Abstract
Background
N6-Methyladenosine (m6A) RNA modification is the most prevalent internal modification pattern in eukaryotic mRNAs and plays critical roles in diverse physiological and pathological processes. However, the expression of m6A regulator YTHDF2, its prognostic value, its biological function, its correlation with tumor microenvironment (TME) immune infiltrates, and related regulatory networks in hepatocellular carcinoma (HCC) remain determined.
Methods
TCGA, GTEx, and GEO databases were used to investigate the expression profile of YTHDF2 in HCC. We performed differentially expressed genes (DEGs) analysis and constructed a PPI network to explore the biological processes of YTHDF2 in HCC. Kaplan-Meier curves and Cox regression analysis were used to assess the prognostic value of YTHDF2 and then a clinical prognostic nomogram was constructed. Additionally, ssGSEA was performed to assess the correlation between YTHDF2 and immune infiltration levels. The TISIDB database was applied to explore the expression of YTHDF2 in immune and molecular subtypes of HCC. GSEA identifies the YTHDF2-related signaling pathways. Finally, we utilized miRNet and starBase database to construct regulatory networks for HCC based on lncRNA-miRNA and miRNA-YTHDF2 interactions.
Results
YTHDF2 was significantly upregulated in HCC tumor tissues compared with the adjacent normal tissues. HCC patients in the high YTHDF2 expression group had poorer survival. Multivariate Cox analysis suggested that YTHDF2 may be a new independent prognostic indicator for HCC patients, with the prognostic nomogram exhibiting satisfactory results. YTHDF2 expression was significantly correlated with TME immune cell-infiltrating characteristics. Strong correlations were also shown in immune subtypes, molecular subtypes and immune checkpoints. Further analysis revealed that the combination of YTHDF2 expression and immune cell score was considerably associated with survival outcome in HCC patients. GESA analysis demonstrated that high YTHDF2 expression is associated with multiple biological processes and oncogenic pathways. Moreover, 14 possible regulatory networks were constructed, which are associated with HCC progression.
Conclusion
Our findings revealed that YTHDF2 may serve as a promising prognostic biomarker for HCC and may regulate the tumor immune microenvironment to provide effective therapeutic strategies.
Keywords: m6A methylation, Hepatocellular carcinoma, Prognostic signature, Immune infiltration, Epitranscriptomics
1. Introduction
As the most abundant RNA modification, N6-Methyladenosine (m6A) RNA modification plays a critical role in biological processes. Dynamical and reversible crosslink among the regulators of m6A (methyltransferase complex, Demethylase and reader protein), is involved in initiation and progression of cancer. More recent studies have revealed that RNA m6A modification is involved in the development of the immune system and the induction of immune response. Cancer drugs targeting m6A methylation are promising in the clinical application, and some have shown satisfactory anticancer effects [[1], [2], [3]].
The m6A reader proteins, including EIF3, YTH family and IGF2BP family, primarily influences gene expression post-transcriptionally through specific recognition or regulating targeted RNA. Aberrantly expressed reader proteins have been correlated with diverse malignant behaviors in multiple cancer types. As the first reported m6A reader, YTHDF2 induces degradation of transcripts by selectively binding m6A-modified mRNA and recruiting them to mRNA decay sites [4]. Previous studies have demonstrated that YTHDF2 plays an oncogenic role in most cancer types. YTHDF2 was upregulated in prostate cancer and its expression can be suppressed by miR-493-3p, thus inhibition of miR-493-3p could abrogate the inhibitory effect of YTHDF2 knockdown on prostate cancer cell proliferation and migration [5]. Paris et al. discovered that YTHDF2 is overexpressed in acute myeloid leukemia and affects homeostasis of hematopoietic stem cells [6]. Overexpression of YTHDF2 is associated with pancreatic progression and shortened patient survival [7]. Nevertheless, YTHDF2 has been reported to play both oncogenic activity and tumor suppressive roles in hepatocellular carcinoma (HCC). The tumor suppressor gene SOCS2 has been identified as the target for METTL3-mediated m6A modification, which is recognized by YTHDF2 leading to its degradation [8]. In contrast, Zhong et al. revealed that YTHDF2 was downregulated in response to hypoxia leading to HCC cell proliferation [9]. In addition, YTHDF2 inhibits the function of innate immunity by binding m6A-modificated circRNA [10]. Winkler et al. 's findings reveal that m6A serves as a negative regulator of interferon response by dictating the fast turnover of interferon mRNAs and thus facilitating viral propagation [11]. However, a comprehensive depiction of YTHDF2 in the prognosis, the tumor microenvironment (TME), and regulatory networks in HCC remains deficiency.
HCC has high relapse and low 5-year survival rates, mainly related to the large heterogeneity among patients. Immune checkpoint blockades (ICBs) have recently exhibited clinical benefits in several types of cancer, although the response rates in HCC were unsatisfactory. The immune microenvironment can affect the effectiveness of ICB therapy. This study aims to explore the role of YTHDF2 in prognosis and TME immune cell infiltration in HCC. Subsequently, we constructed a regulatory network of lncRNAs-miRNAs-m6A regulators in HCC by systematic bioinformatic analysis, including 10 lncRNAs, 27 miRNAs, and YTHDF2, which may represent novel insights into the possible molecular pathways of HCC progression and may provide potential biomarkers for improving therapeutic efficacy.
2. Materials and methods
2.1. Data collection
Gene expression profiles and clinical information of The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) were obtained from the University of California, Santa Cruz (UCSC) Xena database (https://xenabrowser.net/datapages/). The data were further filtered to remove missing results, the RNA sequencing data in Transcripts Per Million (TPM) format and clinical data were retained and further analyzed. To assess YTHDF2 expression profiles in pan-cancer, we obtained tumor tissue from TCGA and normal tissue from the TCGA and GTEx databases.
The study used anonymized, published data requiring no ethics committee approval.
2.2. Differentially expressed analysis
Patients were classified into low and high expression groups according to the median expression value of YTHDF2 in TCGA HCC samples. Expression profiles were compared between low and high YTHDF2 expression groups to identify differentially expressed genes (DEGs) within the limma R package (version 3.6.3). Adjusted P < 0.05 and |log2-fold change (logFC)| >1.5 were considered as thresholds for the DEGs.
The different expression of YTHDF2 between HCC and non-tumor tissue was also analyzed in three RNA-seq datasets (GSE14520, GSE45267, and GSE64041), which were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds).
The YTHDF2 protein expression level in HCC was checked by immunohistochemistry from HPA database (https://www.proteinatlas.org/).
2.3. Survival analysis
Kaplan-Meier survival curves were used to estimate overall survival (OS) and progress free interval (PFI), and the log-rank test was used to assess the survival differences between groups. Univariate and multivariate Cox analyses were performed to identify the independent prognostic factors, hazard ratio (HR) and their 95 % confidence intervals (95 % CI) were computed. Then, the nomogram was constructed to predict 1-, 3- and 5-year OS for patients with HCC. The calibration curve was performed to verify and estimate the prediction accuracy of the nomogram. The performance of the nomograms was evaluated with calibration and clinical practical value by using the calibration curve and decision curve. We also assessed the prognostic performance with time-dependent receiver operating characteristic (ROC) curve analysis at various follow-up times.
2.4. Expression of YTHDF2 in immune and molecular subtypes and its correlation with immune checkpoint genes
The TISIDB database (http://cis.hku.hk/TISIDB/) is an online platform containing a large amount of tumor immune-related data to study tumor and immune system interactions. Correlation between YTHDF2 expression and the immune or molecular subtypes were analyzed using the TISIDB database.
