross-contamination. Moreover, for quantitative measurements, Figure 3o,p shows that yields obtained from the EVs outlets (87.25 ± 3.81%) and LDL outlets (93.83 ± 2.76%) are statistically significant across all experiments. The purity of the mixture fractionation at the EVs outlet was evaluated to be 99.99997 ± 5.77 × 10−6 %, whereas the LDL outlet was evaluated to be 97.32567 ± 3.19564% (Supplementary Figure 11c).
Automated CIF (auto-CIF) Analyzer using Machine Learning.
Although we have calibrated the pH effluent range for the key nanocarriers in spiked buffers, the isoelectric points of the nanocarriers may change in plasma and other physiological fluids. To account for isoelectric point variation and achieve fine fractionation, tedious tuning is required to select the desired pH range from the pH gradient chip, followed by visual inspection and comparison to a pH reference chart. Both procedures are inefficient and can lead to user-dependent variance in pH selection for the separation chip (Supplementary Figure 13a). To reduce user-driven bias and improve device repeatability, we developed the auto-CIF analyzer, a machine learning platform (Supplementary Figure 12) for automatic pH detection. The auto-CIF analyzer’s workflow (Figure 4a) is divided into two modules: an image segmentation module,59 which identifies the ROI using machine learning, and the pH detection module which generates a spatial pH profile of the ROI by assigning a pH value to each pixel using a custom developed calibration curve60 (Supplementary Note 9). The test data prediction results of image segmentation module for xurography-based chip are represented by a confusion matrix (Figure 4d) that shows excellent accuracy (96.30%), sensitivity (94.83%), and specificity (96.73%). Moreover, the area (# of pixels) of the ROI calculated manually correlates linearly with machine learning prediction (Figure 4e). For the pH detection module of the xurography-based chip, the normalized mean RGB values taken from a pH reference chart (Figure 4g) gave poor correlation with the pH (Figure 4i). However, when the data was divided into acidic (pH = [1–6]) and basic pH values (pH = [7–11]), a good calibration curve was obtained (Figure 4i). Subsequently, using this calibration curve, a pH surface plot for the ROI of a test image (shown in Figure 4a) was generated (Figure 4k). The pH plot guides the user regarding in selecting outlets 3 and 4 from the pH gradient chip (Supplementary Figure 13b) to transfer into the separation chip for HDL-LDL mixture fractionation (Supplementary Figure 13c). Line intensity plots were extracted at the indicated four axial locations of a fluorescence image taken at steady-state (Supplementary Figure 13d), indicating clear segregation of peaks in Figure 4l as the sample progresses downstream.
Figure 4.
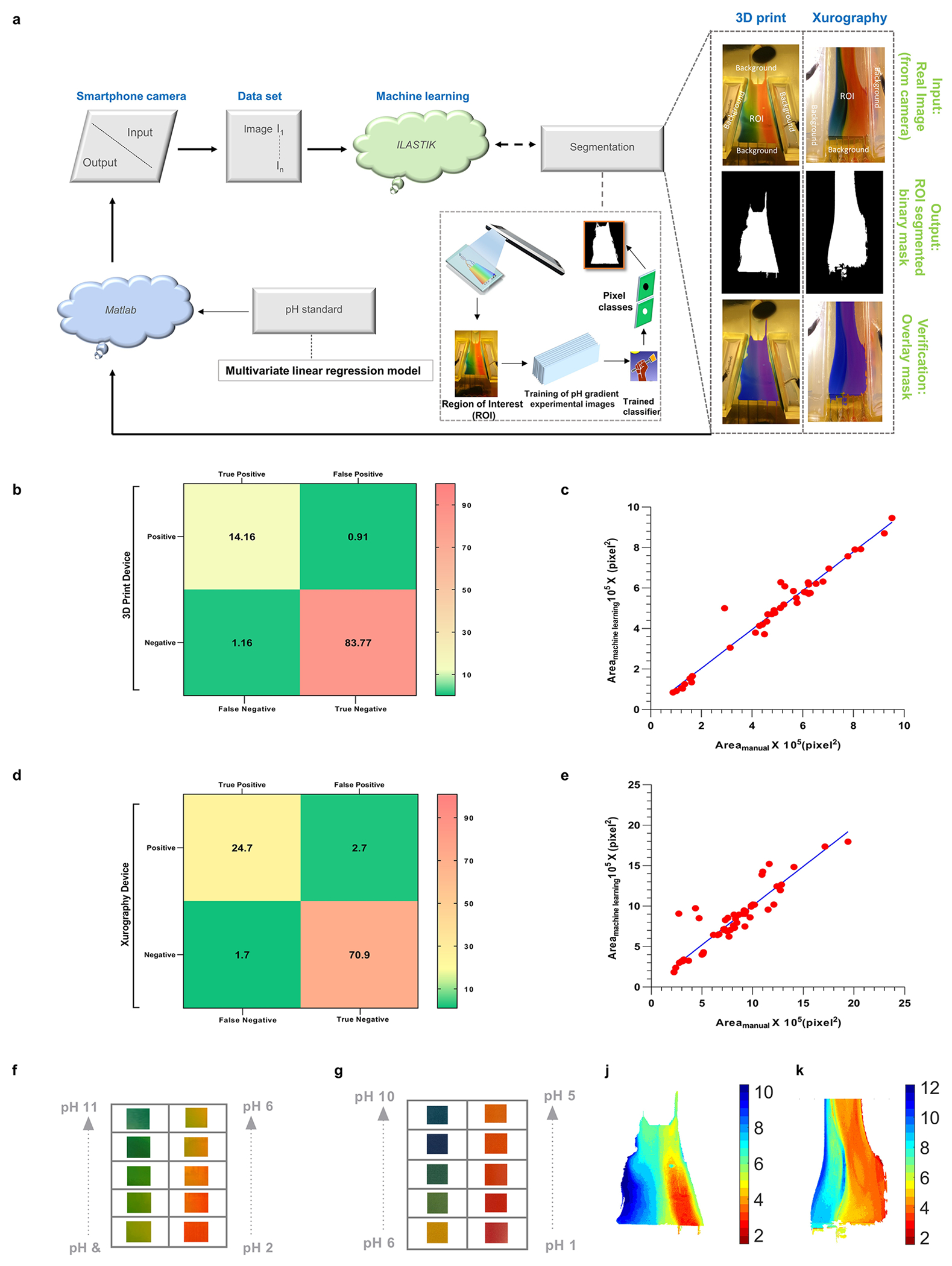
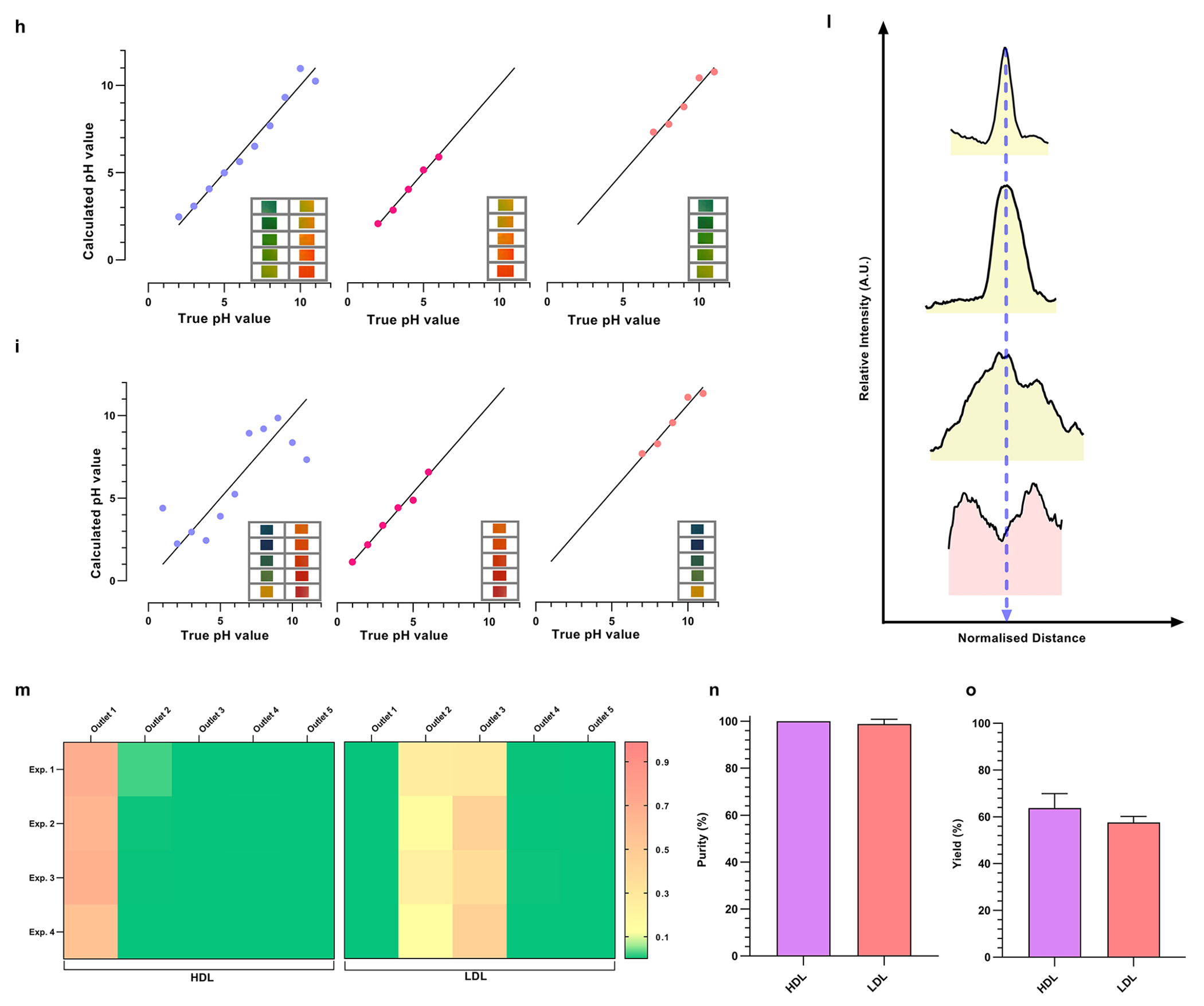
Machine learning based auto-CIF analyzer platform for improved pH judgment and experimental tunability. (a) Complete workflow of the auto-CIF analyzer with image analysis pipeline of xurography and 3D printed chips. (b) Confusion matrix generated for the 3D printed device shows excellent pixel-wise classification accuracy for ilastik classifier trained to identify the ROI. The numbers shown in the boxes represent the percentage prediction accuracy in which a pixel is classified in vertical labels as compared to its true class shown horizontally. (c) ROI area of test images for 3D printed chip predicted by auto-CIF versus measurement performed manually. The slope (0.96) and R2 value (0.96) indicate an excellent performance of the semantic segmentation module. (d) Confusion matrix generated for xurography-based chip also shows excellent pixel-wise classification accuracy for the ilastik classifier trained to identify the ROI. Similarly, as described above, the numbers shown in the boxes represent the percentage prediction accuracy in which a pixel is classified in vertical labels as compared to its true class shown horizontally. (e) Measurements of ROI area of test images for xurography-based chip predicted by auto-CIF versus measurement of ROI area performed manually. The slope (0.97) and R2 value (0.91) indicate excellent performance of the semantic segmentation module. (f) Array of images captured in the 3D printed device corresponding to different pH. (g) Series of images taken from a pH reference chart (Hydrion One Drop Indicator Solution Kit 1–11). (h) Graph of the calculated pH value versus the true pH value for a 3D printed chip obtained by multivariate linear regression using all the normalized mean RGB values as independent variable vectors (MSE = 0.573), only acidic (pH = [2–6]) normalized mean RGB values as independent variables (MSE = 0.175) and only basic (pH = [7–11]) normalized mean RGB values as independent variables (MSE = 0.468). (i) Graph