Abstract
For many viruses, primary infection has been shown to prevent superinfection by a homologous second virus. In this study, we investigated superinfection exclusion of bovine viral diarrhea virus (BVDV), a positive-sense RNA pestivirus. Cells acutely infected with BVDV were protected from superinfection by homologous BVDV but not with heterologous vesicular stomatitis virus. Superinfection exclusion was established within 30 to 60 min but was lost upon passaging of persistently infected cells. Superinfecting BVDV failed to deliver a translatable genome into acutely infected cells, indicating a block in viral entry. Deletion of structural protein E2 from primary infecting BVDV abolished this exclusion. Bypassing the entry block by RNA transfection revealed a second block at the level of replication but not translation. This exclusion did not require structural protein expression and was inversely correlated with the level of primary BVDV RNA replication. These findings suggest dual mechanisms of pestivirus superinfection exclusion, one at the level of viral entry that requires viral glycoprotein E2 and a second at the level of viral RNA replication.
The family Flaviviridae consists of three genera, the classical flaviviruses, the pestiviruses, and the hepatitis C viruses. The pestivirus genus includes bovine viral diarrhea virus (BVDV), classical swine fever virus, and border disease virus, which are important animal pathogens (34, 61). The pestiviruses are enveloped, single-stranded positive-sense RNA viruses (reviewed in reference 41). The RNA genome of BVDV is approximately 12.5 kb in size and contains a single open reading frame flanked by a 5′ nontranslated region and a nonpolyadenylated 3′ nontranslated region. The open reading frame is translated into a nascent polyprotein of approximately 4,000 amino acids via an internal ribosome entry site within the 5′ nontranslated region. The polyprotein is co- and posttranslationally processed by cellular and viral proteases into mature viral proteins, designated from the N terminus as Npro, C, Erns, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B.
Npro, the first viral protein translated in the genome, possesses an autoproteolytic activity responsible for cleavage at its own C terminus. Next encoded in the polyprotein are the viral structural proteins, capsid (C) and three virion-associated glycoproteins, Erns, E1, and E2. Several reports have shown that E2 is a major target of neutralizing antibodies, suggesting a significant role in receptor-mediated viral entry (19, 47, 62). Erns has been reported to have intrinsic RNase activity (50, 68), but its role in viral infection remains ambiguous.
The remainder of the BVDV genome encodes nonstructural (NS) proteins, which are involved in viral replication. NS3 contains a serine protease motif at its N terminus (7, 23) responsible for cleavage between NS3 and NS4A, NS4A and NS4B, NS4B and NS5A, and NS5A and NS5B, (59, 69, 70). NS4A functions as an NS3 protease cofactor for cleavage between NS4B and NS5A and between NS5A and NS5B (59, 70). Additionally, NS3 contains NTPase and helicase sequence motifs, and these enzymatic activities have been demonstrated in vitro (23, 25, 26, 57, 58, 63). NS5B is the viral RNA-dependent RNA polymerase (30, 32, 71). Studies have shown that autonomous BVDV RNA replication in transfected cells does not require viral structural proteins (C, Erns, E1, and E2), Npro, p7, or NS2 (8, 45, 60).
Infections of cattle with BVDV can result in a wide range of clinical symptoms from acute self-limiting disease to a sporadic, fatal mucosal disease (6, 61). Two BVDV biotypes, cytopathic (cp) and noncytopathic (ncp), based on cytopathogenicity in tissue culture cells, can be isolated from animals with mucosal disease. Animals persistently infected with an ncp virus in utero develop mucosal disease due to the emergence of a cp virus, usually by RNA recombination (40). This conclusion is supported by the fact that antigenically and genetically closely related ncp and cp virus pairs can be isolated from animals that have died of mucosal disease (37, 38, 40, 67). Furthermore, experimental superinfection of persistently infected animals with a closely related cp virus can result in mucosal disease (10, 13, 44). Mendez et al. have generated an isogenic pair of infectious cDNA clones, ncp BVDV NADLJiv90− and cp BVDV NADL, the latter having a short in-frame insertion of cellular sequence (Jiv90) in the NS2 gene upstream of the NS2/3 cleavage site (39). These nearly identical but easily distinguishable viruses provide an ideal model pair for the study of superinfection exclusion.
Superinfection exclusion, or homologous interference, is defined as the ability of an established virus infection to interfere with a homologous superinfecting virus. From an evolutionary standpoint, superinfection exclusion can be advantageous for a virus. In a population of cells undergoing viral infection, superinfection exclusion would be beneficial to newly produced virions by favoring entry into uninfected rather than previously infected cells and therefore dissemination. Moreover, a primary virus successfully infecting a cell would be protected from a competing, superinfecting virus. In this regard, it is reasonable to speculate that superinfection exclusion is a powerful strategy for maintaining the genetic diversity of a virus population because it allows replication of variants that range in fitness.
Superinfection exclusion has been described for a number of viruses, including retroviruses, vesicular stomatitis virus, and borna disease virus, as well as several alphaviruses, including Sindbis virus. The mechanisms of superinfection exclusion have been identified at various stages of the viral life cycle, including receptor-mediated attachment (11, 12, 54, 55), penetration of the viral core into infected cells (52, 53, 66), and subsequent replication steps (1, 22, 31).
Superinfection exclusion has been described for pestiviruses (43), but the molecular mechanisms responsible for this phenomenon remain unclear. In this report, we demonstrate that MDBK cells acutely infected with ncp BVDV are protected from cytopathic effect (CPE) following superinfection with cp BVDV but not with an unrelated cp virus, vesicular stomatitis virus. Dual mechanisms of BVDV superinfection exclusion were observed; one occurred at the level of viral entry and another at the level of viral replication. We determined that superinfecting BVDV fails to enter acutely infected cells and that the structural protein E2 was responsible for this block. If the viral entry steps were bypassed by transfection of the viral RNA, the RNA could be translated but failed to replicate in cells acutely infected with BVDV. The second mechanism of superinfection exclusion at viral RNA replication was dependent on the level of primary RNA replication and was not influenced by expression of the viral structural proteins (C, Erns, E1, and E2).
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby bovine kidney (MDBK) cells were obtained from M. Collett (ViroPharma, Inc.) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated horse serum, sodium pyruvate, and penicillin-streptomycin. Media and reagents for cell culture were purchased from Gibco-BRL, Life Technologies Ltd. Stocks of the NADL strain of BVDV were generated by in vitro transcription of pACNR/NADL cDNA, transfection of transcribed RNA into MDBK cells, and collection of cell culture supernatant containing virus (39). Original stocks of other BVDV derivatives, including NADLJiv90−, NADLJiv90−pac, and NADLJiv90−luc viruses, were prepared in the same way from the cDNAs pACNR/NADLJiv90−, pACNR/NADLJiv90−pac, and pACNR/NADLJiv90−luc, respectively. Virus stocks were used without passaging to minimize the possibility of generating variants or defective interfering (DI) RNAs, an extremely rare event in cell culture (42).
Construction of plasmids.
The parental DNA plasmids used in this study were derived from pACNR/NADLJiv90− (formerly called pACNR/cIns−NADL) and pACNR/NADL (39). To generate the pACNR/NADLJiv90−ΔS construct, PCR was performed to amplify the following three fragments. Fragment I (nucleotides 695 to 916, nucleotide numbers are based on the NADLJiv90− sequence) introduced a HindIII site immediately following nucleotides 916. Fragment II (nucleotides 1142 to 1225) introduced a HindIII site immediately preceding nucleotide 1142, and the last two codons (1137CTA CAA1142) were changed to 1137CTG CAG1142 to create a PstI site without changing the amino acid sequence. Fragment III (nucleotides 3440 to 4049) introduced a PstI site immediately preceding nucleotide 3440 and a HindIII site immediately following nucleotide 4049.
The SacI-HindIII, HindIII-PstI, and PstI-BsmI fragments of each PCR-amplified DNA were generated by digestion with the two appropriate restriction endonucleases. Each fragment was cloned into pRS2, a subcloning vector derived from pUC19, and sequenced. These three fragments were ligated to the SacI-BsmI fragment of pACNR/NADLJiv90− generated by digestion with SacI and BsmI. The pACNR/NADLJiv90−ΔS plasmid has a complete BVDV 5′ nontranslated region followed by the Npro gene, 10 N-terminal residues of capsid, an extra Leu codon, 18 C-terminal residues of capsid, nine N-terminal residues of Erns, 48 C-terminal residues of E2, and the remainder of the NADLJiv90− viral genome.
To generate the pACNR/NADLJiv90−ΔS-pac and pACNR/NADLJiv90−pac plasmids, the ubi-pac cassette was subcloned into the pRS2 vector by ligating four pieces generated by PCR amplification. Fragment I (nucleotides 247 to 502) introduced an SphI site upstream of nucleotide 247, and MluI and EcoRI sites downstream of nucleotides 502. The SphI-EcoRI fragment was cloned into the pRS2 vector digested with SphI and EcoRI and sequenced. Fragment II (nucleotides 503 to 727) of the entire ubi gene, flanked by MluI at the 5′ end and SacI at the 3′ end, was cloned into the pRS2 vector digested with MluI and SacI and sequenced. Fragment III, representing the N-terminal portion of the pac gene, was flanked by SacII immediately upstream of the initiation codon AUG for the pac gene and BssHII at the 3′ end. The SacII-BssHII fragment of PCR-amplified DNA was also cloned into the pRS2 vector digested with SacII and BssHII and sequenced. These three fragments were ligated to the pRS2 vector digested with SphI and BssHII (pRS2-SB).
Subsequently, the BssHII-SacI fragment of pRS2-SB was ligated with two fragments, the BssHII-Ecl136II and AclI-SacI fragments of the p5′EMCV/BVDV plasmid described previously (21) digested with AclI, filled in by T4 DNA polymerase, and digested with SacI (pRS2-BS). Next, both the SphI-BssHII fragment of pRS2-SB and the BssHII-SacI fragment of pRS2-BS were ligated to the SphI-SacI fragment of the pRS2 vector (pRS2-SS). Finally, the pACNR/NADLJiv90−ΔS-pac and pACNR/NADLJiv90−pac plasmids were constructed by cloning the SphI-SacI fragment of pRS2-SS into the pACNR/NADLJiv90−ΔS and pACNR/NADLJiv90− plasmids, respectively, which had been digested with SphI and SacI.