The relationship between YTHDF2 expression and immune checkpoint (ICP) genes was explored via the SangerBox website (http://sangerbox.com/Tool), an online tool for analyzing TCGA data.
2.5. Immune infiltration analysis
Immune-cell infiltration levels in tumors were quantified by single-sample gene set enrichment analysis (ssGSEA) in R package gene set variation analysis (GSVA). The SGSEA scores were compared between the YTHDF2 high and low expression group using the Wilcoxon rank-sum test to discover significant differences in specific immune signatures.
Moreover, we computed the Spearman's rank correlation coefficients between YTHDF2 expression with various immune cells, including CD8+/CD4+ T cells, B cells, monocytes, natural killer (NK) cells, dendritic cells (DCs), TAMs, M1macrophages, M2 macrophages, neutrophils, T cells, and related subtypes.
2.6. Functional enrichment analysis
The Metascape database (http://metascape.org/) is a free and credible gene-list analysis online tool for high-throughput functional analysis of genes. It was applied to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. GO analysis is a common method for annotating genes and gene products and for identifying molecular function (MF), biological process (BP) and cellular component (CC) attributes for high throughout genome or transcriptome data. KEGG is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances.
In this study, we assessed the functions of YTHDF2 and their co-expression genes.
2.7. YTHDF2 coexpression network
The protein–protein interaction (PPI) network of co-regulated DEGs and the functional interaction between proteins were analyzed by the STRING database (https://cn.string-db.org/). The combined score threshold of interaction in our study was 0.5. The database has a comprehensive score for each pair of protein relationships ranging from 0 to 1; with higher scores indicating more reliable the PPI relationship.
2.8. Gene set enrichment analysis
The Gene Set Enrichment Analysis (GSEA) was performed using the R package “clusterProfiler” with the following parameters: nPerm = 10000, minGSSize = 10, maxGSSize = 500, and p-value-Cutoff = 0.05. The DEGs between the high and low expression levels of YTHDF2 were selected for analysis. Statistical significance and concordant differences between two biological states were determined with GSEA utilizing a predefined gene set from the MSigDB database (http://www.gsea-msigdb.org/gsea/msigdb/index.jsp). C2.Cp.v7.2.symbols.gmt was selected as the reference gene set of the KEGG and Reactome pathway, C5. All.v7.2.symbols.gmt was selected as the reference gene set of GO term. GSEA was used to generate a gene list with an order based on the correlation between all genes and YTHDF2 expression. The enriched pathways were determined based on the normalized enrichment score (NES), adjusted P-value and false discovery rate (FDR).
2.9. Construction of lncRNA-miRNA-YTHDF2 regulatory networks
All upstream target miRNAs that were correlated with YTHDF2 were obtained from the starBase database (http://starbase.sysu.edu.cn/), containing PITA, miRmap, microT, miRanda, PicTar, and TargetScan prediction programs. Only those miRNAs that appeared in one or more of the prediction procedures were considered for further analysis. Then, miRNet (http://www.mirnet.ca/miRNet/home.xhtml/) and starBase database were utilized to predict the target lncRNA of miRNA. Correlation analysis between lncRNA expression and miRNA expression was performed to screen for lncRNAs that are more consistent with ceRNA conditions. Finally, we constructed regulatory networks for HCC based on lncRNA-miRNA and miRNA-YTHDF2 co-expression patterns.
2.10. Statistical analysis
All analyses were performed using SPSS software version 19.0 (SPSS, Inc., Chicago, IL) and R software version 3.5.0 (Vienna, Austria). Statistical significance was set at 0.05 for two-tailed tests. The normality of the data was tested using the Kolmogorov–Smirnov test. The expression levels of YTHDF2 between normal tissues and tumors were compared by Wilcoxon rank-sum test and Wilcoxon signed-rank test, the stats package (version 4.2.1) and car package (version 3.1.0) of R were employed. Use the ggplot2 software package to visualize the analysis data. The Chi-square test, Fisher exact test, and Wilcoxon rank-sum test were used to compare and analyze the clinical and pathological conditions of the high and low YTHDF2 expression groups.
3. Results
3.1. YTHDF2 expression analysis in pan-cancer
As shown in Fig. 1A, the expression levels of YTHDF2 varied significantly among different cancers, except for kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), and pheochromocytoma and paraganglioma (PCPG). YTHDF2 was down-regulated in adrenocortical carcinoma (ACC), kidney chromophobe (KICH), and acute myeloid leukemia (LAML) while up-regulated in other cancers. Details are shown in Supplementary Table S1 and Table S2.
Fig. 1.
Expression level of YTHDF2 in HCC. (A) The expression of YTHDF2 in pan-cancer data from TCGA and GTEx database. (B) The expression levels of YTHDF2 in all HCC samples from TCGA database. (C) The expression levels of YTHDF2 in paired tumor and adjacent normal tissues in HCC from TCGA database. (D, E and F) showed the expression levels of YTHDF2 in all HCC samples from GSE14520, GSE45267and GSE64041. ns P ≥ 0.05; *P < 0.05; **P < 0.01 and ***P < 0.001.
YTHDF2 was prominently increased in HCC in both unmatched and paired comparative studies based on the TCGA database (Fig. 1B and C). Additionally, we downloaded GSE14520 (Platform: GPL571), GSE45267 (Platform: GPL570), and GSE64041 (Platform: GPL6244) datasets from GEO database to verify YTHDF2 expression in other datasets. The results also confirmed that YTHDF2 was overexpressed in HCC compared with those corresponding normal tissues (Fig. 1D, E, 1F). Immunohistochemical results from the HPA database also indicated that YTHDF2 was significantly overexpressed in HCC samples. HCC tumor tissues had medium or strong YTHDF2 IHC staining, while adjacent normal tissues had weak or medium staining (Fig. 2A). Moreover, YTHDF2 showed high diagnostic accuracy in HCC as shown by its ROC curve with an area under the curve (AUC) value of 0.779 (95 % CI 0.716 to 0.842) (Fig. 2B).
Fig. 2.
Diagnostic and prognostic value of YTHDF2 in HCC. (A) The expression levels of YTHDF2 in normal liver and HCC were visualized by IHC in HPA. (B) Diagnostic ROC curve of YTHDF2 in HCC. (C) Time-dependent ROC curve of YTHDF2 in HCC. (D) OS curves of YTHDF2. (E) PFI curves of YTHDF2.
3.2. Relationship between YTHDF2 expression and clinical features
The characteristics of 374 primary HCC patients with both clinical and gene expression data were downloaded from the TCGA database. Patients were classified into low- and high-expression groups according to the median expression value of YTHDF2 (Supplementary Table S3). The correlation between the YTHDF2 expression level and clinical characteristics was analyzed. We found that YTHDF2 expression was significantly associated with T stage (P = 0.002), pathologic stage (P < 0.001), BMI (P = 0.041), and histologic grade (P = 0.024). No correlation was found between YTHDF2 expression and other clinicopathologic features, as shown in Table 1.
Table 1.
