Abstract
Three Schiff bases were synthesised by the condensation reaction between 2-napthaldehyde and aromatic amines to afford (E)-N-mesityl-1-(naphthalen-2-yl)methanimine (L1), (E)-N-(2,6-dimethylphenyl)-1-(naphthalen-2-yl)methanimine (L2) and (E)-N-(2,6-diisopropylphenyl)-1-(naphthalen-2-yl)methanimine (L3). The synthesised compounds were characterised using UV–visible, NMR (13C & 1H), and Fourier transform infrared spectroscopic methods while their purity was ascertained by elemental analysis. Structural analysis revealed that the naphthalene ring is almost coplanar with the imine functional group as evident by C1–C10–C11–N1 torsion angles of 176.4(2)° and 179.4(1)° in L2 and L3, respectively. Of all the various intermolecular contacts, H⋯H interactions contributed mostly towards the Hirshfeld surfaces of both L2 (58.7 %) and L3 (69.7 %). Quantum chemical descriptors of L1 - L3 were determined using Density Functional Theory (DFT) and the results obtained showed that the energy band gap (ΔE) for L1, L2 and L3 are 3.872, 4.023 and 4.004 eV respectively. The antidiabetic potential of the three compounds were studied using α-amylase and α-glucosidase assay. Compound L1 showed very promising antidiabetic activities with IC50 values of 58.85 μg/mL and 57.60 μg/mL while the reference drug (Acarbose) had 405.84 μg/mL and 35.69 μg/mL for α-amylase and α-glucosidase respectively. In-silico studies showed that L1 docking score as well as binding energies are higher than that of acarbose, which are recognized inhibitors of α-amylase together with α-glucosidase. Further insight from the RMSF, RMSD and RoG analysis predicted that, throughout the simulation L1 showcased evident influence on the structural stability of α-amylase. The antioxidant potential of the compounds was carried out using nitric oxide (NO), ferric reducing ability power (FRAP) and 2,2-diphenyl-1-picrylhydrazyl (DPPH) assays. The compounds exhibited good to fairly antioxidant properties with L1 as well as L3 having IC50 values of 70.91 and 91.21 μg/mL respectively for NO scavenging activities assay, which comparatively outshined acarbose (reference drug) with IC50 value of 109.95 μg/mL. Pharmacology and pharmacokinetics approximations of L1 – L3 showed minimal violation of Lipinski's Ro5 and this projects them to be less toxic and orally bioavailable as potential templates for the design of therapeutics with antioxidant and antidiabetic activities.
Keywords: Schiff bases, Crystal structure, Hirshfeld analysis, Antidiabetes, Antioxidant
1. Introduction
Diabetes mellitus (D.M.) is an endocrinological disorder [1] caused by high blood glucose in animals due to inappropriate secretion of insulin by pancreas in the body. In the 14th century, diabetes mellitus was referred to as black death [2]. It is probably the oldest disease known to man and till today still remains a global health concern [3]. By 2030, researchers are projecting that, about 438 million people might have diabetes in 70 % developing countries if not checked [2]. One of the strategies adopted to combat diabetes mellitus, is to inhibit pancreatic α-amylase and intestinal α-glucosidase (hydrolytic enzymes) which are liable for breaking down carbohydrate into glucose in the body [4]. This will reduce glucose in the blood due to the decrease in the uses of dietary carbohydrates as well as suppresses postprandial glycemia [5]. Current generation of antidiabetic drugs i.e., Acarbose effectively decrease the intestinal absorption of glucose in human body [6]. However, complications such as meteorism, farting, swelling of the abdomen, and perhaps diarrhoea pose as great limitations to their utilisation [7]. Hence, the need to search for new antidiabetic drugs with little or no side effect with excellent activity (see Scheme 1).
Scheme 1.
Preparation of Schiff bases, L1 – L3.
Also, searching for novel antioxidants with excellent properties are of great importance due to their usefulness in slowing down aging, shielding biomolecules from oxidation and fortifying immune system against adverse effect of oxidation stress [[8], [9], [10]]. Reactive oxygen species (R.O.S.) i.e., superoxide radical (O2•-) hydroxyl radical (OH•), hydrogen peroxide (H2O2), are the main cause of oxidative stress [11]. Though at low concentration, R.O.S. has beneficial roles such as defending the body against infectious agents, aid number of cellular signalling systems and induction of mitogenic response [12]. However, at high concentration, R.O.S. damages structures of the cells such as nucleic acids, proteins, membranes, lipids, among others, resulting in cardiovascular, neurodegenerative and cancerous inflammation diseases [13].
Schiff bases together with their metal complexes have been reported for their myriad biological applications [[14], [15], [16]]. They have been utilized as antibacterial [14], antioxidant [17], antituberculosis [18], antidiabetes [19], anticancer [20], among others [21]. The azomethine bond, also known as imine bond (―C N) in Schiff bases are responsible for the crucial role during biological processes which it involves, and this facilitates their wide biological applications [22]. Here in, antidiabetics and antioxidant studies of Schiff bases prepared from 2-naphthaldehyde and aromatic amine is reported. The Schiff bases are designed in such a way to vary the alkyl groups. This would enable us to probe how electronic properties of organic compounds could affect their medicinal properties while we use theoretical approaches (DFT and Molecular docking) to support this assertion. All compounds are characterized by FT-IR, UV–Visible and N.M.R. spectroscopy. We also ran elemental analysis to confirm the purity of these compounds.
2. Experimental section
2.1. Materials
Solvents explored for this study were A.C.S. grade (purity ≥99.5 %) and used without further purification. Reagents used are 2,6-xylidine (99 %), 2,6-diisopropylphenylamine (97 %), mesidine (98 %) and 2-naphthaldehyde (98 %). All chemicals were sourced from Sigma-Aldrich.
2.2. Instrumentation
Bruker AvanceIII 400 MHz spectrometer were used to analyze 1H and 13C NMR spectra of L1 – L3 at 25 °C. Deuterated chloroform was used as a solvent to obtain the 1H N.M.R. and 13C N.M.R. data and signals at δ 7.26 and δ 77.00 ppm are referenced to residual CDCl3. Vario elemental E.L. cube CHNS analyser was used for the elemental analysis and I.R. spectra were obtained on a PerkinElmer Universal A.T.R. spectrum 100 FT-IR spectrometer. Shimadzu UV–Vis–NIR spectrophotometer was used to process the electronic absorption spectra.
2.3. Synthesis of the Schiff bases
The compounds were prepared following a previously reported procedure [23,24]. 2-naphthaldehyde was added to a stirring ethanolic solution of the primary amine (2,6-xylidine, 2,6-diisopropylphenylamine, and mesidine) in a round bottom flask. Four droplets of glacial ethanoic acid were introduced into the mixture, and it was further stirred for 6–7 h at room temperature to afford off-yellow precipitates. The crude products were rinsed thoroughly with hexane to remove unreacted anilines, dried and kept in an air-tight desiccator for further work.
2.3.1. Synthesis of (E)-N-mesityl-1-(naphthalen-2-yl)methanimine
The reaction of 2-naphthaldehyde (0.8 g, 5 mmol) and mesidine (0.7 ml, 5 mmol) in 30 mL of ethanol furnished compound L1 as off-yellow powder. Yield: 82 %, z g. M.p: 162–163 °C. 1H N.M.R. (CDCl3, 600 MHz): δ (ppm) 2.19 (s, 6H, CH3-Ar), 2.33 (s, 3H, CH3-Ar), 6.94 (s, 2H, Ar–H), 7.59 (m, 2H, J.H.H. = 6, Ar–H), 7.93 (d, 1H, J.H.H. = 12, Ar–H), 7.97 (d, 2H, J.H.H. = 12, Ar–H), 8.18 (s, 1H, Ar–H), 8.25 (d, 1H, J.H.H. = 6, Ar–H), 8.39 (s, 1H, CH N). 13C N.M.R. (CDCl3, 150 MHz) δ (ppm): 0.17.23, 19.73, 122.60, 125.62, 126.50, 127.62, 127.71, 127.73, 127.84, 129.84, 132.06, 134.06, 147.70, and 161.73. IR υ (cm−1): 3079(w), 1649(s), 1573(s), 1375(s), 1219(s), 726(m), 590(m). Anal. calcd for C20H19N: C, 87.87; H, 7.01; N, 5.12; Found: C, 87.99; H, 7.27; N, 5.37.
2.3.2. Synthesis of (E)-N-(2,6-dimethylphenyl)-1-(naphthalen-2-yl)methanimine
The reaction of 2-naphthaldehyde (0.8 g, 5 mmol) and 2,6-xylidine (0.6 ml, 5 mmol) in 30 mL of ethanol furnished compound L2 as off-yellow powder. Yield: 87 %, z g. M.p: 174–175 °C. 1H N.M.R. (CDCl3, 600 MHz): δ (ppm) 2.22 (s, 6H, CH3-Ar), 7.01 (t, 1H, J.H.H. = 6, Ar–H), 7.12 (d, 2H, J.H.H. = 6, Ar–H), 7.59 (m, 2H, J.H.H. = 6, Ar–H), 7.94 (d, 1H, J.H.H. = 6, Ar–H), 7.98 (q, 2H, J.H.H. = 6, Ar–H), 8.19 (s, 1H, Ar–H), 8.26 (d, 1H, J.H.H. = 6, Ar–H), 8.41 (s, 1H, CH N). 13C N.M.R. (CDCl3, 150 MHz) δ (ppm): 18.46, 123.71, 123.84, 126.81, 127.30, 127.71, 128.10, 128.20, 128.82, 128.88, 131.07, 133.22, 133.90, 135.25, 151.38, 162.78. IR υ (cm−1): 3057(w), 1632(s), 1575(s), 1377(s), 1217(s), 732(m), 599(m). Anal. calcd for C19H17N: C, 87.99; H, 6.61; N, 5.40; Found: C, 88.09; H, 6.97; N, 5.47.
2.3.3. Synthesis of (E)-N-(2,6-diisopropylphenyl)-1-(naphthalen-2-yl)methanimine
The reaction of 2-naphthaldehyde (0.8 g, 5 mmol) and 2,6-diisopropylphenylamine (0.9 ml, 5 mmol) in 30 mL of ethanol furnished compound L3 as off-yellow powder. Yield: 87 %, z g. M.p: 168–170 °C. 1H NMR (CDCl3, 600 MHz): δ (ppm) 1.22 (d, 12H, CH3–CH), 3.06 (m, 2H, JHH = 6, CH3–CH-Ar), 7.16 (t, 1H, JHH = 6, Ar–H), 7.22 (d, 2H, JHH = 6, Ar–H), 7.59 (m, 2H, JHH = 6, Ar–H), 7.94 (d, 1H, JHH = 12, Ar–H), 7.99 (t, 2H, JHH = 12, Ar–H), 8.20 (s, 1H, Ar–H), 8.25 (d, 1H, JHH = 12, Ar–H), 8.38 (s, 1H, CH N). 13C N.M.R. (CDCl3, 150 MHz) δ (ppm): 23.64, 28.14, 123.20, 123.83, 124.28, 126.84, 127.73, 128.11, 128.89, 131.11, 133.26, 133.87, 135.25, 137.80, 149.50, 162.20. IR υ (cm−1): 3053(w), 1643(s), 1587(s), 1381(s), 1203(s), 743(m), 605(m). Anal. calcd for C23H25N: C, 87.57; H, 7.99; N, 4.44; Found: C, 87.78; H, 7.99; N, 4.71.