of the calculated pH value versus the true pH value for xurography-based chip obtained by multivariate linear regression using all the normalized mean RGB values as independent variable vectors (MSE = 2.17), only acidic (pH = [1–6]) normalized mean RGB values as independent variables (MSE = 0.29) and only basic (pH = [7–11]) normalized mean RGB values as independent variables (MSE = 0.448). (j–k) pH heatmap of a representative 3D printed and xurography based chip after ROI detection using the image segmentation module, respectively. (l) The line intensity plots at different downstream locations are shown confirming the separation of HDL and LDL nanocarriers with auto-CIF analyzer. (m) Heatmaps of yield for the separation of HDL and LDL across 4 different plasma experiments (n = 4) in all 5 outlets of the separation chip. The HDL is coming primarily from outlet 1, whereas LDL is coming out from outlets 2 and 3. (n) Purities of HDL and LDL from human plasma are 99.99904 ± 0.00126% and 98.83900 ± 1.99225% respectively. (o) Yield of HDL and LDL from human plasma are 63.82 ± 6.1% and 57.67 ± 2.53% respectively.
Similarly, we evaluated the feasibility of the CIF device coupled with auto-CIF analyzer by fractionating HDL and LDL nanocarriers present in 20× diluted human plasma samples, using the optimized protocols from spiked buffers, fine-tuned by the auto-CIF analyzer to adjust for the change in isoelectric points. The effluents from all five outlets of separation chip were collected for plasma experiments (n = 4). Figure 4m shows two adjacently placed HDL and LDL heat maps representing their yield from each outlet for all four experiments, where HDL primarily exited outlet 1 and LDL exited outlets 2 and 3. As shown in Figure 4n, the purity of the separation at the HDL outlet was evaluated to be 99.99904 ± 0.00126% whereas for LDL outlets were 98.83900 ± 1.99225%. Figure 4o shows that the yield obtained from the HDL outlet was 63.82 ± 6.10% and 57.67 ± 2.53% from the LDL outlets. Furthermore, to demonstrate the high-throughput fabrication potential of the CIF device, we built a 3D-printed CIF prototype (Supplementary Figure 14), generated a linear pH gradient in the device (Supplementary Figure 2d), and developed an auto-CIF analyzer for this chip type whose workflow is same as described above (Figure 4a). More discussions on Figure 4b,c,f,h,j are provided in Supplementary Note 8.
Separating RNP from HDL, LDL, and EVs from Plasma, Urine, and Saliva Samples.
In the previous section, we demonstrated highly pure fractionation of HDL and LDL ΔpI < 0.5) from human plasma using the CIF coupled with the auto-CIF analyzer. Here, we demonstrate comprehensive, high-purity, high-yield, and probe-free isolation of RNPs from human plasma using the CIF coupled with auto-CIF analyzer. A pH gradient in the range of 2–8 was transferred into the separation chip using the auto-CIF analyzer, as this effluent range worked well previously. We spiked the Cas9-miRNA21 RNP complex into 20× diluted healthy human plasma before injecting the sample into the separation chip. As described earlier, for quantitative assessments, the effluents collected from all the outlets of the chip were analyzed by ELISA, to determine the concentration profiles of RNP, HDL, LDL, and EVs. Figure 5a shows a series of heatmaps of RNP, LDL, EVs and HDL from each outlet of the separation chip for different experiments (n = 3). The heatmaps clearly show high-purity separation of RNP from HDL, LDL, and EVs in plasma with very little crosscontamination. Mostly, RNP is fractionated into outlet 5, while HDL and LDL exit from outlets 1 and 2. EVs on the other hand are obtained in outlets 1–3, indicating that plasma EVs have a wider distribution of isoelectric points as compared to cell culture EVs. Closer data analysis reveals that the RNP yield obtained from outlet 5 (78.06 ± 7.07%) is statistically significant across all experiments as compared to other outlets (Figure 5c). Furthermore, we evaluated the presence of other nanocarriers obtained in the RNP outlet and determined the purity of RNP fractionation to be 93.39 ± 0.88% (Figure 5d). Similarly, we also evaluated the performance of CIF for RNP fractionation using 5× diluted urine and saliva samples (Figure 5e–l). As demonstrated earlier for blood plasma, the CIF platform successfully separated RNP from urine and saliva at high yield (urine = 87.63 ± 6.15% and saliva: 96.82 ± 1.41%) and purity (urine: 95.38 ± 0.79% and saliva: 97.43 ± 1.41%). Therefore, this proof-of-concept study demonstrates the successful fractionation of RNP from HDL, LDL, and EVs nanocarriers in 0.75 mL of various biofluids such as blood plasma, urine, and saliva within 30 min using the difference in their isoelectric point, thus significantly outperforming established gold standard methods such as RIC and CLIP in terms of purity and throughput.