Baseline characteristics of high and low YTHDF2 expression group patients with HCC.
| Characteristic | Low expression of YTHDF2 (n = 187) | High expression of YTHDF2 (n = 187) | P value |
|---|---|---|---|
| Gender | 0.077 | ||
| Female | 52 (27.8 %) | 69 (36.9 %) | |
| Male | 135 (72.2 %) | 118 (63.1 %) | |
| Race | 0.296 | ||
| Asian | 75 (42.1 %) | 85 (46.2 %) | |
| Black or African American | 6 (3.4 %) | 11 (6 %) | |
| White | 97 (54.5 %) | 88 (47.8 %) | |
| Age | 0.055 | ||
| ≤60 | 79 (42.2 %) | 98 (52.7 %) | |
| >60 | 108 (57.8 %) | 88 (47.3 %) | |
| BMI | 0.041 | ||
| ≤25 | 81 (46.8 %) | 96 (58.5 %) | |
| >25 | 92 (53.2 %) | 68 (41.5 %) | |
| AFP(ng/ml) | 0.313 | ||
| ≤400 | 120 (79.5 %) | 95 (73.6 %) | |
| >400 | 31 (20.5 %) | 34 (26.4 %) | |
| Albumin (g/dl) | 0.486 | ||
| <3.5 | 34 (21.1 %) | 35 (25.2 %) | |
| ≥3.5 | 127 (78.9 %) | 104 (74.8 %) | |
| Prothrombin time | 0.950 | ||
| ≤4 | 110 (70.5 %) | 98 (69.5 %) | |
| >4 | 46 (29.5 %) | 43 (30.5 %) | |
| Child-Pugh grade | 0.898 | ||
| A | 124 (91.2 %) | 95 (90.5 %) | |
| B | 11 (8.1 %) | 10 (9.5 %) | |
| C | 1 (0.7 %) | 0 (0 %) | |
| Fibrosis ishak score | 0.442 | ||
| 0 | 43 (37.1 %) | 32 (32.3 %) | |
| 1/2 | 13 (11.2 %) | 18 (18.2 %) | |
| 3/4 | 17 (14.7 %) | 11 (11.1 %) | |
| 5/6 | 43 (37.1 %) | 38 (38.4 %) | |
| Vascular invasion | 0.062 | ||
| No | 117 (70.5 %) | 91 (59.9 %) | |
| Yes | 49 (29.5 %) | 61 (40.1 %) | |
| Residual tumor | 0.903 | ||
| R0 | 166 (94.9 %) | 161 (94.7 %) | |
| R1 | 8 (4.6 %) | 9 (5.3 %) | |
| R2 | 1 (0.6 %) | 0 (0 %) | |
| Histologic grade | 0.024 | ||
| G1 | 30 (16.2 %) | 25 (13.6 %) | |
| G2 | 101 (54.6 %) | 77 (41.8 %) | |
| G3 | 49 (26.5 %) | 75 (40.8 %) | |
| G4 | 5 (2.7 %) | 7 (3.8 %) | |
| Adjacent hepatic tissue inflammation | 0.307 | ||
| None | 66 (54.5 %) | 52 (44.8 %) | |
| Mild | 46 (38 %) | 55 (47.4 %) | |
| Severe | 9 (7.4 %) | 9 (7.8 %) | |
| T stage | 0.002 | ||
| T1 | 109 (58.9 %) | 74 (39.8 %) | |
| T2 | 40 (21.6 %) | 55 (29.6 %) | |
| T3 | 29 (15.7 %) | 51 (27.4 %) | |
| T4 | 7 (3.8 %) | 6 (3.2 %) | |
| N stage | 1.000 | ||
| N0 | 119 (98.3 %) | 135 (98.5 %) | |
| N1 | 2 (1.7 %) | 2 (1.5 %) | |
| M stage | 0.365 | ||
| M0 | 131 (97.8 %) | 137 (99.3 %) | |
| M1 | 3 (2.2 %) | 1 (0.7 %) | |
| Pathologic stage | <0.001 | ||
| Stage I | 104 (58.8 %) | 69 (39.9 %) | |
| Stage II | 39 (22 %) | 48 (27.7 %) | |
| Stage III | 30 (16.9 %) | 55 (31.8 %) | |
| Stage IV | 4 (2.3 %) | 1 (0.6 %) |
3.3. Differential expression and pathway analysis of YTHDF2 in HCC
The DEGs analysis was performed to further explore the functions and pathways affected by YTHDF2 in HCC, patients with HCC were divided into the high expression group and the low expression group according to the expression level of YTHDF2. With |logFC| >1.5 and adjusted P < 0.05 set as the cut-off criteria, a total of 613 DEGs were identified (574 of 613 genes were highly expressed and 39 genes were lowly expressed). DEGs expressions were displayed in a volcano plot (Fig. 3A).
Fig. 3.
DEGs analysis, enrichment pathway analysis and construction of PPI network. (A) The volcano plot of DEGs. (B) PPI network comprising YTHDF2 and its most closely associated genes. Functional enrichment analysis in (C) GO Biological Process, (D) GO Molecular Function, (E) GO Cellular Component, (F) KEGG.
GO and KEGG enrichment analysis of these 613 genes were performed using Metascape. The GO biological processes terms revealed that the co-expressed genes were mainly associated with digestion, neuron projection morphogenesis, chemical synaptic transmission, inorganic ion transmembrane transport, epithelial cell differentiation, etc (Fig. 3C). The GO molecular functions terms were mainly enriched in the ligand-gated ion channel activity, signaling receptor regulator activity, glutamate receptor activity, neurotransmitter receptor activity, neurotransmitter receptor activity, etc (Fig. 3D). The GO cellular component terms were mainly involved in the extracellular matrix, ion channel complex, postsynaptic specialization membrane, mucus layer, axon signaling pathways, etc (Fig. 3E). Regarding to the KEGG pathway analysis as shown in Fig. 3F and Supplementary Table S4. Neuroactive ligand-receptor interaction, taste transduction, gap junction, gastric acid secretion, and cocaine addiction were the significant pathways.
In addition, the PPI network of YTHDF2 and its potential co-expression genes were established based on STRING to further explain the possible downstream reasons (Fig. 3B). The database has an integrated score distributed between 0 and 1 for each pair of protein relationships, and the higher the total score, the more reliable the PPI relationship. The analysis showed that some genes had close relationship with YTHDF2, for instance, METTL14, ALKBH5, CNOT1, METTL3, WTAP, FTO, KIAA1429, ALKBH1, RBM15 and HRSP12.
3.4. The prognostic significance of YTHDF2 in HCC
We evaluated the prognostic significance of YTHDF2 in HCC patients. According to the Kaplan–Meier survival curves, HCC patients with higher YTHDF2 expression exhibited poor OS (HR = 1.74 (1.22–2.47), P = 0.002) and PFI (HR = 1.50 (1.12–2.01), P = 0.006) (Fig. 2D and E). When stratified by clinical features, high YTHDF2 expression was also associated with worse OS in the ≤60 subgroup of age (HR = 1.93 (1.12–3.32), P = 0.016), male subgroup of gender (HR = 1.73 (1.10–2.71), P = 0.016), Asian subgroup of race (HR = 2.84 (1.50–5.36), P<0.001), G3 and G4 subgroup of histologic grade (HR = 2.43 (1.33–4.43), P = 0.003), stage Ⅲ and Ⅳ subgroup of pathologic stage (HR = 1.87 (1.02–3.41), P = 0.039), T2 subgroup of T stage (HR = 2.55 (1.16–5.57), P = 0.015), T3 and T4 subgroup of T stage (HR = 1.99 (1.12–3.53), P = 0.017), N0 subgroup of N stage (HR = 2.17 (1.39–3.40), P<0.001), and M0 subgroup of M stage (HR = 1.96 (1.26–3.05), P = 0.003) (Fig. 4A–I).
Fig. 4.
Association between the prognosis value of YTHDF2 and clinicopathologic characteristics.
Kaplan–Meier survival analysis showed that high expression of YTHDF2 was associated with worse OS in (A) ≤ 60 years old, (B) male, (C) Asian, (D) G3 and G4, (E) stage Ⅲ and Ⅳ, (F) stage T2, (G) stage T3 and T4, (H) stage N0, and (I) stage M0.
