Abstract
Introduction
To compare the performance of droplet digital polymerase chain reaction (ddPCR) and plasma next-generation sequencing (NGS) in detecting clearance of plasma EGFR (pEGFR) mutations.
Methods
Patients with treatment-naive advanced EGFR-mutated lung cancer treated with first-line tyrosine kinase inhibitors (TKIs) were included. pEGFR were measured at baseline and first response assessment using ddPCR and NGS. Clearance of pEGFR was defined as undetectable levels after a positive baseline result. Results were correlated with time-to-treatment failure (TTF). In exploratory analysis, corresponding change in carcinoembryonic antigen (CEA) levels was evaluated.
Results
Between January 1, 2020, and December 31, 2021, 27 patients were recruited. Ex19del comprised 74% (20 of 27) and L858R 26% (seven of 27). Osimertinib was used in 59% (16 of 27), dacomitinib 4% (one of 27), and gefitinib/erlotinib 37% (10 of 27). Sensitivity of ddPCR and NGS in detecting pEGFR mutation at baseline was 70% (19 of 27) and 78% (21 of 27), respectively (p = 0.16). All patients with detectable pEGFR by ddPCR were detected by NGS.
At a median of 8 (range 3–24) weeks post-TKI initiation, clearance of pEGFR was achieved in 68% (13 of 19) and 71% (15 of 21) using ddPCR and NGS, respectively. Concordance between ddPCR and NGS was 79% (kappa = 0.513, p = 0.013). Clearance of pEGFR was associated with longer median TTF (not reached versus 6 months, p = 0.03) and median decrease in CEA levels by 70% from baseline.
In another cohort of 124 patients, decrease in CEA levels by greater than 70% within 90 days of TKI initiation was associated with doubling of both TTF and overall survival.
Conclusions
Plasma NGS trended toward higher sensitivity than ddPCR in detecting pEGFR, although both had similar concordance in detecting pEGFR clearance. Our results support using NGS at diagnosis and interchangeability of NGS and ddPCR for monitoring, whereas CEA could be explored as a surrogate for pEGFR clearance.
Keywords: EGFR-mutated NSCLC, ddPCR, NGS, CEA
Introduction
Clearance of plasma EGFR (pEGFR) mutations after commencement of tyrosine kinase inhibitors (TKIs) has been found to be predictive of improved survival among patients with advanced EGFR-mutated NSCLC.1,2 Given the expanding number of first-line treatment options, there is an urgent need to improve risk stratification to help select patients for combination therapy such as TKI plus chemotherapy,3 and clearance of pEGFR is a potential strategy. Although plasma next-generation sequencing (NGS) is increasingly used at diagnosis and to evaluate treatment resistance among advanced NSCLC, cost-effectiveness remains a significant concern.4 To that end, droplet digital polymerase chain reaction (ddPCR) is well established for both diagnosis and monitoring treatment response in advanced EGFR-mutated NSCLC,1,2 with sensitivity and specificity reaching up to 80% and 100%, respectively,5 at a fraction of the cost of NGS. Nevertheless, head-to-head comparisons between the various assays are scarce and there remains equipoise as to which assay is better suited to assessing pEGFR. We sought to compare the performance of ddPCR and Oncomine Pan-Cancer Cell-Free Assay, a 52-gene NGS panel,6 in detecting clearance of pEGFR as a predictor of outcomes to TKI. In exploratory analysis, corresponding change in carcinoembryonic antigen (CEA) levels was evaluated.
Materials and Methods
Data Collection
This study was conducted under the approval of SingHealth Centralised Institutional Review Board. All participants provided written informed consent. Data collection was performed through manual electronic database review.
Study Population
Patients with treatment-naive, advanced-stage EGFR-mutated NSCLC treated with first-line EGFR TKI at the National Cancer Centre Singapore were included. EGFR mutations were prospectively confirmed by NGS on tumor tissue as per institutional standard of care, and only patients with EGFR exon 19 p.E746_A750del (ex19del) and exon 21 p.L858R (L858R) mutations were included. Exclusion criteria included insufficient plasma volumes for both ddPCR and NGS at baseline pretreatment and at time of first radiological response assessment.
