Abstract
All herpesviruses encode a homolog of glycoprotein M (gM), which appears to function in virion morphogenesis. Despite its conservation, gM is inessential for the lytic replication of alphaherpesviruses. In order to address the importance of gM in gammaherpesviruses, we disrupted it in the murine gammaherpesvirus 68 (MHV-68). The mutant virus completely failed to propagate in normally permissive fibroblasts. The defective genome was rescued by either homologous recombination to restore the wild-type gM in situ or the insertion of an ectopic, intergenic expression cassette encoding gM into the viral genome. Thus, gM was essential for the lytic replication of MHV-68.
All mammalian and avian herpesviruses encode conserved glycoproteins—glycoprotein B (gB), gH, gL, gM, and gN—that mediate the assembly of infectious virions, their exit from the cell, and their subsequent fusion with new cellular membranes. The functions of these glycoproteins are relatively well characterized for the alphaherpesviruses and appear to be qualitatively similar in the beta- and gammaherpesviruses. However, the quantitative contribution each glycoprotein makes to lytic replication in alpha-, beta-, and gammaherpesviruses is not necessarily the same. gM, which forms a complex with gN (14, 17, 18, 20, 29), is one of the most highly conserved of all herpesvirus glycoproteins and has been retained in a recognizable form over hundreds of millions of years of viral evolution. Outside the context of viral infection, alphaherpesvirus gM/gN complexes inhibit membrane fusion by a variety of endogenously synthesized proteins, including those of unrelated viruses (15; C. Crump, personal communication). The Kaposi's sarcoma-associated herpesvirus (KSHV) gM/gN complex has a similar function (17). In infected cells, the pseudorabies virus gM probably has a role in secondary envelopment, since a gM/gE/gI triple mutant has a defect in cytoplasmic virion assembly that is not seen with a gE/gI double mutant (4). However, disrupting gM alone has little effect on viral replication (8). Even a gM/gE double knockout of herpes simplex virus grows fairly normally in vitro (5). There may therefore be considerable functional redundancy in the alphaherpesviruses between gM, gE, gI, and gD (4, 10).
gM is probably an essential component of betaherpesviruses, since in random mutagenesis libraries, transposon insertion into the gM open reading frame (ORF) correlates with viral nonviability (12, 30). However, this phenotype remains to be formally linked to gM function by either viral reversion or complementation.
No gammaherpesvirus gM mutant has been described previously. Indeed, although much is known about cell binding by glycoproteins on gammaherpesvirus virions (2, 13), relatively little is known about the contribution of viral glycoproteins to other aspects of lytic propagation. This reflects the relative difficulty of manipulating the lytic cycle of the Epstein-Barr virus (EBV) and KSHV and of generating and analyzing targeted gene knockouts. EBV lacking gN shows substantial, although not complete, impairment of secondary virion envelopment and egress (19). As the EBV gM is unstable in the absence of gN (18), the gN-deficient phenotype may reflect a partial gM deficiency rather than a lack of gN itself; the contribution of gM instability to the gN-deficient phenotype and the consequences of a complete lack of gM in EBV have yet to be defined.
In contrast to EBV and KSHV, a related murid gammaherpesvirus, MHV-68, readily enters the lytic replication cycle in vitro. It therefore offers an opportunity to address fundamental questions about the function of shared gammaherpesvirus glycoproteins in a more tractable experimental setting. We are using MHV-68 to define the minimum set of glycoproteins required for gammaherpesvirus lytic replication in vitro and in vivo. The ultimate aim is to understand the mechanisms by which a gammaherpesvirus spreads between cells during the colonization of its natural host. Here, we have addressed the role of the MHV-68 gM in viral lytic replication.
MATERIALS AND METHODS
Cell lines BHK-21 cells (American Type Culture Collection [ATCC] CCL-10), NIH-3T3 cells (ATCC CRL-1658), NIH-3T3-CRE cells (6), 293T cells (ATCC CRL-11268), NS0 cells (ATCC CRL-11177), and murine embryonic fibroblasts (MEFs), harvested at 13 to 14 days of gestation, were grown in Dulbecco's modified Eagle medium (Invitrogen, Paisley, United Kingdom) supplemented with 2 mM glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal calf serum (PAA Laboratories, Linz, Austria) (complete medium). Medium for MEFs was further supplemented with 50 μM β-mercaptoethanol. Where indicated, cells were transfected using Fugene-6 (Roche Diagnostics Ltd., Lewes, United Kingdom) according to the manufacturer's instructions.
Plasmids.
The MHV-68 ORF39 (genomic coordinates 56950 through 55799) and ORF53 (genomic coordinates 71701 through 71444) were amplified by PCR (Hi-Fidelity; Roche Diagnostics), including EcoRI and XhoI restriction sites in the respective 5′ and 3′ primers. The PCR products were cloned into the EcoRI and XhoI sites of pMSCV-IRES-NEO and pMSCV-IRES-ZEO (3). To make NIH-3T3-gM/gN cells, each retroviral expression plasmid was cotransfected into 293T cells with the pEQPAM3 packaging plasmid (22). Supernatants were harvested 48 and 72 h posttransfection and used to transduce NIH-3T3 cells in the presence of 6 μg of hexadimethrine bromide (Sigma Chemical Co., Poole, United Kingdom)/ml as previously described (26). Transduced cells were selected with 1 mg of G418/ml plus 600 μg of Zeocin (Invitrogen)/ml.
Viral mutagenesis.
