Abstract
Adenovirus type 5 (Ad5) is one of the most promising vectors for gene therapy applications. Genetic engineering of Ad5 capsid proteins has been employed to redirect vector tropism, to enhance infectivity, or to circumvent preexisting host immunity. As the most abundant capsid protein, hexon modification is particularly attractive. However, genetic modification of hexon often results in failure of rescuing viable viruses. Since hypervariable regions (HVRs) are nonconserved among hexons of different serotypes, we investigated whether the HVRs could be used for genetic modification of hexon by incorporating oligonucleotides encoding six histidine residues (His6) into different HVRs in the Ad5 genome. The modified viruses were successfully rescued, and the yields of viral production were similar to that of unmodified Ad5. A thermostability assay suggested the modified viruses were stable. The His6 epitopes were expressed in all modified hexon proteins as assessed by Western blotting assay, although the intensity of the reactive bands varied. In addition, we examined the binding activity of anti-His tag antibody to the intact virions with the enzyme-linked immunosorbent assay and found the His6 epitopes incorporated in HVR2 and HVR5 could bind to anti-His tag antibody. This suggested the His6 epitopes in HVR2 and HVR5 were exposed on virion surfaces. Finally, we examined the infectivities of the modified Ad vectors. The His6 epitopes did not affect the native infectivity of Ad5 vectors. In addition, the His6 epitopes did not appear to mediate His6-dependent viral infection, as assessed in two His6 artificial receptor systems. Our study provided valuable information for studies involving hexon modification.
The adenovirus type 5 (Ad5)-based gene delivery system has shown great potential for gene therapy applications because Ad5 viruses have a minor pathological effect on humans, can efficiently infect a wide variety of cell types, and can be readily generated to high titers in vitro (3, 26, 42). However, several problems need to be solved before Ad5 vectors become effective in clinics. For example, the receptor for Ad5, coxsackievirus and adenovirus receptor (CAR), is expressed in many cells; thus, infection of Ad5 vectors is not specific to target cells (4, 35). On the other hand, some target tissue express a very low level of CAR and thus are difficult to infect (7, 18, 44). In addition, some patients have preexisting neutralizing antibodies against Ad5 resulting from natural adenovirus infection or previously administered Ad5 vectors (8, 17, 36, 38, 49, 50, 52). This preexisting immunity can prevent subsequent Ad5 infections.
One strategy to address these problems is capsid modification. The capsid of each Ad virion is composed of three major proteins: hexon, fiber, and penton base. Infection of cells by Ad5 is mediated by attachment of fiber protein to the cell surface receptors, CAR (4, 22, 34), followed by interaction of the penton base with αv integrins, which triggers the internalization of the viruses (2, 27, 43). Based on this knowledge, fiber modification has been employed to redirect Ad5 infection by incorporating extra targeting motifs. For example, RGD and polylysine motifs have been incorporated into fiber, and the modified vectors demonstrated CAR-independent Ad5 infection (10, 19, 44, 45, 48). As the largest and most abundant capsid protein, hexon is found to be the major target for host neutralizing antibodies against Ad5 (14-16, 25, 31, 46, 47). Therefore, hexon modification may be used to circumvent the host's preexisting immunity. In this regard, gene therapy investigators have sought both nongenetic and genetic strategies (29).
The nongenetic strategy is to mask the antigens on hexon surface with protective polymers such as polyethylene glycol (PEG), polylactic glycolic acid (PLGA) microspheres, or lipids by in vitro chemical coupling methods (21, 23, 24, 28). These approaches require additional procedures, and their efficacy and safety remain to be tested. The genetic strategy is to modify the hexon gene so that the expressed hexon protein cannot be recognized by the preexisting neutralizing antibodies. For example, since neutralizing antibodies against adenoviruses have serotype specificity and are mostly targeted on the major capsid protein hexon, replacement of Ad5 hexon with hexons from different serotypes may accomplish this goal. Indeed, studies have shown that Ad5 vectors bearing hexons from Ad1, Ad2, Ad3, Ad6, and Ad12 can escape the host's preexisting immunity against Ad5 (16, 31, 47, 51). In addition, Crompton et al. have genetically incorporated an eight-amino-acid sequence from the major antigenic site in the VP1 capsid protein of poliovirus type 3 into the Ad2 hexon and have discovered that this modification can alter the immune response to this virus (6). Hexon modification, however, often results in failure of rescuing viruses or poorly growing viruses (6, 16, 31, 47, 51), suggesting hexon modification may interfere with viral formation. Therefore, it will be very useful to know what regions of hexon can be modified without affecting viability of the Ad5 viruses.
The hexon monomer of Ad5 is composed of 952 amino acid residues. Electron microscopy and X-ray crystallography of human Ad2 and Ad5 hexon revealed that hexon has a dense pedestal base formed by two eight-stranded, antiparallel β barrels stabilized by an internal loop (L3). Three other loops, L1, L2, and L4, project away from the surface of the virion (1, 30, 32, 33, 39, 40). Analysis of the protein sequences of different hexons revealed that, in addition to the conserved regions, there are seven discrete hypervariable regions (HVRs), and these HVRs do not appear to be involved in binding any other viral proteins (5). In addition, previous studies have found that the HVRs of hexon contain the serotype-specific epitopes (5, 32). Therefore, genetic modification of these regions seems to be very promising in circumventing the host immunization response against Ad5. Furthermore, since HVRs are mostly located within the extruding domains of the hexon, genetic incorporation of foreign targeting motifs into these HVRs may allow CAR-independent viral infection. Since hexon has more copies than fiber in the capsid, hexon modification-mediated targeting may be more profound than fiber modification.
In this study, we tested whether foreign peptides can be incorporated into HVRs without affecting the normal function of Ad5 as a gene therapy vector. We genetically incorporated six-histidine (His6) epitopes into different HVRs of Ad5 hexon and characterized the viruses in detail. Our study provided valuable information for Ad5-based gene therapy applications involving hexon modification.
MATERIALS AND METHODS
Antibodies.
Goat polyclonal antibody against Ad2 hexon AB1056, and mouse anti-His-Tag monoclonal antibody (MAB3112) were purchased from Chemicon International, Inc. Alkaline phosphatase (AP)-conjugated donkey anti-goat and goat anti-mouse secondary antibodies were purchased from Jackson ImmunoResearch Laboratories.
Cells and cell culture.
The human embryonic kidney cell line 293, transformed with Ad5-E1 DNA, was purchased from Microbix (Toronto, Ontario, Canada). Human cervix carcinoma HeLa cells and human glioma U118MG cells were obtained from the American Type Culture Collection (ATCC). U118MG.HissFv.Rec cells were obtained by stably transfecting U118MG cells with single-chain antibody against His tag (12). 293, U118MG, and U118MG.HissFv.Rec cells were cultured in Dulbecco's modified Eagle's medium-Ham's F-12 (DMEM/F-12) containing 10% fetal calf serum (FCS) and 2 mM l-glutamine. HeLa cells were grown in Eagle's minimum essential medium (E-MEM) containing 10% FCS and 2 mM l-glutamine. All of the cells were cultured at 37°C in a 5% CO2 atmosphere.