In exploratory analysis, an extended cohort of patients with advanced EGFR-mutated (ex19del or L858R) NSCLC with paired CEA results within 90 days of commencing first-line EGFR TKI and minimum 2 years of follow-up was evaluated to correlate CEA dynamics with treatment outcomes.
Patients were followed up from diagnosis to death or date of last follow-up. The cutoff for data analysis was February 27, 2023.
Outcomes
Primary outcome for this study was clearance of pEGFR, defined as undetectable levels of pEGFR at time of response assessment after a positive baseline result with either NGS or ddPCR. Secondary outcome was time-to-treatment failure (TTF) and overall survival (OS). TTF was defined as time from treatment initiation to treatment discontinuation or death (whichever occurred first); surviving patients with no change in systemic therapy were censored at their date of last follow-up. OS was defined as time from initial diagnosis to date of death, with surviving patients censored at their date of last follow-up.
Preparation and Testing of Plasma Samples
Longitudinal plasma samples were serially collected from each patient before and after initiation of EGFR TKI. Plasma processing protocols, library preparation, and sequencing and variant calling algorithms used for ddPCR and plasma NGS (Oncomine Pan-Cancer Cell-Free Assay) are detailed in the Supplementary Appendix.
Statistical Analysis
Categorical variables were summarized as frequency and percentage, and continuous variables were summarized using median with range or mean with SD and range. McNemar’s test was used to compare sensitivity between ddPCR and NGS in detecting pEGFR at baseline. Concordance between ddPCR and NGS was assessed using the kappa-statistic measure of agreement. Chi-square test or Fisher’s exact test was used to assess the association between categorical variables. Survival curves were estimated using Kaplan-Meier method. Differences in survival curves were assessed using log-rank test. Cox regression analyses were performed to assess the association between CEA with clinicopathologic characteristics and survival outcomes. A two-sided p less than 0.05 was considered statistically significant. All analyses were performed in STATA version 16.0.
Results
Patient Characteristics
A total of 27 patients were recruited from January 1, 2020, to December 31, 2021. Patient characteristics are summarized in Supplementary Table 1. Median age at diagnosis was 63 (range: 45–83) years, 19 (70%) were males, and 17 (63%) were never smokers. Ex19del comprised 74% (20 of 27) and L858R 26% (seven of 27). TP53 co-mutations were detected in 41% (11 of 27), and median programmed death-ligand 1 tumor proportion score was 1% (range: 0%–60%). Osimertinib was used in 59% (16 of 27), dacomitinib in 4% (one of 27), and gefitinib/erlotinib in 37% (10 of 27). Partial response (PR) was achieved in 74% (20 of 27) and stable disease in 26% (seven of 27).
Performance of ddPCR Versus NGS
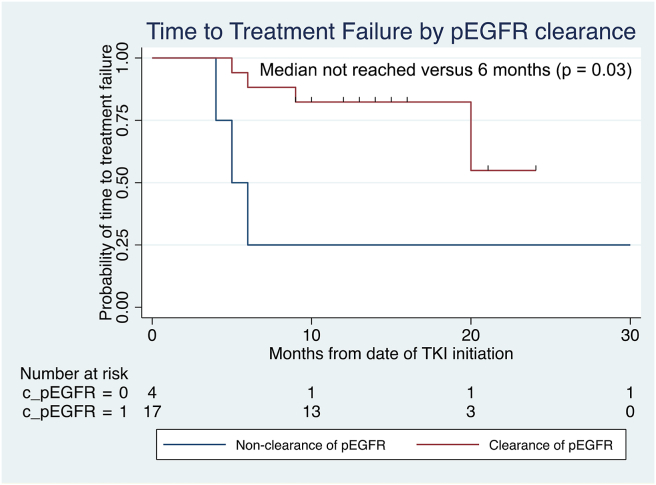
Sensitivity of ddPCR and NGS in detecting pEGFR mutation at baseline was 70% (19 of 27) and 78% (21 of 27), respectively (McNemar chi-square p = 0.16). All patients with detectable pEGFR by ddPCR were detected by NGS. At a median of 8 (range 3–24) weeks post-TKI initiation, clearance of pEGFR was achieved in 68% (13 of 19) and 71% (15 of 21) using ddPCR and NGS, respectively, corresponding to persistence of pEGFR in 22% (six of 27) of patients regardless of assay used. The concordance between ddPCR and NGS was 79% (kappa = 0.513, p = 0.013). At a median follow-up of 19 (range: 13–37) months, clearance of pEGFR by either ddPCR or NGS was associated with longer median TTF (not reached versus 6 months, log-rank p = 0.03), as found in Figure 1.
