Abstract
The full-length sequence of a satellite RNA (sat-RNA) of Beet black scorch virus isolate X (BBSV-X) was determined. This agent is 615 nucleotides long and lacks extensive sequence homology with its helper virus or with other reported viruses. Purified virus particles contained abundant single-stranded plus-sense monomers and smaller amounts of dimers. Single-stranded RNAs from total plant RNA extracts also included primarily monomers and smaller amounts of dimers that could be revealed by hybridization, and preparations of purified double-stranded RNAs also contained monomers and dimers. Coinoculation of in vitro transcripts of sat-RNA to Chenopodium amaranticolor with BBSV RNAs was used to assess the replication and accumulation of various forms of sat-RNA, including monomers, dimers, and tetramers. Dimeric sat-RNAs with 5- or 10-base deletions or 15-base insertions within the junction regions accumulated preferentially. In contrast, the replication of monomeric sat-RNA was severely inhibited by five-nucleotide deletions in either the 5′ or the 3′ termini. Therefore, sequences at both the 5′ and the 3′ ends of the monomers or the presence of intact juxtaposed multimers is essential for the replication of sat-RNA and for the predomination of monomeric progeny. Comparisons of the time courses of replication initiated by in vitro-synthesized monomeric or multimeric sat-RNAs raised the possibility that the dimeric form has an intermediate role in replication. We propose that replication primarily involves multimers, possibly as dimeric forms. These forms may revert to monomers by a termination of replication at 5′ end sequences and/or by internal initiation at the 3′ ends of multimeric junctions.
Beet black scorch virus (BBSV) was first reported in the late 1980s (13, 14). The virus is responsible for serious damage to the sugar beet crop in the Xinjiang, Ningxia, Inner Mongolia, Gansu, and Heilongjiang provinces of China. BBSV infects sugar beet plants systemically through the roots after transmission with Olpidium brassicae zoospores (25) and causes black scorch symptoms on the leaves (6). Mechanical inoculation in a greenhouse resulted in local infections of 15 plant species, including several Chenopodium spp., producing inapparent to extremely mild symptoms or necrotic local lesions (7). Sugar beets on different plantations contaminated by BBSV isolates in China developed similar severe black scorch symptoms on leaves, and most plants died within a week after the appearance of symptoms. Normally, only a single RNA was associated with infections, but a small single-stranded RNA (ssRNA) that is similar to the satellite RNAs (sat-RNA) of other necroviruses (39) has been found in isolates from the Xinjiang province (6). This small RNA multiplied in test plants only when associated with BBSV genomic RNA, either from the same viral isolate or from other sources of BBSV (5). The presence of sat-RNA resulted in more local lesions on the leaves of Chenopodium amaranticolor plants than did inoculation with BBSV RNA alone when the inoculum was highly diluted. Many sat-RNAs of plant viruses such as Turnip crinkle virus (TCV) (9), Cucumber mosaic virus (CMV) (27), Subterranean clover mottle virus (17), and Lucerne transient streak virus (31) have been found to have complicated mechanisms of replication that result in the appearance of different forms of replicating RNAs. BBSV has recently been identified as a new member of the genus Necrovirus based on the sequence of a single 3,644-nucleotide plus-strand RNA (8). The BBSV sequence has a genome organization similar to that of other necroviruses (8) and shares the highest nucleotide sequence identity (61%) with Tobacco necrosis virus D (12). However, the molecular characteristics of the BBSV sat-RNAs have not previously been determined. In this paper, we report a sequence analysis of these sat-RNAs and a description of the multimeric forms involved in replication.
MATERIALS AND METHODS
Virus preparation and RNA extraction.
Four BBSV isolates were collected from the provinces of Xinjiang (BBSV-X), Ningxia (BBSV-N), Inner Mongolia (BBSV-IM), and Heilongjiang (BBSV-H). Among these, only the BBSV-X isolate naturally contained an additional small satellite-like RNA (6). These BBSV isolates were propagated in C. amaranticolor by mechanical inoculation in a greenhouse with a light period of 16 h and a temperature of 20 to 30°C. Virus particles were purified from mechanically inoculated leaves with local lesions at 3 to 4 days postinoculation (dpi) by extraction in 0.2 M phosphate buffer (containing 1% β-mercaptoethanol; pH 7.0) followed by two cycles of sucrose density gradient centrifugation (5). Fifty micrograms of purified BBSV was used for RNA extraction by denaturation in equal volumes of extraction buffer (20 mM Tris-HCl [pH 8.0], 1% sodium dodecyl sulfate, 200 mM NaCl, 5 mM EDTA) and phenol-chloroform (1:1). After phenol extraction and RNA precipitation, the RNAs were subjected to agarose gel electrophoresis, and the single small RNA band revealed by ethidium bromide staining was recovered from the gel by a freeze-thaw procedure (4). Double-stranded RNAs (dsRNAs) were recovered from infected plants on CF-11 cellulose columns followed by a 2 M LiCl2 precipitation step to further separate ssRNAs from dsRNAs as described by Dodds et al. (18).
cDNA synthesis and sequencing.
The small RNAs recovered from BBSV-X were used for cDNA synthesis either directly or after polyadenylation by poly(A) polymerase (GIBCO BRL) at 37°C for 30 min. Using the Universal RiboClone cDNA synthesis system (Promega Biotech), we synthesized cDNAs from 3 μg of RNA template, with either 1 μg of random oligonucleotides or 1.5 μg of oligo(dT)15 as a primer. Double-stranded cDNAs were blunted with T4 DNA polymerase and ligated into the SmaI site of the pGEM-7Zf(+) plasmid (Promega Biotech) for transformation into Escherichia coli strain DH5α. After screening by dot blotting and probing with a sat-RNA labeled with [γ-32P]ATP, positive cDNAs were sequenced in both directions by dideoxynucleotide chain termination (33) in a DNA autosequencer (ABI model 377A). The 5′ sequences of the sat-RNA were obtained by 5′ random amplification of cDNA ends (5′-RACE) and reverse transcription-PCR (RT-PCR) with the BB-S1 and BB-31 primers (Table 1) according to the procedure of Frohman et al. (22). Using templates extracted from purified BBSV-X particles that contained BBSV and sat-RNAs, we amplified products and then ligated them to the plasmid pMD18-T (TaKaRa) for sequencing after propagation in E. coli. The MFOLD web server of M. Zucker (http://bioinfo.rpi.edu/applications/mfold) (version 2.3) was used to analyze the sequences and to determine the secondary structures of the sat-RNAs.
TABLE 1.
List of oligonucleotides used for PCR
| Oligonucleotide | Sequence (5′-3′) | Structure and/or positiona |
|---|---|---|
| BB-S1 | CCTCACCTTCCCTGGCAGAC | nt 303 to 284 |
| BB-S2 | CCCGGGTAGCAATTAATTGC | CCC, nt 615 to 599 |
| BB-S3 | TAATACGACTCACTATAGGAAAGCTACGTGGTCATGGTAG | T7 promoter, G, nt 1 to 22 |
| BB-S4 | GAAAGCTACGTGGTCATGGTAG | nt 1 to 22 |
| BB-S5 | GTTCACATGATAACCGTTGG | nt 460 to 479 |
| BB-S7 | TAATACGACTCACTATAGGGGTAGCAATTAATTGC | T7 promoter, G, nt 615 to 599 |
| BB-S10 | GATATCAAATACTACCTCCC | EcoRV, nt 277 to 258 |
| BB-S11 | TAATACGACTCACTATAGCTACGTGGTCATGGTAGAG | T7 promoter, G, nt 6 to 24 |
| BB-S12 | GCAATTAATTGCTTTATAGC | nt 610 to 591 |
| BB-S13 | CTACGTGGTCATGGTAGAG | nt 6 to 24 |
| BB-31 | GCTCT(A/C)GAGAATTCCC(G)15 | Oligo(dG) anchor |
| MD18-T1 | ATCGTCGACCTGCAGGCATG | nt 425 to 406 in plasmid pMD18-T |
Numbers correspond to the sat-RNA nucleotide positions, and a reverse order of numbers indicates that the primer is complementary to the sat-RNA.
Construction of biologically active cDNAs.
