Abstract
We have reported in the accompanying paper that the BFRF1 protein of Epstein-Barr virus (EBV) is important for efficient primary viral envelopment and egress (A. Farina, R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H.-J. Delecluse, J. Virol. 79:3703-3712). Here we describe the characterization of the product of the EBV BFLF2 gene, which belongs to a family of conserved herpesviral genes which include the UL31 genes of herpes simplex virus and of pseudorabies virus and whose products are known to interact with UL34, the positional homolog of BFRF1. BFLF2 is an early transcript and is expressed in a variety of cell lines upon EBV lytic cycle activation. Western blotting of purified virion preparations showed that BFLF2 is a component of intracellular virions but is absent from mature extracellular virions. Coimmunoprecipitation experiments indicated that BFLF2 interacts with BFRF1, which was confirmed by immunofluorescence confocal microscopy showing that the two proteins colocalize on the nuclear membrane not only upon cotransfection in epithelial cells but also during viral replication. In cells carrying an EBV mutant with the BFRF1 gene deleted (293-BFRF1-KO cells) BFLF2 expression was low, and it was restored to wild-type levels upon treatment of the cells with the proteasome inhibitor MG132. Furthermore, recomplementing the 293-BFRF1-KO cells by BFRF1 transfection restored BFLF2 expression to the wild-type level. In addition, when expressed alone BFLF2 was localized diffusely inside the nucleus, whereas in the presence of BFRF1 the two proteins colocalized at the nuclear rim. Finally, 293 epithelial cells transfected with either protein or cotransfected were analyzed by electron microscopy to investigate potential alterations in the morphology of the nuclear membrane. The ultrastructural analysis revealed that (i) BFRF1 caused duplications of the nuclear membrane, similar to those reported to occur during the course of herpesviral replication, and (ii) while BFLF2 alone did not cause any apparent alteration, coexpression of the two proteins dramatically induced profound convolutions of the duplicated nuclear membrane. Both biochemical and morphological analysis showed association of the BFRF1-BFLF2 complex with a component of the nuclear lamina, lamin B. Taken together, these results and those of the accompanying paper (Farina et al., J. Virol. 79:3703-3712) indicate an important role of BFRF1 and BFLF2 in the early steps of EBV maturation at the nuclear membrane.
Two conserved herpesvirus proteins, designated UL34 and UL31, of herpes simplex virus (HSV) and pseudorabies virus (PrV) are involved in the early steps of viral maturation at the nuclear envelope (reviewed in reference 26). With many similarities and a few differences, accumulating evidence indicates that these proteins and their homologs play similar roles in nuclear egress of both alpha- and betaherpesviruses (9, 17, 23, 30, 33, 34, 36, 37, 40, 48, 50). The physical interaction between the two proteins appears to be important to facilitate virion envelopment at the inner nuclear membrane, a process which probably also involves nuclear lamina disruption, to allow nucleocapsids to gain access at the interior face of the nuclear envelope (28, 39).
We initially identified and characterized the product of the Epstein-Barr virus (EBV) BFRF1 gene, which is the positional homolog of the UL34 gene (14). Based upon its intracellular localization (15) and the functional analysis of a viral mutant with BFRF1 deleted (16), we showed that BFRF1 is involved in viral envelopment and egress, and we suggested that, in analogy with other herpesviruses, it might interact with the EBV homolog of the conserved herpesviral protein UL31, encoded by the BFLF2 gene. Such an interaction has been recently demonstrated by Lake and Hutt-Fletcher (24), who reported that BFRF1 and BFLF2 colocalize in cotransfected cells and that each protein influences the intracellular localization of the other. However, that study was not performed in the context of viral replication. In the present work we confirm and extend their observations by characterizing the BFLF2 protein and showing, using a newly generated monoclonal antibody (MAb) and both during viral replication and by transient-transfection assays, that BFRF1 and BFLF2 form a stable association and colocalize on the nuclear membrane. In addition, using cells transfected with a viral mutant with BFRF1 deleted, we demonstrate that BFRF1 is necessary for BFLF2 intracellular localization, and we suggest that it might be involved in BFLF2 stabilization. Finally, we show that BFRF1, and more strikingly when coexpressed with BFLF2, causes profound morphological alterations of the nuclear membrane and binds to a nuclear lamina component, lamin B.
MATERIALS AND METHODS
Cell culture.
DG75 is a an EBV-negative B-cell line derived from a Burkitt lymphoma (BL) (3). P3HR1 is a human B-cell line derived from an EBV-positive BL that spontaneously produces EBV particles (20). Raji is a human B-cell line derived from a BL harboring a defective EBV genome that is unable to replicate viral DNA and express late viral gene (31). B95-8 is a marmoset B-cell line transformed with EBV in which approximately 5% of cells support spontaneous viral replication (27). The 293 cell line is a human embryonic epithelial kidney cell line that has been transformed by the introduction of the E1a and E1b genes from adenovirus type 5 DNA (19).
293-BFRF1-KO cells, carrying the BFRF1-KO EBV genome, and 293-2089 cells, carrying the wild-type B95-8 EBV genome, were described elsewhere (12, 16).
All cell lines were grown in RPMI 1640 (Euroclone) supplemented with 10% fetal calf serum (Invitrogen). To induce EBV lytic cycle gene expression, cells were treated with 20 ng of 12-tetradecanoylphorbol 13-acetate (TPA) per ml and 3 mM sodium butyrate for 24, 48, and 72 h. Inhibition of viral DNA replication was obtained by addition of phosphonoacetic acid (PAA) to the cell cultures to a final concentration of 1 mM. The proteasome inhibitor MG132 (Calbiochem) was used at a concentration of 5 μM, and lambda phosphatase (New England Biolabs) was used at 1,000 U/ml.
RNA preparation, Northern blot analysis, and 5′ and 3′ rapid amplification of cDNA ends (RACE).
Total RNA was extracted with Trizol (Invitrogen) according to the manufacturer's instructions. Twenty micrograms of RNA was loaded for each sample and resolved by 1.2% agarose-6% formaldehyde gel electrophoresis in 20 mM morpholinepropane sulfonic acid (MOPS) (pH 7.0) (Sigma). After migration, RNA was transferred to a Nytran Plus (Schleicher & Schuell) membrane in 20× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate) and UV cross-linked. The probe for the BFLF2 transcript was the oligonucleotide RG3, whose sequence is 5′-TATGGGGGTGTTCATCTCACGCAG-3′ (genomic coordinates in strain B95-8, 56609 to 56632). The probe was labeled by using [γ-32P]dATP and T4 polynucleotide kinase (Amersham Bioscience) according to the manufacturer's instructions. Hybridizations were carried out in phosphate buffer (0.5 M NaH2PO4 [pH 6.8], 0.5 M Na2HPO4 [pH 6.8], 0.7% sodium dodecyl sulfate [SDS], 1% bovine serum albumin [BSA], 1 mM EDTA) at 42°C overnight. Filters were subsequently washed at 42°C twice in buffer A (0.5% BSA, 5% SDS, 40 mM NaH2PO4 [pH 6.8], 40 mM Na2HPO4 [pH 6.8], 1 mM EDTA) and twice in buffer B (1% SDS, 40 mM NaH2PO4 [pH 6.8], 40 mM Na2HPO4 [pH 6.8], 1 mM EDTA), and bands were traced by autoradiography. As a control for RNA quality and equal loading, membranes were hybridized with a β-actin oligonucleotide probe (5′-TGTTGGCGTACAGGTCTTTGCGGATGTCCA-3′).
In order to map the transcription start site, we used the 5′/3′ RACE second-generation kit (Roche). Briefly, 5 μg of total RNA, extracted from B95-8cells 48 h after TPA and sodium butyrate treatment, was used for cDNA amplification with primer RG2 (5′-CGTGTAGTTTCTGTGGTG-3′; genomic coordinates in B95-8, 56837 to 56854) according to the manufacturer's instructions. The cDNA was subsequently amplified by PCR with primer RGP2 (5′-ATTCACGGCATCTGGGGTG-3′; genomic coordinates in B95-8, 56906 to 56924) and an oligo(dT) primer (supplied with the kit) at 55°C. The 200-bp amplified product was cloned in pGEMT-Easy vector (Promega) and sequenced.
3′-RACE was carried out to map the end of BFLF2 transcription. An oligo(dT) anchor primer (Roche) was used to amplify cDNA, and PCR was performed with primer RSP8 (5′-TGTGGCCCGATCAATATGTTC-3′; genomic coordinates in B95-8, 56747 to 56727) and the PCR anchor primer supplied with the kit. The 800-bp product obtained was cloned in pGEMT-Easy vector (Promega) and finally sequenced.