2.4. Crystal structure determination
Recrystallization of ethanolic solution of L2 and L3 affords a suitable single crystal utilized in this X-ray diffraction study. Bruker Smart APEXII diffractometer with Mo Kα radiation was used for the crystal evaluation and data collection. At different starting angles, reflections were collected, and these were indexed using APEXII program suite [25]. SAINT software was explored for data reduction [26] while SADABS multi-scan technique [27] was used for scaling and absorption corrections. SHELXS program and SHELXL program [28] were used to solve (using direct methods) and refine structures respectively. OLEX2 software [29] was used in designing the graphics of the crystal structures. Non-hydrogen atoms were first refined isotropically and then by anisotropic refinement with the full-matrix least-squares method based on F2 using SHELXL [28]. The refinement parameters and crystallographic data for L2 and L3 are given in Table 1.
Table 1.
Summary of X-ray structure refinement parameters and crystal data collection for L2 and L3.
| Compound | L2 | L3 |
|---|---|---|
| Empirical formula | C19H17N | C23H25N |
| Formula weight | 259.33 | 315.44 |
| Temperature/K | 296.15 | 296.15 |
| Crystal system | Orthorhombic | Monoclinic |
| Space group | P212121 | P21/n |
| a/Å | 7.0306(5) | 13.5344(4) |
| b/Å | 13.743(1) | 11.4699(3) |
| c/Å | 15.381(1) | 13.7414(6) |
| α/° | 90 | 90 |
| β/° | 90 | 119.150(1) |
| γ/° | 90 | 90 |
| Volume/Å3 | 1486.2(2) | 1863.0(1) |
| Z | 4 | 4 |
| ρcalcg/cm3 | 1.159 | 1.125 |
| μ/mm−1 | 0.067 | 0.064 |
| F(000) | 552.0 | 680.0 |
| Crystal size/mm3 | 0.18 × 0.14 × 0.08 | 0.28 × 0.22 × 0.18 |
| 2Θ range for data collection/° | 7.028 to 56.526 | 4.912 to 56.496 |
| Index ranges | −9 ≤ h ≤ 2, −18 ≤ k ≤ 17, −19 ≤ l ≤ 20 | −15 ≤ h ≤ 17, −15 ≤ k ≤ 15, −18 ≤ l ≤ 16 |
| Reflections collected | 8741 | 20119 |
| Independent reflections | 3644 [Rint = 0.0238, Rsigma = 0.0244] | 4582 [Rint = 0.0244, Rsigma = 0.0170] |
| Data/restraints/parameters | 3644/0/183 | 4582/0/221 |
| Goodness-of-fit on F2 | 1.044 | 1.033 |
| Final R indexes [I ≥ 2σ (I)] | R1 = 0.0397, wR2 = 0.1011 | R1 = 0.0457, wR2 = 0.1237 |
| Final R indexes [all data] | R1 = 0.0493, wR2 = 0.1086 | R1 = 0.0597, wR2 = 0.1371 |
| Largest diff. peak/hole/e Å−3 | 0.12/-0.14 | 0.21/-0.14 |
2.5. In vitro antidiabetes studies
2.5.1. α-amylase enzyme inhibitory activity
Using a modified procedure presented by Ibitoye et al. [30], the inhibitory effect of ligands L1 - L3 on α-amylase enzyme activity was evaluated. In a 20,000 μM buffer solution (mono/dibasic sodium phosphate salt, pH 6.8) with 5 U/mL pancreatic α-amylase, increasing doses (63–500 μg/mL) of the ligands or acarbose were incubated for 10 min at 37 °C. The solution was equilibrated further for 20 min at 37 °C after adding 200 μL of 10 mg/mL starch solution. 0.6 mL of DNSA before boiling at 100 °C in a water bath for 10 min. At 540 nm, the optical density of the coloured mixture formed was read in comparison to a control solution devoid of the inhibitors.
2.5.2. α-Glucosidase enzyme inhibitory activity
This activity of ligands L1 - L3 on α-glucosidase enzyme was also elucidated using p-nitrophenol glucopyranoside (pNPG) as the substrate in a slightly modified method from the literature [31]. 0.4 mL of yeast α-glucosidase (1 U/mL) was added to 0.5 mL solution of L1-L3 or acarbose, along with 500 μL of one hundred millimolar buffer solution (mono/dibasic sodium phosphate salt, pH 6.8), and the mixture was incubated at 37 °C for 15 min. The amount of yellow p-nitrophenol emitted from the substrate was then measured at 405 nm after adding 0.2 ml of pNPG (0.005 M).
2.6. In vitro antioxidant activity experiment
2.6.1. Nitric oxide (NO) scavenging capacity
This in vitro assay of the L1 - L3 compounds was done via NO scavenging estimation. The Kurian et al. method [32] was employed in this assay. A 0.50 mL aliquot of various concentrations of L1 - L3 or Trolox was equilibrated at 25 °C for 2 h with 10 mM sodium nitroprusside dispersed in pH 7.4 PBS. Griess reagent (0.20 mL) was added to the solution, before further equilibration at 25 °C for 5 min. Then, using a blank solution devoid of the test samples, the NO scavenging abilities of L1 - L3 and the standard were read at 546 nm.
2.6.2. 2,2′-diphenyl-1-picrylhydrazyl (DPPH) mopping capacity
This assay was done by determining the capacity of the compounds to release electrons to purple-coloured DPPH free radical using Turkoglu et al. [33] previous protocol with minor modification. 400 μL aliquot of 63–500 μg/mL concentrations of the Schiff bases or Trolox and DPPH solution (300 μM) in methanol were maintained away from light for 30 min at ambient laboratory temperature. Then, absorbance was read at 516 nm against a control solution devoid of the samples or Trolox.
2.6.3. Ferric (Fe3+) reducing power capacity
This property of the L1 - L3 Schiff bases was evaluated using potassium ferricyanide K3Fe(C.N.)6 in accordance with Tan and Chan's method [34]. 300 μL of two hundred millimolar phosphate buffer was incubated with 500 μL solution of each L1 - L3 or Trolox (63–500 μg/mL) and 500 μL of 1 % K3Fe(CN)6 solution at 50 °C for 25 min. Afterward, 0.4 mM of 0.1 g/mL TCA solution was added before dilution with 0.3 mL distilled water. The optical density of the resultant mixture was read at 700 nm after adding 0.2 mL FeCl3 (0.1 %). From a standard curve made with gallic acid (y = 0.0243x - 0.0586; R2 = 0.9852), the compounds' capacity to reduce ferric ion was estimated.
2.7. Computational method
2.7.1. DFT calculations
DFT Becke 3 Lee Yang Par (B3LYP) [35] and the 6–31 +G(d,p) basis set for all atoms were employed to optimize the synthesised compounds L1 - L3 to produce the three-dimensional layout using Gaussian 16 module available on the Lengua cluster of the centre for high performance computing (CHPC), Cape Town, South Africa. The frontier molecular orbital (FMO) hypothesis was used to analyze the compounds' chemical reactivity [36]. The part of the molecule with the lowest unoccupied and highest occupied molecular orbitals (LUMO and HOMO) is revealed by FMO studies.
2.7.2. Molecular docking procedure
This docking analysis was done to determine the compounds' affinity for certain carbohydrate-digesting enzymes. In this procedure, α-glucosidase [37] and α-amylase [38] 3D crystallographic structures with the identification codes 1B2Y and 3CTT, respectively, were downloaded from the Protein Data Bank which is an online database (accessed on May 14, 2023). The Chimera software (V.1.16) Dock prep tool's algorithm was employed to delete water molecules and non-protein chemical components that co-crystallized with the enzymes. Then, according to Wang et al. [39], an automated software added hydrogen atoms and gasteiger charges. Prior to 3D conversion and structural optimization with Avogadro software [40], ChemBioDraw Ultra 14.0 was used to create the 2D structures of L1, L2, and L3 (Hanwell et al., 2012). The Schiff bases 3D structures were then independently loaded into UCSF Chimera application (V1.16) and prepared following the same steps as with the digestive enzymes. Each protein's catalytic pocket occupied by their co-crystallized inhibitors (acarbose for α-amylase & casuarine for α-glucosidase), was covered by the search volume while docking. Subsequently, BIOVIA Discovery Studio [41] was utilized to study the 2D interactions of the most stable ligand-protein complexes while their predicted binding energies were also recorded.
2.7.3. Molecular dynamic simulation
The AMBER force field's FF18SB variant [42] was utilized to describe the systems in the GPU style of the AMBER software (AMBER 18), which was used for molecular dynamic simulations. Using the General Amber Force Field (GAFF) and Restrained Electrostatic Potential (RESP) methods, ANTECHAMBER produced atomic partial charges for the ligands. To neutralize all systems, the Leap module of AMBER 18 permitted the inclusion of hydrogen atoms, as well as Na+ and Cl− counterions to α-Amylase. After that, the systems were implicitly suspended within an orthorhombic box made of TIP3P water molecules, with each atom being within 8 Å of any box edge [43]. After heating, an equilibration estimating 200 ps (minimization of 500,000 steps) of each system was conducted; the operating temperature was kept constant at 300 K. With an imposed restraint potential of 500 kcal/mol for both solutes, a preliminary minimization of 2500 steps was completed. The steepest descent method was used for 1000 steps, while conjugate slopes were used for 1500 steps. A further full minimization of 1000 steps was completed using the conjugate gradient algorithm without constraints. A molecular dynamic simulation with gradual heating from 0 K to 300 K was run for 5 ps (minimization of 2500 steps), keeping the systems' volume and atom count constant. The system solutes were imposed with a collision frequency of 1 ps and a potential harmonic constraint of 10 kcal/mol. The pressure and other elements were maintained constant, simulating an isobaric-isothermal ensemble. The Berendsen barostat [44] was used to maintain the system pressure at 1 bar. The SHAKE algorithm was used [45] to constrict hydrogen atom bonds over the course of the molecular dynamic simulation, which took more than 100 ns in total. Each simulation employed an SPFP precision model with a 2 fs step size [46]. The isobaric-isothermal ensemble (N.P.T.), randomized seeding, constant pressure of 1 bar maintained by the Berendsen barostat, pressure-coupling constant of 2 ps, temperature of 300 K, and Langevin thermostat with a collision frequency of 1 ps were all present in the simulations [47].