Based on the full-length sequence of the sat-RNA and the oligonucleotide primers shown in Table 1, 13 cDNA clones were constructed for in vitro transcription of different oligomeric forms of the sat-RNA, as shown in Fig. 7. BB-S2 and BB-S3 primers were designed for RT-PCRs to obtain full-length cDNAs whose in vitro transcription could be driven by a bacteriophage T7 RNA polymerase promoter. Three cytosines (CCC) were fused to the 3′ terminus of the BB-S2 primer (Table 1) to generate a SmaI site by PCR amplification, together with three guanosines (GGG) at the 3′ end of the sat-RNA. The cDNA product was inserted into plasmid pMD18-T (TaKaRa) and introduced into E. coli DH5α for selection of pMBS-M4 (see Fig. 7). A cDNA clone, pUBS-R35, was constructed for minus-sense RNA transcription after PCR amplification with the BB-S4 and BB-S7 primers (see Fig. 7). Using similar strategies, we produced cDNA clones for the in vitro transcription of multimeric sat-RNAs by utilizing the BB-S2 and BB-S4 primers, which lack the T7 RNA polymerase promoter sequences (Table 1). The products were ligated into the SmaI site at the 3′ terminus of the monomeric, dimeric, or trimeric cDNA in order to construct multimers (pMBS-D10, pMBS-T6, and pMBS-Tet7) containing successive monomer units (see Fig. 7).
FIG. 7.
Illustration of cDNA clones used for in vitro transcription of BBSV sat-RNAs. The pMBS-M4 monomeric cDNA plasmid was used for in vitro transcription of the full-length (+) sat-RNA and was modified for transcription of three mutant (Mut) RNAs as follows. Mut-1 contained a 5-nt (5′-TACCC-3′) deletion at the 3′ end of the sat-RNA that was generated by use of the BB-S3 and BB-S12 primers (Table 1) to amplify pMBS-M4. Mut-2 had a 3′ end extension of 39 bases from pMD18-T, produced by digesting pMBS-M4 with EcoRI prior to in vitro transcription. Mut-3 consisted of a three-G extension at the 3′ end of the sat-RNA that was obtained by PCR amplification of pMBS-M4 with the BB-S3 and BB-S2 primers. The pUBS-Md3 monomeric cDNA plasmid contained a 5-nt (5′-GAAAG-3′) deletion from the 5′ end of the sat-RNA produced by amplification with the BB-S11 and BB-S2 primers (Table 1). The plasmid pMBS-M41 lacked the T7 promoter sequence and contained a full-length sat-RNA. The pUBS-R35 plasmid contained the monomeric sat-RNA cDNA in the reverse orientation, produced by amplification with the BB-S7 and BB-S4 primers (Table 1), for in vitro transcription of a negative-sense sat-RNA probe. pMBS-D10 had the full-length dimeric sat-RNA cDNA insert, and the dimeric pMBS-D17 plasmid had 5-nt deletions at the 3′ end (5′-TACCC-3′) and at the 5′ end (5′-GAAAG-3′) of the junction region. The pMBS-D34 dimeric plasmid had a 3′ extension at the junction region consisting of 15 nt (5′-GATCCTCTAGAGATT-3′) derived from the pMD18-T vector. The dimer in pMBS-D6 contained a deletion (5′-GAAAG-3′) at the 5′ end of the junction region. Plasmid pUBS-D21 had a deletion (5′-GAAAG-3′) at the 5′-proximal end of the dimeric cDNA sequence. The dimeric sat-RNA sequence in pMBS-D50 had a deletion (5′-TACCC-3′) at the 3′-proximal end. The plasmid pUBS-D5 contained deletions (5′-TACCC-3′) at the 3′ ends of the cDNAs of each sat-RNA monomer and of the junction region and had an insertion (5′-GGGGAT-3′) from the vector. The pMBS-T6 plasmid contained a trimeric sat-cDNA insert, and pMBS-Tet7 had the tetrameric sequence. pMBS and pUBS nomenclature corresponds to plasmids in the pMD18-T and pUC18 backgrounds, respectively.
For the transcription of mutant sat-RNAs, deletions or insertions were generated in both the monomeric and dimeric forms. Based on the plasmids pMBS-M4 and pUBS-Md3, each of which contains a single cDNA copy of the sat-RNA, four monomeric transcripts with modified 3′ or 5′ ends were synthesized by different strategies (see the scheme in Fig. 7). Similarly, sequence modifications of dimeric cDNAs were amplified by PCRs using different primers, as follows. (i) Five nucleotides (5′-GAAAG-3′) were removed from the 5′ end by use of the MD18-T1 and BB-S13 primers, which are oriented in opposite directions, on pMBS-M4. The deleted cDNA was religated downstream of the fragment amplified with the BB-S12 and BB-S3 primers, in which five nucleotides (5′-TACCC-3′) were deleted from the 3′ terminus. The resulting mutant dimeric pMBS-D17 derivative with two monomers was fused to the truncated ends of the monomers in the head-to-tail direction. (ii) Using the BB-S11 and BB-S2 primers, we deleted five nucleotides (5′-GAAAG-3′) from the 5′ end downstream of the T7 promoter. After ligation into the SmaI site of pUC18, another monomeric cDNA fragment was fused at the 3′ end to form the dimeric pUBS-D21 cDNA. (iii) Similarly, by using the BB-S13 and BB-S2 primers, we excised five nucleotides (5′ GAAAG-3′) from the 5′ end of a monomer that was then fused to a perfect 3′ downstream end (SmaI site) of another monomer in pMBS-M4 to form the pMBS-D6 dimer. (iv) After the formation of blunt ends by the use of T4 DNA polymerase, the fragment released by BamHI and PstI from pMBS-M41 was ligated into the SmaI site of pMBS-M4 in the head-to-tail direction. This manipulation produced an insertion of an additional 15 nucleotides from the pMD18-T plasmid into the junction region of the resulting pMBS-D34 dimer. (v) A mutant monomeric PCR product in which five nucleotides (5′-TACCC-3′) were deleted from the 3′ end was amplified by the BB-S4 and BB-S12 primers and fused downstream of an intact monomer at the SmaI site of pMBS-M4 to form the pMBS-D50 dimer. (vi) Five-nucleotide (5′-TACCC-3′) deletions at the 3′ ends of two monomers were generated separately by use of either the BB-S12 and BB-S3 primers with the T7 promoter or the BB-S12 and BB-S4 primers without the T7 promoter. After ligation of the T7 promoter-driven fragment to the SmaI site of pUC18, the plasmid was redigested with BamHI to insert a second downstream head-to-tail fragment. The resulting dimeric cDNA, pUBS-D5, contained an additional six bases (5′-GGGGAT-3′) from the vector in the junction region.
Plant inoculations with in vitro-synthesized sat-RNAs.
Linearized (at the 3′ end) cDNAs or purified PCR products were used as templates for runoff transcription in vitro at 37°C for 1 h by use of a T7 RNA polymerase kit as described by the manufacturer (Promega). The transcripts contained an additional G residue, and their concentrations were estimated visually on ethidium bromide-stained gels.
In vitro transcripts of the full-length infectious genomic RNA (gRNA) of the BBSV-N isolate were generated from the plasmid pUBF52 (Y.-H. Cao, unpublished data) and used as the helper virus for coinoculations in all experiments with sat-RNAs. For mechanical inoculation, the synthesized BBSV helper RNA was mixed with sat-RNA in a 1:3 or 1:5 proportion and then diluted with an equal volume of inoculation buffer (50 mM glycine, 30 mM K2HPO4, 1% bentonite, 1% celite, pH 9.2). Each young leaf of C. amaranticolor was rubbed with 15 μl of inoculum, and before the development of lesions, total RNAs were extracted at 3 to 4 dpi. The replication of sat-RNAs was assessed with a full-length cDNA probe produced with the DIG High Prime DNA labeling and detection starter kit I (Roche). RNA probes were also prepared by in vitro transcription of plus- or minus-sense sat-RNAs labeled with [α-32P]UTP.
Ribozyme activity analysis.
In order to test whether the in vitro-transcribed multimeric sat-RNAs were capable of self-processing activity, we separated the transcripts in agarose gels immediately after transcription. For these studies, the in vitro tetrameric and dimeric RNA transcripts from the template cDNAs pMBS-Tet7 and pMBS-D10, respectively, were recovered from 0.7% agarose gels by a freeze-thaw procedure as described previously (4). The purified transcripts were incubated in a buffer containing 5 mM MgCl2, 0.5 mM EDTA, and 50 mM Tris-HCl (pH 7.5) at 25°C for 30, 60, 90, or 120 minutes after being heated at 80°C for 1 min in 1 mM EDTA (21) or were incubated in 10 mM MgCl2-50 mM Tris-HCl (pH 7.5) at 37°C for the same time points, but without preheating (36). All reactions were terminated by the addition of an equal volume of 95% formamide-10 mM sodium EDTA and then immediately frozen at −80°C. Before being loading into 1% agarose gels for Northern blot analyses, the samples were thawed and denatured by heating at 80°C for 1 min and then snap cooled on ice.
