Abstract
Recombinant adeno-associated virus (rAAV) vector holds promise for gene therapy. Despite a low frequency of chromosomal integration of vector genomes, recent studies have raised concerns about the risk of rAAV integration because integration occurs preferentially in genes and accompanies chromosomal deletions, which may lead to loss-of-function insertional mutagenesis. Here, by analyzing 347 rAAV integrations in mice, we elucidate novel features of rAAV integration: the presence of hot spots for integration and a strong preference for integrating near gene regulatory sequences. The most prominent hot spot was a harmless chromosomal niche in the rRNA gene repeats, whereas nearly half of the integrations landed near transcription start sites or CpG islands, suggesting the possibility of activating flanking cellular disease genes by vector integration, similar to retroviral gain-of-function insertional mutagenesis. Possible cancer-related genes were hit by rAAV integration at a frequency of 3.5%. In addition, the information about chromosomal changes at 218 integration sites and 602 breakpoints of vector genomes have provided a clue to how vector terminal repeats and host chromosomal DNA are joined in the integration process. Thus, the present study provides new insights into the risk of rAAV-mediated insertional mutagenesis and the mechanisms of rAAV integration.
Recombinant adeno-associated virus serotype 2 (rAAV2) vectors have been considered safe and are widely used for delivering therapeutic genes into experimental animals and human subjects (14). rAAV2 vectors have the ability to integrate into host chromosomes in tissues (18, 21), but extrachromosomal vector genomes dominate over integrated forms and are primarily responsible for persistent expression (4, 25, 28). Despite the low integration efficiency, we need to carefully consider rAAV-mediated insertional mutagenesis because of the following reasons. First, vector-mediated insertional oncogenesis in humans has been reported in a clinical trial for gene therapy for severe combined immune deficiency-X1 (12). Second, Donsante et al. reported that 6 of 12 mucopolysaccharidosis type VII mice treated with a rAAV2 vector developed liver cancers, while none of 24 mucopolysaccharidosis type VII mice treated with bone marrow transplantation did. In their study, detailed analyses of the tumors suggested that rAAV2-mediated insertional mutagenesis was unlikely, but Donsante et al. also pointed out that these data do not exclude rAAV2 as the causative agent for the tumors in rAAV2-treated mice (3, 30). Third, we have recently demonstrated in a small-scale analysis that rAAV2 vectors preferentially integrate into genes (22); we and others have shown that rAAV2 vector integration accompanies chromosomal deletions, although most of the deletions were less than 0.3 kb (20, 22). Fourth, recent advances in rAAV technologies have enabled the transduction of higher numbers of target cells (2, 8, 17), which may increase the absolute number of integration events in target tissues and in theory result in increased risk of insertional mutagenesis. Now that gene therapy-mediated oncogenesis has been proven to occur, we need to thoroughly address this issue.
Human immune deficiency virus type 1 (HIV-1) and murine leukemia virus (MLV) integration sites have been extensively analyzed with high-throughput methods. The results clearly demonstrated that MLV integrations preferentially target transcription start regions, while HIV-1 integrations favor the entire length of genes (29, 32). With rAAV2 vectors, we have just begun to elucidate how vectors integrate into host chromosomal DNA in animals (20, 22). Since rAAV2 vectors have been used for human clinical trials, we need to carefully assess how rAAV2 vector integration affects the host. However, a comprehensive large-scale study with rAAV2-injected animals has been challenging, due to unpredicted complex structures of rAAV2 proviral genomes and the presence of substantial unintegrated extrachromosomal vector genomes in tissues (21, 22, 25), which are not issues in high-throughput analyses for HIV-1 and MLV integrations.
Here, we report the results from a high-throughput analysis of rAAV2 integrations in mouse primary hepatocytes in vivo. The salient feature of the high-throughput method was to use a hereditary tyrosinemia type I (HTI) mouse model and a fumaryl acetoacetate hydrolase (FAH)-expressing rAAV2 shuttle vector (22). This allowed in vivo expansion of hepatocytes that harbored integrated rAAV2 vector genomes and retrieval of rAAV2 proviruses together with flanking genomic sequences on both sides in plasmids. This in vivo selection technique greatly facilitated the high-throughput analysis using rAAV2-injected animal tissues; importantly, an extended analysis of our previous study (22) has indicated that in vivo selection procedures do not substantially alter integration site preference.
MATERIALS AND METHODS
rAAV2 vector production and animal handling.
All animal experiments were performed according to the guidelines for animal care at Stanford University and Oregon Health & Science University. Production and purification of a human FAH-expressing rAAV2 vector (AAV2-EF1α-hFAH.AOS), maintenance of the HTI mice, the procedures for portal vein injection, in vivo selection of hepatocytes transduced with AAV2-EF1α-hFAH.AOS, and hepatocyte transplantation were all described in detail in a previous publication (22). All the animals were treated with 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclohexanedione (NTBC), except during the period of in vivo selection. We injected 12 adult male HTI mice via the portal vein with AAV2-EF1α-hFAH.AOS at a dose of 3.0 × 1011 vector genomes per mouse. The mice were divided into three groups (each group contained four mice). An 8-week in vivo selection was started 3 or 6 weeks postinjection in groups 3 and 1, respectively, by withdrawal of NTBC. The mice in group 2 were kept on NTBC during a period corresponding to the first in vivo selection period for groups 1 and 3. Hepatocytes were then isolated and transplanted into HTI recipient mice and further selected for 7 months. All mice in group 2 died during the second in vivo selection, while all eight recipients from four donors (two recipients per donor) in groups 1 and 3 survived. In our previous study (22), we utilized a recipient from group 1, while in this study, we also analyzed liver DNA from three other recipients from three different donors. In total, we analyzed four recipient mice (mice A and B from group 1 and mice C and D from group 3). Each recipient mouse had a different donor injected with the rAAV2 vector and therefore carried independent rAAV2 integration events. The aim of studying groups 1 and 3 was to investigate whether there was any difference in the forms of integrated vector genomes and integration site preference depending on when in vivo selection started (3 versus 6 weeks postinjection).
Generation of rAAV2 integration libraries.
Detailed strategy for the construction of rAAV2 integration plasmid libraries was summarized in our previous publications (21, 22). Briefly, we treated 20 μg of liver DNA from each recipient mouse with calf intestinal alkaline phosphatase, divided the DNA in half, and digested each DNA preparation with AvrII or KpnI, which does not cleave the vector genome but cuts mouse genomic DNA. We self-ligated the digested DNA with T4 DNA ligase and transformed Escherichia coli (ElectroMAX DH10B; Invitrogen, Carlsbad, Calif.) with 2.5 to 5 μg of ligated products to make a set of AvrII and KpnI digestion-derived libraries. In total, we constructed eight libraries, i.e., four sets of AvrII and KpnI libraries from four recipients.
High-throughput analysis of rAAV2 provirus plasmids.