Plasmids.
The BFLF2 cDNA was first amplified by reverse transcription-PCR performed on total RNA extracted from B95-8 cells at 72 h after induction of virus replication with TPA and sodium butyrate. First-strand cDNA amplification was achieved by using Superscript (Invitrogen) and the primer F2-down (5′-CTGTTTATTTTCCAAAATGAGCTGG-3′) (genomic coordinates in B95-8, 55982 to 56006). Subsequently the primers F2-up (5′-ATGGCCCCGGTCACCCCAGA-3′) (genomic coordinates in B95-8, 56953 to 56916) and F2-down, together with Taq polymerase (Invitrogen), were used to perform PCR amplification. The 954-bp amplified fragment was first cloned in pCR-Script Amp SK(+) vector (Stratagene), and the BFLF2 gene was excised by digestion with BamHI and SacI and subsequently cloned in pet30b(+) (Novagene) cut with BamHI/SacI, in order to obtain a His6-BFLF2 fusion open reading frame (ORF).
A plasmid containing a glutathione S-transferase (GST)-BFLF2 fusion ORF was also generated by cloning a BamHI/EagI filled-in fragment from vector pCR-Script Amp SK(+) in vector pGEX-3X (Pharmacia) digested with SmaI.
The cytomegalovirus (CMV) immediate-early gene promoter-driven eukaryotic expression vector pHD1013 was used to generate plasmids for cell transfection (11). The BFLF2 ORF was excised from plasmid pGEX-3X-BFLF2 by using BamHI and EcoRI and inserted in the pHD1013 BamHI and EcoRI sites.
The CMV-BZLF1 plasmid, whose transfection in eukaryotic cells leads to the expression of ZEBRA protein, was a generous gift of G. Miller. The CMV-BFRF1 construct was described elsewhere (14). One microgram of either the CMV-BFRF1 or CMV-BFLF2 construct was used to transfect 3 × 104 293 cells by using the Fugene 6 kit (Roche) according to the manufacturer's instructions
Production of a MAb against BFLF2.
Escherichia coli BL21 cells were transformed with the His-BFLF2 or GST-BFLF2 plasmid to produce BFLF2 fusion proteins that were successively purified through column chromatography, according to the manufacturer's instructions. Four-week-old BALB/c mice were immunized twice by intraperitoneal injection with 25 μg of purified His-BFLF2 protein emulsified in RIBI adjuvant (RIBI Immunochemical Research). Mice were then given a booster immunization intravenously with 10 μg of the immunogen, and immune splenocytes were removed 3 days later. Somatic cell hybrids were prepared with NS-1 mouse nonsecreting myeloma cells as previously described (29). Hybridoma supernatants were screened for differential immunoreactivity between GST-BFLF2 and GST purified proteins by enzyme-linked immunosorbent assay. Positive hybridoma cell lines were cloned twice by limiting dilution. One MAb, named C1, was selected. Tissue culture supernatant of MAb R4, recognizing the unrelated carcinoembryonic antigen, was used as a negative control (2).
Immunoprecipitation and immunoblotting.
To perform immunoprecipitation assay, cells were harvested by centrifugation, washed once with phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, 1.8 mM KH2PO4, pH 7.4) and lysed in 1× radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM Na2EDTA, 0.5% NP-40, 1% BSA, 1× protease inhibitor cocktail [Roche]) for 30 min on ice (1 × 106 transfected 293 cells were lysed in 300 μl, and 6 × 106 B95-8 cells were lysed in the same volume). After 10 min of centrifugation at 20,000 × g, the supernatants were discarded and the resulting pellets were resuspended in 1× radioimmunoprecipitation assay buffer containing 500 mM NaCl, 0.5% NP-40, 1% BSA, and 1× protease inhibitor cocktail and sonicated for 30 seconds. These cellular extracts were mixed with protein G-Sepharose (Amersham) and the appropriate antibody and incubated overnight at 4°C while rocking gently. Precipitated proteins were collected by centrifugation, washed three times in lysis buffer, and loaded on a polyacrylamide gel for Western blot analysis. Antibody dilutions used for immunoprecipitation analysis were as follows: goat polyclonal anti-lamin B, 1:50; rabbit polyclonal anti-BFRF1, 1:300; and mouse monoclonal anti-BFLF2, 1:50.
To prepare total cell extract, cells were harvested by centrifugation at 1,500 × g for 5 min, washed once with PBS, resuspended in PBS-1% Triton-2 M urea, and lysed by sonication for 1 min. Different amount of cell lysates ranging from 10 to 50 μg were loaded on Nu-Page precast gels (Invitrogen) according to the manufacturer's instructions. Proteins were transferred to nitrocellulose filters (0.45-μm pore size: Schleicher & Schuell) by standard procedures (38). Membranes were incubated for 30 min in blocking solution (5% nonfat dry milk in PBS-0.2% Tween 20) and for 30 min with primary antibody and washed three times with blocking solution. Membranes were successively incubated with horseradish peroxidase-conjugated secondary antibody (Pierce), and after three washings in blocking solution, bands were visualized by enhanced chemiluminescence (ECL system) according to the manufacturer's instructions (Amersham). Antibody dilutions used for Western blot analysis were as follows: goat polyclonal anti-lamin B (Santa Cruz), 1:100; mouse monoclonal anti-BFLF2, 1:50; and mouse monoclonal anti-BFRF1, 1:50.
Virion purification.
Biochemical characterization of extracellular virions was performed by precipitating viruses from infectious supernatants with a polyethylene glycol (PEG)-containing solution (0.5% [wt/vol] PEG 6000 in 5 M NaCl). Viruses were further collected by centrifugation at 9,000 × g for 20 min. To purify intracellular virions, lytically induced cells were extensively washed and sequentially frozen in a dry ice bath and thawed at 37°C three times. Cells were spun down at 5,000 × g for 10 min, and supernatants were filtered with a 0.8-μm-pore-size filter. Viruses present in these supernatants were further PEG precipitated as described for extracellular virions. Purified virions were lysed in LDS sample buffer (Invitrogen), loaded onto Nu-Page gels (Invitrogen), and analyzed by Western blotting.
Immunofluorescence and confocal microscopy.
Chemically induced B95-8 cells, 293-BFRF1-KO cells, and 293-2089 cells were harvested, washed once in PBS, seeded onto multispot microscope slides (ICN), air dried, and fixed for 5 min in acetone-methanol (1:1). 293 cells grown on coverslips covered with 2% gelatin (Sigma) were transfected with CMV-BFRF1, CMV-BFLF2, or vector alone with Fugene 6 (Roche). At 24 h after transfection, the medium was aspirated and the cells were washed once with PBS and fixed in methanol at −20°C for 4 min or in 4% paraformaldehyde in PBS for 30 min at 25°C, followed by treatment with 0.1 M glycine for 20 min at 25°C and with 0.1% Triton X-100 for an additional 5 min at 25°C to allow permeabilization. For double-staining experiments, cells were incubated for 30 min at room temperature with the following primary antibodies: goat polyclonal anti-lamin B, 1:50 in PBS (Santa Cruz); rabbit polyclonal anti-BFRF1, 1:1,000 in PBS; mouse monoclonal anti-BFLF2 C1, 1:20 in PBS; mouse monoclonal anti-BFRF1 E7, 1:20 in PBS; and mouse monoclonal anti-nuclear pore complex, 1:50 in PBS (MAb 414 [Berkeley Antibody Co.], binding to p62, nup155, nup214, and nup358) (10) The cells were then washed in PBS. The primary antibodies were visualized by using fluorescein isothiocyanate (FITC)-conjugated rabbit anti-goat immunoglobulin G (IgG) (Cappel) (1:50), Texas Red-conjugated goat anti-mouse IgG (Jackson) (1:50), Texas Red-conjugated goat anti-rabbit IgG (Jackson) (1:100), FITC-conjugated goat anti-mouse IgG (Cappel) (1:50 in PBS), and FITC-conjugated goat anti-rabbit IgG (Jackson) (1:200 in PBS). After several washes in PBS, coverslips were mounted face down in Mowiol (Calbiochem) with or without 1 μg of DAPI (4′,6′-diamidino-2-phenylindole) per ml for observation on an Axiophot epifluorescence microscope (Zeiss, Oberkochen, Germany). The fluorescence signals were analyzed by recording and merging single-stained images, using a cooled charge-coupled device SPOT-2 color digital camera (Diagnostic Instruments Inc., Sterling Heights, Mich.) and FISH 2000/H1 software (Delta Sistemi, Rome, Italy). Colocalization of fluorescence signals was evaluated with a Zeiss confocal laser scan microscope. To prevent cross talk between the two signals, the multitrack function was used.