Figure 5.
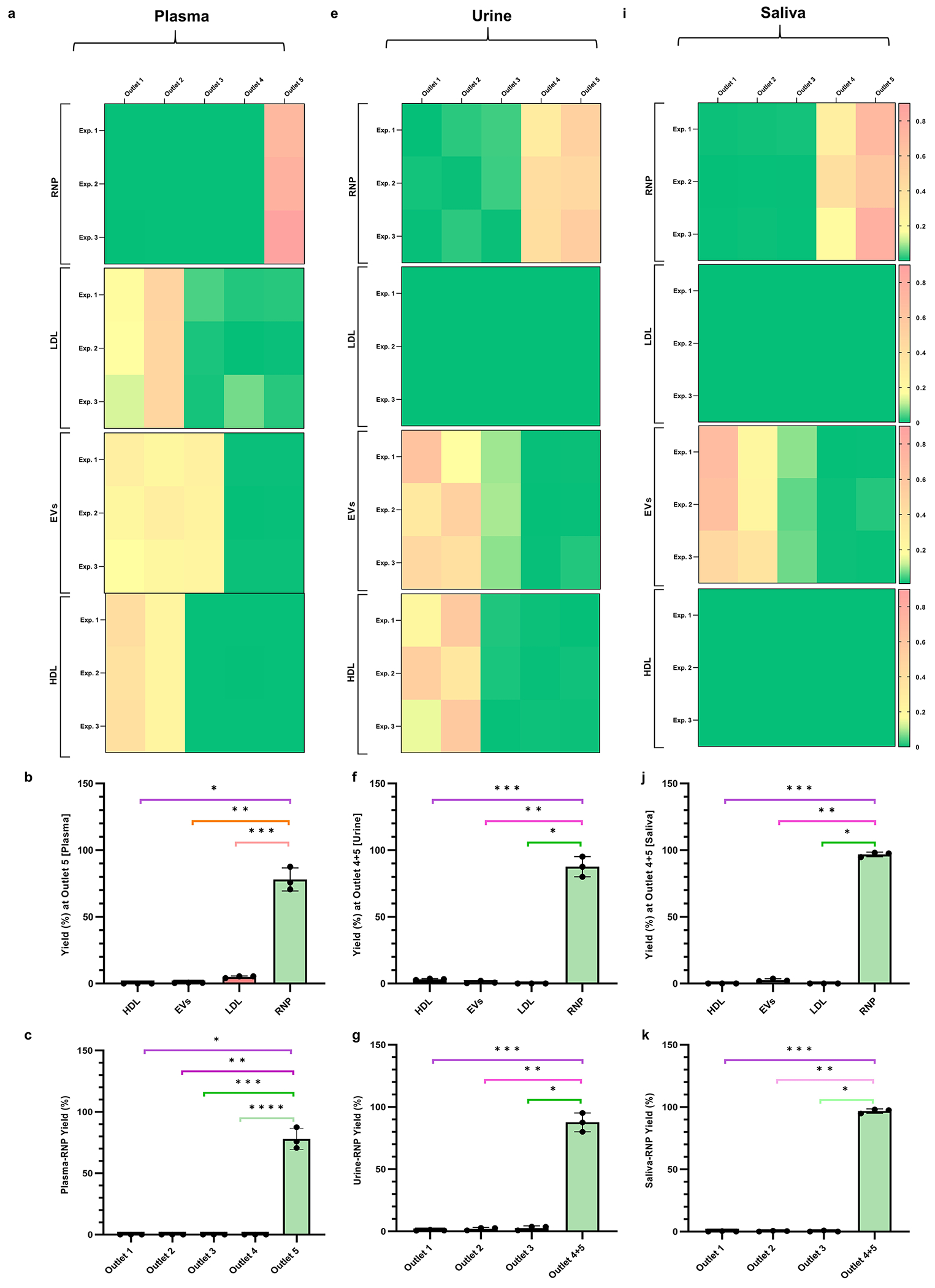
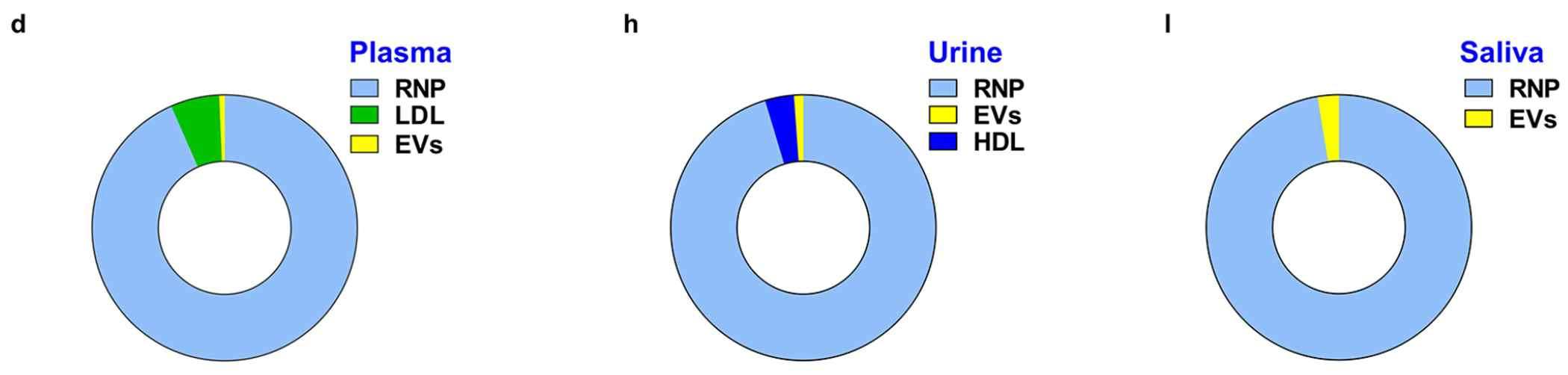
Fractionation of RNP from HDL, LDL and EVs from human plasma, urine and saliva samples. (a) Heatmaps of the yield of RNP, LDL, EVs, and HDL for all 5 outlets of the separation chip (n = 3) for plasma. (b) Column plots showing the yield of RNP, LDL, EVs, and HDL obtained at RNP outlet (outlet 5) (* indicates p = 4.08 × 10−3, ** indicates p = 4.13 × 10−3, and *** indicates p = 4.83 × 10−3). (c) Yield at RNP outlet (outlet 5) as compared with other outlets (* indicates p = 4.08 × 10−3, ** indicates p = 4.08 × 10−3, *** indicates p = 4.08 × 10−3, and **** indicates p = 4.08 × 10−3). (d) Pie chart depicting the purity (93.39 ± 0.88%) of RNP isolation from plasma (n = 3). (e) Heatmaps of the yield of RNP, LDL, EVs, and HDL for all 5 outlets of the separation chip (n = 3) for urine. (f) Column plots showing the yield of RNP, LDL, EVs and HDL obtained at RNP outlet (outlet 4 + 5) (* indicates p < 1 × 10−9, ** indicates p = 2.23 × 10−3, and *** indicates p = 2.55 × 10−3). (g) Yield at RNP outlet (outlet 4 + 5) as compared with other outlets (* indicates p = 1.73 × 10−3, ** indicates p = 2.15 × 10−3, and *** indicates p = 2.43 × 10−3). (h) Pie chart depicting the purity (95.38 ± 0.79%) of RNP isolation from urine (n = 3). (i) Heatmaps of the yield of RNP, LDL, EVs and HDL for all 5 outlets of the separation chip (n = 3) for saliva. (j) Column plots showing the yield of RNP, LDL, EVs and HDL obtained at RNP outlet (outlet 5) (* indicates p < 1 × 10−9, ** indicates p = 1.81 × 10−6, and *** indicates p = < 1 × 10−9). (k) Yield at RNP outlet (outlet 4 + 5) as compared with other outlets (* indicates p = 3.58 × 10−5, ** indicates p = 7.06 × 10−5, *** indicates p = 8.35 × 10−5). (l) Pie chart depicting the purity (97.43 ± 0.79%) of RNP isolation from saliva (n = 3).
