Abstract
Bunyamwera virus (BUNV) is the prototype of the Bunyaviridae family of tri-partite negative-sense RNA viruses. The three BUNV segments possess 3′ and 5′ nontranslated regions (NTRs) that signal two RNA synthesis activities: (i) transcription to generate mRNAs and (ii) replication to generate antigenomes that are replicated to yield further genomes. While the genome acts as a template for synthesis of both transcription and replication products, the antigenome allows synthesis of only replication products, with mRNAs being undetectable. Here, we investigate the basis for the fundamentally different signaling abilities of genomic and antigenomic strands. We show that the identity of only nucleotide position 9 within the genomic 3′ NTR is critical for the different RNA synthesis characteristics of genomic and antigenomic strands, thus identifying this nucleotide as an essential component of the transcription promoter. This nucleotide is distinctive, as it interrupts an unbroken run of conserved complementary nucleotides within the 3′ and 5′ NTRs of all three segments. Our results show that the conserved mismatched arrangement of this nucleotide plays no detectable role in signaling transcription. Instead, we show that the transcription-signaling ability of this position is entirely dependent on its nucleotide identity. We further show that while a U residue at 3′ position 9 is strongly preferred for transcription activity in the context of the genomic promoter, it does not signal transcription in the context of the antigenomic promoter. Therefore, our results show that the identity of 3′ position 9 is crucial for signaling BUNV transcription; however, it is not the sole determinant.
The Bunyaviridae family of segmented negative-sense RNA viruses consists of five genera, namely, Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus. Many members of this family are associated with serious human disease and consequently are described by the Centers for Disease Control and Prevention as category A priority pathogens. Examples include Rift Valley fever phlebovirus, Crimean-Congo hemorrhagic fever nairovirus, and the reassortant Ngari orthobunyavirus (13, 24, 33). In addition, many bunyaviruses are categorized as emerging viral pathogens, also capable of causing human disease with fatal consequences, such as the La Crosse, Cache Valley Fever, and Oropouche orthobunyaviruses and Sin nombre, Hantaan, and Andes hantaviruses.
Bunyamwera virus (BUNV) is the prototype of the genus Orthobunyavirus and the family Bunyaviridae. Many features of BUNV molecular and cellular biology are common to other bunyaviruses, and consequently, BUNV is recognized as a model for studying the many serious human pathogens within the family Bunyaviridae.
The BUNV genome consists of three segments of negative-sense RNA designated small (S), medium (M), and large (L). The S segment encodes the nucleocapsid (N) and nonstructural (NSs) proteins from overlapping reading frames on the same mRNA (12, 20, 21). The M segment encodes the surface glycoproteins G1 and G2 and also the NSm protein of unknown function, again from a single mRNA that encodes a polyprotein precursor that is subsequently cleaved (10, 21, 23). The L segment encodes the RNA-dependent RNA polymerase (RdRp) (11).
The coding regions of each segment are surrounded at both the 3′ and 5′ ends by nontranslated regions (NTRs) that extend for 50 to 85 nucleotides (nt) at the 3′ genomic end and 100 to 174 nt at the genomic 5′ end. The 3′ NTRs of each segment are identical in sequence for the first 11 nt, as are the corresponding 11 nt from each of the 5′ NTRs. In addition, the 3′ and 5′ NTRs of each segment exhibit nucleotide complementarity up to positions 15, 18, and 19 for the S, M, and L segments, respectively. For all three segments, this nucleotide complementarity is unbroken, except at one position, which is a conserved U-G mismatched pairing between nucleotides at 3′ and 5′ position 9.
The BUNV 3′ and 5′ NTRs contain signals that direct the BUNV RdRp to perform two distinct RNA synthesis activities: (i) transcription to generate a single mRNA and (ii) replication to generate a positive-sense antigenome that can in turn be replicated to generate further genomic strands. The BUNV genome and antigenome are complementary in sequence and equal in size. In contrast, the S, M, and L mRNAs of bunyaviruses are different in size from their corresponding templates. This is due to a 3′ truncation of ∼100 (S mRNA) or 40 (M and L mRNAs) nt (6, 8, 14) and an extension of between 12 and 17 nt, which comprises a capped oligoribonucleotide derived from host cell mRNAs (27). These capped 5′ extensions are derived from host-cell mRNAs and are thought to be generated by an endonucleolytic activity of the bunyavirus RdRp (9, 22, 27) in a process similar to that which results in the characteristic 5′ cap structures of influenza virus mRNAs (7, 28, 34).
While the BUNV genome serves as a template for both transcription and replication activities, the BUNV antigenome only signals replication activity (27). This strategy of RNA synthesis, in which transcription occurs only from the negative sense genome, is not universally adopted by all Bunyaviridae family members. Some members of the genera Phlebovirus and Tospovirus exhibit an ambisense strategy in which transcription occurs on both genomic and antigenomic templates of one or more segments, allowing access to open reading frames within both genomic and antigenomic strands. It has also been reported that La Crosse virus, a member of the genus Orthobunyavirus, generates negative-sense transcription products from its antigenomic strand (25), although at a sixfold-reduced abundance compared to mRNAs generated from the genome.
The finding that the BUNV genome is used as a template for transcription whereas transcription by the antigenome is not detected (27) indicates that the BUNV genomic and antigenomic templates are recognized differently by the BUNV RNA synthesis machinery. This disparity is likely due to differences between the nucleotide sequences of the RNA synthesis promoters located within the 3′ and 5′ NTRs on the genomic and antigenomic strands. However, the molecular basis for this fundamental difference is unknown.
The RNA synthesis promoters of bunyavirus members are poorly characterized. Functional analysis of the RNA synthesis promoter of Rift Valley fever virus has shown that the first 13 nt of the 3′ genomic terminus are sufficient for RNA synthesis activity (36). A limited mutagenic analysis of these 13 nt showed that nt 3 to 8 and position 13 were particularly sensitive to alteration. A more extensive study of RNA synthesis promoter activity using Uukuniemi virus (16) showed the importance of nucleotides located at 3′ positions 2, 3, 4, and 8 and 5′ positions 3, 4, 5, and 8 (nucleotide positions are counted from the respective termini). However, these two studies assayed RNA synthesis indirectly by detecting the translation product of a reporter gene. As replication or transcription ultimately affects the accumulation of reporter gene mRNAs, these studies were unable to determine whether these nucleotides were components of the replication or transcription promoters.