Electron microscopy.
293 cells were washed three times in PBS and fixed with 2% glutaraldehyde in the same buffer at 4°C. Samples were postfixed in 1% osmium tetroxide in veronal acetate buffer (pH 7.4) for 1 h at 25°C, stained with 0.1% tannic acid in the same buffer for 30 min at 25°C and with uranyl acetate (5 mg/ml) for 1 h at 25°C, dehydrated in acetone, and embedded in Epon 812. Thin sections were examined either unstained or poststained with uranyl acetate and lead hydroxide, using a Philips Morgagni electron microscope.
RESULTS
Predicted features of the BFLF2 ORF.
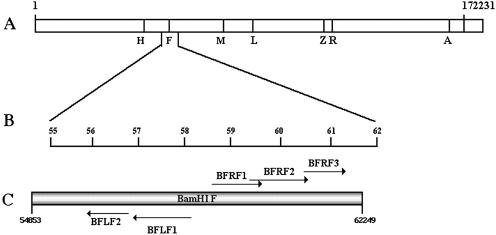
As shown in Fig. 1, BFLF2 is an open reading frame located within BamHI fragment F of the EBV genome. The ATG codon is situated at position 56953 in the B95-8 genome, and the stop codon is at position 55982 (1). Computer analysis showed that BFLF2 potentially encodes a protein of 35.3 kDa, consisting of 318 amino acids and with an isoelectric point of 9.5 BFLF2 is predicted to be a nuclear phosphoprotein. Amino acid analysis of BFLF2 with the program Netphos, version 2.0 (4), predicts 20 possible sites for phosphorylation, 14 on serine, 3 on threonine, and 3 on tyrosine residues. Furthermore, five potential casein kinase II and three protein kinase C phosphorylation sites are present along the amino acid sequence. Although there is no evidence for any N-terminal signal peptide, the high content of basic amino acids could be an indication of nuclear localization.
FIG. 1.
Schematic drawing of the BFLF2 ORF within the EBV genome. (A) BamHI cleavage map of B95-8 EBV. (B) Enlargement of the BamHI region. (C) Positions and transcriptional orientations of the ORFs (indicated by arrows).
Generation of recombinant BFLF2 proteins and of a MAb against BFLF2.
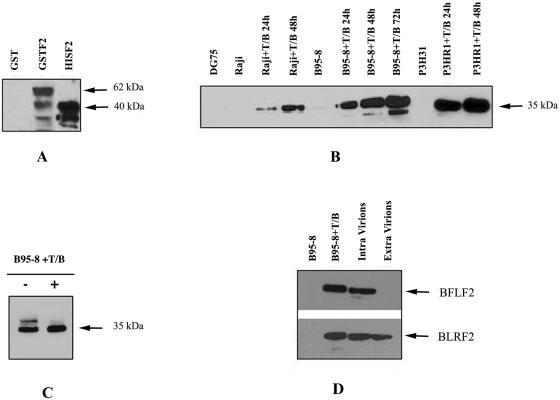
The entire BFLF2 ORF was amplified by reverse transcription-PCR with RNA extracted from B95-8 cells 72 h following chemical induction of the replicative cycle by TPA and sodium butyrate treatment. The DNA fragment was cloned into the vector pCR-Script Amp SK(+) and successively in pet30b(+) in order to obtain a histidine-BFLF2 fusion ORF and subsequently a histidine-BFLF2 fusion protein. This fusion protein was used for mouse immunization, and a MAb, C1, which specifically reacted in Western blot analysis with a GST-BFLF2 fusion protein was selected (Fig. 2A)
FIG. 2.
(A) Western immunoblot showing the specificity of MAb C1. Samples representing His-BFLF2 and GST-BFLF2 fusion proteins together with GST were run, and the immunoblot was probed with the anti-BFLF2 MAb C1. (B) BFLF2 protein expression in different cell lines, either uninduced or following activation with TPA and sodium butyrate (T/B). (C) Phosphorylation of BFLF2 in induced B95-8 cells. Twenty micrograms of cell lysate was treated with λ-phosphatase (+) or left untreated (−) and analyzed by immunoblotting. (D) BFLF2 is present in intracellular virions. Equal amounts of intracellular and extracellular virion lysates were run together with uninduced and induced B95-8 (T/B) cell extracts. The immunoblot probed with the anti-BFLF2 MAb C1 shows positivity only in intracellular virions and in B95-8+T/B cells. As a control for virion preparation, an antibody specific for a viral capsid antigen (BLRF2) was used.
BFLF2 is a lytic phosphoprotein.
To determine whether BFLF2 is expressed in EBV-infected cells, cell extracts obtained from EBV-positive and EBV-negative cell lines were assayed by Western blot analysis with MAb C1 (Fig. 2B). A band of approximately 35 kDa was detected in every cell line following induction of virus replication by treatment with TPA and sodium butyrate, which appeared as a doublet migrating at 35 to 36 kDa in B95-8 cells. A faint band was also observed in uninduced B95-8 cells, consistent with the spontaneous activation of the lytic cycle in approximately 5% of these cells. No signal was detected in the EBV-negative DG75 cells and in uninduced Raji and P3HR-1 cells. In addition, following treatment of the cells with lambda phosphatase, the mobility of one of the two bands of the doublet was altered, indicating that this band represents the phosphorylated product of the BFLF2 protein (Fig. 2C). These data indicate that BFLF2 is a lytic phosphoprotein.
BFLF2 is not detected in extracellular virions.
We next analyzed by Western blotting whether BFLF2 was present in extracellular virions. To this end, cell-free B95-8 viruses from infectious supernatants or intracellular B95-8 viruses obtained from the producing cell line were purified and protein extracts were analyzed by Western blotting. Figure 2D shows that no reactivity for BFLF2 was detectable in extracellular virions with MAb C1, whereas a strong positive signal was visible after immunoblotting with a MAb directed against a known virion component, BLRF2, confirming the presence of virions in the concentrated supernatant.
BFLF2 is expressed as an early transcript.
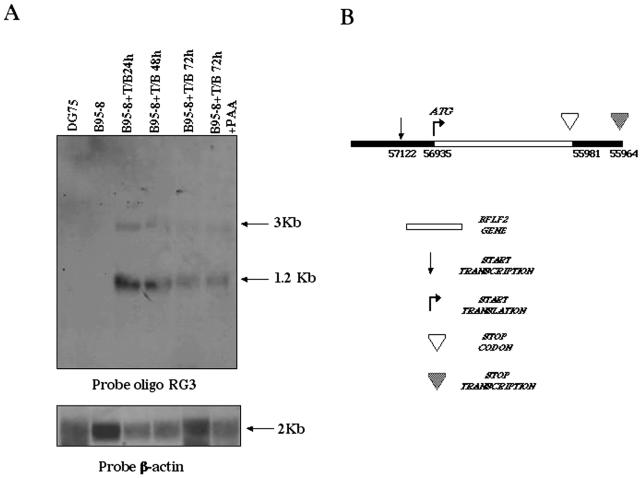
Regulation of BFLF2 transcription was analyzed by Northern blot analysis of RNA extracted from chemically induced B95-8 cells. The DNA oligonucleotide RG3 was used as a probe, and the results are shown in Fig. 3A. Two major transcripts were detected, at 3 and 1.2 kb. The presence of multiple signals is not surprising, since several transcripts encompassing the BamHI F region are known (22). In particular, the 1.2-kb band is consistent with a monocistronic transcript of BFLF2, and the 3-kb signal is consistent with a bicistronic transcript including the BFLF1 and BFLF2 ORFs. As shown in Fig. 1, the BFLF1 ORF is 2 kb long and is located immediately upstream of the BFLF2 ORF. We next investigated the effect of the viral DNA inhibitor PAA on BFLF2 transcription. As shown in Fig. 3A, BFLF2 expression was not affected by PAA. In addition, since BFLF2 is expressed in Raji cells, which harbor a defective viral strain which does not allow the expression of late EBV genes, we conclude that BFLF2, similarly to BFRF1 (14), is expressed as an early gene.