The MHV-68 gM coding sequence occupies genomic coordinates 56950 through 55799 (28). We generated a gM-mutant MHV-68 bacterial artificial chromosome (BAC) by inserting an oligonucleotide with multiple stop codons and a diagnostic EcoRI restriction site (5′-CTAGCTAGCTAGAATTCTAGCTAGCTAG-3′) into a PmlI restriction site at genomic coordinate 56407. The oligonucleotide was first cloned into BamHI genomic clone covering genomic coordinates 49938 through 59884 in pUC9 (9). The vector was digested with PmlI and dephosphorylated with Pandalus borealis alkaline phosphatase (Roche Diagnostics). The oligonucleotide was denatured at 95°C, annealed to itself at 37°C, and phosphorylated with polynucleotide kinase (New England Biolabs, Hitchin, United Kingdom). The vector and oligonucleotide were then joined with T4 DNA ligase (New England Biolabs). A BamHI-SacI fragment incorporating the disrupted ORF39 (genomic coordinates 54271 through 59884) was subcloned into the pST76K-SR shuttle plasmid, and the oligonucleotide insertion was recombined from pST76K-SR into the MHV-68 BAC in DH10B Escherichia coli by using transient RecA expression as previously described (1). A revertant BAC was made by replacing the mutant ORF39 with an intact copy in a similar way. Mutant and wild-type BACs were distinguished by the loss of the PmlI restriction site and the gain of an EcoRI restriction site.
An MHV-68 intergenic expression cassette was constructed in pSP73 (Promega Corporation, Southampton, United Kingdom). First, an SphI fragment containing the bovine growth hormone polyadenylation signal was subcloned from pcDNA3 (Invitrogen) into the SphI site of pSP73 (pSP73-polyA). The M3 promoter (genomic coordinates 7281 through 7780) was then amplified by PCR from MHV-68 genomic DNA, including BglII and EcoRI restriction sites in the respective upstream and downstream primers. This was cloned into the BglII and EcoRI sites of pSP73-polyA (pSP73-proM3-polyA). We then PCR-cloned the gM coding sequence as an EcoRI-XhoI fragment (see paragraph on plasmids, above) and ligated it into the EcoRI and SalI sites of pSP73-proM3-polyA. The enhanced green fluorescent protein (eGFP) coding sequence was separately cloned into the same sites in a similar way. Each expression cassette was then excised from pSP73 with BglII and XhoI and blunted with Klenow fragment DNA polymerase (New England Biolabs). In order to generate suitable recombination flanks for the expression cassette, we subcloned a BglII genomic fragment of MHV-68 (coordinates 75338 through 78717) from an EcoRI-HinDIII genomic clone (9), ligating it into the BglII site of an otherwise unmodified pSP73 vector. This construct was then cut at the MfeI site between the polyadenylation signals of ORF57 and ORF58 (genomic coordinate 77176), blunted with Klenow fragment DNA polymerase, and dephosphorylated. We ligated the blunt-ended expression cassette into the blunted MfeI site. The expression cassette plus flanks was then subcloned as a BglII fragment into pST76K-SR and recombined into the MHV-68 BAC as above. All BACs were reconstituted into infectious virus by transfecting BAC DNA into BHK-21 cells as previously described (7).
Virus growth and titration.
BAC+ viruses were grown in BHK-21 cells and titered by plaque assay on MEFs (7). Where indicated, the loxP-flanked BAC/GFP cassette was removed from viruses by passing it through NIH-3T3-CRE cells. All virus stocks were then prepared in BHK-21 cells. Infected cells and supernatants were sonicated after harvesting. Cell debris was pelleted by low-speed centrifugation (1,000 × g, 3 min) and discarded. Virions were then pelleted by high-speed centrifugation (38,000 × g, 90 min) and stored at −70°C. To prepare MHV-68 virions, the virus pelleted from supernatants was resuspended in phosphate-buffered saline (PBS), sonicated, layered over 5 to 15% Ficoll gradients, and centrifuged again (30,000 × g, 90 min). Virions were recovered as a distinct band, resuspended in PBS, pelleted (30,000 × g, 90 min), sonicated, and stored at −70°C.
Southern blotting.
DNA was extracted from virions by alkaline lysis, phenol-chloroform extraction, and salt-ethanol precipitation as previously described (7). Following restriction enzyme digestion, samples were electrophoresed on 0.8% agarose gels and transferred to positively charged nylon membranes (Roche Diagnostics). A [32P]dCTP-labeled probe (APBiotech, Little Chalfont, United Kingdom) was generated by random primer extension (Nonaprimer kit; Qbiogene, Bingham, United Kingdom) according to the manufacturer's instructions. Membranes were hybridized with probe (65°C, 18 h), washed to a stringency of 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 65°C, and exposed to X-ray film.
Immunoblotting and immunoprecipitation.
Monoclonal antibodies (MAbs) specific for the MHV-68 gM/gN complex were generated according to established protocols (16) by using an NS0 fusion partner and spleen cells of mice previously infected with ORF73-deficient MHV-68 (11). For immune precipitation, MHV-68-infected cells (5 PFU/cell, 18 h) were labeled for 15 min with [35S]cysteine-methionine (PerkinElmer Life Sciences, Cambridge, United Kingdom) and chased for 4 h with 1 mM unlabeled cysteine-methionine as previously described (3). Labeled cells were lysed on ice for 30 min in 1% Triton X-100-50 mM TrisCl (pH 7.4)-150 mM NaCl-5 mM EDTA with 1 mM phenylmethylsulfonyl fluoride and Complete protease inhibitors (Roche Diagnostics). Insoluble debris was removed by centrifugation (13,000 × g, 15 min). Lysates cleared of cell debris were then further cleared by incubation with protein A-Sepharose (AP Biotech). After removing the protein A-Sepharose, specific immune precipitations were performed with MAb 3F7, the gH/L-specific MAb T4C5, or the ORF4-specific MAb T3B8, each followed by further protein A-Sepharose. Sepharose beads with bound antibody were washed five times in lysis buffer. Samples were then dissociated (50°C, 15 min) in Laemmli's buffer, resolved by SDS-polyacrylamide gel elecrtrophoresis (PAGE), and exposed to X-ray film. For immunoblotting, virus stocks or purified virions were lysed directly in Laemmli's buffer, heated (50°C, 15 min) with or without 2-mercaptoethanol, and resolved by Tris-Tricine SDS-PAGE (24). The size-resolved proteins were transferred to polyvinylidene difluoride membranes (Perbio Science, Tattenhall, United Kingdom) and immunoblotted with either the 3F7 MAb or the gB-specific MAb T7H9, followed by horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin G (IgG; Dako Corporation, Ely, United Kingdom). Development was done with enhanced chemiluminescence reagents (AP Biotech).
