Abstract
Retroviral late domains (L domains) are short amino acid sequences in the Gag protein that facilitate the process of budding. L domains act by recruiting the ESCRT complexes, which normally function in the formation of multivesicular bodies. The PTAP late domain of human immunodeficiency virus (HIV) is believed to specifically recruit this machinery by binding the ESCRT protein TSG101. It was recently demonstrated that expression of a C-terminal fragment of TSG101 (TSG-3′) blocked the budding of both PTAP-dependent and PPPY-dependent retroviruses. We show here that TSG-3′ expression leads to the formation of large spherical entities that we call TICS (TSG-3′-induced cellular structures) in the cytoplasm. Rous sarcoma virus (RSV) and murine leukemia virus (MLV) Gag proteins are selectively recruited to these structures, but HIV type 1 Gag is completely excluded. Experiments with various HIV and RSV vector constructs as well as HIV and RSV chimeras suggest that recruitment to the TICS is late domain independent and does not involve recognition of any single amino acid sequence. TICS appear to have no limiting membrane and do not colocalize with markers for any membranous cellular compartment. Wild-type TSG101 is also recruited to TICS, but most other ESCRT proteins are excluded. These structures are similar in nature to aggresomes, colocalize with the aggresome marker GFP-250, and are highly enriched in ubiquitin but in other ways do not fully meet the description of aggresomes. We conclude that the block to retroviral budding by TSG-3′ may be the result of its sequestration of Gag, depletion of free TSG101, or depletion of free ubiquitin.
All retroviruses encode a single structural polyprotein, Gag, which can assemble into a virus-like particle that buds from the plasma membrane independently of all other viral proteins. During or shortly after the process of budding, the virus-encoded protease becomes active and cleaves Gag into its constitutive domains, including the MA (matrix or membrane associated), CA (capsid), and NC (nucleocapsid) domains. Also present in all Gag proteins is a 4-amino-acid sequence known as a late or L domain, which is required at a late stage of budding to facilitate the process of pinching off from the host plasma membrane (6).
Retroviral late domains, first described over 15 years ago, are believed to function by usurping a set of cellular protein complexes known as ESCRTs, the normal task of which is to promote the budding of small vesicles into the lumen of the multivesicular body (MVB, or the vacuole in Saccharomyces cerevisiae). MVBs are specialized endocytic organelles that fuse with or become lysosomes (for reviews, see references 6, 20, and 30). Mono-ubiquitination is one of the signals used to target proteins to this pathway (20). Budding into the MVB, which is topologically and functionally similar to the budding of retroviruses, has been extensively studied in Saccharomyces cerevisiae and requires at least 17 genes for proper formation. Deletion of any one of these 17 genes in yeast results in the formation of a malformed late endosome called a class E compartment (20). The ESCRT proteins, which are encoded by 10 of these genes, form three distinct complexes, ESCRT I, II, and III, that act sequentially to form the budding MVB vesicle.
Three distinct categories of retroviral late domains with apparently distinct mechanisms of recruiting ESCRTs to retroviral budding sites have been described. The equine infectious anemia virus (EIAV) late domain, with core sequence YPDL (29), interacts with the protein AIP1 (37, 41). The yeast homologue of AIP1, Bro1, is also one of the 17 class E compartment genes and interacts with both ESCRT I and ESCRT III (41). Rous sarcoma virus (RSV), murine leukemia virus (MLV), and human T-cell leukemia virus utilize a late domain with the consensus sequence PPXY (43-45), which interacts with WW domains of E3 ubiquitin ligases (14, 21). Though ubiquitination appears to be an important step in ESCRT action, it is currently not clear how the E3 ubiquitin ligase ties in with the ESCRT complexes. The most extensively studied retroviral late domain is the PTAP sequence in human immunodeficiency virus type 1 (HIV-1) Gag p6 (12, 17), which binds the ESCRT I protein TSG101 (Vps 23 in yeast) (9, 40). The N-terminal portion of TSG101 contains a ubiquitin E2 variant domain that is capable of simultaneously binding ubiquitin and PTAP (9, 38). Overexpression of the N-terminal portion of TSG101 effectively blocks the budding of viruses dependent on a PTAP late domain but does not affect other retroviruses (11, 34). In contrast, overexpressing full-length TSG101 or the C-terminal half of TSG101 (TSG-3′) blocks the budding of HIV-1 (referred to hereinafter as HIV) Gag as well as the PPPY L domain-containing MLV Gag but not the YPDL-containing EIAV Gag. It was speculated that TSG-3′ acts by disrupting the cellular endosomal sorting machinery (11), but this hypothesis does not explain why viruses with YPDL late domains appear to be immune to the effects of TSG-3′.
The objective of this study was to determine whether TSG-3′ expression blocks RSV budding and, if so, where that block occurs. We found that TSG-3′ expression decreases RSV budding. Surprisingly, in cells expressing TSG-3′, RSV Gag, but not HIV Gag, was efficiently recruited along with TSG-3′ into nonendocytic aggresome-like structures in the cytoplasm of cells. Although most of the ESCRT machinery was not recruited to these structures, they were highly enriched in wild-type TSG101 as well as ubiquitin. Depletion of either one of these proteins may explain why TSG-3′ blocks only certain retroviruses from budding.
MATERIALS AND METHODS
Cell lines and transfections.
An immortalized line of chicken fibroblast cells (DF-1 [16, 31]) was grown at 37°C in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and 1% heat-inactivated chick serum. Cells to be transfected were grown on glass coverslips in 35-mm-diameter dishes. For each transfection, 1 μg of total DNA was incubated for 15 min with 3 μl of FuGENE6 reagent (Roche) in 100 μl of serum-free medium before the mixture was added to cells. Cells were prepared for imaging between 12 and 20 h posttransfection.
Vectors.