Nucleotide sequence accession number.
The sat-RNA sequence has been submitted to the DDBJ/GenBank/EMBL database and assigned the accession number AY394497.
RESULTS AND DISCUSSION
Plant inoculation and virus purification.
Unlike fungal transmission in the field, the mechanical inoculation of susceptible sugar beet varieties with combinations of BBSV-N and BBSV-X failed to reliably produce symptoms of infection, and virus particles could not be recovered from inoculated plants. Although local lesions appeared on young leaves of C. amaranticolor plants by 3 to 4 dpi (Fig. 1, left and middle) in the greenhouse, no systematic symptoms were evident on the upper leaves (Fig. 1, right), as reported previously for other tested plants (7). In addition, viral or satellite RNAs were not detected by RT-PCR and Northern blotting on the upper leaves of plants that were inoculated with sat-RNA alone or coinoculated with BBSV RNA (data not shown). The BBSV-X isolate containing the sat-RNA caused more necrotic lesions on inoculated leaves than did BBSV-N. This may have been due to a host response induced by the sat-RNA and its helper similar to the indirect pathogenesis which is postulated to result from sat-RNA C associations with TCV in a systemic host (41). Virus particles were purified from the lesions, and each inoculum combination was analyzed for its viral RNA composition. A single coat protein species of approximately 25 kDa was also identified by SDS-polyacrylamide gel electrophoresis in both BBSV-X and BBSV-N purified from infected plants, but not in extracts from healthy plants (data not shown).
FIG. 1.
Symptoms on C. amaranticolor elicited by different BBSV isolates. The images, from left to right, show leaves inoculated with the wild-type BBSV isolate from Xinjiang Province (BBSV-X), which contained sat-RNA, with the BBSV isolate from Ningxia Province (BBSV-N) without a sat-RNA, and with in vitro-synthesized sat-RNA alone. Only the leaves inoculated with isolates developed symptoms.
RNA composition of BBSV preparations.
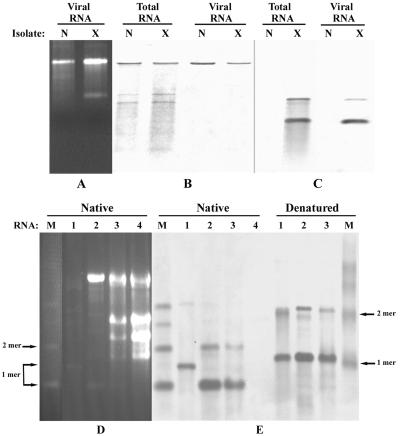
An RNA of ∼3.6 kb corresponding to BBSV gRNA was abundant in ethidium bromide-stained agarose gels (Fig. 2A) and Northern blots of viral RNA preparations (Fig. 2B). In addition to the gRNA present in the viral RNA preparations, two less intense bands which were presumed to be subgenomic mRNAs (sgRNAs) were evident in total RNA extracts from infected plants (Fig. 2B). This suggested that two sgRNAs are produced abundantly in infected tissue but that they are not encapsidated during viral morphogenesis. In addition to the gRNA and sgRNA species, a smaller RNA component was easily detected in ethidium bromide-stained RNAs from BBSV-X isolates, but this species was absent from the BBSV-N isolate (Fig. 2A) and the other two isolates, which were recovered from Inner Mongolia (BBSV-IM) and Heilongjiang (BBSV-H) (data not shown). In addition, Northern blot analysis of the RNAs with a probe specific for the smaller RNA revealed the presence of a minor RNA that migrated at the position expected of a dimer (Fig. 2C). These two smaller RNAs appeared to have little sequence relatedness to the BBSV gRNA because they failed to cross-hybridize reciprocally with probes derived from the gRNA and the smaller RNA species (Fig. 2B and C). Moreover, the small RNAs were present only in BBSV-X particles or in RNA extracts of plants infected by the BBSV-X isolate (Fig. 2C), and hence they had the properties of sat-RNAs.
FIG. 2.
Analysis of RNA components of BBSV isolates. (A) RNAs extracted from virus preparations after electrophoresis in 1.3% agarose gels and ethidium bromide staining. Lanes N and X show the slowly migrating BBSV gRNAs, and lane X shows the rapidly migrating small sat-RNA associated with BBSV-X. (B) Northern blots of BBSV-X and BBSV-N RNAs hybridized with cDNA probes specific for BBSV gRNA. (C) Northern blots of BBSV-X and BBSV-N RNAs probed with a cDNA specific for sat-RNA. Panels B and C were loaded with the same amounts of samples, which were separated in the same gel. (D) Ethidium bromide-stained 1.2% agarose gel showing nondenatured (native) RNAs separated as described below. Lane 1, samples of dsRNAs purified from C. amaranticolor infected with BBSV-X; lane 2, RNAs extracted from purified BBSV-X particles; lane 3, total RNAs from C. amaranticolor leaves inoculated with BBSV-X containing sat-RNA; lane 4, total RNAs from C. amaranticolor leaves infected by in vitro transcripts of BBSV-N RNA without sat-RNA; lane M, plus-sense monomeric (1-mer), dimeric (2-mer), trimeric, and tetrameric marker sat-RNAs. (E) RNAs hybridized with a digoxigenin (DIG)-labeled cDNA probe specific for sat-RNA. RNA samples corresponding to those in panel D were loaded in the same gel as native forms (left) or were denatured first (right) by the addition of 20 μl of RNA buffer (2 μl of 10× MOPS, 3.5 μl of formaldehyde solution, 10 μl of formamide) and heating at 68°C for 10 min. The positions of the monomeric and dimeric sat-RNAs are indicated for both the native and denatured forms.
In further experiments to determine the nature of the replicating intermediates, dsRNAs were extracted from leaves of C. amaranticolor plants infected with BBSV-X isolates and were analyzed by ethidium bromide staining (Fig. 2D). Hybridization analyses suggested that both monomeric and dimeric species were present as dsRNAs in both the native and denatured states (Fig. 2E), suggesting that the larger dimer species may be an independent replicating RNA. We therefore have designated the smaller RNA species a BBSV sat-RNA, and we postulate that the dimer species represents a sat-RNA that replicates independently of the monomeric derivative through a dsRNA dimer.
Sequence analysis and deletions in multimeric sat-RNA.
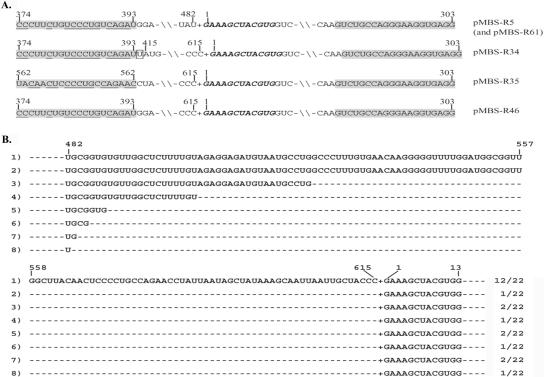
The small RNA component was recovered from agarose gels and used as a template for cDNA synthesis. Six overlapping cDNA clones were selected for sequencing, and a total of 603 nucleotides (nt) were determined by sequencing each of these clones in both directions. Three clones (pGBS-P12, pGBS-P40, and pGBS-P20) with identical 3′ ends, obtained by the use of oligo(dT)15 primers, and three additional cDNA clones (pGBS-R2, pGBS-R27, and pGBS-R19) with internal 3′ sequences amplified by random priming had identical sequences in the overlapping regions of the cDNA clones (Fig. 3A). In order to identify the 5′-proximal sequence, we performed a RACE RT-PCR analysis with a mixture of viral RNAs extracted from the purified BBSV-X isolate, and we detected amplified products in agarose gels with sizes ranging from 350 to 550 bp (data not shown). The sequences of 5 selected cDNAs from a total of 18 revealed a 12-nt sequence (5′-GAAAGCTACGTG-3′) extending beyond the 603-nt sequence of the cloned derivatives (see Fig. 5A). These results thus suggest that the full-length genome of the small BBSV RNA is composed of 615 nt, as illustrated in Fig. 3B.
