Abstract
The 5′ cloverleaf in poliovirus RNA has a direct role in regulating the stability, translation, and replication of viral RNA. In this study, we investigated the role of stem a in the 5′ cloverleaf in regulating the stability and replication of poliovirus RNA in HeLa S10 translation-replication reactions. Our results showed that disrupting the duplex structure of stem a destabilized viral RNA and inhibited efficient negative-strand synthesis. Surprisingly, the duplex structure of stem a was not required for positive-strand synthesis. In contrast, altering the primary sequence at the 5′-terminal end of stem a had little or no effect on negative-strand synthesis but dramatically reduced positive-strand initiation and the formation of infectious virus. The inhibition of positive-strand synthesis observed in these reactions was most likely a consequence of nucleotide alterations in the conserved sequence at the 3′ ends of negative-strand RNA templates. Previous studies suggested that VPgpUpU synthesized on the cre(2C) hairpin was required for positive-strand synthesis. Therefore, these results are consistent with a model in which preformed VPgpUpU serves as the primer for positive-strand initiation on the 3′AAUUUUGUC5′ sequence at the 3′ ends of negative-strand templates. Our results suggest that this sequence is the primary cis-acting element that is required for efficient VPgpUpU-primed positive-strand initiation.
Poliovirus (PV) is a member of the Picornaviridae family of single-stranded positive-sense RNA viruses. The viral genome contains a 3′-terminal poly(A) tail and a large open reading frame that is flanked by nontranslated regions (NTR) at the 5′ and 3′ termini. A small viral protein, VPg (3B), is covalently linked to the 5′-terminal ends of all newly synthesized viral RNAs (1, 30, 43). Poliovirus RNA is first translated in the cytoplasm of the host cell to synthesize the viral structural and replication proteins and is then copied by the viral polymerase (3Dpol) to form full-length negative-strand RNA. The negative-strand RNA then serves as a template for the synthesis of positive-strand progeny RNA. The replication of viral RNA is highly asymmetric, with the ratio of positive-strand to negative-strand RNA synthesis ranging from 3 to 70 in various studies (21, 40, 49).
The 3′ NTR and poly(A) tail, the internal cre(2C) hairpin, and the 5′ cloverleaf are cis-acting elements that are required to replicate the viral genome. The 3′ NTR and poly(A) tail are both required for the efficient initiation of negative-strand RNA synthesis (26, 35, 36, 46, 50). The cre hairpin is a highly conserved structure that is located in various regions in the genomes of different picornaviruses (20, 22, 31, 33, 34, 42, 52) and is required for the synthesis of VPgpUpU (19, 41, 51) and positive-strand RNA (37, 38). The 5′-terminal cloverleaf is another highly conserved structure that is implicated in various aspects of viral RNA replication. The 5′ cloverleaf is organized into stem a, as well as stem-loops b, c, and d (Fig. 1) (2). Stem-loops b and d bind poly(rC) binding protein (PCBP) and viral protein 3CD, respectively, to form a ribonucleoprotein (RNP) complex (2, 26, 39, 41). The binding of PCBP to stem b in the 5′ cloverleaf is required to maintain the stability of poliovirus RNA (39). The binding of 3CD to stem-loop d in the 5′ cloverleaf is also required to stabilize viral RNA and for the initiation of negative-strand RNA synthesis (13). The 5′ cloverleaf bound to both PCBP and 3CD is believed to interact with PABP bound to the 3′ poly(A) tail to form a circular RNP complex that is required to initiate negative-strand RNA synthesis (13, 26, 32, 48). Therefore, the 5′ cloverleaf is a key cis-acting replication element that is required for negative-strand initiation and helps to ensure that virion RNA is the only polyadenylated RNA in infected cells that is copied by the viral polymerase.
FIG. 1.
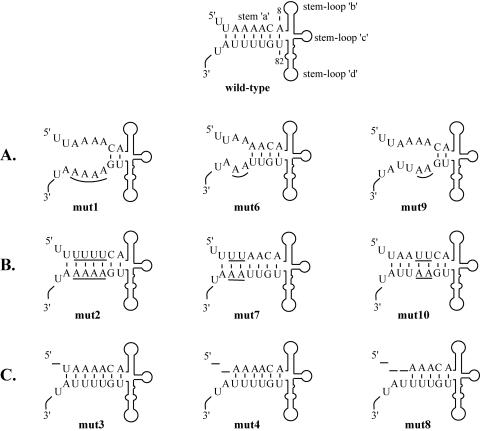
Stem a mutations in the 5′ cloverleaf of poliovirus RNA and predicted secondary structure of the 5′ cloverleaf formed by poliovirus wild-type RNA and the stem a mutants used in this study. The nucleotides that were altered or deleted are underlined. (A) The duplex structure of stem a was disrupted in mut1, mut6, and mut9. (B) By making compensatory nucleotide changes in both strands of stem a, the duplex structure was restored while the primary sequence was changed in mut2, mut7, and mut10. (C) The 5′-terminal nucleotides were sequentially deleted in mut3, mut4, and mut8.
The 3′ end of negative-strand RNA serves as the template for positive-strand initiation. Using a 3′-terminal negative-strand probe, UV cross-linking of 36- and 38-kDa cellular proteins was observed in poliovirus-infected cellular extracts (45). Viral proteins 2C and 2BC were also shown to bind near the 3′-terminal end of negative-strand RNA but not to the 5′ cloverleaf in the positive-strand RNA (5, 6). Similar observations were also reported for the 3′-terminal end of negative-strand RNAs of hepatitis A virus and human rhinovirus (5). Therefore, both cellular and viral proteins are known to bind to the 3′-terminal end of negative-strand RNA, although the functional significance of this binding relative to viral RNA replication has not been determined.
Both the primary sequence and the structure of stem a in the 5′ cloverleaf are highly conserved in enteroviruses and rhinoviruses (29, 54, 55). In this study, we performed an extensive mutational analysis of stem a and determined the effects of these mutations on viral RNA replication in preinitiation RNA replication complexes (PIRCs) and virus formation in HeLa S10 translation-replication reactions. Our results showed that the duplex structure of stem a was required for negative- but not positive-strand initiation. In contrast, the 5′-terminal sequence in stem a, and consequently the 3′-terminal sequence in negative strands, was required for efficient positive-strand initiation and virus formation. This suggests that the 3′-terminal ends of negative-strand templates are the primary sequences that are required in cis for efficient VPgpUpU-primed positive-strand initiation.
MATERIALS AND METHODS
Viral cDNA clones and transcript RNAs.
The viral construct pT7-PV1(A)80 P3 (pT7-P3) was derived from a cDNA clone of the Mahoney strain of poliovirus type 1 (PV1), pT7-PV1(A)80, as previously described (27). Two nucleotides were deleted at positions 652 and 653 in the plasmid pT7-P3 to generate plasmid pT7-PV1(A)80 F3, hereafter referred to as pT7-F3. RNA transcripts obtained from this plasmid were designated F3 RNA (Fig. 2B). The 2-nucleotide deletion in F3 RNA introduced a frameshift mutation that blocked the translation of this RNA after the sixth amino acid. Plasmid pT7-F3 was used as the parental construct for the mutants depicted in Fig. 1. In pT7-F3mut1, 4 nucleotides, T84 to T87, were changed to A84 to A87. pT7-F3mut2 was derived from pT7-F3mut1, in which nucleotides A3 to A6 were altered to T3 to T6. In pT7-F3mut3, the T1 nucleotide was deleted at the 5′-terminal end. In pT7-F3mut4, 2 nucleotides, T1 and T2, were deleted at the 5′-terminal end. In pT7-F3mut6, nucleotides T86-T87 were replaced with nucleotides A86-A87. Plasmid pT7-F3mut7 was derived from pT7-F3mut6, in which nucleotides A3-A4 were replaced with nucleotides T3-T4. Plasmid pT7-F3mut8 contained a 3-nucleotide deletion, T1T2A3, at the 5′-terminal end. Plasmid pT7-F3mut9 contained a 2-nucleotide change from T84-T85 to A84-A85. Plasmid pT7-F3mut10 was derived from pF3mut9, in which nucleotides at positions A5-A6 were replaced with nucleotides T5-T6. Transcript RNAs obtained from these plasmids were designated F3mutN, where N is the mutation number. These transcript RNAs contain two nonviral 5′-terminal G nucleotides that had no effect on negative-strand synthesis but inhibited positive-strand synthesis below detectable levels (12, 25, 37). RNA secondary structures were predicted using the MFOLD program by Zucker and Turner (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi). In the predicted structures, stem-loops b, c, and d in the 5′ cloverleaf were maintained in all the engineered stem a mutations (Fig. 1).
