Abstract
Human papillomaviruses infect stratifying squamous epithelia, causing benign and malignant lesions. Upon differentiation of the host keratinocyte, the virus undergoes a dramatic increase in both DNA replication and transcription from the late promoter, leading to expression of late genes and virion morphogenesis. In human papillomavirus type 31 (HPV31), the late promoter is designated p742 and includes multiple start sites embedded within the E7 gene. In this report, we mapped viral DNA elements that control transcriptional activity from p742. Enhancer elements in the viral upstream regulatory region positively regulate this promoter. The region containing the transcriptional start sites is dispensable for activity, and at least two separate elements in the E6/E7 region are capable of supporting transcription. Of these, we mapped one to a 150-bp region of the E7 open reading frame and designate it the core p742 promoter. Using GF109203X, an inhibitor of protein kinase C signaling, we show that p742 activation is independent of viral genome amplification. Finally, we mapped elements in the region of p742 that confer responsiveness to differentiation and show that the upstream regulatory region does not contribute to the differentiation response of p742. These studies are an important step toward understanding the functioning and regulation of this multiple-start promoter.
Human papillomaviruses (HPVs) are small closed circular DNA viruses that infect the keratinocytes of stratified squamous epithelia, causing benign or malignant hyperproliferative lesions (65, 66). Over 100 types of papillomaviruses have been identified to date, infecting numerous cutaneous or mucosal sites (67). All HPVs have strong species and tissue tropism. Because of their association with malignancy, especially cervical cancer, certain HPV types that infect the anogenital mucosa are of particular concern. Types 16, 18, 31, 33, 39, 45, 51, and 52 are termed high risk because of their frequent presence in cervical carcinomas; whereas the low-risk types 6 and 11 also cause genital lesions, they are rarely associated with malignancy (28, 66).
HPVs are thought to gain access to the basal cells of the epithelium through small traumas (47). After a burst of DNA replication to establish the copy number at 50 to 200 in basal cells, the virus maintains itself in the basal layer, replicating once per cycle as an episome (19). As the cells move from the basal into the spinous layer of the epithelium, the first step in the keratinocyte differentiation program, a substantial increase in viral DNA synthesis occurs (6, 51); this increase is accompanied by a change from theta replication to a rolling-circle mechanism (19). The cells of suprabasal layers normally do not express DNA replication machinery, but the viral oncoproteins E6 and E7 circumvent the normal cellular controls on the cell cycle, causing the cells to continue the expression of replication factors for use by the virus (18, 32, 61). As the infected cells undergo terminal differentiation in the granular and cornified layers, viral late genes, including the major and minor capsid proteins L1 and L2, are expressed and virions are assembled (42).
Both replication of viral DNA and transcription from HPV promoters increase in response to differentiation of host keratinocytes (1, 19, 34, 44-46). In particular, transcripts encoding the capsid proteins L1 and L2 increase dramatically upon differentiation (22, 26, 44, 58), presumably in preparation for virion assembly in the terminally differentiated strata of the epithelium. In HPV31, this increase is largely attributable to an upregulation of transcripts from the viral late promoter p742. This promoter initiates transcription from a family of start sites located near nucleotide 742 of the viral genome in the E7 open reading frame (ORF) (26, 46). Although activity from p742 is detectable in monolayer cultures, suggesting a basal transcriptional activity not dependent on differentiation, the steady-state level of transcripts originating from p742 increases dramatically upon differentiation of infected cells in rafts (44, 46) or upon suspension of the cells in semisolid medium (51). This differentiation responsiveness coupled to its apparent contribution to late gene expression make understanding p742 an important starting point in unraveling the late stages of the productive HPV life cycle.
It is assumed that, as in most promoters, there are elements in the HPV genome that together constitute a “core” p742 promoter, i.e., the information necessary and sufficient for basal transcription. The locations of such elements in the case of p742 or any other HPV late promoter are unknown, as are the roles of any enhancer elements in the upstream regulatory region or elsewhere. Differentiation responsiveness could be conferred by elements either within or in addition to those of the core promoter. The mechanism by which transcripts originating from p742 increase in response to differentiation has not been clearly demonstrated, but three hypotheses may be proposed. First, the increase in transcripts may be due to an increase in transcriptional initiation rate, or transactivation of the promoter, mediated by a combination of positive and negative transcription factors. Second, because all studies to date have measured only steady-state transcript levels, it is possible that increased transcript stability rather than initiation is the key regulatory event. In this case, the relevant genetic elements that would interact with either stabilizing or destabilizing protein factors would function in the RNA rather than DNA. A third and perhaps most mechanistically simple hypothesis is that because viral genome amplification and p742 transcript upregulation occur in the same strata of the tissue, increased gene dosage as a result of amplification may lead to a greater overall level of transcripts even in the absence of changes in initiation or degradation rates. In this case, the major relevant cis element would be the viral origin of replication.