To generate pACNR/NADLJiv90−ΔS-luc, the entire luc gene flanked by SacII and BssHII was PCR amplified from pSP-luc(+) (generously provided by Alexander Kolyhkalov), cloned into pRS2 (pRS2-luc), and sequenced. The SacII site at the 3′ end of the ubi gene was used to fuse the ubi gene in frame. Both the XhoI-SacII fragment of pACNR/NADLJiv90−ΔS and the SacII-BssHII fragment of pRS2-luc were ligated to the XhoI-BssHII fragment of pACNR/NADLJiv90−ΔS-pac. The pACNR/NADLJiv90−luc plasmid was constructed by ligating the XhoI-SacI fragment of pNADLJiv90−ΔS-luc with the XhoI-SacI fragment of pACNR/NADLJiv90−pac.
To generate the pACNR/NADLJiv90−ΔS-luc-pol− plasmid, the PacI-XmaI fragment of pACNR/NADL/AAG (4), which contains mutations at nucleotides 11532 to 11539 (11532GGG ATG AT11539→11532CGG CCG GC11539) to inactivate the catalytic active site (GDD→AAG), was ligated with two fragments, the Pac-SacI and SacI-XmaI fragments of pACNR/NADLJiv90−ΔS-luc.
In order to generate each in-frame deletion mutant in the region of viral structural proteins, we first tried to make each mutant construct in the context of pACNR/NADLJiv90−pac. These efforts were not successful due to the instability of the resulting plasmids. With pBeloBAC11 (New England Biolabs), we generated the pBAC/NADLJiv90−pac, pBAC/NADLJiv90−ΔC-pac, pBAC/NADLJiv90−ΔErns-pac, pBAC/NADLJiv90−ΔE1-pac, and pBAC/NADLJiv90−ΔE2-pac plasmids. Briefly, for pBAC/NADLJiv90−ΔC-pac, nucleotides 2494 to 2718 in the capsid region were deleted and a new HindIII site was introduced by PCR. The primers to generate the downstream region from the introduced HindIII site were 5′-GAT AAG CTT GCA TTG TTG GCG TGG GCA-3′ and 5′-GAT CTC GAG TTC AAT ATT GTA CCA GTT-3′, and the upstream region from the HindIII site was derived from pNADLJiv90−ΔS-pac.
For pBAC/NADLJiv90−ΔErns-pac, the primers to delete nucleotides 2794 to 3384 and introduce a new HindIII site were 5′-GAT CTC GAG TCC GGA TGC TAC AAT AGT-3′, 5′-GAT AAG CTT GTT CCA CTG TGT TAT GTT-3′, 5′-GAT AAG CTT GGC AAG CAG CTC GGG ATA-3′, and 5′-GAT CTC GAG CAG CTG TTG TCA GCT CCA-3′. For pBAC/NADLJiv90−ΔE1-pac, the primers to delete nucleotides 3484 to 3618 and introduce a HindIII site were 5′-GAT CTC GAG GGG CCC CTG CAA CTT TGA-3′, 5′-GAT AAG CTT TTT GCG ATC GAC ATC ACA-3′, 5′-GAT AAG CTT GAG GTA CTA CTA CTT TCT-3′, and 5′-GAT CTC GAG GGC CGG TCC GTT CAG CAG-3′.
For pBAC/NADLJiv90−ΔE2-pac, the primers to delete nucleotides 4072 to 5016 and introduce a HindIII site were 5′-GAT CTC GAG CAG CTG AAG TAA TAC CAG-3′, 5′-GAT AAG CTT GGC ATA CGA GAA TTC AGG-3′, 5-GAT AAG CTT TGG TTT GAC CTG GAG GTG-3′, and 5′-GAT CTC GAG CTT TAC GCT CTC CTC CCT-3′.
Transcription and transfection.
For transcription reactions, plasmids were digested to completion with SdaI (Sse8387I), followed by phenol-chloroform extraction, precipitation with ethanol, and resuspension in distilled H2O. Between 0.5 and 1 μg of linearized DNA plasmid was added to the transcription reaction with the T7 Megascript kit (Ambion) as recommended by the manufacturer. Reactions were incubated at 37°C for 90 min with 1 μCi of [3H]UTP (DuPont) to quantify RNA synthesis. The quality of the synthesized RNAs was monitored by agarose gel electrophoresis. Then the transcription reaction mixtures were treated with DNase I for 30 min at 37°C, aliquoted, and stored at −80°C. Transfection of MDBK cells was performed by electroporation as previously described (39) with a BTX ElectroSquarePorator.
Analysis of superinfection exclusion.
We inoculated 5 × 105 cells per well of a six-well plate with 5 × 106 FFU of ncp BVDV for 1 h with frequent shaking. The inoculum was removed, and the cells were incubated at 37°C for 12 h. After incubation, the cells were washed with medium three times and superinfected with the appropriate virus at 37°C for 1 h. After superinfection, the cells were washed three times with medium. For plaque assays, cells were overlaid with 0.5% LE SeaKem agarose in minimal essential medium containing 5% heat-inactivated horse serum and penicillin-streptomycin and incubated at 37°C for 3 to 5 days. Plaques were visualized by crystal violet staining of formaldehyde-fixed cells as described previously (39). Focus-forming assays were incubated under the same overlay and performed as previously described (48). To monitor CPE in liquid culture, cells were incubated in complete medium at 37°C for 2 to 4 days. After incubation, the cells were fixed and stained with crystal violet.
Luciferase assay.
Transfected MDBK cells (8 × 106), divided equally and seeded in a six-well plate, were washed with phosphate-buffered saline without Ca2+ and Mg2+ and lysed by addition of 0.2 ml of lysis buffer (25 mM Tris-HCl, pH 7.8, 2 mM dithiothreitol, 2 mM EGTA, 1% Triton X-100, vol/vol). Cell lysates were incubated 20 min at room temperature and then cleared of cellular debris by centrifugation. The supernatants were frozen rapidly at −80°C until assaying for luciferase. To measure luciferase activity, 20 μl of cell lysate was added to a luminometer tube which contained 180 μl of luciferase assay reagent [20 mM Tricine, 1.07 mM (MgCO3)4Mg(OH)2. · 5H2O), 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM dithiothreitol, 270 μM coenzyme A (Sigma), 470 μM luciferin (Promega), 530 μM ATP]. The activity was typically measured for 20 s at least in duplicate.
Generation of NADLJiv90−pac, NADLJiv90−ΔS-pac, NADLJiv90−ΔC-pac, NADLJiv90−ΔErns-pac, NADLJiv90−ΔE1-pac, and NADLJiv90−ΔE2-pac MDBK cell lines.
To generate the NADLJiv90−pac, NADLJiv90−ΔS-pac, NADLJiv90−ΔC-pac, NADLJiv90−ΔErns-pac, NADLJiv90−ΔE1-pac, and NADLJiv90−ΔE2-pac cell lines, naïve MDBK cells were electroporated with the appropriate in vitro transcribed RNA. After 12 h of incubation at 37°C, the transfected cells were treated with puromycin (10 μg/ml) for 24 h. Surviving cells were then prepared for superinfection by seeding in a six-well plate (4 × 105) in the presence of 10 μg of puromycin per ml for 24 h prior to superinfection. Established cell lines were maintained in complete medium containing 1 μg of puromycin per ml.
Generation of MDBK cell lines with different levels of BVDV RNA replication.
Two BVDV viral replicon RNAs of NADLJiv90−ΔS-pac and NADLΔS-pac were synthesized by in vitro transcription as described in the text and transfected into naïve MDBK cells. At 24 h postelectroporation, the cells were placed under selection with medium containing 10 μg of puromycin per ml. Puromycin-resistant foci from the selected NADLJiv90−ΔS-pac and NADLΔS-pac cells were expanded individually and maintained for the study. A detailed study of these selected replicons will be presented elsewhere, but as previously found for a study selecting ncp variants of NADL (48), adaptive mutations allowing persistent noncytopathic replication were identified in the NS4A-4B-5A region of the polyprotein for the colonies recovered from the NADLΔS-pac transfectants.
Metabolic labeling of viral RNA.
Cells (106) were cultured in phosphate-free Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated horse serum, 2 μg of dactinomycin, and 20 μCi of [32P]orthophosphate for 6 h. Total RNA was harvested with Trizol reagent. One third of the isolated RNA was denatured with glyoxal and separated by agarose gel electrophoresis as described elsewhere (39). The gel was then fixed with methanol and dried. RNA was visualized and quantified by phosphoimage analysis.
Immunostaining and flow cytometry.
Immunostaining and flow cytometry were performed as described (2). Briefly, cells in a 35-mm2 dish (3 × 105) were trypsinized and fixed with 0.5% paraformaldehyde for 20 min at room temperature. Cells were washed twice with Dulbecco’s phosphate-buffered saline, then blocked and permeabilized with phosphate-buffered saline-5% goat serum-0.1% sodium azide-0.1% saponin for 20 min at room temperature. Cells were then incubated with primary antibody (monoclonal antibody 184) diluted 1:2,000 in SB (phosphate-buffered saline with 0.5% bovine serum albumin, 0.1% sodium azide, and 0.1% saponin) for 30 to 60 min at room temperature. After washing with SB, the cells were incubated with goat anti-mouse immunoglobulin G-Alexa 488 secondary antibody (Molecular Probes, Eugene, Oreg.) diluted 1:1,000 in SB for 30 to 60 min at room temperature. Cells were washed with SB and analyzed by flow cytometry on a FACSCalibur (Becton Dickinson), counting 104 cells.
Real-time quantitative reverse transcription-PCR.
Total RNA was extracted from duplicate wells with Trizol reagent (Invitrogen); 10 ng of total cellular RNA was mixed with primers and probe specific for the BVDV 5′ nontranslated region as well as for bovine beta-actin RNA to normalize total RNA levels. BVDV-specific RNA and bovine beta-actin RNA were amplified with the Platinum Quantitative reverse transcription-PCR ThermoScript One-Step System (Invitrogen) and detected with the ABI Prism 7700 sequence detection system (PE Applied Biosystems, Foster City, Calif.). BVDV and bovine beta-actin cDNAs were generated by reverse transcription at 60°C for 30 min, followed by inactivation of the reverse transcriptase at 95°C for 10 min. cDNAs were then amplified with 40 cycles of 95°C for 15 s and 60°C for 1 min.