2.7.4. Binding free energy calculations of the complexes
The molecular Mechanics/Poisson-Boltzmann surface area approach (MM/PBSA) was used to compute the free binding energy to determine the compounds' binding affinity [48]. The binding free energy was averaged over 100,000 snapshots extracted from the 100 ns trajectory. The following equation can be used to express the free binding energy (ΔG) calculated using this method for each molecular species (complex, ligand, and receptor):
| ΔGbind = Gcomplex − Greceptor − Gligand | [1] |
| ΔGbind = Egas + Gsol − TS | [2] |
| Egas = Eint + Evdw + Eele | [3] |
| Gsol = GGB + GSA | [4] |
| GSA = γ SASA | [5] |
The Egas stands for gas-phase energy, which is made up of the internal energy Eint, Coulomb energy Eele and the van der Waals energies Evdw. The FF14SB force field terms were used to estimate the Egas directly. The energy contribution from the polar, G.G.B. and non-polar, G states was used to compute the solvent-free energy, Gsol. Using a water probe radius of 1.4 Å, the non-polar solvation (S.A.) energy, G.S.A., was calculated from the solvent-accessible surface area (SASA). While the contribution of the polar solvation, G.G.B., was estimated by solving the G.B. equation, S and T represent the total entropy of the solute and temperature, respectively.
2.7.5. Post molecular dynamic analysis
The analysis of Radius of gyration (RoG), Root mean square fluctuation (RMSF) and the Root mean square deviation (RMSD) were done using CPPTRAJ module found in the AMBER 18 suit [49]. The Origin data analysis program was used to generate all average raw data visualizations [50].
3. Results and discussion
3.1. Synthesis
Compounds L1 - L3 were synthesised by introducing 2-naphthaldehyde into a vigorously stirred ethanolic solution containing aromatic amines (namely, 2,6-xylidine, 2,6-diisopropylphenylamine, and mesidine). Following this addition, 2 to 3 drops of glacial acetic acid were incorporated into the resulting solution, and the reaction was allowed to proceed for a duration of 6 h. A pale-yellow solid precipitate formed as a result, which was subsequently filtered and washed with dry hexane three times to eliminate any unreacted amines. The resulting products were left to air dry at room temperature overnight and were then securely stored in a dry vial placed inside a desiccator.
3.2. Spectroscopic studies
3.2.1. 1H and 13C NMR
The 1H and 13C NMR spectra for L1 to L3 were recorded in CDCl3, and their peaks were assigned using 2D NMR techniques. In all compounds, the aliphatic proton signals fell within the range of 1.22–3.06 ppm. Specifically, the methyl proton signals for L1 were detected at 2.19 ppm and 2.33 ppm, whereas those for L2 and L3 were observed at 2.22 ppm and 1.22 ppm, respectively. Within the 1H NMR spectra of L3, quintet peaks between 3.03 and 3.08 ppm were attributed to the methine protons (CH–CH3) within the isopropyl group of L3. Additionally, signals between 6.94 and 8.26 ppm, primarily appearing as singlets, doublets, triplets, and quintets, were assigned to the aromatic protons found in the benzene and naphthalene rings. Distinct peaks at 8.39 ppm for L1, 8.41 ppm for L2, and 8.38 ppm for L3 were also identified in the 1H NMR spectra, and these were attributed to the imine protons (—CH N) within the compounds. In the 13C NMR spectra, the imine carbon (—CH N) resonated at 162 ppm, while the carbon atoms connecting the imine group to the benzene ring (—CH N–C) appeared in the downfield region, specifically between 149 and 152 ppm. Other carbon signals, such as those from aromatic carbons, were observed within the range of 127–136 ppm, while aliphatic carbons exhibited signals around 18–29 ppm. See Fig. S1–S9 in supplementary information for the N.M.R. spectra.
3.2.2. FT-IR and UV–visible spectra of L1 – L3
The Fourier transform infrared spectra of L1 – L3 showcase the major diagnostic peaks. The vibrational frequency band corresponding to the imine (—CH N) group is characterized by a sharp peak at approximately 1649–1632 cm−1, consistent with previously reported findings in the literature [23,51]. Additionally, the Sp3 C–H and Sp2 C–H bands are observed around 2914–2954 cm−1 and 3079–3053 cm−1, respectively. The bending vibrational band associated with ortho-substituted benzene in L1 – L3 is evident around 724–726 cm−1, while peaks around 953–977 cm−1 can be assigned to the trans –CH CH— absorption band, as reported by Kemp [52]. The UV–Visible spectra of L1 – L3 in dichloromethane solvent are provided in the supplementary material (Fig. S13). An intense absorption band appeared in the wavelength range of 235–240 nm, which can be attributed to the π→π* transition induced by the excitation of electrons within the aromatic ring. The second, less intense band, observed around 280–290 nm, is associated with the π→π* transition of the imine functional group, which results from the overlap of two shoulder bands [51].
3.3. Crystal structure analysis for L2 and L3
The crystal structures of L2 and L3 each contain one molecule in their respective asymmetric units as shown in Fig. 1. To best describe and compare the molecular conformation of L2 and L3, C1–C10–C11–N1 and C11–N1–C12–C13 torsion angles were selected. The naphthalene ring is almost coplanar with the imine functional group as evident by C1–C10–C11–N1 torsion angles of 176.4(2)° and 179.4(1)° in L2 and L3, respectively. The geometric orientation of the 2,6-disubstituted phenyl ring appears to be nearly orthogonal to the imine functional group with C11–N1–C12–C13 torsion angles of 73.7(2)° and 78.5(1)° in L2 and L3, respectively. The broadening of the C1–C10–C11–N1 and C11–N1–C12–C13 torsion angles could be ascribed to the availability of sterically demanding iso-propyl substituents in L3 relative to methyl substituents in L2. To the best of our knowledge, we are the first to report the crystal structures of N-(2-naphthylmethylene)-aniline Schiff bases since no hits of closely related compounds were obtained in the Cambridge Structural Database (C.S.D.; version 5.43, November 2022 update) [53]. However, the bond distances and angles in L2 and L3 were found to be comparable with those of N-(1-naphthylmethylene)-aniline Schiff bases (C.S.D. ref. codes: DONVOU [54] and EXIPIL [55].
Fig. 1.
Structures of L2 and L3 with thermal ellipsoids drawn at 50 % probability.
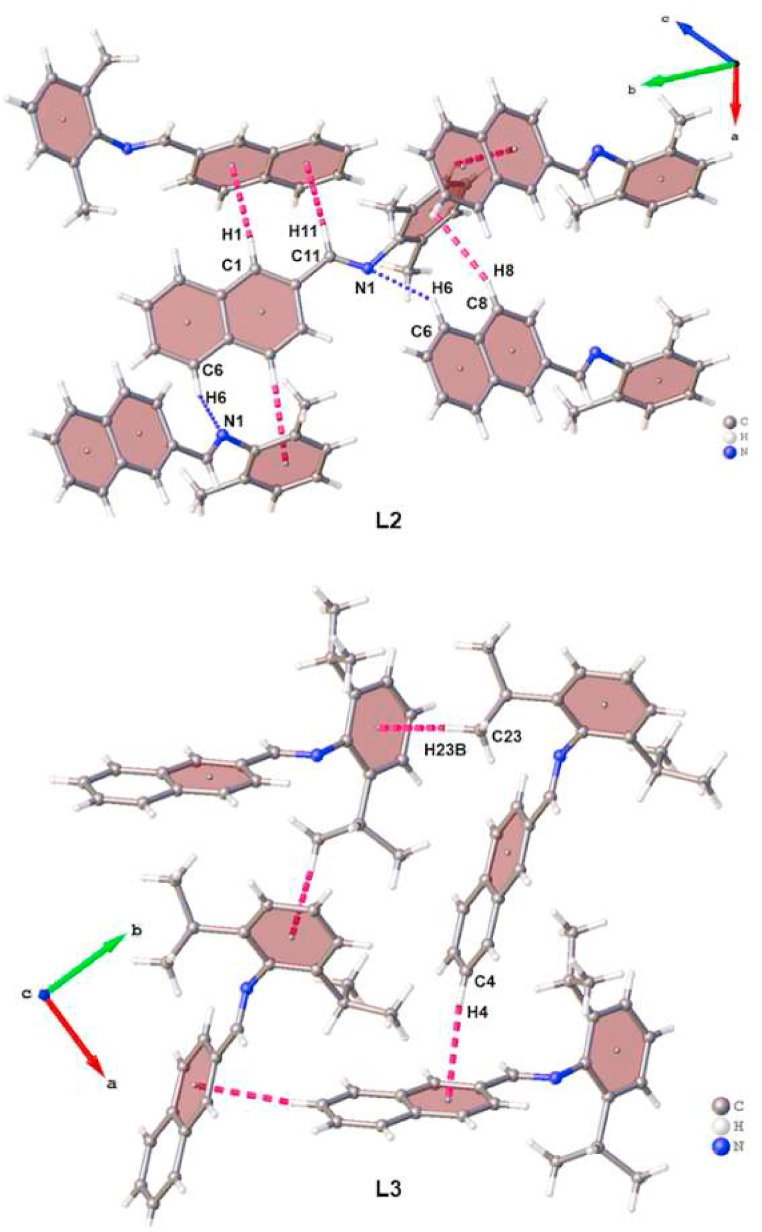
3.3.1. Analysis of intermolecular interactions in L2 and L3
Intermolecular interactions in the crystal packing of L2 and L3 are depicted in Fig. 2 while their geometric parameters are listed in Table 1. In L2, numerous intermolecular C–H … π interactions were observed in the crystal packing. For instance, the H1 and H8 hydrogen atoms of the naphthalene ring exhibits intermolecular C–H … π interactions with the naphthalene (πnaphthalene) and phenyl (πphenyl) rings of two separate neighbouring molecules. The H11 hydrogen atom of the imine carbon forms C11–H11 … πnaphthalene interactions with a naphthalene ring of the same neighbouring molecule as did the H1 hydrogen atom. Collectively, the C–H … πphenyl and C–H … πnaphthalene link together neighbouring molecules of L2 to form chains that extend along the crystallographic a axis. These chain-like structures are linked by intermolecular C6–H6⋯N1 hydrogen bonds to form a two-dimensional supramolecular structure which propagates along the crystallographic ab plane. In L3, the iso-propyl group's H23B atom and the naphthalene ring's H4 atom participate in intermolecular C23–H23B … πphenyl and C4–H4 … πnaphthalene interactions, respectively. These interactions form a 2D supramolecular structure which extends along the crystallographic ab plane.