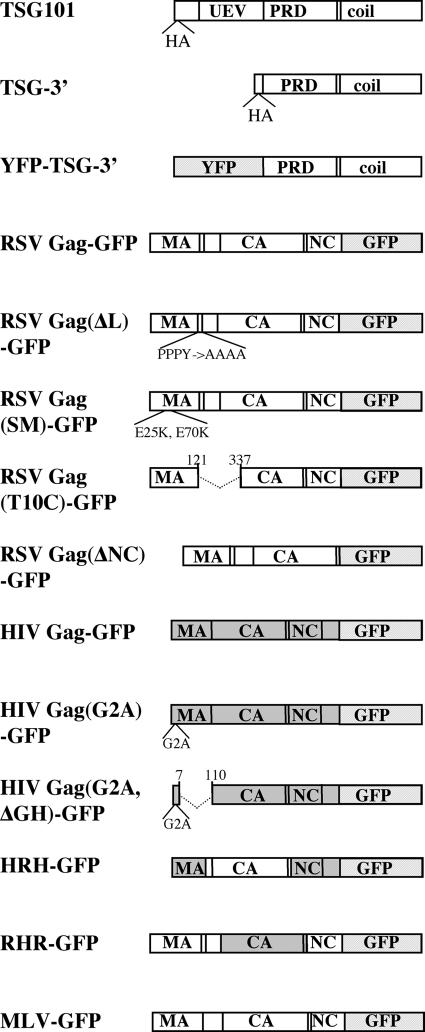
Plasmids expressing N-terminally hemagglutinin (HA)-tagged TSG101 or HA-tagged TSG-3′ were provided by Eric Freed (11). Yellow fluorescent protein (YFP)-TSG-3′ was created by subcloning the SalI/BamHI fragment from TSG-3′ (excluding the HA tag) into the same sites in the vector pEYFP-C1 (Clontech). RSV Gag-green fluorescent protein (GFP), pΔNC-GFP [RSV Gag(ΔNC)-GFP], and Super M-GFP [RSV Gag(SM)-GFP] have been described previously (4, 5). pGag(−) (RSV Gag) and pPPPY-A.GFP [RSV Gag(ΔL)-GFP] also have been described previously (36). RSV Gag(ΔL) (without GFP) was created by introducing the stop codon TAA after the last codon of RSV NC in RSV Gag(ΔL)-GFP. T10C-GFP [RSV Gag(T10C)] was described previously (28). HIV Gag-GFP was provided by Marilyn Resh (15). HIV Gag(G2A)-GFP, HIV Gag(G2A,ΔGH)-YFP, and GFP-VPS28 were provided by Paul Bieniasz. HRH-GFP was created by mutagenic PCR and replaces the CA-SP1 sequence of HIV Gag-GFP (15) with the RSV sequence that includes the last 25 residues of p10, all of CA, and all of SP. The forward primers over the junction sequences were as follows: HIV MA+RSV p10 (ACAGCCAGGTCAGCCAAAATTACCCTGGGCCGGCCCTGACTGACT) and RSV SP+HIV NC (ATCCAGCCCTTAATTATGATGCAGAGAGGCAATTTTAGG). RHR-GFP was created by mutagenic PCR and replaces the CA-SP sequence of RSV Gag-GFP (5) with the CA-SP1 sequence of HIV. The forward primers over the junction sequences were as follows: RSV p10-HIV CA (CTGGTCCGCCCGTGGTGGCCATGCCTATAGTGCAGAACATCCAGG) and HIV SP1+RSV NC (CAAATTCAGCTACCATAATGGCAGTAGTCAATAGAGAGAGGG). MLV Gag-YFP, TGN38-YFP, Lamp1-YFP, and DsRed-CD63 were provided by Walter Mothes (35). Rab5-GFP, Rab7-GFP, and Rab11-GFP were created by Craig Roy and provided by Gary Whittaker (13). Rab9-GFP was created by Suzanne Pfeffer and provided by Gary Whittaker (1). Rab4-GFP was created by Ruth Collins and provided by Gary Whittaker. Golgi-GFP, DsRed-ER, and peroxisome-GFP were purchased from Clontech. GFP-ubiquitin was created by subcloning the NheI/ApaI fragment of pEGFP-C2 (Clontech) into the same sites in Gag-Ub (28) and contains GFP followed by the 76 amino acids of ubiquitin followed by 4 amino acids. The parallel construct GFP-ubiquitin75 contains a stop codon after the 75th amino acid of ubiquitin, thereby deleting the C-terminal glycine residue. GFP-250 was provided by Elizabeth Sztul (7). CHMP1-YFP, CHMP2A-DsRed, CHMP3-YFP, CHMP4B-DsRed, and CHMP6-YFP were provided by Wesley Sundquist (41). GFP-human VPS4 (hVPS4) and GFP-hVPS4(EQ) were provided by Philip Woodman (2).
Budding assays.
The budding of RSV Gag in the presence or absence of TSG-3′ was analyzed by radiolabeling transfected cells for 2.5 h with an l-[35S]methionine-cysteine mix (50 μCi, >1,000 Ci/mmol) 24 h after transfection. Viral proteins were immunoprecipitated from detergent-lysed cells and particles by using a polyclonal rabbit serum against whole RSV, as previously described (42). Immunoprecipitates were separated in sodium dodecyl sulfate-12% polyacrylamide gels, which were subsequently dried and exposed to Kodak X-Omat AR5 X-ray film. Gag proteins were also quantitated by PhosphorImager (Molecular Dynamics) analysis. The budding efficiency was calculated as the amount of Gag in the medium divided by the total amount in the cell lysate and medium.
Reagents and fluorescence imaging.
FM4-64, Lysotracker red, DAPI (4′,6′-diamidino-2-phenylindole), and Mitotracker all were purchased from Molecular Probes. Monoclonal anti-HA (Sigma) was diluted 1:1,000 in phosphate-buffered saline (PBS). Polyclonal anti-GFP (Clontech) was diluted 1:500 in PBS. Tetramethyl rhodamine isothiocyanate- or fluorescein isothiocyanate-conjugated anti-mouse and Cy5-conjugated anti-rabbit (Jackson Laboratories) antibodies were diluted 1:50 in PBS. DF-1 cells were fixed 12 to 20 h posttransfection with 3.7% formaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 15 min, and blocked with 4% bovine serum albumin for 1 h prior to antibody staining. Primary- and secondary-antibody staining were each performed for 1 h at room temperature. Cells were viewed using a Cetus UltraView spinning disk confocal microscope (Perkin Elmer).
Correlative SEM.
DF-1 cells were plated and transfected on coverslips with grids (MatTek Corporation). Cells were fixed 12 to 20 h posttransfection with 4% paraformaldehyde in PBS for 15 min. After fluorescence images were captured and the locations of individual cells on the grid were recorded, cells were fixed with 2.5% glutaraldehyde in 0.2 M sodium phosphate buffer (pH 7.2), postfixed with 2% osmium tetroxide (Electron Microscopy Sciences), dehydrated through a graded series of ethanol washes, critical-point dried, and sputter coated with gold-palladium. The same individual cells were located and visualized by scanning electron microscopy (SEM) by using a model LEO 1550 field emission SEM at 3 kV. In most cases, multiple digital images were captured at the same magnification and pasted together using Adobe Photoshop. The procedure for visualizing TSG-3′ compartments was adapted from a previously described protocol (39). The plasma membranes of live cells were stripped by treating cells with 1% Triton X-100 in PEM (100 mM PIPES [pH 6.9], 0.5 mM MgCl2, 1 mM EGTA, 4% polyethylene glycol [molecular weight, 3,350; Sigma]). After the cells were washed three times with PEM, the actin cytoskeleton was disrupted by incubating cells in a 15-μg/ml concentration of the F-actin-severing protein villin (kindly provided by Anthony Bretscher) in PEM-2 mM CaCl2 for 30 min at room temperature. Cell remains were then fixed with 3.7% formaldehyde in PBS for 15 min, imaged, and processed for SEM as described above.