Construction of adenoviral vectors.
The His6 epitope composed of six histidine residues and flanking Lys-Gly-Ser spacers were genetically incorporated into hexon HVRs at positions marked in Fig. 1. To accomplish this genetic modification, we first obtained hexon fragments containing sequences encoding the His6 epitope in different HVRs via three-step PCR. For instance, to obtain a His6 insertion in HVR2 (HVR2-His6), using Ad5 hexon as template, we first amplified fragment 2L (left to the HVR2 insertion) with primers L (Dra III) and HVR2-His6 (antisense), and fragment 2R (right to the HVR2 insertion) with primers HVR2-His6 (sense) and R (SacI) (Table 1). After purification of fragments 2L and 2R, 25 to 50 ng of each fragment (equal molar ratio) was mixed and used as a template and primers for the second step of PCR, resulting in insertion of sequences encoding His6 and the spacers in HVR2. Next, primers L and R were added into the tubes, and a third step of PCR was used to amplify the HVR2-His6 fragment. Other insertions were obtained in the same way with corresponding HVR-His6 primers, which include HVR3-His6 (antisense), HVR3-His6 (sense), HVR5-His6 (antisense), HVR5-His6 (sense), HVR6-His6 (antisense), HVR6-His6 (sense), HVR7a-His6 (antisense), HVR7a-His6 (sense), HVR7b-His6 (antisense), HVR7b-His6 (sense) (Table 1).
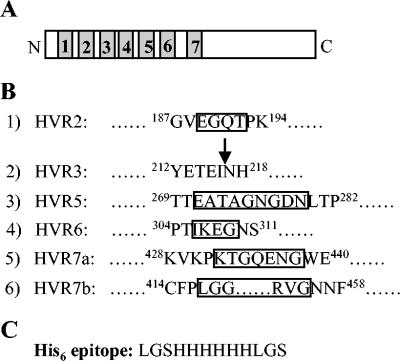
FIG. 1.
Diagram of the His6 incorporation sites in Ad5 hexon. (A) Domain structure of hexon. HVRs (░⃞) are numbered as 1 to 7. (B) Sites in the HVRs that were modified with His6 motif. The amino acid residues marked in the rectangles were replaced with the His6 motif. For HVR3, the arrow marks the position where the His6 motif was inserted. The numbers show the positions of the amino acid residues in the Ad5 hexon. (C) Amino acid residues of the His6 motif that was incorporated into HVRs of Ad5 hexon. LGS sequences flanking the six histidines were included as spacer.
TABLE 1.
Primers used to incorporate the His6 epitope into the HVRs of the Ad5 hexon
| Namea | Sequenceb |
|---|---|
| L (DraIII) | CCTACGCACGACGTGACCACAG |
| R (SacI) | CTAGGGAGCTCTGCAGAACCATG |
| HVR2-His6 (as) | TGAACCTAGGTGATGGTGATGGTGATGGGATCCGAGGACACCTATTTGAATACCCTCCTTTG |
| HVR2-His6 (s) | CTCGGATCCCATCACCATCACCATCACCTAGGTTCACCTAAATATGCCGATAAAACATTTC |
| HVR3-His6 (as) | TGAACCTAGGTGATGGTGATGGTGATGGGATCCGAGTTCGTACCACTGAGATTCTCCTAT |
| HVR3-His6 (s) | CTCGGATCCCATCACCATCACCATCACCTAGGTTCAACTGAAATTAATCATGCAGCTGGG |
| HVR5-His6 (as) | TGAACCTAGGTGATGGTGATGGTGATGGGATCCGAGAGTAGTTGAGAAAAATTGCATTTCC |
| HVR5-His6 (s) | CTCGGATCCCATCACCATCACCATCACCTAGGTTCATTGACTCCTAAAGTGGTATTGTAC |
| HVR6-His6 (as) | TGAACCTAGGTGATGGTGATGGTGATGGGATCCGAGAGTGGGCATGTAAGAAATATGAGTG |
| HVR6-His6 (s) | CTCGGATCCCATCACCATCACCATCACCTAGGTTCAAACTCACGAGAACTAATGGGCC |
| HVR7a-His6 (as) | TGAACCTAGGTGATGGTGATGGTGATGGGATCCGAGAGGTTTTACCTTGGTAAGAGTCTC |
| HVR7a-His6 (s) | CTCGGATCCCATCACCATCACCATCACCTAGGTTCATGGGAAAAAGATGCTACAGAATTTTC |
| HVR7b-His6 (as) | TGAACCTAGGTGATGGTGATGGTGATGGGATCCGAGTGGAAAGCAGTAATTTGGAAGTTC |
| HVR7b-His6 (s) | CTCGGATCCCATCACCATCACCATCACCTAGGTTCAAATAATTTTGCCATGGAAATCAATCTA |
as, antisense; s, sense.
Underlined letters represent the sequences encoding the His6 epitope.
The hexon fragments containing His6 epitope obtained above were purified and subcloned into Ad5 hexon shuttle vector H5/pH5S (described in reference 47) with DraIII and SacI. The resultant shuttle plasmids were named HVR2-His6/pH5S, HVR3-His6/pH5S, HVR5-His6/pH5S, HVR6-His6/pH5S, HVR7a-His6/pH5S, and HVR7b-His6/pH5S, respectively. To create Ad5 vector containing His6 epitopes in the HVRs of hexon, these plasmids were digested with EcoRI and PmeI, and the fragments containing the homologous recombination regions and the hexon genes were purified, then recombined with a SwaI-digested backbone Ad5 vector that lacks the hexon gene, pAd5/ΔH5 (47) in Escherichia coli BJ5183. The resultant clones were designated pAd5/HVR2-His6, pAd5/HVR3-His6, pAd5/HVR5-His6, pAd5/HVR6-His6, pAd5/HVR7a-His6, and pAd5/HVR7b-His6, all of which contain the green fluorescence protein (GFP) gene and firefly luciferase (Luc) gene in the E1 region. The constructs were confirmed by restriction digestions and sequencing.
To rescue viruses, these modified plasmids were digested with PacI, and 2 μg of each purified DNA was transfected into the Ad-E1-expressing 293 cells grown in 60-mm-diameter dishes with Superfect (QIAGEN). After plaques were formed, they were processed for large-scale proliferation in 293 cells, followed by purification by CsCl gradient centrifugation (47).
SDS-PAGE and Western blotting.
A total of 1010 virus particles (VPs) of each CsCl-purified virus were dissolved in Laemmli sample buffer without boiling and separated on 4 to 15% polyacrylamide gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. The gels were either stained with Gelcode blue stain reagent (Pierce) according to the protocol from the manufacturer or transferred to nitrocellulose membrane (Bio-Rad). The membrane was processed to Western blotting with either anti-His-Tag monoclonal antibody or antihexon polyclonal antibody, essentially as described previously (47). In the Western blotting assay of the artificial receptor, cell extracts from both the transient and stable systems were boiled in Laemmli sample buffer, separated by SDS-PAGE on 4 to 15% polyacrylamide gradient gels, and stained with anti-hemagglutinin (HA) antibody.