Fig. 2.
Graphical representation of intermolecular C–H … π interactions (magenta-coloured dashed bonds) and C–H⋯N hydrogen bonds (blue-coloured dotted bonds) in the crystal packing of L2 and L3.
3.3.2. Hirshfeld surface analysis of L2 and L3
The Hirshfeld surface of a crystal is known to be the locations in which the electron density contribution from the molecule within the surface is equal to the contributions from all other molecules. The investigation of crystal structures and the arrangement of molecules offers valuable understanding into this particular phenomenon. The significance of Hirshfeld surface in the field of crystal engineering is paramount [56] as it facilitates the examination of data related to both weak and strong interactions within the crystal system. Fig. 3 depicts the Hirshfeld surfaces of L2 and L3 that have been mapped using the dnorm function including the 2D fingerprint plots of the various intermolecular contacts, were generated using CrystalExplorer21 [57]. The Hirshfeld surface is visually encoded with the colours red, white, and blue, which correspond to the mapped dnorm values [58]. The presence of negative values in the dnorm distribution, as observed in the red regions, signifies that intermolecular distances are comparatively diminutive. The blue regions exhibit positive dnorm values, which signify an increased number of contacts. Conversely, the white regions display dnorm values of zero, indicating contact distances in relation to the van der Waals separation [59]. This section exclusively focuses on the discussion of short intermolecular contacts, as they are believed to play a crucial role in stabilising the crystal lattice. Of all the various intermolecular contacts, H⋯H contacts were found to contribute the most towards the Hirshfeld surfaces of both L2 (58.7 %) and L3 (69.7 %). The large contribution of H⋯H contacts in L3 is attributed due to its higher hydrogen content (7.99 %) than that of L2 (6.61 %). Reciprocal H⋯C contacts were attributed to intermolecular C–H … π interactions and they contributed 38.8 % and 29.0 % towards the Hirshfeld surface of L2 and L3, respectively. Lastly, intermolecular C–H⋯N hydrogen bonds were reciprocal N⋯H contacts and they contribute 1.8 % and 0.7 % towards the Hirshfeld surface of L2 and L3, respectively.
Fig. 3.
dnorm property generated over the Hirshfeld surface and fingerprint plots of the intermolecular contacts with their contribution towards the Hirshfeld surface of L2 and L3.
3.4. Quantum chemical descriptors
The calculated values for relevant quantum descriptors for L1 – L3 are given in Table 2 while their isodensity plots for crucial molecular orbitals are presented in Fig. 4. These quantum descriptor values offer valuable insights into the stability, reactivity, as well as selectivity of organic molecules. Specifically, EHOMO values signify a compound's capacity to give out its most loosely bound electron to a suitable orbital of an acceptor molecule. The higher the EHOMO, the better the electron-donating power of compounds and on the contrary the higher the LHOMO the better the electron-accepting ability [60,61]. Among the studied compounds, it is likely that L1, which possesses an EHOMO value of −5.769 eV, exhibits the highest electron-donating capability. The calculation of energy band gaps (ΔE) through the subtraction of EHOMO from LHOMO serves as a metric for assessing chemical reactivity and stability. It has been reported that a lower ΔE indicates a higher level of both stability and reactivity. Specifically, the calculated ΔE values for L1, L2, and L3 are 3.872 eV, 4.023 eV, and 4.004 eV, respectively. Consequently, it can be inferred that L1, with its lower ΔE value, may possess greater reactivity and stability when compared to L2 and L3.
Table 2.
Quantum chemical descriptors of compounds L1 - L3.
| Parameters | L1 | L2 | L3 |
|---|---|---|---|
| EHOMO (eV) | −5.769 | −5.953 | −5.941 |
| ELUMO (eV) | −1.897 | −1.930 | −1.937 |
| ΔE (eV) | 3.872 | 4.023 | 4.004 |
| η (eV) | 1.936 | 2.012 | 2.002 |
| S (eV−1) | 0.258 | 0.249 | 0.250 |
| IP (eV) | 5.769 | 5.953 | 5.941 |
| A (eV) | 1.897 | 1.930 | 1.937 |
| χ (eV) | −3.833 | −3.941 | −3.939 |
| ω (eV) | 3.794 | 3.861 | 3.875 |
Fig. 4.
HOMO and LUMO plots for studied Schiff bases L1 – L3 at the SMD/B3LYP/6-31+G(d) level of theory.
The global softness (S) characterizes how easily the electron cloud of molecules can undergo deformation or polarization in response to small chemical perturbations. Among the compounds considered, L1 exhibits the highest global softness at 0.258 eV-1. This implies that the electron cloud of L1 readily deforms or polarizes, rendering it more chemically active compared to L2 and L3. Conversely, global hardness (η) is the inverse of global softness, indicating that higher global hardness values correspond to lower chemical reactivity. Examining Tables 2 and it becomes evident that L2 possesses the highest global hardness value, suggesting it is likely the least reactive among the synthesised compounds. The electrophilic (ω) properties for L1, L2, and L3 are determined to be 3.794, 3.861, and 3.875 eV, respectively. These values reveal that L3 exhibits the highest electrophilic ability, while L1 demonstrates the highest nucleophilic ability. Furthermore, the calculated ionization potential (I.P.) values reflect the nucleophilic properties of the synthesised compounds, while the electron affinity (E.A.) values indicate their ability to attract electrons. Notably, L3 possess the highest electron affinity, while L2 has the highest ionization potential.
3.5. Schiff bases L1 – L3 effects on carbohydrate digestive enzyme
3.5.1. α-Amylase inhibition
As displayed in Fig. 5, it is evident that the % α-amylase inhibitory activities increase with increasing concentration. While the percentage inhibition of both sample L1 and sample L2, as well as Acarbose, was indistinguishable at 63 μg/mL, Table 3 reveals that sample L1 exhibited the highest suppressive effect on the carbohydrate digestive enzyme, as indicated by its IC50 value of 58.85 μg/mL, which is significantly lower than that of Acarbose (405.84 μg/mL) and the other compounds (L2 > 500 μg/mL; L3 > 500 μg/mL).
Fig. 5.
Effect of L1 – L3 synthesised Schiff bases on α-amylase enzyme inhibition. The mean activity ± standard deviation of each compound represented by bars at each concentration with different alphabets (a–c) are significantly different at p < 0.05.
Table 3.
Antidiabetic potential of L1 – L3 at different concentration using α-glucosidase and α-amylase assays.
| Compounds | α-Amylase (μg/mL) | α-Glucosidase (μg/mL) |
|---|---|---|
| L1 | 58.85 | 57. 60 |
| L2 | >500 | 147.42 |
| L3 | >500 | 206.02 |
| Acarbose (Standard) | 405.84 | 35.67 |
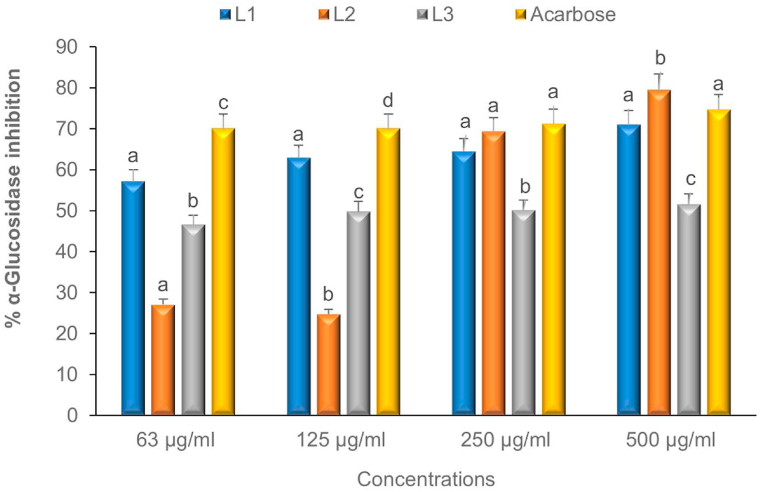
3.5.2. α-Glucosidase inhibition
The results illustrating the inhibitory capacity of the Schiff bases on α-glucosidase are presented in Fig. 6. Among the synthesised samples, L1 exhibited a significantly higher inhibitory effect at concentrations of 63–125 μg/mL. Furthermore, when comparing L1, L2, and acarbose, their percentage inhibition at 250 μg/mL did not exhibit statistically significant differences, however, at a concentration of 500 μg/mL, L2 displayed significantly greater activity. Intrestingly, the IC50 values for these Schiff bases, as shown in Table 3, were higher compared to acarbose (IC50 = 35.67 μg/mL). More importantly, L1 demonstrated the most potent enzyme inhibition amongst the compounds, with an IC50 value of 57.60 μg/mL. This suggests that L1 holds promise as a potential template in the development of antidiabetic therapeutics.
Fig. 6.
Effect of L1 – L3 synthesised Schiff bases on α-glucosidase enzyme inhibition. The mean activity ± standard deviation of each compound represented by bars at each concentration with different alphabets (a–d) are significantly different at p < 0.05.
3.6. Computational techniques
3.6.1. Molecular dynamic simulation
Molecular docking experiments were employed in order to anticipate the binding energies of compounds L1 – L3 and acarbose, which served as a reference drug. These studies were conducted with specific target enzymes, namely alpha-amylase and alpha-glucosidase. The binding energies obtained are summarized in Table 4, while comprehensive insights into the interactions between the receptors and ligands can be found in the supplementary material (Fig. S14–S18). Remarkably, among all the compounds tested, L1 exhibited the highest binding scores, surpassing even acarbose in terms of binding affinity. This finding is in excellent agreement with experimental results, where L1 demonstrated greater promise than the reference drug. To comprehend L1's binding affinity and its potential as an anti-diabetic agent against α-amylase in comparison to acarbose, an extensive analysis involving molecular dynamic simulations (MDS) was carried out. This analysis focused on L1, which exhibited notable docking scores, and acarbose (the reference drug).
Table 4.