FIG. 2.
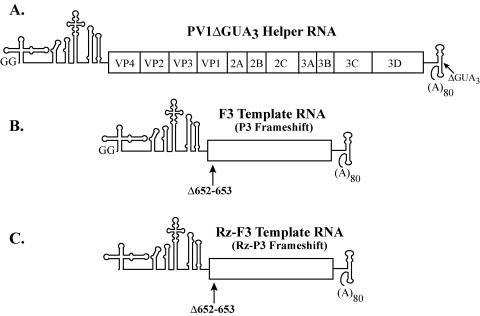
Schematic of poliovirus RNAs utilized in this study. (A) Diagram of the nonreplicating helper RNA, PV1ΔGUA3, which encodes all of the viral proteins. (B) Diagram of F3 template RNA. F3 RNA contains a frameshift mutation, as indicated in the diagram, and does not encode any viral proteins. This RNA contains two 5′-terminal nonviral G nucleotides and was used in assays for negative-strand RNA synthesis. (C) Diagram of Rz-F3 template RNA. This RNA contains a self-cleaving 5′-terminal hammerhead ribozyme that results in the formation of viral transcripts with an authentic 5′ end. Rz-F3 RNA supports the synthesis of both negative- and positive-strand product RNAs.
All the mutations described above were also engineered into plasmids containing a hammerhead ribozyme (Rz) between the T7 promoter and the 5′ end of the poliovirus sequence (12, 37). For each mutation that was engineered at the 5′ end of the poliovirus sequence, the complementary sequence in the hammerhead ribozyme was altered to ensure efficient cleavage. Transcripts obtained from these plasmids are indicated with an Rz prefix and contain an authentic 5′-terminal end (Fig. 2C). The Rz RNA transcripts support the synthesis of both negative- and positive-strand RNAs (12, 25, 37). Some of the stem a mutations were also engineered into full-length pT7-Rz-PV1(A)80 and designated pT7-Rz-PV1(A)80mutN, where N represents the mutations 1 through 10, as described above. The RNAs transcribed from these plasmids were able to encode all the viral replication proteins and were designated Rz-PV1mutN RNA. pT7-PV1(A)80ΔG7418-A7422 (pPV1ΔGUA3) contained a GUAAA deletion in the 3′ NTR (Fig. 2A) (13). RNA transcripts obtained from this plasmid were designated helper RNAs in this study and were used to synthesize the viral replication proteins in reactions containing F3 RNA templates (37).
All plasmid DNAs were linearized with Mlu1 and transcribed in vitro in reactions containing bacteriophage T7 RNA polymerase and 500 μM each nucleoside triphosphate, as previously described (37). Where indicated, transcript RNAs were synthesized with a 5′ cap using a 7-methyl guanosine cap analog (13). RNA transcripts were purified by Sephadex G-50 gel filtration chromatography as described previously and stored in ethanol at −20°C (10).
RNA stability assays.
Labeled viral RNAs were transcribed in reactions containing 50 μCi of [α-32P]CTP (400 Ci/mmol) for RNA stability assays. Equimolar amounts of a labeled viral RNA and the helper RNA were added to HeLa S10 translation-replication reactions at a total concentration of 50 μg/ml. The composition of HeLa S10 translation-replication reactions has been described previously (9, 11). The reaction mixtures were incubated at 34°C for 4 h. At the indicated time points, 20 μl of the translation reaction mixture was removed and added to 400 μl of 0.5% sodium dodecyl sulfate buffer (10 mM Tris HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl). A 50-μl portion of this reaction mixture was removed in duplicate, and labeled RNA was precipitated in 1 ml of 5% trichloroacetic acid-2% sodium pyrophosphate-5 μl of yeast tRNA (50 μg/ml). The labeled RNA was collected on filters and quantitated by liquid scintillation counting. The amount of labeled RNA recovered at each time point was calculated as a percentage of the amount of input RNA. From the remaining portion of the reaction mixture, the labeled RNA was recovered by phenol-chloroform extraction and ethanol precipitation. The labeled RNA was then analyzed by CH3HgOH-agarose gel electrophoresis and detected by autoradiography of the dried gel.
RNA replication assays.
HeLa S10 translation-replication reactions containing 50 μg of RNA transcripts/ml were prepared as previously described (9). In trans-replication assays, the helper and template RNAs were added in equimolar concentrations. After incubation at 34°C for the appropriate time, PIRCs were isolated by centrifugation. The synthesis of labeled viral RNA in PIRCs was measured by the two methods described below.
(i) Method 1: negative-strand RNA synthesis.
PIRCs isolated from HeLa S10 translation-replication reactions containing F3 template RNA (wild type or mutant) were resuspended in a reaction mixture containing 35.5 mM HEPES (pH 8.0), 120 mM KCH3CO2, 2.75 mM Mg(CH3CO2)2, 5 mM KCl, 3 mM dithiothreitol, 0.5 mM CaCl2, 1 mM EGTA, 30 mM creatine phosphate, 0.4 mg of creatine kinase/ml, 1 mM ATP, 250 μM (each) GTP and UTP, 125 μM CTP, 30 μCi of [α-32P]CTP (400 Ci/mmol), and 50 μg of puromycin/ml. The reaction mixtures were incubated at 37°C for 8 min. Labeled viral RNA products were recovered by phenol extraction and ethanol precipitation and were analyzed by CH3HgOH-agarose gel electrophoresis and autoradiography. The amount of labeled RNA was quantitated using a Molecular Dynamics PhosphorImager and was defined in terms of phosphorimager (PI) units.
(ii) Method 2: positive-strand RNA synthesis.
F3 template RNA or Rz-F3 template RNA (wild type or mutant) was resuspended in HeLa S10 translation-replication reaction mixtures containing the helper RNA and incubated at 34°C for 1 to 4 h as indicated in the figure legends. Where indicated, the helper RNA was translated in the reactions prior to the addition of the template RNA. PIRCs were resuspended in the reaction mixture described above and incubated for 1 h at 37°C. The labeled viral RNA products were recovered and analyzed as described above.
PIRCs formed with normal transcript RNAs synthesize only labeled negative-strand RNA (12, 25, 37). In contrast, PIRCs formed with Rz-RNAs support the synthesis of both negative- and positive-strand labeled product RNAs as described previously (12, 25, 37). To calculate the total amount (in PI units) of labeled positive-strand RNA synthesized (y), the amount of labeled negative-strand RNA synthesized (x) was subtracted from the total amount of labeled RNA synthesized in reactions containing Rz-RNA, which includes both negative- and positive-strand RNAs (x + y). Therefore, the amount of labeled positive-strand RNA synthesized in a reaction was calculated as follows: y = (x + y) − x. The ratio between positive- and negative-strand RNAs was calculated by dividing the PI units obtained for positive-strand RNA by the PI units obtained for negative-strand RNA (i.e., y/x).
Virus production in HeLa S10 translation-replication reactions.
Infectious poliovirus was synthesized in HeLa S10 translation-replication reactions as previously described (7, 8). Briefly, reaction mixtures that contained 50 μg of Rz-PV1 RNA (wild type or mutant)/ml were incubated at 34°C for 18 h. The reaction mixtures were then treated with 20 μg of RNase A/ml and 8 μg of RNase T1/ml for 25 min at 25°C. The amount of infectious virus synthesized in each reaction was determined by using a plaque assay on BSC40 cells as described previously (7, 8).