To more accurately investigate BUNV RNA synthesis, we recently described the use of a BHK cell-based assay that allows direct detection of BUNV replication and mRNA transcription products either by direct visualization of metabolically labeled RNAs separated by gel electrophoresis or by using primer extension analysis (3, 4). This assay offers a fundamental advantage over methods that use reporter gene analysis to measure RNA synthesis in that it allows unambiguous distinction among BUNV genomes, BUNV antigenomes, and BUNV mRNAs. We can therefore functionally dissect the role of each nucleotide within the BUNV promoter in promoting either replication or transcription activities.
Previously, we showed that the RNA replication abilities of the three BUNV segments were not equal, with the relative replication abilities being M > L > S (3). We further showed that these different RNA replication abilities were signaled by segment-specific sequences located between nucleotide positions 17 and 23 within both 3′ and 5′ NTRs, thus for the first time identifying nucleotides specific to the BUNV replication promoter. More recently, we used this assay to show that the promoter for BUNV RNA replication required cooperation between the 3′ and 5′ NTRs, and we provided evidence showing that this cooperation was likely due to basepairing interactions driven by the extensive degree of nucleotide complementarity present within 3′ and 5′ NTRs (4).
Here, we extend these studies by investigating the nucleotide composition of the BUNV mRNA transcription promoter. Our results show that a single conserved nucleotide difference between genomic and antigenomic strands is a major determinant in signaling transcription. Therefore, for the first time, we identify a critical component of the BUNV transcription promoter.
MATERIALS AND METHODS
Plasmid constructions.
The previously described plasmid pBUN-S(ren) contained the entire 3′ and 5′ NTR sequences of the BUNV S segment (3). These sequences surrounded a 936-nt cDNA encoding the Renilla luciferase protein and were flanked by sequences of the T7 RNA polymerase promoter and the hepatitis delta virus self-cleaving ribozyme. The T7 RNA polymerase product generated from this construct was subsequently cleaved by the ribozyme generating an antigenomic sense RNA having two additional non-BUNV residues at the 5′ end and an authentic 3′ end. All other plasmids used in this study were derived from pBUN-S(ren) using Quikchange mutagenesis (Stratagene, La Jolla, Calif.) with mutagenic oligonucleotides (Operon, Inc., Valencia, Calif.). All nucleotide changes were confirmed as correct by DNA sequence analysis.
Transfections.
BUNV RNAs were generated from cDNA-derived BUNV-specific genome analogs as previously described (3). Briefly, 2-μg quantities of plasmids expressing the BUNV S and L segment coding regions were transfected along with 6 μg of genome analog-expressing plasmid into BHK-21 cells previously infected with the vaccinia virus recombinant vTF7-3. BUNV-specific RNAs were metabolically labeled by incubating monolayers with [3H]uridine (33 μCi/ml; Perkin-Elmer Life Sciences Inc., Boston, Mass.) in the presence of actinomycin D (10 mg/ml; Sigma Chemical Co., St. Louis, Mo.) 12 h after transfection. After a 6-h labeling period, total cellular RNAs were harvested using the RNeasy procedure (QIAGEN Inc., Valencia, Calif.).
RNA analysis.
Metabolically labeled RNAs were separated using agarose-urea gel electrophoresis and visualized by fluorography and autoradiography. The RNA synthesis activity of genomic or antigenomic strands was also determined using primer extension analysis as described previously (3). Briefly, positive-sense RNAs were detected using the 33P end-labeled polyacrylamide gel electrophoresis-purified oligodeoxynucleotide primer BUNMRNA (5′-GAGCCTTTAATGACCTTCTGTTGG-3′), whereas negative-sense RNAs were detected using the 33P end-labeled polyacrylamide gel electrophoresis-purified oligodeoxynucleotide RENSEQ (5′-ATCAAATCGTTCGTTGAGCGAG-3′) or minus-80 (5′-GGTTGGGGACAGAAAGACAGCGGGC-3′). The labeled primer extension products were separated using a standard 6% sequencing gel and visualized by autoradiography. Sequence ladders were generated by modified T7 DNA polymerase (Sequenase 2.0; U.S. Biochemical Corp., Cleveland, Ohio) using the appropriate end-labeled primer in combination with plasmid pBUN-S(ren). Densitometric analysis was performed using a Howtek Scanmaster 3 scanner controlled by Pdi Quantity One software. For each RNA band, boundaries were manually defined, and a local lane background density was subtracted. The transcription ability of a template was normalized to the abundance of its corresponding genomic RNA strand.
RESULTS
The BUNV antigenomic RNA synthesis promoter does not signal transcription.
A previous study suggested that the BUNV antigenome does not signal transcription, as shown by the inability to detect negative-sense RNAs having the characteristic 5′ BUNV mRNA extensions (27). However, this conclusion was based on sequencing a limited number of reverse transcription-PCR-generated cDNAs, so the existence of so-called “anti-mRNAs” at low abundance could not be ruled out. We wanted to reexamine the ability of the BUNV antigenome strand to signal transcription using the alternative approach of primer extension analysis. This approach allowed us to analyze a more numerous RNA population and thus to provide a more accurate assessment of the transcription-signaling ability of the antigenomic strand. To achieve this aim, we analyzed the RNA synthesis activity of the previously described genome analog template BUN-S(ren) (3), which contained the entire S segment 3′ and 5′ NTRs surrounding a heterologous sequence (Renilla luciferase).
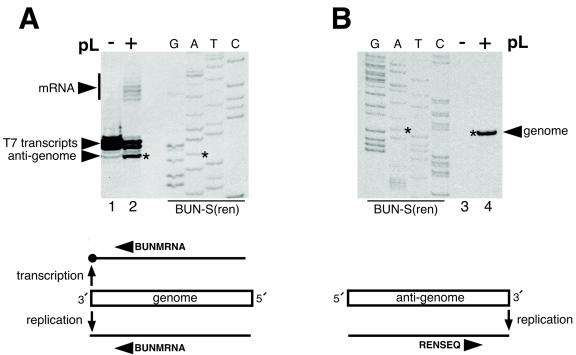
RNAs generated from the negative-sense genomic strand of BUN-S(ren) were detected by primer extension using the negative-sense oligonucleotide BUNMRNA. This analysis detected the BUNV antigenome replication product and the characteristic 5′-extended BUNV mRNAs (Fig. 1A, lane 2) and, in addition, detected the primary T7 RNA polymerase transcription products from plasmid pBUN-S(ren). Primer extension detects two major T7 RNA polymerase products that differ in size by a single nucleotide, which is likely due to the addition of a 5′ cap by the recombinant vTF7-3 that is present in all transfections. These findings showed that the genomic RNA template contained a promoter that was capable of signaling both RNA replication and mRNA transcription (Fig. 1A, schematic).