Because the elements that control the basal or differentiation-inducible function of any HPV late promoter have not been systematically mapped, we have initiated studies aimed at identifying the elements that control transcription from HPV31 p742 and, in particular, the element(s) that controls responsiveness to differentiation. In this report, we show that the viral upstream regulatory region contains enhancer elements that influence p742 activity but not responsiveness to differentiation. We also map the core p742 promoter to a 150-bp region in the E7 open reading frame and show that p742 activation is not dependent on genome amplification. Finally, we show that although the region containing the start sites is not needed for strong promoter activity, it is necessary for responsiveness to differentiation.
MATERIALS AND METHODS
Plasmid constructs.
The oligonucleotides and primers (Integrated DNA Technologies, Coralville, Iowa) used for cloning the constructs described in this study are shown in Table 1. Primers 1 to 14 were designed to introduce a KpnI site at the 5′ end of the resulting PCR product. Similarly, primers 17 to 21 were designed to introduce a BglII site at the 3′ end. PCR was performed with the Expand High Fidelity PCR System (Roche, Indianapolis, Ind.) with pBS-HPV31 as a template. The PCR products were precipitated with ethanol, resuspended in water, digested with KpnI and BglII, purified with the Geneclean or Mermaid kits (Qbiogene, Carlsbad, California), and ligated into KpnI- and BglII-digested pGL2B (Promega, Madison, Wis.). Alternatively, the PCR products were cloned into the pCRII vector with the TA cloning kit (Invitrogen, Carlsbad, Calif.), and then subsequently freed by digestion with KpnI and BglII and ligated into digested pGL2B vector.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Name | Positions | Sequencea |
|---|---|---|---|
| 1 | CM742 1706 5 | 7067-7084 | ccgggtaccTGGATGTGTATGTAATAC |
| 2 | CM742 1530 5 | 7243-7260 | ccgggtaccCCTGTGTGTGTTGTGTAT |
| 3 | CM742 1380 5 | 7393-7410 | ccgggtaccCCATAGTAAAAGTTGTAC |
| 4 | CM742 1230 5 | 7543-7560 | ccgggtaccTTGGTTTCCTGAATACTA |
| 5 | CM742 1080 5 | 7693-7710 | ccgggtaccACATATATTATATTATCC |
| 6 | CM742 930 5 | 7843-7860 | ccgggtaccATAATTAATTGCATATAG |
| 7 | CM742 780 5 | 81-98 | ccgggtaccAGTATTTTGTGCAAACCT |
| 8 | CM742 630 5 | 231-248 | ccgggtaccGTATTAGATTTTGCATTT |
| 9 | CM742 480 5 | 381-398 | ccgggtaccACAAACAAAGGTATATGT |
| 10 | CM742 330 5 | 531-548 | ccgggtaccAGACCTCGTACTGAAACC |
| 11 | CM742 280 5 | 581-598 | ccgggtaccTTGCAAGACTATGTGTTA |
| 12 | CM742 180 5 | 681-698 | ccgggtaccCAGCTGGACAAGCAGAAC |
| 13 | CM742 130 5 | 731-748 | ccgggtaccTGTTGTCAGTGTAAGTCT |
| 14 | CM742 80 5 | 781-798 | ccgggtaccAGATATTCGCATATTGCA |
| 15 | CM742 30 5 | 831-861 | cGCCCCAACTGTTCTACTAGACTGTAACTACAa |
| 16 | CM742 30 3 | 862-831 | gatcTTGTAGTTACAGTCTAGTAGAACAGTTGGGGCggtac |
| 17 | CM742 3 | 861-844 | gcgagatctTGTAGTTACAGTCTAGTA |
| 18 | CM742 1520 3 | 680-663 | cgcgagatctGACTGTCT ATGACATCCT |
| 19 | CM742 1370 3 | 530-513 | cgcgagatctTCTCCAAC ATGCTATGCA |
| 20 | CM742 1220 3 | 380-363 | cgcgagatctCAATTTTT CTAATGTTGT |
| 21 | CM742 230 3 | 630-613 | cgcgagatctCAGTGGAGGTCAGTTGCC |
| 22 | CM31enhd 5 | 7413-7391 | GGTGTACAACTTTTACTATGGCG |
| 23 | CM31enhd 3 | 7790-7815 | TTTAAACTGCCAAGGTTGTGTCATGC |
| 24 | CM31KEd 5 | 7594-7569 | GCAGGAAACTACAAGCCAGAATGTTG |
| 25 | CM31AEd 3 | 7595-7621 | CTAACACACCTTGCCAACATATAATCC |
| 26 | 330Δ280 5 | 531-580 | cAGACCTCGTACTGAAACCCAAGTGTAAACATGCGTGGAGAAACACCTACGa |
| 27 | 330Δ280 3 | 580-531 | gatcTCGTAGGTGTTTCTCCACGCATGTTTACACTTGGGTTTCAGTACGAGGTCTggtac |
| 28 | 280Δ230 5 | 581-630 | cTTGCAAGACTATGTGTTAGATTTGCAACCTGAGGCAACTGACCTCCACTGa |
| 29 | 280Δ230 3 | 630-581 | gatcTCAGTGGAGGTCAGTTGCCTCAGGTTGCAAATCTAACACATAGTCTTGCAAggtac |
| 30 | 230Δ180 5 | 631-680 | cTTATGAGCAATTACCCGACAGCTCAGATGAGGAGGATGTCATAGACAGTCa |
| 31 | 230Δ180 3 | 680-631 | gatcTGACTGTCTATGACATCCTCCTCATCTGAGCTGTCGGGTAATTGCTCATAAggtac |
| 32 | CMp99TATA5 | 53-89 | CCGAAAAC GGTTGGgAgA cAAAGCACAT AGTATTTTG |
| 33 | CMp99TATA3 | 89-53 | CAAAATACTA TGTGCTTTgT cTcCCAACCG TTTTCGG |
Nucleotides not found in the HPV31 sequence are in lowercase.