We also did a comprehensive study to evaluate the potential adverse effect of CIF on the separated nanocarriers due to shear and pH. Shear can be easily ruled out as widely used techniques in the field, such as ultracentrifugation and ultrafiltration apply shear several orders of magnitude higher than what is applied during CIF, and have shown that the structure, integrity, and biological function of extracellular nanocarriers remains intact. To study the impact of pH, all the nanocarriers were first subjected in different pH conditions from 3 to 7.4 and then were analyzed using various techniques (Supplementary Figures 20–23) including the Bradford assay, cholesterol tests, RT-qPCR, and transmission electron microscopy (TEM) imaging. The physical and chemical properties of the nanocarriers remained unchanged, indicating that they will also be unaffected during short-lived immersion in a different pH during CIF separation.
CONCLUSION
In this study, we report a bias-free, high-throughput (~mL/h) and continuous free-flow isoelectric separation device that allows binary fractionation and isolation of nanocarriers such as LLPs, RNP, and EVs as well as fractionation of RNP from LLPs and EVs in 20× diluted healthy human plasma, 5× diluted urine, and 5× diluted saliva at high yield and purity. This technology is based on the distinct isoelectric points of individual RNA nanocarriers (instead of size and density where significant overlap exists) allowing their fractionation within a residence time of 10 s and with a minimum ΔpI resolution of 0.3. In the pH gradient chip, the applied voltage bias across two BiMs splits water into hydronium and hydroxide ions in a single microfluidic channel, thus overcoming the need for ampholytes or multiple acid/base buffer inflows. The linear pH profile results from the rapid equilibration of the inert ions to a no-flux Boltzmann equilibrium and reversible water dissociation at each location to maintain electroneutrality. We have demonstrated versatile amplification of the spatial pH resolution by extracting one portion of the full pH profile and expanding it in subsequent trapezoidal separation chip(s), further optimized by a machine learning procedure, as necessary for nanocarrier separation. Furthermore, we propose that this efficient, yet low-cost device has the potential to be used in a star topological configuration, with the pH gradient chip being the central node and different separation chips acting as connected nodes (Supplementary Figure 18) for high-throughput separation of multiple nanocarriers from a single 2 mL plasma sample. As multiplexed detection of different biomarkers is challenging due to wild-type/inhibiting molecules, spectral overlap, and low sensitivity of biosensors, high-throughput CIF-based purification can help enable bias-free and label-free separation, leading to more accurate detection and quantification. This device represents a significant advancement as it overcomes the various limitations described earlier of commonly utilized traditional isolation technologies including ultracentrifugation,61,62 immunocapture, and ultrafiltration techniques.
Although there is enormous interest in extracellular RNA-carriers spanning basic laboratory research, the pharmaceutical industry, and the clinic, the lack of carrier purification as well as unequivocally demonstrating the specific RNA pool as causative in physiological effects on target cells are among the most significant challenges in this field. Since non-EV carriers fractionate with EVs during common isolation procedures, it is possible that while RNAs are contained within specific subtypes of EVs as reported in the literature, others are present in other carriers that co-isolate. The CIF-based purification described here alleviates these issues. Additionally, we envision that CIF platform’s future application in exploring the heterogeneity of EVs such as fractionating different EV types with cargoes derived from cancer cells (e.g., GPC-1, Active EGFR, AR-V7)63,64 will require a fine pH gradient, which will likely require multiple separation devices to achieve sufficient pH resolution. We also anticipate that this technology can be used to purify other biological nanoparticles including virus vaccines,65 exosome drug carriers,66 amyloid-beta aggregates,67 and peptide assemblies.68
METHODS
Reagents and Biosamples.
1× PBS (Fischer Scientific, USA) and 10× TAE buffer (Sigma-Aldrich, USA) consisting of 400 mM Tris-acetate and 10 mM ethylenediaminetetraacetic acid (EDTA) were purchased and used as received. HDL and LDL were purchased from MyBiosource, USA. CFSE Cell Proliferation Kit (ThermoFischer Scientifics, USA) and Atto 550 NHS ester (Sigma-Aldrich, USA) were purchased and used for the labeling of LPPs, RNP, and commercial EVs. MicroRNA 21 (miR21) (Integrated DNA Technologies, USA) with a sequence of TAGCTTATCAGACTGATGTTGA and Cas 9 (New England Biolabs, USA) were purchased and used for RNP complex formation. Human preadipocyte (mesenchymal stem cell) EVs (concentration > billion particles/vial) were purchased from a commercial vendor (cat # EXP-F100, Zen-Bio, USA) and stored at −80 °C upon receipt. 10 mL of 3 healthy uninfected urine samples was purchased from Biochemed Services, USA. Bradford Reagent (Sigma-Aldrich, USA) and BSA standards (Thermo Fischer Scientific, USA) were purchased and stored at 4 °C until use. Amplex Red Cholesterol Assay Kit (Invitrogen, USA), miRCURY LNA RT Kit(Qiagen, USA), and miScript SYBR Green PCR Kit (Qiagen, USA) were stored at −20 °C until further use.
Xurography-Based Fabrication and Operation of CIF Device.
Xurography-based pH gradient chip was fabricated by sandwiching together two polycarbonate sheets (300 μm thickness) between a double-sided Kapton tape (PPTDE-3, Katontape.com) containing the microfluidic structure using a plotter cutter (Graphtec Pro FC7000). Upstream from the pH generation chip comprises of an IEM-based water splitting slot followed by a tree-like microchannel (splitter-mixer) connected to a trapezoidal-shaped microfluidic channel downstream. The width and length of the splitter-mixer channels are 0.5 mm × 26 mm, while the trapezoidal-shaped chamber has a dimension of 20 mm × 26 mm × 37 mm (b1 × b2 × l). Bipolar membranes were fabricated by stacking together a cation-exchange membrane (CEM) and an anion-exchange membrane (AEM) (Mega a.s., Czech Republic) using UV-based glue (Acryfix, USA). Several 5 mm pieces of polycarbonate clear tubing (McMaster-Carr, USA) were cut and glued at inlets/outlets. 5 mL of deionized water was injected using Multi-Syringe Programmable Syringe Pumps (Braintree Scientific, USA) under 80 V (Keithley 2400 SourceMeter, USA) unless otherwise specified. For smooth operation of the CIF chip, the working range of the flow rate in the pH gradient chip lies between 9 mL/h to 12 mL/h. For all experiments, we selected a flow rate of 12 mL/h in the pH gradient chip. During the experiments, to minimize pressure imbalance between outlets, the key outlets of the pH generation chip were connected to a separation chip, and others were connected to a dummy separation chip using silicone tubing (1/32″, Cole-Parmer, USA). Similarly, the separation chip was also fabricated and operated as described above. The dimensions of the trapezoidal channel were 9 mm × 26 mm × 37 mm (b1 × b2 × l). Both sides of the trapezoidal microfluidic channel of the separation chip were exposed to two reservoirs that were filled with 4% agarose gel after covering it with a filter paper from the reservoir side. The combination of filter paper and the agarose gel acts as a salt bridge to ensure a reliable electrical connection between the trapezoidal channel and the voltage source while avoiding any bubble escape into the channel. 200 V was applied transverse to the sample flow unless specified otherwise, and the sample was injected at a flow rate of 3 mL/h.
3D Printed Chip.
A 3D model was designed in Solidworks 2019 (Supplementary Figure 14), and a stereolithography (.stl) file was sent to a commercial vendor (Boston Micro Fabrication, Maynard, MA, USA) to fabricate a 3D printed chip with HTL resin.
Sample Preparation: HDL, LDL, RNP, and EVs Preparation.