ELISA.
The enzyme-linked immunosorbent assay (ELISA) binding assay was performed essentially as described previously (48). In brief, different amounts of viruses ranging from 4 × 106 to 9 × 109 VPs were immobilized on wells of a 96-well plate (Nunc Maxisorp) by overnight incubation in (per well) 100 μl of 100 mM carbonate buffer (pH 9.5) at 4°C. After extensive washes with 0.05% Tween 20 in Tris-buffered saline (TBS) and blocking with blocking solution (2% bovine serum albumin and 0.05% Tween 20 in TBS), the immobilized viruses were incubated with anti-His tag monoclonal antibody (Chemicon) for 2 h at room temperature, followed by AP-conjugated goat anti-mouse antibody incubation. Color reaction was performed with p-nitrophenyl phosphate (Sigma) as recommended by the manufacturer, and optical density at 405 nm (OD405) was obtained with a microplate reader (Molecular Devices).
Gene transfer assay. (i) Gene transfer assay in HeLa cells, U118MG cells, and U118MG.HissFv.Rec cells.
Gene transfer efficacy of the viruses was evaluated by luciferase activity essentially as described previously (47). In brief, HeLa, U118MG, and U118MG.HissFv.Rec cells were plated in 24-well plates at a density of 105 cells per well the day before infection. Cells were infected at multiplicities of infection (MOI) of 1, 10, and 100 VPs/cell in triplicates. Twenty-four hours later, the cells were lysed in (per well) 250 μl of reporter lysis buffer (Promega), followed by one freeze/thaw cycle. Five microliters of each sample was used to measure the luciferase activity with a luciferase assay kit (Promega) and a luminometer (Berthold, Gaithersburg, Md.).
(ii) Gene transfer assay in transient artificial system.
To establish a transient system expressing an artificial receptor (AR) for His-Tag, U118MG cells were infected with Ad5.MK.AR, which encodes anti-His-Tag single-chain antibody, the AR for His tag, in the E1 region under the control of a cytomegalovirus (CMV) promoter with an MOI of 300, and cultured for 3 days to allow the AR to express. A vector with E1 deleted, Ad5.E1dd, was used as a control. These cells were then infected with the His6-containing viruses at an MOI of 100, and their gene transfer efficacy was measured 24 h later as described above.
Thermostability assay.
To test the thermostability of these viruses, viruses equivalent to an MOI of 100 were diluted in E-MEM containing 2% FCS and incubated at 45°C for different time intervals before infecting HeLa cells (11). Luciferase activity in infected cells was analyzed 24 h postinfection as described above.
RESULTS
Construction of hexon-modified viruses.
The HVRs of hexon are less conservative among different serotypes of adenoviruses and do not appear to interact with other viral proteins. Therefore, hexon modification may be made at these regions without affecting virus viability. To test this hypothesis, we explored the ability of the HVRs to accommodate foreign motifs. According to the X-ray crystallographic structure of adenovirus hexon, HVR1 and HVR4 contain internal β-sheet structure and are without a major ectodomain as predicted. In contrast, HVR2, HVR3, HVR5, HVR6, and HVR7 extrude from the capsid surface and contain flexible loop structures. We therefore focused on these HVRs and incorporated a His6 epitope into each of them (Fig. 1). Two versions of HVR7 modification were created because HVR7 contains a large loop. Thus, we replaced a small fragment (HVR7a) and a large fragment (HVR7b) in HVR7. The His6 epitope was chosen as a marker because antibody against His6 is commercially available, which facilitates detection of the genetic incorporation and its effect. The amino acid residues marked in the box were replaced with the His6 motif LGSHHHHHHLGS (Fig. 1), in which the LGS sequences acted as a spacer to present the six histidines (41). Genetic incorporation of the His6 motif into Ad5 hexon was achieved by bacterium-based homologous recombination. The resultant HVR-His6-containing vectors encoded luciferase and GFP double reporters and were replication deficient. In the study, all of the viruses but Ad5/HVR7b-His6 were successfully rescued in 293 cells stably expressing Ad5-E1 proteins. The inability to rescue Ad5/HVR7b-His6 suggested that replacement of a large fragment of HVR7 interfered with viral formation. This will be discussed in detail later. Nonetheless, incorporation of His6 in other HVRs resulted in similar virus yields to unmodified viruses, which ranged from 3 × 1012 to 5 × 1012 VPs in a culture of 3 × 108 cells, suggesting that the hexon modifications did not affect virus formation and the HVRs were able to accommodate foreign peptides.
Expression and presentation of His6 epitopes in hexons of modified viruses.
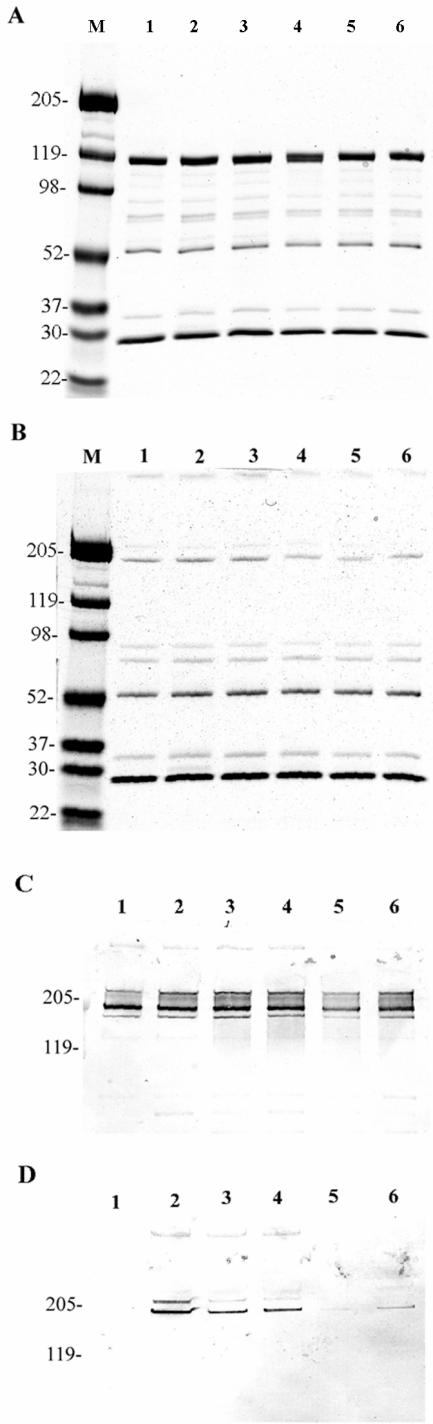
Next we examined whether the incorporated His6 epitopes were indeed expressed in hexon proteins and how they were presented. In this regard, we performed SDS-PAGE and Western blotting analysis after lysis of purified virions in Laemmli sample buffer. Since several antihexon antibodies that we tested before do not recognize denatured (boiled) hexon protein (47), the viral samples for Western blotting assays were not boiled. In the experiments, 1010 VPs per well were lysed and subjected to SDS-PAGE. The gels were either stained with Gelcode blue stain reagent (Pierce) or transferred to nitrocellulose membrane for Western blotting. Gelcode blue staining demonstrated that the modified viruses contained proper viral proteins, as compared to the unmodified Ad5, and the quantitation of the viruses is consistent (Fig. 2A and B). Western blotting assays with antihexon and anti-His tag antibody suggested the His6 epitopes were indeed expressed in hexons of modified viruses but not in unmodified Ad5 hexon (Fig. 2C and D). However, His6 epitopes incorporated in different HVRs exhibited differential antibody accessibility in the hexon protein, as reflected by the intensity of the bands in the anti-His tag Western blotting assay. Hexons of Ad5/HVR2-His6, Ad5/HVR3-His6, and Ad5/HVR5-His6 appeared to be readily accessible to anti-His tag antibody, while hexons of Ad5/HVR6-His6 and Ad5/HVR7a-His6 were much less accessible. These data indicated that the His6 epitopes inserted in HVR2, HVR3, and HVR5 were better presented on the surface of hexon protein than in HVR6 and HVR7.