FIG. 3.
cDNA synthesis and sequencing of BBSV satellite RNA. (A) Sequencing strategy using cDNA plasmids (pGBS-P12, pGBS-P40, and pGBS-P20) that were amplified with oligo(dT)15 primers and plasmids (pGBS-R2, pGBS-R27, and pGBS-R19) that were recovered after priming with random primers. (B) Complete nucleotide sequence of the 615-nt BBSV sat-RNA. The 12 underlined bases at the 5′ end were determined by RACE RT-RCR as shown in Fig. 5A.
FIG. 5.
Junction sequences of multimeric sat-RNAs. (A) Sequences of cDNA clones obtained by 5′ RACE RT-PCR. The extra 12 nt beyond the 603-nt sequence determined from the cloned plasmids are highlighted with bold italic letters. Imperfect inverted repeats of primer BB-S1 are shaded, and nucleotides with reverse complementarity to the 303- to 284-nt region are underlined. The numbers above each sequence indicate the nucleotide positions. An extra U residue in the gap in the pMBS-R34 insert is boxed. −\\−, common internal nucleotides; +, head-to-tail fusion of the two monomers. (B) Partial sequences of cDNAs obtained by RT-PCR with the primers BB-S5 and BB-S10. The intact junction sequence from the 3′ to 5′ end is shown in row 1, and variable deletions at the 3′ end are shown in rows 2 to 8. The numbers at the top represent the nucleotide positions, and the numbers on the right indicate the number of cDNA clones with a particular sequence among the 22 cDNAs that were sequenced. +, head-to-tail fusion of the two monomers. The dashed lines between nucleotides indicate deleted nucleotides.
The results of sequence alignments revealed no extensive sequence homology between the small monomeric sat-RNA and those of other viruses deposited in the EMBL databases, including the BBSV gRNA (8). Only four small open reading frames were found in the small RNA (data not shown). These open reading frames may encode predicted proteins with a maximum molecular mass of <2.8 kDa on the sense strand and <5.5 kDa on the antisense strand. Moreover, in contrast to the capsid proteins encoded by satellite viruses of Tobacco necrosis virus, Tobacco mosaic virus, Panicum mosaic virus, and Maize white line mosaic virus (2), no protein was produced from transcripts of the small RNA by in vitro translation in a reticulocyte lysate system (Promega) (data not shown). These results thus clearly show that the sat-RNA does not encode a protein capable of encapsidating the 615-nt RNA, and this observation, combined with the encapsidation of the RNA by the 25-kDa BBSV coat protein, provides additional evidence that it is a satellite RNA. Therefore, all of the available evidence agrees with our previous suggestion that the small monomeric RNA is a sat-RNA whose replication and encapsidation depend on the presence of the BBSV helper virus (7).
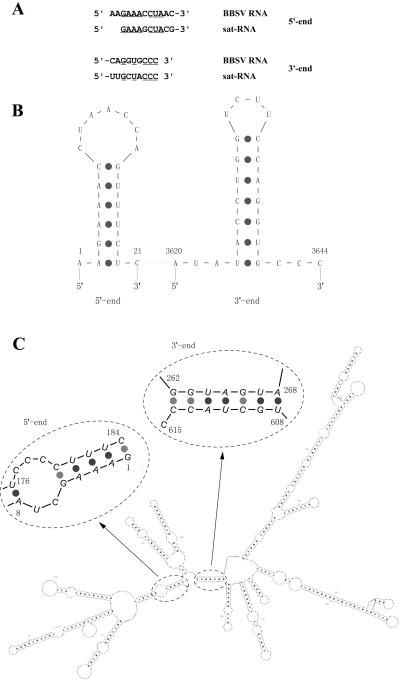
Sequence analyses of the 5′- and 3′-proximal ends of the sat-RNA and of BBSV gRNA (8) revealed several common nucleotides (Fig. 4A) and indicated that both the sat-RNA and gRNA contain potential stem-loop structures at their 5′ and 3′ termini (Fig. 4B). However, a comprehensive analysis of the entire sat-RNA sequence revealed more energetically favorable long-distance stem-loop interactions of both the 5′- and 3′-terminal regions (−237.5 kcal/mol at 25°C) (Fig. 4C). This analysis indicated that the first eight 5′-terminal residues of the sat-RNA have the potential to form a five-residue paired stem-loop in which nt 1 to 5 are engaged in long-distance base pairing with nt 180 to 184 (Fig. 4C). This pentanucleotide stem is interrupted by an unpaired bulge formed by nt 6 and 7 and nt 177 to 179 followed by a second hexanucleotide stem formed by the pairing of nt 8 to 13 with nt 171 to 176 (Fig. 4C). Interestingly, four paired nucleotides of the pentanucleotide sat-RNA stem (Fig. 4C) are identical to those in the six-residue stem-loop at the 5′ end of the BBSV gRNA (Fig. 4B). Moreover, although the long-distance 5′ sat-RNA stem-loop structure differs from that of the helper virus loop, five of seven residues in both of the loops (5′-CUAXCCX-3′) have sequence similarity (Fig. 4B and C). In the 3′ end region of the sat-RNA, long-distance pairing of residues 608 to 614 also forms a heptanucleotide stem with nt 262 to 268 (Fig. 4C), but this sequence has no obvious similarity to sequences surrounding the heptanucleotide stem-loop formed at the 3′ terminus of the BBSV gRNA (Fig. 4B). The highly ordered secondary structure formed by the 615-nt sat-RNA has approximately 70% base pairing, and the sequence consists of 26 short inverted repeats of 3 to 17 nt throughout the entire genome (Fig. 4C).
FIG. 4.
Comparison of structures of BBSV satellite and genomic RNAs. (A) Terminal sequences of BBSV RNA and satellite RNA. (B) Predicted stem-loop structures at the terminal regions of BBSV genomic RNA. (C) Computer-generated structure of the entire 615-nt sat-RNA. The 5′- and 3′-terminal regions are enlarged to show details of the stem-loops formed by long-distance pairing. The numbers in the circles indicate the nucleotide positions of the sat-RNA.
A potentially important observation is that the long-distance pairing brings the 5′- and 3′-proximal regions of the sat-RNA into close proximity (Fig. 4C), which might promote efficient replicase reinitiation during amplification of the sat-RNA. This highly organized structure consists of several double-stranded regions and stem-loops similar to those in other small subviral RNAs, such as the TCV sat-RNA C, which also forms dimers and other multimeric forms (10, 31, 34), and the CMV sat-RNA D4 (24, 30). These features may contribute to the high stability and survivability generally exhibited by sat-RNAs, which were first reported by Mossop and Francki (29). Similar structures have also been postulated to facilitate template switching of the RNA replicase of TCV and other viruses by reinitiation at base-paired regions (9). Interestingly, these structural features are also similar to those of the CMV Y satellite RNA and the potato spindle tuber viroid, which are both noncoding RNAs involved in RNA silencing (40).
In addition to the six cDNAs (Fig. 3A) that were amplified from the purified BBSV sat-RNA components recovered from agarose gels, five additional cDNA clones (Fig. 5A) were obtained from viral RNAs extracted from purified BBSV-X. The BB-S1 RT-PCR primer was designed to initiate reactions at positions 303 to 284 (Fig. 3B) and to extend to the 5′ end of the sat-RNA, whose sequences were flanked by oligo(dG) sequences derived from the anchor primer BB-31 (Table 1). However, of the five cDNAs recovered, four were amplified by pairing of BB-S1 at the designated site and at an imperfect inverted upstream repeat between positions 374 and 393, whereas the fifth was amplified by spurious pairing at another imperfect inverted repeat between positions 562 and 581 (Fig. 5A). Nevertheless, the results revealed that all five of these clones contained fusions of sequences of different lengths from the 3′ end of the monomeric sequence to the exact 5′ end of the next sat-RNA monomeric unit.
The clone pMBS-R46 differed from pMBS-R35 in that it had a longer 3′ end sequence than the first sat-RNA monomer that started at nt 374, but otherwise it was similar to pMBS-R35 in extending to the BB-S1 binding region of the next unit without any sequence gaps. The pMBS-R34, pMBS-R5, and pMBS-R61 inserts contained the same 5′ end sequences as pMBS-R46, but internal deletions were also found in these three clones. These results thus revealed that fused cDNAs of the multimeric sat-RNAs were amplified by priming of the BB-S1 primer but that the primer hybridized imperfectly between different short inverted repeats in the sat-RNA. In addition, the multimeric forms may be RNA intermediates formed during sat-RNA replication, which contain high-frequency deletions at the 3′ ends of the junction regions.