A known concentration (0.28 μg/mL) of HDL and LDL in 1× PBS was labeled with CFSE dye and Atto dye, respectively, using manufacturer’s protocols and purified using Amicon Ultra 0.5 centrifugal filter kit (Sigma-Aldrich, USA). The conjugation of HDL and LDL with their respective dyes was then verified with on-chip gel electrophoresis (Supplementary Figure 4). For RNP complex formation, 1 μL of miRNA was mixed with 10 μL of Cas9 protein from their original vial and then left at room temperature for 30 min. The complex was further diluted 100 times with 1× PBS before being stored at 4 °C until use. Cas9 was labeled with Atto dye using the manufacturer’s labeling protocol, and then the RNP was purified from unbound free-floating dye molecule using Amicon Ultra 0.5 centrifugal filter kit. RNP formation was verified by on-chip gel electrophoresis (Supplementary Figure 9). For EVs, prior to use, the cryovial was thawed by placing it in a 37 °C water bath for 1 h and later centrifuged at 17000g for 10 min. The labeling of EVs was done by adding 10 μL of commercial EVs and 2 μL CFSE dye into 1× PBS to a final volume of 2 mL.
ζ Potential Measurement.
The ζ potential of HDL, LDL, RNP, and EVs was measured in 1× PBS (pH 7.4) buffer using a NanoBrook ZetaPALS potential analyzer (Brookhaven Instruments Corporation, USA).
Healthy Human Plasma, Urine, and Saliva.
Healthy human plasma samples were purchased (Zen-Bio Inc., USA) and collected in tubes of 10 mL with EDTA coagulant. Plasma from two different donors was used in which the first donor is a white female (age = 39) with Hispanic ethnicity, while the other donor was an African American male (age = 62). The samples were obtained following FDA-mandated testing for pathogens. Every urine sample was centrifuged for 10 min at 2000g at 4 °C. Following this, the supernatant was filtered through a Millipore 0.22 μm membrane filter and kept at 80 °C until use. For RNP fractionation from human plasma, urine, and saliva, 1.5 mL of sample was injected into the separation chip at a flow rate of 1.5 mL/h.
Fractionation of Binary Mixture of Nanocarriers Spiked in Buffer.
After selecting the appropriate outlets from pH gradient chip, a 3 mL of nanocarrier binary mixture for three different cases (HDL-LDL, HDL-RNP, and LDL-EVs) spiked in PBS was injected into the separation chip at a flow rate of 3 mL/h except for LDL-EVs fractionation which ran at 1.5 mL/h. For human plasma experiments, 1.5 mL of 20× diluted plasma was injected into the separation chip at a flow rate of 1.5 mL/h. A voltage bias of 150 V was applied for HDL-LDL fractionation, whereas 200 V was applied for HDL-RNP and LDL-EVs fractionation.
HDL, LDL, RNP, and EVs ELISA.
Apolipoprotein A1 Human ELISA Kit (ThermoFisher Scientific, USA) and Human ApoB ELISA Kit (ThermoFisher Scientific, USA) were used for the quantification of HDL and LDL samples following the manufacturer’s protocols. Briefly, 100 μL of protein samples was added to each of the 96-well plates and incubated overnight at 4 °C with gentle shaking. The samples were discarded and washed 4 times with 1× wash buffer. 100 μL of biotin conjugate was added, incubated for 1 h with gentle shaking, and washed four times. 100 μL of Streptavidin-HRP was then added, incubated for 45 min with gentle shaking, and washed four times. Finally, 100 μL of TMB substrate was added, incubated for 30 min, and a 50 μL of stop solution to each well. For RNP complex quantification, EpiQuik CRISPR/Cas9 Assay ELISA Kit (Epigentek, USA) was used following the manufacturer’s protocol. 100 μL of Cas9 Binding Buffer (CBB) and 1 μL of the sample were added to each blank well and was incubated at 37 °C for 120 min. After 3 times washing with 150 μL wash buffer, 50 μL of the Cas9 Detection Complex Solution (made by mixing 1 mL of diluted wash buffer, 1 μL of detection antibody, 1 μL of signal indicator, and 1 μL of enhancer solution) to each well and incubated for 50 min at room temperature. After 3 times washing, 100 μL of developer solution was added and incubated at room temperature for 10 min, and 100 μL of stop solution was added to each well.
For EVs quantification, Human CD63 ELISA Kit (ThermoFischer Scientific, USA) was used following the manufacturer’s protocol with the kit. In short, 100 μL of the EVs samples was added individually to the 96-well plates and incubated overnight at 4 °C with gentle shaking. The solution was washed 4 times with wash buffer before adding 100 μL of biotin conjugate for each well and incubated for 1 h with gentle shaking. Then, 100 μL Streptavidin-HRP was added to each well and incubated for 45 min before discarding and further washing the solution 4 times. TMB substrate (100 μL) was then added and incubated for 30 min, and 50 μL of Stop Solution was added to each well. For all ELISA experiments, the 96-well plates were imaged by a microplate reader (Tecan Infinite M200 Pro) at 450 nm and were compared with the calibration curve (obtained by 4 parameter logistic regression (4PL)) of the protein standards.
Calculation of Separation Performance Metrics.
The yield of an experiment is determined by the mass of protein obtained at each outlet as compared to the initial mass used for the separation experiment. Numerically, the yield for the ith outlet for each sample is calculated as
where the protein sample concentration is obtained by ELISA and Volumei is the volume collected from the ith outlet.
Recovery for a sample from the fractionation experiment is calculated using the values of the intensity peaks obtained from the spectrophotometer fluorescence scan for both inlet and sample outlet. For example, in the HDL and LDL separation experiment, recovery is calculated as
where IHDL is the HDL intensity peak value obtained from the denoted outlet, IHDL inlet is the HDL intensity peak value of the sample in the inlet mixture, ILDL is the LDL intensity peak value obtained from the denoted outlet, and ILDL inlet is the LDL intensity peak value of the sample in the inlet mixture
The purity for HDL and LDL separation was determined using
For HDL and RNP separation experiments, purity was determined using
For LDL and EVs separation experiments, purity was determined using
For RNP separation from plasma, urine and saliva experiments, purity was determined using
Bradford Assay.
A 96-well plate Bradford assay was performed by following the manufacturer’s protocol. Briefly, 5 μL of the protein standards, controls, and samples were injected into separate wells. 250 μL of the Bradford reagent was added to each well and incubated at room temperature for 30 min. The absorbance was measured at 595 nm using a microplate reader, and the concentration of unknowns was determined using the standard curve.
Cholesterol Assay.
The cholesterol assay was performed according to the manufactures protocol. In brief, 50 μL of the diluted samples, standards, and controls were pipetted into separate wells of a microplate. 50 μL of 300 μM Amplex Red reagent containing 2 U/mL HRP, 2 U/mL cholesterol oxidase, and 0.2 U/mL cholesterol esterase was added to each microplate well containing the samples and controls and incubated for 30 min at 37 °C. The fluorescence was measured using a microplate reader using excitation at 545 nm and emission detection at ~590 nm.
RT-qPCR.
Reverse transcription was carried out using a miRCURY LNA RT Kit following the kit protocol. Briefly, 10 μL of reverse transcription reaction was prepared using 5× miRCURY RT reaction buffer (2 μL), RNase-free water (5 μL), 10× miRCURY RT enzyme mix (1 μL), and miR21 (2 μL). The PCR plate was incubated for 60 min at 42 °C followed by incubation for 5 min at 95 °C to inactivate the reverse transcriptase enzyme. Then, the cDNA obtained from RT was diluted 10 times using RNase-free water, and the qPCR was carried out using miRCURY LNA SYBR Green PCR Kit (Qiagen) on a StepOnePlus Real-Time PCR System (Applied Biosystems) following the manufacturers protocols. In brief, 10 μL of the reaction mixture contained 2× miRCURY SYBR Green Master Mix (5 μL), ROX reference dye (0.05 μL), PCR primer mix (1 μL), cDNA (3 μL), and RNase-free water (1 μL). The PCR plate was incubated for 2 min at 95 °C, followed by 40 cycles of 95 °C for 10 s and 56 °C for 60 s. Standard curves were generated from a series of dilutions and were used to estimate the copy number. All experiments were performed in triplicates.
Numerical Simulation.