Thin-section EM.
DF-1 cells were transfected with a 1:1 ratio of YFP-TSG-3′-RSV Gag. Fourteen hours posttransfection, cells were trypsinized, fixed with 4% paraformaldehyde in PBS, and sorted by fluorescence-activated cell sorting into fluorescent and nonfluorescent cells. After being sorted, both populations of cells were fixed and prepared for electron microscopy (EM) as described above through the ethanol washes and then embedded in pure SPURR plastic (Electron Microscopy Sciences). Both populations of cells were thin sectioned and viewed on a Morgagni transmission electron microscope (TEM).
RESULTS
TSG101-3′ expression blocks RSV budding.
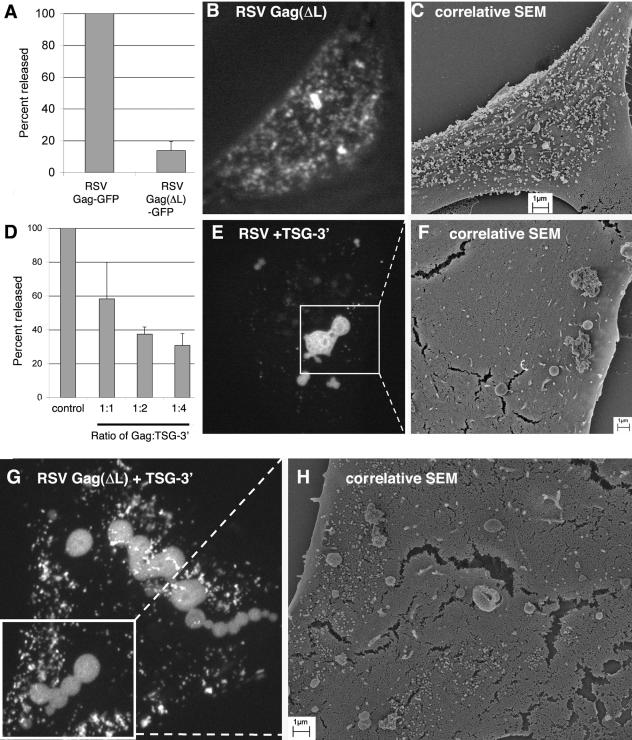
It was previously reported that expression of a truncated form of TSG101 (TSG-3′) (Fig. 1) blocks budding of both PTAP-containing and PPPY-containing retroviruses (11, 34). In order to determine whether TSG-3′ would block RSV Gag budding, we coexpressed TSG-3′ with a previously described GFP-tagged RSV Gag protein (5). As a positive control, we first performed budding assays using RSV Gag-GFP with or without its native PPPY late domain [RSV Gag(ΔL)] (Fig. 1). As expected, the amount of Gag-GFP(ΔL) released from transfected cells was only about 10% of that of Gag-GFP (Fig. 2A). To determine where this block was occurring, cells were transfected with a 9:1 molar ratio of Gag(ΔL)-Gag(ΔL)-GFP and the same individual cells were visualized first by confocal microscopy and then by SEM (Fig. 2B and C). Although Gag-GFP alone buds at a rate similar to that of wild-type Gag (5, 23), we have found that it forms morphologically aberrant virus particles if it is not coexpressed with excess wild-type Gag protein (unpublished data). The majority of Gag(ΔL)-GFP appeared by confocal microscopy to have accumulated on the plasma membranes of transfected cells. SEM images of the same cells verified that the fluorescent spots observed by confocal microscopy directly correlated with virus particles accumulated on the surfaces of transfected cells.
FIG. 1.
Vectors used in this study. HRH-GFP and RHR-GFP are chimeras of RSV Gag and HIV Gag. Light-gray domains represent fluorescent tags. Dark-gray boxes represent HIV sequence. A full description of all vectors is in Materials and Methods. UEV, ubiquitin E2 variant domain; PRD, proline-rich domain; coil, coiled-coil domain.
FIG. 2.
RSV Gag sequestration by TSG-3′. (A) Budding efficiency of RSV Gag-GFP(ΔL). Gag-GFP was normalized to 100%. (B, E, G) Fluorescence-merged Z sections of cells transfected with a mixture of DNAs at the ratios indicated. (B) Nine-to-one ratio of RSV Gag(ΔL) to RSV Gag(ΔL)-GFP; (E) 9:1:10 DNA ratio of RSV Gag to RSV Gag-GFP to TSG-3′; (G) 9:1:10 ratio of RSV Gag(ΔL) to RSV Gag(ΔL)-GFP to TSG-3′. (C, F, H) Corresponding SEM images of the surfaces of the same individual cells as shown in panels B, E, and G, respectively. (D) Budding efficiency of Gag in cells cotransfected with RSV Gag and TSG-3′ at the ratios indicated. Percentages released are expressed relative to the release of parallel cells transfected with an equal amount of Gag.
When parallel experiments were performed by coexpressing RSV Gag (or Gag plus Gag-GFP for the imaging experiments) with TSG-3′, we found that Gag release was blocked by TSG-3′ in a dose-dependent manner (Fig. 2D). Surprisingly, when Gag-GFP was viewed by confocal microscopy, the majority of the protein was found to have accumulated in large, round structures in the cytoplasm of transfected cells (Fig. 2E). SEM analysis of the same cells revealed few virus particles on the cell surface (Fig. 2F), verifying that most of the Gag was inside the cell. We refer to these TSG101-3′-induced cellular structures as TICS.
Because retroviral late domains are believed to interact with the ESCRT machinery, which includes TSG101, we hypothesized that recruitment of RSV Gag to TICS occurred through its PPPY late domain. However, when Gag(ΔL)-GFP [plus RSV Gag(ΔL)] was cotransfected with TSG-3′, Gag(ΔL)-GFP still accumulated in the TICS (Fig. 2G). In these cells a greater proportion of the Gag(ΔL)-GFP protein was found on the plasma membrane, and the Gag(ΔL)-GFP in the plasma membrane directly corresponded with properly budding or budding-arrested virus (Fig. 2H). Thus, recruitment to TICS is not dependent upon a functional L domain.
HIV Gag does not accumulate in TICS.