FIG. 1.
Primer extension analysis of RNA synthesis activities of genomic and antigenomic promoters. RNAs were harvested from vTF7-3-infected BHK-21 cells transfected with a cDNA expressing the BUN-S(ren) genome analog and either BUNV S and L support plasmids (pL) (+) or BUNV S support plasmid alone (−). (A) Positive-sense RNAs generated by the genomic strand of BUN-S(ren) were analyzed by primer extension analysis using the negative-sense oligonucleotide BUNMRNA. The variously sized mRNA 5′ extensions are marked with a vertical bar. (B) Negative-sense RNAs generated by the antigenomic strand of BUN-S(ren) were analyzed using the positive-sense oligonucleotide RENSEQ. For each panel, the cDNA expressing BUN-S(ren) was sequenced using the corresponding end-labeled primer to act as a size marker, and the genomic terminal nucleotide is marked with an asterisk. The abilities of genome and antigenome templates to transcribe and replicate are schematically summarized below each corresponding autoradiograph.
We next performed primer extension analysis on RNAs generated from the positive-sense antigenomic RNA strand of BUN-S(ren) using the positive-sense oligonucleotide RENSEQ. This analysis detected a single product that corresponded to the negative-sense genomic RNA (Fig. 1B, lane 4) but did not detect products corresponding to negative-sense transcripts. This analysis indicated that the antigenomic strand of BUN-S(ren) contains a promoter only for RNA replication and not for mRNA transcription (Fig. 1B, schematic), thus confirming previous findings by others (27).
Identification of nucleotides critical for transcription-signaling ability.
The results described above showed that the BUNV genomic strand contains an RNA synthesis promoter active for transcription and replication whereas the antigenomic RNA synthesis promoter only signals replication. The nucleotide differences that exist between the promoter regions of these two strands must therefore be involved in signaling transcription. The goal of this study was to identify these nucleotides and thus to define the BUNV transcription promoter.
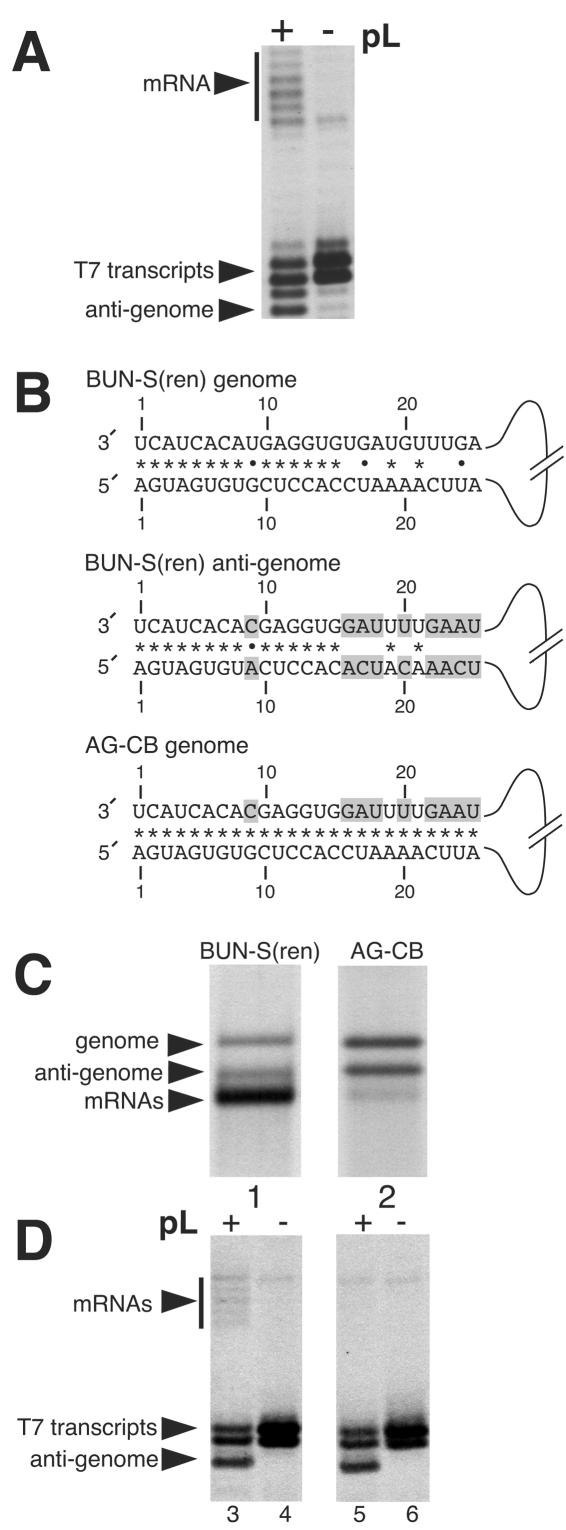
We previously showed that the BUNV RNA replication promoter comprised nucleotides located within both the 3′ and 5′ NTRs (4), and thus we wanted to examine the possibility that nucleotides from both the 3′ and 5′ ends may also build up the transcription promoter. A comparison of the genomic and antigenomic RNAs of template BUN-S(ren) shows that 18 nucleotide differences exist between the first 25 nt of the 3′ and 5′ NTRs, 9 nucleotide differences in the 3′ NTR and 9 nucleotide differences in the 5′ NTR (Fig. 2B). We hypothesized that these nucleotide differences were responsible for the different transcription-signaling abilities of genomic and antigenomic RNAs. We chose to consider only the first 25 nt of the 3′ and 5′ NTRs, as these nucleotides alone are sufficient to signal transcription abilities indistinguishable between the genomic strand and template BUN-S(ren) (Fig. 2A).
FIG. 2.