Oligonucleotides 15 to 16 and 26 to 31 were designed so that when annealed to the complementary oligonucleotide, the ends would resemble KpnI- and BglII-digested overhangs and could be ligated directly into the digested vector. The primers used to create each plasmid construct were as follows, with the name of the plasmid followed by the letter designation of the HPV fragment in parentheses and the numbers of the primers used in PCR (Table 1): pGL2B-1706 (fragment A): 1 and 17; pGL2B-1530 (fragment B): 2 and 17; pGL2B-1380 (fragment C): 3 and 17; pGL2B-1230 (fragment D): 4 and 17; pGL2B-1080 (fragment E): 5 and 17; pGL2B-930 (fragment F): 6 and 17; pGL2B-780 (fragment G): 7 and 17; pGL2B-630 (fragment H): 8 and 17; pGL2B-480 (fragment I): 9 and 17; pGL2B-330 (fragment J): 10 and 17; pGL2B-180 (fragment K): 12 and 17; pGL2B-130 (fragment L): 13 and 17; pGL2B-80 (fragment M): 14 and 17; pGL2B-30 (fragment N): 15 and 16; pGL2B-630Δ180 (fragment O): 8 and 18; pGL2B-630Δ330 (fragment P): 8 and 19; pGL2B-630Δ480 (fragment Q): 8 and 20; pGL2B-480Δ180 (fragment R): 9, and 18; pGL2B-480Δ330 (fragment S): 9 and 19; pGL2B-330Δ180 (fragment T): 10 and 18; pGL2B-330Δ230 (fragment T1,2): 10 and 21; pGL2B-280Δ180 (fragment T2,3): 11 and 18; pGL2B-330Δ280 (fragment T1): 26 and 27; pGL2B-280Δ230 (fragment T2): 28 and 29; and pGL2B-230Δ180 (fragment T3): 30 and 31.
The p99 TATA mutants were made with primers 32 and 33, with the corresponding wild-type plasmids as templates, and the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Constructs containing internal deletions of the auxiliary and/or keratinocyte enhancers were made by PCR with pGL2B-1706 as a template with primers that diverge away from the region to be deleted, leaving a linearized fragment lacking the deleted region. The linearized fragment was recircularized to create a plasmid with the deletion. The enhancer deletions were made with the following primers: pGL2B-1706 ΔAE, 22 and 25; pGL2B-1706 ΔKE, 23 and 24; and pGL2B-1706 ΔAEKE, 22 and 23.
Cell culture and transfections.
CIN-612 9E primary human foreskin keratinocytes (HFK), and C33A cells were maintained with standard cell culture techniques as described (53). Human foreskin keratinocytes were isolated from neonatal foreskins and maintained in medium 154 (Cascade Biologics) as described (40). The HFK31a:1 cell line was derived from electroporation of human foreskin keratinocytes with HPV31a genomic DNA as described (43) and maintains HPV31 genomes episomally (39). Transfection followed by monolayer culture or differentiation by suspension in 1.6% methylcellulose was performed as reported previously (10, 11, 53), except that cells were plated directly in KGM overnight before transfection.
For the experiments in Fig. 4 to 6, 2 μg of DNA/well (monolayer) or 8 μg of DNA/plate (methylcellulose) was transfected with 10 μl/well (monolayer) or 40 μl/plate (methylcellulose) of Lipofectamine reagent. Lysates from transfected cells were prepared and assayed for luciferase activity as described (10, 11, 53). Experiments were repeated three to six times (monolayer) or four to eight times (methylcellulose). The luciferase activity for each experimental construct was normalized to the activity of pGL2B-1706 (Fig. 2 and 3) or pGL2B-630 (Fig. 4 to 6) for the individual experiment, and the normalized values were averaged. Error bars represent one standard error of the mean.
FIG. 4.
Localization of the core p742 promoter. Reporter constructs containing 5′ or 3′ deletions of the E6/E7 region were transfected into CIN-612 9E cells and cultured under (a) monolayer or (b) methylcellulose conditions. Numbers to the left and right of the bars in the schematic diagram represent the nucleotide positions of the 5′ and 3′ termini of the fragments in the HPV31 genome. The raw luciferase values were normalized to the value of fragment H within each experiment, and then the normalized values were averaged. Bars represent ± 1 standard error of the mean.
FIG. 6.
Activity of segments of the proximal element (fragment T). Numbers to the right and left of the bars in the schematic diagram represent the nucleotide positions of the 5′ and 3′ termini of the fragments in the HPV31 genome. Reporter constructs were transfected into CIN-612 9E cells and cultured in monolayer (a) or methylcellulose (b). The raw luciferase values were normalized to the value of fragment H within each experiment, and then the normalized values were averaged. Bars represent ± 1 standard error of the mean.