Figure 1.
TTF by clearance of pEGFR status, revealing significantly improved TTF among patients with clearance of pEGFR. pEGFR, plasma EGFR mutation; TKI, tyrosine kinase inhibitor; TTF, time-to-treatment failure.
Correlation With CEA Levels
Paired CEA results were available for 15 of 27 (56%) patients, of which 12 of 15 (80%) were raised at baseline (defined as >4.8 ng/mL as per institutional standards). Median baseline CEA was 15.7 ng/mL (range <1.8–5974). All three patients with undetectable baseline pEGFR by NGS had CEA levels less than 20 ng/mL. There was no correlation between raised baseline CEA levels and EGFR mutation allele frequency by NGS or ddPCR (p = 0.52 and p = 0.64, respectively).
Clearance of pEGFR was achieved in nine patients with paired CEA results, which was associated with median decrease in CEA levels by 70% from baseline (range 57%–81%). Of the three patients who had persistently detected pEGFR, corresponding change in CEA levels was 31.8 ng/mL to 9 ng/mL (75% decrease), 9.8 ng/mL to 3.7 ng/mL (62% decrease), and 1.8 ng/mL to 2.0 ng/mL (11% increase), respectively.
Survival Outcomes Analyzed by Change in CEA Levels
In exploratory analysis, change in CEA levels was correlated with survival outcomes to first-line EGFR TKI in a separate cohort of patients. A total of 124 patients diagnosed between May 12, 2008, and December 1, 2020, had paired CEA results at baseline and within 90 days of starting EGFR TKI. For patients with multiple CEA results, the nadir reading was used for analysis.
Patient characteristics and treatment responses are summarized in Supplementary Table 2. CEA was raised at baseline in 74% (92 of 124), with median levels of 32.5 (range 1.2–1952) ng/mL. Baseline CEA levels greater than 30 ng/mL were significantly associated with age above 65 years (64% versus 43%, p = 0.02), liver metastases (93% versus 48%, p < 0.01), and bone metastases (77% versus 30%, p < 0.01). There was no significant association between baseline CEA levels and smoking status (p = 0.85), sex (p = 0.12), EGFR mutation subtype (p = 0.57), or brain metastases (p = 0.20).
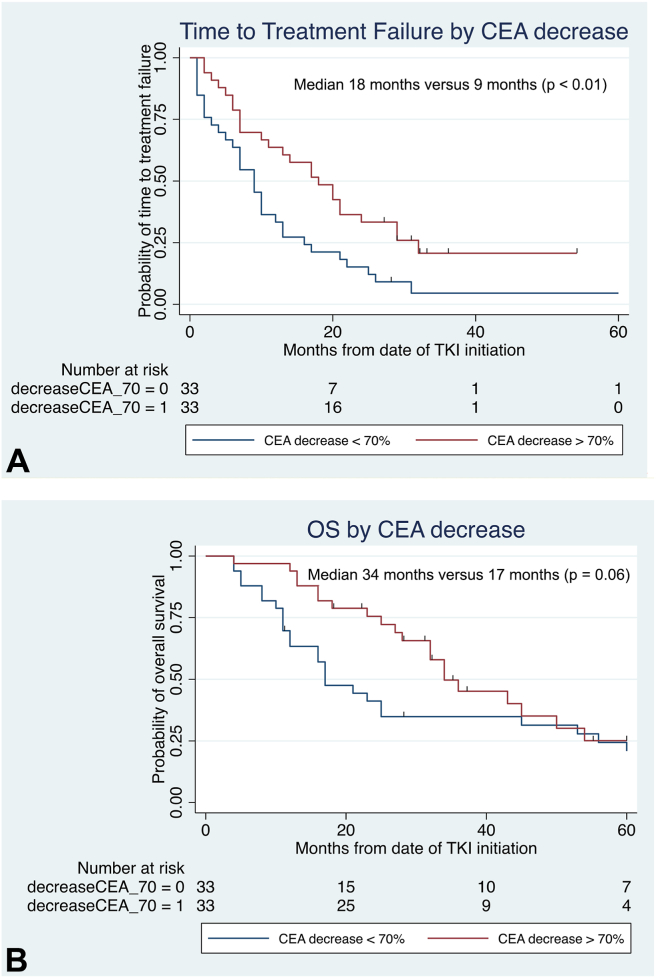
Median time from TKI initiation to CEA nadir was 54 (range 14–90) days. Among 66 of 124 (53%) patients with baseline CEA levels greater than 30 ng/mL, decrease in CEA levels by greater than 70% within 90 days of TKI initiation was significantly correlated with improved TTF (median 18 months versus 9 months, p < 0.01) as found in Figure 2A. These patients were more likely to achieve PR compared with patients who did not have decrease in CEA level by greater than 70% (58% versus 25%, Fisher’s exact p = 0.02). OS was similarly doubled among patients with decrease in CEA levels by greater than 70% as found in Figure 2B, although this was of borderline statistical significance (median 34 mo versus 17 mo, p = 0.06). In contrast, seven of 66 (11%) patients whose CEA levels increased after commencing TKI experienced shorter TTF (median 10 mo versus 13 mo, p = 0.06) and significantly worse OS (median 11 mo versus 34 mo, p < 0.01) as compared with patients with decrease in CEA levels post-TKI.