Schematic representation of RNA templates BUN-S(ren) and AG-CB and analysis of their RNA synthesis activities. (A) The first 25 nt of both 3′ and 5′ NTRs of BUN-S(ren) are sufficient to signal transcription. RNAs were generated in vTF7-3-infected BHK cells transfected with the corresponding genome analog expressing cDNA and either BUNV S and L support plasmids (pL) (+) or S support plasmid alone (−). Bands representing mRNAs (vertical bar), T7 RNA polymerase primary transcripts, and BUNV-specific antigenomic RNAs (arrowheads) are marked. Template BUN-S(ren)-25/25 is identical to template BUN-S(ren), except that it contains only the terminal 25 nt of 3′ and 5′ NTRs. (B) Representation of the first 25 nt of both 3′ and 5′ NTRs of the genomic and antigenomic strands ofBUN-S(ren) and the genomic strand of AG-CB. The AG-CB antigenome is not shown, as it is identical in sequence to the AG-CB genomic strand for the nucleotides shown. Nucleotide differences from the genomic strand of BUN-S(ren) are shaded, and the potential to form Watson-Crick base pairs (*) or noncanonical U-G pairings (•) are also shown. Template AG-CB was derived from BUN-S(ren) by making nine nucleotide changes to the corresponding cDNA. (C) The RNA synthesis characteristics of BUN-S(ren) and AG-CB were compared by direct visualization of metabolically labeled actinomycin D-resistant RNAs using agarose-urea gel electrophoresis. This gel system allows separation of the three BUNV-specific RNAs, due to its ability to resolve RNAs based on both size and nucleotide composition. (D) RNAs generated by the genomic strands of BUN-S(ren) and AG-CB were also detected using primer extension analysis with the negative-sense oligonucleotide BUNMRNA. These RNAs were generated in vTF7-3-infected BHK cells transfected with the corresponding genome analog expressing cDNA and either BUNV S and L support plasmids (+) or S support plasmid alone (−). Bands representing mRNAs (vertical bar), T7 RNA polymerase primary transcripts, and BUNV-specific antigenomic RNAs (arrowheads) are marked.
As an initial step in locating the BUNV transcription signal, we first wanted to analyze the roles of the nine nucleotide differences that existed between the 3′ ends of genomic and antigenomic strands. To achieve this, we altered template BUN-S(ren) to generate template AG-CB (for antigenome, copy back), named from the fact that the 3′ ends of both its genomic and antigenomic strands were identical to the 3′ NTR of the wild-type BUN-S(ren) antigenome for the first 25 nt (Fig. 2B). Analysis of the RNA synthesis activity of this template allowed us to investigate whether the 3′ genomic NTR contained an essential transcription signal.
Agarose-urea gel electrophoresis of metabolically labeled and actinomycin D-resistant RNAs showed that template AG-CB was able to generate both positive- and negative-sense replication products with an abundance equal to that of BUN-S(ren) (Fig. 2C, lane 2). In contrast, the transcription activity of template AG-CB was inhibited, so that the abundance of transcription products from this template was barely detectable above background signal (Fig. 2C, lane 2). Densitometric analysis of autoradiographs indicated that the residual transcription ability of AG-CB was reduced by >100-fold compared to that of the wild-type template BUN-S(ren) (results not shown). These findings were supported by primer extension analysis, in which the negative-sense oligonucleotide BUNMRNA detected positive-sense replication products but failed to detect synthesis of mRNAs from the genomic RNA template (Fig. 2D, lane 5). Together, these results show that the nine nucleotide differences between templates BUN-S(ren) and AG-CB contain an essential component of the BUNV transcription promoter. This group of nine nucleotide differences includes positions 16, 17, 18, 20, 22, 23, 24, and 25 and also the conserved mismatched nucleotide at position 9.
A single nucleotide difference between the BUNV genome and antigenome abolishes transcription-signaling ability.
The results described above showed that, in contrast to the wild-type template BUN-S(ren), template AG-CB was unable to transcribe from the genomic strand. As the genomic strands of templates BUN-S(ren) and AG-CB differ at only 9 nt, we next wanted to determine which of these positions were responsible for transcription-signaling ability (Fig. 3A).
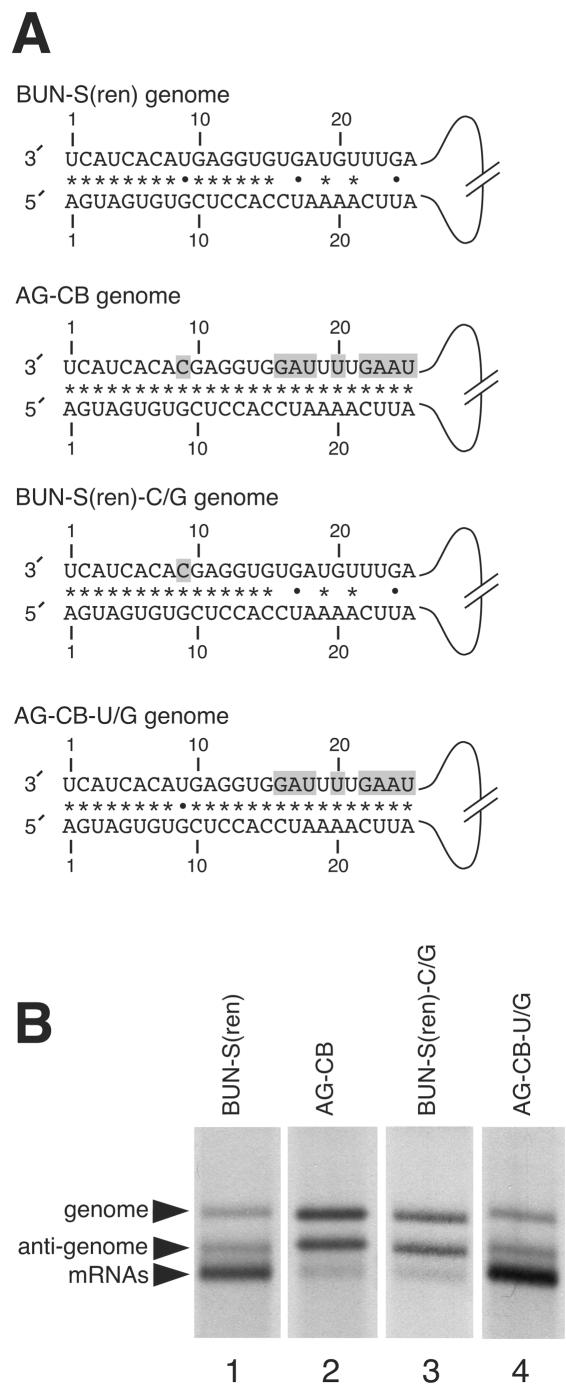
FIG. 3.