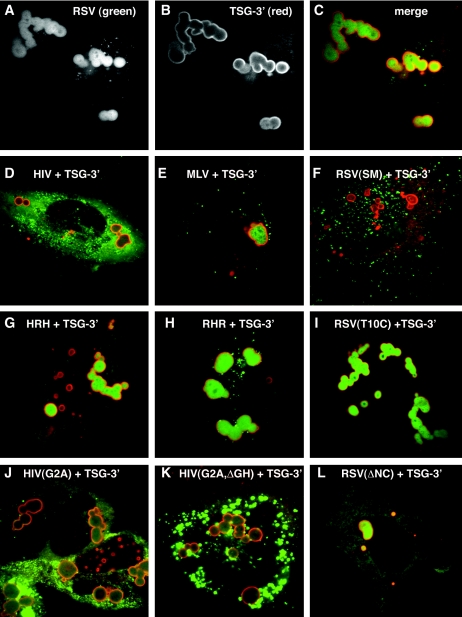
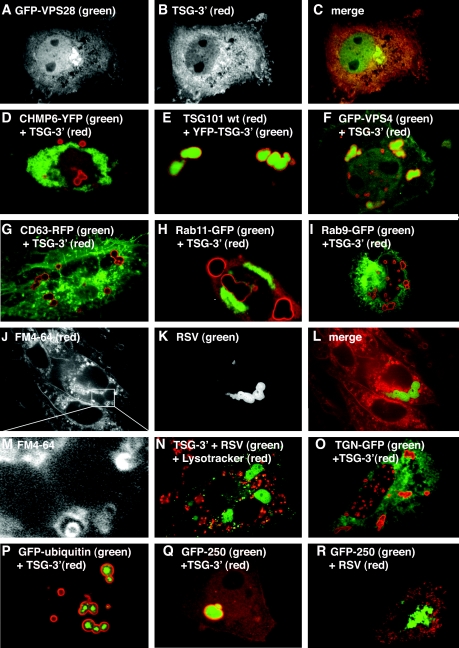
It has been reported previously that overexpression of TSG-3′ forms large cytoplasmic structures, but Gag proteins were not observed inside them (10, 11). Immunofluorescence staining against TSG-3′ yielded the same ring-like staining of the compartments as has been previously described, and the staining verified that RSV Gag-GFP was in the same compartment as the TSG-3′ protein (Fig. 3A to C). To determine whether other retroviral Gag proteins are recruited to these compartments, cotransfections of TSG-3′ with RSV Gag-GFP, HIV Gag-GFP, or MLV Gag-YFP were performed. HIV Gag-GFP was entirely excluded from the TICS (Fig. 3D). MLV Gag-GFP was recruited to the TICS, though the amount of Gag in the TICS varied more than with RSV.
FIG. 3.
Recruitment of Gag proteins by TSG-3′. (A-C) Cells transfected with a 1:1 ratio of RSV Gag-GFP to TSG-3′. (A) RSV Gag-GFP. (B) Immunofluorescence visualization of the HA tag of TSG-3′. (C) Merge of panels A and B. (D-L) Cells cotransfected with a 1:1 ratio of TSG-3′ to individual GFP or YFP-tagged Gag vectors. Each image is a merge of the TSG-3′ immunofluorescence image (red) and the fluorescent Gag image (green). (D) HIV Gag-GFP. (E) MLV Gag-YFP. (F) RSV Gag(SM)-GFP. (G) HRH-GFP. (H) RHR-GFP. (I) RSV Gag(T10C)-GFP. (J) HIV Gag(G2A)-GFP. (K) HIV Gag(G2A,ΔGH)-YFP. (L) RSV Gag(ΔNC)-GFP.
A rapidly budding RSV Gag mutant does not go to TICS.
RSV Gag, unlike HIV Gag, quickly accumulates in the nucleus when cells are treated with the nuclear export inhibitor leptomycin B (4, 32), indicating that the trafficking pathway of RSV may be distinct from that of HIV. A mutant form of RSV Gag with two extra positive charges in the MA domain [RSV Gag(SM)] (Fig. 1) buds faster than wild-type Gag and is not affected by leptomycin B treatment (4), presumably by having an increased affinity for plasma membrane. When we cotransfected RSV Gag-GFP(SM) with TSG-3′, the amount of Gag accumulating in TICS was dramatically decreased (Fig. 3F). This experiment demonstrates that recruitment to TICS can be modulated by very small genetic changes and suggests a possible link between membrane association and recruitment to TICS.
No single domain in Gag modulates recruitment to TICS.
Recruitment to TICS probably involves protein-protein interactions, and therefore we sought to determine what sequence in RSV participated in these interactions. To address this question, we made two chimeric proteins in which the CA domains of RSV and HIV were exchanged (Fig. 1). HRH-GFP replaces CA-SP1 of HIV with CA-SP of RSV plus the 25 amino acids of p10 adjacent to CA, which are known to be required for proper spherical assembly of RSV (18, 19). RHR-GFP replaces CA-SP of RSV with CA-SP1 of HIV (Fig. 1). Both RHR and HRH can assemble into proper spherical particles and both RHR-GFP and HRH-GFP produce a punctate pattern in transfected cells similar to that of wild-type Gag-GFPs (data not shown). When cotransfected with TSG-3′, HRH was efficiently recruited to TICS (Fig. 3G), initially suggesting that a sequence in RSV CA was important for this effect. However, RHR-GFP also was efficiently recruited to the TICS (Fig. 3H). The only sequence shared between RHR and HRH is the 25 amino acids of RSV p10 adjacent to CA, a sequence which also contains the Crm1-dependent nuclear export sequence (32). To determine whether this stretch of amino acid residues is responsible for the recruitment to TICS, we cotransfected TSG-3′ with RSV Gag(T10C)-GFP, a deletion construct missing the entire region surrounding the 25 amino acids (Fig. 1 and 3I). RSV Gag(T10C)-GFP also accumulated in TICS. Taken together, these experiments demonstrate that no single amino acid sequence in Gag controls recruitment to, or exclusion from, TICS.
Recruitment to TICS does not require membrane binding.
RSV and HIV Gag proteins differ in their interactions with membranes. Whereas HIV Gag must be N-terminally myristylated for membrane association (3), RSV Gag appears to interact with membranes only electrostatically (5). This difference, combined with the observation that rapidly budding RSV Gag(SM) is not found in TICS, led us to hypothesize that recruitment of Gag proteins to TICS is a consequence of slower membrane association. To test this hypothesis, we cotransfected TSG-3′ with three different vectors encoding Gag proteins with weakened membrane associations. The first two were nonmyristylated forms of HIV Gag-GFP, one containing a point mutation eliminating the myristylation site [HIV Gag(G2A)-GFP] and the other containing this mutation plus a deletion of the globular head of MA [HIV Gag(G2A,ΔGH)-YFP] (Fig. 1 and 3J to K). Both of these proteins formed cytoplasmic aggregates, but neither was recruited to TICS. The third Gag construct was RSV Gag(ΔNC)-GFP, which does not multimerize or associate with membranes (Fig. 3L) (4). This protein was recruited to TICS but less efficiently than the wild type, since a portion of Gag-GFP remained diffuse in the cytoplasm. We interpret these experiments to mean that recruitment to TICS is not a consequence of delayed membrane association.