The BVDV forward and reverse primers are 5′-GGTGACTGCAGGTCGGGTAG-3′ and 5′-GGTAAAATAGTGGCCCTGGCTT-3′, respectively. The probe sequence is 5′-6FAM-CAGAGGACCTGTGAGCGGGATCTACCT-TAMRA-3′ (Eli Lilly and Co.). The forward and reverse primers for bovine beta-actin are 5′-ATGTGGATCAGCAAGCAGGAGTA-3′ and 5′-AAGCATTTGCGGTGGACAA-3′, respectively. The probe sequence is 5′-VIC-CGAGTCTGGCCCCTC-MGBNFQ-3′ (ABI). BVDV RNA levels were normalized to levels of bovine beta-actin RNA.
RESULTS
MDBK cells acutely infected with ncp BVDV are protected from CPE when superinfected with cp BVDV but not with vesicular stomatitis virus.
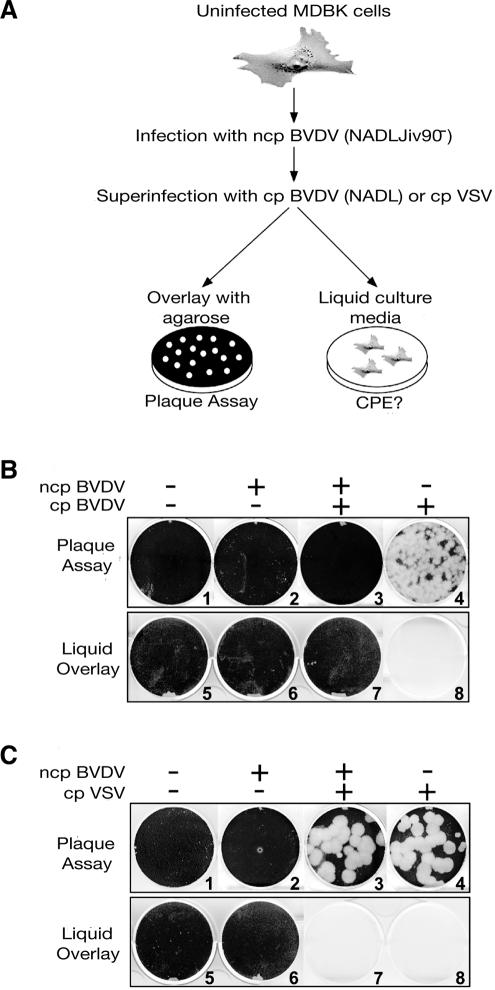
To examine whether ncp BVDV-infected MDBK cells are protected from subsequent BVDV infection, MDBK cells were infected with ncp NADLJiv90− at a multiplicity of infection of 10 and challenged at 12 h postinfection with cp NADL (Fig. 1A). As a control, naïve MDBK cells were infected in parallel with cp BVDV. CPE caused by cp BVDV replication was monitored quantitatively by plaque assay or qualitatively by incubating infected monolayers in liquid medium.
FIG. 1.
MDBK cells acutely infected with ncp BVDV are protected from CPE when superinfected with homologous cp BVDV but not with vesicular stomatitis virus. (A) Experimental procedures to examine superinfection exclusion in ncp BVDV-infected MDBK cells. (B and C) Naïve MDBK cells were first mock infected or infected with ncp NADLJiv90− BVDV. Subsequently, the cells were either mock infected or infected with only cp NADL BVDV or cp vesicular stomatitis virus (VSV). At 12 h postinfection, ncp NADLJiv90−-infected cells were washed three times with medium and either mock superinfected or superinfected with cp NADL BVDV or cp vesicular stomatitis virus for 1 h. The + and − above each column of plates indicate infection and mock infection, respectively, with the particular virus shown at the top left. The cells were overlaid with agarose (B and C, plates 1 to 4) or incubated in liquid culture medium (B and C, plates 5 to 8). The cells were fixed and stained with crystal violet to visualize live cells surviving CPE after 5 days (plaque assay) or 3 days (liquid overlay).
Plaques were formed on cell monolayers in plaque assays and no viable cells were observed for cultures incubated in liquid medium following infection of naïve MDBK cells with cp BVDV (Fig. 1B, plates 4 and 8). In contrast, neither plaques nor recognizable CPE resulted from superinfection of acutely ncp BVDV-infected cells with cp BVDV (Fig. 1B, plates 3 and 7), even 5 days postsuperinfection, when naïve MDBK cells began to die due to overgrowth. Primary ncp BVDV-infected cells did not show any signs of CPE (Fig. 1B, plates 2 and 6). In addition to ncp NADLJiv90− virus, MDBK cells infected with a heterologous ncp strain of BVDV, SD-1 (18), were also protected from CPE upon superinfection with cp NADL (data not shown).
Next, we investigated if ncp BVDV-infected MDBK cells are generally protected from virus-induced CPE, including that induced by a heterologous virus. To do so, vesicular stomatitis virus, a negative-sense interferon-sensitive rhabdovirus, was used to challenge the ncp BVDV-infected MDBK cells. CPE induced by vesicular stomatitis virus infection of BVDV-infected cells was indistinguishable from that of vesicular stomatitis virus-infected naïve MDBK cells, as determined by plaque assays (Fig. 1C, compare plates 3 and 4) and observing infected monolayers overlaid with liquid medium (Fig. 1C, compare plates 7 and 8). Thus, MDBK cells infected with ncp BVDV, either ncp NADLJiv90− or ncp SD-1 BVDV, were protected from CPE when superinfected with homologous cp BVDV but not with heterologous cp vesicular stomatitis virus. This suggests that the block may be virus specific and not mediated by a general cellular antiviral defense pathway such as interferon or a block in the ability of the cells to undergo virus-induced cell death.
To address whether the lack of CPE was due to a block in the ability of superinfecting virus to replicate in acutely infected cells, MDBK cells acutely infected with ncp NADLJiv90− were superinfected with ncp NADLJiv90−pac, which expresses a dominant selectable marker, the puromycin N-acetyltransferase (pac) gene. After selection with puromycin, neither foci nor surviving cells were observed in superinfected monolayers, whereas naïve MDBK cells infected with ncp NADLJiv90−pac were transduced to puromycin resistance with high efficiency (data not shown). These results indicate that the lack of CPE observed in the first experiments was likely due to the failure of the superinfecting cp BVDV to replicate efficiently in cells acutely infected with ncp BVDV (rather than inhibition of CPE despite replication of the cp virus).
Superinfection exclusion is established rapidly.
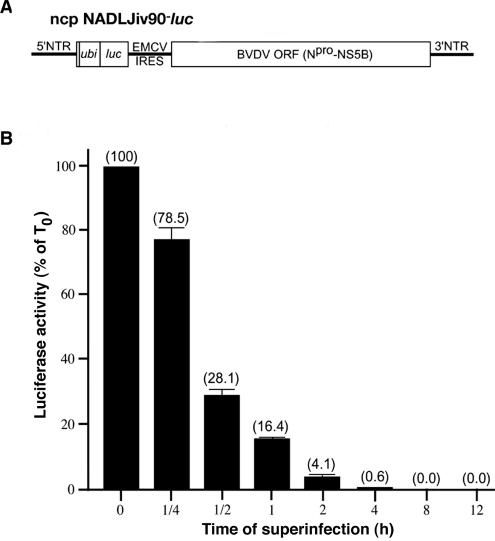
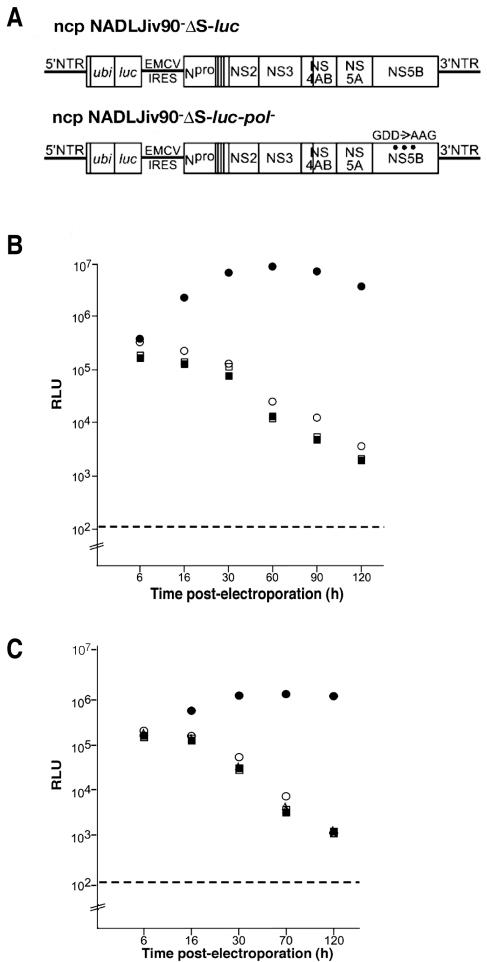
To determine the kinetics with which superinfection exclusion is established, an ncp virus expressing luciferase, NADLJiv90−luc, was generated (Fig. 2A). The ubiquitin gene (ubi) was inserted adjacent to the N terminus of the luc gene to create the correct N terminus of luciferase upon cleavage of Ubi. Expression of luc was driven by the BVDV 5′ nontranslated region, and expression of the BVDV proteins Npro to NS5B was driven by the encephalomyocarditis virus internal ribosome entry site. The level of luciferase activity from ncp NADLJiv90−luc after superinfection of ncp NADLJiv90−-infected cells was used as a measure of the degree of superinfection exclusion. Naïve MDBK cells were infected with ncp NADLJiv90− virus for 0, 0.25, 0.50, 1, 2, 4, 8, or 12 h prior to superinfection with ncp NADLJiv90−luc. At 24 h postsuperinfection, the luciferase activity was determined. Cells incubated with ncp NADLJiv90− for at least 30 to 60 min prior to superinfection with ncp NADLJiv90−luc had dramatically reduced luciferase activity compared to cells simultaneously coinfected with the two viruses (Fig. 2B). Beyond 4 h of incubation, little luciferase activity was detected. Thus, superinfection exclusion of BVDV is efficiently established within 30 to 60 min after primary BVDV infection.