Schematic representations of the genomic strands of BUN-S(ren), AG-CB, BUN-S(ren)-C/G, and AG-CB-U/G templates and analysis of their RNA synthesis activities. (A) Templates AG-CB-U/G and BUN-S(ren)-C/G were derived from template AG-CB, and their nucleotide differences from the genomic strand of wild-type template BUN-S(ren) are shaded. The potential to form Watson-Crick base-pairs (*) or noncanonical U-G pairings (•) are also shown. (B) The RNA synthesis activities of BUN-S(ren), AG-CB, AG-CB-U/G, and BUN-S(ren)-C/G were analyzed by direct visualization of metabolically labeled actinomycin D-resistant RNAs using agarose-urea gel electrophoresis. BUNV-specific RNAs are marked with arrowheads.
Due to their clustered location within the 3′ NTR of AG-CB, and for ease of description, we separated this group of 9 nt into the distal group containing nt 16, 17, 18, 20, 22, 23, 24, and 25 (Fig. 3A) and the nucleotide at 3′ position 9 that is normally mismatched in the wild-type sequence. We first changed the distal nucleotides of template AG-CB back to their wild-type identities to generate template BUN-S(ren)-C/G (Fig. 3A). This template differed from BUN-S(ren) only at the mismatched 3′ position 9. Analysis of the RNA synthesis activity of this template using agarose-urea gel electrophoresis revealed that it possessed a phenotype indistinguishable from that of the parental template, AG-CB, in that it was able to signal replication but was deficient at signaling transcription (Fig. 3B, lane 3). This showed that these eight 3′ nucleotides did not contain the essential transcription signal and consequently implied that the remaining mismatched nucleotide 9 was critical for signaling transcription.
This was confirmed by altering the 3′ nucleotide position 9 of template AG-CB to the wild-type identity to generate the template AG-CB-U/G. This template possessed the wild-type 3′ nucleotide position 9 but still maintained the eight nucleotide differences at the distal group positions. Analysis of the RNA synthesis activity of this template by agarose-urea gel electrophoresis revealed that restoration of nucleotide position 9 to its wild-type identity correspondingly restored the transcription ability of template AG-CB-U/G to levels exhibited by the wild-type template BUN-S(ren) (Fig. 3B, lane 4). Taken together, these results reveal two important features of the BUNV transcription promoter. First, the 3′ nucleotide position 9 is critical for transcription-signaling ability, and second, the distal-group nucleotides perform little, if any, role in building up the BUNV transcription promoter.
Template abilities of BUNV RNAs with alternative nucleotides at 3′ and 5′ position 9.
Above, we showed that the identity of 3′ nucleotide position 9 of the BUN-S(ren) genomic RNA was critical for transcription activity. Interestingly, 3′ position 9 occupies a distinctive and conserved location within the BUNV NTRs in that it interrupts an unbroken run of 15, 18, or 19 complementary nucleotides in the BUNV S, M, and L segments, respectively. For all three segments, the nucleotide pairing of position 9 would allow a noncanonical U-G pairing to form in the genomic RNA with an incompatible A-C pairing in the antigenomic strand. This arrangement allows the possibility that differential association of genomic and antigenomic strands at 3′ and 5′ nucleotide position 9 could provide the molecular basis for their different RNA synthesis activities.
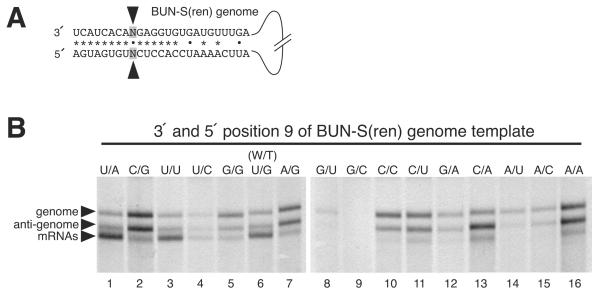
To test this possibility, we incorporated all possible nucleotide changes to both 3′ and 5′ position 9 of BUN-S(ren) (Fig. 4A) to generate 15 new templates. We analyzed the RNA synthesis abilities of the resulting panel of templates by agarose-urea gel electrophoresis (Fig. 4B). This analysis revealed a wide variety of RNA synthesis abilities, with many templates exhibiting low or undetectable transcription activities and others exhibiting low levels of RNA synthesis in general. However, two important trends emerged. First, it was evident that only 2 of the 15 templates (U/A and U/U; lanes 1 and 3) exhibited a level of transcription signaling ability that was comparable to that of the wild-type BUN-S(ren) template (U/G; lane 6). This finding shows that a U residue at 3′ position 9 of the genomic strand is a critical component of the BUNV transcription promoter that cannot be replaced by any other residue without loss of activity. The low level of mRNAs generated by template U/C (lane 4), which also has a U residue at 3′ position 9, is likely due to poor replication activity of this template, reducing the supply of transcriptionally active genomic strands. Second, our finding that templates U/U, U/G, and U/A have very similar phenotypes shows that the 5′ position 9 is not critical for signaling transcription from the genomic strand in a nucleotide-specific manner. Furthermore, as these three templates exhibit different potentials for association at 3′ and 5′ nucleotide position 9 through either canonical or noncanonical basepairing, these results show that the transcription-signaling ability of 3′ nucleotide 9 is not affected by its potential to interact with the corresponding nucleotide within the 5′ NTR. Taken together, our findings suggest that the 5′ nucleotide at position 9 does not build up the BUNV transcription promoter, and consequently, the noncomplementary arrangement of nucleotide position 9 is not critical for its transcription-signaling ability.
FIG. 4.