We propagated each E. coli colony in 96-well plates and purified plasmid DNA with a Perfectprep Plasmid 96 Vac Direct Bind system (Eppendorf, Hamburg, Germany). We digested each plasmid with a corresponding restriction enzyme (either AvrII or KpnI) and ran the digests on a 0.8% agarose gel together with undigested counterparts to check the quantity and quality of the DNA and screen for bona fide rAAV2 proviral plasmids. Bona fide plasmid clones must be digestible with the corresponding restriction enzyme used for self circularization and be larger than the 1-U length of the vector (5.0 kb). Some plasmids did not propagate sufficiently for screening and sequencing. In this case, we manually prepared individual plasmid DNA. We sequenced plasmid DNA with the 3730xl DNA Analyzer (Applied Biosystems, Foster City, Calif.). The sequence primers we used were as follows: EF1αP8, 5′-CAGTACACGACATCACTTTCCCAG-3′; EF1αP11, 5′-GGCTAGAGACTTATCGAAAG-3′; OriP1, 5′-CGCACGAGGGAGCTTCCAGG-3′; and OriP2, 5′-CAGCAACGCGGCCTTTTTACGGT-3′.
We designed EF1αP8 and EF1αP11 for 5′ vector-cellular DNA junction sequencing, while OriP1 and OriP2 were for 3′ junction sequencing. We sequenced plasmid DNA with a set of primers (EF1αP8 and OriP2). EF1αP11 and OriP1 were occasionally used to further investigate the junctions. Finally, the sequence reads were clustered to identify duplicate clones and determine the total number of independent rAAV2 integration plasmids. We considered an integration event independent only when the structures of rAAV2 provirus and vector-cellular DNA junction sequences were unique.
Mapping of the integration sites.
We first searched the isolated vector-flanking DNA sequences against the public mouse genome database (October 2003 freeze) using both the University of California—Santa Cruz (UCSC) BLAT program and the National Center for Biotechnology Information (NCBI) BLAST program. We considered sequence matches to be authentic only if a sequence match extended over the length of the high-quality sequence with high sequence identity and yielded no more than one best hit with ≥95% identity. When there was more than one best hit with ≥95% identity in a sequence read, we carried out extended sequencing as described below to unambiguously map to a unique genomic locus. Unlike PCR-based approaches, which can only provide flanking genomic sequence information of a couple of hundred base pairs at most (28, 32), the plasmid rescue approach we applied in the present study allowed us to analyze thousands of base pairs around the integration sites. When the sequence search did not yield a match that met all the above criteria, we also compared vector-flanking DNA sequences against human genome DNA using the same programs described above or against DNA databases of all organisms. In addition, we compared the vector-flanking DNA sequence with the sequence of the plasmids used for the rAAV2 vector productions, pAAV-EF1α-hFAH.AOS2 (AAV2-EF1α-hFAH.AOS vector plasmid) (22), pHLP19 (AAV2 helper plasmid) (10), and pladeno5 (adenovirus type 5 helper plasmid) (10). This was necessary because these nonvector DNAs, including human genomic DNA derived from cells used for vector production, can often be incorporated in recombinant viral genomes (13, 20-22).
Extended sequence analysis of rAAV2 provirus plasmids.
For mapping of the two hot spots for rAAV2 integration, the rRNA gene repeat and ubiquitin C (Ubc) gene, we extensively sequenced vector-flanking mouse genomic DNA sequences to solidify the mapping results. In addition, we performed restriction enzyme mapping of the rAAV2 provirus plasmids as previously described (22) to support the mapping results.
We set up multiple forward and reverse sequence primers based on the information obtained from our sequence analysis and the mouse genome database. We always included a set of primers that could read through the AvrII or KpnI sites in the rAAV2 provirus plasmids so that we could obtain a contiguous sequence read from one vector-flanking mouse genomic sequence end to the other. This could give us sequence information remote from the integration site within the same AvrII-AvrII or KpnI-KpnI genomic fragment and was particularly useful for unambiguous mapping.
The following primers were used for the Ubc gene: Ubc1 Rev, 5′-GCTTTCACTCTGCTGTGTCTAGCC-3′; Ubc2 For, 5′-CCAGTAGGAACAGGTCTTTTTCCAG-3′; and Ubc3 Rev, 5′-CCTTGATAGTTTTAGCCTGTCGCTT-3′.
Sequencing of rAAV2 integration plasmids with these primers could allow us to unambiguously map 3 integrations to the Ubc gene locus.
The following three primer sets were used for the rRNA gene repeat. Set 1 consisted of the following: rDNAIGS1 Rev, 5′-CCATCTCTCAGGACCGACTGAC-3′; rDNAIGS2 For, 5′-GCAGTCAGGTGCTCTTACCCAC-3′; rDNAIGS3 For, 5′-CGGGGGAGAGGGTGTAAATCT-3′; rDNAIGS4 For, 5′-TGGACCAATTAGTTGGCTGGTTT-3′; rDNAIGS5 For, 5′-TGAACCAGAGAGTTTGGATGTCAA-3′; rDNAIGS6 For, 5′-CGCGCGCTCGTTTTATAAATACT-3′; rDNAIGS7 For, 5′-GGATCGTCTTCTCCTCCGTCTC-3′; and rDNAIGS8 For, 5′-TCTGTGGGATTATGACTGAACGC-3′. Set 2 consisted of the following: rDNAIGS9 Rev, 5′-TGTTACACAGAGAAACTGCATCATGA-3′; rDNAIGS10 For, 5′-AAGCCTTAAAAAGCACTCTGACAGC-3′; rDNAIGS11 For, 5′-GCCCGGACTAATTTTATTTGTTTGA-3′; rDNAIGS12 For, 5′-TAGTTTCTTAGTGTAAGCAGCCCTGG-3′; and rDNAIGS13 Rev, 5′-GACCTATTGTTTCAGGTCGCTTTG-3′. Set 3 consisted of the following: rDNAIGS14 Rev, 5′-TTGGTAGCCTCAAACTCAGAGAGG-3′; rDNAIGS15 For, 5′-GCACGCGCTGTTTCTTGTAAG-3′; rDNAIGS16 For, 5′-CTGGCCTTGAACACATTAATCTGTC-3′; rDNAIGS17 For, 5′-GCCTCTCAGGTTGGTGACACA-3′; rDNAIGS18 Rev, 5′-ACTGATAAGACCGACAGGTCAATGA-3′; rDNAIGS19 Rev, 5′-GTACTCGGGACTCTCCACCTCC-3′; rDNAIGS20 For, 5′-GGTGGCAACGTTACTAGGTCGA-3′; rDNAIGS21 For, 5′-GCGCGGTTTTCTTTCATTGAC-3′; rDNAIGS22 Rev, 5′-TTTTCTGTGTAGCCCTATCGGACTT-3′; and rDNAIGS23 For, 5′-GTTAAAGGTGTGCTCCACAATTGC-3′.
Sequencing with primer sets 1, 2, and 3 covered up to 4.4-, 1.7-, and 4-kb flanking mouse genomic sequences, respectively. For eight rAAV integration plasmids, we mapped them to the rRNA gene repeats because none of the mouse genomic regions except the rRNA gene repeat (GenBank accession numbers X82564 and AF441173; Third Party Annotation accession number BK000964) matched the entire sequence queries with high sequence identity. We could map the other two integrations to the rRNA gene repeat without the extended sequence analysis.