FIG. 3.
Analysis of BFLF2 RNA expression. (A) Northern blot analysis of RNA extracted from B95-8 cells at different times after treatment with TPA and sodium butyrate (T/B), with and without treatment with phosphonoacetic acid (T/B+PAA). Twenty micrograms of total RNA was separated on a 1.2% agarose-6% formaldehyde gel, and the blot was subsequently probed with the oligonucleotide RG3 as described in Materials and Methods. Equal RNA loading was assessed by β-actin hybridization. (B) 5′ and 3′ RACE analysis of BFLF2 transcripts. The transcription start and stop sites of the BFLF2 ORF as they were mapped by 5′-3′ RACE assay are shown. Genomic coordinates are given with respect the B95-8 viral strain.
In order to map the 5′ end of the BFLF2 transcript, we next performed 5′ RACE analysis on RNA extracted from chemically induced B95-8 cells. The oligonucleotides RGP2 and RSP8 were used to amplify the region which lies upstream of the BFLF2 ORF in RNA molecules, and the results are shown in Fig. 3B. Sequence analysis of the fragment amplified by 5′ RACE maps the transcription start at position −167 (with respect to the ATG), in proximity to a hypothetical TATA box situated within the BFLF1 ORF. This is consistent with the presence of a monocistronic transcript for BFLF2 which gives rise to the 1.2-kb band in the Northern blot probed with the RG3 oligonucleotide. We also performed 3′ RACE experiments to map the end of BFLF2 transcription, as shown in Fig. 3B. An oligo(dT) anchor primer was first used to amplify cDNA, followed by PCR amplification with oligonucleotide RSP8 and a PCR anchor primer. The 800-bp amplified fragment was sequenced, and the result maps the transcription end at position 55965 (in the B95-8 genome), 14 nucleotides downstream from the stop codon of the BFLF2 gene.
BFLF2 interaction with BFRF1.
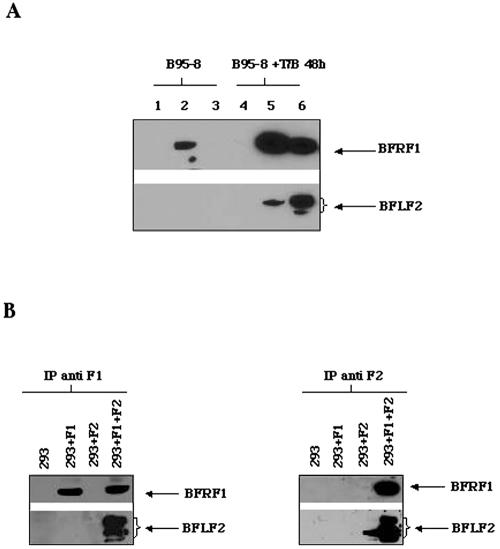
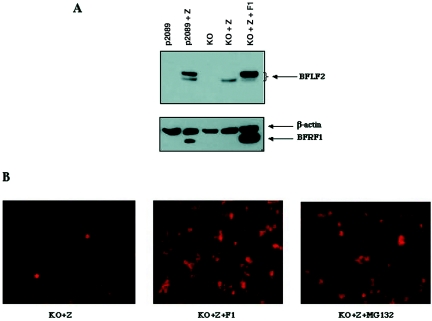
To assess physical interaction between BFRF1 and BFLF2, coimmunoprecipitation experiments were performed with cell lysates from both chemically induced B95-8 cells and 293 cells transiently transfected with a plasmid expressing either BFRF1 or BFLF2 or cotransfected with both genes. Coprecipitation (Fig. 4) demonstrated a direct interaction of the two proteins. Having shown a direct physical interaction between BFRF1 and BFLF2, we next analyzed the expression of BFLF2 in 293 cells infected with an EBV mutant carrying a deletion of the BFRF1 gene, which was obtained through bacterial artificial chromosomal technology and described in the accompanying paper (16). Control 293 cells carrying wild-type virus (p2089) were described previously (12). Figure 5 shows an immunoblot with the anti-BFLF2 MAb in 293 cells carrying wild-type virus (p2089), either uninduced or transfected with CMV-BFRF1 (p2089+Z), and in uninduced 293 cells carrying the mutant virus (KO), induced by CMV-BZLF1 transfection (KO+Z) or induced by CMV-BZLF1 transfection and recomplemented by CMV-BFRF1 transfection (KO+Z+F1). As shown in Fig. 5, BFLF2 was undetectable in both uninduced 293-2089 and 293-BFRF1-KO cells. Following lytic cycle activation, BFLF2 was visible as a doublet, with the higher band more intense than the lower one, in the cells infected with wild-type virus and transfected with CMV-BFRF1 (p2089+Z), whereas in the induced 293-BFRF1-KO cells only a weak band was detectable, corresponding to the lower band of the BFLF2 doublet. These data indicate that in absence of BFRF1, the expression of BFLF2 is strongly limited. In addition, since as shown above (Fig. 2C), the higher band of the doublet represents the phosphorylated product of the protein, the absence of the higher band of the doublet in KO+Z cells indicates that the protein is not phosphorylated. Finally, BFLF2 expression and phosphorylation were both restored to the higher levels in induced KO cells recomplemented with BFRF1.
FIG. 4.
(A) BFLF2 and BFRF1 coprecipitate in B95-8 cells. Cellular extracts from B95-8 cells and B95-8 cells activated with TPA and sodium butyrate (T/B) for 48 h were immunoprecipitated with rabbit anti-BFRF1 polyclonal antibody and with MAb C1 against BFLF2. Immunoprecipitated proteins were subjected to electrophoresis and transferred to nitrocellulose filters. The resulting immunoblots were probed with anti-BFRF1 MAb E7 and anti-BFLF2 MAb C1. Lanes 1 and 4: extracts from uninduced and induced B95-8 cells immunoprecipitated with protein G-conjugated beads without antibody. Lanes 2 and 5: extracts immunoprecipitated with anti-BFRF1 polyclonal antibody. Lanes 3 and 6: extracts immunoprecipitated with anti-BFLF2 MAb C1. (B) BFLF2 and BFRF1 coprecipitate in 293 transfected cells. Cellular extracts from 293 cells transiently transfected with CMV-BFRF1 (F1) or CMV-BFLF2 (F2) or cotransfected with CMV-BFRF1 and CMV-BFLF2 (F1+F2) were immunoprecipitated with anti-BFRF1 polyclonal antibody (IP anti F1) and with the anti-BFLF2 MAb C1 (IP anti F2). After electrophoresis, the resulting immunoblots were probed with anti-BFRF1 MAb E7 and with anti-BFLF2 MAb C1.
FIG. 5.
(A) BFLF2 expression in the presence or absence of BFRF1. Equal amounts of cellular extracts from 293 cells carrying the wild-type virus (p2089) or a virus with BFRF1 deleted (KO) were loaded on a polyacrylamide gel, and the resulting immunoblot was probed with the anti-BFLF2 MAb C1. The BFRF1-KO virus is characterized in the accompanying paper (16), and 293-2089 cells were described previously (12). Lane 1, 293 cells carrying the wild-type genome; lane 2, p2089 cells transiently transfected with CMV-BZLF1; lane 3, 293-BFRF1-KO cells; lane 4, 293-BFRF1-KO cells transiently transfected with CMV-BZLF1; lane 5, 293-BFRF1-KO cells transiently transfected with CMV-BZLF1 and CMV-BFRF1. The same blot was stripped and reprobed with anti-β-actin and anti-BFRF1 antibodies to assess equal loading and presence of BFRF1. (B) Indirect immunofluorescence of BFLF2 expression in 293-BFRF1-KO cells transiently transfected with CMV-BZLF1, in 293-BFRF1-KO cells transiently cotransfected with CMV-BZLF1 and CMV-BFRF1, and in 293-BFRF1-KO cells transiently transfected with CMV-BZLF1 and treated with the proteasome inhibitor MG132.
To assess whether the minimal expression of BFLF2 in cells lacking BFRF1 might be due to degradation of the protein, 293-BFRF1-KO cells were induced to undergo lytic cycle activation by BZLF1 transfection in the presence of the proteasome inhibitor MG132. MG132 treatment fully restored BFLF2 expression to levels similar to those of BFRF1-recomplemented cells (Fig. 5B) without affecting BFRF1 expression, suggesting that the ubiquitin-proteasome system probably plays a role in BFLF2 degradation and that BFRF1 is involved in BFLF2 stabilization. These results are consistent with a similar previous observation for HSV type 1 (HSV-1) (49), and additional studies to formally prove this hypothesis and to further investigate the mechanisms involved in the regulation of BFLF2 expression are ongoing in our laboratory.