Isolation and sequencing of poliovirion RNA.
The virus produced in a HeLa S10 translation-replication reaction was used to prepare a first-passage virus stock. A 50-μl HeLa S10 reaction mixture that contained 50 μg of Rz-PV1 RNA, Rz-PV1mut3 RNA, or Rz-PV1mut4 RNA/ml was incubated at 34°C for 18 h. The reaction mixtures were treated with RNase A and RNase T1, as described above. The reaction mixtures were diluted to 200 μl and used to infect a BSC40 monolayer (4 × 106 cells) in a T-25 flask. The virus was adsorbed for 30 min at 37°C (with rocking every 10 min). Five milliliters of Eagle's minimum essential medium was added, and incubation was continued until complete cytopathic effect was observed. The T-25 flasks containing the infected cells in Eagle's minimum essential medium were freeze-thawed three times. The medium was transferred to 15-ml conical tubes, and cellular debris was centrifuged at 800 × g for 5 min. Virus was pelleted from the supernatant by centrifugation at 147,000 × g for 2 h at room temperature. The virus pellet was resuspended in 0.5% sodium dodecyl sulfate buffer. Viral RNA was isolated by phenol-chloroform extraction and ethanol precipitation as described previously (10). The sequence at the 5′-terminal end of the isolated virion RNA was amplified using a 5′ rapid amplification of cDNA ends (RACE) procedure (Invitrogen) according to the manufacturer's instructions. Briefly, first-strand cDNA was synthesized using a virus-specific primer (5′CAGGTTTCATCACAGAAAGTGGG3′) and SuperScriptTM II reverse transcriptase. A homopolymeric tail was added to the 3′ end of the cDNA by using terminal transferase (Invitrogen). The tailed cDNA was amplified by PCR using the abridged anchor primer (Invitrogen) and a virus-specific primer (5′ATTGTCACCATAAGCAGCCA3′) to obtain a PCR product ∼600 nucleotides long. The 5′-terminal nucleotides of the PCR products were sequenced by both automated sequencing (ABI Prism; Perkin-Elmer) and cycle sequencing (Promega) using an internal virus-specific primer (5′CGGGAAGGGAGTATAAAACAGG3′). The sequence of the first 140 nucleotides at the 5′-terminal end of the PCR products was compared with the sequence determined for wild-type poliovirion RNA using the same procedure.
RESULTS
Effects of stem a mutations on viral RNA stability.
Previous studies have shown that the 5′ cloverleaf plays an important role in regulating the translation, replication, and stability of poliovirus RNA (13, 26, 32, 41, 48). In this study, we determined how mutations that altered the primary sequence and structure of stem a in the 5′ cloverleaf affected the replication of poliovirus RNA. Previous studies have shown that some of the mutations in stem-loop b and stem-loop d of the 5′ cloverleaf destabilize viral RNA in HeLa S10 translation-replication reactions (13, 39). Therefore, it was essential to first ascertain the effects of stem a mutations on RNA stability before testing them in replication assays. In addition, to dissociate viral protein synthesis from stability and replication, we used trans-replication assays that contained a nontranslating template RNA (F3 RNA) and a nonreplicating helper RNA (PV1ΔGUA3), which provided equivalent amounts of the viral replication proteins in trans independently of the template RNA used in each reaction (Fig. 2).
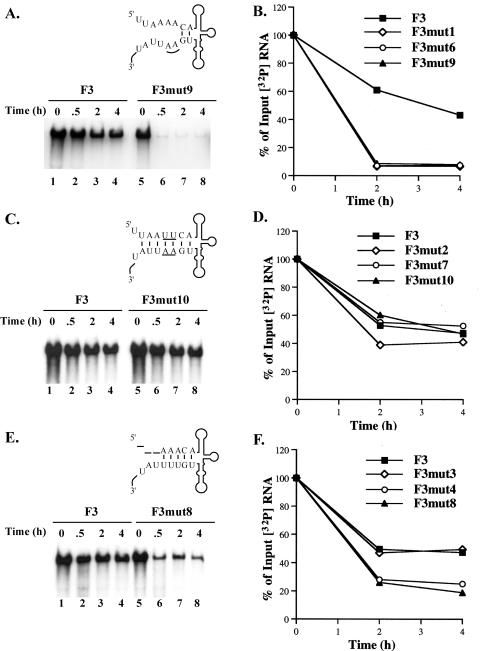
The stability of 32P-labeled F3 RNA was measured in the presence of helper RNA in HeLa S10 translation-replication reactions. The amount of labeled RNA remaining at each time point was calculated as a percentage of the amount of input RNA. F3 RNA was relatively stable, and ∼45% of the input RNA remained intact after 4 h (Fig. 3A, lanes 1 to 4). F3mut1 RNA, F3mut6 RNA, and F3mut9 RNA contained mutations that disrupted the duplex structure of stem a (Fig. 1A). F3mut9 RNA was very unstable compared to F3 RNA, and most of this RNA was degraded after 30 min (Fig. 3A). F3mut6 RNA and F3mut1 RNA were also unstable and were completely degraded by 2 h (Fig. 3B). These results indicate that the duplex structure of stem a is required to maintain viral RNA stability.
FIG. 3.
Effects of stem a mutations on viral RNA stability. HeLa S10 translation-replication reaction mixtures (150 μl) containing equimolar amounts of helper RNA and the indicated 32P-labeled viral RNA were incubated for 4 h at 34°C, and 20-μl samples of the reaction mixture were removed at the indicated time points. The 32P-labeled viral RNA remaining at each time point was determined by gel electrophoresis (A, C, and E) or by precipitation in trichloroacetic acid (B, D, and F) as described in Materials and Methods.
To investigate this observation further, we engineered compensatory mutations in the mutant RNAs described above to restore the duplex structure of stem a (F3mut 2, F3mut7, and F3mut10 [Fig. 1B]). There was no significant difference between the stability of F3mut10 RNA and that of F3 RNA (Fig. 3C), and after 4 h, ∼45% of the input RNA was recovered with both RNAs (Fig. 3D). Likewise, the stabilities of F3mut2 RNA and F3mut7 RNA were similar to the stability of F3 RNA (Fig. 3D). Therefore, restoring the duplex structure of stem a restored the stability of these RNAs to wild-type levels, whereas changing the primary sequence of stem a had no significant effect on viral RNA stability.
Additional stem a mutations, in which the 5′-terminal nucleotides were sequentially deleted (F3mut3, F3mut4, and F3mut8 [Fig. 1C]), were also analyzed for stability. Deleting the 5′-terminal U from stem a in F3mut3 RNA had no significant effect on RNA stability (Fig. 3F). This deletion did not affect the number of basepaired nucleotides in stem a and therefore maintained the duplex structure of stem a. In contrast, deleting 2 or 3 nucleotides at the 5′-terminal end (F3mut4 and F3mut8 [Fig. 1C]) produced RNAs that exhibited reduced stability (Fig. 3E and 3F). In this case, only 20 to 25% of the input RNA was recovered at 4 h. It is important to note that in both of these mutations the number of basepaired nucleotides in stem a was reduced (Fig. 1C). Therefore, these results also indicated that the duplex structure of stem a was required to maintain the stability of viral RNA in HeLa S10 translation-replication reactions.
Effects of stem a mutations on negative-strand RNA synthesis.