FIG. 2.
Deletion analysis of the p742 upstream region. A 1,706-bp fragment of the HPV31 genome extending from the 3′ end of the L1 gene through the upstream regulatory region (URR) and E6 and E7 genes to the 5′ end of the E1 gene was cloned into the pGL2B luciferase reporter vector (fragment A). AE, auxiliary enhancer; KE, keratinocyte enhancer. 5′ deletions of this fragment with wild-type sequence (black bars) or containing a mutated p99 TATA box (grey bars) were also generated and cloned. Numbers to the left of the bars in the schematic diagram represent the nucleotide position of the 5′ end of the fragment in the HPV31 genome. The constructs were transfected into CIN-612 9E cells (a and d), primary HFKs (b and e), or C33A cells (c) and cultured in monolayer (a, b, and c) or in methylcellulose (d and e) as described in Materials and Methods. After 48 h, the cells were washed in phosphate-buffered saline, lysed, and assayed for luciferase activity. The raw luciferase values were normalized to the value of fragment A within each experiment, and then the normalized values were averaged. Bars represent ± 1 standard error of the mean. Bent arrows represent transcriptional start sites.
FIG. 3.
Deletion of upstream regulatory region enhancer elements. Reporter constructs lacking the auxiliary enhancer (ΔAE), keratinocyte enhancer (ΔKE), or both (ΔAE/KE) were transfected into CIN-612 9E cells, cultured under (a) monolayer or (b) methylcellulose conditions, and assayed as described in the legend to Fig. 1. Numbers to the left or under the bars in the schematic diagram represent the nucleotide positions in the HPV31 genome of the 5′ and 3′ termini or of the deleted regions within the fragments. The raw luciferase values were normalized to the value of pGL2B-1706 within each experiment, and then the normalized values were averaged. Bars represent ± 1 standard error of the mean.
To assay differentiation responsiveness, we performed duplicate transfection in 100-mm cell culture dishes as described above for methylcellulose transfections. Following the transfection, the cells were washed with phosphate-buffered saline and detached from the plate by trypsin treatment. Duplicate transfections were pooled and centrifuged, and half of the cells were suspended in methylcellulose and half were plated in 100-mm dishes. Following 48 h of culture, the methylcellulose was washed by two to three rounds of dilution in phosphate-buffered saline followed by centrifugation, the monolayer cultures were detached by trypsin treatment, and both samples were resuspended in 1 ml of phosphate-buffered saline. The cells in each sample were counted with a Coulter Z1 particle counter (Coulter Corp.), and then equal numbers of cells were added to a microcentrifuge tube, centrifuged briefly, resuspended in passive lysis buffer, and assayed as described (10, 11, 53). Induction was calculated by subtracting the value of the empty vector from the raw monolayer or methylcellulose luciferase values for each condition and then dividing the resulting methylcellulose value by the resulting monolayer value. Data represent the mean of 6 to 12 experiments. Error bars represent one standard error of the mean.
Analysis of replication and transcript expression.
GF109203X hydrochloride (62) was obtained from Alexis Biochemicals, dissolved in water at 3 mg/ml, and stored at −20°C in small aliquots. CIN-612 9E cells were plated overnight at a density of 1 × 106 to 2 × 106 cells/100-mm plate. The next day, cells were trypsinized and grown either in monolayer cultures at 106 cells/100-mm plate or suspended at 106 cells/10 ml of methylcellulose. GF109203X was diluted in E medium (monolayer) or in methylcellulose before the addition of cells. Following 48 h of culture, cells were harvested by washing (methylcellulose) or by treatment with trypsin (monolayer), and the cultures were divided for either DNA or RNA analysis. DNA extraction and Southern analysis were performed as described (42, 43). The probe for the RNase protection assay was prepared by digesting pGL2B-330 with KpnI and BglII and ligating the resulting HPV fragment (J) into pGEM-7Zf(+) (Promega) which had been digested with KpnI and BamHI. The resulting plasmid (pGEM-330) was linearized with KpnI, treated with protease K, phenol-chloroform-isoamyl alcohol extracted, precipitated with ethanol, and resuspended in Tris-EDTA. RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer's instructions, followed by treatment with DNase as described (44); 10 μg of total RNA was analyzed by RNase protection assay as described (44) with the RPA III kit (Ambion) replacing the RPA II kit. A probe against cyclophilin (Ambion) was included as an internal control (57).
To detect the start sites used on reporter plasmid templates, we transfected 9.6 × 106 HFKs with 48 μg of reporter plasmid or empty vector DNA with Lipofectamine reagent as described (10, 11, 53). Following 48 h of incubation, the cells were trypsinized and pelleted by centrifugation, and total RNA was harvested with Trizol reagent. mRNA was purified from total RNA with the Oligotex mRNA mini kit (Qiagen); 1 μg of total RNA from the HFK31a:1 cell line or 1 μg of poly(A)-selected mRNA from transfected HFKs were mixed with riboprobe, coprecipitated with 5 μg of yeast RNA, and then subjected to hybridization and RNase protection assay analysis as described above.