BFLF2 colocalizes with BFRF1 in both infected and transfected cells.
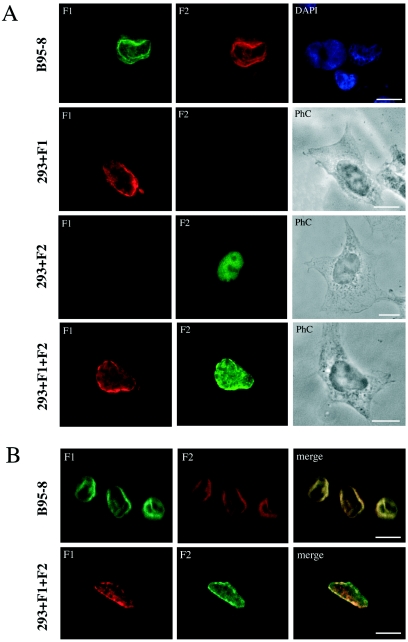
Having shown that there is a physical interaction between BFRF1 and BFLF2 during viral infection as well as in transfected cells, we next analyzed the intracellular localization of the two proteins by immunofluorescence and confocal microscopy. In chemically induced B95-8 cells (Fig. 6), both proteins were detectable on the nuclear rim. Confocal analysis showed colocalization of the two proteins, as shown by the yellow color in merged images. Following transient transfection on 293 epithelial cells, BFRF1 appeared to be localized on the nuclear membrane, sometimes concentrated on thickened portions of the membrane. Occasional cytoplasmic staining in proximity to the nuclear membrane was also observed. In contrast, BFLF2 appeared to be diffusely localized inside the nucleus. In some experiments, equal amounts of cells singly transfected with BFRF1 and BFLF2 were mixed and spotted onto a glass slide and analyzed by immunofluorescence with antibodies directed against BFRF1 and BFLF2. As expected, approximately half of the cells reacted with the MAb against BFRF1 and half reacted with the second antibody, ruling out potential cross-reactions between primary or secondary antibodies in double-fluorescence experiments. In cotransfected cells, both proteins localized at the nuclear rim, occasionally concentrated over irregular fragmentations of the membrane, and confocal analysis showed a full colocalization of BFRF1 with BFLF2.
FIG. 6.
Indirect immunofluorescence of B95-8 and 293 cells expressing BFLF2 and BFRF1. (A) Double immunofluorescence of chemically induced B95-8 cells and of 293 cells transfected with CMV-BFRF1 (293+ F1) or CMV-BFLF2 (293+F2) or cotransfected with CMV-BFRF1 and CMV-BFLF2 (293+F1+F2). Bars, 10 μm. PhC, phase contrast. (B) Confocal immunofluorescence of induced B95-8 cells and cotransfected 293 cells, as described for panel A. Yellow staining in the merged images shows colocalization of the two signals. Bars, 10 μm.
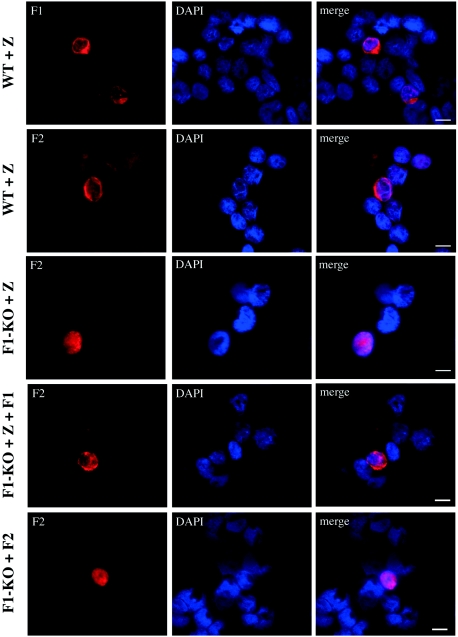
We next examined BFLF2 localization in 293 cells carrying the wild-type virus or the BFRF1-KO virus. In uninduced wild-type 293 cells, neither BFRF1- nor BFLF2-positive cells were visible at immunofluorescence (not shown). Upon lytic cycle activation by transfection with ZEBRA, approximately 10% of the cells expressed BFRF1 and BFLF2, and both proteins appeared to be localized at the nuclear rim (Fig. 7, WT+Z). As expected, the percentage of BFRF1-positive cells increased in cells cotransfected with ZEBRA plus BFRF1, whereas the percentage of BFLF2-positive cell was not affected (data not shown).
FIG. 7.
Indirect immunofluorescence of 293 cells carrying the wild-type virus and transfected with CMV-BZLF1 (WT+Z) and of 293 cells carrying the BFRF1-KO virus and transfected with CMV-BZLF1 (F1-KO+Z), cotransfected with CMV-BZLF1 and CMV-BFRF1 (F1-KO+Z+F1), or transfected with CMV-BFLF2 (F1-KO+F2). In the absence of BFRF1, BFLF2 is localized inside the nucleus, while in the presence of BFRF1, it is localized on the nuclear rim. Bars, 10 μm.
In 293 cells in which the gene encoding BFRF1 has been deleted (293-BFRF1-KO cells) (16), again neither BFRF1 nor BFLF2 was detectable. As expected, they remained negative for BFRF1 upon transfection with ZEBRA, whereas approximately 30% of the cells became positive for BFRF1, with the typical fluorescence pattern on the nuclear rim, following BFRF1 recomplementation (not shown). Very few BFLF2-positive cells (fewer than 1%) were visible following lytic cycle activation, confirming the previous biochemical data indicating that BFLF2 expression is dependent upon BFRF1. BFLF2 was localized diffusely in the nucleus (Fig. 7, F1-KO+Z). Conversely, when 293-BFRF1-KO cells were recomplemented with BFRF1, the number of BFLF2-positive cells increased to approximately 10%, and BFLF2 localization became similar to that of BFRF1, concentrated around the nuclear rim (Fig. 7, F1-KO+Z+F1). Finally, as expected, BFLF2 transfection in 293-BFRF1-KO cells resulted in a high percentage of cells expressing BFLF2, with the typical diffuse nuclear staining, whereas BFRF1 expression was not influenced, and no BFRF1-positive cells were visible (not shown).
In conclusion, the results with 293-BFRF1-KO cells confirmed the previous results with EBV producer lymphoid cells or transfected 293 cells, indicating that in the absence of BFRF1, BFLF2 is localized inside the nucleus, whereas it colocalizes with BFRF1 on the nuclear membrane when BFRF1 is expressed.
The nuclear membrane structure of 293 cells is altered by BFRF1 and BFLF2 expression.
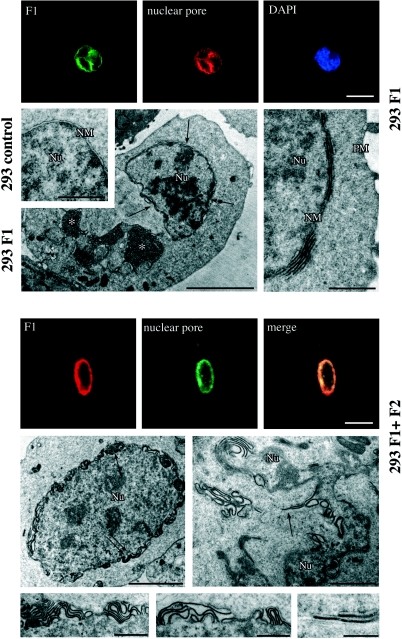
Since by immunofluorescence, and previously by immunoelectron microscopy for BFRF1 (15), we have demonstrated the localization of BFRF1 on the nuclear membrane and since we show here that BFLF2, when coexpressed with BFRF1, acquires a similar nuclear membrane localization, we analyzed whether the single or double expression of the two proteins might induce structural alterations of the nuclear membrane. To this end, we used untransfected 293 cells and 293 cells transfected with either BFRF1, BFLF2, BFRF1 and BFLF2, or a vector control. At 24 h following transfection, approximately 20% of the cells were positive for the transfected proteins, and in the BFRF1- and BFLF2-cotransfected cells, the vast majority (over 90%) of the cells reacted with both anti-BFRF1 and anti-BFLF2 antibodies as revealed by immunofluorescence analysis (not shown). First, we performed double-immunofluorescence experiments with a MAb directed against the nuclear pore complex (10), as a marker for the nuclear membrane, and the anti-BFRF1 rabbit polyclonal antibody, and we observed that staining of BFRF1 in both singly and doubly transfected 293 cells fully corresponded to the nuclear membrane labeling and that both signals were not uniform but were concentrated in portions of the nuclear rim (Fig. 8). Parallel ultrastructural analysis, performed by conventional transmission electron microscopy, revealed that in 293 cells transfected with BFRF1 alone, well demarcated areas of the nuclear membranes appeared multilayered and characterized by narrow packaging of multiple cisternal layers strictly adhering to each other and giving rise to dense osmiophylic membrane domains at the linear contacts between the stacked cisternae (Fig. 8). Occasionally, perinuclear aggregates of endoplasmic reticulum cisternae, frequently in continuity with the outer nuclear membrane, were detected (Fig. 8), which may correspond to the cytoplasmic structures positively stained for BFRF1 by immunofluorescence (Fig. 6A). Interestingly, in 293 cells coexpressing both BFRF1 and BFLF2, these multilayered portions of the nuclear membranes were more irregularly organized; in fact, the packaged cisternae appeared dilated, not linear, and occasionally were in concentric whorls. Neither in untransfected 293 cells nor in cells transfected with BFLF2 or with vector alone were similar nuclear membrane alterations observed. These results indicate that BFRF1 alone is responsible for the membrane duplications frequently observed during EBV replication (13, 43) and that BFLF2 influences this effect, possibly rendering these areas more suitable for viral envelopment.