A 2D finite element simulation model of the ionic behavior in both straight and trapezoidal channels was developed using COMSOL Multiphysics 4.4. The steady-state model solved the Poisson–Nernst–Planck (PNP) equations coupled with Poisson–Boltzmann (PB) and Navier–Stokes (NS) equations. For fluid flow, a fully developed flow was assumed with no-slip boundary condition on the walls. For PNP and PB equations, the walls of the channel were assigned a DC voltage bias of −V/2 and +V/2 from left to right with a no net ionic flux boundary condition. A constant concentration boundary condition was applied at the inlet, whereas at the outlet, a zero diffusive boundary condition was specified.
Image Acquisition.
All colorimetric images were captured by a smartphone (Apple iPhone 11 Pro) that was mounted on a tripod in a dark chamber with an external white light source to maintain constant illumination condition. For capturing fluorescence images, the separation chip was placed on a Dark Reader blue transilluminator (DR89 Mid-Size Transilluminator, Clare Chemical Research, USA) that used pure visible blue light as the excitation source. The amber screen was adjusted on top of the chip using a clamp.
Image Analysis for Line Intensity Plots.
Matlab was used to split the separation fluorescence images into red, green, and blue channels. The contrast of the green channel image was enhanced using imadjust function, and line intensity plots were made.
auto-CIF Analyzer.
A Matlab application was developed that integrates an image segmentation module (by running ilastik in headless mode) and a pH detection module. The app enables the user to load a captured digital image, view the segmented image of the ROI alongside the overlaid image using machine learning, and determines the pH surface plot across the ROI. Please check Supplementary Note 9 for more details.
Quantification and Statistical Analysis.
We used OriginPro 8.6 and GraphPad Prism for graphical representation along with statistical analysis. The data in this study have been presented as mean ± standard deviation unless otherwise specified. For statistical analysis, GraphPad Prism was used to perform Student’s two-tailed independent t test with Welch correction, and a p-value <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work has been supported by the National Institute of Health (NIH) Common Fund, through the Office of Strategic Coordination/Office of the NIH Director, grant number 4UH3CA241684-03. We acknowledge Mike Brueseke at the Center for Environmental Science and Technology (CEST) for helping in ζ potential measurements. We also acknowledge Boston Micro Fabrication (BMF), Maynard, MA, USA for supplying the 3D printed CIF chip.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.3c01340.
Water splitting module (Supplementary Figure 1); finite element simulation setup (Supplementary Figure 2); separation chip optimization using 50 nm carboxylated and aminated polystyrene nanoparticles (Supplementary Figure 3); agarose gel electrophoresis of the labeled HDL and LDL (Supplementary Figure 4); chip optimization for HDL and LDL (Supplementary Figure 5); experimental setup for fractionation of HDL and LDL (Supplementary Figure 6); optimization of the IEF separation of a mixture of HDL and RNP (Supplementary Figure 7); fractionation of HDL and RNP mixture and gel electrophoresis (Supplementary Figure 8); the agarose gel electrophoresis of tagged HDL, LDL, RNP and commercial EVs (Supplementary Figure 9); separation of LDL and EVs mixture in the separation chip under a very high-resolution pH gradient (Supplementary Figure 10); recovery and purity data for binary fractionation of nanocarriers (Supplementary Figure 11); graphical user interface for the auto-CIF analyzer (Supplementary Figure 12); auto CIF analyzer platform for fractionating HDL and LDL mixture (Supplementary Figure 13); Solidworks model and the 3D printed separation device (Supplementary Figure 14); histograms of the red, green and blue intensities of an image of a pH reference chart and 3D printed chip corresponding to different pH (Supplementary Figures 15 and 16); results of image segmentation module for xurography and 3D printed chips (Supplementary Figure 17); star topological configuration with pH gradient chip (Supplementary Figure 18); yield and purity of RNP fractionation across various biofluids to demonstrate interday and intraday performance of CIF (Supplementary Figure 19); Bradford assay results for HDL and LDL after incubation in different pH (Supplementary Figure 20); cholesterol assay results for HDL and LDL after incubation in different pH (Supplementary Figure 21); RT qPCR results for mir21 after incubation in different pH (Supplementary Figure 22); TEM images of commercial EVs after incubation in different pH (Supplementary Figure 23); simulated pH profiles to depict insensitivity of linear pH gradient to flow (Supplementary Figure 24); pH gradient chip (Supplementary Note 1); theoretical explanation of pH profile (Supplementary Note 2); mathematical calculation of linear pH profile in both trapezoidal and straight channels (Supplementary Note 3); insensitivity of linear pH gradient to flow (Supplementary Note 4); optimized conditions for separating HDL and LDL (Supplementary Note 5); optimized conditions for carboxylated and aminated latex particles in the separation chip (Supplementary Note 6); optimized conditions for separating RNP and HDL (Supplementary Note 7); use of auto-CIF analyzer for 3D printed CIF microfluidic chip (Supplementary Note 8); image segmentation module and pH detection module of auto-CIF analyzer (Supplementary Note 9); and on-chip gel electrophoresis and gel electrophoresis experimental details (Supplementary Note 10) (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsnano.3c01340
The authors declare no competing financial interest.
Contributor Information
Himani Sharma, Department of Chemical and Biomolecular Engineering, University of Notre Dame, Notre Dame, Indiana 46556, United States.
Vivek Yadav, Department of Chemical and Biomolecular Engineering, University of Notre Dame, Notre Dame, Indiana 46556, United States.
Crislyn D’Souza-Schorey, Harper Cancer Research Institute and Department of Biological Sciences, University of Notre Dame, Notre Dame, Indiana 46556, United States.
David B. Go, Department of Chemical and Biomolecular Engineering and Department of Aerospace and Mechanical Engineering, University of Notre Dame, Notre Dame, Indiana 46556, United States
Satyajyoti Senapati, Department of Chemical and Biomolecular Engineering, University of Notre Dame, Notre Dame, Indiana 46556, United States.
Hsueh-Chia Chang, Department of Chemical and Biomolecular Engineering, Department of Aerospace and Mechanical Engineering, and Harper Cancer Research Institute, University of Notre Dame, Notre Dame, Indiana 46556, United States.