FIG. 2.
Superinfection exclusion is established within 30 to 60 min of primary BVDV infection. (A) Schematic diagram of ncp NADLJiv90−luc viral RNA. (B) Naïve MDBK cells were infected with ncp NADLJiv90− for 0.25, 0.5, 1, 2, 4, 8, or 12 h prior to superinfection. The cells were then superinfected with ncp NADLJiv90−luc virus. In parallel, naïve MDBK cells were coinfected with the ncp NADLJiv90− and ncp NADLJiv90−luc viruses, indicated as time point 0. After 24 h of incubation, the cells were lysed and assayed for luciferase activity.
Superinfection exclusion is transient and lost upon passaging.
To determine if superinfection exclusion was maintained during passaging of persistently infected cell cultures, naïve MDBK cells were infected with ncp NADLJiv90− and maintained by passaging every 2 to 3 days. At each passage, the ncp NADLJiv90−-infected cells were superinfected with cp NADL and monitored for the ability to undergo CPE.
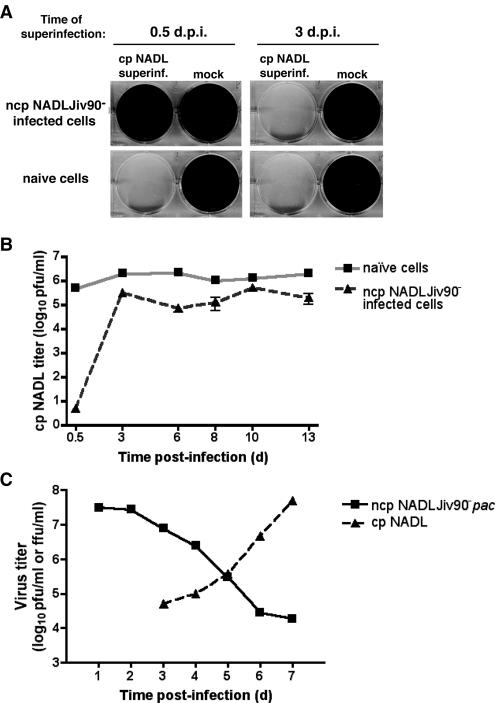
At passage 0, neither CPE nor visible plaques (Fig. 3A and data not shown) were found in the ncp NADLJiv90−-infected cells upon superinfection with cp NADL. In contrast, upon superinfection of passage 1 cells, persistently infected for 3 days, major CPE and visible plaques were observed (Fig. 3A and data not shown). There was, however, an approximately sixfold decrease in titer compared to cp NADL infection of naïve passage 1 cells (Fig. 3B). The extent of CPE and the size of the plaques were also diminished compared to infection of the naïve passage 1 cells (Fig. 3A and data not shown). Superinfection of additional passages of persistently infected cells did not result in further changes to cp NADL titer, plaque size, or the extent of CPE (Fig. 3B and data not shown). Thus, while persistently infected cells quickly lost the majority of the superinfection exclusion phenotype, their infectibility was not fully restored even out to passage 20 and greater (data not shown). Nonetheless, the ability to efficiently exclude a superinfecting virus is primarily a transient phenotype associated with acutely infected cells.
FIG. 3.
MDBK cells persistently infected with ncp NADLJiv90− are not protected from cp NADL-induced CPE. (A) MDBK cells were infected with ncp NADLJiv90−. At 0.5 and 3 days postinfection (d.p.i.) the cells were superinfected with cp NADL or mock superinfected as indicated above each column of plates. Naïve cells were also infected or mock infected for comparison. The cells were incubated in liquid medium for 3 days, at which time they were fixed and stained with crystal violet to visualize live cells surviving CPE. (B) ncp NADLJiv90−-infected and uninfected cells were maintained by passaging for a period of 13 days. Every 2 to 3 days, the cells were superinfected with cp NADL. Naïve cells were maintained over the same time period and infected in parallel. After superinfection, monolayers were overlaid with agarose, incubated for 3 days, and then fixed and stained with crystal violet. Plaques were enumerated and cp NADL titer on naïve cells or ncp NADLJiv90−-infected cells was determined. (C) MDBK cells were infected with ncp NADLJiv90−pac and maintained under puromycin selection. At 1, 2, 3, 4, and 5 days after acute infection, supernatants were harvested from the infected cells to determine ncp NADLJiv90−pac titer by focus-forming assay and also superinfected with cp NADL. Two days later, supernatants were harvested and titered by plaque assay to measure the yield of cp NADL. The plaque and focus-forming assay plates were incubated for 3 days under overlaid agarose and then fixed and stained with crystal violet or anti-BVDV antibody, respectively. The titers of ncp NADLJiv90−pac and cp NADL are expressed as focus-forming units (FFU) or PFU per milliliter.
To see if changes in the persisting ncp virus had caused the loss of superinfection exclusion, ncp NADLJiv90− released from persistently infected cells (passage 23) was used to acutely infect naïve MDBK cells. These cells were then competent to exclude superinfecting cp NADL (data not shown), suggesting that no significant changes to the persisting virus had occurred. To see if cellular changes were involved in the loss of superinfection exclusion over time, persistently ncp NADLJiv90−-infected cells were acutely reinfected with ncp NADLJiv90− and subsequently challenged with cp NADL. Plaques were seen after superinfection with cp NADL despite the acute reinfection (data not shown), indicating that superinfection exclusion had not been restored. These results suggest that a cellular alteration or adaptation occurred during prolonged persistent infection leading to a loss of superinfection exclusion.
It was possible that the loss of superinfection exclusion was due to a mixed population of cells, with only some infected by ncp NADLJiv90−. To rigorously rule out this possibility, we examined the percentage of BVDV-positive cells at each passage by flow cytometry with an NS3-specific BVDV antibody. Over a period of five passages, the majority of cells, 75.1 ± 13.0%, remained positive for BVDV antigen, although the ability to exclude superinfecting cp NADL was lost after 3 days of infection (passage 1) when at least 91% of cells were NS3 positive.
To further verify that individual persistently infected cells lost the ability to exclude superinfecting virus, we infected cells with ncp NADLJiv90−pac and maintained infected monolayers under puromycin selection. At 1, 2, 3, 4, and 5 days after acute infection, cells were superinfected with cp NADL. Two days later, supernatants were harvested and titered by plaque assay to measure the yield of cp NADL. Over the 5-day period, the decrease in virus production from the puromycin-selected cells directly correlated with the increase in cp NADL titer from these cells (Fig. 3C). The fact that the cells were maintained under puromycin selection throughout the experiment ensured that cells superinfected with cp BVDV harbored replicating ncp BVDV, albeit at decreasing titers. These experiments strongly suggest that the loss of superinfection exclusion over time seen in persistently infected cells is due to an alteration occurring in the cells that remained persistently infected with BVDV rather than to a subpopulation of uninfected cells.
We also examined whether cells persistently infected with ncp BVDV lose the ability to exclude superinfecting ncp BVDV as well as cp BVDV. Persistently infected MDBK cells were infected with ncp NADLJiv90−luc every 2 to 3 days over a period of five passages. At 24 h postsuperinfection, the cells were harvested to assay for luciferase activity. Although a small increase in luciferase activity was observed after superinfection of persistently infected cells at passage 2 to 5, the average luciferase activity on the persistently infected cells was only 0.69 ± 1.2% of that on the naïve cells and never rose to more than 3% of the naïve cell luciferase activity (data not shown). Therefore, cells persistently infected with ncp BVDV remain efficient at excluding ncp BVDV over time, as measured by this assay.
Superinfecting BVDV is blocked at the level of viral entry.
The lack of BVDV replication could be due to the inability of superinfecting BVDV particles to efficiently enter a BVDV-infected cell and/or the inability of the superinfecting viral genome to be translated and replicated after entry. To examine these two options, ncp NADLJiv90−-infected cells were challenged with NADLJiv90−luc. To assay primary translation of incoming genome RNA, luciferase activity was measured at 6 h postinfection with ncp NADLJiv90−luc. The 6-h time point was chosen by two criteria. First, infection with NADLJiv90−luc in the absence or presence of a potent BVDV-specific RNA-dependent RNA polymerase inhibitor (4), resulted in luciferase activities differing by approximately 2.5-fold at 6 h, compared to a greater than 500-fold difference by 15 h due to replication in the absence of the inhibitor (data not shown). Second, transfection experiments showed no significant difference in luciferase levels between replication-competent and replication-defective BVDV replicons at the 6 h time point (see below).
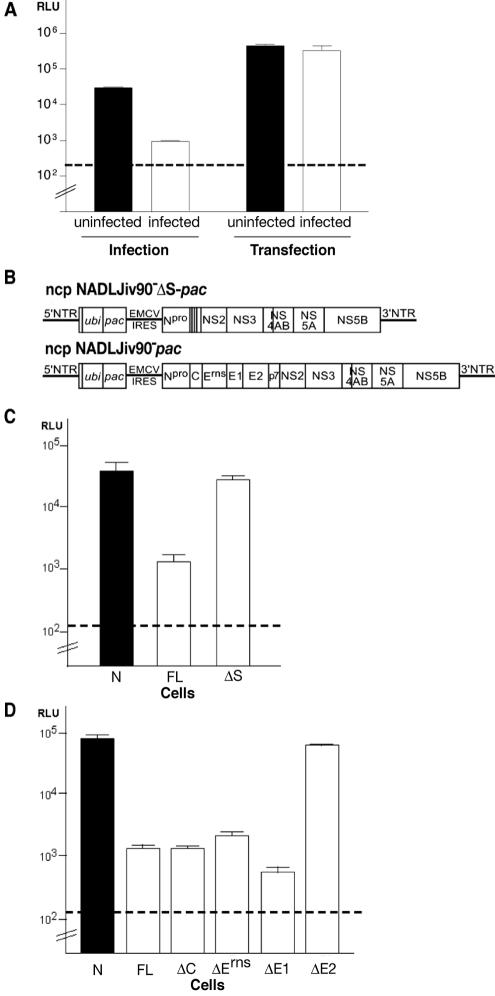
Superinfection of ncp NADLJiv90−-infected cells with ncp NADLJiv90−luc virus resulted in luciferase activity almost 2 logs lower than that from naïve MDBK cells infected with ncp NADLJiv90−luc (Fig. 4A, infection). Transfection of this construct into ncp NADLJiv90−-infected cells showed that the superinfecting genome could be efficiently translated (Fig. 4A, transfection; also see below). Therefore, superinfecting ncp NADLJiv90−luc was unable to deliver its translation-competent RNA genome into ncp NADLJiv90−-infected MDBK cells, indicating a block at the level of entry. Entry is loosely defined here to encompass all of the pretranslation steps.