FIG. 8.
Immunofluorescence and electron microscopic analyses of the nuclear membrane structure in transfected 293 cells. Upper panels: CMV-BFRF1-transfected cells (293 F1). Lower panels: CMV-BFRF1- and CMV-BFLF2-cotransfected cells (293 F1+F2). Double-immunofluorescence staining with anti-nuclear pore complex and anti-BFRF1 antibodies reveals an irregular concentration of the two signals in both transfected and cotransfected cells (bar, 10 μm). Electron microscopic analysis of 293 F1 cells reveals focal multilayering of the nuclear membrane (NM) (arrows in the left panel and detail in the right panel), which is not observed in untransfected cells (293 control). Asterisks indicate cytoplasmic membrane structures possibly corresponding to aggregates of endoplasmic reticulum cisternae (asterisks in the left panel). Nu, nucleus; PM, plasma membrane. (Bars, in sequential order: 1, 5, and 1 μm.) Parallel electron microscopic analysis of 293 F1+F2 cells shows a highly irregular structure of the nuclear membrane, with more pronounced multilayering (enlargements of the areas indicated by arrows are shown in the lower panels). (Bars, in sequential order: 5, 2, 1, 1, and 0.5 μm.)
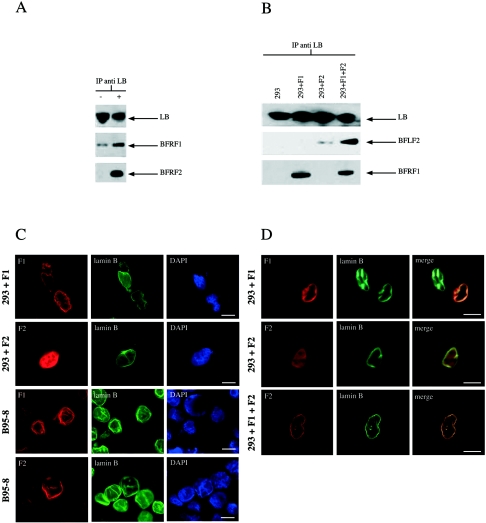
BFRF1 and BFLF2 interact with lamin B.
Finally, given the dramatic alterations induced on the nuclear membrane upon cotransfection with BFRF1 and BFLF2, we analyzed the possible interactions of either protein with lamin B, a component of the nuclear lamina which is known to directly interact with its receptor on the nuclear membrane. Coimmunoprecipitation experiments (Fig. 9) indicated that in chemically induced B95-8 cells both BFRF1 and BFLF2 interact with lamin B, whereas in transiently transfected cells BFLF2 interacts with lamin B only when BFRF1 is expressed. The interaction between BFRF1, BFLF2, and lamin B was also examined by immunofluorescence, both in chemically induced B95-8 cells and upon BFRF1 and BFLF2 transfection in 293 cells. The results of these experiments are shown in Fig. 9C and D. In B95-8 cells, in which we showed in the previous experiments (Fig. 6) that BFRF1 and BFLF2 colocalize, both proteins, as well as lamin B, were uniformly detected on the nuclear membrane. In BFRF1-transfected 293 cells, BFRF1 and lamin B signals were visible on the nuclear rim (Fig. 9C), and confocal analysis showed colocalization of the signals (Fig. 9D). In addition, BFRF1 was also visible on cytoplasmic structures in proximity to the nuclear membrane (upper cell on the left in the first row of Fig. 9C), which may correspond to the structures visualized by electron microscopy and indicated in Fig. 8. As expected, these cytoplasmic structures were not labeled with the anti-lamin B antibody. Conversely, in BFLF2-transfected 293 cells, the lamin B was evenly localized on the nuclear membrane, whereas BFLF2 maintained its usual diffuse localization inside the nucleus. Confocal analysis confirmed the lack of colocalization between BFLF2 and lamin B (Fig. 9D, middle panels). Furthermore, in cells cotransfected with BFRF1 and BFLF2, BFLF2 localization on the nuclear membrane, as well as its colocalization with lamin B, was rescued (Fig. 9D, lower panels). These results indicate that BFRF1 colocalizes with lamin B and recruits BFLF2 on the nuclear membrane. BFLF2, in the absence of BFRF1, maintains its nuclear localization and does not interact with lamin B.
FIG. 9.
(A) Lamin B (LB) coprecipitates with BFRF1 and BFLF2 in B95-8 cells. Cellular extracts from untreated B95-8 cells (−) and B95-8 cells induced with TPA plus butyrate (+) were immunoprecipitated (IP) with anti-lamin B polyclonal antibody. Immunoprecipitated proteins were run on polyacrylamide gels, and the resulting immunoblots were probed with the anti-lamin B antibody and with MAbs directed against BFRF1 and BFLF2. (B) Lamin B coprecipitates with BFRF1 and BFLF2 in 293 cells. 293 cells were transfected with CMV-BFRF1 (293+F1) and CMV-BFLF2 (293+F2) and cotransfected with CMV-BFRF1 and CMV-BFLF2 (293+F1+F2). At 24 h following transfection, cells were lysed and the resulting extracts were immunoprecipitated with anti-lamin B antibody. After electrophoresis, proteins were transferred to nitrocellulose membranes and analyzed by Western blotting as described for panel A. (C) Indirect immunofluorescence of BFRF1, BFLF2, and lamin B expression in induced B95-8 cells and in 293 cells transfected with CMV-BFRF1 (293+F1) or with CMV-BFLF2 (293+F2). (D) Confocal microscopy showing that BFRF1 colocalizes with lamin B, whereas BFLF2, in the absence of BFRF1, does not colocalize with lamin B. Bars, 10 μm.
DISCUSSION
The experiments presented in the present paper were aimed to identify and characterize the product of the BFLF2 gene of EBV, as well as to investigate the interaction of BFLF2 with an additional viral protein, BFRF1, and the modifications induced by this interaction in terms of intracellular localization of the two proteins and of alterations of the cellular nuclear membrane in the context of viral replication.
The interest in BFLF2 stems from the fact that it belongs to a conserved family of herpesvirus proteins which has been widely studied in HSV-1, HSV-2, and PrV and appears to play an important role in virus envelopment. Its homolog in HSV-1, designated UL31, is a nuclear matrix phosphoprotein of 37 kDa (6). UL31 homologs in other herpesviruses, with molecular masses varying from 42 kDa (UL53 in human CMV) to 34 kDa (in HSV-2), to 29 kDa (in PrV), have also been described (9, 17, 28, 33, 51). While CMV UL53 is a component of extracellular virions, UL31 of HSV-1 and PrV are not. Our present results indicate that BFLF2 is a nuclear phosphoprotein of approximately 35 kDa, expressed early during the viral replicative cycle, which is not detectable in extracellular virions. The kinetics of BFLF2 expression differs from those of all other herpesvirus homologs, which have been reported as true late proteins.