Figure 2.
(A) TTF by decrease in CEA levels and (B) OS by decrease in CEA levels, both revealing improved survival among patients with CEA level decrease greater than 70% from baseline. CEA, carcinoembryonic antigen; OS, overall survival; TTF, time-to-treatment failure.
Discussion
We found that plasma NGS trended toward higher sensitivity than ddPCR in detecting EGFR mutations at baseline among patients with advanced EGFR-mutated NSCLC, although both had similar concordance in detecting clearance of pEGFR. Our results are consistent with previous studies in the treatment resistance setting, revealing that NGS has slightly higher sensitivity than ddPCR although both tests are highly concordant and correlate well with tissue-based results.7,8 Given the paucity of data comparing the performance of ddPCR against NGS for detecting pEGFR in the treatment-naive setting, relevant considerations in the real-world include cost, turnaround time, and accessibility. Although NGS has the potential to provide more information such as co-mutations, it is disadvantaged by higher costs, longer turnaround time, and more complex bioinformatics analyses. ddPCR, however, is limited by the ability to detect limited genomic aberrations.7 Our results support performing NGS at diagnosis and the interchangeability of NGS and ddPCR in monitoring response to EGFR TKI, for which ddPCR may be preferred given lower cost and shorter turnaround time.
Clearance of pEGFR by either ddPCR or NGS was significantly predictive of improved TTF, similar to prior reports.1, 2 Although FLAURA2 revealed improved progression-free survival with the addition of chemotherapy to first-line osimertinib, this is at the expense of added toxicities and the need for 3-weekly hospital visits for intravenous infusions.3 Whether persistence of pEGFR after osimertinib initiation can be used as a biomarker to predict the benefit of adding chemotherapy to osimertinib is currently being evaluated prospectively in a randomized phase 2 trial (NCT04410796). Besides the potential to adapt treatment strategies on the basis of clearance of pEGFR, longitudinal plasma profiling also avails the opportunity to detect T790M among patients treated with first-/second-generation EGFR TKI before radiological progression and hence facilitate earlier switch to osimertinib. This strategy was evaluated in the APPLE trial, with promising results albeit limited by small patient numbers.9 Considering that both ddPCR and NGS may not be widely available and the challenges of real-time longitudinal monitoring in the clinical setting, we explored the role of CEA as a surrogate measure of pEGFR clearance.