Immunofluorescence.
Cells were washed in PBS, fixed in methanol (−20°C, 5 min), and blocked for 30 min in PBS-10% fetal calf serum-0.1% Tween. For monoclonal antibody staining, hybridoma supernatants were diluted 1/2 in PBS-0.1% Tween and incubated with cells for 1 h at 4°C. Detection was carried out with Alexa 594-coupled goat anti-mouse IgG (1 h, 4°C). Cells were washed three times in PBS-0.1% Tween 20 after each antibody incubation. In some experiments, nuclei were counterstained with DRAQ5 (Alexis Corporation, San Diego, Calif.). Fluorescence was visualized on a Leica confocal microscope. For GFP detection, unfixed cells were visualized under UV excitation on an Olympus microscope, and images were captured with a Hamamatsu digital camera.
Flow cytometry.
Cells infected with GFP+ viruses were washed in PBS and analyzed directly for green channel fluorescence. For specific staining, cells were incubated for 30 min on ice for 1 h with MAb 3F7, washed two times, and incubated with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG polyclonal antibody (PAb; Dako Corporation). After washing two times, cells were analyzed on FACSCalibur using Cellquest software (Becton-Dickinson, Oxford, United Kingdom). Graphs were plotted with FCSPress version 1.3 (http://www.fcspress.com).
RESULTS AND DISCUSSION
Characterization of MHV-68 gM and gN.
Since there is little information available on MHV-68 gM, we first undertook a preliminary characterization of this protein and its likely binding partner, gN. The MHV-68 gM homolog is encoded by ORF39; its gN homolog is encoded by ORF53. We identified a gM/gN-specific monoclonal antibody (MAb 3F7) in a panel of MHV-68-specific monoclonal antibodies by immune fluorescence of BHK-21 fibroblasts cotransfected with ORF39 and ORF53 (Fig. 1A). Both genes were required for detectable protein expression, suggesting that the epitope-bearing protein was unstable in the absence of its binding partner. When both gM and gN were present, the transfected cells showed perinuclear staining with the 3F7 MAb, which was therefore specific for either a gN-dependent gM epitope or a gM-dependent gN epitope.
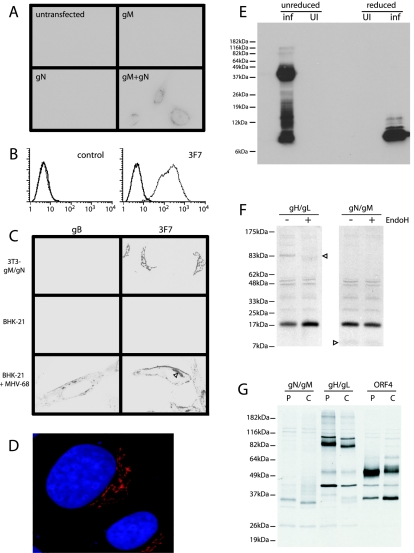
FIG. 1.
Characterization of the MHV-68 gM/gN complex. (A) BHK-21 cells were transfected with gN, gM, or gM plus gN or left untransfected. All cells were then stained with 3F7 MAb and Alexa 594-coupled goat anti-mouse IgG PAb. Three representative transfected cells are seen in the gM+gN panel. No staining was seen with gM or gN alone. (B) Uninfected (solid lines) or MHV-68-infected (dotted lines, 3 PFU/cell, 18 h) BHK-21 cells were stained with secondary antibody alone (control) or with MAb 3F7 plus FITC-conjugated rabbit anti-mouse IgG PAb. Samples were analyzed by flow cytometry. These cells were not fixed or permeabilized, so staining was restricted to epitopes accessible at the cell surface, presumably on virions. (C) NIH-3T3 cells stably transduced with retroviral vectors expressing gM and gN (3T3-gM/gN), uninfected BHK-21 cells, and MHV-68-infected BHK-21 cells (2 PFU/cell, 18 h) were each fixed with methanol, permeabilized with 0.1% Tween 20, and stained either with MAb 3F7 or with the isotype-matched, gB-specific MAb T7H9. Detection was done with Alexa 594-coupled goat anti-mouse IgG PAb. 3T3-gM/gN cells showed only a discrete perinuclear cap of 3F7 staining. The equivalent perinuclear cap of staining in MHV-68-infected BHK-21 cells is indicated by an arrowhead. (D) 3T3-gM/gN cells were stained with MAb 3F7 (red) and counterstained with DRAQ-5 (blue) to show the perinuclear distribution of gM/gN. (E) MHV-68 virions (inf) were purified from the supernatants of MHV-68-infected BHK-21 cells byultracentrifugation on Ficoll density gradients. Lysates of uninfected BHK-21 cells (UI) were used as a control. All samples were denatured (50°C, 15 min) in Laemmli's buffer with (reduced) or without (unreduced) 2-mercaptoethanol. Samples were resolved by Tris-Tricine SDS-PAGE, transferred to polyvinylidene difluoride membranes, and immunoblotted with MAb 3F7. (F) BHK-21 cells were infected with MHV-68 (5 PFU/cell, 18 h) and then labeled for 1 h with [35S]cysteine-methionine. We immunoprecipitated gM/gN with MAb 3F7. For comparison, we immunoprecipitated gH/gL with MAb T4C5. Immunoprecipitates were dissociated at 95°C, treated (+) or not (−) with endoglycosidase H (EndoH), and resolved by Tris-Tricine SDS-PAGE. Bands corresponding to gH and gN are indicated by arrowheads. (G) BHK-21 cells were infected with MHV-68 (5 PFU/cell, 18 h) and then pulse labeled (P, 15 min) with [35S]cysteine-methionine followed by a 4-h chase (C) with excess unlabeled cysteine-methionine. gM/gN was precipitated with MAb 3F7. For comparison, we immunoprecipitated from the same cells gH/gL with MAb T4C5 and ORF4 with MAb T3B8. Immunoprecipitates were dissociated at 50°C and resolved by SDS-PAGE.