FIG. 2.
SDS-PAGE and Western blotting confirmed the HVR-His6-containing viruses. In the assays, 1010 VPs of purified viruses were lysed in Laemmli sample buffer, followed by boiling for 5 min (A) or without boiling (B to D), and separated on 4 to 15% polyacrylamide gradient SDS-PAGE gels. The proteins were either stained with Gelcode blue stain reagent (Pierce) (A and B) or transferred to nitrocellulose membrane and then stained with anti-hexon polyclonal antibody AB1056 (C) or anti-His tag monoclonal antibody (D). Lane M, molecular weight marker; lane 1, Ad5 (unmodified); lane 2, Ad5/HVR2-His6; lane 3, Ad5/HVR3-His6; lane 4, Ad5/HVR5-His6; lane 5, Ad5/HVR6-His6; lane 6, Ad5/HVR7a-His6. The weak anti-His tag staining of hexons of Ad5/HVR6-His6 and Ad5/HVR7a-His6 suggested that the His6 epitopes incorporated in HVR6 and HVR7a were not well accessible to the anti-His antibody at the protein level.
Presentation of His6 epitope on virion surface.
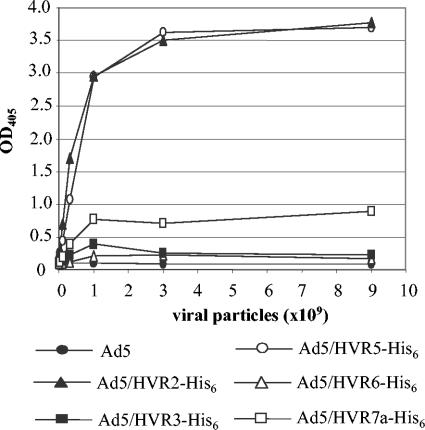
Exposure of an epitope on the surface of a protein does not necessarily mean it is exposed on an intact virion. Therefore, we next examined whether the His6 epitopes were presented on the surface of virions using an ELISA. In the assay, different amount of purified viruses were immobilized on the wells of ELISA plates and incubated with anti-His tag antibody. The binding of anti-His antibody to the intact virions was detected with AP-conjugated secondary antibody, followed by AP color reaction. The results showed that Ad5/HVR2-His6 and Ad5/HVR5-His6 bound anti-His tag antibody most strongly and Ad5/HVR7-His6 showed weak binding, while Ad5/HVR3-His6 and Ad5/HVR6-His6 showed minimal binding (similar to unmodified Ad5) (Fig. 3). This suggested that the His6 epitopes were exposed on the virion surfaces when incorporated in HVR2 and HVR5, partially exposed in HVR7, while not exposed in HVR3 and HVR6. As noted, the His6 epitope in HVR3 was readily accessible to anti-His tag antibody at the protein level but not in the context of intact virion. The discrepancy may be due to the complicated surroundings of the epitopes in the virions. In particular, hexon interacts with many other proteins in a virion, which include polypeptides VI, VIII, and IX (13, 37). These interactions can affect the exposure of the His6 epitopes by affecting the conformation of hexon or by simply covering the His6 epitopes.
FIG. 3.
Results of an ELISA binding assay suggesting that the His6 epitopes incorporated in different HVRs exhibited differential accessibility to anti-His tag antibody in the context of intact virion. In the assay, various amounts of purified viruses were immobilized on the wells of 96-well ELISA plates and incubated with anti-His tag antibody. The binding was detected with AP-conjugated secondary antibody. The results suggested that the His6 epitopes incorporated into HVR2 and HVR5 were accessible to anti-His tag antibody at the virion level, indicating they were exposed on the virion surface.
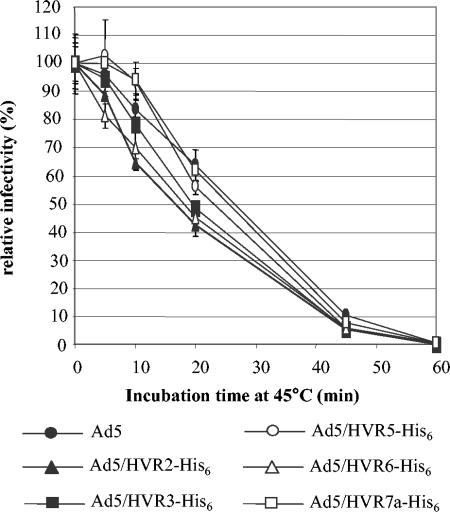
Thermostability of the modified viruses.
One problem often associated with capsid modification of adenoviruses is instability of the modified viruses. This is especially evident with hexon modification, since hexon constitutes the major part of adenovirus capsid (16, 31, 47, 51). We therefore tested whether incorporation of His6 epitopes in HVRs could affect the structural integrity of the virions by comparing their thermostability with that of unmodified Ad5. The viruses were incubated at 45°C in infection media (E-MEM containing 2% FCS) for different time intervals before infecting HeLa cells, and their relative gene transfer efficiencies were obtained by comparing to that of unheated viruses with luciferase reporter. We found that all of the modified viruses showed similar stability to the unmodified viruses (Fig. 4). Although Ad5/HVR2-His6 and Ad5/HVR6-His6 appeared to be less stable than unmodified and other modified viruses, the differences were not statistically significant. This suggested that incorporation of His6 epitopes in HVRs of hexon did not significantly affect the stability of Ad5, consistent with our results on virus production showing the modified viruses had similar yields to unmodified Ad5.
FIG. 4.
Thermostability of HVR-His6-containing viruses. The viruses equivalent to an MOI of 100 were diluted in infection media (E-MEM plus 2% FCS) and incubated at 45°C for different time intervals before infecting HeLa cells in triplicates. The cell lysates were processed for luciferase activity assay. The relative light units (RLU) showing the activity of luciferase were detected with a luciferase assay kit (Promega) and a luminometer. The relative infectivity was obtained by changing the RLU readings of the heat-treated viruses to percentages of the readings of corresponding untreated viruses.