Elevated CEA levels are associated with EGFR-mutated NSCLC, and dynamic changes have been found to correlate with TKI response, although factors that predict for elevated baseline CEA levels remain poorly understood.10 We found that age above 65 years, liver metastases, and bone metastases were significantly associated with baseline CEA levels greater than 30 ng/mL. Notably, decrease in CEA level from baseline by greater than 70% within 90 days of TKI initiation was associated with doubling of both TTF and OS and achieving PR in these patients. Conversely, patients with increase in CEA levels post-TKI experienced inferior survival outcomes. A study of 51 patients found that patients with CEA level decrease of greater than or equal to 23% from baseline post-EGFR TKI had a higher rate of pEGFR clearance on NGS compared with patients with less than 23% decrease (70.6% versus 35.3%, p = 0.016), although survival outcomes by CEA levels were not analyzed.11 Taken together, our results support exploring the role of decrease in CEA levels as a surrogate measure for clearance of pEGFR, particularly when access to NGS or ddPCR is a concern.
Limitations of our study include the small sample size, retrospective nature, and not all patients had corresponding NGS, ddPCR, and CEA data available at the same time points. Uncommon EGFR mutations were not evaluated, and being a single-center study could also limit the generalizability of the data, although reassuringly our results are generally consistent with what has been reported in the literature.
Conclusions
In conclusion, we found that plasma NGS trended toward higher sensitivity than ddPCR in detecting pEGFR at baseline, although both had similar concordance in detecting clearance of pEGFR. Clearance of pEGFR by either ddPCR or NGS was predictive of improved TTF. Among patients with baseline CEA greater than 30 ng/mL, decrease in CEA levels from baseline by greater than 70% within 90 days of TKI initiation was associated with doubling of both TTF and OS. Our results support using NGS at diagnosis and the interchangeability of NGS and ddPCR in monitoring response to EGFR TKI, whereas decrease in CEA levels could be explored as a surrogate for pEGFR clearance.
CRediT Authorship Contribution Statement
Stephanie P. L. Saw: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Writing—original draft, Writing—review and editing.
Gek San Tan: Investigation, Data curation, Formal analysis, Software, Writing—original draft.
Wei Chong Tan: Data curation, Investigation, Writing—review and editing.
Aaron C. Tan: Investigation, Writing—review and editing.
Gillianne G. Y. Lai: Investigation, Writing—review and editing.
Darren W. T. Lim: Investigation, Writing—review and editing.
Ravindran Kanesvaran: Investigation, Writing—review and editing.
Wan Ling Tan: Investigation, Writing—review and editing.
Sze Huey Tan: Formal analysis, Supervision, Visualization, Writing—review and editing.
Tony Kiat Hon Lim: Investigation, Writing—review and editing.
Anders J. Skanderup: Data curation, Writing—original draft, Writing—review and editing.
Daniel S. W. Tan: Conceptualization, Data curation, Funding acquisition, Supervision, Writing—original draft, Writing—review and editing.
Acknowledgments
Data collection for this study was conducted through the Lung Cancer Consortium Singapore. This work was supported by grants from the National Medical Research Council of Singapore (NMRC/TCR/007-NCC/2013 and NMRC/OFLCG/002-2018).
Footnotes
Disclosure: Dr. Saw reports receiving grant support from AstraZeneca and Guardant Health; consulting fees from Pfizer and Bayer; honoraria from AstraZeneca and Merck Sharp & Dohme; and travel support from Merck Sharp & Dohme and AstraZeneca, outside the submitted work. Dr. A. Tan reported receiving consulting fees from Amgen, Bayer, and Pfizer and honoraria from Amgen, Janssen, Pfizer, Juniper Biologics, and Guardant Health AMEA, outside the submitted work. Dr. Lai reported receiving honoraria from AstraZeneca; receiving travel support from DKSH; serving on the advisory board for Amgen; and receiving grants from Amgen, Merck, AstraZeneca, Pfizer, and Roche, outside the submitted work. Dr. D.W.T. Lim reported receiving grants from Bristol-Myers Squibb and travel support from Taiho Pharmaceuticals and Pfizer, outside the submitted work. Dr. Kanesvaran reported receiving honoraria from AstraZeneca, Astellas, and Bristol-Myers Squibb and travel support from Merck and AstraZeneca, outside the submitted work. Dr. W.L. Tan reported receiving grants from AstraZeneca; honoraria from Amgen, Merck, and Novartis; and travel support from AstraZeneca, Ipsen, Boehringer Ingelheim, Bristol-Myers Squibb, and DKSH, outside the submitted work. Dr. T.K.H. Lim reported receiving honoraria from Bayer, Amgen Oncology, AstraZeneca, Merck Sharp & Dohme, Johnson and Johnson, Novartis, and Amoy Diagnostics, outside the submitted work. Dr. D.S.W. Tan reported receiving grants from ACM Biolabs, Bayer, Pfizer, AstraZeneca, and Amgen and consulting fees from Amgen, AstraZeneca, DKSH, Novartis, Boehringer Ingelheim, Bayer, GlaxoSmithKline, Merck, Pfizer, Roche, and Takeda, outside of the submitted work. The remaining authors declare no conflict of interest.