Flow cytometry (Fig. 1B) established that the 3F7 epitope was present on the surfaces of MHV-68-infected fibroblasts. Immune fluorescence (Fig. 1C) of permeabilized, infected cells with the 3F7 MAb showed staining in a well-defined perinuclear cap and at the plasma membrane. Fibroblasts stably expressing recombinant gM and gN (Fig. 1C and D) showed a much more restricted staining pattern, with the 3F7 epitope expressed only in a well-defined perinuclear cap. This was consistent with gM/gN localization to Golgi membranes, a pattern typical of gM/gN expression outside the context of viral infection (20; C. Crump, personal communication). The more restricted staining in Fig. 1C compared to Fig. 1A probably reflects lower expression levels in stable cell lines. Surface staining was not observed by flow cytometry of either BHK-21 cells cotransfected with gM and gN or of NIH-3T3 cells stably transduced with retroviral vectors expressing gM and gN (data not shown). The cell surface staining seen after MHV-68 infection was therefore likely to be due to gM and gN being incorporated into virions and then exported to the cell surface. It is also possible that gM/gN heterodimers were independently exported to the cell surface when another viral protein was expressed.
We used immunoblots to distinguish whether the 3F7 MAb recognized gM or gN (Fig. 1E). The predicted sizes of unglycosylated gM and gN are 44 and 7 kDa (after signal peptide cleavage), respectively. The 3F7 MAb detected an 8-kDa band in reduced lysates of purified virions and was therefore specific for gN. Immunoblots of unreduced virion lysates identified not only the 8-kDa band but also higher-molecular-mass bands, with the predominant one at 40 kDa. It therefore seemed likely that, as in other herpesviruses, the MHV-68 gN is disulphide linked to gM and both proteins are incorporated into virions. The 10- to 20-kDa bands detected with MAb 3F7 in unreduced virion lysates may be gN oligomers.
It was just possible to discern an 8-kDa band corresponding to gN by immunoprecipitation with the 3F7 MAb from virus-infected cells (Fig. 1F). When immunoprecipitates were dissociated at 50°C rather than 95°C prior to loading, we could also identify a 35-kDa protein precipitated by MAb 3F7 from pulse-labeled cells (Fig. 1G), which was likely to be gM. After a 4-h chase, there were both a smaller (33-kDa) band, suggestive of protein cleavage, and more diffuse bands in the 40- to 45-kDa range, suggestive of glycan modification. Why it was smaller than the predicted molecular mass of 44 kDa is not clear. The 35-kDa protein was prone to aggregation after denaturation, since it entered SDS-PAGE gels after heating to 50°C but not after boiling, consistent with the highly hydrophobic nature of gM.
A targeted disruption of ORF39.
In order to disrupt ORF39 (genomic coordinates 56950 through 55799), we inserted an oligonucleotide with multiple stop codons in all reading frames into a PmlI restriction site at genomic coordinate 56407. (Fig. 2A). Southern blots confirmed the predicted genomic structure of the mutant BAC and its revertant (Fig. 2B and C).
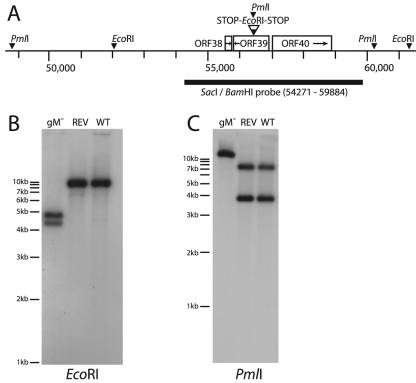
FIG. 2.
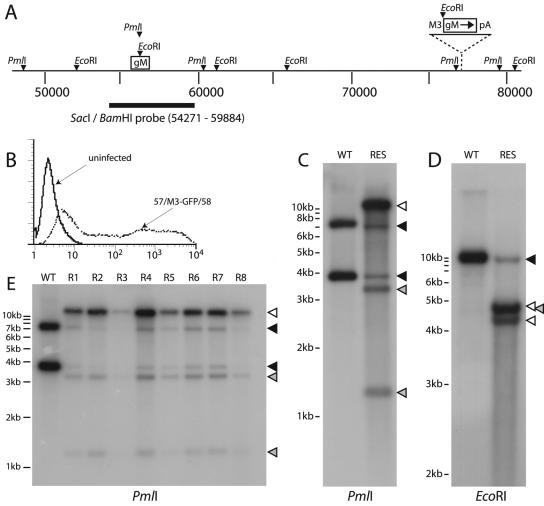
Disruption of ORF39 in the MHV-68 BAC. (A) We generated a gM-deficient MHV-68 BAC by inserting an oligonucleotide with multiple stop codons and an internal EcoRI restriction site into a PmlI restriction site midway through ORF39. (B) BAC DNA was digested with EcoRI, electrophoresed, transferred to nylon membranes, and probed with a BamHI-SacI genomic fragment as indicated in panel A. The predicted bands were 9,215 bp for the wild type (WT) and revertant (REV) and 4,343 and 4,872 bp for the gM mutant (gM−). (C) The same samples were digested with PmlI. Because the oligonucleotide insertion disrupted the PmlI site at genomic coordinate 56407, the predicted band for the mutant BAC was 11,420 bp, with 7,542 and 3,878 bp for the wild type and revertant.