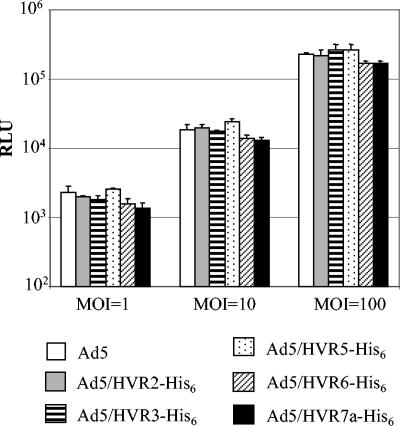
Gene transfer efficacies of the viruses in HeLa cells.
Since capsid proteins play critical roles in Ad5 infection, insertion of foreign peptide into hexon may interfere with the native infectivity of Ad5. We thus investigated whether incorporation of His6 epitopes in HVRs affected gene transfer efficacies of the viruses in HeLa cells. HeLa cells were chosen because they express a high level of CAR, the native Ad5 receptor, and can be effectively infected by unmodified Ad5 vector. Viruses equivalent to MOI of 1, 10, and 100 were used to infect the HeLa cells, and the gene transfer efficacies were assessed with the reporter firefly luciferase. All of the modified viruses exhibited similar luciferase activity compared to unmodified Ad5 (Fig. 5). Ad5/HVR6-His6 and Ad5/HVR7a-His6 appeared to have less efficiency than unmodified and other modified viruses, but the differences were not statistically significant (Fig. 5). These data suggested that His6 incorporation in HVRs of hexon did not interfere with native infectivity of Ad5.
FIG. 5.
Gene transfer efficacies of the HVR-His6-containing viruses in HeLa cells. In the assay, HeLa cells were infected with these viruses at three different MOI (MOI of 1, 10, and 100) in triplicate, and the cell lysates were processed for the luciferase activity assay as described in the legend to Fig. 4.
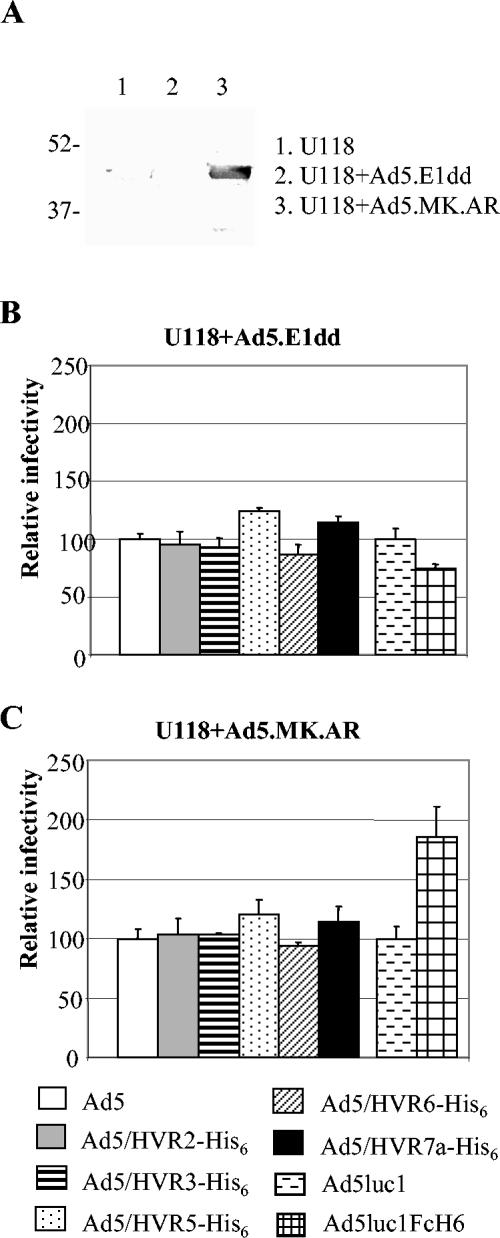
Gene transfer efficacies of the viruses in His6 AR systems.
We further investigated whether the His6 epitope incorporated in HVRs of Ad5 hexon could mediate independent viral infection. Two AR systems were employed in this regard. In the first one, we established a transient AR system by infecting CAR-negative U118MG cells with Ad5.MK.AR, an Ad5 vector encoding anti-His tag single-chain antibody (the AR for His6 epitopes) in the E1 region (12; P. J. Mahasreshti et al., unpublished data). The AR was driven by CMV promoter and was composed of an immunoglobulin κ-chain signal peptide, an HA epitope, the single-chain antibody against His6 epitope, and the transmembrane domain of platelet-derived growth factor (PDGF) recetor (12). As demonstrated previously (12), the Ig κ-chain leader directs the AR to the cell surface, the PDGF receptor transmembrane domain anchors the AR in the plasma membrane, and the HA epitope facilitates AR detection. As shown in Fig. 6A, the AR was expressed in the U118MG cells preinfected with Ad5.MK.AR, but not in uninfected cells or the cells preinfected with a negative control, Ad5.E1dd, an E1-deleted Ad5 vector. To test the gene transfer efficacies of the hexon-modified viruses in this AR system, the U118MG cells were preinfected with either Ad5.MK.AR or Ad5.E1dd and cultured at 37°C in 5% CO2 for 3 days to allow expression of the AR. Then the preinfected U118MG cells were infected with the HVR-His6-containing viruses at an MOI of 100, and their gene transfer efficacies were examined with the luciferase assay. A fiber-modified vector containing His6 motif in fiber, Ad5luc1FcH6, has been shown to be able to infect cells via the His6 AR expressed on the targeting cells (12) and thus was used as a positive control. Compared to the control vector Ad5luc1, Ad5luc1FcH6 showed enhanced gene transfer efficacy in the transient AR system, but not in the control cells, indicating the transient AR system was functional (Fig. 6B and C). In contrast, all of the hexon-modified viruses showed similar infectivity compared to unmodified Ad5 in U118MG preinfected with both Ad5.MK.AR and Ad5.E1dd (Fig. 6B and C). These data suggested that His6 epitope incorporated in the hexon HVRs did not mediate independent viral infection in the transient system.
FIG. 6.
Gene transfer efficacies of the modified viruses in a transient artificial receptor system. (A) Western blotting assay showing AR expression in Ad5.MK.AR-infected U118MG cells, but not in uninfected cells or Ad5.E1dd-infected cells. (B and C) Gene transfer efficacies of the modified viruses in the transient AR system. In the assay, U118MG cells were infected with Ad5.E1dd (B) or Ad5.MK.AR (C) at an MOI of 300. Three days later, the cells were infected with the His6-containing viruses at an MOI of 100, and their gene transfer efficacies were measured 24 h later as described in the legend to Fig. 4. A fiber-modified virus, Ad5luc1FcH6, which contains the His6 motif at the C-terminal end of fiber, and its unmodified control, Ad5luc1, were used as positive controls. The relative infectivity was obtained by changing the RLU readings of the modified viruses to a percentage of the readings of unmodified viruses. The results suggested that the His6 epitopes incorporated in HVRs did not confer the viruses' His6-mediated gene transfer ability.