Analysis of the RNA synthesis-signaling abilities of 3′ and 5′ nucleotide position 9 of the genomic template. (A) The nucleotide identity of 3′ and 5′ genomic position 9 was changed from the wild-type combination (U-G) to all other possible permutations. (B) The RNA synthesis characteristics of these altered templates were analyzed by direct visualization of metabolically labeled actinomycin D-resistant RNAs using agarose-urea gel electrophoresis. BUNV-specific RNAs are marked with arrowheads.
Transcription signaling from the antigenomic strand.
The results described above show that a U residue at 3′ position 9 is a critical requirement for transcription from the genomic strand.
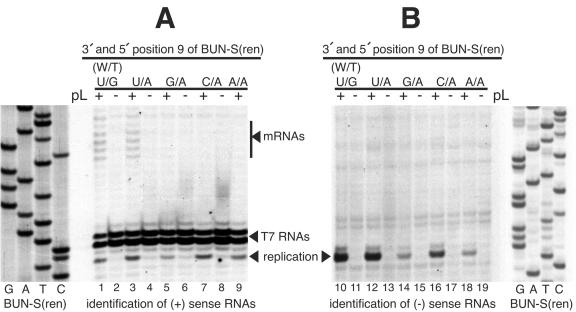
We next wanted to determine whether the different identities of this nucleotide in genomic and antigenomic RNAs comprised the sole factor that determined whether the strands were transcriptionally active. If no other nucleotide signals were required for transcription, then embedding a U nucleotide at 3′ position 9 within the replication-competent antigenomic strand would be expected to confer transcription-signaling ability on this template. To test this, we used primer extension analysis to determine the RNA synthesis activities of both the genomic (Fig. 5A) and antigenomic (Fig. 5B) strands of templates having position 9 nucleotide pairings of U-A, C-A, A-A, and G-A, as described above, all of which have the critical U residue at antigenomic 3′ position 9.
FIG. 5.
Primer extension analysis of the RNA synthesis-signaling abilities of BUNV templates having 3′ and 5′ position 9 combinations of U-A, C-A, G-A, and A-A to detect transcription from the antigenomic strand. RNAs were harvested from vTF7-3-infected BHK-21 cells transfected with a cDNA expressing a genome analog and either BUNV S and L support plasmids (pL) (+) or BUNV S support plasmid alone (−). (A) Positive-sense RNAs generated from genomic strands were detected using the negative-sense oligonucleotide BUNMRNA. The variously sized mRNA 5′ extensions are marked with a vertical bar. (B) Negative-sense RNAs generated from the antigenomic strand were detected using oligonucleotide minus-80. In each panel, the cDNA expressing template BUN-S(ren) was sequenced with the corresponding end-labeled oligonucleotide to act as a size marker.
As expected from the results described above, primer extension using the negative-sense oligonucleotide BUNMRNA detected antigenomic strands generated by all the tested templates (Fig. 5A, lanes 1, 3, 5, 7, and 9) and also detected positive-sense transcription products from templates BUN-S(ren) and U/A, as demonstrated by detection of the characteristic laddered banding pattern corresponding to the extended 5′ terminus of the BUNV mRNAs (Fig. 5A, lanes 1 and 3). In contrast, primer extension analysis using the positive-sense primer minus-80 detected the negative-sense genomic RNAs generated from each template (Fig. 5B, lanes 10, 12, 14, 16, and 18) but did not detect transcription products, even after long exposure of the autoradiograph (Fig. 5B). These findings suggest that while a U residue at 3′ nucleotide 9 is critical for transcription activity in the context of the genomic promoter, this nucleotide does not signal transcription in the context of the antigenomic promoter. Therefore, our results suggest that the identity of 3′ position 9 is critical for determining transcription activity; however, it is not the sole determinant.
DISCUSSION
The three negative-sense genomic strands of Bunyamwera virus serve as templates for both mRNA transcription and RNA replication. In contrast, each antigenomic strand is the template only for RNA replication, with mRNA synthesis being undetectable. In this report, we showed that the transcription activity of BUNV genomic RNAs is governed by the presence of a U residue at 3′ position 9. As the identity of this nucleotide differs in genomic and antigenomic RNAs, our results suggest that it plays a crucial role in determining the fundamentally different RNA synthesis promoter activities of genomic and antigenomic strands.
The nucleotides at 3′ and 5′ position 9 occupy distinctive locations within the BUNV NTRs in that they interrupt an unbroken run of complementarity in all three BUNV segments. These nucleotides would potentially allow a noncanonical U-G pairing to form in the BUNV genome with an incompatible A-C pairing in the antigenome. This arrangement allows the possibility that differential association of genomic and antigenomic strands at nucleotide position 9 could contribute to their different RNA synthesis activities. This signaling strategy would require that both 3′ and 5′ position 9 nucleotides be involved in building up the transcription promoter.
We investigated this possibility and showed that of all possible nucleotide combinations only templates U/A, U/U, and U/G (wild type) exhibited abundant transcription (Fig. 4B, lanes 1, 3, and 6, respectively). This observation revealed two important findings: first, that a U at position 9 is required to signal abundant transcription and, second, that the identity of 5′ position 9 is not critical for signaling transcription activity. These results suggest that the signaling ability of 3′ position 9 is not dependent on its ability to associate with the corresponding 5′ nucleotide, and consequently, we suggest that the 5′ position 9 does not build up the BUNV transcription promoter. Rather, the 3′ position 9 alone signals transcription in a nucleotide-specific manner. We previously showed that nt 12 to 15 of both 3′ and 5′ NTRs built up the BUNV RNA replication promoter (4), and therefore, our results show that both paired and unpaired nucleotides are likely involved in signaling BUNV RNA synthesis, although within different regions of the BUNV NTRs.
While nucleotide-specific signaling of transcription can account for the strict conservation of the U residue at 3′ position 9, it does not explain why the G residue at the corresponding 5′ position 9 is also strictly conserved in all three BUNV segments. A possible explanation is that alternative nucleotides at 5′ position 9 may result in subtle differences in RNA synthesis activity that are beyond the sensitivity of our detection methods. Such mutations may alter the fitness of BUNV in the context of a natural infection. Alternatively, we cannot rule out the possibility that the conserved G residue at 5′ position 9 is required for an alternative function unrelated to signaling abundance of mRNA synthesis, such as selection of host cell mRNAs during the cap-snatching process or even packaging of genomic RNAs into the virus particle.