These BACs were then transfected into BHK-21 cells. Virus reconstituted from the wild-type and revertant BACs multiplied rapidly, whereas no infectivity was recovered from cells transfected with gM-negative BACs (Fig. 3A). Because the MHV-68 BAC cassette includes eGFP expressed from a human cytomegalovirus IE1 promoter (1), fibroblasts transfected with BAC DNA or infected with virus containing the BAC cassette show green fluorescence. By 3 days after transfection, green fluorescence had spread to all cells in cultures transfected with wild-type or revertant BACs. Single green fluorescent cells were also observed after transfection with ORF39-disrupted BACs, but there was no spread to neighboring cells, consistent with the lack of recoverable infectivity. eGFP+ cells had disintegrated by 4 days after transfection (Fig. 3B) and were lost completely from cultures upon further passage. The same pattern was observed with three independently derived ORF39-disrupted BAC mutants, in multiple transfections, and in NIH-3T3 cells as well as BHK-21 cells. Thus, MHV-68 unable to express gM completely failed to spread.
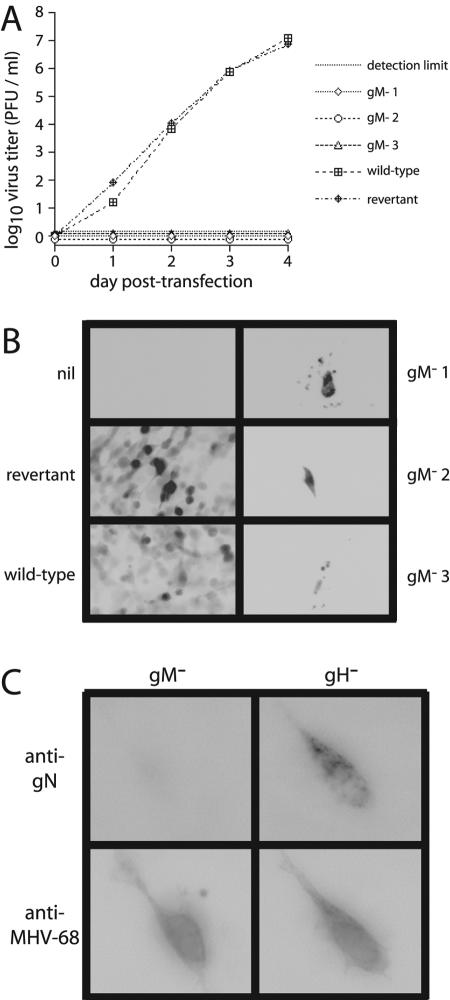
FIG. 3.
Disrupting gM completely abolishes MHV-68 lytic spread. (A) BHK-21 cells were transfected with 5 μg of BAC DNA. Supernatants were harvested for infectious virus after transfection as indicated and titered by plaque assay. gM−1, gM−2, and gM−3 are three independently derived BAC mutants. (B) The spread of infection from transfected cells was also assayed by the expression of eGFP, which is transcribed from a human cytomegalovirus IE1 promoter as part of the MHV-68 BAC cassette (1). Representative areas of cultures 4 days after transfection are shown as reverse images such that dark areas in the picture correspond to green fluorescence. By this time, cells transfected with ORF39-disrupted BACs had become fragmented, while the surrounding cells appeared entirely normal. Nil, cotransfected. (C) At 24 h after transfection, cells were costained for MHV-68 antigens with an immune rabbit serum and for gN with MAb 3F7 by using FITC-coupled goat anti-mouse IgG and Alexa 594-coupled goat anti-rabbit IgG PAbs for detection. To control for the possible effects of viral spread, we compared the gM-disrupted BAC with a gH-disrupted BAC, which also failed to generate infectious virus (J. S. May and P. G. Stevenson, unpublished data). Representative transfected cells are shown.
Cells transfected with gM-negative BACs showed evidence of viral protein synthesis in that they could be stained with an MHV-68-specific immune serum (Fig. 3C) or with a gH/gL-specific MAb (data not shown). However, they showed no staining with the 3F7 MAb. This implied that virus-expressed gN was unstable in the absence of gM or that expression of the 3F7 gN epitope was gM dependent, in agreement with the results of transfections with gN and gM expression plasmids (Fig. 1A).
Rescue of the gM-deficient phenotype by repair of ORF39.
We then tested whether complementation in trans could rescue the gM-deficient BACs. Thus, we cotransfected BHK-21 cells with a gM-negative BAC and a gM expression plasmid. This led to spreading green fluorescence in the transfected cells (data not shown), but a proportion of the virus recovered also spread without further complementation, suggesting that homologous recombination had restored ORF39 in the viral genome and thereby generated revertant viruses (Fig. 4A). This was confirmed by PCR analysis of DNA recovered from infectious clones isolated by limiting dilution: all showed evidence of an intact ORF39 (Fig. 4B). DNA sequence analysis of PCR products from three clones confirmed the presence of wild-type ORF39 (data not shown). Southern blots of the same DNA samples showed three distinct patterns (Fig. 4C). These were explained by the expression plasmid integrating into the MHV-68 genome by a single crossover in ORF39, either 5′ or 3′ of the inserted oligonucleotide, and then resolving this intermediate to restore the native gM locus.