TICS cannot be stained internally with antibodies.
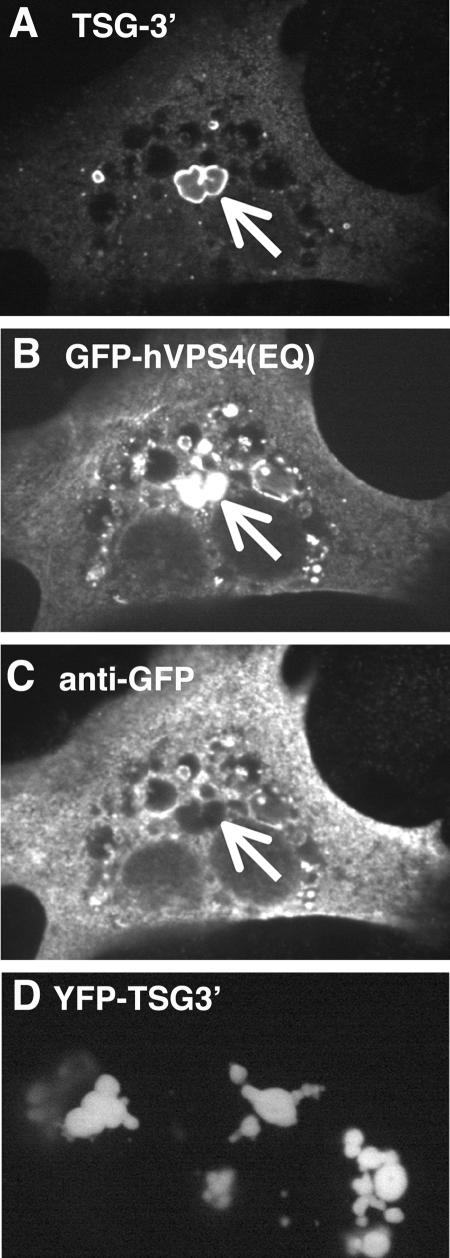
Although multiple GFP-tagged proteins were excluded from TICS, we wanted to rule out the possibility that the recruitment of RSV Gag-GFP to TICS was caused by the GFP tag itself. Surprisingly, diverse polyclonal antibodies to RSV Gag and Gag-GFP failed to stain either protein inside the TICS (data not shown). Because RSV Gag-GFP could be seen inside the TICS, it appeared that the antibodies could not penetrate these structures even after permeablization with Triton X-100. Antibody exclusion was most apparent when immunofluorescence staining was performed against the protein GFP-hVPS(EQ) (a dominant negative class E protein, discussed below) which localizes both to the interior of TICS and to the outside of other non-TSG-3′ vesicles. A large portion of GFP-hVPS4(EQ) was found to accumulate in the interior of TICS (Fig. 4A and B). However, when the same cells were stained with an antibody against GFP, only the GFP excluded from the TICS was labeled with antibody (Fig. 4C). In this instance the antibody did not even stain GFP at the outer rim of the TICS, which is delineated by TSG-3′. Only GFP-expressing cells were recognized by the GFP antibody, eliminating background fluorescence as an explanation. It therefore appears that some TICS-associated proteins (such as TSG-3′) are present throughout the TICS and that other TICS-associated proteins [such as VPS4(EQ)-GFP] are found only at the interior of the structure. In support of this notion, an N-terminally YFP-tagged TSG-3′ protein efficiently induced the formation of TICS, but the YFP tag was distributed equally through the structures and not just at the outer edge (Fig. 4D). We therefore hypothesize that antibodies are fully excluded from the interior of TICS.
FIG. 4.
Antibody staining of TSG-3′ compartments. (A-C) Cell transfected with a 1:1 ratio of TSG-3′ to GFP-hVPS4(EQ). (A) Immunofluorescent stain against the HA tag of TSG-3′. (B) GFP-hVPS4(EQ) fluorescence. (C) Immunofluorescent stain against GFP. Arrows point to the GFP-hVPS4(EQ)-enriched TICS. (D) Image of YFP-TSG-3′ fluorescence.
Not all ESCRT proteins associate with TICS.
TSG101 is a member of the ESCRT complex and interacts with all other members of this complex either directly or indirectly. To determine whether the rest of the ESCRT complex is recruited to the TICS along with TSG-3′, we performed cotransfections with TSG-3′ and several ESCRT proteins (Fig. 5). Cotransfection of TSG-3′ with its binding partner, GFP-VPS28, caused a dramatic relocalization of the TSG-3′ protein from TICS to a diffuse cytoplasmic distribution with some punctate appearance (Fig. 5A to C). Five other ESCRT proteins did not colocalize with TICS: CHMP1-YFP, CHMP2A-DsRed, CHMP3-YFP, CHMP4B-DsRed, and CHMP6-YFP (Fig. 5D, Table 1; see Fig. S1 in the supplemental material). The only ESCRT protein found to associate with TICS was wild-type TSG101 (Fig. 5E). Another class E compartment protein which acts downstream of the ESCRT complexes is the AAA-ATPase protein VPS4, whose function is to facilitate the disassembly of the final ESCRT complex. A GFP-tagged hVPS4 (GFP-hVPS4), which has been reported to be distributed throughout the cytoplasm (2), was efficiently recruited into TICS (Fig. 5F). A dominant negative VPS4 with a point mutation in its ATPase domain [GFP-hVPS4(EQ)] displayed a mixed phenotype when it was cotransfected with TSG-3′ (Fig. 4), with a portion of GFP-hVPS4(EQ) appearing to line the outer edge of endocytic compartments, as has been previously described (2), while the remainder of GFP-hVPS4 localized strongly within the TICS. These experiments demonstrate that TICS recruit some, but not most, of the ESCRT and ESCRT-associated proteins.
FIG. 5.