Determination of host genomic deletions, duplications, and translocations.
We determined the length of host genomic deletions and duplications at integration sites by aligning sequences of rAAV2 provirus plasmid clones, the rAAV2 vector genome, and the mouse genome. When there was a microhomology at a junction, we took the maximum possible size of the deletion as the host chromosomal deletion and the minimum size of the duplication as the target site duplication. We confirmed the duplication only if the entire duplicated sequence was observed in both 5′ and 3′ sequence reads and was not in simple repeats of the genome. When 5′ and 3′ sequence reads did not land on the same chromosome, we analyzed physical linkage of the 5′ and 3′ flanking mouse genomic sequences at the AvrII or KpnI site by sequencing. We confirmed the translocation only if we observed a contiguous sequence read spanning the AvrII or KpnI site that consisted of 5′- and 3′-flanking mouse genomic sequences derived from two different chromosomes.
Determination of breakpoints of the vector genome.
We determined the nucleotide positions where rAAV2 vector and mouse genomes were recombined in the same way as described above. When there were microhomologies around the recombination sites, we took a nucleotide position furthest from the center of the vector genome as the breakpoint.
Bioinformatics.
We downloaded coordinates of RefSeq genes, CpG islands, and other annotation tables for the October 2003 mouse genome freeze from the UCSC genome project website. In the present study, we defined a genomic region between transcriptional start and stop boundaries of one of the 17,724 RefSeq genes as a “gene,” as reported by Wu et al. (32). When we identified two recombination sites at an integration site, resulting in deletions or duplications, we considered an integration as “hitting a gene” if (i) both recombination sites were in a gene(s), (ii) one of the two recombination sites was in a gene, or (iii) none of the recombination sites were in a gene but a gene(s) resided in the deleted chromosomal region. We should mention that the criteria for rAAV2 integration into regions of interest were different from those for MLV and HIV-1 integrations (29, 32). MLV and HIV-1 integrations do not accompany host chromosomal deletions at integration sites, while rAAV2 integration normally accompanies host chromosomal deletions, which were taken into account in the criteria. Such criteria are particularly important when we consider functional loss of cellular genes by rAAV2 integration, because disruption of genes can occur even when rAAV2 vector genomes recombine with intergenic sequences of host chromosomal DNA.
We also analyzed integrations around transcription start sites, transcription stop sites, and CpG islands with various-sized windows. We also scored 5′ and 3′ recombination sites separately without considering deletions or duplications.
We assessed transcriptional activity of each rAAV2-targeted gene using a publicly available web-based microarray gene expression database, as previously described by Wu et al. (32). We used mouse liver expression database entries GSM4659, GSM4661, and GSM4669 in the Gene Expression Omnibus (GEO) data repository and GNF Gene Expression Atlas 2 from the Genomics Institute of the Novartis Research Foundation (31).
In the GNF data set, expression data from 77 RefSeq genes that had rAAV2 integration within genes and 59 RefSeq genes that had rAAV2 integration within ±5 kb of transcription start sites were available. For the GEO datasets, we filtered all the spots by criteria (more than two times the standard deviation of the background; not saturated or irregular) and obtained 9,527 spots. A total of 2,272 of 9,527 spots could be linked to RefSeq genes and were used as a reference for statistical analysis. A total of 20 RefSeq genes integrated within genes and 22 RefSeq genes integrated within ±5 kb of transcription start sites were available for the analysis.
We analyzed gene expression data of RefSeq genes hit by rAAV2 integration in two different ways. First, we compared expression level of each rAAV2-integrating gene with the median expression level from all the 61 tissues analyzed (GNF data set) (31) or the value from universal control RNA (GEO database). Second, we compared the expression level of each rAAV2-integrating gene with the median expression value from all the genes analyzed in the liver. The second analysis was possible only when we used the GEO database.
We searched cancer-related genes hit by rAAV2 integrations by using web-based public databases, i.e., the Retrovirus Tagged Cancer Gene Database (RTCGD) (1) and the Tumor Gene Database (TGDB) from Baylor College of Medicine.
Statistical analyses.
We investigated the bias for or against preferred integration in RefSeq genes, near transcription start sites, and in or near CpG islands by comparing the observed frequency with that from computer-simulated 10,000 random integrations and assessing statistical significance of the bias with the χ2 test. Since rAAV2 integrations accompany host chromosomal deletions of various sizes, which may affect the integration frequency towards increasing the probability of hitting genes, we also generated computer-simulated 10,000 random integrations with a window of various sizes (200 bp, 1 kb, and 10 kb) to mimic chromosomal deletions at integration sites. Ten integrations landing in the rRNA repeats were excluded from this statistical analysis because the rRNA gene repeats have not been assembled in the mouse genome database and the computer-simulated random integration data did not take into account this redundant gene.
We compared the observed frequency of the integration into the rRNA gene repeats with that from a random integration model by a one-tailed binomial test. Since the size of a haploid mouse genome is 3 × 109 bp and 200 copies of 45.3-kb rRNA gene repeats form a target of 9 × 106 bp in length, the expected probability of an integration landing in the rRNA gene repeats by a random integration model is (9 × 106)/(3 × 109) = 0.003. Therefore, the probability of having (n) integrations into the rRNA gene repeat in 347 integration events follows the equation P(n) = 347Cn(0.003)n(0.997)347−n.
Preferential integration in a particular set of genes was assessed by binomial testing as well. We made the assumption that each of the 17,724 RefSeq genes has the same probability of being hit by rAAV2 vector. When the total number of integration events in RefSeq genes is 179, the probability that a gene has (n) times integration in a random integration model follows the equation P(n) = 179Cn(1/17,724)n(17,723/17,724)179−n; i.e., P(0) = 9.90 × 10−1, P(1) = 1.00 × 10−2, P(2) = 5.02 × 10−5, and P(3) = 1.67 × 10−7. We compared these values with the observed values by a χ2 test.
To statistically analyze the transcriptional status of rAAV2-targeted genes in the liver, we compared the median expression level for rAAV2-targeted genes with that for all the genes represented in the database by a two-tailed Mann-Whitney test.
URLs.
The URLs for sites consulted for this study are as follows: UCSC Genome Informatics, http://www.genome.ucsc.edu; NCBI Mouse Genome Resources, http://www.ncbi.nlm.nih.gov/genome/guide/mouse/; NCBI Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/; RTCGD, http://RTCGD.ncifcrf.gov/; TGDB, http://Condor.bcm.tmc.edu/oncogene.html; and the Gene Expression Atlas of the Genomics Institute of the Novartis Research Foundation, http://expression.gnf.org/cgi-bin/index.cgi.
RESULTS
rAAV2 provirus integration libraries.