Docked energies (kcal mol−1) of compounds L1 — L3 against selected α-amylase and α-glucosidase enzymes.
| α-Amylase |
α-Glucosidase |
|
|---|---|---|
| Binding energy (kcal mol−1) | ||
| Sample L1 | −9.0 | −7.7 |
| Sample L2 | −8.5 | −7.5 |
| Sample L3 | −8.8 | −7.4 |
| Acarbose | −7.3 | −6.1 |
Molecular dynamics (M.D.) simulations are of great importance in this study because they encompass intricate molecular structural motions, enabling a comprehensive analysis of structural shades at specific protein sites. This includes identifying crucial binding regions (hot spots) and eliminating structural anomalies that may arise due to the conditions used in MD characterization, as emphasized in the work done by Hernández-Rodríguez et al. [62]. Moreover, MD simulations provide a robust foundation for improving the accuracy of free energy estimations related to interactions between proteins and ligands, a valuable complement to experimental procedures [62]. After conducting extensive 100 ns molecular dynamics (MD) simulations in our current study, we employed the molecular mechanics Poisson-Boltzmann surface area (MMPBSA) technique. The MMPBSA approach is highly regarded for its robustness and reliability in estimating binding free energies (ΔGbind) in various molecular recognition scenarios, with a notable example being the research conducted by Wang and colleagues in 2018 [63]. MMPBSA was employed in a meticulous examination of the interactions between molecules involving L1 and α-Amylase, as well as the reference standard (acarbose). The outcome of these simulations, as shown in Table 5, showed the binding free energy score of L1 complexes to be −22.627 kcalmol-1, whereas the standard reference, acarbose, exhibited a binding free energy of −22.206 kcalmol-1, determined through the integration of various energy components. This insightful analysis sheds light on the favourable binding affinity of L1 compared to the reference standard acarbose, providing valuable molecular-level insights into their interactions with α-Amylase.
Table 5.
The complex's thermodynamic binding free energy profile.
| Energy Components (kcalmol−1) | |||||
|---|---|---|---|---|---|
| Complex | ΔEvdW | ΔEelec | ΔGgas | ΔGsolv | ΔGbind |
| Acarbose + α-Amylase | −54.154 | 15.846 | −38.307 | 16.101 | −22.206 |
| L1 + α-Amylase | −35.064 | 2.939 | −32.125 | 9.497 | −22.627 |
By examining the binding free energy, we gain a subtle perspective on the interplay of energy components. Notably, the energy contribution of van der Waals surpasses that of electrostatic energy in the standard reference Acarbose and compound L1, elucidating the predominant effect of van der Waals forces in shaping the overall binding energy.
3.6.2. Protein-ligand interaction after molecular dynamic simulation
The exploration of protein-ligand interactions has emerged as a pivotal approach for dissecting the intricate molecular connections that transpire within the protein's active sites when bound to ligands, as underscored by the research of Obakachi et al. [64]. A paramount aspect of this investigation is ascertaining whether the ligand or drug engages with the indispensable amino acids critical for inhibition. The nature and abundance of these interactions often wield significant influence over the resultant binding energy values when a molecule becomes intricately bound to an enzyme within a complex. In our quest to fathom the diverse array of interactions between amino acids and the ligand L1, alongside the reference compound Acarbose (A.C.B.), we crafted a compelling depiction of protein-ligand interactions, vividly illustrated in Fig. 7, Fig. 8.
Fig. 7.
Interactive structures of the α-Amylase in complex with Acarbose in 3D and 2D.
Fig. 8.
Interactive structures of the α-Amylase in complex with L1 in 3D and 2D.
Protein structures rely on a multifaceted network of non-covalent interactions, encompassing hydrophobic forces, electrostatic attractions, hydrogen bonds, and van der Waals interactions, as elucidated by Obakachi et al. in their 2021 research [64]. Notably, the interaction plot shows that the L1 compound and the reference drug A.C.B both contain van der Waals interactions, which significantly contributes to protein stability. While hydrogen bonding is discernible in the case of the reference drug A.C.B., it is notably absent in the L1 compound. Instead, a spectrum of other interactions, including Pi-sigma, Pi-Pi T-shaped, Pi-alkyl, and Alkyl interactions, is observed for compound L1. Drawing from existing literature on proteins, it's well-established that certain amino acids, namely Phe, Trp, and Tyr, often participate in π-interactions, such as π–π, cation-π, and π-alkyl bonds. The identified interactions play pivotal roles in shaping protein structure, governing protein-ligand interactions, and contributing substantively to protein stability, catalytic processes, and self-assembly dynamics [65]. Furthermore, an extensive exploration of π–π interactions' function in the interaction dynamics between aromatic drugs and proteins reveals their profound influence on drug binding to proteins, as illuminated by Shao et al. [66].
3.6.3. Molecular dynamics studies of L1 and A.C.B. With α-AMYLASE protein
An in-depth molecular dynamic simulation (M.D.S.) investigation was conducted, encompassing 100 nano-second simulations. During this extensive simulation period, a meticulous analysis was undertaken, tracking and calculating critical structural parameters, including the Root Mean Square Deviation (RMSD), Radius of Gyration (RoG), and Root Mean Square Fluctuation (RMSF). These parameters were evaluated for both the ligand-bound and free (apo-enzyme) trajectories, yielding insightful data that was further visualised through plots and graphical representations.
A gauge of structural stability and system convergence is the Root Mean Square Deviation (RMSD), with lower values denoting greater stability [64]. In the context of the α-Amylase systems, including the bound L1, reference Acarbose (A.C.B.), and unbound α-Amylase (Apo), both L1 and A.C.B. converge nearly immediately, akin to the unbound system, which converges in less than 2ns (Fig. 9a). The C-alpha RMSD backbone atoms of all configurations exhibit relative stability, albeit with subtle differences. Notably, the Apo system displays a slightly lower RMSD (≈1.4 Å) than A.C.B., with an RMSD of (≈1.6 Å), while the L1 bound system exhibits a higher RMSD (≈1.2 Å). The average RMSD values for α-Amylase (Apo), A.C.B., and L1 are 1.235 Å, 1.285 Å, and 1.161 Å, respectively. These results highlight that the L1 system, characterised by lower RMSD values, enhances enzyme stability to a greater extent than the reference drug, which exhibits a comparatively higher RMSD than the unbound Apo system. Notably, the plots reveal that the binding of L1 and A.C.B. does not compromise protein stability or integrity of the structure. In addition to determining the RMSD, we examined the Radius of Gyration (RoG) plots for the α-Amylase protein system (Fig. 9b). RoG provides insights into structural compactness, with lower values indicating greater stability [67]. The RoG plots echo the RMSD findings, with the L1 compound binding leading to enhanced structural compactness and heightened system stability compared to the apo-enzyme and the standard drug. The average RoG values for A.P.O., A.C.B., and L1 are 23.204 Å, 23.143 Å, and 23.115 Å, respectively, further underscoring the stability conferred by the L1 compound1. Furthermore, we delved into the Root Mean Square Fluctuation (RMSF) plot for the α-Amylase protein system (Fig. 9c). RMSF illuminates the impact of ligand binding on active site residue behaviour, with high RMSF values signifying greater flexibility and movement. Remarkably, the standard A.C.B. and the L1 compound exhibited the maximum levels of fluctuation values, suggesting increased residue flexibility. This observation contrasts with the high RoG and RMSD values observed for the apo-enzyme and the reference compound, emphasising the multifaceted dynamics in the system.
Fig. 9.
showcases (a) RMSD, (b) RoG, and (c) RMSF plots in comparison for α-Amylase systems (A.P.O., A.C.B., and L1) during 100 ns MD simulations.
3.6.4. Snapshot analysis of protein-ligand interactions using MD simulations
Using molecular dynamics simulations and analysing snapshots at regular intervals provide deep insights into the stability and consistency of interaction between protein and ligand [68]. These insights help researchers to understand the mechanisms of drug action better and to design more effective drugs with improved stability and efficacy. Overall, analysing the snapshots at regular intervals can provide a powerful tool for studying protein-ligand interactions and developing new drugs with improved therapeutic properties [69]. The snapshots were taken within the range of 25–100 ns at 25 ns intervals for L1 and analysed to affirm the consistencies between α-amylase-L1 interactions (Fig. 10). The analysis showed that, most of the L1-residue interactions were stable throughout the simulation period, predicting that L1 forms a very stable complex with the protein. This finding indicates that the protein-ligand interaction of the compound L1 is consistent and stable over time, which is an important factor in drug design and development.
Fig. 10.
Snapshot Images captured at 25 ns, 50 ns, 75 ns, and 100 ns intervals, depicting essential site residues and their interactions with L1 within the active pocket of α-amylase.
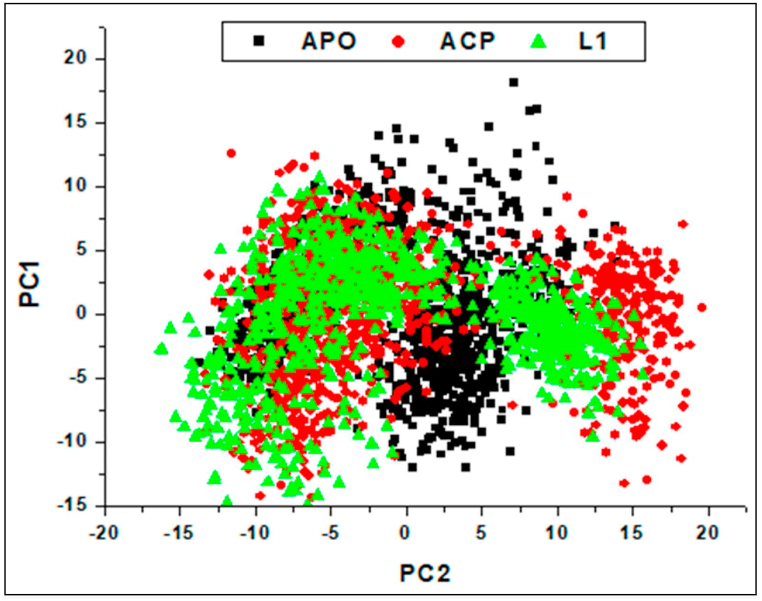
3.6.5. Principle component analysis (P.C.A.)
Several articles discuss the use of Principal Component Analysis (PCA) to elucidate MD simulations of biological macromolecules [70,71]. This analysis (PCA) provides insights into the displacement patterns of Amylase C-α atoms, a revelation unveiled through Molecular Dynamics (MD) simulations conducted using CPPTRAJ in AMBER18 GPU [72]. It was conducted to facilitate the integration of in vitro and computational studies of compound L1 in relation to the reference compound acarbose. The primary objective of employing PCA was to evaluate the structural similarity or dissimilarity between the synthesised compound L1 and the reference compound acarbose. The outcomes of the PCA analysis, as depicted in Fig. 11, revealed a notable structural resemblance between these compounds. This structural similarity observed between L1 and acarbose is instrumental in understanding the remarkably close binding affinities observed in Table 5, which stand at −22.206 kcal/mol for acarbose and −22.627 kcal/mol for L1. This synergy between computational and in vitro findings underscores the potential of L1 as a candidate with comparable binding affinity to acarbose, a significant result with implications for further drug development and therapeutic applications. Furthermore, the PCA analysis of Amylase protein dynamics during interactions with inhibitors, specifically PC1 and PC2 components, unveiled key attributes of magnitude. It clarified the patterns of atomic displacements, showcasing their unidirectional and uniquely oriented nature, which can be attributed to distinct vibrational modes. The PCA scatter plot in Fig. 11 visually represents the unbound Amylase, Amylase-ACB complex, and Amylase-L1 complex.