REFERENCES
- (1).Mathieu M; Martin-Jaular L; Lavieu G; Théry C Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol 2019, 21 (1), 9–17. [DOI] [PubMed] [Google Scholar]
- (2).Gruner HN; McManus MT Examining the Evidence for Extracellular RNA Function in Mammals. Nat. Rev. Genet 2021, 22 (7), 448–458. [DOI] [PubMed] [Google Scholar]
- (3).Das S; Ansel KM; et al. The Extracellular RNA Communication Consortium: Establishing Foundational Knowledge and Technologies for Extracellular RNA Research. Cell 2019, 177 (2), 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ainsztein AM; Brooks PJ; Dugan VG; Ganguly A; Guo M; Howcroft TK; Kelley CA; Kuo LS; Labosky PA; Lenzi R; et al. The NIH Extracellular RNA Communication Consortium. J. Extracell. Vesicles 2015, 4 (1), 27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Skog J; Würdinger T; Van Rijn S; Meijer DH; Gainche L; Curry WT; Carter BS; Krichevsky AM; Breakefield XO Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cell Biol 2008, 10 (12), 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Li K; Wong DK; Luk FS; Kim RY; Raffai RL Isolation of Plasma Lipoproteins as a Source of Extracellular RNA. In Extracellular RNA; Springer, 2018; pp 139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).McMahon KM; Thaxton CS High-Density Lipoproteins for the Systemic Delivery of Short Interfering RNA. Expert Opin. Drug Delivery 2014, 11 (2), 231–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wei Z; Batagov AO; Schinelli S; Wang J; Wang Y; El Fatimy R; Rabinovsky R; Balaj L; Chen CC; Hochberg F; et al. Coding and Noncoding Landscape of Extracellular RNA Released by Human Glioma Stem Cells. Nat. Commun 2017, 8 (1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Tauber D; Tauber G; Parker R Mechanisms and Regulation of RNA Condensation in RNP Granule Formation. Trends Biochem. Sci 2020, 45 (9), 764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jan AT; Rahman S; Khan S; Tasduq SA; Choi I Biology, Pathophysiological Role, and Clinical Implications of Exosomes: A Critical Appraisal. Cells 2019, 8 (2), 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhang Q; Higginbotham JN; Jeppesen DK; Yang Y-P; Li W; McKinley ET; Graves-Deal R; Ping J; Britain CM; Dorsett KA; et al. Transfer of Functional Cargo in Exomeres. Cell Rep. 2019, 27 (3), 940–954. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Thompson AG; Gray E; Heman-Ackah SM; Mäger I; Talbot K; El Andaloussi S; Wood MJ; Turner MR Extracellular Vesicles in Neurodegenerative Disease—Pathogenesis to Biomarkers. Nat. Rev. Neurol 2016, 12 (6), 346–357. [DOI] [PubMed] [Google Scholar]
- (13).Zernecke A; Preissner KT Extracellular Ribonucleic Acids (RNA) Enter the Stage in Cardiovascular Disease. Circ. Res 2016, 118 (3), 469–479. [DOI] [PubMed] [Google Scholar]
- (14).Quinn JF; Patel T; Wong D; Das S; Freedman JE; Laurent LC; Carter BS; Hochberg F; Keuren-Jensen KV; Huentelman M; et al. Extracellular RNAs: Development as Biomarkers of Human Disease. J. Extracell. Vesicles 2015, 4 (1), 27495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhang Q; Jeppesen DK; Higginbotham JN; Graves-Deal R; Trinh VQ; Ramirez MA; Sohn Y; Neininger AC; Taneja N; McKinley ET; et al. Supermeres Are Functional Extracellular Nanoparticles Replete with Disease Biomarkers and Therapeutic Targets. Nat. Cell Biol 2021, 23 (12), 1240–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Li K; Rodosthenous RS; Kashanchi F; Gingeras T; Gould SJ; Kuo LS; Kurre P; Lee H; Leonard JN; Liu H Advances, Challenges, and Opportunities in Extracellular RNA Biology: Insights from the NIH ExRNA Strategic Workshop. JCI Insight 2018, 3 (7), e98942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Arroyo JD; Chevillet JR; Kroh EM; Ruf IK; Pritchard CC; Gibson DF; Mitchell PS; Bennett CF; Pogosova-Agadjanyan EL; Stirewalt DL; et al. Argonaute2 Complexes Carry a Population of Circulating MicroRNAs Independent of Vesicles in Human Plasma. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (12), 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Geekiyanage H; Rayatpisheh S; Wohlschlegel JA; Brown R; Ambros V Extracellular MicroRNAs in Human Circulation Are Associated with MiRISC Complexes That Are Accessible to Anti-AGO2 Antibody and Can Bind Target Mimic Oligonucleotides. Proc. Natl. Acad. Sci. U. S. A 2020, 117 (39), 24213–24223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mateescu B; Kowal EJK; van Balkom BWM; Bartel S; Bhattacharyya SN; Buzás EI; Buck AH; de Candia P; Chow FWN; Das S; Driedonks TAP; Fernández-Messina L; Haderk F; Hill AF; Jones JC; Van Keuren-Jensen KR; Lai CP; Lässer C; di Liegro I; Lunavat TR; Lorenowicz MJ; Maas SLN; Mäger I; Mittelbrunn M; Momma S; Mukherjee K; Nawaz M; Pegtel DM; Pfaffl MW; Schiffelers RM; Tahara H; Théry C; Tosar JP; Wauben MHM; Witwer KW; Nolte-‘t Hoen ENM Obstacles and Opportunities in the Functional Analysis of Extracellular Vesicle RNA – an ISEV Position Paper. J. Extracell. Vesicles 2017, 6 (1), 1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Gyuris A; Navarrete-Perea J; Jo A; Cristea S; Zhou S; Fraser K; Wei Z; Krichevsky AM; Weissleder R; Lee H; Gygi SP; Charest A Physical and Molecular Landscapes of Mouse Glioma Extracellular Vesicles Define Heterogeneity. Cell Rep. 2019, 27 (13), 3972–3987.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Driedonks TAP; Nijen Twilhaar MK; Nolte-‘t Hoen ENM Technical Approaches to Reduce Interference of Fetal Calf Serum Derived RNA in the Analysis of Extracellular Vesicle RNA from Cultured Cells. J. Extracell. Vesicles 2019, 8 (1), 1552059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Galvanin A; Dostert G; Ayadi L; Marchand V; Velot É; Motorin Y Diversity and Heterogeneity of Extracellular RNA in Human Plasma. Biochimie 2019, 164, 22–36. [DOI] [PubMed] [Google Scholar]
- (23).Hentze MW; Castello A; Schwarzl T; Preiss T A Brave New World of RNA-Binding Proteins. Nat. Rev. Mol. Cell Biol 2018, 19 (5), 327–341. [DOI] [PubMed] [Google Scholar]
- (24).Gebauer F; Schwarzl T; Valcárcel J; Hentze MW RNA-Binding Proteins in Human Genetic Disease. Nat. Rev. Genet 2021, 22 (3), 185–198. [DOI] [PubMed] [Google Scholar]
- (25).Kang D; Lee Y; Lee J-S RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers 2020, 12 (9), 2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hong S. RNA Binding Protein as an Emerging Therapeutic Target for Cancer Prevention and Treatment. J. Cancer Prev 2017, 22 (4), 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Cao X; Jin X; Liu B The Involvement of Stress Granules in Aging and Aging-Associated Diseases. Aging Cell 2020, 19 (4), e13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).An H; de Meritens CR; Shelkovnikova TA Connecting the “Dots”: RNP Granule Network in Health and Disease. Biochim. Biophys. Acta BBA - Mol. Cell Res 2021, 1868 (8), 119058. [DOI] [PubMed] [Google Scholar]
- (29).Markmiller S; Soltanieh S; Server KL; Mak R; Jin W; Fang MY; Luo E-C; Krach F; Yang D; Sen A; Fulzele A; Wozniak JM; Gonzalez DJ; Kankel MW; Gao F-B; Bennett EJ; Lécuyer E; Yeo GW Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172 (3), 590–604.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ramanathan M; Porter DF; Khavari PA Methods to Study RNA–Protein Interactions. Nat. Methods 2019, 16 (3), 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Hafner M; Katsantoni M; Köster T; Marks J; Mukherjee J; Staiger D; Ule J; Zavolan M CLIP and Complementary Methods. Nat. Rev. Methods Primer 2021, 1 (1), 20. [Google Scholar]