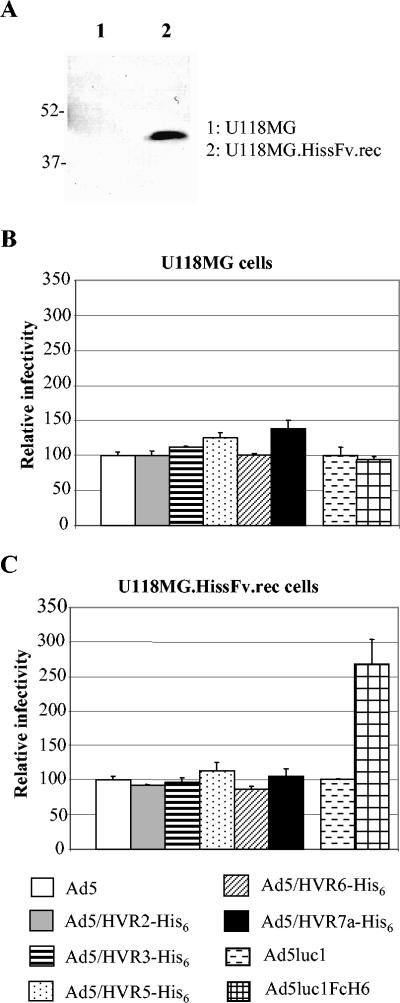
To confirm this conclusion, we employed a second AR system, the U118MG.HissFv.rec cells—cell line that stably expressed the AR for His tag on cell surface (12). Expression of the AR was confirmed by Western blotting assay with anti-HA antibody (Fig. 7A). We next infected the stable AR cells and the control U118MG cells with the hexon-modified viruses and compared their infectivity with that of unmodified Ad5 vector in both cell lines. Similarly, Ad5luc1 and Ad5luc1FcH6 were used as a positive control in this system. Consistent with studies in the transient system, hexon-modified viruses did not show enhanced gene transfer efficacy in the stable system, while the positive control did (Fig. 7B and C). Taken together, these data demonstrated that His6 epitopes incorporated in HVRs of hexon did not mediate cell infection.
FIG. 7.
Gene transfer efficacies of the hexon-modified viruses in a stable AR system. (A) Expression of the AR in the stable U118MG.HissFv.Rec cells. Cell extracts from U118MG cells or U118MG.HissFv.Rec cells were subjected to Western blotting analysis with anti-HA antibody which recognizes the HA epitope fused to the AR. (B and C) Gene transfer efficacies of the modified viruses in the stable AR system. In this assay, U118MG cells (B) and U118MG.HissFv.Rec cells (C) were infected with the modified viruses at an MOI of 100. Ad5luc1 and Ad5luc1FcH6 were used as positive controls. The cell lysates were subjected to luciferase assay, and the relative infectivity was obtained as described in the legend to Fig. 6. The data confirmed that the His6 epitopes incorporated in HVRs did not confer the viruses' His6-mediated gene transfer ability.
DISCUSSION
In this study, we identified several HVRs in Ad5 hexon as proper sites for foreign peptide incorporation, using the His6 motif as a marker. We found that the His6 motif could be genetically inserted into HVR2, HVR3, HVR5, HVR6, and HVR7 of the Ad5 hexon without affecting virus formation, virion stability, and CAR-mediated Ad5 native infectivity. However, different HVRs appeared to present the His6 epitopes differentially. The His6 epitopes incorporated in HVR2, HVR3, and HVR5 appeared to be exposed on the protein surface because they were readily detected in Western blotting assays with anti-His tag antibody. Nonetheless, in the context of intact virion, only the His6 epitopes inserted in HVR2 and HVR5 appeared to be exposed, as assessed by ELISA, suggesting protein interactions in the intact virion have some effect on epitope exposure. Furthermore, we found that in all of the modified adenovirus vectors, the His6 epitopes did not mediate CAR-independent cell infection in the presence of fiber.
This study demonstrated that HVRs of hexon can accommodate insertions of foreign peptides. The definition of HVRs used in the study is based on analysis performed by Crawford-Miksza and Schnurr (5). While we were preparing the manuscript for this report, another study performed by Rux et al. was published, in which the HVRs of hexon were redefined (33). Using high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods, Rux and his colleagues identified nine HVRs. In general, the positions of the newly defined HVRs are in agreement with what Crawford-Miksza and Schnurr have defined. The exceptions are HVR3 and HVR7. The authors assigned HVR3 to a different region, and HVR7 was defined more precisely as HVR7, HVR8, and HVR9. According to the new definition, the previously defined HVR3 is in a well-conserved region. We successfully rescued Ad5.HVR3-His6, suggesting some regions outside HVRs may also accommodate certain modifications. As noted, we were able to rescue Ad5.HVR7a-His6 viruses, but not Ad5.HVR7b-His6. This may be explained with the newly identified HVR7, HVR8, and HVR9. According to the sequence information, our HVR7a-His6 modification is in HVR8, whereas the HVR7b-His6 modification involved sequences ranging from part of HVR7 to part of HVR9. These results indicate that HVR8 is able to accommodate modifications. However, replacement of sequences ranging from HVR7 to HVR9 interferes with virion assembly since it involves the conserved sequences in this region. Our results are in agreement with an early study, in which 8-amino-acid sequences from the poliovirus type 3 VP1 capsid protein were used to replace residues 442 to 449 of the Ad2 hexon, resulting in viruses that grow poorly (6). The position is in HVR7 according to Crawford-Miksza and Schnurr's definition. However, the insertion extends between HVR7 and HVR8, which is a largely conserved region in the redefinition. Therefore, the redefinition of HVR7-HVR9 appears to explain the results better and thus may be more accurate.
Although His6 epitopes inserted into HVR2 and HVR5 are exposed on the surface of the Ad5 virion, they do not appear to mediate viral infection of cells. This was demonstrated in both transient and stable AR systems, suggesting peptides incorporated into hexon are not able to mediate cell targeting. However, a previous study has shown that RGD peptide incorporated into HVR5 is able to mediate CAR-independent cell infection (41). The controversy may arise from the nature and length of the incorporated peptides. Longer and well-protruding ligands have better accessibility to their receptors. In addition, the ligand-receptor binding affinity can also have an effect on the ligand-mediated cell infection. It is possible that the AR for the His6 epitope, the single-chain antibody against His tag, has weaker binding affinity to His6 peptide inserted in the hexon than anti-His tag antibody; thus, even though it is readily exposed on the virion surface, it cannot mediate cell infection. Furthermore, the presence of fiber, the major cell targeting protein of Ad5, may spatially block access of the incorporated ligands to the receptors expressed on the cell surface. In this regard, our future study involving employment of fiberless Ad will help clarify whether the epitopes incorporated in hexon can mediate additional cell targeting.