FIG. 4.
Homologous BVDV superinfecting particles are blocked at the level of viral entry. (A) Naïve (uninfected) and ncp NADLJiv90−-infected (infected) cells were superinfected (Infection) or transfected (Transfection) with ncp NADLJiv90−luc. At 6 h postsuperinfection or transfection, the cells were lysed and luciferase activity was determined. (B) Schematic diagram of ncp NADLJiv90−pac and ncp NADLJiv90−ΔS-pac viral RNAs. (C) ncp NADLJiv90−pac (FL), ncp NADLJiv90−ΔS-pac (ΔS), and naïve (N) cells were superinfected with ncp NADLJiv90−luc. At 6 h postinfection, the cells were lysed and luciferase activity was determined. (D) Naive (N), ncp NADLJiv90−pac (FL), ncp NADLJiv90−ΔC-pac (ΔC), ncp NADLJiv90−ΔErns-pac (ΔErns), ncp NADLJiv90−ΔE1-pac (ΔE1), and ncp NADLJiv90−ΔE2-pac (ΔE2) selected MDBK cells were infected with ncp NADLJiv90−luc. At 6 h postinfection, the cells were lysed and luciferase activity was determined. The dashed lines represent the background level of the assay from naïve MDBK cells.
To investigate the importance of the BVDV structural proteins, an MDBK cell line expressing only the BVDV nonstructural proteins was generated by transfection of a subgenomic ncp NADLJiv90−ΔS-pac replicon and selection with puromycin (Fig. 4B). An MDBK cell line expressing all of the viral proteins was also generated with full-length ncp NADLJiv90−pac RNA (Fig. 4B). Both puromycin-selected cell populations were challenged with ncp NADLJiv90−luc, and the luciferase activity was determined at 6 h postinfection. In ncp NADLJiv90−pac selected cells (Fig. 4C, FL) the luciferase activity after superinfection was more than 50-fold less than in naïve cells (Fig. 4C, N). In contrast, the ncp NADLJiv90−ΔS-pac selected cells were as susceptible as naïve cells for delivery and early translation of luciferase-expressing virion RNA (Fig. 4C, ΔS). These results demonstrated that expression of one or more BVDV structural proteins was required for superinfection exclusion at the level of viral entry.
To investigate which viral structural protein was responsible for exclusion during entry, we generated four puromycin-selected MDBK cell lines. Each harbored a recombinant viral RNA containing an in-frame deletion of each viral structural gene, designated NADLJiv90−ΔC-pac, NADLJiv90−ΔErns-pac, NADLJiv90−ΔE1-pac, and NADLJiv90−ΔE2-pac. The selected cells appeared to express similar amounts of NS2-3 protein, measured by immunoblotting or radioimmunoprecipitation with an anti-NS3-specific antiserum (data not shown). The MDBK cell lines were infected with ncp NADLJiv90−luc viral particles, and luciferase activity at 6 h postinfection was determined.
The luciferase activities obtained from ncp NADLJiv90−ΔC-pac, ncp NADLJiv90−ΔErns-pac, and ncp NADLJiv90−ΔE1-pac mutant cell lines upon superinfection with ncp NADLJiv90−luc virus (Fig. 4D, ΔC, ΔErns, and ΔE1) were similar to that of cells expressing a full complement of structural proteins (Fig. 4D, FL). These results suggest that the viral structural proteins C, Erns, and E1 do not play a major role in exclusion at the level of viral entry. In contrast, ncp NADLJiv90−luc infection of the ncp NADLJiv90−ΔE2-pac selected cells resulted in luciferase activity (Fig. 4D, ΔE2) similar to that obtained from infection of naïve MDBK cells (Fig. 4D, N). Hence, these findings show that deletion of viral glycoprotein E2 was sufficient to abolish the exclusion at the level of viral entry. Interestingly, we observed that although ncp NADLJiv90−luc could be efficiently translated in the ncp NADLJiv90−ΔE2-pac selected cells, there was no amplification of this genome, suggesting that a second, internal block to superinfection might exist.
Transfection of BVDV RNA reveals a second block at replication but not translation.
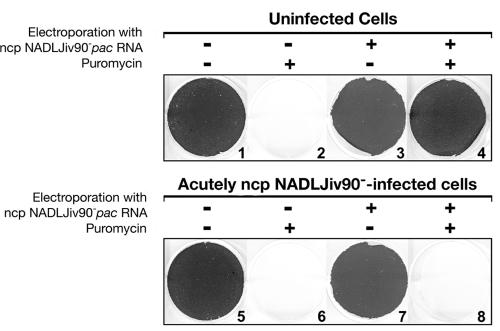
To determine if superinfecting viral RNA would be competent for replication if it were allowed to bypass the viral entry steps, MDBK cells acutely infected with ncp NADLJiv90− were transfected with ncp NADLJiv90−pac RNA. These cells did not survive puromycin selection (Fig. 5, plate 8), indicating that the transfected ncp NADLJiv90−pac RNA had failed to replicate. Naïve MDBK cells transfected with ncp NADLJiv90−pac RNA transcripts were able to grow in medium containing puromycin and eventually became confluent (Fig. 5, plate 4), whereas MDBK cells infected with only ncp NADLJiv90− were unable to survive puromycin selection (Fig. 5, plate 6). Transfection of the alphavirus-based expression vector SINrep3-lacZ (20) indicated that there was no significant difference in transfection efficiency of naïve versus BVDV-infected MDBK cells (data not shown). These findings demonstrate that the ncp NADLJiv90−pac RNA transfected into MDBK cells acutely infected with ncp NADLJiv90− was unable to replicate to a level required for puromycin selection and suggest an additional postentry block to superinfection.
FIG. 5.
Transfected ncp NADLJiv90−pac RNA fails to replicate in ncp NADLJiv90− infected cells. Naïve MDBK cells were either mock infected (plates 1 to 4) or acutely infected with ncp NADLJiv90− (plates 5 to 8). The cells were then mock transfected or transfected with in vitro-transcribed ncp NADLJiv90−pac RNA. The + and − above each column of plates indicate transfection and mock transfection with ncp NADLJiv90−pac RNA, respectively. Following transfection, the cells were incubated in culture medium with and without 5 μg of puromycin per ml as indicated, and viable cells were fixed and stained with crystal violet after 5 days.
Since translation of the incoming genome is a prerequisite for RNA replication, we undertook additional experiments (besides that in Fig. 4A) to examine translation of BVDV RNA transfected into acutely BVDV-infected MDBK cells. To more clearly distinguish between translation of input RNA versus signal due to productive replication, we compared a subgenomic ncp BVDV replicon, ncp NADLJiv90−ΔS-luc, encoding the luc reporter, with its isogenic replication-incompetent derivative, ncp NADLJiv90−ΔS-luc-pol− (Fig. 6A). In naïve MDBK cells transfected with ncp NADLJiv90−ΔS-luc RNA (Fig. 6B, solid circles), the luciferase activity at 6 h posttransfection was similar to that observed with the pol− replicon (Fig. 6B, open circles), indicating (as mentioned earlier) that detectable RNA replication had not taken place at this early time point. Transfection of either ncp replicon into ncp NADLJiv90−-infected MDBK cells resulted in similar luciferase levels at the 6-h time point (Fig. 6B, solid and open squares). Therefore, in ncp NADLJiv90−-infected MDBK cells, transfected ncp NADLJiv90−ΔS-luc viral RNA can be translated as efficiently as in uninfected cells.
FIG. 6.
ncp NADLJiv90−ΔS-luc viral RNA transfected into ncp NADLJiv90−-infected MDBK cells is competent for translation but not for replication. (A) Schematic diagram of ncp NADLJiv90−ΔS-luc and ncp NADLJiv90−ΔS-luc-pol− viral RNAs. (B) We electroporated 8 × 106 naïve (solid circles) or ncp NADLJiv90−-infected MDBK cells (solid squares) with 5 μg of ncp NADLJiv90−ΔS-luc RNA. In parallel, 5 μg of ncp NADLJiv90−ΔS-luc-pol− RNA was electroporated into naïve (open circles) and ncp NADLJiv90−-infected (open squares) cells. The luciferase activity was measured at the indicated time. The dashed line indicates the background level of the assay from naïve MDBK cells. (C) Naïve MDBK cells were transfected with ncp NADLJiv90−ΔS-pac or ncp NADLJiv90−pac RNA and then selected with 5 μg of puromycin per ml. Both ncp NADLJiv90−ΔS-pac selected (solid and open triangles) and ncp NADLJiv90−pac selected cells (solid and open squares) were electroporated with either ncp NADLJiv90−ΔS-luc RNA (solid triangles and squares) or ncp NADLJiv90−ΔS-luc-pol− RNA (open triangles and squares). Naïve cells (solid and open circles) were also electroporated with ncp NADLJiv90−ΔS-luc RNA (solid circles) or ncp NADLJiv90−ΔS-luc-pol− RNA (open circles). After incubation for the indicated times, cell lysates were prepared and luciferase activity was determined.
After 6 h, the luciferase activity in naïve MDBK cells transfected with ncp NADLJiv90−ΔS-luc-pol− RNA gradually decreased over time (Fig. 6B, open circles). In contrast, the luciferase activity from naïve cells transfected with ncp NADLJiv90−ΔS-luc RNA increased dramatically at 16 h posttransfection and was maintained until the final time point at 120 h posttransfection (Fig. 6B, solid circles). In contrast, in ncp NADLJiv90−-infected MDBK cells, both initial activities decreased over time, similarly to naïve MDBK cells transfected with ncp NADLJiv90−ΔS-luc-pol− RNA (Fig. 6B, open circles). Therefore, in ncp NADLJiv90−-infected MDBK cells, viral RNA can be translated, but this RNA fails to replicate. These results demonstrate a second block at the level of BVDV RNA replication. The fact that there is no amplification of luciferase expression from the pol− genome after transfection of ncp NADLJiv90−-infected cells indicated that lethal mutations in the BVDV RNA-dependent RNA polymerase, NS5B, could not be trans-complemented, consistent with previous results (24).