By injecting rAAV2 vector into HTI mouse livers at 3.0 × 1011 vector genomes per animal and performing in vivo hepatocyte selection, we generated eight rAAV2 integration libraries from four mice (mice A, B, C, and D). Libraries from each mouse represented independent rAAV2 integration events. Mice A and B belonged to group 1, while mice C and D belonged to group 3. These two groups were different in that in vivo selection was started 6 and 3 weeks postinjection in groups 1 and 3, respectively (see Materials and Methods). Since there was no difference in the forms of rAAV2 vector genomes and integration site preference between these two groups (unpublished data), we combined them together for the analysis. The 14 integration events previously isolated from mouse A (22) were included in the present study. We picked 733 plasmid colonies and identified 699 bona fide clones by restriction enzyme digestion screening of recovered plasmids. For 34 of 733 colonies, plasmid DNAs either were not recovered or did not meet the criteria for bona fide clones. We determined vector DNA-flanking unknown sequences in 699 plasmid clones and identified 393 integration events from four mice (56, 68, 116, and 153 events from mouse A, B, C, and D, respectively). All 393 integration events exhibited different proviral structures or occurred at different locations of the mouse genome and therefore were considered independent integration events. A majority of the 393 independent integration events were sequenced only once (280 of 393 integrations; 71%) or twice (60 of 393 integrations; 15%) in the 699 bona fide plasmid clones. Of these integration events, we could unambiguously map 347 different integrations in the mouse genome.
Preferential rAAV2 integration near gene regulatory sequences.
In the present study, observed frequencies of integrating into regions of interest (i.e., in or near RefSeq genes, near transcription start sites, and in or near CpG islands) were compared to those from computer-simulated 10,000 random integrations with various sized windows (Table 1). Windows in this case mimicked host chromosomal deletions at each random integration site.
TABLE 1.
rAAV2 integration site preference in primary mouse hepatocytes
| Targeted genomic region | % of integrations
|
||||||
|---|---|---|---|---|---|---|---|
| rAAV2 (no. of events analyzed)a
|
Random (10,000 events)
|
||||||
| Totalb (337) | 5′ Junctionc (280) | 3′ Junctionc (270) | 0-bp window | 200-bp window | 1-kb window | 10-kb window | |
| Within RefSeq genes | 53.1 | 46.4 | 49.3 | 26.0 | 26.1 | 26.6 | 31.2 |
| Upstream of genes (≤1 kb) | 15.4 | 11.4 | 12.2 | 0.6 | 0.8 | 1.3 | 1.3 |
| (≤5 kb) | 25.8 | 21.8 | 22.2 | 3.2 | 3.4 | 3.8 | 6.1 |
| Downstream of genes (≤1 kb) | 3.6 | 3.6 | 3.3 | 0.8 | 0.9 | 1.2 | 1.4 |
| (≤5 kb) | 12.5 | 12.5 | 12.6 | 3.4 | 3.8 | 3.9 | 6.0 |
| Near transcriptional start site (±1 kb) | 27.3 | 24.6 | 24.1 | 1.2 | 1.4 | 1.9 | 2.8 |
| (±5 kb) | 43.9 | 42.9 | 44.1 | 6.4 | 6.5 | 7.0 | 11.8 |
| In CpG island | 24.9 | 23.2 | 23.0 | 0.5 | 0.6 | 0.8 | 0.9 |
| Near CpG island (±1 kb) | 37.1 | 35.7 | 35.6 | 1.7 | 1.8 | 2.3 | 3.4 |
| (±5 kb) | 49.3 | 48.6 | 48.5 | 6.4 | 6.5 | 6.9 | 11.1 |
All values were statistically significant compared to any of the random integration models (χ2 test; P < 0.002).
Host chromosomal deletions at integration sites were considered. Ten integrations into the rDNA repeats were excluded.
Host chromosomal deletions were not considered.
We first investigated whether rAAV2 integration preferred intergenic regions or intragenic regions in the present large-scale analysis. Consistent with our previous study (22), rAAV2 preferentially integrated into RefSeq genes at a frequency of 53% (Table 1). This was significantly higher than the computer-simulated 10,000 random integrations with windows of any size (26.0 to 31.2%, χ2 test, P < 0.000000001). The frequency of 53% was lower than our previous observations from 14 integration sites (64%) (22), but this was presumably because the definitions of a gene used in our previous and present studies were different. When we applied the definition used for our previous study, the frequency of integration into genes was 62%.
We next investigated whether rAAV2 integration was similar to MLV integration in its preference for regulatory sequences. Surprisingly, 27 and 37% of total integrations landed within the region ±1 kb from the transcription start sites and CpG islands, respectively (Table 1). These frequencies were over 10 times higher than the predicted frequency of random integration (χ2 test; P < 0.000000001). With a window of ±5 kb, approximately half of the integrations occurred in this vicinity.
To assess whether the same preference was observed in rAAV2 vector integration sites isolated directly from rAAV2-injected mice without any selection, we reanalyzed our previous data with the criteria used for the present study. In our previous study, we only investigated whether rAAV2 integration landed in genes or intergenic regions but did not analyze the data in terms of integration near transcription start sites or in or near CpG islands (22). As a result, we demonstrated the same integration site preference even in the absence of selective pressure (Table 2). In particular, a strong preference for integrating near transcription start sites (±1 kb) and near CpG islands (±1 kb) was demonstrated with P values of <0.00001 and <0.00000001, respectively (χ2 test), under nonselective conditions.
TABLE 2.
rAAV2 integration sites isolated from in vivo-selected and nonselected primary mouse hepatocytes
| Targeted genomic region | % of integrations
|
|||
|---|---|---|---|---|
| rAAV2 (no. of events)a
|
Random (10,000 events) | |||
| No selection (total, 13) | In vivo selection (total, 14) | Total (total, 27b) | ||
| Within RefSeq genes | 61.5 (8)* | 57.1 (8)* | 59.3 (16)* | 26.0 |
| Upstream of genes (≤1 kb) | 7.7 (1)* | 7.1 (1)* | 7.4 (2)* | 0.6 |
| (≤5 kb) | 7.7 (1) | 7.1 (1) | 7.4 (2) | 3.2 |
| Downstream of genes (≤1 kb) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.8 |
| (≤5 kb) | 0.0 (0) | 7.1 (1) | 3.7 (1) | 3.4 |
| Near transcriptional start site (±1 kb) | 15.4 (2)* | 14.3 (2)* | 14.8 (4)* | 1.2 |
| (±5 kb) | 15.4 (2) | 35.7 (5)* | 25.9 (7)* | 6.4 |
| In CpG island | 7.7 (1)* | 7.1 (1)* | 7.4 (2)* | 0.5 |
| Near CpG island (±1 kb) | 23.1 (3)* | 14.3 (2)* | 18.5 (5)* | 1.7 |
| (±5 kb) | 30.8 (4)* | 28.6 (4)* | 29.6 (8)* | 6.4 |
All values were statistically compared to the computer-simulated 10,000 random integration frequency. Values in parentheses indicate the number of events. Values with an asterisk are significantly higher than the random integration models (χ2 test; P < 0.008).
The 27 integration events were reported previously (22).