Cite this article as: Saw SPL, Tan GS, Tan WC, et al. Brief report: droplet digital polymerase chain reaction versus plasma next-generation sequencing in detecting clearance of plasma EGFR mutations and carcinoembryonic antigen levels as a surrogate measure. JTO Clin Res Rep. 2023;4:2023;4:100599.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2023.100599.
Supplementary Data
References
- 1.Zhou C., Imamura F., Cheng Y., et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR-TKIs in the FLAURA trial. J Clin Oncol. 2019;37(suppl 15) 9020–9020. [Google Scholar]
- 2.Buder A., Hochmair M.J., Setinek U., Pirker R., Filipits M. EGFR mutation tracking predicts survival in advanced EGFR- mutated non-small cell lung cancer patients treated with osimertinib. Transl Lung Cancer Res. 2020;9:239–245. doi: 10.21037/tlcr.2020.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janne P., Planchard D., Cheng Y., et al. PL03.13 osimertinib with/without platinum-based chemotherapy as first-line treatment in patients with EGFRm advanced NSCLC (FLAURA2) J Thorac Oncol. 2023;18:S36–S37. [Google Scholar]
- 4.Yang S.C., Lin C.C., Chen Y.L., Su W.C. Economic analysis of Tissue-First, plasma-first, and complementary NGS approaches for treatment-naïve metastatic lung adenocarcinoma. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oxnard G.R., Paweletz C.P., Kuang Y., et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.So M.K., Park J.H., Kim J.W., Jang J.H. Analytical validation of a pan-cancer panel for cell-free assay for the detection of EGFR mutations. Diagn Basel Switz. 2021;11:1022. doi: 10.3390/diagnostics11061022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding P.N., Becker T., Bray V., et al. Plasma next generation sequencing and droplet digital PCR-based detection of epidermal growth factor receptor (EGFR) mutations in patients with advanced lung cancer treated with subsequent-line osimertinib. Thorac Cancer. 2019;10:1879–1884. doi: 10.1111/1759-7714.13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steendam C.M.J., Atmodimedjo P., de Jonge E., et al. Plasma cell-free DNA testing of patients with EGFR mutant non–small-cell lung cancer: droplet digital PCR versus next-generation sequencing compared with tissue-based results. JCO Precis Oncol. 2019;3:1–9. doi: 10.1200/PO.18.00401. [DOI] [PubMed] [Google Scholar]
- 9.Remon J., Besse B., Aix S.P., et al. Osimertinib treatment based on plasma T790M monitoring in patients with EGFR-mutant non-small cell lung cancer (NSCLC): EORTC Lung Cancer Group 1613 APPLE phase II randomized clinical trial. Ann Oncol. 2023;34:468–476. doi: 10.1016/j.annonc.2023.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y., Song P., Li H., Jia H., Zhang B. Elevated serum CEA levels are associated with the explosive progression of lung adenocarcinoma harboring EGFR mutations. BMC Cancer. 2017;17:484. doi: 10.1186/s12885-017-3474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng J., Wang Y., Hu C., et al. Predictive value of early kinetics of ctDNA combined with cfDNA and serum CEA for EGFR-TKI treatment in advanced non-small cell lung cancer. Thorac Cancer. 2022;13:3162–3173. doi: 10.1111/1759-7714.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.