The generation of a specific MAb has allowed us to study the intracellular localization of the protein in virus-infected cells and its interaction with the recently described BFRF1 gene product (14), the UL34 positional homolog of EBV. Although the function of BFLF2 is still unknown, clues come from its intracellular localization and from its interaction with BFRF1, as well from analogies with other herpesviral homologs. Pull-down experiments (50) and yeast two-hybrid studies (17) have previously demonstrated a physical interaction between UL34 and UL31. Muranyi et al. also showed that M50/p35 and M53/p38, their respective homologs in mouse cytomegalovirus, coprecipitate (28). Accumulating evidence from several herpesvirus systems suggests that the physical interaction of UL31 with UL34 is a necessary event for their colocalization on the nuclear rim and for correct viral envelopment at the nuclear membrane. Our demonstration that BFRF1 also is essential for efficient envelopment of EBV (16) and the results presented in the present paper, showing that BFRF1 and BFLF2 coprecipitate and colocalize in both infected and cotransfected cells, are all consistent with the model proposed for other herpesviruses (reviewed in reference 26).
Concerning UL34 localization, our present confocal and previous immunoelectron microscopic analyses (15) have now clearly defined the unequivocal predominant nuclear membrane localization of BFRF1. Previous apparently conflicting results regarding the localization of BFRF1 homologs in other herpesviruses might simply reflect differences in the visualization methods (conventional versus confocal fluorescence) or be due to different viral strains or various target cell lines used. For example, UL34 in HSV-2 appears to be preferentially localized on the endoplasmic reticulum, and the presence of UL31 relocates the protein from the endoplasmic reticulum to the internal nuclear membrane (48).
Different results have also been reported regarding the presence of UL34 in extracellular virions. In the case of HSV-2, UL34 was detected on mature virions (40), while both the presence and absence of HSV-1 UL34 in the virion have been documented (36, 50). EHV-1 UL34 was not detectable in extracellular virions (30), and in PrV it was visualized on intracellular virions but not on the mature extracellular virions (17). The latter behavior was considered further evidence in favor of the envelopment-de-envelopment-reenvelopment process of alphaherpesviruses. Similarly, our recent results (16) and our previous observation by immunoelectron microscopy showing BFRF1 labeled intracellular virions but unlabeled extracellular virions (15) suggest that BFRF1 is not a component of the mature virions.
The reduced BFLF2 expression in cells not expressing BFRF1 is also similar to previous data reported for HSV-1, where immunoblot analysis indicated a reduction of UL31 protein in cells infected with a UL34 mutant virus (49), suggesting that some UL31 was mistargeted and degraded through the proteasome pathway in the absence of UL34. Our preliminary observations with the proteasome inhibitor MG132 suggest a similar degradation of BFLF2. In PrV, in contrast, UL34 does not seem to be required for stable expression of UL31 (17).
Viruses have often been used as a tool to investigate the cell biology of intracellular organelles and of the nuclear membrane, and the occurrence of morphological nuclear alterations in the course of viral infection is a well-known phenomenon. These modifications have been particularly documented in the process of viral entry, where incoming virions need to modify the nuclear membrane to gain access to the nucleus (46). For herpesviruses, binding of HSV-1 capsids to microtubules to reach the nuclear pore complex has been shown (42). In addition, HSV-1 UL34 and UL31 were shown to form complexes with dynein intermediate chains (50), but the significance of this interaction for virus entry is unclear. Since we showed here and in the accompanying paper (16) that both BFLF2 and BFRF1 are not components of the extracellular virions, it is unlikely that they participate in the process of viral entry.
One common feature shared by several viruses during the envelopment process to exit from the nucleus is the modification of the nuclear envelope structure. The nuclear envelope is composed of the outer and inner nuclear membranes and a complex network of stable, filamentous proteins called lamins, which are attached to the nuclear membrane by a large number of integral membrane proteins and which collectively form the nuclear lamina. For herpesviruses, dismantling of the nuclear lamina during HSV-1 infection has been reported by Scott and O'Hare (39), and, remarkably, a similar effect has been attributed to protein kinase C (PKC)-mediated phosphorylation of the nuclear lamina induced by the BFRF1 and BFLF2 homologs during mouse cytomegalovirus infection (28). Our results, showing the association of the BFRF1-BFLF2 complex with lamin B at the nuclear membrane, are consistent with the possible involvement of these proteins in affecting nuclear lamina structure and function. In addition, an interesting observation is represented by the profound nuclear alterations induced by expression of BFRF1, which is more strikingly observed when it is coexpressed with BFLF2, which are reminiscent of the nuclear membrane reduplications described previously as a peculiar, although not specific, feature of EBV replication (13, 43). These observations, together with our previous immunoelectron microscopic findings (15) on the preferential localization of BFRF1 over duplicated domains of the nuclear membrane in infected cells, strongly suggest that BFRF1, in conjunction with BFLF2, plays a key role in viral envelopment. These nuclear membrane duplications differ from the effects induced by HSV-1 UL34, which consist of dissociation of the inner and outer nuclear membranes (50).
The nuclear envelope alterations observed in the course of herpesviral infection can be also reminiscent of some features of nuclear membrane reconstruction occurring after mitotic division (for recent reviews, see references 25 and 47). One of the factors that is required for nuclear envelope assembly events is the small GTPase Ran. It has been proposed that chromatin-associated Ran-GTP recruits importin-β, which then binds to nucleoporins, to begin the assembly of the nuclear envelope (reviewed in reference 32). In addition, it has been shown that an excess of Ran-GTP or removal of importin-β induces formation of nuclear pore-containing membrane structures, known as annulate lamellae (45). Interestingly, while studying the intracellular maturation pathway of a human betaherpesvirus, human herpesvirus 6, we observed (5, 44) that annulate lamellae are also formed following human herpesvirus 6 infection of T cells and that they might represent a storage compartment of the viral glycoprotein gp116. However, we never observed annulate lamellae in the course of EBV replication, and we can exclude the possibility, by their ultrastructural characteristics as well as by the lack of staining with the antibody directed against the nuclear pore complex, that the cytoplasmic perinuclear organelles observed in the present work (Fig. 8) are annulate lamellae.
Furthermore, the nuclear membrane duplications induced by BFRF1 suggest that the BFRF1-BFLF2 complex might act, either directly or through binding to some of the nuclear proteins (such as emerin, LAP2, or MAN1) involved in the bridge between the nuclear membrane and the lamina, in the process of nuclear membrane reorganization. Of great interest is that very recent reports have demonstrated the role of UL34 and UL31 in altering the distribution of LAP2 and lamin A/C following HSV-1 infection (35, 41).
The relevant differences between the effects caused by BFRF1 alone and those caused by BFRF1 plus BFLF2 on the structure of the nuclear membrane are in accordance with the scenario proposed by Muranyi et al. (28) for mouse CMV, where the BFRF1 homolog strongly modifies the nuclear membrane, whereas the BFLF2 homolog qualitatively and quantitatively modulates this effect.
Further work to elucidate the role of BFRF1-BFLF2 complex in nuclear membrane disruption and reconstitution and to identify potential nuclear membrane proteins which might participate to this process will most likely represent a fruitful area for future studies.
In HSV-1 and in PrV an additional viral protein, a viral kinase encoded by the US3 gene, is also essential for viral envelopment by interacting with the UL34-UL31 complex (26). A US3 homolog is not present in the EBV genome, and preliminary results from Lake and Hutt-Fletcher (24) suggest that BGLF4, the only known EBV-encoded kinase (7, 18), is not responsible for phosphorylation of BFRF1 and BFLF2, which are thus probably phosphorylated by a cellular kinase. Of interest is that in mouse CMV, the BFRF1 homolog has been shown to recruit cellular PKC to dissolve the nuclear lamina, allowing virions to gain access to the nuclear membrane for primary envelopment (28). Since it has been shown that PKC-βII is directly responsible for lamin B phosphorylation and solubilization (21) and that the selective PKC-β inhibitor hispidin reduces lamin B phosphorylation and totally prevents lamin B proteolysis (8), it might be of interest to investigate whether the use of selective PKC inhibitors might interfere with the correct intracellular localization of lamin B or with its interaction with BFRF1, ultimately leading to the lamina disassembly that is needed for the breakdown of the nuclear envelope.
In conclusion, our results confirm the preliminary observations of Lake and Hutt-Fletcher (24) and provide, in conjunction with the data reported in the accompanying paper (16), a mechanistic explanation of the BFRF1-BFLF2 interaction in infected cells. Our experiments with the BFRF1-KO virus, where BFLF2 expression also is drastically reduced, indicate that the two proteins are necessary for efficient viral envelopment at the nuclear membrane. Whether the lack of BFLF2, in the presence of BFRF1, would also affect the process of viral envelopment will be analyzed in future studies by constructing, with the use of bacterial artificial chromosomal technology, a viral mutant with the BFLF2 gene deleted.