Our results show that of the nine nucleotide differences between templates BUN-S(ren) and AG-CB analyzed in this report, only position 9 exerts an influence on mRNA synthesis. When the distal group of nucleotides (positions 16, 17, 18, 20, 22, 23, 24, and 25) of BUN-S(ren) were altered to give template AG-CB-U/G, mRNA synthesis showed no detectable reduction. However, when 3′ position 9 alone was altered, as in template BUN-S(ren)-C/G, mRNA synthesis was inhibited. Changes to the distal-group nucleotides within the 3′ NTR may reduce transcription, but only indirectly, as a consequence of affecting replication and thus reducing the generation of transcriptionally active genomes. The roles of nucleotides 1 to 8 and 10 to 15 of 3′ and 5′ NTRs in signaling transcription remain to be determined. However, as these nucleotides are conserved between genomic and antigenomic RNAs, they cannot be responsible for the different activities of the two RNA strands. It is possible that these conserved nucleotides may be involved in signaling RNA replication, which is a common activity of both genomic and antigenomic strands.
In this study, we showed that a further signal is required to form the BUNV transcription promoter in addition to 3′ nucleotide 9. The finding that the terminal-proximal 25 nt from both 3′ and 5′ NTRs are sufficient to signal transcription (Fig. 2A) leaves two possible regions within the BUNV genome analog in which the additional signal may reside. The first is the Renilla luciferase coding region, and the second is within the 5′ NTR. As the Renilla luciferase open reading frame is heterologous to that of BUNV, we suggest that it is unlikely to contain a signal that is required for BUNV transcription. It is more likely that there is an additional signal within the 5′ end of the genomic strand. To agree with our finding that a U residue at 3′ position 9 does not confer transcription-signaling ability on the antigenome, this signal must be present within the BUN-S(ren) genomic strand and absent from the BUN-S(ren) antigenomic strand. We are presently engaged in locating this additional signal.
In an attempt to derive a working model for how BUNV transcription is signaled, we have drawn heavily on the well-studied RNA synthesis strategy of the related influenza virus. A current model for influenza virus mRNA synthesis proposes that the RdRp associates with the 5′ genomic termini and subsequently recruits the 3′ terminus of the same RNA molecule (19, 39). Following cleavage of a capped host cell oligoribonucleotide to act as a primer, the RdRp migrates away from the 3′ genomic end while still tethered to the 5′ terminus and continues until it approaches its own 5′ binding site, at which point the sterically hindered RdRp terminates transcription (19, 26). Termination occurs on a short U tract that is reiteratively transcribed by the RdRp to yield the 3′ mRNA poly(A) tail (32, 35). It is now well established that influenza virus transcription involves nucleotides within both the 3′ and 5′ termini, and recently, the roles of individual nucleotides in the transcription process have been determined (1, 2, 18, 29-31, 37, 38, 40).
The findings of the present study suggest that sequences required for BUNV transcription activity are also distributed within both the 3′ and 5′ termini of the BUNV genomic strand. Our working model proposes that the BUNV transcription promoter includes the U at 3′ position 9 in addition to presently unidentified nucleotides within the 5′ terminus of the genomic strand. If the BUNV transcription promoter shares structural features with the influenza virus promoter, these nucleotides may represent an initial binding site for the viral transcriptase.
It is interesting that a mismatched nucleotide interrupting otherwise complementary termini is a conserved feature of many members of the family Bunyaviridae. The level of conservation is strongest in members of the genera Orthobunyavirus, Nairovirus, and Hantavirus, in which a mismatch is a consistent feature of all three segments (Fig. 6). Our finding that the identity of the mismatched 3′ nucleotide position 9 is a critical component of the BUNV transcription promoter may also apply to other bunyaviruses that possess similar terminal structures. The recent development of RNA synthesis assays for Hantaan (15), La Crosse (5), and Crimean Congo hemorrhagic fever (17) viruses will allow this hypothesis to be tested.
FIG. 6.

Schematics of conserved 3′ and 5′ NTR sequences of Orthobunyavirus, Hantavirus, and Nairovirus members. Complementary nucleotide pairs (*) and mismatched pairs (shaded and numbered) are shown. The nucleotides represented are conserved in S, M, and L segments. The Orthobunyavirus sequence was compiled from Bunyamwera virus and La Crosse virus. The Hantavirus sequence was compiled from Hantaan, Andes, Puumula, and Sin Nombre viruses. The Nairovirus sequence was compiled from Crimean Congo hemorrhagic fever virus and Dugbe virus.
Acknowledgments
We acknowledge members of the Gail W. Wertz and L. Andrew Ball laboratories for helpful discussion during the course of this study. We also thank Rodney Harris for expert technical assistance and R. M Elliott (Glasgow, United Kingdom) for the gift of plasmids expressing BUNV S and L segment open reading frames and genome analog BUN-S(ren).
This work was supported by an unrestricted infectious diseases research award to G.W.W. and by NIH grant AI 59174 to J.N.B.