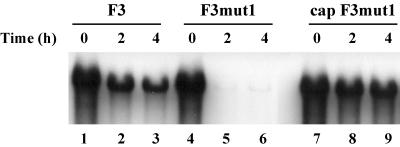
Since some of the stem a mutations affected the stability of the template RNAs, we added a 7-methyl guanosine cap to restore the stability of the template RNA to assay for negative-strand synthesis. It was previously shown that the addition of the 5′ cap has no effect on the initiation of negative-strand RNA synthesis (13). F3mut1 RNA was very unstable compared to F3 RNA (Fig. 3B and Fig. 4, compare lanes 1 to 3 with lanes 4 to 6). However, the addition of a 5′ cap to F3mut1 RNA restored its stability to levels equivalent to that of F3 RNA (Fig. 4, compare lanes 1 to 3 with lanes 7 to 9). Similarly, the addition of a 5′ cap fully restored the stability of F3mut6 RNA and F3mut9 RNA (data not shown). Therefore, the use of capped RNAs in negative-strand synthesis assays eliminated any effect that viral RNA instability might have on RNA replication.
FIG. 4.
Effect of adding a 5′ cap on the stability of F3mut1 RNA. HeLa S10 translation-replication reaction mixtures (150 μl) containing equimolar amounts of helper RNA and 32P-labeled F3 RNA, F3mut1 RNA, or F3mut1 RNA with a 5′ cap (cap F3mut1) were incubated for 4 h at 34°C, and 20-μl samples of the reaction mixture were removed at the indicated time points. The 32P-labeled viral RNA remaining at each time point was determined by gel electrophoresis as described in Materials and Methods.
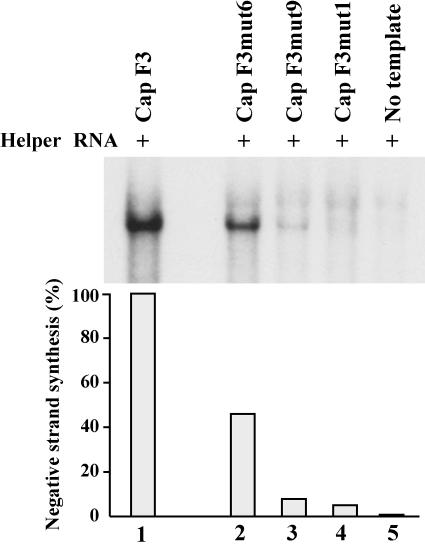
To investigate the effects of mutations that disrupted the duplex structure of stem a on negative-strand synthesis, we used F3mut1 RNA, F3mut6 RNA, and F3mut9 RNA in HeLa S10 translation-replication reactions. The level of negative-strand RNA synthesis observed in PIRCs isolated from reactions containing F3mut6 RNA, in which the duplex structure of stem a was partially disrupted, was 47% of the level observed with F3 RNA (Fig. 5, lanes 1 to 2). In reactions containing F3mut9 RNA or F3mut1 RNA, however, negative-strand synthesis was only 9 or 6% of the level observed with F3 RNA, respectively (Fig. 5, lanes 3 to 4). These results suggested that the primary sequence and/or the duplex structure of stem a was required for negative-strand initiation.
FIG. 5.
Effect of disrupting the duplex structure of stem a on negative-strand RNA synthesis. PIRCs were isolated from HeLa S10 translation-replication reactions containing the nonreplicating helper RNA and the indicated template RNAs and were resuspended in reaction mixtures containing [32P]CTP and incubated at 37°C for 8 min. The 32P-labeled product RNAs synthesized in these reactions were characterized by denaturing agarose gel electrophoresis and quantitated using a PhosphorImager as described in Materials and Methods. Labeled RNA synthesized in the reactions containing the mutant RNAs (lanes 2 to 4) was expressed as a percentage of the labeled RNA synthesized with F3 RNA (lane 1).
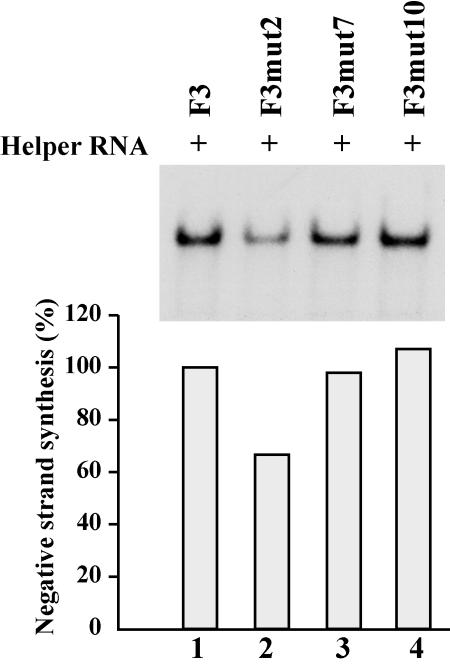
To address this question, we used stem a mutants in which the duplex structure was maintained but the primary sequence was altered (F3mut2, F3mut7, and F3mut10 [Fig. 1B]). These RNAs were stable in HeLa S10 translation-replication reactions, as described above. Equivalent levels of negative-strand synthesis were observed in PIRCs containing F3 RNA, F3mut7 RNA, or F3mut10 RNA (Fig. 6, lanes 1, 3, and 4). These results suggest that the deficiency in initiation of negative-strand synthesis observed with F3mut6 and F3mut9 RNAs was not due to the change in the primary sequence of stem a but was primarily a result of disrupting the duplex structure of stem a. With F3mut2 RNA, negative-strand synthesis was ∼65% of the level observed with F3RNA (Fig. 6, lanes 1 to 2). Although the duplex structure of stem a was maintained in F3mut2 RNA, 8 nucleotides were changed in this mutant, which may have caused the small decrease in negative-strand synthesis observed. Therefore, we concluded that maintaining the duplex structure of stem a was of primary importance for negative-strand initiation.
FIG. 6.
Effect of altering the 5′-terminal sequence on negative-strand RNA synthesis. PIRCs isolated from HeLa S10 reactions containing helper RNA and the indicated template RNA were resuspended in reaction mixtures containing [32P]CTP and incubated at 37°C for 8 min. The 32P-labeled product RNAs synthesized in these reactions were characterized by gel electrophoresis and quantitated as described for Fig. 5.
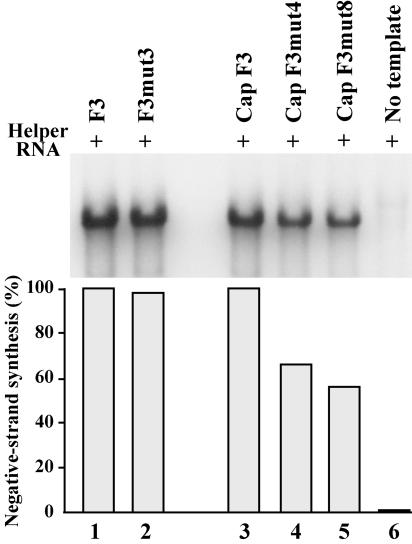
We next determined how the sequential deletion of nucleotides at the 5′-terminal ends of viral RNA templates affected negative-strand synthesis. Deleting the 5′-terminal U in F3mut3 RNA had no effect on negative-strand synthesis (Fig. 7, lanes 1 to 2). In contrast, removing 2 or 3 nucleotides from the 5′ terminus resulted in a measurable decrease in negative-strand synthesis. In reactions containing F3mut4 RNA and F3mut8 RNA, negative-strand synthesis was reduced to 66 and 56% of the amount observed with F3 RNA, respectively (Fig. 7, lanes 3 to 5). Based on the results described above, the reduction in negative-strand synthesis observed with F3mut4 RNA and F3mut8 RNA was most likely a result of shortening the length of stem a by 1 or 2 bp, respectively. These results were also consistent with the results with F3mut6, where the length of stem a was reduced by 3 bp and negative-strand synthesis was reduced to 47% of the levels observed with F3 RNA (Fig. 5). Therefore, the duplex structure of stem a appears to be essential for efficient negative-strand initiation.
FIG. 7.
Effect of the sequential deletion of the 5′-terminal nucleotides on negative-strand RNA synthesis. PIRCs isolated from HeLa S10 reactions containing helper RNA and the indicated template RNA were resuspended in reaction mixtures containing [32P]CTP and incubated at 37°C for 8 min. The 32P-labeled product RNAs synthesized in these reactions were characterized by gel electrophoresis and were quantitated as described for Fig. 5.