Furthermore, we performed univariate and multivariate Cox analyses of OS in HCC patients, and the results are shown in Table 2. Multivariate Cox analyses revealed that YTHDF2 remained independently associated with OS (HR = 1.495 (1.043–2.142), P = 0.028), along with T stage. We subsequently integrated the aforementioned variables to construct a clinical prognostic nomogram for HCC. The nomogram assigned risk scores to each variable according to their contributions, as shown in Fig. 5A. By adding up the total scores shown on the bottom scale, we can predict the probability of survival for the individual. The C-index of the prognostic model was 0.648 (95%CI: 0.623–0.674). The calibration curves were plotted with bootstrapping (cycle = 1000) (Fig. 5B–D). There was a favorable agreement between observed and predicted probabilities. Additionally, the clinical usefulness of the nomogram was assessed using decision curve analysis by quantifying the net benefits (Fig. 5E–G).
Table 2.
Univariate and multivariate Cox proportional hazard analyses of YTHDF2 expression.
| Variable | Number | Univariate analysis |
Multivariate analysis |
|||
|---|---|---|---|---|---|---|
| HR (95 % CI) | P value | HR (95 % CI) | P value | |||
| Gender | ||||||
| Male | 252 | Reference | ||||
| Female | 121 | 1.261 (0.885–1.796) | 0.200 | |||
| Age | ||||||
| ≤60 | 177 | Reference | ||||
| >60 | 196 | 1.205 (0.850–1.708) | 0.295 | |||
| Race | ||||||
| White | 185 | Reference | ||||
| Black or African American | 17 | 1.198 (0.520–2.760) | 0.672 | |||
| Asian | 159 | 0.756 (0.519–1.101) | 0.144 | |||
| YTHDF2 | ||||||
| Low | 187 | Reference | ||||
| High | 186 | 1.643 (1.160–2.328) | 0.005 | 1.495 (1.043–2.142) | 0.028 | |
| T stage | ||||||
| T1 | 183 | Reference | ||||
| T2 | 94 | 1.431 (0.902–2.268) | 0.128 | 1.320 (0.828–2.105) | 0.243 | |
| T3 | 80 | 2.674 (1.761–4.060) | <0.001 | 2.442 (1.596–3.737) | <0.001 | |
| T4 | 13 | 5.386 (2.690–10.784) | <0.001 | 5.572 (2.779–11.175) | <0.001 | |
| Histologic grade | ||||||
| G1 | 55 | Reference | ||||
| G2 | 178 | 1.162 (0.686–1.969) | 0.576 | |||
| G3 | 123 | 1.185 (0.683–2.057) | 0.545 | |||
| G4 | 12 | 1.681 (0.621–4.549) | 0.307 | |||
| Adjacent hepatic tissue inflammation | ||||||
| None | 118 | Reference | ||||
| Mild | 101 | 1.204 (0.723–2.007) | 0.476 | |||
| Severe | 17 | 1.144 (0.447–2.930) | 0.779 | |||
| AFP(ng/ml) | ||||||
| ≤400 | 215 | Reference | ||||
| >400 | 64 | 1.075 (0.658–1.759) | 0.772 | |||
| Vascular invasion | ||||||
| No | 208 | Reference | ||||
| Yes | 109 | 1.344 (0.887–2.035) | 0.163 | |||
| Child-Pugh grade | ||||||
| A | 218 | Reference | ||||
| B&C | 22 | 1.643 (0.811–3.330) | 0.168 | |||
| Fibrosis ishak score | ||||||
| 0 | 75 | Reference | ||||
| 1/2 | 31 | 0.935 (0.437–2.002) | 0.864 | |||
| 3/4 | 28 | 0.698 (0.288–1.695) | 0.428 | |||
| 5/6 | 80 | 0.737 (0.410–1.325) | 0.308 | |||
| Residual tumor | ||||||
| R0 | 326 | Reference | ||||
| R1&R2 | 18 | 1.604 (0.812–3.169) | 0.174 | |||
Fig. 5.
Construction and validation of nomogram based on YTHDF2 expression. (A) The nomogram for predicting survival probability at 1-, 3- and 5-years in HCC patients. (B–D) Calibration curves of the nomogram. (E–G) Decision curve analysis of the nomogram.
We also evaluated the prognostic accuracy of YTHDF2 using time-dependent ROC analysis at various follow-up times. ROC analysis showed satisfactory sensitivity and specificity with YTHDF2 in predicting OS at 1, 3, and 5 years, as shown in Fig. 2C. Taken together, all these results suggested that the high YTHDF2 expression level is associated with poor prognosis in patients with HCC.
3.5. Immune cell infiltration analysis
The tumor immune microenvironment is a key factor associated with tumor progression and can affect the prognosis of patients with HCC. To further explore the potential role of YTHDF2 in the tumor immune microenvironment, we calculated immune cell infiltration scores in HCC using ssGSEA from the R package. The correlation coefficients between YTHDF2 expression and the abundances of immune cell infiltration were analyzed using Spearman tests.
We found that YTHDF2 expression was positively correlated with infiltration levels of aDC (r = 0.124, P = 0.016), Eosinophils (r = 0.114, P = 0.027), Macrophages (r = 0.153, P = 0.003), NK CD56bright cells (r = 0.147, P = 0.004), T helper cells (r = 0.320, P < 0.001), Tcm (r = 0.139, P = 0.007), TFH (r = 0.141, P = 0.006), Th1 cells (r = 0.104, P = 0.045), and Th2 cells (r = 0.284, P < 0.001). In contrast, YTHDF2 expression was negatively correlated with that of Cytotoxic cells (r = −0.243, P < 0.001), DC (r = −0.241, P < 0.001), pDC (r = −0.307, P < 0.001) and Th17 cells (r = −0.120, P = 0.020) (Fig. 6A and B).
Fig. 6.
Correlation between YTHDF2 expression and immune infiltration. (A–B) Correlation between 24 immune cell infiltration. (C) The immune cell infiltration in the high- and low-expression groups of YTHDF2.
Further research showed significant differences in infiltrating immune cell levels, including Cytotoxic cells, DC, Eosinophils, NK CD56bright cells, pDC, T helper cells, TFH, and Th2 cells (P < 0.05), when YTHDF2 expression was categorized into high and low groups (Fig. 6C). High expression of YTHDF2 may promote intratumoral accumulation of eosinophils, NK CD56bright cells, TFH and Th2 cells, especially T helper cells. As expected, these results indicate that YTHDF2 plays a key role in immune infiltration of HCC.
Furthermore, since YTHDF2 expression is significantly correlated with immune infiltration and poor prognosis in HCC, we finally performed prognosis analyses using combinations of YTHDF2 expression and Immune cells score to assess whether YTHDF2 expression affects the prognosis of HCC due to immune infiltration. Patients with low YTHDF2 expression had better OS than those with high YTHDF2 expression, regardless of T helper cells, NK CD56bright cells, iDC, macrophages, eosinophils, Tem, B cells, NK CD56dim cells, Th17 cells, Tgd, CD8 T cells, cytotoxic cells, mast cells, neutrophils, TReg, NK cells, T cells, Tcm, or pDC scores. Meanwhile, HCC patients with high YTHDF2 expression and high CD8 T cells score or cytotoxic cells score had better OS compared to those with high YTHDF2 expression and low CD8 T cells score or cytotoxic cells score (Fig. 7A–S).
Fig. 7.
Impact of immune cell infiltration on prognosis in HCC patients. Kaplan–Meier survival analysis using combinations YTHDF2 expression and Immune cells score, including (A) B cells, (B) CD8 T cells, (C) cytotoxic cells, (D) eosinophils cells, (E) iDC cells, (F) macrophages cells, (G) mast cells, (H) neutrophils cells, (I) NK CD56bright cells, (J) NK CD56dim cells, (K) NK cells, (L) pDC cells, (M) T cells, (N) T helper cells, (O) Tcm cells, (P) Tem cells, (Q) Tgd cells, (R) Th17 cells, and (S) TReg cells.