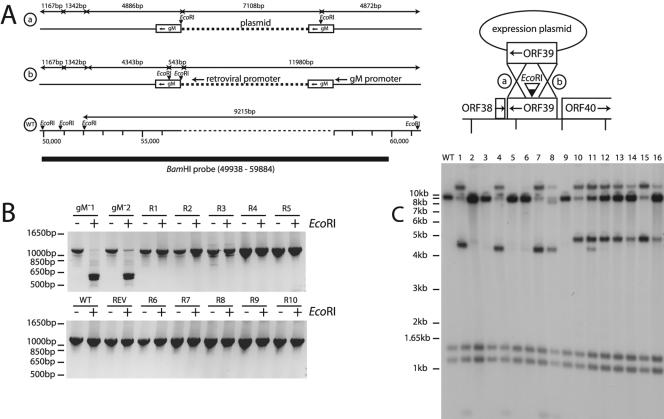
FIG. 4.
Restoration of infectivity during complementation correlates with the repair of ORF39. (A) Virus recovered from BHK-21 cells cotransfected with ORF39-disrupted BACs and a retroviral gM expression plasmid were cloned by limiting dilution. There was the potential for gM to be reconstituted by recombination between BAC and expression plasmid DNA either 3′ (pattern a) or 5′ (pattern b) of the oligonucleotide insertion, thereby integrating the expression plasmid into the BAC. There could then be further recombination between the gM copies to excise the plasmid sequence and reconstitute the wild-type (WT) genome. (B) ORF39 was amplified by PCR of BAC DNA (gM−1, gM−2, WT, REV) or of DNA from the infectious viral clones recovered after cotransfection (R1 through R5). PCR products were then digested (+) or not (−) with EcoRI. Digestion of the 1,150-bp gM band into 608- and 543-bp fragments indicated the presence of the inserted oligonucleotide. The proportions of mutant and intact ORF39 cannot be compared, since the oligonucleotide insertion may have reduced the efficiency of PCR amplification. The key point is that appreciable amounts of wild-type viral DNA were always present in infectious viral clones. (C) Viral DNA was digested with EcoRI, electrophoresed, blotted, and probed with a labeled BamHI genomic clone as indicated for panel A. The numbers 1 through 16 refer to individual infectious virus clones recovered after cotransfection. The 1,167- and 1,342-bp fragments were unaffected by gM reconstitution. A 5′ crossover in ORF39 to integrate the expression plasmid (1,150 bp of ORF39 plus 6.5 kb of plasmid) should have given 4,343-, 543-, and 11,980-bp fragments. A 3′ crossover in ORF39 should have given 4,886-, 7,108-, and 4,872-bp fragments. The latter pattern was never seen complete, indicating that there was selective pressure to lose the EcoRI site from the right-hand copy of ORF39. A double crossover to excise all plasmid sequences gave the 9,215-bp wild-type band that was present to some degree in all samples.
Clones 1, 4, 7, and 8 (crossover 5′ of the oligonucleotide insertion [Fig. 4A, pattern b]) showed only a small amount of the 9.2-kb wild-type band. Plasmid insertion downstream of an intact ORF39 was therefore a relatively stable arrangement, without strong selection for a second recombination event. In contrast, clones 10 through 16 (clone 11 was mixed) contained significant amounts of the 9.2-kb wild-type band. The 4.8-kb band in these clones was consistent with recombination between the retroviral and BAC ORF39 copies 3′ of the oligonucleotide insertion (Fig. 4A, pattern a). However, a 7.2-kb band corresponding to the retroviral plasmid plus the 3′ half of the mutant ORF39 was never observed. Instead, there was a 12-kb band, consistent with repair of the oligonucleotide insertion in the upstream copy of gM. This suggested that either the retroviral promoter was inadequate to drive gM expression or the oligonucleotide insertion in the middle of gM compromised a function to the right of ORF39 in the viral genome. After homologous recombination 5′ of the oligonucleotide insertion, there was clearly strong selective pressure for a second recombination event to give a wild-type ORF39 locus (clones 2, 3, 5, 6, and 9).
Rescue of the gM-deficient phenotype by ectopic gM expression.
The rescue of gM-deficient BACs by homologous recombination with plasmid-based gM confirmed that disruption of the gM locus was responsible for the nonviability of the gM-mutant BACs. However, because complementation involved plasmid integration and integration of plasmid 5′ of the intact gM copy was unstable, we could not be sure that gM itself was the crucial function, rather than an overlapping function, such as an ORF40 promoter. In order to link definitively the nonviable virus phenotype to gM expression, we rescued the ORF39-deficient BAC by expressing ORF39 from an ectopic site in the viral genome.
We first established a viable intergenic expression system for MHV-68 by using the intergenic region between ORF57 and ORF58 (Fig. 5A). ORFs 57 and 58 meet at their 3′ ends, so the intervening sequence is unlikely to contain crucial viral promoter functions; the equivalent intergenic region in KSHV contains multiple vIRF genes (23). As a strong lytic promoter, we used the 500 bp upstream of the transcriptional start site of the MHV-68 M3, an abundant early-late lytic gene (27). Preliminary experiments using eGFP expression from the M3 promoter region established that the ORF57/ORF58 intergenic expression cassette was actively transcribed in lytically infected cells (Fig. 5B). We then generated an equivalent gM expression cassette and recombined it into the gM-deficient BAC. This restored viral viability, and the gM rescue virus propagated efficiently after the transfection of BAC DNA into BHK-21 cells.
FIG. 5.