Effects of stem a mutations on positive-strand RNA synthesis.
We next investigated the effects of stem a mutations on the initiation of positive-strand RNA synthesis. Poliovirus transcript RNAs containing an authentic 5′-terminal end (Rz-RNA) synthesize both negative- and positive-strand product RNAs (10, 12, 25, 37). The abilities of PV1 RNA to synthesize negative-strand RNA and of Rz-PV1 RNA to synthesize both negative- and positive-strand product RNAs were previously confirmed using an RNase T1 fingerprint analysis (9, 37). Since poliovirus replication is highly asymmetric, an excess of positive-strand RNA over negative-strand RNA is synthesized in reactions containing Rz-RNA templates. In reactions containing F3 RNA, the labeled product RNA represents newly synthesized negative strands, whereas in reactions containing Rz-F3 RNA, the labeled RNA represents both negative- and positive-strand RNAs (Fig. 8, lanes 1 and 2). We quantitated the amount of labeled negative-strand RNA, the amount of labeled positive-strand RNA, and the ratio of positive/negative-strand synthesis (see Materials and Methods). In trans-replication assays with Rz-F3RNA and F3 RNA, the positive/negative-strand ratio ranged from 7.0 to 10.0 in five separate experiments (Fig. 8 to 10). Therefore, stem a mutations that specifically inhibit positive-strand synthesis were expected to reduce the positive/negative-strand ratio below 7. In contrast, mutations that inhibit negative-strand synthesis would inhibit the total amount of RNA synthesized but not lower the positive/negative-strand ratio. Therefore, the positive/negative-strand ratio determined in these experiments was used as a measure of the efficiency of positive-strand RNA synthesis.
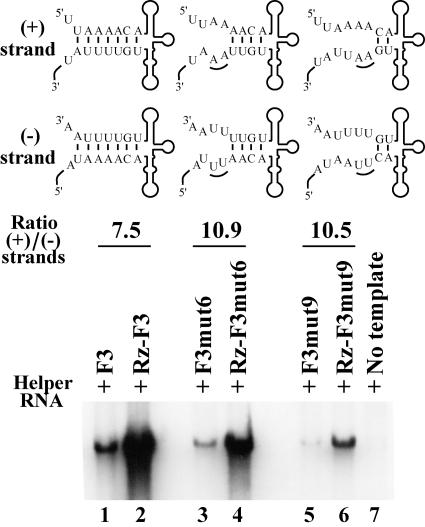
FIG. 8.
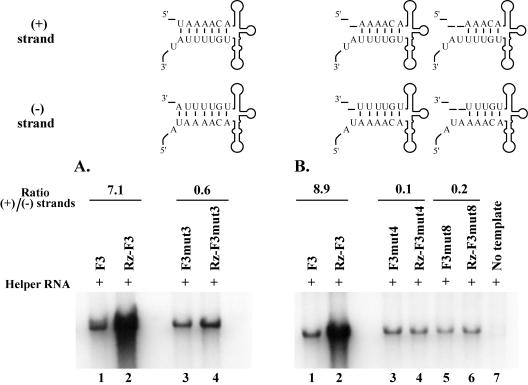
Effect of disrupting the duplex structure of stem a on positive-strand RNA synthesis. HeLa S10 translation-replication reaction mixtures that contained 2 mM guanidine HCl and helper RNA were incubated at 34°C for 1 h. The indicated template RNA was added and allowed to remain in the reaction for 1 h. PIRCs were isolated from these reactions, resuspended in reaction mixtures containing [32P]CTP, and incubated for 1 h at 37°C. The resulting 32P-labeled RNA products were then analyzed by gel electrophoresis and quantitated using a PhosphorImager. The ratios of positive/negative-strand synthesis shown were calculated for the wild-type and mutant RNAs as described in Materials and Methods. The predicted secondary structures for the 5′ end of the positive strand and the 3′ end of the negative strand are shown for each RNA. The altered nucleotides are underlined.
FIG. 10.
Effect of sequential deletion of 5′-terminal nucleotides on positive-strand RNA synthesis. (A) HeLa S10 translation-replication reaction mixtures that contained 2 mM guanidine HCl, helper RNA, and the indicated template RNA were incubated at 34°C for 4 h. (B) HeLa S10 translation-replication reaction mixtures that contained 2 mM guanidine HCl and helper RNA were incubated at 34°C for 1 h. The indicated template RNA was added and incubated for 1 h. PIRCs were isolated from the reactions shown in panels A and B, resuspended in reaction mixtures containing [32P]CTP, and incubated for 1 h at 37°C. The resulting 32P-labeled product RNA was analyzed by gel electrophoresis and quantitated as described for Fig. 8. The ratios of positive/negative-strand synthesis shown were calculated for the wild-type and mutant RNAs as explained in Materials and Methods. The predicted secondary structures for the 5′ end of the positive strand and the 3′ end of the negative strand are shown for mutant RNAs. The deleted nucleotides are underlined.
As shown earlier, some of the RNAs containing mutations in stem a were unstable in the HeLa S10 translation-replication reactions. However, it was not possible to stabilize the Rz-RNAs by adding a 5′ cap to the original transcript RNAs, since the 5′ cap is removed when the ribozyme undergoes autocatalytic cleavage. Therefore, to reduce the degradation of the template RNA, we presynthesized the viral replication proteins and reduced the time the template RNA was incubated in the HeLa S10 translation-replication reactions. In these experiments, HeLa S10 translation-replication reactions containing the helper RNA were incubated at 34°C for 1 h, the template RNA was then added, and the reaction mixtures were incubated for one additional hour. PIRCs were isolated from these reactions, and RNA replication was measured as described above. In PIRCs containing Rz-F3 RNA and F3 RNA, the positive/negative-strand ratio was 7.5 (Fig. 8, lanes 1 to 2). In the same experiment, a positive/negative-strand ratio of 10.9 was obtained for Rz-F3mut6 RNA and F3mut6 RNA (Fig. 8, lanes 3 to 4). Similar results were also obtained with Rz-F3mut9 RNA and F3mut9 RNA, where the positive/negative-strand ratio was 10.5 (Fig. 8, lanes 5 to 6). Note that in both these mutations, the duplex structure of stem a was disrupted (Fig. 1A). Therefore, these results suggest that the duplex structure of stem a was not directly required for positive-strand initiation.
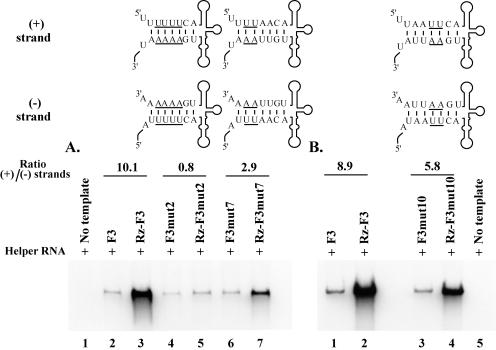
To study the effect of altering the primary sequence at the 5′-terminal ends of poliovirus RNA transcripts on positive-strand RNA synthesis, we used stem a mutations in which the duplex structure of stem a was maintained but the primary sequence was changed (Fig. 1B). In reactions containing Rz-F3mut7 RNA and F3mut7 RNA, the positive/negative-strand ratio was 2.9 (Fig. 9A, lanes 6 to 7). In the same experiment, the positive/negative-strand ratio was 10.1 with Rz-F3 and F3 RNAs (Fig. 9A, lanes 2 to 3). Similarly, in reactions containing Rz-F3mut10 RNA and F3mut10 RNA, the positive/negative-strand ratio was 5.8 compared to a positive/negative-strand ratio of ∼8.9 obtained with Rz-F3 RNA and F3 RNA (Fig. 9B, lanes 1 to 4). When 4 nucleotides in the primary sequence were changed, positive-strand synthesis was severely inhibited. A positive/negative-strand ratio of 0.8 was obtained in reactions containing Rz-F3mut2 RNA and F3mut2 RNA (Fig. 9A, lanes 4 to 5). Taken together, these results demonstrated that altering 2 to 4 nucleotides at the 5′ terminus had a moderate to severe inhibitory effect on positive-strand initiation.