In order to ensure that the BB-S1 results were not artifacts arising during PCR amplification and to confirm the head-to-tail nature of the multimeric sat-RNAs in the wild-type virus, we designed the BB-S5 and BB-S10 primers (Table 1), which do not correspond to any inverted repeated sequences in other regions of the sat-RNA, for RT-PCR amplification. Amplification using the mixed viral RNAs from the purified BBSV-X isolate and sequencing results revealed that of the 22 sequenced cDNAs, all contained intact 5′ end sequences fused to the 3′ regions of adjacent satellite sequences. These results confirmed the junction sequences of multimeric molecules (Fig. 5B). Among these clones, 12 cDNAs had perfect 3′-terminal ends fused to the intact 5′ ends of the next monomeric unit, whereas 10 clones had variable deletions ranging from 58 to 133 bases at the 3′ termini. The largest deletion, which occurred in clone 8, was exactly the same as those appearing in pMBS-R5 and pMBS-R61 (Fig. 5A). These results thus indicate that in addition to the monomers shown in Fig. 2C, multimeric forms occur frequently within the BBSV sat-RNAs in vivo, and that 3′-terminal deletions in the junction regions are also present within the multimeric population. These multimers may be produced either by a rolling circle mechanism or by replicase-driven copy-choice recombination, with the abnormal forms subsequently being selected during replication, as suggested by Mayo et al. (28) and Cascone et al. (11).
Our results indicate, however, that rolling circle mechanisms are not involved in the replication of sat-RNAs because similar relative mobilities were observed in agarose gels before and after formaldehyde and formamide denaturation of various sat-RNAs. These sat-RNAs included those identified in total RNA extracts, RNAs extracted from purified BBSV-X particles, dsRNAs purified from infected plants, and marker RNAs consisting of the in vitro-transcribed linear oligomers shown in Fig. 2E (right panel). These results thus indicate that circular sat-RNA intermediates that might form rolling circle templates are not prominent components of the sat-RNAs. In addition, the sat-RNAs appeared not to form hammerhead structures typical of those found in other sat-RNAs (21), and the results of in vitro cleavage experiments indicated that dimeric or tetrameric sat-RNA transcripts are relatively stable and do not produce specific self-processing products (data not shown).
Analysis of proportions of plus- and minus-strand sat-RNAs.
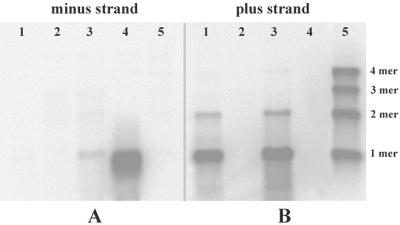
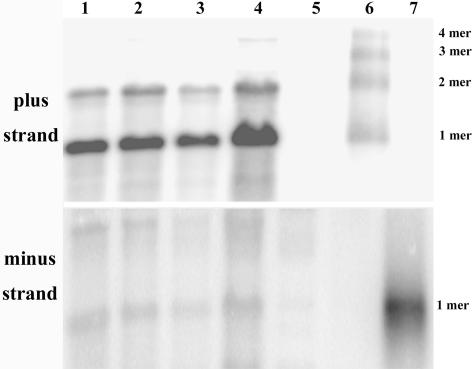
Because both monomeric and dimeric dsRNA forms of the sat-RNA were present in total RNA extracts (Fig. 2D and E), the levels of plus- and minus-strand sat-RNAs accumulating during infection were determined. Total RNAs extracted from infected plants at 3 to 4 days postinfection and RNAs from virus preparations were separated in agarose gels and blotted onto Hybond-H+ filters (Amersham Pharmacia) for hybridization. For these experiments, a plus-sense probe designed to detect minus-strand sat-RNAs failed to hybridize to RNAs purified from BBSV-X particles (Fig. 6A, lane 1), to total RNAs from lesions appearing on leaves inoculated with the BBSV-N isolate, which lacks the sat-RNA (Fig. 6A, lane 2), or to marker RNAs (Fig. 6A, lane 5), which consisted of 20 ng each of the monomer, dimer, trimer, and tetramer plus-sense sat-RNA transcripts derived from the plasmids pMBS-M4, pMBS-D10, pMBS-T6, and pMBS-Tet7, respectively (Fig. 7). However, a faint minus-strand sat-RNA band was present in the total RNAs extracted from lesions produced by BBSV-X infections (Fig. 6A, lane 3), and the intensity of hybridization can be compared with that in the adjacent positive control lane (Fig. 6A, lane 4), which was loaded with 30 ng of the negative-sense satellite RNA transcript derived from the pUBS-R35 clone (Fig. 7).
FIG. 6.
Northern blot analyses of BBSV sat-RNAs. RNA samples were loaded in the same agarose gel and probed with the plus-sense (A) or minus-sense (B) transcript of the sat-RNA. Lanes 1, viral RNAs extracted from purified BBSV-X particles; lanes 2, total RNAs from C. amaranticolor infected by in vitro-transcribed BBSV-N genomic RNA; lanes 3, total RNAs from C. amaranticolor infected by the BBSV-X isolate; lanes 4, in vitro transcripts of the minus-sense sat-RNA control; lanes 5 (from top to bottom), tetramer (4 mer), trimer (3 mer), dimer (2 mer), and monomer (1 mer) plus-sense sat-RNA transcripts.
In contrast, hybridizations designed to detect positive-strand sat-RNAs (Fig. 6B) revealed the presence of intense monomers and lower levels of dimers (Fig. 6B, lane 1), indicating that both species were encapsidated in purified virions (Fig. 6B, lane 1). The same relative sat-RNA proportions were also present in total RNAs extracted from developing local lesions (Fig. 6B, lane 3). All four species of plus-strand marker RNAs hybridized with about the same intensity to the minus-strand probe (Fig. 6B, lane 5).
Thus, the results of these experiments show that minus-strand derivatives are not encapsidated in virions, and their presence in the total RNA extracts from infected tissue suggests that minus-strand RNAs may have a role in replication. In contrast, monomers preferentially accumulated in both minus- and plus-sense forms in BBSV-X-infected tissue, and positive-strand dimers similar to those seen for wild-type BBSV-X (Fig. 2C) were also present. These results, in combination with the monomers and the fused 3′ and 5′ sequences shown in Fig. 5, suggest that multimeric forms of the satellite RNAs may accumulate as intermediates during replication. Therefore, we hypothesize that the BBSV sat-RNA species may use mechanisms of replicase reinitiation or template switching similar to the models described in previous reports for TCV (9) and Flock house virus (FHV) (1). The results also suggest that the minus-sense forms, at least as monomers, are components of dsRNA molecules in vivo, as indicated from the results in Fig. 2D and E, and that they act as intermediates in the replication of the sat-RNA (38).
Infection and replication of in vitro-transcribed sat-RNA.
In order to obtain more information about the properties of the sat-RNA and the mechanisms employed in replication, we constructed 13 cDNA clones for in vitro transcription of the sat-RNAs to be used for plant infections (Fig. 7). The linearized plasmid pMBS-M4, which contains the 615-nt sat-RNA cDNA under control of the T7 promoter, was used as a template for runoff transcription of the intact sat-RNA. When the in vitro-transcribed sat-RNA and BBSV gRNA were coinoculated mechanically into C. amaranticolor, local lesions appeared on the inoculated leaves. These lesions appeared to be identical to those of wild-type BBSV infections, but no symptoms were detected when the sat-RNA alone was inoculated (Fig. 1, right panel).
Multimeric forms have been widely reported previously for different subviral RNAs, mostly as intermediates or by-products of replication (1). To investigate the properties of the multimeric BBSV sat-RNA templates more fully, we joined monomeric cDNAs in tandem to create pMBS-D10, pMBS-T6, and pMBS-Tet7 (Fig. 7) for the transcription of dimeric, trimeric, and tetrameric sat-RNAs, respectively. Young leaves of C. amaranticolor were inoculated with in vitro transcripts of multimeric sat-RNAs in the presence of BBSV-N RNA transcripts, and total RNAs extracted from the leaves at 3 to 4 dpi were used for Northern hybridizations.