RESULTS
To analyze the functional elements of p742, we used the pGL2B luciferase reporter plasmid (10, 11, 53). By cloning various wild-type and mutant HPV sequences into this plasmid, we could determine their transcriptional activities with luciferase activity as a readout. This strategy required isolating a region of the HPV31 genome to drive reporter activity. Because the boundaries of p742 have not been defined, any selection of a subset of HPV sequences involved some risk that a relevant element would be excluded. We reasoned that the promoter was likely to contain the region encompassing the start sites. From work in our laboratory (44, 46) and others (26), we knew that p742 drives transcription from a family of start sites in the E7 open reading frame between nucleotides 700 and 800 of the HPV31 genome. We assumed that most or all of the control elements for this promoter would be located 5′ of this region. Because of the large number of start sites, to ensure that all would be included, we set the nucleotide just before the ATG of E1 (nucleotide 861) as the 3′ boundary of our region of study. This position also serves as the reference point (+1) for locations within the promoter. Because the upstream regulatory region of HPV31, like that of other HPVs, contains enhancer elements that control the early promoter(s), and considering that enhancers can work at some distance from a promoter, we reasoned that some elements in the upstream regulatory region may be important for p742 regulation as well. We therefore included the entire upstream regulatory region in our cloned fragment, setting the 5′ boundary just downstream of the stop codon of L1 (nucleotide 7066). This region, extending from the end of L1 through the upstream regulatory region and the E6 and E7 open reading frames to just before the beginning of the E1 open reading frame, totaled 1,706 bp. This fragment (A) was subcloned into pGL2B to create pGL2B-1706.
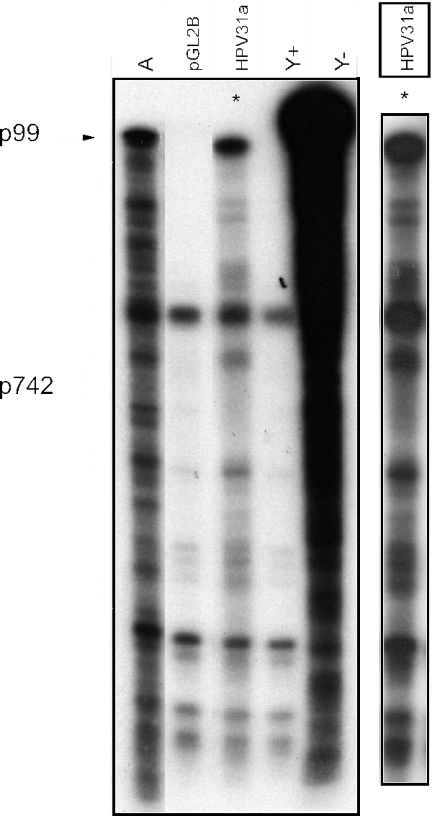
To determine whether the pattern of transcriptional activity from a transiently transfected fragment A reporter resembles the pattern from episomally replicating whole virus genomes, we transfected either the empty vector or the fragment A reporter plasmid into HFK cells. After 48 h of incubation in monolayers, we isolated mRNA and analyzed the transcripts by RNase protection assay. Total RNA from a cell line maintaining HPV31a genomes episomally was also analyzed for comparison. The results are shown in Fig. 1. Some nonspecific bands are visible, probably as a result of the vast excess of probe relative to target RNA in this experiment. However, the transcriptional pattern from the transfected reporter largely resembles that of the infected cell RNAs. The larger size of the top band is probably due to additional protection by polylinker- and vector-derived sequences in the riboprobe by vector sequences present in reporter RNAs. Minor alterations in the positions of the bands are seen but are probably not significant for the purposes of measuring overall activity, given that even extensive alterations of start site patterns often do not alter the activity or regulation of multiple start promoters (4, 7, 12, 36, 48, 56). It is evident that no essential information for generally wild-type p742 transcription is present downstream of nucleotide 861.
FIG. 1.
RNase protection assay of transcripts from transfected reporter template. A 1,706-bp fragment of the HPV31 genome extending from the 3′ end of the L1 gene through the upstream regulatory region (URR) and E6 and E7 genes to the 5′ end of the E1 gene was cloned into the pGL2B luciferase reporter vector to create pGL2B-1706 (fragment A). This plasmid or the empty vector was transfected into HFK cells and incubated in monolayer culture. After 48 h, mRNA was isolated and RNase protection analysis was performed with a riboprobe specific for transcripts in the p742 region. The RNA analyzed in each lane is as follows: A, mRNA from HFKs transected with the fragment A reporter; pGL2B, mRNA from HFKs transfected with the empty vector; HPV31a, total RNA from monolayer HFK31a:1 cells; Y+ and Y−, total yeast RNA with and without nuclease digestion, respectively. A darker exposure of the HPV31a lane (*) from the same gel is shown to the right.
Mapping the enhancer.