We also assessed the transcriptional status of rAAV2-integrated genes in mouse liver with publicly available microarray databases. We did not observe any preference for integration into genes up- or down-regulated in the liver compared to the median expression values from all the tissues analyzed (31) or the values from universal control RNA. However, the genes with rAAV2 integration near the transcription start sites (±5 kb) were more actively transcribed in the liver than all RefSeq genes available in the GEO database. Median values of the expression signals of targeted genes were twofold higher than that for all the genes with a marginal statistical significance (Mann-Whitney test; P = 0.016 to 0.036).
These observations demonstrate that rAAV2 vectors have a strong preference for integrating in or near gene regulatory sequences. Importantly, this preference did not change when we analyzed 5′ and 3′ vector genome integration sites separately and did not consider host chromosomal deletions at integration sites (Table 1). In addition, our results implied that genes with higher transcriptional activity have gene regulatory regions more susceptible to rAAV2 integration.
Genomic regions recurrently hit by rAAV2 integration.
It is of interest that we found 14 genomic regions where the rAAV2 vector recurrently integrated (Table. 3). In the present study, 179 integrations landed in RefSeq genes, and 164 of 17724 RefSeq genes had rAAV integrations within genes. Of the 164 RefSeq genes hit by rAAV2 integration, 151, 11, and 2 RefSeq genes were hit once, twice, and three times, respectively. In a random integration model based on the assumption described in the Methods section, 177, 0.9, and 0.002 RefSeq genes would be hit once, twice, and three times, respectively. This indicates that rAAV2 integration is biased toward integration into a particular set of genomic regions, including the ones in Table 3 (χ2 test; P = 0.0053). The average and median sizes of 17724 RefSeq genes are 40 and 14 kb, respectively; therefore, the size of rAAV2-targeted genes itself cannot explain the recurrent hits. Because in a random integration model there would be no genes targeted three times in a sample size of 179 genes, three repeatedly hit genomic regions would be considered hot spots for rAAV2 integration. If we include rAAV2 integrations landing near transcription start sites of up to −1.4 kb, four genes were targeted three times in the present study (Table 3). They were serologically defined colon cancer antigen 33 (Sdccag33), Ubc, upstream binding transcription factor, RNA polymerase I (Ubft), and Riken cDNA 1300002F13 (130002F13Rik) genes. Significantly, all four targeting events were observed independently in different mice (Table 3).
TABLE 3.
Genes or genomic regions recurrently hit by rAAV2 integration
| Target gene or genomic region
|
No. of integration eventsa
|
||||||
|---|---|---|---|---|---|---|---|
| Symbol | Name | Size (kb) | Mouse
|
Total (no. in RefSeq gene)b | |||
| A | B | C | D | ||||
| Bhmt | C betaine-homocysteine methyltransferase | 20.7 | 1 | 1 | 2 (2) | ||
| Cmas | Cytidine monophospho-N-acetylneuraminic acid synthetase | 28.7 | 1 | 1 | 2 (2) | ||
| Gsk3b | Glycogen synthase kinase 3 beta | 198.1 | 2 | 2 (2) | |||
| Gtf2i | General transcription factor III | 76.9 | 1 | 1 | 2 (2) | ||
| LOC381596c | LOC381596 | 13.8 | 2 | 2 | |||
| Myo6 | Myosin IV | 91.9 | 1 | 1 | 2 (2) | ||
| Nedd4I | Neural precursor cell expressed, developmentally down-regulated gene 4 like | 28.1 | 1 | 1 | 2 (2) | ||
| Sdccag33c | Serologically defined colon cancer antigen 33 | 2.0 | 1 | 2d | 3 | ||
| Spag9 | Sperm-associated antigen 9 | 127.3 | 1 | 1 | 2 (2) | ||
| Ubc | Ubiquitin C | 1.5-26.9 | 2 | 1 | 3 (3) | ||
| Ubtf | Upstream binding transcription factor; RNA polymerase I | 10.5 | 2e | 1 | 3 (2) | ||
| Usp10 | Ubiquitin specific protease 10 | 46.7 | 2 | 2 (2) | |||
| 1300002F13Rik | Riken cDNA 1300002F13 gene | 13.8 | 2 | 1 | 3 (3) | ||
| 45S pre-rRNA gene repeatc | 45.3 × 200f | 2 | 2 | 3 | 3 | 10 | |
Integrations into genes (with no footnote) and into upstream regulatory regions (indicated by footnote d or e) are listed.
The numbers in parentheses represent integrations in RefSeq genes.
These are not RefSeq genes. LOC381596 and Sdccag33 are identified as genes in the NCBI Map Viewer. The Sdccag33 gene is classified as a known gene according to the definition by UCSC Genome Bioinformatics.
One of the two integrations found in mouse C landed 1.4 kb upstream of the Sdccag33 gene, as defined by NCBI.
One of the two integrations found in mouse B landed 0.3 kb upstream of the Ubtf gene.
There are approximately 200 copies of the 45S pre-rRNA repeats per haploid mouse genome.
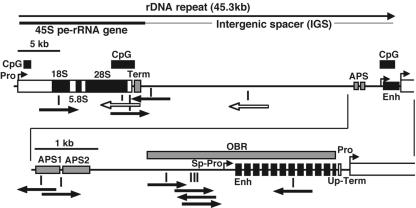
The most interesting finding is that 10 integrations (3% of total integrations) landed in the rRNA gene repeats (Fig. 1). Each rRNA gene repeat unit consists of the 45S pre-rRNA gene and an intergenic spacer. In mouse cells, there are ∼200 rRNA gene repeats of 45.3 kb in length per haploid genome in the nucleolus organizer regions (NORs) (11). Although they form a large target of approximately 9 Mb in length per haploid (0.3% of the mouse genome), 10 hits out of 347 integrations is 10 times higher than the frequency expected from a random integration model (binominal test; P < 0.000001). In addition, seven integrations were clustered within a short DNA stretch of ∼4 kb in length, the 5′ regulatory elements of the 45S pre-rRNA gene, including the enhancer repeats, spacer promoter, origin of bidirectional replication, and amplification-promoting sequences (9, 26). This region also coincides with DNase I hypersensitive sites and CpG islands (16), consistent with the general nature of integration site selection by rAAV2, as described above. The other three integrations also landed in or near CpG islands in the rRNA gene repeats. Two of the 15 rAAV2 integrations previously isolated from nonselected mouse liver (21) landed in the rRNA gene repeats as well.
FIG. 1.
Distribution of integration sites in the rRNA gene repeats, the hottest spot for rAAV2 integration. One complete rRNA gene repeat is depicted. Ten independent integrations among 347 integrations isolated from in vivo-selected primary mouse hepatocytes were mapped in the rRNA gene repeats and are shown together in a rRNA gene repeat with closed arrows. Orientation of each arrow represents vector genome orientation relative to that for the transgene expression cassette. Most of the integrations landed on narrow regions containing the regulatory elements for the 45S pre-rRNA gene. Such elements include the enhancer repeats (Enh), spacer promoter (Sp-Pro), origin of bidirectional replication (OBR), amplification-promoting sequences (APS), and terminator (Term). These elements coincide with CpG islands. Open arrows show the 2 of 15 integration sites we previously isolated from nonselected mouse primary hepatocytes (21). Up-Term, upstream terminator.