FIG. 9.
Effect of changing the primary sequence of stem a on positive-strand RNA synthesis. PIRCs were isolated from HeLa S10 reactions containing helper RNA and the indicated template RNA and were resuspended in reaction mixtures containing [32P]CTP and incubated for 1 h at 37°C. The resulting 32P-labeled product RNAs were analyzed by gel electrophoresis and quantitated as described for Fig. 8. Results for separate experiments are shown in panels A and B. The ratios of positive/negative-strand synthesis shown were calculated for the wild-type and mutant RNAs as described in Materials and Methods. The predicted secondary structures for the 5′ end of the positive strand and the 3′ end of the negative strand are shown for mutant RNAs. The altered nucleotides are underlined.
We then determined how deletion of the 5′-terminal nucleotides from viral RNA templates affected plus-strand RNA synthesis. A positive/negative-strand ratio of 0.6 was observed in reactions containing Rz-F3mut3 RNA and F3mut3 RNA (Fig. 10A, lanes 3 to 4). In reactions containing Rz-F3mut4 RNA and F3mut4 RNA or Rz-F3mut8 RNA and F3mut8 RNA, positive/negative-strand ratios of 0.1 and 0.2, respectively, were obtained (Fig. 10B, lanes 3 to 6). These results suggested that even a single-nucleotide deletion at the 5′-terminal end resulted in a severe inhibition of positive-strand initiation. The most likely explanation for these results is that the negative-strand RNA templates synthesized with these mutations do not serve as efficient templates for the initiation of positive-strand RNA synthesis. Taken together with earlier observations, these results indicate that the primary sequence, but not the duplex structure, of stem a is required for positive-strand initiation.
Effects of stem a mutations on virus production in HeLa S10 translation-replication reactions.
HeLa S10 translation-replication reactions support the complete replication of poliovirus, including the formation of infectious virus (7, 8). To determine the effects of the stem a mutations on the production of infectious virus, we engineered these mutations in full-length Rz-RNA transcripts. As expected, Rz-PV1 RNA supported the efficient production of infectious virus in the HeLa S10 translation-replication reactions (Table 1 and Fig. 11). With Rz-PV1 RNA, the virus titer ranged from 5.0 × 105 to 1.3 × 106 PFU/ml (Table 1). The mutant RNAs in which stem a was disrupted (mut1, mut6, and mut9) were unstable in the HeLa S10 translation-replication reactions (Fig. 3B) and did not support efficient negative-strand RNA synthesis (Fig. 5). Therefore, full-length RNAs containing these mutations were not characterized for virus formation. In the cases where the 5′-terminal sequence was changed but the duplex structure of stem a was maintained (mut2, mut7, and mut10), virus production was either partially or completely inhibited (Table 1 and Fig. 11). With mut2 RNA, virus production was completely inhibited, and with mut7 RNA, the virus titer was reduced by ∼4 log units (Table 1 and Fig. 11). These results were consistent with the previous observation that positive-strand synthesis was strongly inhibited in reactions containing Rz-F3mut2 RNA and Rz-F3mut7 RNA (Fig. 9A). With mut10 RNA, the virus titer was reduced by 1 log unit and the average plaque size was only slightly reduced compared to wild-type virus (Table 1 and Fig. 11). This was also consistent with the small decrease in positive-strand synthesis that was observed with Rz-F3mut10 RNA (Fig. 9B). Therefore, stem a mutations in which the 5′-terminal sequence was mutated inhibited both positive-strand RNA synthesis and virus production.
TABLE 1.
Effects of stem a mutations on virus production in HeLa S10 translation-replication reactions
| RNA transcript | PFU/ml | Plaque diam (mm)a |
|---|---|---|
| Rz-PV1 | 1.3 × 106 | 2.1 ± 0.4 |
| Rz-PV1mut2 | NDb | ND |
| Rz-PV1mut7 | 2.5 × 101 | 1.0 ± 0.5 |
| Rz-PV1 | 5.0 × 105 | 2.3 ± 0.4 |
| Rz-PV1mut10 | 5.0 × 104 | 1.9 ± 0.3 |
| Rz-PV1 | 1.3 × 106 | 2.1 ± 0.4 |
| Rz-PV1mut3 | 9.6 × 104 | 2.0 ± 0.3 |
| Rz-PV1mut4 | 4.3 × 104 | 2.1 ± 0.4 |
| Rz-PV1mut8 | 8.8 × 102 | 0.6 ± 0.2 |
Plaque diameter was calculated as an average of at least 15 plaques, except Rz-PV1mut7 RNA, which produced only 5 plaques.
ND, no detectable plaques were obtained.
FIG. 11.
Plaque morphology of virus produced in HeLa S10 reactions. Monolayers of BSC40 cells were infected with the virus produced in the HeLa S10 translation replication reactions containing the indicated RNAs. The cells were overlaid with Eagle's minimum essential medium containing 1% methyl cellulose and incubated at 37°C for 2 days. Plaques were visualized by staining the monolayers with 0.02% crystal violet. Representative wells for the virus produced in the reactions containing Rz-PV1 RNA (Wild-type) and each of the indicated mutant RNAs are shown.
Restoration of the 5′-terminal sequence in progeny virion RNA from virus produced in reactions containing mut3 and mut4 RNAs.
In mutant RNAs in which the 5′-terminal nucleotides were sequentially deleted (mut3, mut4, and mut8), virus production was strongly inhibited and the virus titer was reduced by 1 to 3 log units (Table 1). Therefore, these results confirmed that the sequence at the 5′-terminal end of poliovirus RNA or, more likely, the sequence at the 3′-terminal end of the negative-strand RNA templates was required for the efficient initiation of positive-strand synthesis and the formation of infectious virus. Surprisingly, the virus synthesized in reactions containing the mut3 and mut4 RNAs produced plaques identical in size to those produced by wild-type virus (Table 1 and Fig. 11). This suggested that the virus produced in these reactions contained virion RNA with a wild-type 5′-terminal sequence. This was confirmed by sequencing the 5′ end of the virion RNA using a 5′ RACE procedure, as described in Materials and Methods. The 5′-terminal sequence obtained in both cases (mut3 and mut4) was identical to that observed for wild-type virion RNA (Table 2). These results indicated that the 5′-terminal UU sequence was fully restored in virion RNAs synthesized in the HeLa S10 reactions containing either mut3 or mut4 input RNA.
TABLE 2.
Sequences of 5′-terminal ends of input transcript RNA and recovered progeny virion RNA
| RNA | Sequence of 5′-terminal enda |
|---|---|
| Wild-type virion RNA | UUAAAACAG |
| Rz-PV1 mut3 input RNA | _UAAAACAG |
| Mut3 progeny virion RNA | UUAAAACAG |
| Rz-PV1 mut4 input RNA | __AAAACAG |
| Mut4 progeny virion RNA | UUAAAACAG |
The sequences for wild-type poliovirion RNA and for the virion RNAs from virus produced in HeLa S10 reactions containing the indicated input RNAs were determined using a 5′ RACE procedure as described in Materials and Methods.
DISCUSSION
In this study, we showed that mutations in stem a in the 5′ cloverleaf have specific effects on various steps in the replication of poliovirus RNA. The duplex structure of stem a, as part of the 5′ cloverleaf, was essential for viral RNA stability and negative-strand synthesis in HeLa S10 translation-replication reactions. In contrast, altering the 5′-terminal sequence in poliovirus RNA had little or no effect on negative-strand synthesis but dramatically inhibited positive-strand synthesis. This was most likely a consequence of inhibiting positive-strand initiation at the 3′-terminal ends of negative-strand RNA templates. Therefore, our results suggest that the conserved sequence found at the 3′-terminal end of negative-strand RNA is an important cis-acting element that is required for positive-strand initiation.