Fig. 11.
Illustration of the Principal Component Analysis (PCA) of C-α backbone atoms, tracking Apo Amylase, L1, and A.C.B. bound Amylase conformations across the trajectories PC1 and PC2 throughout the 100 ns MD simulation.
The plot depicted unique conformations and movements within the critical subspace defined by the two principal components. While the Apo system exhibited inter-residue displacement along the eigenvectors, the bound systems demonstrated pronounced separation in motion patterns between the A.C.B. and L1 complex systems. The α-amylase-L1 complex displayed a dispersed good motion along the two eigenvectors, similar to the α-amylase-ACB complex. The binding of L1 molecules to the Amylase enzyme could potentially lead to increased structural activity similar to the compact motions exhibited by the A.C.B. molecule. These findings align with the earlier discussed results, collectively supporting the notion that the binding of the proposed molecules stabilised a highly active conformation that promotes α-amylase enzymatic activity inhibition.
The results from the in silico studies (molecular docking, molecular dynamic simulation and PCA) indicated that L1 bound to the same pocket as acarbose and had very close binding affinities. Hence, the need to carry out kinetic studies in future experiments before we could conclude if they both have the same mode of inhibition (competitive or non-competitive inhibition). Knowing these will enhance our understanding of how they could constitute a better alternative to acarbose. Should it be discovered subsequently that this compound could bind to other allosteric sites on the enzyme, they could be modified chemically to possibly display lesser adverse effects compared to acarbose without compromising their excellent antidiabetic activity.
3.7. In vitro antioxidant properties of L1 – L3
3.7.1. Nitric oxide (NO) scavenging activity
Fig. 12 displays the NO scavenging effects of the compounds. At 63–250 μg/mL, sample L1 activities were significantly higher than those of the other compounds investigated and Trolox. Although sample L2 activity at 500 μg/mL was lower compared with the other test compounds and standard, the 70.91 μg/mL and 91.21 μg/mL IC50 values (Table 6) respectively indicates sample L1 and L3 have better NO scavenging antioxidant potency compared with Trolox (IC50 = 109.59 μg/mL).
Fig. 12.
L1 – L3 synthesised Schiff bases NO• radical scavenging activtities compared with Trolox at increasing concentration. The mean activity ± standard deviation of each compound represented by bars at each concentration with different alphabets (a–c) are significantly different at p < 0.05.
Table 6.
Antioxidant potentials of L1 – L3 Schiff bases in vitro.
| Compounds | FRAP (μg/mL) | DPPH (μg/mL) | NO (μg/mL) |
|---|---|---|---|
| L1 | >500 | 25,44 | 70,91 |
| L2 | >500 | 35,69 | 119,68 |
| L3 | >500 | 67,81 | 91,21 |
| Acarbose (Standard) | 292,51 | 13,62 | 109,59 |
3.7.2. DPPH radical mopping activity
The amount to which L1 - L3 could donate and pair with the odd electrons in the DPPH free radical is shown by the data in Fig. 13. L1 - L3 antioxidant capacities at the test concentrations were significantly lower than those of Trolox (IC50 = 13.62 μg/mL) despite the fact that at 500 μg/mL, the compound's DPPH scavenging capabilities were statistically equivalent. Sample L1 has the least IC50 value of 25.44 μg/mL. It has the most superior Trolox equivalent antioxidant capacity (TACE) value, followed by L2 and L3, respectively (Table 6). Using quantum chemical descriptor values, particularly EHOMO and ionization potential, it is possible to explain further why L1 has better antioxidant activity than L2 and L3. These two variables express how well a substance can donate electrons. The ability to donate electrons declines with decreasing EHOMO (and increases with increasing ionization potential). Thus, L1, which had a greater EHOMO and a lower ionization potential than the other compounds, produced the best DPPH radical scavenging ability, making it a better choice for developing therapeutic antioxidants [73].
Fig. 13.
L1 – L3 synthesised Schiff bases DPPH radical scavenging activities compared with Trolox at increasing concentration. The mean activity ± standard deviation of each compound represented by bars at each concentration with different alphabets (a–c) are significantly different at p < 0.05.
3.7.3. Ferric reducing power of L1 – L3
The result in Fig. 14 compares the reducing power of the synthesised Schiff bases with Trolox at different concentrations. At 63–250 μg/mL, the Fe3+ reduction properties of the compounds were significantly lower than those of the standard antioxidant. However, at these concentrations, the F.R.P. of the compounds were not statistically different from each other. Although samples L1 and L2 showed slightly better abilities at 500 μg/mL, the 292.51 μg/mL IC50 of Trolox in Table 6 indicates its superior potency relative to the synthesised compounds.
Fig. 14.
L1 – L3 synthesised Schiff bases Fe3+ reducing power activities compared with Trolox at increasing concentration. The mean activity ± standard deviation of each compound represented by bars at each concentration with different alphabets (a–c) are significantly different at p < 0.05.
3.8. Druglikeness and pharmacokinetics properties of L1 – L3
Compounds L1 – L3 displayed promising antidiabetes and antioxidant properties. Therefore, the need to predict their drug-likeness properties using appropriate web tools. SwissADME and pKCMS are web-based analytical tools used to approximate the drug-like nature, ADME parameters, pharmacokinetic properties and toxicity of potential drug candidates [74,75]. The physicochemical and pharmacokinetic parameters of L1 – L3 are given in Table 7 and related with the acceptable threshold of Lipinski's Ro5. Reports indicate that promising drug molecules should comply with the acceptable threshold of Lipinski's Ro5 by not exceeding it or perhaps have a minimal violation [76]. We predicted physicochemical properties such as molecular weight (M.W.), aqueous solubility (Log S), lipophilicity (Log P), hydrogen bond donor (HBD), and acceptor (H.B.A.) ability, rotatable bonds (RotB), topological polar surface area (TPSA), and skin permeation (LogKp). We also estimated the toxicity and the pharmacokinetic parameters which include P-glycoprotein (P-gp) substrate, blood-brain barrier (BBB) permeant and gastrointestinal (GI) absorption.
Table 7.
Estimated pharmacokinetics and physicochemical parameters of L1 – L3.
| L1 | L2 | L3 | Acceptable threshold (Ro5) | |
|---|---|---|---|---|
| Physicochemical properties | ||||
| Molecular weight (Da) | 259.34 | 273.37 | 315.45 | <500Da |
| LogP | 4.83 | 5.16 | 6.02 | <5 |
| LogS (mol/L) | −5.18 | −5.48 | −6.26 | 0 → −6 |
| TPSA (A2) | 12.36 | 12.36 | 12.36 | ≤140 |
| HBA | 1 | 1 | 1 | ≤10 |
| HBD | 0 | 0 | 0 | ≤5 |
| RotBs | 2 | 2 | 4 | <10 |
| Pharmacokinetics properties | ||||
| G.I. absorption | High | High | Low | |
| B.B.B. Permeant | Yes | No | No | |
| P-gp Substrate | No | No | Yes | |
| LogKp (skin permeation) | −4.19 | −4.01 | −3.45 | |
Molecular weight of the compounds does not violate Lipinski's Ro5, which ascribes 500 g/mol as the M.W. of a promising drug molecule. Drug molecules with M.W. within the threshold of Lipinski's Ro5 find it easier to be transported to biomolecules target [77]. The LogS value of L1 and L2 fell within the range of acceptable standard of 0 → −6 mol/L while L3 deviates minimally. These depict that, L1 and L2 can easily permeate through the intestinal epithelium surface because they are lipophilic in nature. The predicted LogP values for L2 and L3 have minimal violation for Lipinski's Ro5 (<5) while L1 falls under the acceptable range. We could deduce that L1 is more lipophilic than L2 and L3, perhaps the reason why it displayed better antidiabetes potential relative to other compounds and even reference drug. The predicted values for TPSA are within the accepted values of (≤140) for L1 – L3. This implies that the compounds could be transported through a lipid bilayer that is densely packed, i.e., the gastrointestinal tract (G.T.) [78]. Drugs with small TPSA values tend to penetrate through the cells easily when compared with the ones having high values and this validates their bioavailability together with using them as therapeutics in the future. RotBs, HBDs, and HBAs also pinpoint the tendency of the compounds to be promising drug candidates. Rotatable bonds could be defined as typical single bonds, joined to a nonterminal heavy atom and not present in the ring [79]. They reveal the molecular flexibility of drug compounds. In Table 7, the estimated values for RotBs, HBD.s, and H.B.A.s fell within the acceptable threshold of Lipinski's Ro5.
Intestinal absorption is one of the critical stages that is very crucial in the discovery of orally bioavailable drugs [80]. In the intestine, a promising drug candidate should be absorbed easily. All the compounds displayed high GI absorption except L3, and only L1 has the tendency to penetrate through the brain-blood barrier (BBB). P-glycoprotein in the intestine reduces the bioavailability of a wide range of drugs [81]. L1 and L2 are not P-gp substrates, while L3 does, and this indicates their promising bioavailability. The rate at which the compounds penetrate through the skin is in the order of L3 > L2 > L1.
4. Conclusion
Three Schiff bases were synthesised from 2-naphthaldehyde and aromatic amines and subsequently characterized using UV–vis, FT-IR, and NMR spectroscopic techniques. X-ray structural analysis of compounds L2 and L3 revealed that the naphthalene ring in both molecules is almost coplanar with the imine functional group, as shown by their torsion angles. Notably, the crystal packing of L2 and L3 displayed prominent C–H … π interactions. Among various intermolecular contacts, H⋯H interactions were found to contribute significantly to the Hirshfeld surfaces of both compounds. Calculated quantum chemical parameters provided insights into their chemical reactivity and stability, with the order being L1 > L3 > L2. In vitro antidiabetes indicated that L1 exhibited superior activity compared to its counterparts and even outperformed the acarbose. Computational investigations such as molecular docking, MD simulation, post-MD and PCA were conducted to corroborate the favourable in vitro results of L1's efficacy. The binding score and binding energies of L1 outshined the one for acarbose (reference drug). The RMSD, RMSF and RoG results from post-MD analysis predicted that, L1 will actively bind the targets and significantly impose structural stability on α-amylase relative to acarbose. The outcome of the PCA analysis revealed a structural resemblance between L1 and acarbose. The convergence and consistency in all the computational parameters substantiate the reliability of L1's interaction with α-amylase protein, aligning with its potent in vitro activity. Furthermore, L1 – L3 demonstrated moderate to good antioxidant properties, with both L1 and L3 displaying better NO free radical scavenging activities than L2 and the reference drug. Remarkably, all these compounds exhibited minimal violations of Lipinski's Rule of Five (Ro5), suggesting their potential as orally less toxic and bioavailable templates for the design of antidiabetic therapeutic agents.