Host chromosomal changes at integration sites.
Vector-flanking mouse genomic sequences were determined in both sides and unambiguously mapped in 218 integration sites. With these 218 integration sites, we could determine host chromosomal changes at integration sites (Table 4). Contrary to the prediction from our previous small-scale study that the majority of genomic DNA deletions at integration sites are less than 0.3 kb in length (22), our results demonstrated that deletions over 0.3 kb were relatively frequent (33%) and that deletions over 1 kb were observed in 17% of the integrations. The median size of deletions was 205 bp, while a deletion as large as 5.5 Mb was also observed. Although target site deletion is a hallmark for rAAV2 integration (20, 22), we confirmed target site duplications ranging from 1 to 277 bp in 13 of 218 integrations (6% of the integration events) (Table 4). In addition, we confirmed translocations associated with integration in five cases (2% of the integrations). At vector-cellular DNA junctions in two integration events, we observed interpositions of a stretch of mouse chromosomal DNA jumping from a region remote from the integration sites. The interposed DNA fragments at recombination sites were a 296-bp satellite DNA fragment from an unidentified chromosome and a 539-bp DNA fragment residing on the same chromosome but 60 Mb away from the integration site. These observations indicate that host chromosomal changes are unpredictable and more complex than the previous presumptions from small-scale studies (20, 22).
TABLE 4.
Host chromosomal changes associated with rAAV2 integration
| Target site deletion
|
Target site duplication
|
Translocation
|
|||
|---|---|---|---|---|---|
| Size of deletion (bp) | No. of events | Size of duplication (bp) | No. of events | Chromosome | No. of events |
| 0-10 | 11 | 1 | 1 | t(3:12) | 1 |
| 11-30 | 27 | 2 | 2 | t(3:14) | 1 |
| 31-100 | 40 | 3 | 3 | t(6:15) | 1 |
| 101-300 | 51 | 4 | 1 | t(9:17) | 1 |
| 301-1,000 | 34 | 12-13 | 2 | t(11:19) | 1 |
| 1,001-3,000 | 27 | 40-79 | 3 | ||
| >3,000 | 10 | 277 | 1 | ||
| Total | 200/218 (91.7%)a | 13/218 (6%) | 5/218 (2%) | ||
Vector-flanking mouse genomic sequences were determined in both sides and unambiguously mapped in 218 integration events. The frequencies of deletions, duplications, and translocations are calculated based on this number. Median target site deletion size, 205 bp; maximum, 5.5 Mb.
rAAV2 integrations in or near cancer-related genes.
We investigated at what frequency and how rAAV2 vectors integrate in or near (within ±5 kb upstream or downstream of) cancer-related genes with Web-based cancer and tumor gene databases, i.e., RTCGD and TGDB. We found 12 integrations (3.5% of 347 integrations) landing within or near cancer-related genes (Table 5). Nine of these have been identified as common integration sites (CISs) isolated from various MLV-tagged but exclusively hematologic tumor types (1). CISs represent targets of retroviral integration in more than one tumor and are thus likely to encode a disease gene. In particular, Zfp36, Hmga1, Vegfb, and Hic1, which were hit by rAAV2 integration, were isolated from eight, four, three, and three independent tumors, respectively. Although the significance is not clear, many retrovirus integration sites isolated from mouse tumors but not classified as CISs were also hit by rAAV2 integration. These retrovirus integration sites included Calr, Camk1g, Dhrs8, Dpp8, Epb4.1, Hps3, Lrdd, Nedd4, Nedd4l, Nfib, Nfyc, Rbm3, Rdx, Siat4a, Syt12, Tcte3, Topors, Tpm3, Ubc, Ubtf, Usp10, 1200010C09Rik, 1600019O04Rik, 1700027M01Rik, 2700094L05Rik, 4932417H02Rik, and C530046K17Rik. It may be noted that Nedd4l, Ubc, Ubtf, and Usp10 are among the genes recurrently hit by rAAV2 integration (Table 3). The other three cancer-related genes listed in Table 5 are found in the Baylor College of Medicine TGD. Importantly, 4 of these 12 integrations in or near cancer-related genes occurred within ±1 kb of the transcription start site and upstream of the coding sequences, leaving open reading frames and intervening introns intact (Table 5).
TABLE 5.
Cancer-related genes hit by rAAV2 integration
| RIS namea | Gene hit by rAAV2 integration
|
Location of integration sites
|
|||
|---|---|---|---|---|---|
| Symbol | Name | Relative to gene (up, in, or down)b | Distance from transcription start site (kb) | Relative to ORF (up, in, or down) | |
| Coro2a | Coro2a | Coronin actin binding protein 2A | In | Down | |
| Dkmi28 | Vegfb | Vascular endothelial growth factor B | Down | Down | |
| Evi24 | Zfp36 | Zinc finger protein 36 | Up-in | −1.1 to + 0.6 | In |
| Evi41 | Crry | Complement receptor related protein | In | Down | |
| Evi63 | Epha2 | Eph receptor A2 | In | In | |
| Evi99 | Stag1 | Stromal antigen 1 | Up-in | −0.1 to +0.4 | Up |
| Evi130 | Hmga1 | High mobility group AT-hook 1 | In | +0.9 | Up |
| Evi132 | Nfkbie | Nuclear factor of kappa light polypeptide gene | Down | Down | |
| Hic1 | Hic1 | Hypermethylated in cancer 1 | Up | −0.8 | Up |
| N/Ac | Cdc25a | Cell division cycle 25 homolog A | In | In | |
| N/Ac | Fosl2 | Fos-like antigen 2 | In | In | |
| N/Ac | Pten | Phosphatase and tensin homolog | In | +0.5 | Up |
RISs, retroviral integration sites. Listed are only CISs that likely encode a cancer gene.
Up, integration within 5 kb upstream of the transcription start site of RefSeq genes; in, integration between start and stop sites of genes; down, integration within 5 kb downstream of the transcription stop sites of genes.
N/A, not applicable. They are cancer-related genes listed in the web-based TGDB provided by Baylor College of Medicine.
To investigate whether cancer-related genes are preferential targets for rAAV2 integration, we performed a statistical analysis. We analyzed 9,329 computer simulated random integrations that landed in or near (within ±5 kb upstream or downstream of) RefSeq genes and found that 212 CISs were hit among 9,329 random integrations in or near RefSeq genes, which accounts for 2.3% of total integrations in or near genes. This frequency was statistically compared to the observed frequency in the present study, i.e., 9 CIS integrations among 209 integrations in or near RefSeq genes. As a result, the frequency of rAAV integration in or near CISs among all the integrations landing in or near RefSeq genes was not different from that of random integration (χ2 test; P = 0.056). Thus, our study demonstrates that cancer-related genes are not preferential targets for rAAV2 integration.
Structures of rAAV2 proviral genomes.