Colocalization of TSG-3′ with ESCRT and other cellular markers. (A-C) Cell transfected with a 1:1 ratio of GFP-VPS28 to TSG-3′. (A) Fluorescence image of GFP-VPS28 expression. (B) Immunofluorescence image of TSG-3′ expression. (C) Merge of panels A and B. (D, F-I, O-Q) Cells cotransfected with a 1:1 ratio of TSG-3′ and vectors expressing various fluorescently tagged cellular proteins. Each image is a merge of the TSG-3′ immunofluorescent stain image (red) and the fluorescent protein image (green). (D) CHMP6-YFP (ESCRT 3 protein). (F) GFP-hVPS4. (G) DsRed-CD63 (MVB marker). (H) Rab11-GFP (recycling endosome marker). (I) Rab9-GFP (late endosome marker). (O) TGN38-GFP (trans-Golgi marker). (P) GFP-ubiquitin (ubiquitin marker). (Q) GFP-250 (aggresome marker). (E) Cell transfected with a 1:1 ratio of YFP-TSG-3′ (green) to wild-type TSG101 (TSG101 wt) (red). (J-M) Cell transfected with a 1:1 ratio of RSV Gag-GFP to TSG-3′ treated with FM4-64 for the duration of the transfection (20 h). (J) Image of FM4-64 staining. (K) Image of RSV Gag-GFP expression (in TSG-3′ compartments). (L) Merge of panels J and K. (M) Blow up of the boxed location of panel J. (N) Cell transfected with a 1:1 ratio of RSV Gag-GFP (green) to TSG-3′ and stained live for 10 min with Lysotracker (red, acidic compartment marker). (R) Cell transfected with a 1:1 ratio of GFP-250 (green, aggresome marker) to RSV Gag-DsRed (red).
TABLE 1.
Summary of colocalization with TICS
| Cellular compartments or protein | Marker | Phenotypea |
|---|---|---|
| Cellular compartments | ||
| Early endosome | Rab4-GFP | + |
| Early endosome | Rab5-GFP | − |
| Late endosomes | Rab7-GFP | − |
| Late endosomes | Rab9-GFP | + |
| Recycling endosomes | Rab11-GFP | − |
| MVBs | DsRed-CD63 | − |
| All endosomes | FM4-64 | − |
| Lysosomes | Lamp1-YFP | − |
| Acidic compartments | Lysotracker | − |
| Nucleus | DAP1 | − |
| Mitochondria | Mitotracker | − |
| Trans-Golgi network | TGN-YFP | − |
| Golgi | Golgi-YFP | − |
| Endoplasmic reticulum | DsRed-ER | − |
| Peroxisome | Peroxi-GFP | − |
| Cytoplasm | DsRed | + |
| Ubiquitin | GFP-ubiquitin | +++ |
| Aggresomes | GFP-250 | +++ |
| Gag proteins | ||
| RSV Gag-GFP | +++ | |
| RSV Gag(ΔL)-GFP | ++ | |
| RSV Gag(SM)-GFP | +/− | |
| RSV Gag(T10C)-GFP | +++ | |
| RSV Gag(ΔNC)-GFP | ++ | |
| HIV Gag-GFP | − | |
| HIV Gag(G2A)-GFP | − | |
| HIV Gag(G2A,ΔGH)-GFP | − | |
| HRH-GFP | +++ | |
| RHR-GFP | +++ | |
| MLV-GFP | ++ | |
| MVB proteins | ||
| GFP-VPS28 | NT | |
| CHMP1-YFP | − | |
| CHMP2A-DsRed | − | |
| CHMP3-YFP | − | |
| CHMP4B-DsRed | − | |
| CHMP6-YFP | − | |
| VPS4-GFP | ++ | |
| VPS4(EQ)-GFP | ++ | |
| TSG101 wild type | +++ |
+++, mostly in compartments; ++, enriched in compartments; +, not excluded; −, excluded; NT, no TICS formed.
TICS are not endocytic compartments.
We initially assumed that TICS represent some form of late endocytic vesicle or MVB, as this is the normal location of TSG101 action. We were, therefore, surprised to find that the TICS did not colocalize with typical markers for the MVB (DsRed-CD63), the late endosome (Rab11-GFP), or the recycling endosome (Rab9-GFP) (Fig. 5G to I; Table 1). Early endosome markers (Rab4-GFP and Rab5-GFP) and another late endosome marker (Rab7-GFP) also did not colocalize with TICS (Table 1; see Fig. S2 in the supplemental material). To verify that TICS are not endocytic vesicles, we treated live TSG-3′-transfected cells for 1 h with the lipophilic dye FM4-64 to stain the membranes of all endocytic compartments, and the results showed no fluorescence at the boundaries of the TICS (data not shown). Finally, to eliminate the possibility that the TICS start out as endocytic compartments which mature and stop fusing with new vesicles, thereby preventing FM4-64 from reaching the structures, we stained live cells with FM4-64 for the entire duration of the a transfection (20 h) (Fig. 5J to M). This effectively stained most membranes in the cell, yet it did not stain the TICS. A blown-up image of the FM4-64 staining near the TICS reveals efficient staining of vesicles near the TICS but no staining of the TICS themselves (Fig. 5M). Furthermore, TICS did not colocalize with markers for acidic compartments (Lysotracker and Lamp1-YFP) (Fig. 3N; see Fig. S2 in the supplemental material), nuclei (DAPI) (see Fig. S2 in the supplemental material), the trans-Golgi network (TGN38-GFP) (Fig. 5O), mitochondria (mitotracker) (see Fig. S2 in the supplemental material), the endoplasmic reticulum (DsRed-ER) (see Fig. S2 in the supplemental material), or peroxisomes (persoxisome-GFP) (Table 1). Collectively, these experiments demonstrate that TICS are not standard cellular structures.
TICS are similar to aggresomes.
We next asked if TICS are specialized protein aggregates, such as aggresomes (for reviews, see references 8 and 19). A property of some aggresomes is enrichment in ubiquitin. To determine whether TICS are enriched in ubiquitin, cells were cotransfected with plasmids expressing TSG-3′ and GFP-ubiquitin, which has been used as a live-cell marker for ubiquitin (24). GFP-ubiquitin was found exclusively inside the TICS with a nonregular or compact staining pattern towards the centers of the structures (Fig. 5P). The significance of this distribution is unknown. A different ubiquitin construct missing the terminal conjugating glycine residue (GFP-ubiquitin 75) was not as highly enriched in TICS, suggesting that the ubiquitin in the TICS exists largely in a form that is conjugated to other proteins (see Fig. S2 in the supplemental material). A GFP-tagged protein that forms aggresomes (GFP-250) (7) was also found to be highly enriched inside TICS (Fig. 5Q). These experiments suggest that cells may recognize some GFP-tagged proteins, such as RSV Gag-GFP, as misfolded proteins when they are coexpressed with TSG-3′ and sequester them into aggresome-like compartments. If this is true, RSV Gag-GFP would likely also be recruited into classic aggresomes. To test this hypothesis, we cotransfected cells with GFP-250 along with DsRed-tagged RSV Gag (Fig. 5R). This Gag construct, which is efficiently recruited into TICS (data not shown), was fully excluded from GFP-250-induced aggresomes. These results imply, first, that although they share some properties, TICS are not identical to aggresomes and, second, that recruitment of Gag to TICS is probably not just a consequence of misfolding and/or aggregation.