Since hexon is the major target for the neutralizing antibodies against adenovirus vectors, previous studies involving hexon modifications mainly focused on circumventing the host neutralization response to Ad vectors (16, 31, 47, 51). A hexon switch strategy has been employed in this regard. In particular, the hexons of Ad5 vectors were replaced with hexons from different serotypes, including Ad1, Ad2, Ad3, Ad6, and Ad12. These hexon switch modifications demonstrated their ability to escape the preexisting neutralizing antibodies when administrated sequentially (16, 31, 47, 51). However, a major problem associated with this strategy is the inability to rescue viable viruses with most alternate hexons. In this regard, hexons from more than 20 adenovirus serotypes have been investigated, but only the aforementioned five hexons were able to form viable viruses after replacement of the Ad5 hexon (16, 31, 47, 51). In addition, the yields of the hexon-chimeric Ad5 vectors are lower than that of native Ad5 vector, and they are more susceptible to heat inactivation, suggesting the foreign hexons do not encapsidate the Ad5 genome as efficiently as the native Ad5 hexon. These properties of the hexon-chimeric Ad5 vectors may result from the incompatibility of the hexons with other Ad5 capsid proteins that are involved in virion assembly (9, 37). Since HVRs of hexons contain the serotype-specific epitopes and they do not appear to be involved in binding any other viral proteins, it is speculated that switching only the HVR regions of hexon may allow better rescue of hexon chimeras while retaining their ability to circumvent the host neutralizing response. Our current study suggest that the HVRs of the Ad5 hexon can accommodate modifications without affecting viral formation and stability, thus providing support for the possibility of switching HVRs to escape host neutralization response. An alternative strategy in this regard is to genetically incorporate some ligands to mask the antigenic epitopes of HVRs in a similar way to the nongenetic strategy to overcome host immunity.
In addition to potential utility of hexon modifcations in cell targeting and in escaping host neutralization response, hexon modification may be used for other purposes, such as in vivo imaging of Ad virions. Some imaging reagents such as GFP have been incorporated in Ad5 capsid protein pIX, which allowed live imaging of the Ad5 virion (20). Hexon is the most abundant capsid protein, and each virion contains 240 copies of hexon homotrimer. Therefore, incorporation of imaging reagents in hexon may provide strong signals for live and in vivo imaging purposes. An unsolved problem, however, is the size limit for insertions in hexon. This will be investigated in our future studies.
Acknowledgments
We thank Joanne T. Douglas for providing us with U118MG.HissFv.rec cells.
This work was supported by National Institutes of Health and National Cancer Institute grants R01 HL67962, R01 CA86881, R01 CA090547, R01 AG021875, and P50 CA89019, and a research grant from the Muscular Dystrophy Association (United States).
REFERENCES
- 1.Athappilly, F. K., R. Murali, J. J. Rux, Z. Cai, and R. M. Burnett. 1994. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 A resolution. J. Mol. Biol. 242:430-455. [DOI] [PubMed] [Google Scholar]
- 2.Bai, M., B. Harfe, and P. Freimuth. 1993. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J. Virol. 67:5198-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, B. G., C. J. Crews, and J. T. Douglas. 2002. Targeted adenoviral vectors. Biochim. Biophys. Acta 1575:1-14. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 5.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crompton, J., C. I. Toogood, N. Wallis, and R. T. Hay. 1994. Expression of a foreign epitope on the surface of the adenovirus hexon. J. Gen. Virol. 75:133-139. [DOI] [PubMed] [Google Scholar]
- 7.Curiel, D. T., E. Wagner, M. Cotten, M. L. Birnstiel, S. Agarwal, C. M. Li, S. Loechel, and P. C. Hu. 1992. High-efficiency gene transfer mediated by adenovirus coupled to DNA-polylysine complexes. Hum. Gene Ther. 3:147-154. [DOI] [PubMed] [Google Scholar]
- 8.Dai, Y., E. M. Schwarz, D. Gu, W. W. Zhang, N. Sarvetnick, and I. M. Verma. 1995. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proc. Natl. Acad. Sci. USA 92:1401-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Halluin, J. C. 1995. Virus assembly. Curr. Top. Microbiol. Immunol. 199:47-66. [PubMed] [Google Scholar]
- 10.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 72:9706-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dmitriev, I. P., E. A. Kashentseva, and D. T. Curiel. 2002. Engineering of adenovirus vectors containing heterologous peptide sequences in the C terminus of capsid protein IX. J. Virol. 76:6893-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas, J. T., C. R. Miller, M. Kim, I. Dmitriev, G. Mikheeva, V. Krasnykh, and D. T. Curiel. 1999. A system for the propagation of adenoviral vectors with genetically modified receptor specificities. Nat. Biotechnol. 17:470-475. [DOI] [PubMed] [Google Scholar]
- 13.Everitt, E., B. Sundquist, U. Pettersson, and L. Philipson. 1973. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology 52:130-147. [DOI] [PubMed] [Google Scholar]
- 14.Gahéry-Ségard, H., F. Farace, D. Godfrin, J. Gaston, R. Lengagne, T. Tursz, P. Boulanger, and J.-G. Guillet. 1998. Immune response to recombinant capsid proteins of adenovirus in humans: antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J. Virol. 72:2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gall, J., A. Kass-Eisler, L. Leinwand, and E. Falck-Pedersen. 1996. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 70:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gall, J. G. D., R. G. Crystal, and E. Falck-Pedersen. 1998. Construction and characterization of hexon-chimeric adenoviruses: specification of adenovirus serotype. J. Virol. 72:10260-10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey, B.-G., N. R. Hackett, T. El-Sawy, T. K. Rosengart, E. A. Hirschowitz, M. D. Lieberman, M. L. Lesser, and R. G. Crystal. 1999. Variability of human systemic humoral immune responses to adenovirus gene transfer vectors administered to different organs. J. Virol. 73:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, M., K. R. Zinn, B. G. Barnett, L. A. Sumerel, V. Krasnykh, D. T. Curiel, and J. T. Douglas. 2002. The therapeutic efficacy of adenoviral vectors for cancer gene therapy is limited by a low level of primary adenovirus receptors on tumour cells. Eur. J. Cancer 38:1917-1926. [DOI] [PubMed] [Google Scholar]
- 19.Koizumi, N., H. Mizuguchi, N. Utoguchi, Y. Watanabe, and T. Hayakawa. 2003. Generation of fiber-modified adenovirus vectors containing heterologous peptides in both the HI loop and C terminus of the fiber knob. J. Gene Med. 5:267-276. [DOI] [PubMed] [Google Scholar]