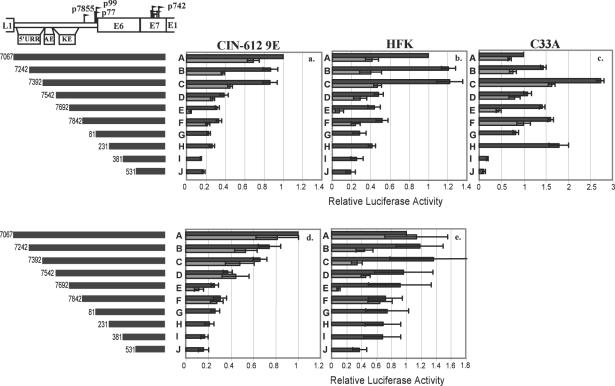
To determine the 5′ boundary of the promoter region as well as any enhancer elements that may be present, we created a series of 5′ deletion constructs that differed by150 bp (Fig. 2). The HPV fragments represented by these deletions are designated with the letters A to J and have the 5′ ends indicated, with a common 3′ end at nucleotide 861. The reporter plasmids were transfected into three different cell types. The CIN-612 9E cell line is derived from a low-grade cervical lesion and maintains HPV31b episomally (26). This cell line is capable of producing infectious virions when grown in organotypic culture and therefore possesses all of the viral and cellular factors needed for the virus to complete its life cycle (42). We used this cell line to represent the situation in a normal infected cell with all of the necessary cellular and viral gene products present. Normal human foreskin keratinocytes (HFKs) are primary cells derived from neonatal foreskins. These cells are not immortalized, contain no viral gene products, and represent the cell environment as it would be found initially upon infection. The C33A cell line is derived from a cervical carcinoma but is not known to harbor any HPV sequences (15). Although not representative of either the infected or the normal uninfected situations, C33A cells are often used in transcriptional studies of HPV and were therefore initially included for comparison.
Each of these cell types was transfected with the deletion constructs described and grown in monolayer culture, which represents the undifferentiated condition. The results are shown in Fig. 2a, b, and c (black bars). A number of findings are apparent. First, the pattern of activities of these constructs in C33A cells differed from that seen in CIN-612 9E cells or HFKs. Several elements appeared to be repressive in C33A cells but not in the other cell types, for example, between nucleotides 7242 and 7392 (−1530 to −1380) and between 81 and 231 (−780 to −630). Second, in both CIN-612 9E cells and HFKs, there was a loss in transcriptional activity relative to the activity of fragment A as sequences were deleted from the 5′ end. In either cell type, the bulk of the loss of activity was between nucleotides 7392 and 7542 (−1380 to −1230) bp upstream of E1 (compare fragments C and D). This region was previously identified as an enhancer and called the auxiliary enhancer (30). Third, there also was only a minor loss of activity upon subsequent deletions up to within 330 bp of the E1 start codon.
Fragment A contains not only p742 but also p99, the major viral early promoter (46). It is likely that p99 also contributes to reporter activity in the context of these constructs. To test this possibility, we created variants of the deletion mutants containing a mutation in the p99 TATA box. This mutation was predicted to abolish p99 promoter activity (49) and was found by RNase protection assay to abolish the start site at nucleotide 99 and by luciferase assay to eliminate transcriptional activity from the upstream regulatory region (data not shown). These TATA mutant constructs were transfected into CIN-612 9E, HFK, and C33A cells as described, and the results are also shown in Fig. 2a, b, and c (grey bars; note that although fragment G contains the start site for p99, the TATA box is absent.) These data indicate that roughly half of the activity of each construct was attributable to p99 in monolayer culture. Furthermore, the loss of activity seen with deletion of nucleotides 7392 to 7542 was no longer apparent without p99 activity, indicating that the contribution of the auxiliary enhancer to reporter activity is mediated through p99 in monolayer culture. Finally, there was little consistent effect of even extensive 5′ deletion on reporter activity in the absence of p99, indicating that in monolayer culture, the elements that drive the activity of p742 must be located very close to the start sites. Our laboratory has found that studies of HPV in C33A cells frequently yield results that differ substantially from those obtained in cell types that more closely resemble infected or uninfected cells in vivo (Meyers et al., unpublished observations). Because of these disparities, we did not pursue further studies with C33A cells.
Because p742 transcripts increase upon differentiation, it was important to examine the activity of p742 in differentiating conditions in order to understand how this promoter is regulated. Suspension of keratinocytes in medium containing 1.6% methylcellulose causes at least a population of cells to differentiate (51), providing a convenient method to test numerous transfected DNAs under differentiating conditions. CIN-612 9E and HFK cells were transfected and suspended in methylcellulose for 48 h, followed by washing and measurement of luciferase activity. Figures 2d and e show the data from these experiments and permit several conclusions. First, although a significant portion of the activity of the reporters in monolayer was attributable to p99, the effect of the p99 TATA mutation was less apparent in methylcellulose culture in CIN-612 9E cells. This is expected since p99 is constitutive and p742 is inducible (46). As p742 is induced by differentiation, the relative importance of p742 in driving the reporter should be increased, and so the loss of a p99 component should have less of an impact. The reason for the more complex pattern of activities seen in HFKs is not understood.