- (32).Zheng JJ; Agus JK; Hong BV; Tang X; Rhodes CH; Houts HE; Zhu C; Kang JW; Wong M; Xie Y; et al. Isolation of HDL by Sequential Flotation Ultracentrifugation Followed by Size Exclusion Chromatography Reveals Size-Based Enrichment of HDL-Associated Proteins. Sci. Rep 2021, 11 (1), 16086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhang J; Nguyen LT; Hickey R; Walters N; Wang X; Kwak KJ; Lee LJ; Palmer AF; Reátegui E Immunomagnetic Sequential Ultrafiltration (ISUF) Platform for Enrichment and Purification of Extracellular Vesicles from Biofluids. Sci. Rep 2021, 11 (1), 8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Li X; Corbett AL; Taatizadeh E; Tasnim N; Little JP; Garnis C; Daugaard M; Guns E; Hoorfar M; Li ITS Challenges and Opportunities in Exosome Research—Perspectives from Biology, Engineering, and Cancer Therapy. APL Bioeng. 2019, 3 (1), 011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhang H; Freitas D; Kim HS; Fabijanic K; Li Z; Chen H; Mark MT; Molina H; Martin AB; Bojmar L; et al. Identification of Distinct Nanoparticles and Subsets of Extracellular Vesicles by Asymmetric Flow Field-Flow Fractionation. Nat. Cell Biol 2018, 20 (3), 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zhang H; Lyden D Asymmetric-Flow Field-Flow Fractionation Technology for Exomere and Small Extracellular Vesicle Separation and Characterization. Nat. Protoc 2019, 14 (4), 1027–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wunsch BH; Smith JT; Gifford SM; Wang C; Brink M; Bruce RL; Austin RH; Stolovitzky G; Astier Y Nanoscale Lateral Displacement Arrays for the Separation of Exosomes and Colloids down to 20 Nm. Nat. Nanotechnol 2016, 11 (11), 936–940. [DOI] [PubMed] [Google Scholar]
- (38).Wu M; Chen C; Wang Z; Bachman H; Ouyang Y; Huang P-H; Sadovsky Y; Huang TJ Separating Extracellular Vesicles and Lipoproteins via Acoustofluidics. Lab. Chip 2019, 19 (7), 1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hattori Y; Shimada T; Yasui T; Kaji N; Baba Y Micro-and Nanopillar Chips for Continuous Separation of Extracellular Vesicles. Anal. Chem 2019, 91 (10), 6514–6521. [DOI] [PubMed] [Google Scholar]
- (40).Gámez-Valero A; Monguió-Tortajada M; Carreras-Planella L; Franquesa M; Beyer K; Borràs FE Size-Exclusion Chromatography-Based Isolation Minimally Alters Extracellular Vesicles’ Characteristics Compared to Precipitating Agents. Sci. Rep 2016, 6 (1), 33641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Kohlheyer D; Eijkel JC; Schlautmann S; Van Den Berg A; Schasfoort RB Microfluidic High-Resolution Free-Flow Isoelectric Focusing. Anal. Chem 2007, 79 (21), 8190–8198. [DOI] [PubMed] [Google Scholar]
- (42).Jezierski S; Belder D; Nagl S Microfluidic Free-Flow Electrophoresis Chips with an Integrated Fluorescent Sensor Layer for Real Time PH Imaging in Isoelectric Focusing. Chem. Commun 2013, 49 (9), 904–906. [DOI] [PubMed] [Google Scholar]
- (43).Cheng L-J; Chang H-C Microscale PH Regulation by Splitting Water. Biomicrofluidics 2011, 5 (4), 046502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Schaumburg F; Urteaga R; Kler PA; Berli CL Design Keys for Paper-Based Concentration Gradient Generators. J. Chromatogr. A 2018, 1561, 83–91. [DOI] [PubMed] [Google Scholar]
- (45).Yoo K; Shim J; Liu J; Dutta P Mathematical and Numerical Model to Study Two-Dimensional Free Flow Isoelectric Focusing. Biomicrofluidics 2014, 8 (3), 034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Rader DJ; Hovingh GK HDL and Cardiovascular Disease. Lancet 2014, 384 (9943), 618–625. [DOI] [PubMed] [Google Scholar]
- (47).Fernandez ML; Webb D The LDL to HDL Cholesterol Ratio as a Valuable Tool to Evaluate Coronary Heart Disease Risk. J. Am. Coll. Nutr 2008, 27 (1), 1–5. [DOI] [PubMed] [Google Scholar]
- (48).Linsel-Nitschke P; Tall AR HDL as a Target in the Treatment of Atherosclerotic Cardiovascular Disease. Nat. Rev. Drug Discovery 2005, 4 (3), 193–205. [DOI] [PubMed] [Google Scholar]
- (49).Vickers KC; Palmisano BT; Shoucri BM; Shamburek RD; Remaley AT MicroRNAs Are Transported in Plasma and Delivered to Recipient Cells by High-Density Lipoproteins. Nat. Cell Biol 2011, 13 (4), 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Goldstein JL; Brown MS A Century of Cholesterol and Coronaries: From Plaques to Genes to Statins. Cell 2015, 161 (1), 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Gofman JW; Lindgren F; Elliott H; Mantz W; Hewitt J; Strisower B; Herring V; Lyon TP The Role of Lipids and Lipoproteins in Atherosclerosis. Science 1950, 111 (2877), 166–186. [DOI] [PubMed] [Google Scholar]
- (52).Zhang Y; Deng Z; Lou D; Wang Y; Wang R; Hu R; Zhang X; Zhu Q; Chen Y; Liu F High-Efficiency Separation of Extracellular Vesicles from Lipoproteins in Plasma by Agarose Gel Electrophoresis. Anal. Chem 2020, 92 (11), 7493–7499. [DOI] [PubMed] [Google Scholar]
- (53).Chistiakov DA; Orekhov AN; Bobryshev YV ApoA1 and ApoA1-Specific Self-Antibodies in Cardiovascular Disease. Lab. Invest 2016, 96 (7), 708–718. [DOI] [PubMed] [Google Scholar]
- (54).Packard CJ; Demant T; Stewart JP; Bedford D; Caslake MJ; Schwertfeger G; Bedynek A; Shepherd J; Seidel D Apolipoprotein B Metabolism and the Distribution of VLDL and LDL Subfractions. J. Lipid Res 2000, 41 (2), 305–317. [PubMed] [Google Scholar]
- (55).Sadik N; Cruz L; Gurtner A; Rodosthenous RS; Dusoswa SA; Ziegler O; Solinge TSV; Wei Z; Salvador-Garicano AM; Gyorgy B Extracellular RNAs: A New Awareness of Old Perspectives. Extracellular RNA; Springer: New York, 2018; pp 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Zhao C; Sun X; Li L Biogenesis and Function of Extracellular MiRNAs. ExRNA 2019, 1 (1), 1–9.34171007 [Google Scholar]
- (57).Yuana Y; Koning RI; Kuil ME; Rensen PC; Koster AJ; Bertina RM; Osanto S Cryo-Electron Microscopy of Extracellular Vesicles in Fresh Plasma. J. Extracell. Vesicles 2013, 2 (1), 21494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Sódar BW; Kittel Á; Pálóczi K; Vukman KV; Osteikoetxea X; Szabó-Taylor K; Németh A; Sperlágh B; Baranyai T; Giricz Z; et al. Low-Density Lipoprotein Mimics Blood Plasma-Derived Exosomes and Microvesicles during Isolation and Detection. Sci. Rep 2016, 6 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Berg S; Kutra D; Kroeger T; Straehle CN; Kausler BX; Haubold C; Schiegg M; Ales J; Beier T; Rudy M; et al. Ilastik: Interactive Machine Learning for (Bio) Image Analysis. Nat. Methods 2019, 16 (12), 1226–1232. [DOI] [PubMed] [Google Scholar]
- (60).Li G; Su H; Ma N; Zheng G; Kuhn U; Li M; Klimach T; Pöschl U; Cheng Y Multifactor Colorimetric Analysis on PH-Indicator Papers: An Optimized Approach for Direct Determination of Ambient Aerosol PH. Atmospheric Meas. Technol 2020, 13 (11), 6053–6065. [Google Scholar]
- (61).Chen Y; Zhu Q; Cheng L; Wang Y; Li M; Yang Q; Hu L; Lou D; Li J; Dong X; Lee LP; Liu F Exosome Detection via the Ultrafast-Isolation System: EXODUS. Nat. Methods 2021, 18, 212–218. [DOI] [PubMed] [Google Scholar]
- (62).Kanwar SS; Dunlay CJ; Simeone DM; Nagrath S Microfluidic Device (ExoChip) for on-Chip Isolation, Quantification and Characterization of Circulating Exosomes. Lab. Chip 2014, 14 (11), 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Xu R; Rai A; Chen M; Suwakulsiri W; Greening DW; Simpson RJ Extracellular Vesicles in Cancer—Implications for Future Improvements in Cancer Care. Nat. Rev. Clin. Oncol 2018, 15 (10), 617–638. [DOI] [PubMed] [Google Scholar]
- (64).Minciacchi VR; Zijlstra A; Rubin MA; Di Vizio D Extracellular Vesicles for Liquid Biopsy in Prostate Cancer: Where Are We and Where Are We Headed? Prostate Cancer Prostatic Dis. 2017, 20 (3), 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).van Riel D; de Wit E Next-Generation Vaccine Platforms for COVID-19. Nat. Mater 2020, 19 (8), 810–812. [DOI] [PubMed] [Google Scholar]
- (66).Luan X; Sansanaphongpricha K; Myers I; Chen H; Yuan H; Sun D Engineering Exosomesas Refined Biological Nanoplatforms for Drug Delivery. Acta Pharmacol. Sin 2017, 38 (6), 754–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Chen G; Xu T; Yan Y; Zhou Y; Jiang Y; Melcher K; Xu HE Amyloid Beta: Structure, Biology and Structure-Based Therapeutic Development. Acta Pharmacol. Sin 2017, 38 (9), 1205–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Levin A; Hakala TA; Schnaider L; Bernardes GJL; Gazit E; Knowles TPJ Biomimetic Peptide Self-Assembly for Functional Materials. Nat. Rev. Chem 2020, 4 (11), 615–634. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