Duplex structure of stem a is required for viral RNA stability and negative-strand synthesis.
Our results indicate that mutations in stem a that disrupted its duplex structure resulted in the rapid degradation of viral RNA in HeLa S10 translation-replication reactions. In mutations where the duplex structure of stem a was restored by inserting compensatory nucleotide changes, stability was restored to the viral RNA. Therefore, even though the primary sequence in stem a was altered, viral RNA was stable, provided that the duplex structure of stem a was maintained. These results were consistent with those of previous studies in which stem-loops b and d in the 5′ cloverleaf were also shown to be required to maintain viral RNA stability (13, 39). Therefore, the duplex structure of stem a, as part of the 5′ cloverleaf, was required to maintain the stability of viral RNA in the HeLa S10 translation-replication reactions.
To eliminate stability as a factor in studying the effects of stem a mutations on negative-strand synthesis, we added a 5′ cap to unstable mutant RNAs. As shown here and previously reported (13), the presence of a 5′ cap on wild-type viral RNA has no effect on negative-strand synthesis. Therefore, the addition of a 5′ cap stabilized mutant viral RNAs without affecting their ability to serve as templates for negative-strand synthesis. The results of this study indicated that when stem a was disrupted, negative-strand synthesis was markedly reduced. Therefore, we concluded that the duplex structure of stem a was required for negative-strand initiation. In cases where the duplex structure of stem a was maintained but the primary sequence was altered, there was little or no effect on negative-strand RNA synthesis. In addition, since negative-strand synthesis was also inhibited when stem a was shortened in mut4 and mut8 RNAs, it appears that the length of stem a is also important for negative-strand initiation. These findings were also consistent with the results of a previous study in which it was shown that a mutation (pDNC-91) which completely disrupted the structure of stem a but maintained the 5′-terminal sequence was lethal in transfected cells (2). Therefore, we can now predict that the pDNC-91 mutation severely inhibited negative-strand synthesis, which would explain why this was a lethal mutation. Our results with stem a mutations are also consistent with previous studies that suggest that the 5′ cloverleaf, as part of a circular RNP complex, is required in cis for the initiation of negative-strand synthesis (13, 26, 32, 48). Similar models have now been proposed for other positive-strand RNA viruses in which both the 5′ and 3′-terminal ends are required to initiate negative-strand RNA synthesis (18, 19, 28, 53).
5′-terminal sequence of stem a is required for positive-strand synthesis.
In this study, mutations that altered the 5′-terminal sequence in the viral RNA (mut2 and mut7 RNAs) produced little or no virus (Table 1) and resulted in a large decrease in the ratio of positive/negative-strand synthesis. In direct contrast, mutations that disrupted the duplex structure of stem a but did not alter the 5′-terminal sequence (mut6 and mut9 RNAs) had no effect on the ratio of positive/negative-strand synthesis. Taken together, these results indicate that the 5′-terminal sequence in viral RNA, or more likely the 3′-terminal sequence in negative-strand RNA templates, is required in cis to initiate positive-strand synthesis. These results are consistent with those of previous studies with poliovirus and other positive-strand RNA viruses. A mutation in stem a similar to mut2 in this study (pDNC-902) was lethal in transfected cells (2). Although not characterized in the previous study, this mutation would be expected to severely inhibit positive-strand initiation and the formation of infectious virus. In Sindbis virus RNA, the deletion of nucleotide 5 or nucleotides 2 to 4 at the 5′ end has no effect on negative-strand synthesis. These mutations, however, inhibit viral replication, which suggests that positive-strand synthesis is specifically inhibited by the mutations (19). Similarly, previous studies showed that flock house virus requires an authentic 3′ end on negative-strand templates to initiate positive-strand RNA synthesis (3).
It has been suggested that newly synthesized negative-strand RNA is associated with input positive-strand viral RNA in the form of a double-stranded RNA intermediate (RF RNA) (4, 51). Multiple rounds of positive-strand initiation on negative-strand templates are known to result in the formation of replicative-intermediate RNAs. It is likely that both viral and cellular proteins are needed to destabilize the duplex structure at the 3′ end of the RF RNA to initiate positive-strand synthesis. Previous studies suggested that poliovirus protein 2C binds to the sequence at the 3′ end of negative-strand RNA (5). In addition, a cellular protein, p36, has been identified which binds to a sequence near the 3′ end of poliovirus negative-strand RNA (44, 45). The 36-kDa protein was recently reported to be hnRNP C (14, 17), which is also one of several nuclear proteins that are selectively redistributed to the cytoplasm upon poliovirus infection (15, 23). Interestingly, examination of the 3′ terminus in negative-strand RNA suggests that this sequence is similar to the RNA sequence recognized by the hnRNP C proteins (C1/C2). The hnRNP C proteins bind to (U)n motifs that contain >3 consecutive Us (47). Therefore, the 3′-terminal sequence in poliovirus negative-strand RNA, 3′AAUUUUGUC5′, appears to be a candidate sequence for binding hnRNP C1. Although the precise role of hnRNP C1, as well as other cellular and viral proteins, is not known, it is possible that the binding of these proteins could play a role in partially denaturing the RF RNA, thereby facilitating the initiation of positive-strand RNA synthesis.
Proposed model for VPgpUpU-primed positive-strand synthesis.
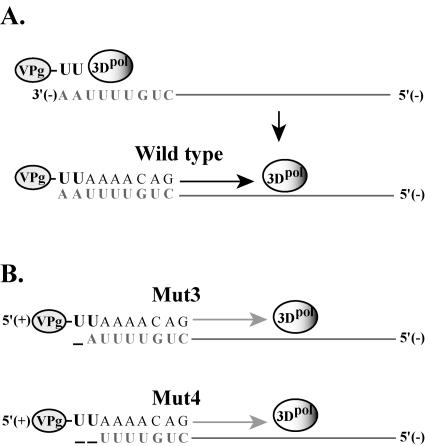
Recently, we and others reported that VPgpUpU, which is synthesized on the cre(2C) hairpin, is specifically required as a primer to initiate positive-strand RNA synthesis (37, 38). The severe reduction in the ratio of plus-strand to minus-strand synthesis that was observed in reactions with mut3 and mut4 RNAs suggested that the negative-strand templates in which either one or both of the 3′-terminal A nucleotides were deleted did not serve as efficient templates for positive-strand initiation. These results are consistent with a model in which preformed VPgpUpU base pairs with the two 3′-terminal A nucleotides in the negative-strand RNA and acts as a primer for the initiation of positive-strand RNA synthesis (Fig. 12A). The deletion of either one or both of the 3′-terminal A nucleotides would be expected to interfere with the ability of VPgpUpU to base pair with the 3′ end of the negative-strand templates and to function as a primer for positive-strand initiation (Fig. 12B). This would explain the dramatic reduction in the ratio of plus-strand to minus-strand synthesis and the drop in the virus titer observed with mut3 and mut4 RNAs. Interestingly, the virus synthesized in reactions containing Rz-PV1mut3 and Rz-PV1mut4 RNAs produced wild-type-size plaques. We confirmed that the progeny virus recovered from these reactions contained a wild-type 5′-terminal sequence. Therefore, these results suggest that even in the absence of the 3′-terminal A nucleotides in negative-strand RNA, preformed VPgpUpU was able to initiate positive-strand synthesis and restore the 5′-terminal sequence in the progeny virion RNA, although very inefficiently (Fig. 12B). These results are consistent with those of previous studies in which virus with wild-type plaque morphology and sequence was recovered from cells transfected with coxsackievirus B3 or poliovirus RNA transcripts lacking the two 5′-terminal U nucleotides (24, 29). Overall, these results strongly support a model in which preformed VPgpUpU is used to initiate positive-strand RNA synthesis (Fig. 12B).
FIG. 12.