To examine if the BVDV structural proteins were required for superinfection exclusion at the level of RNA replication, subgenomic ncp NADLJiv90−ΔS-pac and full-length ncp NADLJiv90−pac selected cells were transfected with NADLJiv90−ΔS-luc (Fig. 6C, solid triangles) or NADLJiv90−ΔS-luc-pol− (Fig. 6C, open triangles). In both cases, the luciferase activity decreased over time and mirrored that of ncp NADLJiv90−pac selected cells transfected with the luc replicons (Fig. 6C, solid and open squares). These results show that expression of the BVDV structural proteins is not required for exclusion at the level of viral RNA replication.
Level of RNA replication correlates with the extent of superinfection exclusion.
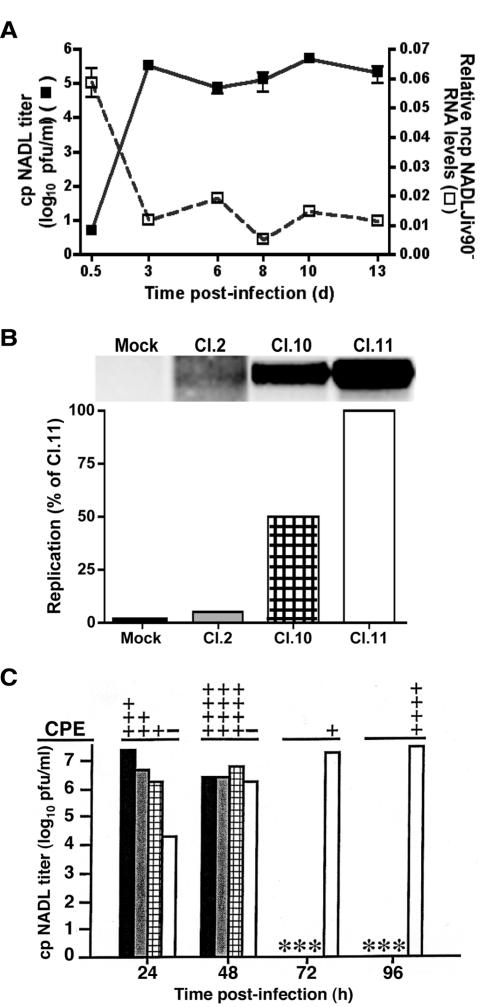
A possible factor in the block at RNA replication could be the level of RNA replication in the infected cell. This hypothesis was supported by the observation that when puromycin-selected cells infected with ncp NADLJiv90−pac were superinfected with cp NADL at various time points post-primary infection, the increase in cp NADL titer over time on the persistently infected cells directly correlated with a decrease in the ncp NADLJiv90−pac titer (Fig. 3C). We also looked directly at RNA levels in persistently infected cells. As superinfection exclusion disappeared in persistently infected cells approximately 3 days postinfection, the appearance of high titers of cp NADL correlated with a sixfold decrease in ncp NADLJiv90− RNA levels as quantitated by real-time quantitative reverse transcription-PCR (Fig. 7A). In later passages the level of ncp NADLJiv90− RNA remained relatively constant, as did the titer of cp NADL on the persistently infected cell monolayers (Fig. 7A).
FIG. 7.
Level of primary viral RNA replication correlates with the degree of superinfection exclusion at viral RNA replication. (A) ncp NADLJiv90−-infected cells were maintained by passaging for a period of 13 days. Every 2 to 3 days, the cells were superinfected with cp NADL and titers were determined by plaque assay, as indicated on the left. The ncp NADLJiv90−-infected cells were also analyzed for levels of ncp NADLJiv90− RNA at each time point. Total cellular RNA was isolated, and BVDV-specific RNA was quantitated with real-time quantitative reverse transcription-PCR. The relative levels of ncp NADLJiv90− RNA, indicated on the right, are expressed as the ratio of copies of BVDV RNA to copies of bovine beta-actin RNA. (B) Cell clones harboring different levels of BVDV RNA were generated by transfection of naïve MDBK cells with ncp NADLJiv90−ΔS-pac or cp NADLΔS-pac RNAs and selection with puromycin. Cl.2 was derived from ncp NADLJiv90−ΔS-pac selected cells, and Cl.10 and Cl.11 were derived from cp NADLΔS-pac selected cells. The three cell clones were metabolically labeled for 6 h with [32P]orthophosphate in the presence of dactinomycin. The viral RNAs were analyzed by gel electrophoresis and visualized by autoradiography. Labeled viral RNA wasquantitated by phosphoimage analysis. The relative levels of RNA replication are expressed in the bar graph as a percentage of the replication in Cl.11, which was set at 100%. (C) Naïve MDBK cells (solid bar), Cl.2 (gray bar), Cl.10 (patterned bar), and Cl.11 (open bar) were infected with cp NADL. At a given time point, culture supernatants were collected and used to determine the virus titer. The extent of CPE caused by cp NADL viral replication was estimated under light microscopy and indicated by +/-. * represents the absence of attached viable cells on culture dishes.
To further examine this possible correlation, we generated MDBK cell lines supporting different levels of viral RNA replication by transfecting puromycin-selectable replicons lacking the structural region to exclude structural protein effects. Since RNA replication of cp NADL has been shown to be higher than that of ncp NADLJiv90− (39), both cp and ncp replicons were used. Naïve MDBK cells were transfected with either ncp NADLJiv90−ΔS-pac or cp NADLΔS-pac viral RNAs and yielded puromycin-resistant colonies that were picked and expanded into cell lines. As might be expected from previous work (48), the replicons within clones Cl.2, Cl.10, and Cl.11 were sequenced and found to contain adaptive mutations in the NS4B-5A region, allowing persistent ncp replication. Some of these mutations have been characterized previously (48); others are the subject of ongoing work (M. Paulson, personal communication). Metabolic labeling of viral RNA with [32P]orthophosphate showed the relatively low level of viral RNA replication in Cl.2 (<5%), harboring the ncp NADLJiv90−ΔS-pac replicon, and higher levels in Cl.10 (50%) and Cl.11 (100%), harboring cp NADLΔS-pac replicons (Fig. 7B).
The cell clones were infected with cp NADL and observed for the extent of CPE and virus yield. At 24 h postinfection, maximum virus titer and clear CPE were observed in naïve MDBK cells (Fig. 7C, solid bar). In contrast, only Cl.2 (Fig. 7C, gray bar) and Cl.10 (Fig. 7C, patterned bar) displayed extensive CPE and reached maximum titer by 48 h postinfection. Cl.11, which had the highest level of ncp BVDV RNA replication, was protected from significant CPE until 72 h postinfection (Fig. 7C, open bar). Therefore, the level of ncp BVDV primary RNA replication strongly correlates with the extent of superinfection exclusion at the RNA replication level.
DISCUSSION
This report examines the phenomenon of superinfection exclusion in BVDV and examines the viral factors involved in this process. Our results indicate that cells acutely infected with BVDV are protected against superinfection with homologous BVDV. These cells, however, remain susceptible to a heterologous virus, vesicular stomatitis virus, suggesting a BVDV-specific exclusion. The ability to exclude superinfecting BVDV is established quickly, within 30 to 60 min after primary infection, but is transient, as persistently infected passaged cells are not efficiently protected. The results of this study show that BVDV superinfection exclusion is mediated by dual mechanisms, one at the level of viral entry and a second at the level of viral RNA replication.
The first mechanism of superinfection exclusion in cells acutely infected with BVDV is a block at the level of entry. This block was dependent upon expression of the BVDV structural proteins. The importance of the BVDV structural proteins in superinfection exclusion is supported by recent studies by Reimann et al., in which cells constitutively expressing the BVDV structural proteins Npro to E2 cannot be efficiently infected with BVDV (49). Our study indicates that expression of BVDV glycoprotein E2 appears to play an essential role in the entry block. The importance of E2 in superinfection exclusion is also consistent with the description of an MDBK cell line expressing E2, called MDBK-E2IRESp7, which was shown to be nonpermissive for BVDV infection (27).
Exclusion at an early stage of viral infection has been shown for a variety of viruses. One well-characterized mechanism is downregulation of the cell surface receptor after primary viral infection. In human immunodeficiency virus infection, the viral Env precursor gp160 binds to CD4 in the endoplasmic reticulum, thereby preventing transport of the viral receptor to the cell surface (17, 28, 56). Additionally, the Env protein of the retrovirus foamy virus was shown to be sufficient for mediating superinfection exclusion in foamy virus-infected cells (9). The influenza virus enzyme neuraminidase removes sialic acid from glycoproteins in the secretory pathway and from the surface of infected cells (3, 33, 46), thereby preventing entry of superinfecting influenza viruses.
Exclusion of a homologous virus can also be due to interference with receptor-mediated surface binding, as shown for respiratory syncytial virus with pseudotyped antigenically related respiratory syncytial virus (54, 55). Furthermore, steps postbinding may be blocked. Multiple mechanisms of vesicular stomatitis virus superinfection exclusion exist at the level of entry, including a reduced rate of endocytic vesicle formation, decreased internalization of bound ligands, and competition between virions for coated pits (52, 66). Based on our findings, it is possible that the block at BVDV entry might be due to a defect in receptor-mediated binding on the plasma membrane, virion internalization, fusion, or nucleocapsid uncoating. The recent identification by Maurer et al. of CD46 as a cellular receptor for BVDV should allow us to assess whether E2-CD46 interactions play a role in superinfection exclusion (36).
We also demonstrate a second mechanism of superinfection exclusion at the level of viral RNA replication. Transfected viral RNAs, although fully competent for translation, were not able to replicate in the ncp NADLJiv90−-infected MDBK cells. This mechanism is consistent with previous observations in other viral systems. For example, temperature-sensitive Sindbis virus mutants in two different RNA-negative complementation groups failed to exclude superinfecting viruses at the nonpermissive temperature, although complementation could be observed (1, 29). These experiments demonstrate that replication of the superinfecting virus was blocked after attachment, penetration, and translation of the superinfecting viral RNA.