3.6. Relationship between YTHDF2 expression and immune subtype and molecular subtype
To further explore the underlying mechanism of the relationship between YTHDF2 expression and the tumor immune microenvironment, we performed analysis by TISDB. The result suggested that YTHDF2 expression was significantly associated with immune subtype including C1 (wound healing), C2 (IFN-gamma dominant), C3 (inflammatory), C4 (lymphocyte depleted), C5 (immunologically quiet), and C6 (TGF-b dominant) (Fig. 8A). Similar results were also shown in molecular subtype (iCluster1, iCluster2, iCluster3) (Fig. 8B).
Fig. 8.
Expression of YTHDF2 in immune and molecular subtypes and its correlation with immune checkpoint genes. (A) The relationship between YTHDF2 expression and HCC immune subtypes and molecular subtypes. (B) The heat map of the 50 immune checkpoint genes correlated to YTHDF2. *P < 0.05; **P < 0.01 and ***P < 0.001.
3.7. Relationship between YTHDF2 expression and immune checkpoints genes
Given the crucial role of immune checkpoint genes in controlling immune system function and modulating tumor immune escape, we further investigated the interrelationship between YTHDF2 and several well-known immune checkpoint genes in HCC. YTHDF2 expression was significantly correlated with these immune checkpoint genes (Fig. 8C). These results further demonstrated the findings that YTHDF2 is intimately engaged in the regulation of the immune interactions and suggest that YTHDF2 plays an important role in immune escape in the tumor microenvironment, which may help anticancer immunotherapy.
3.8. GSEA in YTHDF2 expression phenotype
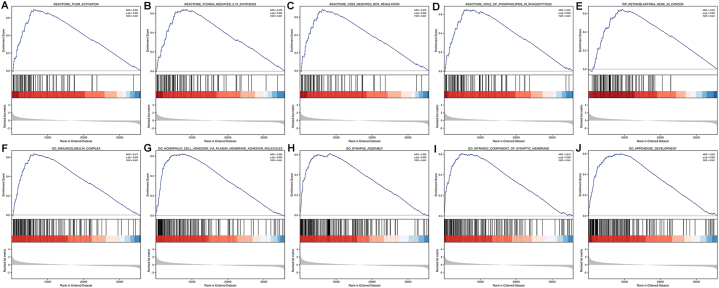
To further explore the functions and biological pathways of YTHDF2 in the progression of HCC, the GSEA analysis of differences between low and high YTHDF2 expression group was conducted to determine the GO term, Reactome pathway and KEGG pathway associated with YTHDF2. Using MSigDB Collection (C2.Cp.v7.2.symbols.gmt and C5.All.v7.2.symbols.gmt), we selected the most significantly enriched signaling pathways (FDR <0.05, adjusted P < 0.05) based on their normalized enrichment scores (NESs). GSEA analysis revealed that the high YTHDF2 expression phenotype was concentrated principally in immunoglobulin complex, homophilic cell adhesion via plasma membrane adhesion molecules, synapse assembly, intrinsic component of synaptic membrane, appendage development, FCGR activation, FCGR3a mediated IL10 synthesis, CD22 mediated BCR regulation, role of phospholipids in phagocytosis, and retinoblastoma gene in cancer (Fig. 9A–J and Table 3). These results indicate that high YTHDF2 expression is associated with hyperactivation of multiple oncogenic pathways in HCC.
Fig. 9.
Enrichment plots from GSEA. (A) FCGR activation, (B) FCGR3a mediated IL10 synthesis, (C) CD22 mediated BCR regulation, (D) role of phospholipids in phagocytosis, (E) retinoblastoma gene in cancer, (F) immunoglobulin complex, (G) homophilic cell adhesion via plasma membrane adhesion molecules synapse assembly, (H) intrinsic component of synaptic membrane, and (I) appendage development. ES, Enrichment score; FDR, false discovery rate; NES, normalized ES.
Table 3.
Gene set enrichment analysis.
| Description | FDR | NES | P.adjust |
|---|---|---|---|
| C5.All.v7.2.symbols.gmt | |||
| GO_IMMUNOGLOBULIN_COMPLEX | 0.001 | 2.617 | 0.002 |
| GO_HOMOPHILIC_CELL_ADHESION_VIA_PLASMA_MEMBRANE_ADHESION_MOLECULES | 0.001 | 2.595 | 0.002 |
| GO_SYNAPSE_ASSEMBLY | 0.001 | 2.555 | 0.002 |
| GO_INTRINSIC_COMPONENT_OF_SYNAPTIC_MEMBRANE | 0.001 | 2.512 | 0.002 |
| GO_APPENDAGE_DEVELOPMENT | 0.001 | 2.505 | 0.002 |
| C2.Cp.v7.2.symbols.gmt | |||
| REACTOME_FCGR_ACTIVATION | 0.001 | 2.561 | 0.002 |
| REACTOME_FCGR3A_MEDIATED_IL10_SYNTHESIS | 0.001 | 2.476 | 0.002 |
| REACTOME_CD22_MEDIATED_BCR_REGULATION | 0.001 | 2.475 | 0.002 |
| REACTOME_ROLE_OF_PHOSPHOLIPIDS_IN_PHAGOCYTOSIS | 0.001 | 2.467 | 0.002 |
| WP_RETINOBLASTOMA_GENE_IN_CANCER | 0.001 | 2.455 | 0.002 |
3.9. Construction of lncRNA-miRNA-YTHDF2 regulatory networks
Indeed, growing evidences have suggested an important role of the m6A RNA methylation regulators and their related regulatory networks in tumor progression and growth [12,13]. We attempted to construct lncRNA-miRNA-m6A regulator regulatory networks in HCC. PITA, miRmap, microT, miRanda, PicTar, and TargetScan databases were first applied to jointly predict upstream miRNAs that could potentially bind to YTHDF2. Twenty-seven target miRNAs were further filtered out based on their correlation with YTHDF2 expression, and their correlation coefficients were presented in Supplementary Table S5.
Next, the upstream lncRNAs of the 27 target miRNAs were predicted using miRNet and starBase database (Supplementary Fig. S1). According to the competing endogenous RNA (ceRNA) hypothesis, there is an inverse relationship between lncRNA and miRNA. As shown in Fig. 10A and lncRNAs (PRKAG2-AS1, PVT1, KCNQ1OT1, SNHG1, MALAT1, MIR155HG, MCM3AP-AS1, ZSCAN16-AS1, LINC00667 and DANT2) that were negatively correlated with the expression of 27 target miRNAs were obtained to further identify the potential upstream regulators of YTHDF2.
Fig. 10.
Prediction of the upstream lncRNAs potentially binding to YTHDF2 and lncRNA-miRNA-m6A regulator (YTHDF2) regulatory networks construction in HCC. (A) The correlation between miRNA and the target lncRNA. (B) The Sankey diagram shows the lncRNA-miRNA-m6A regulator (YTHDF2) regulatory network based on lncRNA-miRNA and miRNA-YTHDF2 co-expression patterns.