Second, deletion from the 5′ end resulted in gradually reduced activity in methylcellulose culture, especially in CIN-612 9E cells, indicating that the upstream regulatory region contributes to p742 activity under differentiating conditions. Because in methylcellulose most of the activity was attributable to p742, at least in CIN-612 9E cells, these elements appear to be authentic regulators of p742 activity. We do not understand the consistently low activity of the TATA mutant fragment E relative to fragments D and F.
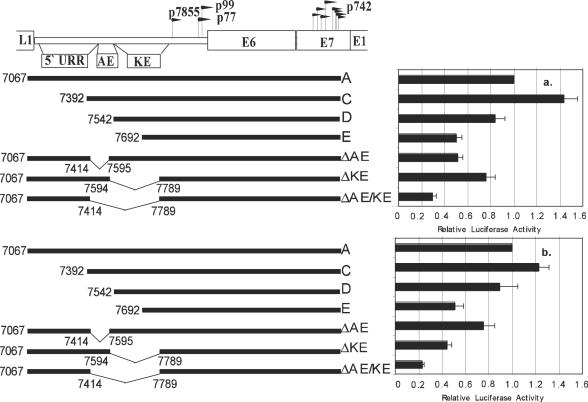
To test whether the auxiliary enhancer, the neighboring keratinocyte enhancer (35), or both regions contribute to p742 activity, we made internal deletions of each region either alone or in combination in the context of pGL2B-1706 (fragment A) and studied their activities in monolayer and methylcellulose cultures as described above. In both monolayer and methylcellulose culture, deletion of the auxiliary enhancer, the keratinocyte enhancer, or both led to a reduction of activity relative to the wild type (Fig. 3). Although the differences between deletion of the auxiliary enhancer and deletion of the keratinocyte enhancer were not sufficient to unambiguously determine which is the more important element, it is clear that both contributed to activity under both monolayer and methylcellulose conditions in CIN-612 9E cells. Because activity in methylcellulose is attributable mostly to p742 in CIN-612 9E cells, we can conclude that both the auxiliary enhancer and keratinocyte enhancer contribute to p742 activity, at least in methylcellulose. We therefore suggest that the auxiliary enhancer and keratinocyte enhancer act as enhancers of p742 activity under differentiating conditions.
Mapping the core promoter.
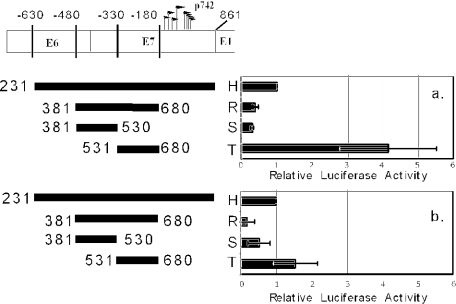
To disentangle the contribution of p99 from that of p742, we chose to focus our attention on fragment H, which includes the region surrounding and upstream of the p742 region but does not contain p99, the other upstream regulatory region promoters (46), or any of their reported control elements. It also lacks the upstream regulatory region, meaning that its activity should represent that of the core p742 promoter, the minimal region necessary and sufficient to drive basal transcription without the contribution of enhancers. To map the minimal region sufficient to drive transcription, we made a series of deletions of fragment H and tested them for activity in the CIN-612 9E cell line, under either monolayer or methylcellulose culture conditions (Fig. 4).
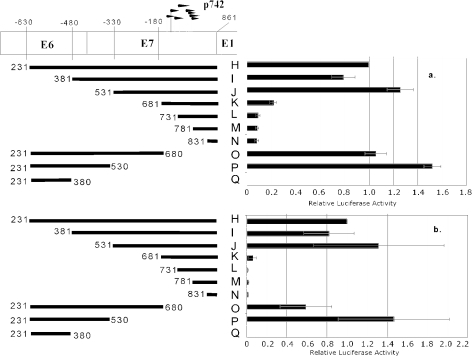
5′ deletion through the promoter revealed that information essential to transcriptional activity in both monolayer and methylcellulose culture maps to between 330 and 180 bp upstream of the start codon of E1 (fragment J versus K, corresponding to nucleotides 531 to 681 of the genome), indicating that the 5′ boundary of a major transcriptional element resides in this region. In contrast, by 3′ deletion, most activity was lost by deletion of the segment between 330 and 480 bp upstream of E1, corresponding to nucleotides 381 to 530 of the genome (fragment I versus J). It is important to note that fragment J and fragment P have no sequence in common. Because both fragments had activity comparable to that of the full-length fragment H, there must be at least two elements in this region, one with a 3′ boundary between nucleotides 380 to 530 and one with a 5′ boundary between nucleotides 531 to 681, either of which is necessary for driving transcription in the absence of the other. We also note that the two elements must not be additive, because deletion of either did not lead to a loss in activity as long as the other was present (fragments H and I versus J and H and O versus P). We designated the element mapping downstream of 531 the proximal element and the element mapping upstream of 530 the distal element. Finally, we observe that the known major transcriptional start sites for p742 (Fig. 1) are located within 180 bp of the E1 start codon (16, 44, 46). Because neither fragment O nor fragment P contains this region but both retain robust transcriptional activity, the DNA surrounding the transcriptional initiation sites does not play a role in controlling the strength of p742 activity.