Rescue of the ORF39-deficient BAC by intergenic ORF39 expression. (A) An intergenic expression cassette, designed to transcribe gM from an ectopic viral M3 promoter, was recombined into the gM mutant MHV-68 BAC between ORF57 and ORF58. (B) An equivalent intergenic eGFP expression cassette (57/M3-GFP/58) showed strong green fluorescence in lytically infected BHK-21 cells (0.5 PFU/cell, 18 h) by flow cytometry. The BAC cassette had been removed from this virus, so eGFP expression was driven solely by the ectopic M3 promoter. This established that the intergenic expression cassette was functional. (C) Southern blots of the rescued gM mutant virus (RES) showed with PmlI digestion a 11,420-bp band due to loss of the PmlI site in ORF39 (open arrowhead). However, there was also a low level of the 7,542- and 3,878-bp wild-type (WT) bands (black arrowheads). Hybridization of the probe to the intergenic ORF39 expression cassette gave the predicted bands of 1,254 and 3,337 bp (gray arrowheads). These were weaker due to less overlap with the probe. (D) EcoRI digestion also showed evidence of some wild-type DNA (9,215-bp band) (black arrowhead). The 4,343- and 4,872-bp bands seen with the rescued virus correspond to the oligonucleotide insertion (open arrowheads). There was also a 4,862-bp band (gray arrowhead) which ran as a doublet with the 4,872-bp band, corresponding to the intergenic ORF39 expression cassette. (E) All clones of the rescued viruses (R1 through R8) showed an pattern identical to that of the parent. The rescued pattern (11,420 bp) was always dominant, but each clone also showed a low level of wild-type ORF39 (7,542 and 3,878 bp). The proportion of wild-type DNA appeared to be constant. The open arrowhead shows the gM mutant band, the black arrowheads show wild-type bands, and the gray arrowheads show intergenic expression cassette bands.
Southern blots of viral DNA confirmed the predicted genomic structure of the gM rescue virus but also showed a small proportion of wild-type DNA in the complemented virus stocks (Fig. 5C and D). There was no sign of wild-type genomes in digests of the gM rescue BAC DNA (data not shown), so the appearance of wild-type DNA presumably reflected a double recombination between the inserted and endogenous copies of ORF39, restoring an intact ORF39 to its normal site. Recombination could not occur within a single viral genome without deleting an essential central region but could potentially occur between genomes. We then cloned this mixed-virus stock by limiting dilution to remove any contaminating wild-type virus. However, all clones of the mixed virus showed the same pattern of low-level wild-type DNA (Fig. 5E). There was no obvious change in the overall proportion of wild-type DNA with passage, and no wild-type—or even predominantly wild-type—clones were identified.
This was very different to the pattern of clones obtained after recombination of the gM expression plasmid into the viral genome (Fig. 4C). There, there was strong selective pressure for the flank 5′ of gM to be repaired. With intergenic gM expression, recombination to repair the endogenous gM locus did not appear to confer a significant increase in viral fitness, since there was no evidence of selection for wild-type virus with passage. Thus, the M3 promoter, unlike the integrated retroviral promoter, expressed sufficient gM to rescue the mutant virus.
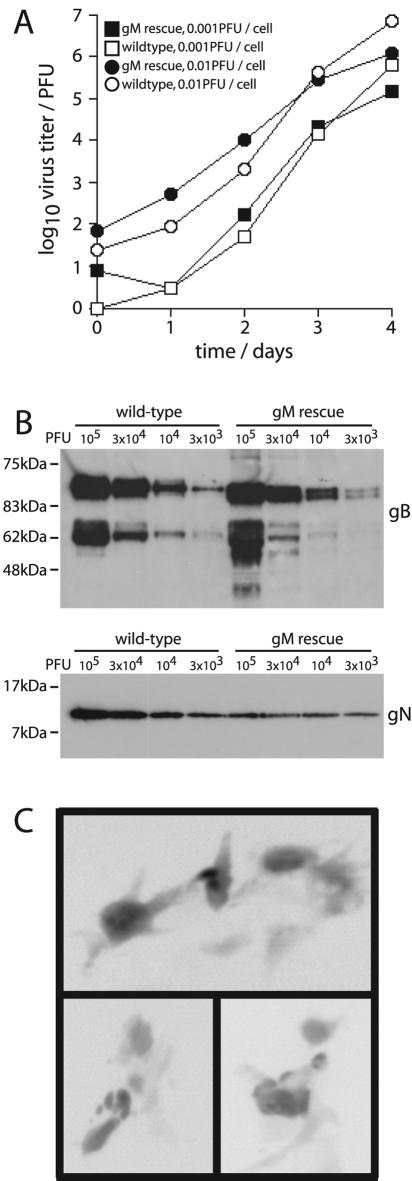
Growth curves (Fig. 6A) established that the complemented virus grew comparably to wild-type virus. Thus, there was rescue from complete nonviability to wild-type levels of lytic replication. It was formally possible that the normal growth of the gM rescue virus was due to a small proportion of replication-competent wild-type genomes in otherwise replication-defective virus stocks, with stock titers measuring only the wild-type virus. This would predict considerably more viral protein for a given plaque titer in stocks of the gM rescue virus. However, despite wild-type DNA being <10% of total viral DNA in gM rescue virus stocks (Fig. 5), the gB content of stocks was equivalent between wild-type and gM rescue viruses (Fig. 6B). More gN was present in the wild-type stocks, presumably reflecting that gM rescue still gave suboptimal reconstitution of the gM/gN heterodimer, but this appeared to be tolerated. The intergenic expression cassette was therefore responsible for restoring viral fitness, establishing that gM expression was the essential viral function compromised by the oligonucleotide insertion.
FIG. 6.