Therefore, we can construct 14 pairs of regulatory networks (PRKAG2-AS1 - hsa-miR-141-3p - YTHDF2, PRKAG2-AS1 - hsa-miR-200a-3p - YTHDF2, PVT1 - hsa-miR-194-5p - YTHDF2, KCNQ1OT1 - hsa-miR-194-5p - YTHDF2, SNHG1 - hsa-miR-194-5p - YTHDF2, MALAT1 - hsa-miR-194-5p - YTHDF2, MIR155HG - hsa-miR-194-5p - YTHDF2, MCM3AP-AS1 - hsa-miR-194-5p - YTHDF2, ZSCAN16-AS1 - hsa-miR-181c-5p - YTHDF2, LINC00667 - hsa-miR-181c-5p - YTHDF2, DANT2 - hsa-miR-181c-5p - YTHDF2, ZSCAN16-AS1 - hsa-miR-181d-5p - YTHDF2, LINC00667 - hsa-miR-181d-5p - YTHDF2 and DANT2 - hsa-miR-181d-5p - YTHDF2) based on the above results (Fig. 10B).
4. Discussion
M6A RNA methylation as one of the most common types of RNA modification in eukaryotes has been reported to be functional in many biological processes [14]. The m6A reader YTHDF2 played an important role in modulating the immune response, which could suppress innate immunity by binding m6A-modificated circRNA [10,11]. Additionally, aberrantly expressed YTHDF2 has been correlated with progression in multiple cancer. YTHDF2 was found upregulated in pancreatic cancer which promoted cancer cell proliferation via activation of Akt/GSK3β/Cyclin D1 pathway [7]. In endometrial tumors, the m6A level is lower, leading to inhibited YTHDF1-mediated-tranlation of tumor suppressor gene PHLPP2 and impaired YTHDF2-directed-degradation to oncogene mTORC2 [15]. Recent studies indicate that YTHDF2 has a dual role in HCC. YTHDF2 expression can inhibit expression of tumor suppressor genes. High m6A RNA methylation level promotes the development of HCC through increasing the expression of METTL3 and YTHDF2 while inhibiting SOCS2 expression [16,17]. By comparison, Zhong et al. showed that YTHDF2 overexpression inhibited the MEK/ERK pathway which was activated by hypoxia and act as a tumor suppressor with inhibitory effect on tumor growth [9]. Overall, research on the detailed mechanism by which YTHDF2 affects HCC pathogenesis remains controversial.
In this research, we comprehensively assessed the prognostic value of YTHDF2 expression and its correlation with TME immune infiltrates in patients with HCC. We used the TCGA, GTEx and GEO databases to determine the expression level of YTHDF2 in cancers and normal tissues. The results indicated that YTHDF2 had significantly higher expression in most cancer types including HCC, and correlated with several clinical features (T stage, pathologic stage, histologic grade, and BMI). Then the relationship between YTHDF2 expression and prognosis was explored. High YTHDF2 expression patients had a worse prognosis in HCC. Similar results have been obtained when stratified by clinical features. Multivariate Cox model and time-dependent ROC analysis also suggested that YTHDF2 was strongly associated with prognosis. We further developed and validated a clinical prognostic nomogram incorporating YTHDF2 expression and T stage, which showed satisfactory discrimination, calibration and clinical practical value. GO and KEGG pathway enrichment analysis showed that the DEGs were mainly enriched in ion channels and neuronal activity. The transports of ions across the cell membrane is a fundamental process in maintaining normal cellular function and activity [18,19]. The transport of ions across the membrane is critical in both normal and tumour cell survival and may be a factor in progression from normal to malignant state [20]. Several studies suggest that ion channel and neurotransmitter signaling in malignant cells and the tumor microenvironment may facilitate metastatic progression [21,22]. Neurons and their axonal projections have been implicated as a common, functionally enabling component of heterozygous cellular microenvironments in tumors. Signaling between innervation and cancer cells enables multiple hallmarks of cancer, including sustained proliferative signaling, resistance to cell death, activation of invasion and metastasis, and tumor-promoting inflammation. At the same time, the reciprocal effects of cancer cells on the nervous system result in the remodeling of neural form and function that contributes to neurological complications of cancers and amplifies the consequences of neurons on cancer pathophysiology, including encouraging nerve ingrowth, increasing neuronal excitability, reinforcing nerve-cancer interactions. Autocrine/paracrine signaling mediated by glutamate, synaptic transmission, and neurotransmitters is strongly associated with cell proliferation and tumorigenesis [23].
Based on the STRING database, a PPI network of YTHDF2 was constructed. The top ten functional partner genes were selected with a high degree of connectivity. Aberrant expression of m6A regulators such as METTL14, ALKBH5, METTL3, WTAP, FTO, KIAA1429 and RBM15 has been observed in various diseases, including cancer, obesity, infertility, autoimmune diseases and neurological diseases [11,[24], [25], [26], [27]]. The AlkB family such as ALKBH1, ALKBH5 and FTO are involved in the development of cancers [28]. CCR4–NOT complex plays critical roles in liver homeostasis, from gene transcription, protein translation, and the epigenetic modification of RNA [29]. Liver-specific disruption of CNOT1, encoding a scaffold subunit of the CCR4–NOT complex, leads to aberrant gene expression that is associated with lethal hepatitis [30]. The m6A modification might regulate the function of circRNAs in various tumors [10,31], and m6A-modified circRNAs were found to be endoribonuclease-cleaved via a YTHDF2-HRSP12-RNase P/MRP axis [32].
To further explore the function of YTHDF2 in HCC, we performed GSEA using TCGA data. GSEA showed that the immunoglobulin complex, homophilic cell adhesion via plasma membrane adhesion molecules, synapse assembly, intrinsic component of synaptic membrane, appendage development, FCGR activation, FCGR3a mediated IL10 synthesis, CD22 mediated BCR regulation, role of phospholipids in phagocytosis, and retinoblastoma gene pathway were positively enriched when YTHDF2 was highly expressed. It is indicated that YTHDF2 inhibitors promote tumor cell differentiation and immune response. Immunotherapy is a new cancer therapy strategy that stimulates and improves the natural ability of the immune system to attack cancer cells. Immunotherapy resistance has become one of the main obstacles to cancer remission [33]. Anti-inflammatory cytokines such as IL-10 can suppress effector T cell responses and play an important role in maintaining self-tolerance [34]. Depletion of IL-10 in the TME can enhance or restore antitumor immune effects. Taken together, these results indicated that YTHDF2 was a potential prognostic biomarker in HCC that required further clinical validation.
Comprehensive analysis of TME immune cell infiltration to develop personalized immunotherapy for patients is of great significance for the effective evaluation and prediction of the dynamics of tumour response to immunotherapeutics and the adoption of combined therapy strategy [35,36]. The microenvironment of HCC contributes to immunological changes during the progression of HCC [37]. ssGSEA combined with Spearman correlation was adopted to investigate the relationship between YTHDF2 expression and immune infiltration levels in HCC. Our results indicate that YTHDF2 expression has a significantly positive correlation with aDC, Eosinophils, Macrophages, NK CD56bright cells, T helper cells, Tcm, TFH, Th1 cells, and Th2 cells, while the inverse correlation was found in Cytotoxic cells, DC, pDC, Th17 cells. We also found that immune cell infiltration in HCC is related to prognosis. As one type of T helper cell, Th2 cell can lead to the inhibition of the host immune system, and produce IL-4, consequently resulting in activation of several cancer-related pathways [38,39]. Tfh cells can differentiate to become Th1 and Th2 cells and are essential for generating protective humoral immunity [40,41]. NK cells play an important role in providing an antiviral immune response and immune surveillance against tumors [42]. Macrophage infiltration has been proposed to be pivotal for tumor immune evasion, angiogenesis, growth and metastasis [43]. DCs and Cytotoxic cells were essential contributors to anti-tumor immunity [44]. All these findings exhibited the crucial role of YTHDF2 in immune infiltration in HCC.