- 20.Le, L. P., M. Everts, I. P. Dmitriev, J. G. Davydova, M. Yamamoto, and D. T. Curiel. 2004. Fluorescently labeled adenovirus with pIX-EGFP for vector detection. Mol. Imaging 3:105-116. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. G., S. J. Yoon, C. D. Kim, K. Kim, D. S. Lim, Y. I. Yeom, M. W. Sung, D. S. Heo, and N. K. Kim. 2000. Enhancement of adenoviral transduction with polycationic liposomes in vivo. Cancer Gene Ther. 7:1329-1335. [DOI] [PubMed] [Google Scholar]
- 22.Louis, N., P. Fender, A. Barge, P. Kitts, and J. Chroboczek. 1994. Cell-binding domain of adenovirus serotype 2 fiber. J. Virol. 68:4104-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews, C., G. Jenkins, J. Hilfinger, and B. Davidson. 1999. Poly-L-lysine improves gene transfer with adenovirus formulated in PLGA microspheres. Gene Ther. 6:1558-1564. [DOI] [PubMed] [Google Scholar]
- 24.Meunier-Durmort, C., R. Picart, T. Ragot, M. Perricaudet, B. Hainque, and C. Forest. 1997. Mechanism of adenovirus improvement of cationic liposome-mediated gene transfer. Biochim. Biophys. Acta 1330:8-16. [DOI] [PubMed] [Google Scholar]
- 25.Molinier-Frenkel, V., R. Lengagne, F. Gaden, S.-S. Hong, J. Choppin, H. Gahery-Ségard, P. Boulanger, and J.-G. Guillet. 2002. Adenovirus hexon protein is a potent adjuvant for activation of a cellular immune response. J. Virol. 76:127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemerow, G. R. 2000. Adenoviral vectors—new insights. Trends Microbiol. 8:391-394. [DOI] [PubMed] [Google Scholar]
- 27.Nemerow, G. R., and P. L. Stewart. 1999. Role of αv integrins in adenovirus cell entry and gene delivery. Microbiol. Mol. Biol. Rev. 63:725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Riordan, C. R., A. Lachapelle, C. Delgado, V. Parkes, S. C. Wadsworth, A. E. Smith, and G. E. Francis. 1999. PEGylation of adenovirus with retention of infectivity and protection from neutralizing antibody in vitro and in vivo. Hum. Gene Ther. 10:1349-1358. [DOI] [PubMed] [Google Scholar]
- 29.Ritter, T., M. Lehmann, and H. D. Volk. 2002. Improvements in gene therapy: averting the immune response to adenoviral vectors. Biodrugs 16:3-10. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, M. M., J. L. White, M. G. Grutter, and R. M. Burnett. 1986. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232:1148-1151. [DOI] [PubMed] [Google Scholar]
- 31.Roy, S., P. S. Shirley, A. McClelland, and M. Kaleko. 1998. Circumvention of immunity to the adenovirus major coat protein hexon. J. Virol. 72:6875-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rux, J. J., and R. M. Burnett. 2000. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol. Ther. 1:18-30. [DOI] [PubMed] [Google Scholar]
- 33.Rux, J. J., P. R. Kuser, and R. M. Burnett. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 77:9553-9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santis, G., V. Legrand, S. S. Hong, E. Davison, I. Kirby, J. L. Imler, R. W. Finberg, J. M. Bergelson, M. Mehtali, and P. Boulanger. 1999. Molecular determinants of adenovirus serotype 5 fibre binding to its cellular receptor CAR. J. Gen. Virol. 80:1519-1527. [DOI] [PubMed] [Google Scholar]
- 35.Seki, T., I. Dmitriev, K. Suzuki, E. Kashentseva, K. Takayama, M. Rots, T. Uil, H. Wu, M. Wang, and D. T. Curiel. 2002. Fiber shaft extension in combination with HI loop ligands augments infectivity for CAR-negative tumor targets but does not enhance hepatotropism in vivo. Gene Ther. 9:1101-1108. [DOI] [PubMed] [Google Scholar]
- 36.Sharpe, S., A. Fooks, J. Lee, K. Hayes, C. Clegg, and M. Cranage. 2002. Single oral immunization with replication deficient recombinant adenovirus elicits long-lived transgene-specific cellular and humoral immune responses. Virology 293:210-216. [DOI] [PubMed] [Google Scholar]
- 37.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 2111-2148. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 38.Smith, T. A., B. D. White, J. M. Gardner, M. Kaleko, and A. McClelland. 1996. Transient immunosuppression permits successful repetitive intravenous administration of an adenovirus vector. Gene Ther. 3:496-502. [PubMed] [Google Scholar]
- 39.Stewart, P. L., and R. M. Burnett. 1995. Adenovirus structure by X-ray crystallography and electron microscopy. Curr. Top. Microbiol Immunol. 199:25-38. [DOI] [PubMed] [Google Scholar]
- 40.van Oostrum, J., P. R. Smith, M. Mohraz, and R. M. Burnett. 1987. The structure of the adenovirus capsid. III. Hexon packing determined from electron micrographs of capsid fragments. J. Mol. Biol. 198:73-89. [DOI] [PubMed] [Google Scholar]
- 41.Vigne, E., I. Mahfouz, J.-F. Dedieu, A. Brie, M. Perricaudet, and P. Yeh. 1999. RGD inclusion in the hexon monomer provides adenovirus type 5-based vectors with a fiber knob-independent pathway for infection. J. Virol. 73:5156-5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vorburger, S. A., and K. K. Hunt. 2002. Adenoviral gene therapy. Oncologist 7:46-59. [DOI] [PubMed] [Google Scholar]
- 43.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 44.Wickham, T. J., P. W. Roelvink, D. E. Brough, and I. Kovesdi. 1996. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 14:1570-1573. [DOI] [PubMed] [Google Scholar]
- 45.Wickham, T. J., E. Tzeng, L. L. Shears II, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wohlfart, C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 62:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, H., I. Dmitriev, E. Kashentseva, T. Seki, M. Wang, and D. T. Curiel. 2002. Construction and characterization of adenovirus serotype 5 packaged by serotype 3 hexon. J. Virol. 76:12775-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, H., T. Seki, I. Dmitriev, T. Uil, E. Kashentseva, T. Han, and D. T. Curiel. 2002. Double modification of adenovirus fiber with RGD and polylysine motifs improves coxsackievirus-adenovirus receptor-independent gene transfer efficiency. Hum. Gene Ther. 13:1647-1653. [DOI] [PubMed] [Google Scholar]
- 49.Yang, Y., S. E. Haecker, Q. Su, and J. M. Wilson. 1996. Immunology of gene therapy with adenoviral vectors in mouse skeletal muscle. Hum. Mol. Genet. 5:1703-1712. [DOI] [PubMed] [Google Scholar]
- 50.Yang, Y., Q. Li, H. C. J. Ertl, and J. M. Wilson. 1995. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J. Virol. 69:2004-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Youil, R., T. J. Toner, Q. Su, M. Chen, A. Tang, A. J. Bett, and D. Casimiro. 2002. Hexon gene switch strategy for the generation of chimeric recombinant adenovirus. Hum. Gene Ther. 13:311-320. [DOI] [PubMed] [Google Scholar]
- 52.Zaiss, A.-K., Q. Liu, G. P. Bowen, N. C. W. Wong, J. S. Bartlett, and D. A. Muruve. 2002. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J. Virol. 76:4580-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]