REFERENCES
- 1.Azzeh, M., R. Flick, and G. Hobom. 2001. Functional analysis of the influenza A virus cRNA promoter and construction of an ambisense transcription system. Virology 289:400-410. [DOI] [PubMed] [Google Scholar]
- 2.Bae, S. H., H. K. Cheong, J. H. Lee, C. Cheong, M. Kainosho, and B. S. Choi. 2001. Structural features of an influenza virus promoter and their implications for viral RNA synthesis. Proc. Natl. Acad. Sci. USA 98:10602-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr, J. N., R. M. Elliott, E. F. Dunn, and G. W. Wertz. 2003. Segment-specific terminal sequences of Bunyamwera bunyavirus regulate genome replication. Virology 311:326-338. [DOI] [PubMed] [Google Scholar]
- 4.Barr, J. N., and G. W. Wertz. 2004. Bunyamwera bunyavirus RNA synthesis requires cooperation of 3′- and 5′-terminal sequences. J. Virol. 78:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blakqori, G., G. Kochs, O. Haller, and F. Weber. 2003. Functional L polymerase of La Crosse virus allows in vivo reconstitution of recombinant nucleocapsids. J. Gen. Virol. 84:1207-1214. [DOI] [PubMed] [Google Scholar]
- 6.Bouloy, M., N. Pardigon, P. Vialat, S. Gerbaud, and M. Girard. 1990. Characterization of the 5′ and 3′ ends of viral messenger RNAs isolated from BHK21 cells infected with Germiston virus (Bunyavirus). Virology 175:50-58. [DOI] [PubMed] [Google Scholar]
- 7.Bouloy, M., S. J. Plotch, and R. M. Krug. 1978. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc. Natl. Acad. Sci. USA 75:4886-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham, C., and J. F. Szilagyi. 1987. Viral RNAs synthesized in cells infected with Germiston Bunyavirus. Virology 157:431-439. [DOI] [PubMed] [Google Scholar]
- 9.Duijsings, D., R. Kormelink, and R. Goldbach. 2001. In vivo analysis of the TSWV cap-snatching mechanism: single base complementarity and primer length requirements. EMBO J. 20:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, R. M. 1985. Identification of nonstructural proteins encoded by viruses of the Bunyamwera serogroup (family Bunyaviridae). Virology 143:119-126. [DOI] [PubMed] [Google Scholar]
- 11.Elliott, R. M. 1989. Nucleotide sequence analysis of the large (L) genomic RNA segment of Bunyamwera virus, the prototype of the family Bunyaviridae. Virology 173:426-436. [DOI] [PubMed] [Google Scholar]
- 12.Elliott, R. M. 1989. Nucleotide sequence analysis of the small (S) RNA segment of Bunyamwera virus, the prototype of the family Bunyaviridae. J. Gen. Virol. 70:1281-1285. [DOI] [PubMed] [Google Scholar]
- 13.Elliott, R. M., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. Pettersson, A. Plyusnin, and C. S. Schmaljohn. 2000. Family Bunyaviridae, p. 599-621. In M. H. V. van Regenmortel et al. (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of viruses. Academic Press, San Diego, Calif.
- 14.Eshita, Y., B. Ericson, V. Romanowski, and D. H. Bishop. 1985. Analyses of the mRNA transcription processes of snowshoe hare bunyavirus S and M RNA species. J. Virol. 55:681-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flick, K., J. W. Hooper, C. S. Schmaljohn, R. F. Pettersson, H. Feldmann, and R. Flick. 2003. Rescue of Hantaan virus minigenomes. Virology 306:219-224. [DOI] [PubMed] [Google Scholar]
- 16.Flick, R., F. Elgh, and R. F. Pettersson. 2002. Mutational analysis of the Uukuniemi virus (Bunyaviridae family) promoter reveals two elements of functional importance. J. Virol. 76:10849-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flick, R., K. Flick, H. Feldmann, and F. Elgh. 2003. Reverse genetics for Crimean-Congo hemorrhagic fever virus. J. Virol. 77:5997-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flick, R., and G. Hobom. 1999. Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew’ conformation. J. Gen. Virol. 80:2565-2572. [DOI] [PubMed] [Google Scholar]
- 19.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller, F., A. S. Bhown, and D. H. Bishop. 1983. Bunyavirus nucleoprotein, N, and a non-structural protein, NSS, are coded by overlapping reading frames in the S RNA. J. Gen. Virol. 64:1705-1714. [DOI] [PubMed] [Google Scholar]
- 21.Fuller, F., and D. H. Bishop. 1982. Identification of virus-coded nonstructural polypeptides in bunyavirus-infected cells. J. Virol. 41:643-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcin, D., M. Lezzi, M. Dobbs, R. M. Elliott, C. Schmaljohn, C. Y. Kang, and D. Kolakofsky. 1995. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol. 69:5754-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentsch, J. R., and D. L. Bishop. 1979. M viral RNA segment of bunyaviruses codes for two glycoproteins, G1 and G2. J. Virol. 30:767-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerrard, S. R., L. Li, A. D. Barrett, and S. T. Nichol. 2004. Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa. J. Virol. 78:8922-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacker, D., S. Rochat, and D. Kolakofsky. 1990. Anti-mRNAs in La Crosse bunyavirus-infected cells. J. Virol. 64:5051-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagen, M., T. D. Chung, J. A. Butcher, and M. Krystal. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol. 68:1509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin, H., and R. M. Elliott. 1993. Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J. Virol. 67:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krug, R. M., B. A. Broni, and M. Bouloy. 1979. Are the 5′ ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell 18:329-334. [DOI] [PubMed] [Google Scholar]
- 29.Leahy, M. B., H. C. Dobbyn, and G. G. Brownlee. 2001. Hairpin loop structure in the 3′ arm of the influenza A virus virion RNA promoter is required for endonuclease activity. J. Virol. 75:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leahy, M. B., G. Zecchin, and G. G. Brownlee. 2002. Differential activation of influenza A virus endonuclease activity is dependent on multiple sequence differences between the virion RNA and cRNA promoters. J. Virol. 76:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, M. T., K. Klumpp, P. Digard, and L. Tiley. 2003. Activation of influenza virus RNA polymerase by the 5′ and 3′ terminal duplex of genomic RNA. Nucleic Acids Res. 31:1624-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, X., and P. Palese. 1994. Characterization of the polyadenylation signal of influenza virus RNA. J. Virol. 68:1245-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichol, S. T. 2001. Bunyaviruses, p. 1603-1633. In D. M. Knipe, B. N. Fields, P. M. Howley, and D. E. Griffin (ed.), Fields' virology. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 34.Plotch, S. J., M. Bouloy, I. Ulmanen, and R. M. Krug. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23:847-858. [DOI] [PubMed] [Google Scholar]
- 35.Poon, L. L., D. C. Pritlove, E. Fodor, and G. G. Brownlee. 1999. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 73:3473-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prehaud, C., N. Lopez, M. J. Blok, V. Obry, and M. Bouloy. 1997. Analysis of the 3′ terminal sequence recognized by the Rift Valley fever virus transcription complex in its ambisense S segment. Virology 227:189-197. [DOI] [PubMed] [Google Scholar]
- 37.Pritlove, D. C., L. L. Poon, E. Fodor, J. Sharps, and G. G. Brownlee. 1998. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J. Virol. 72:1280-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao, P., W. Yuan, and R. M. Krug. 2003. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 22:1188-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiley, L. S., M. Hagen, J. T. Matthews, and M. Krystal. 1994. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 68:5108-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng, H., P. Palese, and A. Garcia-Sastre. 1996. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication. Virology 217:242-251.8599209 [Google Scholar]