Previous studies have shown that in addition to single-copy integration with various terminal deletions, rAAV2 proviral structures often exhibit unpredictable complex structures. In Southern blot analysis of integrated rAAV2 vector genomes, smear signals with no discrete bands were observed with single cutter-digested liver DNA, demonstrating that there was no detectable vector genome concatemers as observed with extrachromosomal rAAV2 vector genomes (unpublished data). Although we could not determine the exact frequency of integrations of complex proviral genomes in the present study, we found that at least 95 (24%) of 393 proviral genomes exhibited some complexity. In addition to head-to-head, head-to-tail, and tail-to-tail configurations with various deletions and rearrangements (66 events), we observed interpositions of various nonvector sequences, i.e., rAAV vector plasmid backbone sequences in 22 proviruses, human genome sequences in 6 proviruses, or AAV helper plasmid sequences in 2 proviruses. This demonstrates that nonvector DNA, including human genomic DNA from cells used for vector production, could hitchhike with considerable frequency to host genomes in transduced tissues via viral particles.
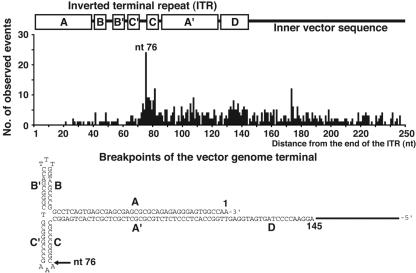
Vector genome terminal deletion occurs when rAAV2 integrates. In the present high-throughput analysis, we mapped 602 breakpoints from 358 independent integration events. In total, 365 (60%) of 602 recombinations occurred within the inverted terminal repeat (ITR) sequences. There was no preference for flip- or flop-oriented ITRs (68 versus 73). Both 5′ and 3′ breakpoints within 250 nucleotides from the vector end (total, 571 breakpoints) were combined together, and their distribution was analyzed with a histogram (Fig. 2). The result clearly showed nonrandom distribution of the recombination sites, which previous small-scale studies failed to elucidate (21, 27). The distal portion of the ITR is a nonpreferential substrate for recombination and clearly bordered by a recombinational hot spot around the proximal three-base loop of the T-shaped ITR (Fig. 2).
FIG. 2.
Distribution of the breakpoints of the vector genome terminal of rAAV2 proviruses. A total of 571 breakpoints within the 250 nucleotides near the vector ends are shown with 5′ and 3′ breakpoints combined. A hot spot for recombination (nucleotide 76) bordering two distinct regions preferred and nonpreferred for recombination is shown on a flip-oriented ITR with secondary structure. A to D, subregions of the ITR.
DISCUSSION
The present study significantly extended our recent observations of rAAV2 vector integrations in animal tissues, by the performance of a high-throughput characterization and statistical analyses of a large number of integration events. We demonstrated that rAAV2 has a very strong preference for integrating not only into genes (22) but also in or near gene regulatory sequences, similar to MLV integration. In addition, we found several hot spots for rAAV2 integration. The most prominent hot spot was the rRNA gene repeats, in particular in or near the regulatory elements of the 45S pre-rRNA gene.
Preferential integration into the rRNA gene units has not been reported in any other integrating vector currently explored for human gene therapy. The significance of this observation is threefold. First, disruption of a rRNA gene unit among hundreds of repeats should not be disadvantageous to host cells, as proven by a long evolutional history of insect retrotransposons that reside in this vicinity as parasites (5). Second, our results indicated that RNA polymerase II (Pol II)-driven transgene could be expressed in the vicinity of NORs where RNA Pol I dominates. We cannot totally exclude the possibility that clonally expanded hepatocytes expressing FAH had both a silent rAAV2 integration in the NOR and another integrated rAAV2 genome transcribed by Pol II outside the NOR. However, it is unlikely that a hepatocyte had multiple integrations, considering that the average vector copy number in the livers was less than 0.03 double-stranded (ds) vector genome copy number per diploid genomic equivalent before in vivo selection (22), and a majority of vector genomes of less than 0.03 ds vector genome copy number per diploid genomic equivalent should be extrachromosomal (25). In addition, we found that rAAV2 provirus DNA integrated in the rRNA gene repeats retained the ability to produce active FAH enzyme in mammalian cells when introduced into in vitro cultured cells by DNA transfection, in all of the seven rAAV2 provirus DNA we tested (data not shown). Third, integrated genetic materials in the rRNA gene repeats could be stably maintained over a number of cell divisions at least under a selective pressure. Thus, our study may imply that the rRNA gene repeats can be engineered as a harmless platform for stable Pol II-driven transgene expression.
The risk of rAAV2-mediated insertional mutagenesis has been believed to be negligible, on the basis of low integration frequency in vivo and no direct evidence of tumorigenesis by rAAV2 integration in a number of preclinical studies (15). Perhaps this is true in most gene therapy settings that target postmitotic quiescent tissues. In addition, rAAV2's strong preference for integrating into the rRNA gene region may contribute to reducing the risk of insertional mutagenesis. However, this does not prove that rAAV2 integration never results in insertional oncogenesis. It may become an issue of concern when rAAV2 vectors target proliferating cells such as hematopoietic cells or tissues undergoing regeneration, such as chronic liver inflammation.
In this regard, the following observations in the present study may draw further attention to the risk of rAAV2 vector integration. First, large host chromosomal deletions at integration sites, which had been considered rare in our previous study (22), were found to be relatively common. In addition, translocations were occasionally found. These chromosomal changes may result in the disruption of genes; therefore, functional loss of cellular genes should be more carefully considered. Second, rAAV2 vectors preferentially integrated near gene regulatory sequences. This nature of rAAV2 integration may not disrupt genes but in some cases may allow proviruses to drive flanking cellular genes, similar to retroviral long terminal repeats. This is because unpredictable complex structures of rAAV2 proviruses with various deletions and rearrangements (22) may delete poly(A) sequences or place functional enhancer/promoter sequences next to open reading frames of flanking cellular genes. Third, cancer-related genes were found to be hit by rAAV2 integration at a frequency of 3.5%. A majority of these cancer-related genes have been identified in hematologic malignancies, and the significance of these in liver remains to be addressed. However, we need to keep in mind that one third of integrations occurred within ±1 kb of the transcription start sites and upstream of the coding sequences of cancer-related genes; therefore, not only loss of function but also gain of function of these genes may be possible.
In addition to the implications regarding the risk of rAAV2-mediated insertional mutagenesis, the present study has provided new insights into the mechanisms of rAAV integration. First, the presence of interposed nonvector DNA sequences in rAAV2 proviral genomes suggests that ds proviral genomes found in chromosomes may have been formed from single-stranded vector genomes by leading-strand synthesis (6, 7). This is because duplex DNA formation by annealing of complementary single-stranded plus and minus genomes (24) does not easily explain the presence of ds proviral genomes with various interpositions of nonviral DNA, even though annealing may be the major mechanism for extrachromosomal vector genomes (24). Second, the presence of a recombination hot spot at the proximal three-base loop of the vector genome ITR that separates two distinct regions that are nonpreferential and preferential for recombination indicates that ITRs with secondary structure are substrates for rAAV2 integration and that initial resolution of the vector genome occurs at the three base loops of the T-shaped ITRs, although the ITR sequences themselves may not be prerequisite for vector genome integration (23). Third, both target site deletions and duplications were observed, suggesting the presence of at least two pathways for rAAV2 integration. Most recently, Miller et al. have demonstrated that rAAV vectors integrate at chromosomal DNA breaks (19).