Model for the VPgpUpU-primed initiation of positive-strand RNA synthesis at the 3′ end of negative-strand RNA templates. (A) The poliovirus polymerase, 3Dpol, utilizes preformed VPgpUpU as a primer to initiate positive-strand RNA synthesis at the 3′ ends of poliovirus negative-strand RNA templates. VPgpUpU pairs with the two complementary A nucleotides at the 3′-terminal end of a negative-strand RNA template to facilitate the efficient initiation of positive-strand synthesis. (B) mut3 and mut4 RNAs contain either a 5′-terminal U or UU deletion, respectively (Fig. 1). Therefore, mut3 and mut4 negative-strand RNAs would contain a 3′-terminal A or AA deletion. As shown in panel B, VPgpUpU is able to function as a primer for positive-strand initiation on both of the mutant negative-strand templates. In this case, however, initiation would be inefficient (depicted by light-gray arrows) due to the absence of either one or both of the 3′-terminal A nucleotides. This mechanism would restore the wild-type sequence at the 5′ ends of nascent positive strands and would explain why only wild-type progeny virus was recovered from reactions containing either mut3 or mut4 input RNAs. For clarity, the model was simplified to focus on the roles of 3Dpol, VPgpUpU, and the 3′ termini of negative-strand templates during positive-strand initiation. Not depicted in this model are the cellular membranes, the cellular proteins, and the precursor forms of VPg, 3Dpol, and the other viral proteins that are most likely part of functional RNA replication complexes. In addition, this model is not meant to imply that other sequences in negative-strand RNA, including the 5′ end, might play a role in positive-strand initiation in some host cells (16).
A characteristic of positive-strand RNA viruses is that viral replication is highly asymmetric, with numerous positive strands being synthesized on a single negative-strand template. This asymmetry is dependent on highly efficient positive-strand RNA synthesis on negative-strand templates. Recent findings suggest that an excess of the primer VPgpUpU preformed on the cre(2C) hairpin could potentially promote efficient positive-strand initiation (37, 38). The results of the present study indicate that the primary sequence at the 3′-terminal ends of negative-strand templates is also required for efficient positive-strand RNA initiation. Therefore, the conserved 3′-terminal sequence in negative-strand templates and the synthesis of excess VPgpUpU appear to work together to promote efficient positive-strand initiation and the asymmetric replication of poliovirus RNA.
Acknowledgments
This work was supported by Public Health Service grants AI15539 and AI32123 from the National Institute of Allergy and Infectious Diseases.
We thank Joan Morasco for excellent technical assistance and Sushma Ogram and Jessica Parilla for critically reading the manuscript.
REFERENCES
- 1.Ambros, V., and D. Baltimore. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263-5266. [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 3.Ball, L. A. 1994. Replication of the genomic RNA of a positive-strand RNA animal virus from negative-sense transcripts. Proc. Natl. Acad. Sci. USA 91:12443-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltimore, D. 1969. The replication of poliovirus, p. 101-176. In The biochemistry of viruses. Marcel Dekker, New York, N.Y.
- 5.Banerjee, R., and A. Dasgupta. 2001. Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J. Gen. Virol. 82:2621-2627. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee, R., W. Tsai, W. Kim, and A. Dasgupta. 2001. Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′ terminus negative-strand cloverleaf requires an intact stem-loop b. Virology 280:41-51. [DOI] [PubMed] [Google Scholar]
- 7.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton, D. J., and J. B. Flanegan. 1993. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J. Virol. 67:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3Dpol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 11.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barton, D. J., B. J. Morasco, L. E. Smerage, and J. B. Flanegan. 2002. Poliovirus RNA replication and genetic complementation in cell-free reactions, p. 461-469. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 13.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6:702-713. [DOI] [PubMed] [Google Scholar]
- 15.Belov, G. A., A. G. Evstafieva, Y. P. Rubtsov, O. V. Mikitas, A. B. Vartapetian, and V. I. Agol. 2000. Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology 275:244-248. [DOI] [PubMed] [Google Scholar]
- 16.Brown, D. M., S. E. Kauder, C. T. Cornell, G. M. Jang, V. R. Racaniello, and B. L. Semler. 2004. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 78:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunner, J. E., J. H. C. Nguyen, H. H. Roehl, T. V. Ho, K. M. Swiderek, and B. L. Semler. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 79:3254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corver, J., E. Lenches, K. Smith, R. A. Robison, T. Sando, E. G. Strauss, and J. H. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 77:2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frolov, I., R. Hardy, and C. M. Rice. 2001. cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2A(pro). J. Virol. 75:10979-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giachetti, C., and B. L. Semler. 1991. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J. Virol. 65:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustin, K. E. 2003. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex. Virus Res. 95:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmon, S. A., O. C. Richards, D. F. Summers, and E. Ehrenfeld. 1991. The 5′-terminal nucleotides of hepatitis A virus RNA, but not poliovirus RNA, are required for infectivity. J. Virol. 65:2757-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurgens, C., and J. B. Flanegan. 2003. Initiation of poliovirus negative-strand RNA synthesis requires precursor forms of p2 proteins. J. Virol. 77:1075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klump, W. M., I. Bergmann, B. C. Muller, D. Ameis, and R. Kandolf. 1990. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J. Virol. 64:1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, Y. F., A. Nomoto, B. M. Detjen, and E. Wimmer. 1977. A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. USA 74:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mason, P. W., S. V. Bezborodova, and T. M. Henry. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76:9686-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melchers, W. J. G., J. G. J. Hoenderop, H. J. Bruins Slot, C. W. A. Pleij, E. V. Pilipenko, V. I. Agol, and J. M. D. Galama. 1997. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 71:686-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirmomeni, M. H., P. J. Hughes, and G. Stanway. 1997. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 71:2363-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morasco, B. J., N. Sharma, J. Parilla, and J. B. Flanegan. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 77:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray, K. E., A. W. Roberts, and D. J. Barton. 2001. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA 7:1126-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 42.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersson, R. F., J. B. Flanegan, J. K. Rose, and D. Baltimore. 1977. 5′-terminal nucleotide sequences of poliovirus polyribosomal RNA and virion RNA are identical. Nature 268:270-272. [DOI] [PubMed] [Google Scholar]
- 44.Roehl, H. H., T. B. Parsley, T. V. Ho, and B. L. Semler. 1997. Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation. J. Virol. 71:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roehl, H. H., and B. L. Semler. 1995. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J. Virol. 69:2954-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohll, J. B., D. H. Moon, D. J. Evans, and J. W. Almond. 1995. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 69:7835-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sokolowski, M., and S. Schwartz. 2001. Heterogeneous nuclear ribonucleoprotein C binds exclusively to the functionally important UUUUU-motifs in the human papillomavirus type-1 AU-rich inhibitory element. Virus Res. 73:163-175. [DOI] [PubMed] [Google Scholar]
- 48.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verheyden, B., S. Lauwers, and B. Rombaut. 2003. Quantitative RT-PCR ELISA to determine the amount and ratio of positive- and negative strand viral RNA synthesis and the effect of guanidine in poliovirus infected cells. J. Pharm. Biomed. Anal. 33:303-308. [DOI] [PubMed] [Google Scholar]
- 50.Wang, J., J. M. Bakkers, J. M. Galama, H. J. Bruins Slot, E. V. Pilipenko, V. I. Agol, and W. J. Melchers. 1999. Structural requirements of the higher order RNA kissing element in the enteroviral 3′UTR. Nucleic Acids Res. 27:485-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wimmer, E., C. U. T. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 52.Yang, Y., R. Rijnbrand, K. L. McKnight, E. Wimmer, A. Paul, A. Martin, and S. M. Lemon. 2002. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J. Virol. 76:7485-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 54.Zell, R., K. Sidigi, E. Bucci, A. Stelzner, and M. Gorlach. 2002. Determinants of the recognition of enteroviral cloverleaf RNA by coxsackievirus B3 proteinase 3C. RNA 8:188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zell, R., and A. Stelzner. 1997. Application of genome sequence information to the classification of bovine enteroviruses: the importance of 5′- and 3′-nontranslated regions. Virus Res. 51:213-229. [DOI] [PubMed] [Google Scholar]