Exclusion of BVDV at the level of viral RNA replication did not require expression of the viral structural proteins. Instead, the degree of exclusion correlated with the level of primary viral RNA replication. The mechanism by which existing BVDV RNA replication inhibits superinfecting RNA replication remains to be determined. One possibility is that viral factors from the primary virus might affect replication of the superinfecting virus. For Sindbis virus, production of trans-acting nsP2 protease by the primary virus might process precursors required for replication of the superinfecting virus (31). Alternatively, exclusion might be due to a limiting cellular factor(s). BVDV RNA elements or proteins could interact with limiting cellular factors essential for replication and sequester them during the course of infection, thus reducing their availability for an incoming homologous virus. Studies of hepatitis C virus replication in cell culture show that replication efficiency is decreased in the presence of increasing amounts of transfected replication-defective viral RNA (35). This effect is dependent upon translation of hepatitis C virus replicase proteins and suggests that one or more cellular factors are limiting for hepatitis C virus replication.
Interference at the level of RNA replication may also be related to the availability of subcellular sites for replication. Positive-strand RNA viruses replicate their RNAs in association with existing or virus-modified subcellular membrane organelles. Flavivirus RNA replication, for example, occurs in a specialized membrane compartment surrounding the nucleus (14 to 16, 64, 65). Bromegrass mosaic virus induces spherule-like invaginations into the lumen of the endoplasmic reticulum that are still connected to the cytosol by a narrow channel. Negative-strand RNA molecules are sequestered in these compartments, where they are used as templates for viral genome amplification (51). During acute primary infection, these specialized areas might become saturated with replication complexes, blocking access of the superinfecting viral RNA and preventing its entry into functional replication complexes. Limiting host components and sites of replication may well be virus specific, explaining exclusion of homologous but not heterologous viruses.
The notion that cp BVDV arises from ncp BVDV by a rare RNA recombination event leading to fatal mucosal disease prompts the question of whether persistently ncp BVDV-infected cells might not exclude superinfecting cp BVDV in vivo. If not, how would a cp BVDV variant that arose be propagated? One possibility is that cp BVDV might have a different cell tropism than ncp BVDV. While cp and ncp BVDVs may well have different replication efficiencies in a given cell type, it seems unlikely that these variants would infect different target cells, since they have identical or nearly identical structural proteins. Alternatively, infection may well be dynamic in persistently infected animals, with not all susceptible target cells preinfected with ncp BVDV. Our study showed a loss of superinfection exclusion in ncp BVDV-infected cells after one to two passages. This shows that acutely infected cells that are initially refractory to superinfection may eventually become permissive, providing a reservoir of susceptible cells. This would give the founder virus an initial replication advantage over a superinfecting variant, but this advantage would diminish over time. Another possibility is that superinfection exclusion may not be efficiently established in all cell types.
Besides the studies discussed above, two other papers have examined pestivirus superinfection exclusion. The recent paper by Baigent et al. reported that calf testis cells could be productively superinfected with cp BVDV 48 h after acute infection with ncp BVDV (5). NS3 and CPE became detectable after an additional 48 h, or 4 days post-acute infection (significant delay compared to cp BVDV infection of naïve cells). These data are consistent with the observations reported here. Another earlier study was conducted by Mittelholzer and colleagues for classical swine fever virus (43). Classical swine fever virus is generally noncytopathic and readily establishes persistent infections in cell culture. Persistently infected cultures, serially passaged more than 100 times yielded cp defective interfering RNAs that spread through some of the cultures and killed the majority of the cells. The few surviving cells were still infected but had either lost the cp defective interfering RNA or contained an ncp classical swine fever virus variant that was able to “control” the replication of the remaining defective interfering RNA to a level that was no longer cytopathic. Interestingly, virus production in the persistently infected classical swine fever virus cultures prior to the emergence of the cp defective interfering RNAs was about 100-fold lower than after acute infection, analogous to our results for MDBK cells persistently infected with ncp BVDV. Thus, the emergence of a packaged cp defective interfering RNA variant, like superinfection with cp BVDV, was able to spread through the culture, replicate to high levels, and kill the majority of the cells.
Interesting questions are raised by both of these studies. For example, what is the mechanism that downregulates the replication level of the ncp virus after the acute phase of infection? Our data suggest that cellular changes or alterations contribute to the loss of superinfection exclusion for cp BVDV. Superinfection exclusion was not restored by reinfection of the persistently ncp BVDV-infected cells with the original ncp BVDV. In addition, ncp BVDV from persistently infected cells was capable of establishing superinfection exclusion in naïve MDBK cells. Persistent BVDV infection thus appears to induce a cellular alteration that downregulates ncp virus replication and allows superinfection to occur. The nature of these cellular alterations remains unknown, but it will be interesting to see if this state requires continuing ncp pestivirus replication or if stable epigenetic changes have occurred. Notably, persistently infected cells remained relatively resistant to superinfection by homologous ncp BVDV, in contrast to cp BVDV, over the time tested. It is interesting that the same mechanism does not apply to a superinfecting cp virus (in the case of our BVDV studies) or to the cp classical swine fever virus defective interfering RNA characterized by Mittelholzer et al. In persistently infected cells as opposed to acute infection, both cp and ncp BVDVs may be able to enter the cell more efficiently, encountering lower levels of the E2-mediated block. At the replication level, however, the ncp BVDV remains blocked, whereas the cp BVDV can somehow enter productive replication, possibly due to fundamental differences in its replication efficiency or mechanism.
Acknowledgments
We thank Matthew Evans, Richard Hardy, Margaret MacDonald, and Tina Myers for comments on drafts of the manuscript and Rebecca Moran for excellent technical assistance.
This work was supported by PHS grants CA57973 and AI40034, the Greenberg Medical Research Institute, and a Korea Research Foundation grant (KRF-2001-042-D00071). D.M.T. was supported in part through a Women and Science Fellowship.
REFERENCES
- 1.Adams, R. H., and D. T. Brown. 1985. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J. Virol. 54:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agapov, E. V., C. L. Murray, I. Frolov, L. Qu, T. M. Myers, and C. M. Rice. 2004. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J. Virol. 78:2414-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Air, G. M., and W. G. Laver. 1989. The neuraminidase of influenza virus. Proteins 6:341-356. [DOI] [PubMed] [Google Scholar]
- 4.Baginski, S. G., D. C. Pevear, M. Seipel, S. C. Sun, C. A. Benetatos, S. K. Chunduru, C. M. Rice, and M. S. Collett. 2000. Mechanism of action of a pestivirus antiviral compound. Proc. Natl. Acad. Sci. USA 97:7981-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, J. C. 1987. Bovine viral diarrhea virus: a review. J. Am. Vet. Med. Assoc. 190:1449-1458. [PubMed] [Google Scholar]
- 7.Bazan, J. F., and R. J. Fletterick. 1989. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology 171:637-639. [DOI] [PubMed] [Google Scholar]
- 8.Behrens, S.-E., C. W. Grassmann, H.-J. Thiel, G. Meyers, and N. Tautz. 1998. Characterization of an autonomous subgenomic pestivirus RNA replicon. J. Virol. 72:2364-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg, A., T. Pietschmann, A. Rethwilm, and D. Lindemann. 2003. Determinants of foamy virus envelope glycoprotein mediated resistance to superinfection. Virology 314:243-252. [DOI] [PubMed] [Google Scholar]
- 10.Bolin, S. R., A. W. McClurkin, R. C. Cutlip, and M. F. Coria. 1985. Severe clinical disease induced in cattle persistently infected with noncytopathic bovine viral diarrhea virus by superinfection with cytopathic bovine viral diarrhea virus. Am. J. Vet. Res. 46:573-576. [PubMed] [Google Scholar]
- 11.Bratt, M. A., and H. Rubin. 1968. Specific interference among strains of Newcastle disease virus. II. Comparison of interference by active and inactive virus. Virology 35:381-394. [DOI] [PubMed] [Google Scholar]
- 12.Bratt, M. A., and H. Rubin. 1968. Specific interference among strains of Newcastle disease virus, III Mechanisms of interference. Virology 35:395-407. [DOI] [PubMed] [Google Scholar]
- 13.Brownlie, J., M. C. Clarke, and C. J. Howard. 1984. Experimental production of fatal mucosal disease in cattle. Vet. Rec. 114:535-536. [DOI] [PubMed] [Google Scholar]
- 14.Cauchi, M. R., E. A. Henchal, and P. J. Wright. 1991. The sensitivity of cell-associated dengue virus proteins to trypsin and the detection of trypsin-resistant fragments of the nonstructural protein NS1. Virology 180:659-667. [DOI] [PubMed] [Google Scholar]
- 15.Chu, P. W., and E. G. Westaway. 1987. Characterization of Kunjin virus RNA-dependent RNA polymerase: reinitiation of synthesis in vitro. Virology 157:330-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu, P. W. G., and E. G. Westaway. 1985. Replication strategy of Kunjin virus: Evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology 140:68-79. [DOI] [PubMed] [Google Scholar]
- 17.Crise, B., L. Buonocore, and J. K. Rose. 1990. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J. Virol. 64:5585-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng, R., and K. V. Brock. 1992. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathic bovine viral diarrhea virus strain SD-1. Virology 191:867-869. [DOI] [PubMed] [Google Scholar]
- 19.Deregt, D., Bolin, S. R., van den Hurk, J., Ridpath, J. F., and S. A. Gilbert. 1998. Mapping of a type 1-specific and a type-common epitope on the E2 (gp53) protein of bovine viral diarrhea virus with neutralization escape mutants. Virus Res. 53:81-90. [DOI] [PubMed] [Google Scholar]