The results shown in Fig. 8 indicate that each of the satellite RNA multimeric forms is able to replicate and that in each case, monomeric plus-strand forms predominate along with smaller amounts of dimers (Fig. 8, top panel). A faint band of tetramers was also observed when RNA samples from the tetrameric inoculations were overloaded (Fig. 8, top panel, lane 4), but trimers were not detected in the hybridizations. In additional experiments, minus-sense monomeric and dimeric sat-RNAs were present at similar ratios to those of the corresponding plus-sense species, albeit with much lower abundances than those of the plus strands (Fig. 8, bottom panel). Thus, the monomeric and dimeric dsRNA forms appear to be replicative intermediates, and both species may serve as templates for the preferential accumulation of positive-sense monomeric sat-RNA. Based on these results, it seems likely that internal termination and/or reinitiation of RNA replicase on multimeric templates is involved in forming monomers. Previously proposed mechanisms of reinitiation to produce multimers from monomeric species (9) can also be evoked to account for the sat-RNA dimers, trimers, and tetramers noted in our studies. According to these models, retention of the replicase at the 3′ end of the nascent strand, followed by reinitiation at the 3′ end of the template strand, may lead to the generation of multimers during subsequent rounds of replication. In addition, since plus-sense monomeric molecules are the predominant products of various types of input templates, including integrated cDNAs (3), perhaps monomeric forms are the most highly selected biologically active forms, whereas dimers (and possibly trimers and tetramers) may have important roles in sat-RNA evolution. Such multimeric replicative forms may have obvious advantages in template variability and in generating diversity among sat-RNAs that might result in novel forms for strain selection.
FIG. 8.
Infectivities of multimeric sat-RNAs transcribed in vitro. Total RNAs of C. amaranticolor inoculated with in vitro transcripts of BBSV-N genomic RNA plus the following transcripts: lane 1, monomeric sat-RNA transcripts; lane 2, dimeric sat-RNA transcripts; lane 3, trimeric sat-RNA transcripts; lane 4, tetrameric sat-RNA transcripts; lane 5, only BBSV gRNA transcripts (negative control); lane 6, a mixture of monomeric, dimeric, trimeric, and tetrameric sat-RNA in vitro transcripts for use as molecular size markers; lane 7, minus-sense sat-RNA in vitro transcripts to serve as a control for the plus-sense probe. RNA samples were probed with a minus-sense (top) or plus-sense (bottom) sat-RNA probe, respectively.
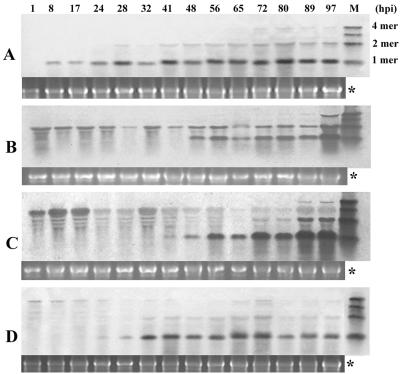
In order to understand the relationships among the molecular forms of the BBSV sat-RNAs and to monitor the time course of their replication, we extracted total RNAs at different times after the inoculation of C. amaranticolor leaves with monomeric, dimeric, trimeric, and tetrameric in vitro transcripts. Compared to the monomeric sat-RNA (Fig. 9A), the inoculated multimeric sat-RNAs degraded gradually after infection. Replication products of the multimers consisted primarily of the monomeric form, which appeared between 24 and 48 h postinoculation (hpi) and remained intact up to 97 hpi (Fig. 9B, C, and D). In agreement with the results shown in Fig. 8, the dimeric forms accumulated at lower levels than those of monomers, and the ratios of the monomers and dimers remained constant irrespective of whether the inoculum consisted of monomers, trimers, or tetramers (Fig. 9A, C, and D). However, the dimeric forms normally became evident 12 to 24 h later than the monomers, but the timing of their appearance varied with the source of the inoculum. Therefore, it seems that the dimeric form is an important template for the replication of monomeric sat-RNA, and as indicated above, dimers may have different biological roles from those of monomers.
FIG. 9.
Time course of replication of sat-RNA derivatives in C. amaranticolor leaves. Northern blots of leaf extracts after coinoculation with in vitro-synthesized monomer, dimer, trimer, and tetramer sat-RNA transcripts are shown in panels A, B, C, and D, respectively. A full-length DIG-labeled sat-cDNA was used as a probe. The panel marked with “*” shows a 28S rRNA loading control for each lane. M, mixture of monomeric, dimeric, trimeric, and tetrameric sat-RNA transcripts.
Mutagenesis of end sequences related to sat-RNA replication.
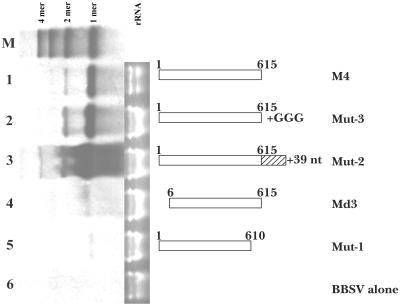
Since multimeric sat-RNAs occur widely in vivo and can be used as templates for the replication of other unit-sized molecules, the proximal nucleotides at either the 3′ or 5′ end must be very important for the initiation or termination of RNA replicase. To determine the nature of this requirement, we designed an additional set of clones for in vitro transcription of sat-RNAs with variable termini as outlined in the diagram in Fig. 10. Plants were coinoculated with BBSV gRNA and sat-RNA transcripts, and at 3 to 4 dpi, RNAs were recovered from the lesions for Northern blot analyses. These analyses were compared to those of inocula containing the 615-nt wild-type sat-RNA (Fig. 10, lane 1) and suggest that extensions consisting of three G residues or an extra 39 bases at the 3′ end of the monomer (Fig. 10, lanes 2 and 3) were removed from the transcripts during replication and that their presence in the initial inoculum did not reduce the accumulation of sat-RNAs. These results agree with those reported for FHV, although heterologous terminal extensions did inhibit FHV RNA replication to some extent (1). However, a 5-nt deletion at the 5′ (5′-GAAAG-3′) or 3′ (5′-TACCC-3′) terminus of the sat-RNA almost completely eliminated replication (Fig. 10, lanes 4 and 5). Therefore, although the 3′ terminus will tolerate extensions, both the 5′- and 3′-terminal regions appear to be essential for sat-RNA replication.
FIG. 10.
Northern blot of infections with sat-RNA mutant transcripts. Lane 1, total RNAs extracted from C. amaranticolor inoculated with in vitro transcripts of BBSV RNA plus full-length sat-RNA from pMBS-M4 (see Fig. 7); lane 2, Mut-3 sat-RNA transcripts; lane 3, Mut-2 sat-RNAs; lane 4, pUBS-Md3 sat-RNAs; lane 5, Mut-1 sat-RNA transcripts; lane 6, extracts from leaves inoculated with an in vitro transcript of BBSV-N genomic RNA alone. The diagrams at the right represent each mutated sat-RNA used for inoculation. M, mixture of monomeric, dimeric, trimeric, and tetrameric sat-RNAs used as markers. The numbers on the diagrams represent the terminal nucleotides of the transcript or the number of extra nucleotides at the 3′ end. Note that the rRNA loading lanes reveal that the total RNAs in lane 2, 3, 4, and 5 were twice as concentrated as those in lanes 1 and 6.
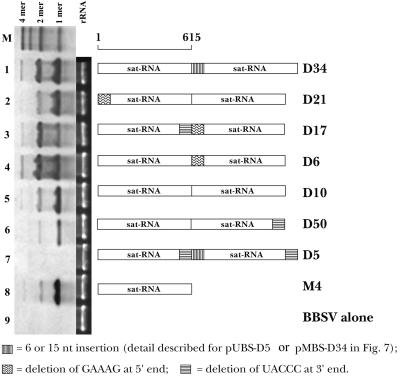
To provide additional information about the roles of the terminal sequences in the replication of the BBSV sat-RNA, we modified the dimeric sequences proximally and/or internally and used them for in vitro transcription. Northern blot analyses of the total RNAs recovered from C. amaranticolor lesions at 3 to 4 dpi indicated that the monomers were about fivefold more abundant than the dimers when the inoculum consisted of dimer transcripts (Fig. 11, lane 5). Similar ratios were noted when a 5-nt deletion was made at either the 5′ end (pUBS-D21) or the 3′ end (pMBS-D50) (Fig. 11, lanes 2 and 6) or when the monomer was used for sat-RNA inoculation (Fig. 11, lane 8). However, the dimeric forms accumulated to about the same levels as the monomers when internal deletions were made at the 5′ end of the second monomer (5′-GAAAG-3′; pMBS-D6) or when 15 nt from the vector were inserted at the junction region between the monomers (pMBS-D34) (Fig. 11, lanes 1 and 4). The abundance of the dimers also increased substantially and exceeded those of the monomers when five nucleotides were deleted from the 3′ (5′-TACCC-3′) and 5′ (5′-GAAAG-3′) ends of the junction regions formed by the head-to-tail monomers (Fig. 11, lane 3) (pMBS-D17). Three repetitions of these experiments provided similar results and indicated that the junction sequences of the dimeric sat-RNAs can have substantial effects on the normal ratio of dimers to monomers. This supports the hypothesis that the release of RNA replicase from templates produces unit-sized progeny species. As was the case with the monomeric mutants (Fig. 10, lanes 4 and 5), double deletions (5′-TACCC-3′) at the internal and proximal 3′ ends of the dimer (pUBS-D5) totally eliminated replication of the sat-RNA (Fig. 11, lane 7). Therefore, the terminal nucleotides are clearly essential for the initiation of replication of sat-RNA and for the production of both monomers and dimers. These results also indicate that monomeric molecules can result from either internal initiation or premature termination of RNA transcription on dimeric RNA templates, as assumed previously for other RNAs (16).