CRediT authorship contribution statement
Segun D. Oladipo: Writing - review & editing, Writing - original draft, Visualization, Resources, Methodology, Funding acquisition, Formal analysis, Data curation, Conceptualization. Robert C. Luckay: Writing - review & editing, Writing - original draft, Visualization, Validation, Supervision, Resources, Project administration, Funding acquisition, Conceptualization. Kolawole A. Olofinsan: Writing - review & editing, Writing - original draft, Software, Methodology, Formal analysis, Data curation. Vincent A. Obakachi: Writing - review & editing, Writing - original draft, Software, Methodology, Data curation. Sizwe J. Zamisa: Writing - review & editing, Writing - original draft, Software, Methodology, Formal analysis, Data curation. Adesola A. Adeleke: Writing - review & editing, Software, Resources, Methodology, Formal analysis, Data curation. Abosede A. Badeji: Writing - review & editing, Software, Methodology, Formal analysis, Data curation. Segun A. Ogundare: Writing - review & editing, Formal analysis, Data curation. Blassan P. George: Writing - review & editing, Methodology, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors acknowledged the funding from the National Research Foundation (NRF), Stellenbosch University, Matieland, 7602, South Africa. The first author is a recipient of the postdoctoral fellowship award of the NRF at Stellenbosch University, Matieland, South Africa. The authors are also grateful to the South Africa Centre for High-Performance Computing, CHPC [www.chpc.ac.za] for resources.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23174.
Contributor Information
Segun D. Oladipo, Email: segun.oladipo@oouagoiwoye.edu.ng.
Robert C. Luckay, Email: rcluckay@sun.ac.za.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Ford E.S., Li C., Zhao G. Prevalence and correlates of metabolic syndrome based on a harmonious definition among adults in the U.S. J. Diabetes. 2010;2:180–193. doi: 10.1111/j.1753-0407.2010.00078.x. [DOI] [PubMed] [Google Scholar]
- 2.Deepthi B., Sowjanya K., Lidiya B., Bhargavi R.S., Babu P.S. A modern review of diabetes mellitus: an annihilatory metabolic disorder. J. In Silico In Vitro Pharmacol. 2017;3:1. [Google Scholar]
- 3.Ahmed A.M. History of diabetes mellitus. Saudi Med. J. 2002;23:373–378. [PubMed] [Google Scholar]
- 4.Prpa E.J., Bajka B.H., Ellis P.R., Butterworth P.J., Corpe C.P., Hall W.L. A systematic review of in vitro studies evaluating the inhibitory effects of polyphenol-rich fruit extracts on carbohydrate digestive enzymes activity: a focus on culinary fruits consumed in Europe. Crit. Rev. Food Sci. Nutr. 2021;61:3783–3803. doi: 10.1080/10408398.2020.1808585. [DOI] [PubMed] [Google Scholar]
- 5.Omoregie H.O., Yusuf T.L., Oladipo S.D., Olofinsan K.A., Kassim M.B., Yousuf S. Antidiabetes, antimicrobial and antioxidant studies of mixed β-diketone and diimine copper (II) complexes. Polyhedron. 2022;217 [Google Scholar]
- 6.Zhang M., Feng R., Yang M., Qian C., Wang Z., Liu W., Ma J. Effects of metformin, Acarbose, and sitagliptin monotherapy on gut microbiota in Zucker diabetic fatty rats. B.M.J. Open Diabetes Res. Care. 2019;7 doi: 10.1136/bmjdrc-2019-000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishoff H. Pharmacological properties of the novel glucosidase inhibitors BAY m 1099 (miglitol) and BAY o 1248. Diabetes Res. Clin. Pract. 1985;1:S53. [Google Scholar]
- 8.Brewer M.S. Natural antioxidants: sources, compounds, mechanisms of action, and potential applications. Compr. Rev. Food Sci. Food Saf. 2011;4:221–247. [Google Scholar]
- 9.Sies H. Oxidative stress: oxidants and antioxidants. Exp. Physiol. 1997;82:291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- 10.Oladipo S.D., Omondi B., Mocktar C. Co (III) N, N′‐diarylformamidine dithiocarbamate complexes: synthesis, characterisation, crystal structures and biological studies. Appl. Organomet. Chem. 2020;34 [Google Scholar]
- 11.Kostova I., Saso L. Advances in research of Schiff-base metal complexes as potent antioxidants. Curr. Med. Chem. 2013;20:4609–4632. doi: 10.2174/09298673113209990149. [DOI] [PubMed] [Google Scholar]
- 12.Valko M., Rhodes C.J.B., Moncol J., Izakovic M.M., Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Halliwell B. Antioxidants in human health and disease. Annu. Rev. Nutr. 1996;16:33–50. doi: 10.1146/annurev.nu.16.070196.000341. [DOI] [PubMed] [Google Scholar]
- 14.Yusuf T.L., Oladipo S.D., Olagboye S.A., Zamisa S.J., Tolufashe G.F. Solvent-free synthesis of nitrobenzyl Schiff bases: characterisation, antibacterial studies, density functional theory and molecular docking studies. J. Mol. Struct. 2020;1222 [Google Scholar]
- 15.Ceramella J., Iacopetta D., Catalano A., Cirillo F., Lappano R., Sinicropi M.S. A review on the antimicrobial activity of schiff bases: data collection and recent studies. Antibiotics. 2022;11:191. doi: 10.3390/antibiotics11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adeleke A.A., Zamisa S.J., Omondi B. Ag(I) complexes of imine derivatives of unexpected 2-thiophenemethylamine homo-coupling and bis-(E)-N-(furan-2-ylmethyl)-1-(quinolin-2-yl)methanimine. Molbank. 2021;2021(2):M1235. [Google Scholar]
- 17.Yusuf T.L., Akintayo D.C., Oladipo S.D., Adeleke A.A., Olofisan K., Vatsha B., Mabuba N. The effect of structural configuration on the D.N.A. binding and in vitro antioxidant properties of new copper (ii) N2O2 Schiff base complexes. New J. Chem. 2022;46:12968–12980. [Google Scholar]
- 18.Prasad S.R., Satyanarayan N.D., Shetty A.S.K., Thippeswamy B., Thippeswamy B. Synthesis, antimicrobial, and antitubercular evaluation of new Schiff bases with in silico ADMET and molecular docking studies. Eur. J. Chem. 2022;13:109–116. [Google Scholar]
- 19.Afzal H.R., Khan N.U.H., Sultana K., Mobashar A., Lareb A., Khan A., Gull A., Afzaal H., Khan M.T., Rizwan M., Imran M. Schiff bases of pioglitazone provide better antidiabetic and potent antioxidant effect in a streptozotocin–nicotinamide-induced diabetic rodent model. ACS Omega. 2021;6:4470–4479. doi: 10.1021/acsomega.0c06064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matela G. Schiff bases and complexes: a review on anti-cancer activity. Anti Cancer Agents Med. Chem. 2020;20:1908–1917. doi: 10.2174/1871520620666200507091207. [DOI] [PubMed] [Google Scholar]
- 21.Hameed A., Al-Rashida M., Uroos M., Abid Ali S., Khan K.M. Schiff bases in medicinal chemistry: a patent review (2010-2015) Expert Opin. Ther. Pat. 2017;27:63–79. doi: 10.1080/13543776.2017.1252752. [DOI] [PubMed] [Google Scholar]
- 22.Olagboye S.A., Yusuf T.L., Oladipo S.D., Zamisa S.J. Crystal structure of bis (2-hydroxy-6-((phenylimino) methyl) phenolato-κ2N, O) copper (II), C26H20CuN2O4. Z. fur. Krist – New Cryst. 2020;235:689–692. [Google Scholar]
- 23.Oladipo S.D., Yusuf T.L., Zamisa S.J., Tolufashe G.F., Olofinsan K.A., Tywabi-Ngeva Z., Mabuba N. Synthesis, crystal structure with free radical scavenging activity and theoretical studies of Schiff bases derived from 1-naphthylamine, 2, 6-diisopropylaniline, and substituted benzaldehyde. Eur. J. Chem. 2021;12:204–215. [Google Scholar]
- 24.Adesola A.A., Zamisa S.J., Omondi B. Crystal structure of 4-(1-phenylimidazo[1,5-a]pyridin-3-yl)benzoic acid (C20H14N2O2) Z. fur Krist. - New Cryst. Struct. 2019;234:1157–1159. [Google Scholar]
- 25.Bruker APEX II, Bruker AXS: Madison, Wisconsin, USA, 2009.
- 26.Bruker SAINT, Bruker AXS: Madison, Wisconsin, USA, 2009.
- 27.Bruker SADABS, Bruker AXS: Madison, Wisconsin, USA, 2009.