TICS are amorphous structures when visualized by EM.
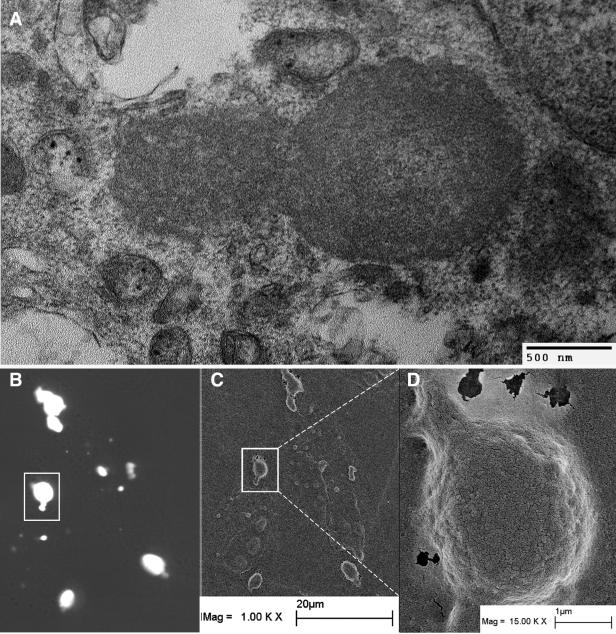
To obtain thin-section TEM images of TICS, cells were transfected with a 1:1 ratio of YFP-TSG-3′ to RSV Gag and sorted into fluorescent and nonfluorescent cells by fluorescence-activated cell sorting. Both populations of cells were then embedded and thin sectioned. Large amorphous aggregates were frequently observed in the fluorescent population (Fig. 6A) but not in the nonfluorescent population (data not shown). These structures were dense and round but showed no signs of either assembled virus particles or a limiting lipid bilayer. To obtain SEM images of the TICS, cells were again transfected with a 1:1 ratio of YFP-TSG-3′ to RSV Gag on coverslips with grids. The membranes of the unfixed cells were stripped, and the actin cytoskeleton was disrupted prior to fixation as described in Materials and Methods. Individual cellular remnants were viewed first by confocal microscopy and then processed and viewed by SEM. The TICS were not affected by the treatment and were easily discernible by SEM (Fig. 6B to D).
FIG. 6.
TEM and SEM analysis of TSG-3′ compartments. (A) Thin-section TEM of cell transfected with YFP-TSG-3′ and RSV Gag. (B-D) Cell transfected with a 1:1 ratio of YFP-TSG-3′ to RSV Gag. The cell was stripped of its membrane and actin cytoskeleton prior to imaging. (B) Fluorescence image of cellular remains and TSG-3′ compartments. (C-D) Low- and high-magnification SEM images of the same cellular remains.
In summary, we found that expression of TSG-3′ blocks the budding of RSV and causes an accumulation of RSV Gag into large nonendocytic structures that we refer to as TICS. Not all retroviral Gag proteins are equally sequestered into these structures, but there does not appear to be any single amino acid sequence that triggers recruitment. Most ESCRT proteins are not recruited to TICS, but the structures do appear to recruit cellular TSG101 and ubiquitin. TICS do not appear to have a limiting membrane and do not comply with the characteristics of any traditional cellular compartments but appear to be unique structures similar to aggresomes.
DISCUSSION
What are TICS?
The lack of colocalization of TICS with known cellular markers suggests that TICS do not represent a typical cellular compartment. Perhaps the most surprising feature about these structures is their apparent lack of a limiting membrane, despite the fact that they appear strikingly spherical. Though we cannot formally exclude the possibility of an external lipid monolayer or bilayer, we could not detect a membrane with fluorescent lipid probes or by thin-section TEM. Because TICS remain intact even after the rest of the cell is stripped away, if a lipid membrane did surround the structures, it is not needed to hold the structures together. Most likely TICS are masses of protein held together primarily through protein-protein interactions.
TICS are similar but not identical to classic aggresomes. Whereas aggresomes are found near the microtubule-organizing center and there is typically only one aggresome per cell (8, 22), cells expressing TSG-3′ commonly contain 10 or more TICS, which are distributed throughout the cell. Thin sections of classic aggresomes display dense but rather disperse aggregations of proteins in comparison to thin sections of TICS, which are solid round masses (Fig. 6A). Fluorescence images of cells transfected with YFP-TSG-3′ (which induces and labels TICS) display a morphology distinct from that of GFP-250 (which induces and labels aggresomes). In general, the TICS are smaller and brighter and have a more regular spherical shape than aggresomes. Finally, TICS efficiently recruit RSV Gag-GFP and exclude antibodies, whereas aggresomes are routinely stained with antibodies and do not recruit RSV Gag-GFP.
Given the differences between aggresomes and TICS, it is curious that TSG-3′ and GFP-250 strongly colocalize. Is TSG-3′ recruited to aggresomes, or is GFP-250 recruited to TICS? Judging solely by fluorescence images, it appears that both events occur, as structures formed with GFP-250 and TSG-3′ display both morphologies when they are observed by confocal microscopy (data not shown). Despite their differences, both TICS and aggresomes are composed of protein aggregates, which may amalgamate.
Another cellular entity composed of misfolded proteins is the dendritic cell aggresome-like induced structures (DALIS) (24, 25). DALIS form in dendritic cells following inflammatory stimulation or induced protein misfolding. Like TICS, DALIS do not necessarily form at the microtubule-organizing center, and the structures are highly enriched in ubiquitin. However, the DALIS are transient structures and are described as being unique to dendritic cells (24, 25).
What modulates recruitment to TICS?
Why some retroviral Gag proteins are recruited to TICS and others are not largely remains a mystery. The observation that both RSV and HIV chimeric proteins with exchanged CA domains are recruited to TICS demonstrates that there is not one single domain of RSV Gag that causes recruitment or one single domain of HIV Gag that causes exclusion. However, it remains possible that there are redundant domains in RSV that trigger recruitment. It is tempting to hypothesize that recruitment is a consequence of protein tertiary structure. One could postulate that TSG-3′ nucleates and maintains TICS but that they are filled with proteins that are unstable or are recognized by cellular proteins as being misfolded. This would explain why the artificial chimeric proteins, which might be less stable than their parent proteins, would be recruited to the TICS. Contrary to this hypothesis is the observation that the fast-budding RSV Gag(SM) (with only two point mutations in MA) is excluded from TICS. This exclusion suggests that recruitment to TICS can at least be circumvented. This circumvention may be a result of Gag reaching the plasma membrane before it can be sequestered or a result of Gag not trafficking by the pathway that leads to TICS.