FIG. 11.
Effects of terminal sequences on the replication of dimeric sat-RNAs. Lane 1, total RNAs of C. amaranticolor leaves inoculated with BBSV RNA transcripts plus dimeric sat-RNAs from pMBS-D34; lane 2, BBSV RNA plus pUBS-D21 sat-RNA; lane 3, BBSV RNA plus pMBS-D17 RNA; lane 4, BBSV RNA plus pMBS-D6 RNA; lane 5, BBSV RNA plus pMBS-D10 RNA; lane 6, BBSV RNA plus pMBS-D50 RNA; lane 7, BBSV RNA plus pUBS-D5 RNA; lane 8, BBSV RNA plus intact monomeric sat-RNA from pMBS-M4; lane 9, BBSV RNA alone. M, mixture of in vitro-transcribed monomeric, dimeric, trimeric, and tetrameric sat-RNAs used as molecular markers. The diagrams at the right represent each mutated sat-RNA constructed as described in the legend to Fig. 7. A 28S rRNA loading control is shown in the middle. The cDNA of the sat-RNA was labeled with DIG for use as a probe.
The secondary structure of the multimeric sat-RNA species was investigated further by a structural analysis with MFOLD. The results indicated that the 17-base stem-loop (including bases forming bulges) and the seven-residue paired stem in the 5′- and 3′-terminal regions of the monomer (Fig. 4C, enlarged regions) are fully conserved in the dimer, trimer, and tetramer derivatives and that, as indicated in the two-dimensional depiction, the 5′ and 3′ termini are located close to each other. However, a twofold rotational symmetry based on the monomeric structure (Fig. 4C) was found only for the dimer and tetramer species, not for the trimer structure (data not shown). These predictions may provide an explanation for the results of sat-RNA mutagenesis. Since the 5′ or 3′ sequence essential for maintaining the long-distance stem-loop structures was deleted in the Md3 or Mut-1 mutant, respectively, replication of the mutant monomeric sat-RNAs was almost completely eliminated (Fig. 10, lanes 4 and 5). In contrast, when 5 nt at the 3′- and 5′-proximal ends of the junction regions of the dimeric mutant pMBS-D17 were deleted (Fig. 7), dimeric forms of the sat-RNA were replicated preferentially (Fig. 11, lane 3). However, replication was not evident with the mutant pUBS-D5, in which the 5′-terminal semi-stem-loop in the junction region and the two 3′-terminal stems in the proximal and junction regions were disrupted (Fig. 11, lane 7).
According to the Seventh Report of the International Committee on Taxonomy of Viruses (39), the sat-RNA of Beet black scorch necrovirus belongs to the small linear satellite RNA subgroup, which contains several subviral RNAs associated with helper viruses in the Tombusviridae family, including Turnip crinkle carmovirus (35), Cymbidium ringspot tombusvirus (CyRSV) (32), Tomato bushy stunt tombusvirus (TBSV) (23), and Cucumber necrosis tombusvirus (CuNV) (19). The BBSV sat-RNA has a 615-nt noncoding genome that is similar in size to the satellite RNAs of CyRSV (∼0.7 kb) and TBSV (∼0.7 kb). However, defective interfering (DI) RNAs similar to those found in CyRSV (16), TBSV (26), TCV (35), and CuNV (20) or chimeric satellite RNA forms have not been identified either under field conditions or after multiple passages in plants coinfected with BBSV genomic and satellite RNAs.
As reported for many subviral RNAs (9, 20, 27, 31), multimeric molecules were also associated with BBSV sat-RNA infections. Our analyses of multimer junctions of sat-RNAs in the population of wild-type subviral RNAs demonstrated that nucleotide deletions often occur in the junction regions of head-to-tail repeats of multimeric species. In addition, the precise fusions of monomer units amplified by perfect pairing of the cDNA primers and those obtained by imperfect pairing to different regions of the dimeric sat-RNAs (Fig. 5A and B) revealed 3′-terminal deletions ranging up to 133 nt in the junction regions. These results contrasted with those for TCV (9) and CuNV (20), in which the junction regions contain deletions at both the 3′ and 5′ ends of the dimeric sat-RNAs. However, in vitro transcripts of monomeric mutants containing deletions of 156 or 169 bases around the same region were not infectious when coinoculated with the helper virus (data not shown). Considering the results of Northern blot analysis, in which only small proportions of short variable length sat-RNAs were detected, we conclude that deletions of the sat-RNAs occur randomly during replication to produce aberrant species that are later eliminated by selection in vivo.
Multimeric sat-RNAs of CyRSV have been postulated to be by-products of replication or stable aggregates of dsRNA monomers (15). However, an alternative model whereby replication is reinitiated without release of the replicase from nascent strands (9) has been proposed to explain the formation of multimeric sat-RNA molecules from monomeric templates. In addition, the possibility that dimeric RNAs may represent intermediates in the formation of monomers has been suggested for the DI RNAs of CuNV (20). Our present research suggests that these models can be extended to include additional steps in BBSV sat-RNA replication and also provides evidence that monomers are produced efficiently from multimeric templates. Our results also indicate that the intact 3′- and 5′-proximal sequences of BBSV sat-RNA may have a direct role in maintaining normal monomer-to-dimer ratios, but they contrast with the results obtained by Carpenter et al. (9), in which the accumulation of TCV sat-RNA C monomers containing a 22-base internal deletion (79 to 100 nt) in a total of 356 nucleotides was not affected by replacement or relocation of the nucleotides. Future investigations to elucidate important steps that occur during BBSV sat-RNA replication will focus on the factors in the junction regions that control the relative accumulation of monomers and dimers and their inactivation by deletions in the junction region. Other topics for study will involve potential cell-to-cell movements of the BBSV sat-RNA dimers and their similarity to movement processes reported for CyRSV DI RNAs (37).
Acknowledgments
We thank Andrew O. Jackson (Department of Plant and Microbial Biology, University of California at Berkeley) and Sek-Man Wong (Department of Biological Science, National University of Singapore, Singapore) for their helpful suggestions and constructive criticism.
This work was supported by grants 30270063 and 30325001 from the National Nature Science Foundation of China.
REFERENCES
- 1.Albariño, C. G., B. D. Price, L. D. Eckerle, and L. A. Ball. 2001. Characterization and template properties of RNA dimers generated during flock house virus RNA replication. Virology 289:269-282. [DOI] [PubMed] [Google Scholar]
- 2.Ban, N., S. B. Larson, and A. Mcpherson. 1995. Structural comparison of the plant satellite viruses. Virology 214:571-583. [DOI] [PubMed] [Google Scholar]
- 3.Baulcombe, D. C., G. R. Saunders, M. W. Bevan, M. A. Mayo, and B. D. Harrison. 1986. Expression of biologically active viral satellite RNA from the nuclear genome of transformed plants. Nature 321:446-449. [Google Scholar]
- 4.Benson, S. A. 1984. A rapid procedure for the isolation of DNA fragments from agarose gels. Bio/Technology 1984(March/April):66-67. [Google Scholar]
- 5.Bo, Y. X., Z. N. Cai, Q. Ding, J. L. Yu, and Y. Liu. 1996. Characterization of beet black scorch virus RNA, partial cDNA cloning and sequencing of the small component. J. Agric. Biotechnol. 4:269-276. [Google Scholar]
- 6.Cai, Z. N., D. H. Chen, M. S. Wu, X. M. Cui, J. L. Yu, and Y. Liu. 1993. Identification of pathogenic virus of beet black scorch disease and detection by synthesized cDNA probes. J. Beijing Agric. Univ. 19:112. [Google Scholar]
- 7.Cai, Z. N., Q. Ding, Y. H. Cao, Y. X. Bo, D. E. Lesemann, J. X. Jiang, R. Koenig, J. L. Yu, and Y. Liu. 1999. Characterization of a sugar beet (Beta vulgaris L.) virus causing black scorch symptom in China, a possible member of Necrovirus, p. 9-12. In J. L. Sherwood and C. M. Rush (ed.), Proceedings of the Fourth International Working Group of Plant Viruses with Fungal Vectors. American Society of Sugar Beet Technologists, Denver, Colo.