Then we found that YTHDF2 expression was significantly different in different immune subtypes and molecular subtypes, which might demonstrate that YTHDF2 has significant potential to serve as biomarkers. In addition, immune checkpoint genes play a major role in controlling the function of the immune system and modulating tumor immune escape [34,35,37]. A strong relationship was discovered between YTHDF2 and immune checkpoint genes, which provides opportunities for developing effective immunotherapies. These results provide evidence suggesting that targeting YTHDF2 is a novel strategy for anticancer immunotherapy.
We hypothesized that ion channel and neuronal activities obtained from our enrichment analyses might be associated with immune infiltration in the tumor microenvironment of hepatocellular carcinoma, as they have been shown to be related to with tumor immunity in previous studies. The value of ion channels in tumor immunomodulation has received widespread attention [45]. Under normal conditions, ion channels are abundantly expressed in immune cells and play an essential role in maintaining immune cell activity, regulating lymphocyte development, and modulating immune response [46]. In the tumor immune microenvironment, ion channel dysfunction and inactivation of ion channels that recognize tumor antigens affect tumor cell anti-cancer immunity, and tumor cell hyperpolarization increases the possibility of immune evasion and promotes tumor progression [47,48]. Direct paracrine and electrochemical communication between neurons and cancer cells, as well as indirect interactions across a wide range of malignancies via neural effects on the immune system and stromal cells in the tumor microenvironment. Crosstalk between neurons, cancer cells and immune cells can regulate treatment resistance, anti-cancer immunity, and pro-cancer inflammation [49]. It has been shown that neurons in the NF1-associated low-grade gliomas (LGGs) TME secreted the paracrine factor mid-kine, which stimulated the recruitment and activation of CD8+ lym-phocytes to secrete the chemokine CCL4, thereby inducing expression in microglial/myeloid cells of CCL5, which triggers the cell cycle (proliferative signaling), suppresses apoptotic cell death in the cancer cells, and evading immune destruction [50]. Therefore, we speculated that YTHDF2 may play a critical role in immune infiltration in HCC.
Previous studies on ceRNA have demonstrated that ncRNAs, including miRNAs and lncRNAs, participate in the regulation of cancer-related biological processes via cross-talk [51]. We utilized the starBase and miRNet database to investigate the upstream regulatory miRNAs and lncRNAs of YTHDF2. As a result, 27 miRNAs and 10 lncRNAs were identified based on the ceRNA hypothesis. Since lncRNAs can interact with miRNAs through their response elements within a ceRNA network, we constructed regulatory networks based on co-expressed lncRNA-miRNA and miRNA-YTHDF2. By performing a comprehensive correlation analysis, 14 pairs of potential regulatory networks were eventually identified, which were potentially related to the progression of HCC. Numerous studies supported that these ncRNAs functioned as a crucial regulator in HCC. For instance, lncRNA MCM3AP-AS1 exerted an oncogenic role in HCC via targeting miR-194-5p and subsequently facilitated FOXA1 expression [52]. ZSCAN16-AS1 expedited HCC progression via modulating the miR-181 c-5p/SPAG9 axis to activate the JNK pathway [53]. Hepatoma cell-derived extracellular vesicles promote tumor metastasis by inducing the differentiation of bone marrow stem cells through microRNA-181d-5p and the FAK/Src pathway [54]. Targeting PRKAG2-AS1 distinctly inhibited proliferation, migration, and invasion in HCC cells [55]. LINC00667 acts as a tumor promotor in promoting HCC progression through targeting miR-130a-3p/AR axis [56]. Taken together, these reports further supported the feasibility of our bioinformatics analysis.
Although we have conducted a comprehensive and systematic analysis of YTHDF2, our study has several potential limitations. First, we conducted the study based on public databases without verification in vivo/in vitro experiments. Second, the function of the upstream regulatory mechanism and downstream target genes of YTHDF2 were not fully investigated in this study. Though the ceRNA regulatory network of YTHDF2 was obtained through bioinformatics analysis, more functional experiments and clinical translational studies are required to elucidate the biological function of these predicted molecular mechanisms in HCC. It remains to be discussed whether other ncRNAs, such as circRNAs, also play an important role in HCC. We expect to discover more efficient regulatory networks to improve patient outcomes. Third, the mechanisms by which YTHDF2 participates in immune regulation remain unknown and more investigations are needed to further elucidate the underlying biological mechanisms and the clinical significance of YTHDF2 in HCC progression. Finally, the sample size in our study was relatively small, and multicenter, multicenter collaborative prospective studies with larger sample sizes are needed to support our results.
5. Conclusions
In summary, our study revealed that YTHDF2 could serve as a promising prognostic biomarker for individual patients, and might play an important role in HCC progression by regulating the tumor immune microenvironment. Specific molecular mechanisms awaited further exploration.
Data availability
The datasets presented in this study can be found in online repositories including TCGA and GTEx (https://xenabrowser.net/datapages/), GEO (https://www.ncbi.nlm.nih.gov/gds), HPA (https://www.proteinatlas.org/), TISIDB (http://cis.hku.hk/TISIDB/), Metascape (http://metascape.org/), STRING (https://cn.string-db.org/), MSigDB (http://www.gsea-msigdb.org/gsea/msigdb/index.jsp), starBase (http://starbase.sysu.edu.cn/) and miRNet (http://www.mirnet.ca/miRNet/home.xhtml/). The accession numbers can be found in the article. Further inquiries can be directed to the corresponding author.
Ethics declarations
Review and/or approval by an ethics committee was not needed for this study because the study used anonymized, published data.
Funding statement
This work was supported by the Startup Fund for scientific research, Fujian Medical University (2019QH1186).