Acknowledgments
This work was partially supported by grants from the MIUR, Ministero della Sanità, Progetto AIDS, Associazione Italiana di ricerca sul Cancro (AIRC), and Istituto-Cenci-Bolognetti Foundation.
REFERENCES
- 1.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Bei, R., A. Moretti, V. Visco, R. De Filippi, K. Y. Tsang, L. Frati, and R. Muraro. 1996. Cell mediated cytotoxicity of human colon carcinoma cells by a monoclonal antibody (R4) recognizing the carcinoembryonic antigen (CEA) and CEA-related molecules. Int. J. Oncol. 8:1127-1135. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Bassat, H., N. Goldblum, S. Mitrani, T. Goldblum, J. M. Yoffey, M. M. Cohen, Z. Bentwich, B. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a ‘Burkitt-like’ malignant lymphoma (line DG75). Int. J. Cancer 19:27-33. [DOI] [PubMed] [Google Scholar]
- 4.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351-1362. [DOI] [PubMed] [Google Scholar]
- 5.Cardinali, G., M. Gentile, M. Cirone, C. Zompetta, L. Frati, A. Faggioni, and M. R. Torrisi. 1998. Viral glycoproteins accumulate in newly formed annulate lamellae following infection of lymphoid cells by human herpesvirus 6. J. Virol. 72:9738-9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. E., and B. Roizman. 1993. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J. Virol. 67:6348-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, M. R., S. J. Chang, H. Huang, and J. Y. Chen. 2000. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J. Virol. 74:3093-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiarini, A., J. F. Whitfield, U. Armato, and I. Dal Pra. 2002. Protein kinase C-beta II Is an apoptotic lamin kinase in polyomavirus-transformed, etoposide-treated pyF111 rat fibroblasts. J. Biol. Chem. 277:18827-18839. [DOI] [PubMed] [Google Scholar]
- 9.Dal Monte, P., S. Pignatelli, N. Zini, N. M. Maraldi, E. Perret, M. C. Prevost, and M. P. Landini. 2002. Analysis of intracellular and intraviral localization of the human cytomegalovirus UL53 protein. J. Gen. Virol. 83:1005-1012. [DOI] [PubMed] [Google Scholar]
- 10.Davis, L. I., and G. Blobel. 1987. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA 84:7552-7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, M. G., and E. S. Huang. 1988. Transfer and expression of plasmids containing human cytomegalovirus immediate-early gene 1 promoter-enhancer sequences in eukaryotic and prokaryotic cells. Biotechnol. Appl. Biochem. 10:6-12. [PubMed] [Google Scholar]
- 12.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epstein, M. A., and B. C. Achong. 1979. Morphology of the virus and virus-induced cytopathologic changes, p. 23-27. In M. A. Epstein and B. G. Achong (ed.), The Epstein-Barr virus. Springer Verlag, New York, N.Y.
- 14.Farina, A., R. Santarelli, R. Gonnella, R. Bei, R. Muraro, G. Cardinali, S. Uccini, G. Ragona, L. Frati, A. Faggioni, and A. Angeloni. 2000. The BFRF1 gene of Epstein-Barr virus encodes a novel protein. J. Virol. 74:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farina, A., G. Cardinali, R. Santarelli, R. Gonnella, J. Webster-Cyriaque, R. Bei, R. Muraro, L. Frati, A. Angeloni, M. R. Torrisi, and A. Faggioni. 2004. Intracellular localization of the Epstein-Barr virus BFRF1 gene product in lymphoid cell lines and oral hairy leukoplakia lesions. J. Med. Virol. 72:102-111. [DOI] [PubMed] [Google Scholar]
- 16.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H.-J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 79:3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gershburg, E., and J. S. Pagano. 2002. Phosphorylation of the Epstein-Barr virus (EBV) DNA polymerase processivity factor EA-D by the EBV-encoded protein kinase and effects of the l-riboside benzimidazole 1263W94. J. Virol. 76:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, F. L., W. C. Smiley, W. Russell, and R. Nairn. 1977. Characteristics of human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 20.Hinuma, Y., M. Konn, J. Yamaguchi, D. J. Wudarski, J. R. Blakeslee, and J. T. Grace. 1967. Immunofluorescence and herpes-like particles in the P3HR-1 Burkitt lymphoma cell line. J. Virol. 1:1045-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hocevar, B. A., D. J. Burns, and A. P. Fields. 1993. Identification of protein kinase C (PKC) phosphorylation sites on human lamin B. Potential role of PKC in nuclear lamina structural dynamics. J. Biol. Chem. 268:7545-7552. [PubMed] [Google Scholar]
- 22.Hudson, G. S., T. J. Gibson, and B. G. Barrell. 1985. The BamHI F region of the B95-8 Epstein-Barr Virus genome. Virology 147:99-109. [DOI] [PubMed] [Google Scholar]
- 23.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lake, C. M., L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99-106. [DOI] [PubMed] [Google Scholar]
- 25.Mattaj, I. W. 2004. Sorting out the nuclear envelope from the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 5:65-69. [DOI] [PubMed] [Google Scholar]
- 26.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, G., and M. Lipman. 1973. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc. Natl. Acad. Sci. USA 70:190-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 29.Muraro, R., D. Wunderlich, A. Thor, J. Lundy, P. Noguchi, R. Cunningham, and J. Schlom. 1985. Definition by monoclonal antibodies of a repertoire of epitopes on carcinoembryonic antigen differentially expressed in human colon carcinomas versus normal adult tissues. Cancer Res. 45:5769-5780. [PubMed] [Google Scholar]
- 30.Neubauer. A., J. Rudolph, C. Brandmuller, F. T. Just, and N. Osterrieder. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology. 300:189-204. [DOI] [PubMed] [Google Scholar]
- 31.Pulvertaft, R. J. V. 1964. Cytology of Burkitt's tumour (African lymphoma). Lancet i:238-240. [DOI] [PubMed]
- 32.Quimby, B. B., and M. Dasso. 2003. The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol. 15:338-344. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78:399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Scott, E. S., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 75:8818-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiba, C., T. Daikoku, F. Goshima, H. Takakuwa, Y. Yamauchi, O. Koiwai, and Y. Nishiyama. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 81:2397-2405. [DOI] [PubMed] [Google Scholar]
- 41.Simpson-Holley, M., J. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the viral periphery. J. Virol. 78:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus I capsids to the nucleus. J. Cell Biol. 136:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torrisi, M. R., M. Cirone, A. Pavan, C. Zompetta, G. Barile, L. Frati, and A. Faggioni. 1989. Localization of Epstein-Barr virus envelope glycoproteins on the inner nuclear membrane of virus-producing cells. J. Virol. 63:828-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torrisi, M. R., M. Gentile, G. Cardinali, M. Cirone, C. Zompetta, L. V. Lotti, L. Frati, and A. Faggioni. 1999. Intracellular transport and maturation pathway of human herpesvirus 6. Virology 257:460-471. [DOI] [PubMed] [Google Scholar]
- 45.Walther, T. C., P. Askjaer, M. Gentzel, A. Habermann, G. Griffiths, M. Wilm, I. W. Mattaj, and M. Hetzer. 2003. RanGTP mediates nuclear pore complex assembly. Nature 424:689-694. [DOI] [PubMed] [Google Scholar]
- 46.Whittaker, G. R., M. Kann, and A. Helenius. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627-651. [DOI] [PubMed] [Google Scholar]
- 47.Wozniak, R., and P. C. Clarke. 2003. Nuclear pores: sowing the seeds of assembly on the chromatin landscape. Curr. Biol. 13:R970-R972. [DOI] [PubMed] [Google Scholar]
- 48.Yamauchi, Y., C. Shiba, F. Goshima, A. Nawa, T. Murata, and Y. Nishiyama. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 82:1423-1428. [DOI] [PubMed] [Google Scholar]
- 49.Ye, G. J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. USA 97:11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu, H. Y., H. Yamada, Y. M. Jiang, M. Yamada, and Y. Nishiyama. 1999. Intracellular localization of the UL31 protein of herpes simplex virus type 2. Arch Virol. 144:1923-1935. [DOI] [PubMed] [Google Scholar]