- 8.Cao, Y. H., Z. N. Cai, Q. Ding, D. W. Li, C. G. Han, J. L. Yu, and Y. Liu. 2002. The complete nucleotide sequence of beet black scorch virus (BBSV), a new member of the genus Necrovirus. Arch. Virol. 147:2431-2435. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, C. D., P. J. Cascone, and A. E. Simon. 1991. Formation of multimers of linear satellite RNAs. Virology 183:586-594. [DOI] [PubMed] [Google Scholar]
- 10.Carpenter, C. D., and A. E. Simon. 1998. Analysis of sequences and predicted structures required for viral satellite RNA accumulation by in vivo genetic selection. Nucleic Acids Res. 26:2426-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascone, P. J., C. D. Carpenter, X. H. Li, and A. E. Simon. 1990. Recombination between satellite RNAs of turnip crinkle virus. EMBO J. 9:1709-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coutts, R. H. A., J. E. Rigden, A. R. Slabas, G. P. Lomonossoff, and P. J. Wise. 1991. The complete nucleotide sequence of tobacco necrosis virus strain D. J. Gen. Virol. 72:1521-1529. [DOI] [PubMed] [Google Scholar]
- 13.Cui, X. M. 1988. An icosahedral virus found in sugar beet. J. Xinjiang Shihezi Agric. College 10:73-78. [Google Scholar]
- 14.Cui, X. M., Z. N. Cai, J. Wu, and Y. Liu. 1991. Study on symptom pattern of sugarbeet rhizomania disease. Plant Protect. 17:5-7. [Google Scholar]
- 15.Dalmay, T., and L. Rubino. 1994. The nature of multimeric forms of cymbidium ringspot tombusvirus satellite RNA. Arch. Virol. 138:161-167. [DOI] [PubMed] [Google Scholar]
- 16.Dalmay, T., G. Szittya, and J. Burgyan. 1995. Generation of defective interfering RNA dimers of cymbidium ringspot tombusvirus. Virology 207:510-517. [DOI] [PubMed] [Google Scholar]
- 17.Davies, C., J. Haseloff, and R. H. Symons. 1990. Structure, self-cleavage, and replication of two viroid-like satellite RNAs (virusoids) of subterranean clover mottle virus. Virology 177:216-224. [DOI] [PubMed] [Google Scholar]
- 18.Dodds, J. A., T. J. Morris, and R. L. Jordan. 1984. Plant viral double-stranded RNA. Annu. Rev. Phytopathol. 22:151-168. [Google Scholar]
- 19.Finnen, R. L., and D. M. Rochon. 1993. Sequence and structure of defective interfering RNAs associated with cucumber necrosis virus infections. J. Gen. Virol. 74:1715-1720. [DOI] [PubMed] [Google Scholar]
- 20.Finnen, R. L., and D. M. Rochon. 1995. Characterization and biological activity of DI RNA dimers formed during cucumber necrosis virus coinfections. Virology 207:282-286. [DOI] [PubMed] [Google Scholar]
- 21.Forster, A. C., and R. H. Symons. 1987. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49:211-220. [DOI] [PubMed] [Google Scholar]
- 22.Frohman, M. A., M. K. Dush, and G. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallitelli, D., and R. Hull. 1985. Characterization of satellite RNAs associated with tomato bushy stunt virus and five other defective tombusviruses. J. Gen. Virol. 66:1533-1543. [Google Scholar]
- 24.Garcia-Arenal, F., and P. Palukaitis. 1999. Structure and functional relationships of satellite RNAs of cucumber mosaic virus. Curr. Top. Microbiol. Immunol. 239:37-64. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, J. X., J. F. Zhang, S. C. Che, D. J. Yang, J. L. Yu, Z. N. Cai, and Y. Liu. 1999. Transmission of beet black scorch virus by Olpidium brassicae. J. Jiangxi Agric. Univ. 21:525-528. [Google Scholar]
- 26.Jones, R. W., A. O. Jackson, and T. J. Morris. 1990. Defective-interfering RNAs and elevated temperatures inhibit replication of tomato bushy stunt virus in inoculated protoplasts. Virology 176:539-545. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda, T., T. Natsuaki, W.-Q. Wang, and S. Okuda. 1997. Formation of multimers of cucumber mosaic virus satellite RNA. J. Gen. Virol. 78:941-946. [DOI] [PubMed] [Google Scholar]
- 28.Mayo, M. A., K. I. Berns, C. Fritsch, J. M. Kaper, A. O. Jackson, M. J. Leibowitz, and J. M. Taylor. 1995. Subviral agents: satellite, p. 487-492. In F. A. Murphy, C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers (ed.), Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, New York, N.Y.
- 29.Mossop, D. W., and R. I. B. Francki. 1979. The stability of satellite RNA in vivo and in vitro. Virology 94:243-253. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Alvarado, G., and M. J. Roossinck. 1997. Structural analysis of a necrogenic strain of cucumber mosaic cucumovirus satellite RNA in planta. Virology 236:155-166. [DOI] [PubMed] [Google Scholar]
- 31.Roossinck, M. J., D. Sleat, and P. Palukaitis. 1992. Satellite RNAs of plant viruses: structures and biological effects. Microbiol. Rev. 56:256-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubino, L., J. Burgyan, F. Grieco, and M. Russo. 1990. Sequence analysis of cymbidium ringspot virus satellite and defective interfering RNAs. J. Gen. Virol. 71:1655-1660. [DOI] [PubMed] [Google Scholar]
- 33.Sanger, G., S. Nicklen, and A. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon, A. E. 1999. Replication, recombination, and symptom-modulation properties of the satellite RNA of turnip crinkle virus. Curr. Top. Microbiol. Immunol. 239:19-36. [DOI] [PubMed] [Google Scholar]
- 35.Simon, A. E., and S. H. Howell. 1986. The virulent satellite RNA of turnip crinkle virus has a major domain homologous to the 3′-end of the helper virus genome. EMBO J. 5:3423-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song, S. I., S. L. Silver, M. A. Aulik, L. Rasochova, B. R. Mohan, and W. A. Miller. 1999. Satellite cereal yellow dwarf virus-RPV (satRPV) RNA requires a double hammerhead for self-cleavage and an alternative structure for replication. J. Mol. Biol. 293:781-793. [DOI] [PubMed] [Google Scholar]
- 37.Szittya, G., D. Silhavy, T. Dalmay, and J. Burgyán. 2002. Size-dependent cell-to-cell movement of defective interfering RNAs of cymbidium ringspot virus. J. Gen. Virol. 83:1505-1510. [DOI] [PubMed] [Google Scholar]
- 38.Tousch, D., M. Jacquemond, and M. Tepfer. 1994. Replication of cucumber mosaic virus satellite RNA from negative-sense transcripts produced either in vitro or in transgenic plants. J. Gen. Virol. 75:1009-1014. [DOI] [PubMed] [Google Scholar]
- 39.Van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. Carstens, M. K. Estes, S. Lemon, D. McGeoch, R. B. Wickner, M. A. Mayo, C. R. Pringle, and J. Maniloff (ed.). 2000. Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses, p. 1025-1032. Academic Press, New York, N.Y.
- 40.Wang, M. B., X. Y. Bian, L. M. Wu, L. X. Liu, N. A. Smith, D. Isenegger, R. M. Wu, C. Masuta, V. B. Vance, J. M. Watson, A. Rezaian, E. S. Dennis, and P. M. Waterhouse. 2004. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 101:3275-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, F., and A. E. Simon. 2003. Enhanced viral pathogenesis associated with a virulent mutant virus or a virulent satellite RNA correlates with reduced virion accumulation and abundance of free coat protein. Virology 312:8-13. [DOI] [PubMed] [Google Scholar]