- 20.Frolov, I., T. A. Hoffman, B. M. Prágai, S. A. Dryga, H. V. Huang, S. Schlesinger, and C. M. Rice. 1996. Alphavirus-based expression systems: strategies and applications. Proc. Natl. Acad. Sci. USA 93:11371-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frolov, I., M. S. McBride, and C. M. Rice. 1998. cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA 4:1418-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geib, T., C. Sauder, D. Venturelli, C. Hässler, P. Staeheli, and M. Schwemmle. 2003. Selective virus resistance conferred by expression of borna disease virus nucleocapsid components. J. Virol. 77:4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbalenya, A. E., A. P. Donchenko, E. V. Koonin, and V. M. Blinov. 1989. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 17:3889-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grassman, C. W., O. Isken, N. Tautz, and S. E. Behrens. 2001. Genetic analysis of the pestivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J. Virol. 75:7791-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grassmann, C. W., O. Isken, and S. E. Behrens. 1999. Assignment of the multifunctional NS3 protein of bovine viral diarrhea virus during RNA replication: an in vivo and in vitro study. J. Virol. 73:9196-9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, B., C. Liu, J. Lin-Goerke, D. R. Maley, L. L. Gutshall, C. A. Feltenberger, and A. M. Del Vecchio. 2000. The RNA helicase and nucleotide triphosphatase activities of the bovine viral diarrhea virus NS3 protein are essential for viral replication. J. Virol. 74:1794-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada, T., N. Tautz, and H. J. Thiel. 2000. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 74:9498-9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jabbar, M. A., and D. P. Nayak. 1990. Intracellular interaction of human immunodeficiency type 1 (ARV-2) envelope glycoprotein gp160 with CD4 blocks the movement and maturation of CD4 to the plasma membrane. J. Virol. 64:6297-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnston, R. E., K. Wan, and J. H. R. Bose. 1974. Homologous interference induced by Sindbis virus. J. Virol. 14:1076-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao, C. C., A. M. Del Vecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Karpf, A., E. Lenches, E. G. Strauss, J. H. Strauss, and D. T. Brown. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 71:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai, V. C., C. C. Kao, E. Ferrari, J. Park, A. S. Uss, J. Wright-Minogue, Z. Hong, and J. Y. Lau. 1999. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 73:10129-10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamb, R. A., and P. W. Choppin. 1983. The gene structure and replication of influenza virus. Annu. Rev. Biochem. 52:467-506. [DOI] [PubMed] [Google Scholar]
- 34.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 35.Lohmann, V., S. Hoffmann, U. Herian, F. P. F., and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurer, K., T. Krey, V. Moennig, H.-J. Thiel, and T. Rümenapf. 2004. CD46 is a cellular receptor for bovine viral diarrhea virus. J. Virol. 78:1792-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClurkin, A. W., S. R. Bolin, and M. F. Coria. 1985. Isolation of cytopathic and noncytopathic bovine viral diarrhea virus from the spleen of cattle acutely and chronically affected with bovine viral diarrhea. J. Am. Vet. Med. Assoc. 186:568-569. [PubMed] [Google Scholar]
- 38.McKercher, D. G., J. K. Saito, G. L. Crenshaw, and R. B. Bushnell. 1968. Complications in cattle following vaccination with a combined bovine viral diarrhea-infectious bovine rhinotracheitis vaccine. J. Am. Vet. Med. Assoc. 152:1621-1624. [PubMed] [Google Scholar]
- 39.Mendez, E., N. Ruggli, M. S. Collett, and C. M. Rice. 1998. Infectious bovine viral diarrhea virus (Strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J. Virol. 72:4737-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyers, G., N. Tautz, P. Becher, H.-J. Thiel, and B. M. Kümmerer. 1996. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J. Virol. 70:8606-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyers, G., and H.-J. Thiel. 1996. Molecular characterization of pestiviruses. Adv. Virus Res. 47:53-118. [DOI] [PubMed] [Google Scholar]
- 42.Mittelholzer, C., C. Moser, J.-D. Tratschin, and M. A. Hoffman. 1997. Generation of cytopathogenic subgenomic RNA of classical swine fever virus in persistently infected porcine cell culture. Virus Res. 51:125-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mittelholzer, C., C. Moser, J.-D. Tratschin, and M. A. Hofmann. 1998. Porcine cells persistently infected with classical swine fever virus protected from pestivirus-induced cytopathic effect. J. Gen. Virol. 79:2981-2987. [DOI] [PubMed] [Google Scholar]
- 44.Moennig, V., H.-R. Frey, E. Liebler, P. Pohlenz, and B. Liess. 1990. Reproduction of mucosal disease with cytopathogenic bovine viral diarrhea virus selected in vitro. Vet. Rec. 127:200-203. [PubMed] [Google Scholar]
- 45.Myers, T. M., V. G. Kolupaeva, E. Mendez, B. S. G., I. Frolov, C. U. T. Hellen, and C. M. Rice. 2001. Efficient translation initiation is required for replication of bovine viral diarrhea virus subgenomic replicons. J. Virol. 75:4226-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palese, P., K. Tobita, M. Ueda, and R. W. Compans. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397-410. [DOI] [PubMed] [Google Scholar]
- 47.Paton, D. J., J. P. Lowings, and A. D. Barrett. 1992. Epitope mapping of the gp53 envelope protein of bovine viral diarrhea virus. Virology 190:763-772. [DOI] [PubMed] [Google Scholar]
- 48.Qu, L., L. K. McMullan, and C. M. Rice. 2001. Isolation and characterization of noncytopathic pestivirus mutants reveals a role for nonstructural protein NS4B in cytopathogenicity. J. Virol. 75:10651-10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reimann, I., G. Meyers, and M. Beer. 2003. Trans-complementation of autonomously replicating bovine viral diarrhea virus replicons with deletions in the E2 coding region. Virology 307:213-227. [DOI] [PubMed] [Google Scholar]
- 50.Schneider, R., G. Unger, R. Stark, E. Schneider-Scherzer, and H.-J. Thiel. 1993. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science 261:1169-1171. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 52.Simon, K. O., J. J. C. Jr., P. A. Whitaker-Dowling, J. S. Youngner, and C. C. Widnell. 1990. Cellular mechanisms in the superinfection exclusion of vesicular stomatitis virus. Virology 177:375-379. [DOI] [PubMed] [Google Scholar]
- 53.Singh, I. R., M. Suomalainen, S. Varadarajan, H. Garoff, and A. Helenius. 1997. Multiple mechanisms for the inhibition of entry and uncoating of superinfecting Semliki Forest virus. Virology 231:59-71. [DOI] [PubMed] [Google Scholar]
- 54.Steck, F. T., and H. Rubin. 1966. The mechanism of interference between an Avian Leukosis Virus and Rous Sarcoma virus, I Establishment of interference. Virology 29:628-641. [DOI] [PubMed] [Google Scholar]
- 55.Steck, F. T., and H. Rubin. 1966. The mechanism of interference between an Avian Leukosis Virus and Rous Sarcoma virus, II. Early steps of infection by RSV of cells under conditions of interference. Virology 29:642-653. [DOI] [PubMed] [Google Scholar]
- 56.Stevenson, M., C. Meier, A. M. Mann, N. Chapman, and A. Wasiak. 1988. Envelope glycoprotein of HIV induces interference and cytolysis resistance in CD4+ cells: mechanism for persistence in AIDS. Cell 53:483-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzich, J. A., J. K. Tamura, F. Palmer-Hill, P. Warrener, A. Grakoui, C. M. Rice, S. M. Feinstone, and M. S. Collett. 1993. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 67:6152-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamura, J. K., P. Warrener, and M. S. Collett. 1993. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology 193:1-10. [DOI] [PubMed] [Google Scholar]
- 59.Tautz, N., K. Elbers, D. Stoll, G. Meyers, and H.-J. Thiel. 1997. Serine protease of pestiviruses: determination of cleavage sites. J. Virol. 71:5415-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tautz, N., T. Harada, A. Kaiser, G. Rinck, S. Behrens, and H. J. Thiel. 1999. Establishment and characterization of cytopathogenic and noncytopathogenic pestivirus replicons. J. Virol. 73:9422-9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thiel, H.-J., P. G. W. Plagemann, and V. Moennig. 1996. Pestiviruses, p. 1059-1073. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Raven Press, New York, N.Y. [Google Scholar]
- 62.Toth, R. L., P. F. Nettleton, and M. A. McCrae. 1999. Expression of the E2 envelope glycoprotein of bovine viral diarrhea virus (BVDV) elicits virus-type specific neutralizing antibodies. Vet. Microbiol. 65:87-101. [DOI] [PubMed] [Google Scholar]
- 63.Warrener, P., J. K. Tamura, and M. S. Collett. 1993. An RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 67:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wengler, G., G. Wengler, T. Nowak, and E. Castle. 1990. Description of a procedure which allows isolation of viral nonstructural proteins from BHK vertebrate cells infected with the West Nile flavivirus in a state which allows their direct chemical characterization. Virology 177:795-801. [DOI] [PubMed] [Google Scholar]
- 65.Westaway, E. G. 1987. Flavivirus replication strategy. Adv. Virus Res. 33:45-90. [DOI] [PubMed] [Google Scholar]
- 66.Whitaker-Dowling, P. A., J. S. Youngner, C. C. Widnell, and D. K. Wilcox. 1983. Superinfection exclusion by vesicular stomatitis virus. Virology 131:137-143. [DOI] [PubMed] [Google Scholar]
- 67.Wilhelmsen, C. L., S. R. Bolin, J. F. Ridpath, N. F. Cheville, and J. P. Kluge. 1991. Lesions and localization of viral antigen in tissues of cattle with experimentally induced or naturally acquired mucosal disease, or with naturally acquired chronic bovine viral diarrhea. Am. J. Vet. Res. 52:269-275. [PubMed] [Google Scholar]
- 68.Windisch, J. M., R. Schneider, R. Stark, E. Weiland, G. Meyers, and H. J. Thiel. 1996. RNase of classical swine fever virus: biochemical characterization and inhibition by virus-neutralizing monoclonal antibodies. J. Virol. 70:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiskerchen, M., and M. S. Collett. 1991. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology 184:341-350. [DOI] [PubMed] [Google Scholar]
- 70.Xu, J., E. Mendez, P. R. Caron, C. Lin, M. A. Murcko, M. S. Collett, and C. M. Rice. 1997. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J. Virol. 71:5312-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong, W., L. L. Gutshall, and A. M. Del Vecchio. 1998. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J. Virol. 72:9365-9369. [DOI] [PMC free article] [PubMed] [Google Scholar]