- 28.Sheldrick G.M. A short history of Shelx. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 29.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A., Puschmann H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009;42:339–341. [Google Scholar]
- 30.Ibitoye O.B., Olofinsan K., Teralı K., Ghali U.M., Ajiboye T.O. Bioactivity‐guided isolation of antidiabetic principles from the methanolic leaf extract of Bryophyllum pinnatum. J. Food Biochem. 2018;42 [Google Scholar]
- 31.Olofinsan K.A., Erukainure O.L., Brian B.K., Islam M.S. Harpephyllum caffrum stimulates glucose uptake, abates redox imbalance and modulates purinergic and glucogenic enzyme activities in oxidative hepatic injury. Asian Pac. J. Trop. Biomed. 2022;12:9. [Google Scholar]
- 32.Kurian G.A., Suryanarayanan S., Raman A., Padikkala J. Antioxidant effects of ethyl acetate extract of Desmodium gangeticum root on myocardial ischemia reperfusion injury in rat hearts. Chin. Med. 2010;5:1–7. doi: 10.1186/1749-8546-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turkoglu A., Duru M.E., Mercan N., Kivrak I., Gezer K. Antioxidant and antimicrobial activities of Laetiporus sulphureus. Bull.) Murrill. Food Chem. 2007;101:267–273. [Google Scholar]
- 34.Tan Y.P., Chan E.W.C. Antioxidant, antityrosinase and antibacterial properties of fresh and processed leaves of Anacardium occidentale and Piper betle. Food Biosci. 2014;6:17–23. [Google Scholar]
- 35.Becke A.D. Density‐functional thermochemistry. I. The effect of the exchange‐only gradient correction. J. Chem. Phys. 1992;96:2155–2160. [Google Scholar]
- 36.Martínez J. Local reactivity descriptors from degenerate frontier molecular orbitals. Chem. Phys. Lett. 2009;478:310–322. [Google Scholar]
- 37.Nahoum V., Roux G., Anton V., Rouge P., Puigserver A., Bischoff H., Henrissat B., Payan F. Crystal structures of human pancreatic α-amylase in complex with carbohydrate and proteinaceous inhibitors. Biochem. J. 2000;346:201–208. [PMC free article] [PubMed] [Google Scholar]
- 38.Cardona F., Parmeggiani C., Faggi E., Bonaccini C., Gratteri P., Sim L., Gloster T.M., Roberts S., Davies G.J., Rose D.R., Goti A. Total syntheses of casuarine and its 6‐O‐α‐glucoside: complementary inhibition towards glycoside hydrolases of the GH31 and GH37 families. Chem. Eur J. 2009;15:1627–1636. doi: 10.1002/chem.200801578. [DOI] [PubMed] [Google Scholar]
- 39.Wang J., Wang W., Kollman P.A., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualisation, and analysis platform. J. Cheminf. 2012;4:1–17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Systèmes D. Dassault Systèmes Biovia; San Diego, CA, USA: 2016. Biovia, Discovery Studio Modeling Environment. [Google Scholar]
- 42.Nair P.C., Miners J.O. Molecular dynamics simulations: from structure function relationships to drug discovery. silico Pharmacol. 2014;2:1–4. doi: 10.1186/s40203-014-0004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryckaert J.P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 44.Berendsen H.J., Postma J.V., Van Gunsteren W.F., DiNola A.R.H.J., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 45.Gonnet P., Shake P.- A quadratically convergent SHAKE in O (n2) J. Comp. Physiol. 2007;220:740–750. [Google Scholar]
- 46.Le Grand S., Götz A.W., Walker R.C. SPFP: speed without compromise—a mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013;184:374–380. [Google Scholar]
- 47.Larini L., Mannella R., Leporini D. Langevin stabilisation of molecular-dynamics simulations of polymers by means of quasisymplectic algorithms. J. Chem. Phys. 2007;126 doi: 10.1063/1.2464095. [DOI] [PubMed] [Google Scholar]
- 48.Ylilauri M., Pentikäinen O.T. MMGBSA as a tool to understand the binding affinities of filamin–peptide interactions. J. Chem. Inf. Model. 2013;53:2626–2633. doi: 10.1021/ci4002475. [DOI] [PubMed] [Google Scholar]
- 49.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 50.Seifert E. OriginPro 9.1: scientific data analysis and graphing software-software review. J. Chem. Inf. Model. 2014;54:1552. doi: 10.1021/ci500161d. [DOI] [PubMed] [Google Scholar]
- 51.Oladipo S.D., Yusuf T.L., Zamisa S.J., Shapi M., Ajayi T.J. Synthesis, crystal structure, Hirshfeld surface analysis and DFT studies of N-(2, 6-diisopropylphenyl)-1-(4-methoxyphenyl) methanimine. J. Mol. Struct. 2021;1241 [Google Scholar]
- 52.Kemp W. Bloomsbury Publishing; 2017. Organic Spectroscopy. [Google Scholar]
- 53.Ferrence G.M., Tovee C.A., Holgate S.J., Johnson N.T., Lightfoot M.P., Ward K.L., Nowakowska-Orzechowska S.C. C.S.D. Communications of the Cambridge structural database. IUCrJ. 2023;10:6–15. doi: 10.1107/S2052252522010545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moghadam F.N., Amirnasr M., Eskandari K., Meghdadi S. A new disulfide Schiff base as a versatile "OFF–ON–OFF" fluorescent–colorimetric chemosensor for sequential detection of C.N.− and Fe3+ ions: combined experimental and theoretical studies. New J. Chem. 2019;43:13536–13544. [Google Scholar]
- 55.Orona G., Molinar V., Fronczek F.R., Isovitsch R. 2-[(Naphthalen-1-ylmethylidene) amino]-5-methylphenol. Acta Crystallogr. E. 2011;67:o2505–o2506. doi: 10.1107/S1600536811034556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spackman M.A., Jayatilaka D. Hirshfeld surface analysis. CrystEngComm. 2009;11:19–32. [Google Scholar]
- 57.Spackman P.R., Turner M.J., McKinnon J.J., Wolff S.K., Grimwood D.J., Jayatilaka D., Spackman M.A. CrystalExplorer: a program for Hirshfeld surface analysis, visualisation and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021;54:1006–1011. doi: 10.1107/S1600576721002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spackman M.A., Byrom P.G. A novel definition of a molecule in a crystal. Chem. Phys. Lett. 1997;267:215–220. [Google Scholar]
- 59.Mudsainiyan R.K., Pandey S.K. A combined theoretical calculation and Hirshfeld surface analysis of cooperative non‐covalent interactions in the crystal packing in [Cu (L1) 2 (E.D.A.)] Z. Anorg. Allg. Chem. 2017;643:1245–1252. [Google Scholar]
- 60.Oladipo S.D., Zamisa S.J., Badeji A.A., Ejalonibu M.A., Lawal I.A., Henni A., Lawal M.M. Ni2+ and Cu2+ complexes of N-(2,6-dichlorophenyl)-N-mesityl formamidine dithiocarbamate structural and functional properties of CYP3A4. Sci. Rep. 2023;13 doi: 10.1038/s41598-023-39502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akpan E.D., Oladipo S.D., Quadri T.W., Olasunkanmi L.O., Nwanna E.E., Omondi B., Ebenso E.E. Formamidine-based thiuram disulfides as efficient inhibitors of acid corrosion of mild steel; electrochemical, surface and density functional theory/Monte Carlo simulation studies. ACS Omega. 2022;7:26076–26091. doi: 10.1021/acsomega.2c00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hernández-Rodríguez M.C., Rosales-Hernández M., E Mendieta-Wejebe J., Martinez-Archundia M., Correa Basurto J. Current tools and methods in molecular dynamics (M.D.) simulations for drug design. Curr. Med. Chem. 2016;23:3909–3924. doi: 10.2174/0929867323666160530144742. [DOI] [PubMed] [Google Scholar]
- 63.Wang C., Greene D.A., Xiao L., Qi R., Luo R. Recent developments and applications of the MMPBSA method. Front. Mol. Biosci. 2018;4:87. doi: 10.3389/fmolb.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Obakachi V.A., Kushwaha N.D., Kushwaha B., Mahlalela M.C., Shinde S.R., Kehinde I., Karpoormath R. Design and synthesis of pyrazolone-based compounds as potent blockers of SARS-CoV-2 viral entry into the host cells. J. Mol. Struct. 2021;1241 doi: 10.1016/j.molstruc.2021.130665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burley S.K., Petsko G.A. Aromatic-aromatic interaction: a mechanism of protein structure stabilisation. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 66.Shao J., Kuiper B.P., Thunnissen A.M.W., Cool R.H., Zhou L., Huang C., Dijkstra B.W., Broos J. The role of tryptophan in π interactions in proteins: an experimental approach. J. Am. Chem. Soc. 2022;144:13815–13822. doi: 10.1021/jacs.2c04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akinpelu O.I., Lawal M.M., Kumalo H.M., Mhlongo N.N. Computational studies of the properties and activities of selected trisubstituted benzimidazoles as potential antitubercular drugs inhibiting MTB-FtsZ polymerisation. J. Biomol. Struct. Dyn. 2022;40:1558–1570. doi: 10.1080/07391102.2020.1830176. [DOI] [PubMed] [Google Scholar]
- 68.Nichols S.E., Baron R., Ivetac A., McCammon J.A. Predictive power of molecular dynamics receptor structures in virtual screening. J. Chem. Inf. Model. 2011;6:1439–1446. doi: 10.1021/ci200117n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palma J., Pierdominici‐Sottile G. On the uses of PCA to characterise molecular dynamics simulations of biological macromolecules: basics and tips for an effective use. ChemPhysChem. 2023;24 doi: 10.1002/cphc.202200491. [DOI] [PubMed] [Google Scholar]
- 71.Adewumi A.T., Oluyemi W.M., Adekunle Y.A., Adewumi N., Alahmdi M.I., Soliman M.E., Abo-Dya N.E. Propitious indazole compounds as β‐ketoacyl‐ACP synthase inhibitors and mechanisms unfolded for TB cure: integrated rational design and MD simulations. ChemistrySelect. 2023;8 [Google Scholar]
- 72.Galindo-Murillo R., Roe D.R., Cheatham T.E., III Convergence and reproducibility in molecular dynamics simulations of the DNA duplex d (GCACGAACGAACGAACGC) Biochim. Biophys. Acta Gen. Subj. 2015;5:1041–1058. doi: 10.1016/j.bbagen.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oladipo S.D., Tolufashe G.F., Mocktar C., Omondi B. Ag (I) symmetrical N, N′-diarylformamidine dithiocarbamate PPh3 complexes: synthesis, structural characterization, quantum chemical calculations and in vitro biological studies. Inorganica. Chim. Acta. 2021;520 [Google Scholar]
- 74.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pires D.E., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oladipo S.D., Olotu F.A., Soliman M., Mocktar C., Omondi B. Formamidine-based thiuram disulfides: synthesis, structural characterisation, biological studies, and preliminary cheminformatics evaluation. J. Mol. Struct. 2020;1219 [Google Scholar]
- 77.Olotu F.A., Munsamy G., Soliman M.E. Does size really matter? Probing the efficacy of structural reduction in the optimisation of bioderived compounds–a computational "proof-of-concept". Comput. Struct. Biotechnol. J. 2018;16:573–586. doi: 10.1016/j.csbj.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shityakov S., Neuhaus W., Dandekar T., Förster C. Analysing molecular polar surface descriptors to predict blood-brain barrier permeation. Int. J. Comput. Biol. Drug Des. 2013;6:146–156. doi: 10.1504/IJCBDD.2013.052195. [DOI] [PubMed] [Google Scholar]
- 79.Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 80.Sjögren E., Westergren J., Grant I., Hanisch G., Lindfors L., Lennernäs H., Abrahamsson B., Tannergren C. In silico predictions of gastrointestinal drug absorption in pharmaceutical product development: application of the mechanistic absorption model GI-Sim. Eur. J. Pharmaceut. Sci. 2013;49:679–698. doi: 10.1016/j.ejps.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 81.Cornaire G., Woodley J., Hermann P., Cloarec A., Arellano C., Houin G. Impact of excipients on the absorption of P-glycoprotein substrates in vitro and in vivo. Int. J. Pham. 2004;278:119–131. doi: 10.1016/j.ijpharm.2004.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.