Regardless of what mechanism triggers the recruitment of Gag to TICS, to some extent TICS prevent Gag from following its normal pathway, which is to exit the cell by budding. The observation that cells expressing RSV Gag(ΔL)-GFP plus TSG-3′ have Gag(ΔL)-GFP both in virus particles at the plasma membrane and in TICS implies that there is a point of no return after which Gag in the process of assembly can no longer be sequestered into TICS. In fact, the observation that cells expressing RSV Gag(ΔL)-GFP appear to have less Gag in the TICS (Table 1) than wild-type Gag is probably just a consequence of there being a higher proportion of Gag on the plasma membrane. In cells expressing wild-type Gag plus TSG-3′, the Gag-GFP that does not go to TICS probably buds away from the cell, effectively reducing the background and making the TICS appear brighter. But what is the point of no return at which Gag-GFP can no longer be sequestered into TICS? Is it the point when Gag begins assembling into a viral particle or the point at which Gag reaches the membrane? A possible unifying explanation of the results reported here would be that Gag assembly or Gag binding the plasma membrane circumvents recruitment to TICS. Perhaps RSV Gag-GFP(SM) is excluded from TICS because it rapidly binds the plasma membrane and all of the HIV Gag constructs are excluded from TICS (including the constructs which never go to a membrane) because they assemble faster than their RSV counterparts or the chimeric Gags.
Why does TSG-3′ block retroviral budding?
It is not surprising that expression of TSG-3′ causes a decrease in MLV and RSV budding when there is a competition between budding and sequestration into TICS. However, TSG-3′ also blocks the release of HIV Gag (11, 34) despite not sequestering HIV Gag into the TICS. The results reported here provide two possible explanations for why HIV budding is blocked. First, wild-type TSG101 is sequestered into TICS, and because depletion of this protein blocks HIV budding (9), overexpression of TSG-3′ might have the same effect as depleting the cell of free TSG101. Second, ubiquitin is highly enriched in TICS. Since most retroviruses, including HIV and RSV, are blocked by depletion of free ubiquitin within cells by proteasome inhibitors (28, 33, 34), expression of TSG-3′ may have similar effects on the levels of free ubiquitin in the cell and thereby inhibit budding indirectly. In support of the latter hypothesis, the retrovirus EIAV, which contains a YPDL late domain, is insensitive to proteasome inhibitors (26, 27) and also is not affected by TSG-3′ expression (34).
In summary, we have shown that the expression of TSG-3′ causes the formation of large unique cellular structures that selectively incorporate particular proteins, including VPS4, ubiquitin, and RSV Gag-GFP. TICS are similar in nature to aggresomes but differ in both action and appearance and represent unique cellular structures.
Supplementary Material
Acknowledgments
We thank Walter Mothes, Paul Bieniasz, Eric Freed, Alan Rein, Anthony Bretscher, Gary Whittaker, John Parker, Uta vonSchwedler, Wesley Sundquist, William Brown, and Elizabeth Sztul for providing reagents and Dimiter Demirov for useful discussions.
This work was supported by NIH grants CA20081 and CA47482.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Barbero, P., L. Bittova, and S. R. Pfeffer. 2002. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 156:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, N., and P. Woodman. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell 11:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callahan, E. M., and J. W. Wills. 2003. Link between genome packaging and rate of budding for Rous sarcoma virus. J. Virol. 77:9388-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan, E. M., and J. W. Wills. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus gag protein. J. Virol. 74:11222-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Mata, R., Z. Bebok, E. J. Sorscher, and E. S. Sztul. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146:1239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Mata, R., Y. S. Gao, and E. Sztul. 2002. Hassles with taking out the garbage: aggravating aggresomes. Traffic 3:388-396. [DOI] [PubMed] [Google Scholar]
- 9.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 10.Goff, A., L. S. Ehrlich, S. N. Cohen, and C. A. Carter. 2003. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J. Virol. 77:9173-9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goila-Gaur, R., D. G. Demirov, J. M. Orenstein, A. Ono, and E. O. Freed. 2003. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol. 77:6507-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Göttlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guignot, J., E. Caron, C. Beuzon, C. Bucci, J. Kagan, C. Roy, and D. W. Holden. 2004. Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J. Cell Sci. 117:1033-1045. [DOI] [PubMed] [Google Scholar]
- 14.Heidecker, G., P. A. Lloyd, K. Fox, K. Nagashima, and D. Derse. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78:6636-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 17.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi, S. M., and V. M. Vogt. 2000. Role of the Rous sarcoma virus p10 domain in shape determination of gag virus-like particles assembled in vitro and within Escherichia coli. J. Virol. 74:10260-10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 21.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 23.Larson, D. R., Y. M. Ma, V. M. Vogt, and W. W. Webb. 2003. Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy. J. Cell Biol. 162:1233-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lelouard, H., V. Ferrand, D. Marguet, J. Bania, V. Camosseto, A. David, E. Gatti, and P. Pierre. 2004. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J. Cell Biol. 164:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lelouard, H., E. Gatti, F. Cappello, O. Gresser, V. Camosseto, and P. Pierre. 2002. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature 417:177-182. [DOI] [PubMed] [Google Scholar]
- 26.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raiborg, C., T. E. Rusten, and H. Stenmark. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15:446-455. [DOI] [PubMed] [Google Scholar]
- 31.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 32.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 99:3944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Kräusslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shehu-Xhilaga, M., S. Ablan, D. G. Demirov, C. Chen, R. C. Montelaro, and E. O. Freed. 2004. Late domain-dependent inhibition of equine infectious anemia virus budding. J. Virol. 78:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785-801. [DOI] [PubMed] [Google Scholar]
- 36.Spidel, J. L., R. C. Craven, C. B. Wilson, A. Patnaik, H. Wang, L. M. Mansky, and J. W. Wills. 2004. Lysines close to the Rous sarcoma virus late domain critical for budding. J. Virol. 78:10606-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Göttlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 38.Sundquist, W. I., H. L. Schubert, B. N. Kelly, G. C. Hill, J. M. Holton, and C. P. Hill. 2004. Ubiquitin recognition by the human TSG101 protein. Mol. Cell 13:783-789. [DOI] [PubMed] [Google Scholar]
- 39.Svitkina, T. M., and G. G. Borisy. 1998. Correlative light and electron microscopy of the cytoskeleton of cultured cells. Methods Enzymol. 298:570-592. [DOI] [PubMed] [Google Scholar]
- 40.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Schwedler, U. K., M. Stuchell, B. Müller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Kräusslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 42.Weldon, R. A., Jr., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric gag protein into retroviral particles. J. Virol. 64:4169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.