In summary, the present study has provided new insights into the risk of rAAV2 vector integration. The issue of the risk of vector integration should not be limited to rAAV2 vectors and will have to be considered for rAAV vectors derived from other serotypes. Further investigations toward fully understanding the mechanisms of rAAV integration and host cellular responses are imperative for the development of safer and more effective vector systems.
Acknowledgments
We thank Jeffery Giering for helpful discussions and Patrice Held for performing FAH enzyme activity assay.
This work was supported by a National Hemophilia Foundation Career Development Award to H.N., a grant from the National Heart, Lung, and Blood Institute of the U.S. National Institutes of Health (NIH) to M.A.K. (HL66948), two grants from the National Institute of Diabetes and Digestive and Kidney Diseases, NIH, to H.N. (DK068636) and M.G. (DK18252), and at least in part by the National Cancer Institute, NIH, Department of Health and Human Services (DHHS), under contract N01-CO-12400 with SAIC-Frederick, Inc.
The contents of this publication do not necessarily reflect the views or policies of DHHS, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Akagi, K., T. Suzuki, R. M. Stephens, N. A. Jenkins, and N. G. Copeland. 2004. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 32:D523-D527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chao, H., Y. Liu, J. Rabinowitz, C. Li, R. J. Samulski, and C. E. Walsh. 2000. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol. Ther. 2:619-623. [DOI] [PubMed] [Google Scholar]
- 3.Donsante, A., C. Vogler, N. Muzyczka, J. M. Crawford, J. Barker, T. Flotte, M. Campbell-Thompson, T. Daly, and M. S. Sands. 2001. Observed incidence of tumorigenesis in long-term rodent studies of rAAV vectors. Gene Ther. 8:1343-1346. [DOI] [PubMed] [Google Scholar]
- 4.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 72:8568-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eickbush, T. H. 2002. R2 and related site-specific non-long terminal repeat retrotransposons, p. 813-835. In N. L. Craig, R. C. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 6.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher, K. J., G. P. Gao, M. D. Weitzman, R. DeMatteo, J. F. Burda, and J. M. Wilson. 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol. 70:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gogel, E., G. Langst, I. Grummt, E. Kunkel, and F. Grummt. 1996. Mapping of replication initiation sites in the mouse ribosomal gene cluster. Chromosoma 104:511-518. [DOI] [PubMed] [Google Scholar]
- 10.Grimm, D., S. Zhou, H. Nakai, C. E. Thomas, T. A. Storm, S. Fuess, T. Matsushita, J. Allen, R. Surosky, M. Lochrie, L. Meuse, A. McClelland, P. Colosi, and M. A. Kay. 2003. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood 102:2412-2419. [DOI] [PubMed] [Google Scholar]
- 11.Grozdanov, P., O. Georgiev, and L. Karagyozov. 2003. Complete sequence of the 45-kb mouse ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 82:637-643. [DOI] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 13.Huser, D., S. Weger, and R. Heilbronn. 2003. Packaging of human chromosome 19-specific adeno-associated virus (AAV) integration sites in AAV virions during AAV wild-type and recombinant AAV vector production. J. Virol. 77:4881-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kay, M. A., C. S. Manno, M. V. Ragni, P. J. Larson, L. B. Couto, A. McClelland, B. Glader, A. J. Chew, S. J. Tai, R. W. Herzog, V. Arruda, F. Johnson, C. Scallan, E. Skarsgard, A. W. Flake, and K. A. High. 2000. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat. Genet. 24:257-261. [DOI] [PubMed] [Google Scholar]
- 15.Kay, M. A., and H. Nakai. 2003. Looking into the safety of AAV vectors. Nature 424:251. [DOI] [PubMed] [Google Scholar]
- 16.Langst, G., T. Schatz, J. Langowski, and I. Grummt. 1997. Structural analysis of mouse rDNA: coincidence between nuclease hypersensitive sites, DNA curvature and regulatory elements in the intergenic spacer. Nucleic Acids Res. 25:511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarty, D. M., P. E. Monahan, and R. J. Samulski. 2001. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 8:1248-1254. [DOI] [PubMed] [Google Scholar]
- 18.Miao, C. H., R. O. Snyder, D. B. Schowalter, G. A. Patijn, B. Donahue, B. Winther, and M. A. Kay. 1998. The kinetics of rAAV integration in the liver. Nat. Genet. 19:13-15. [DOI] [PubMed] [Google Scholar]
- 19.Miller, D. G., L. M. Petek, and D. W. Russell. 2004. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 36:767-773. [DOI] [PubMed] [Google Scholar]
- 20.Miller, D. G., E. A. Rutledge, and D. W. Russell. 2002. Chromosomal effects of adeno-associated virus vector integration. Nat. Genet. 30:147-148. [DOI] [PubMed] [Google Scholar]
- 21.Nakai, H., Y. Iwaki, M. A. Kay, and L. B. Couto. 1999. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J. Virol. 73:5438-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakai, H., E. Montini, S. Fuess, T. A. Storm, M. Grompe, and M. A. Kay. 2003. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 34:297-302. [DOI] [PubMed] [Google Scholar]
- 23.Nakai, H., E. Montini, S. Fuess, T. A. Storm, L. Meuse, M. Finegold, M. Grompe, and M. A. Kay. 2003. Helper-independent and AAV-ITR-independent chromosomal integration of double-stranded linear DNA vectors in mice. Mol. Ther. 7:101-111. [DOI] [PubMed] [Google Scholar]
- 24.Nakai, H., T. A. Storm, and M. A. Kay. 2000. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J. Virol. 74:9451-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai, H., S. R. Yant, T. A. Storm, S. Fuess, L. Meuse, and M. A. Kay. 2001. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 75:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pikaard, C. S., L. K. Pape, S. L. Henderson, K. Ryan, M. H. Paalman, M. A. Lopata, R. H. Reeder, and B. Sollner-Webb. 1990. Enhancers for RNA polymerase I in mouse ribosomal DNA. Mol. Cell. Biol. 10:4816-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutledge, E. A., and D. W. Russell. 1997. Adeno-associated virus vector integration junctions. J. Virol. 71:8429-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnepp, B. C., K. R. Clark, D. L. Klemanski, C. A. Pacak, and P. R. Johnson. 2003. Genetic fate of recombinant adeno-associated virus vector genomes in muscle. J. Virol. 77:3495-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 30.Senior, K. 2002. Adeno-associated virus vectors under scrutiny. Lancet 359:1216. [DOI] [PubMed] [Google Scholar]
- 31.Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching, D. Block, J. Zhang, R. Soden, M. Hayakawa, G. Kreiman, M. P. Cooke, J. R. Walker, and J. B. Hogenesch. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]