Replication of complemented ORF39 mutant virus. (A) BHK-21 cell monolayers were infected with either wild-type or gM rescue viruses as indicated. Viral titers with time were measured by plaque assay. (B) Virus stocks were lysed in Laemmli's buffer and analyzed for viral protein content by SDS-PAGE and immunoblotting for gB and gN. The number of viral PFU equivalents in each lane is indicated. The two main bands for gB represent the 120-kDa full-length protein and a 65-kDa C-terminal cleavage product; each exists as two or three different glycoforms. (C) Examples of the spread of GFP expression observed after the transfection of gM-disrupted BAC DNA into 3T3-gM/gN cells, with groups of 5 to 10 GFP+ cells, are shown. Figure 3B shows equivalent BACs transfected into noncomplementing BHK-21 cells.
Some complementation of the gM-disrupted BAC was also possible by transfecting BAC DNA into the NIH-3T3 cells stably expressing gM and gN (Fig. 6C). In contrast to the single eGFP+ cells seen after the transfection of unmodified fibroblasts (Fig. 3B), there was a definite spread of eGFP expression between adjacent gM and gN+ fibroblasts. Further passage of these cultures on BHK-21 cells showed no evidence of recombination between cellular and viral gM to generate revertant viruses (data not shown). This complementation by cellular gM/gN expression therefore supported further the idea that gM expression was the crucial viral function lost with oligonucleotide insertion into ORF39. The fact that the only limited complementation was achieved—viral titers remained very low—implied that cellular gM/gN expression was well below optimal levels for viral replication. This was not surprising, since MHV-68 causes host shutoff (25) and the accumulation of gN/gM in NIH-3T3-gM/gN cells was always well below that seen in infected fibroblasts (Fig. 1). The key point is that some viability was restored to the gM-deficient BAC.
In conclusion, gM was found to be essential for MHV-68 lytic replication in fibroblasts. Multiple, independently derived BACs with an oligonucleotide inserted into ORF39 were completely replication defective, and this phenotype could be rescued by reversion of the BAC, repair of ORF39, or ectopic gM expression from the viral genome or from cellular DNA. The absolute requirement of MHV-68 for gM contrasted markedly with herpes simplex virus and pseudorabies virus, in which gM is inessential, but was consistent with an essential role for gM in human cytomegalovirus. The function of the MHV-68 gM/gN remains unknown. However, the evidence from other viruses suggests that a role in secondary envelopment is likely (15, 17, 19). Whether this function of gM/gN is essential or not presumably depends on whether other viral glycoproteins, such as gE/gI in alphaherpesviruses, have evolved sufficient interaction with tegument proteins to substitute for gM/gN (21). Alternatively, gM/gN in MHV-68 may fulfil a further function that is essential for viral lytic replication.
Acknowledgments
Thanks are due to Colin Crump for helpful discussions.
This work was supported by Medical Research Council grants G108/462 and G9800903. P.G.S. is an MRC/Academy of Medical Sciences Clinician Scientist.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2002. Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 3.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. [DOI] [PubMed] [Google Scholar]
- 4.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browne, H., S. Bell, and T. Minson. 2004. Analysis of the requirement for glycoprotein M in herpes simplex virus type 1 morphogenesis. J. Virol. 78:1039-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, H. M., B. de Lima, V. Morton, and P. G. Stevenson. 2003. Murine gammaherpesvirus 68 lacking thymidine kinase shows severe attenuation of lytic cycle replication in vivo but still establishes latency. J. Virol. 77:2410-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lima, B. D., J. S. May, and P. G. Stevenson. 2004. Murine gammaherpesvirus 68 lacking gp150 shows defective virion release but establishes normal latency in vivo. J. Virol. 78:5103-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dijkstra, J. M., N. Visser, T. C. Mettenleiter, and B. G. Klupp. 1996. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J. Virol. 70:5684-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efstathiou, S., Y. M. Ho, and A. C. Minson. 1990. Cloning and molecular characterization of the murine herpesvirus 68 genome. J. Gen. Virol. 71:1355-1364. [DOI] [PubMed] [Google Scholar]
- 10.Farnsworth, A., K. Goldsmith, and D. C. Johnson. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 77:8481-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler, P., S. Marques, J. P. Simas, and S. Efstathiou. 2003. ORF73 of murine herpesvirus-68 is critical for the establishment and maintenance of latency. J. Gen. Virol. 84:3405-3416. [DOI] [PubMed] [Google Scholar]
- 12.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutt-Fletcher, L. M., and C. M. Lake. 2001. Two Epstein-Barr virus glycoprotein complexes. Curr. Top. Microbiol. Immunol. 258:51-64. [DOI] [PubMed] [Google Scholar]
- 14.Jöns, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Köhler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495-497. [DOI] [PubMed] [Google Scholar]
- 17.Koyano, S., E. C. Mar, F. R. Stamey, and N. Inoue. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485-1491. [DOI] [PubMed] [Google Scholar]
- 18.Lake. C. M., S. J. Molesworth, and L. M. Hutt-Fletcher. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 72:5559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lake, C. M., and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74:11162-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Persons, D. A., M. G. Mehaffey, M. Kaleko, A. W. Nienhuis, and E. F. Vanin. 1998. An improved method for generating retroviral producer clones for vectors lacking a selectable marker gene. Blood Cells Mol. Dis. 24:167-182. [DOI] [PubMed] [Google Scholar]
- 23.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 25.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevenson, P. G., J. S. May, X. G. Smith, S. Marques, H. Adler, U. H. Koszinowski, J. P. Simas, and S. Efstathiou. 2002. K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3:733-740. [DOI] [PubMed] [Google Scholar]
- 27.van Berkel, V., K. Preiter, H. W. Virgin IV, and S. H. Speck. 1999. Identification and initial characterization of the murine gammaherpesvirus 68 gene M3, encoding an abundantly secreted protein. J. Virol. 73:4524-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Virgin, H. W., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, S. X., X. P. Zhu, and G. J. Letchworth. 1998. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the UL49.5 protein. J. Virol. 72:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]