CRediT authorship contribution statement
Hang Wang: Writing - review & editing, Writing - original draft, Visualization, Methodology, Formal analysis, Conceptualization. Hui Cai: Software, Resources, Investigation, Data curation. Li Li: Writing - review & editing, Validation, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23204.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Visvanathan A., et al. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 2.Yue H., et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Research. 2015;43(1):373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixit D., et al. The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 2021;11:480–499. doi: 10.1158/2159-8290.CD-20-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L., et al. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer. 2019;18(1):176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J., et al. Downregulation of N6-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N6-methyladenosine levels. Oncotarget. 2018;9(3):3752–3764. doi: 10.18632/oncotarget.23365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paris, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–148.e6. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J., et al. Cell Cycle; 2017. YTH Domain Family 2 Orchestrates Epithelial-Mesenchymal Transition/proliferation Dichotomy in Pancreatic Cancer Cells; pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhe, et al. MicroRNA-145 ModulatesN6-methyladenosine levels by targeting the 3′-untranslated mRNA region of theN6-methyladenosine binding YTH domain family 2 protein. J. Biol. Chem. 2017;292(9):3614–3623. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y., et al. N6-Methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76(1):96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler R., et al. mA modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 2019;20(2):173–182. doi: 10.1038/s41590-018-0275-z. [DOI] [PubMed] [Google Scholar]
- 12.Lin C., et al. The N-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol. Cancer. 2022;21(1):80. doi: 10.1186/s12943-022-01560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang D., et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46(8):3906–3920. doi: 10.1093/nar/gky130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uddin M.B., Wang Z., Yang C. The m6A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol. Cancer. 2021;20(1):61. doi: 10.1186/s12943-021-01356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., et al. mA mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20(9):1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamoto A., et al. A designed cell-penetrating human SOCS2 protein suppresses GH-dependent cancer cell proliferation. Bioence Biotechnology and Biochemistry. 2018;83:1–9. doi: 10.1080/09168451.2018.1536516. [DOI] [PubMed] [Google Scholar]
- 17.Shi L., et al. MiR-492 exerts tumor-promoting function in prostate cancer through repressing SOCS2 expression. Eur. Rev. Med. Pharmacol. Sci. 2019;23(3):992–1001. doi: 10.26355/eurrev_201902_16986. [DOI] [PubMed] [Google Scholar]
- 18.Kulbacka J., et al. Cell membrane transport mechanisms: ion channels and electrical properties of cell membranes. Adv. Anat. Embryol. Cell Biol. 2017;227:39–58. doi: 10.1007/978-3-319-56895-9_3. [DOI] [PubMed] [Google Scholar]
- 19.Jentsch T. VRACs and other ion channels and transporters in the regulation of cell volume and beyond. Nat. Rev. Mol. Cell Biol. 2016;17(5):293–307. doi: 10.1038/nrm.2016.29. [DOI] [PubMed] [Google Scholar]
- 20.Lang F., Stournaras C. Ion channels in cancer: future perspectives and clinical potential. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 2014;369(1638) doi: 10.1098/rstb.2013.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnon C., et al. Autonomic nerve development contributes to prostate cancer progression. Science (New York, N.Y.) 2013;341(6142) doi: 10.1126/science.1236361. [DOI] [PubMed] [Google Scholar]
- 22.Rahrmann E., et al. The NALCN channel regulates metastasis and nonmalignant cell dissemination. Nat. Genet. 2022;54(12):1827–1838. doi: 10.1038/s41588-022-01182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D., Monje M. Cancer hallmarks intersect with neuroscience in the tumor microenvironment. Cancer Cell. 2023;41(3):573–580. doi: 10.1016/j.ccell.2023.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J., Du B. Novel positioning from obesity to cancer: FTO, an m6A RNA demethylase, regulates tumour progression. J. Cancer Res. Clin. Oncol. 2019;145(1):19–29. doi: 10.1007/s00432-018-2796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng Y., et al. Epitranscriptomic mA regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018;97(2):313–325.e6. doi: 10.1016/j.neuron.2017.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B., et al. mA regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer. 2020;19(1):53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., et al. Molecular characterization and clinical relevance of mA regulators across 33 cancer types. Mol. Cancer. 2019;18(1):137. doi: 10.1186/s12943-019-1066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geng Y., et al. Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival. Ann. Transl. Med. 2020;8(6):387. doi: 10.21037/atm.2020.03.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collart M. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley interdisciplinary reviews. RNA. 2016;7(4):438–454. doi: 10.1002/wrna.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi A., et al. The CCR4-NOT complex maintains liver homeostasis through mRNA deadenylation. Life Sci. Alliance. 2020;3(5) doi: 10.26508/lsa.201900494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R., et al. N-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019;10(1):4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L., et al. The role of N-methyladenosine (mA) modification in the regulation of circRNAs. Mol. Cancer. 2020;19(1):105. doi: 10.1186/s12943-020-01224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., et al. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat. Rev. Cancer. 2021;21(9):541–557. doi: 10.1038/s41568-021-00383-9. [DOI] [PubMed] [Google Scholar]
- 34.Ringelhan, et al. The immunology of hepatocellular carcinoma. Nat. Immunol. 2018;19:222–232. doi: 10.1038/s41590-018-0044-z. [DOI] [PubMed] [Google Scholar]
- 35.Hui L., Chen Y. Tumor microenvironment: sanctuary of the devil. Cancer Lett. 2015;368(1):7–13. doi: 10.1016/j.canlet.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 36.Petitprez F., et al. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front. Immunol. 2020;11:784. doi: 10.3389/fimmu.2020.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y., et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184(2):404–421.e16. doi: 10.1016/j.cell.2020.11.041. [DOI] [PubMed] [Google Scholar]
- 38.Zhao P., et al. Dendritic cell immunotherapy combined with cytokine-induced killer cells promotes skewing toward Th2 cytokine profile in patients with metastatic non-small cell lung cancer. Int. Immunopharm. 2015;25(2):450–456. doi: 10.1016/j.intimp.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 39.Dey P., et al. Oncogenic KRAS-driven metabolic reprogramming in pancreatic cancer cells utilizes cytokines from the tumor microenvironment. Cancer Discov. 2020;10(4):608–625. doi: 10.1158/2159-8290.CD-19-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rezende R., et al. γδ T cells control humoral immune response by inducing T follicular helper cell differentiation. Nat. Commun. 2018;9(1):3151. doi: 10.1038/s41467-018-05487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu C., et al. Prognostic biomarker DDOST and its correlation with immune infiltrates in hepatocellular carcinoma. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.819520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanier L.L. NK cell recognition. Annu. Rev. Immunol. 2005;23(1):225. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 43.Gajewski T.F., Schreiber H., Fu Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saulep-Easton D., et al. Cytokine-driven loss of plasmacytoid dendritic cell function in chronic lymphocytic leukemia. Leukemia. 2014;28(10):2005–2015. doi: 10.1038/leu.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panyi G., Beeton C., Felipe A. Ion channels and anti-cancer immunity. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 2014;369(1638) doi: 10.1098/rstb.2013.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamza A., et al. Ion channel mediated mechanotransduction in immune cells. Curr. Opin. Solid State Mater. Sci. 2021;25(6) doi: 10.1016/j.cossms.2021.100951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prevarskaya N., Skryma R., Shuba Y. Ion channels and the hallmarks of cancer. Trends Mol. Med. 2010;16(3):107–121. doi: 10.1016/j.molmed.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Bose T., Cieślar-Pobuda A., Wiechec E. Role of ion channels in regulating Ca2⁺ homeostasis during the interplay between immune and cancer cells. Cell Death Dis. 2015;6(2):e1648. doi: 10.1038/cddis.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler F., et al. Cancer neuroscience: state of the field, emerging directions. Cell. 2023;186(8):1689–1707. doi: 10.1016/j.cell.2023.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo X., et al. Midkine activation of CD8 T cells establishes a neuron-immune-cancer axis responsible for low-grade glioma growth. Nat. Commun. 2020;11(1):2177. doi: 10.1038/s41467-020-15770-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karreth F., Pandolfi P. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol. Cancer. 2019;18(1):28. doi: 10.1186/s12943-019-0957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J., et al. ZSCAN16-AS1 expedites hepatocellular carcinoma progression via modulating the miR-181c-5p/SPAG9 axis to activate the JNK pathway. Cell Cycle. 2021;20(12):1134–1146. doi: 10.1080/15384101.2021.1919828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei H., et al. Hepatoma cell-derived extracellular vesicles promote liver cancer metastasis by inducing the differentiation of bone marrow stem cells through microRNA-181d-5p and the FAK/src pathway. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.607001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Ou Y., et al. Targeting antisense lncRNA PRKAG2-AS1, as a therapeutic target, suppresses malignant behaviors of hepatocellular carcinoma cells. Front. Med. 2021;8 doi: 10.3389/fmed.2021.649279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin Z., et al. LncRNA LINC00667 aggravates the progression of hepatocellular carcinoma by regulating androgen receptor expression as a miRNA-130a-3p sponge. Cell death discovery. 2021;7(1):387. doi: 10.1038/s41420-021-00787-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories including TCGA and GTEx (https://xenabrowser.net/datapages/), GEO (https://www.ncbi.nlm.nih.gov/gds), HPA (https://www.proteinatlas.org/), TISIDB (http://cis.hku.hk/TISIDB/), Metascape (http://metascape.org/), STRING (https://cn.string-db.org/), MSigDB (http://www.gsea-msigdb.org/gsea/msigdb/index.jsp), starBase (http://starbase.sysu.edu.cn/) and miRNet (http://www.mirnet.ca/miRNet/home.xhtml/). The accession numbers can be found in the article. Further inquiries can be directed to the corresponding author.
Ethics declarations
Review and/or approval by an ethics committee was not needed for this study because the study used anonymized, published data.