The experiments above show that the presence of either the proximal element or distal element is necessary for the transcriptional activity of the reporter constructs. Information necessary for the activity of the distal element is located between nucleotides 380 and 530, and information necessary for the activity of the proximal element is located between nucleotides 531 and 681. To determine whether either of these necessary regions is sufficient to drive the activity of the proximal element or the distal element, we created and tested fragments that isolate the two regions from surrounding sequences, either separately or in combination (Fig. 5). In contrast to fragment P (see Fig. 4), the nucleotide 381 to 530 segment alone (fragment S) was unable to support transcription to a level comparable to that of fragment H, indicating that some information between nucleotides 231 and 380 is necessary for distal element activity but is not sufficient (see fragment Q, Fig. 4). The segment between nucleotides 531 and 680 in isolation (fragment T), however, was sufficient for robust activity, suggesting that this fragment contains the entire proximal element.
FIG. 5.
Activities of the proximal and distal elements. Numbers to the right and left of the bars in the schematic diagram represent the nucleotide positions of the 5′ and 3′ termini of the fragments in the HPV31 genome. Reporter constructs were transfected into CIN-612 9E cells and cultured in monolayer (a) or methylcellulose (b). The raw luciferase values were normalized to the value of fragment H within each experiment, and then the normalized values were averaged. Bars represent ± 1 standard error of the mean.
When the information between nucleotides 381 and 530 is added to the segment containing proximal element activity, the activity was low (fragment R), suggesting that although some information between nucleotides 381 and 530 is necessary for the distal element, these elements inhibited the activity of the proximal element. This inhibition was not seen when upstream sequences were present (see fragment O, Fig. 4), suggesting that the observed repression of activity was counteracted by elements between nucleotides 231 and 380 (−630 to −480). Relative to fragment H, the proximal element (fragment T) had substantially higher activity in monolayer than in methylcellulose. This result suggests either that the proximal element is repressed by differentiation in methylcellulose or that fragment H is activated by differentiation.
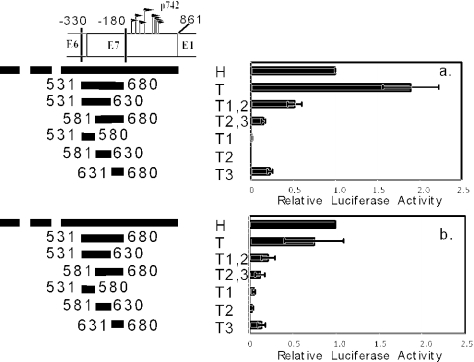
To determine whether the 150-bp fragment containing the proximal element (fragment T) can be further subdivided, we created constructs in which 50-bp segments of fragment T were placed upstream of luciferase, either alone or in combination, to test their activities (Fig. 6). Again, fragment T drove transcription at a level comparable to that of fragment H, and the relative activity was higher in monolayer than in methylcellulose. None of the three 50-bp segments (T1, T2, or T3) could drive transcription as well as fragment T in either monolayer or methylcellulose. Additionally, 100-bp fragments consisting of the upstream two (T1 and T2) or downstream two (T2 and T3) 50-bp segments also did not support transcription to a degree comparable to fragment T. This indicates that information essential for the function of the proximal element is present in each of these three 50-bp segments. Based on these results, we conclude that fragment T contains necessary and sufficient information for promoter activity and designated it the core p742 promoter.
Relationship of amplification to upregulation of p742 transcripts.
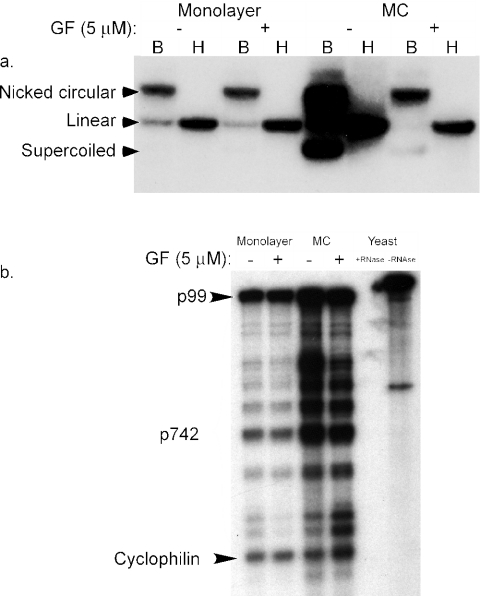
Having mapped the enhancer and basal promoter elements of p742, we turned our attention to the regulation of the promoter by differentiation. As described above, several hypotheses could explain the increase in the steady-state level of transcripts from p742 when cells differentiate. A simple possibility is that differentiation-dependent amplification of viral genomes provides an increased number of templates from which transcription can occur. Template amplification would lead to an increase in the steady-state level of transcripts even in the absence of changes in rates of transcription initiation or degradation. To attempt to prevent differentiation of CIN-612 9E cells suspended in methylcellulose, we treated cells with the general protein kinase C inhibitor GF109203X (62). This compound has been reported to block the cell cycle withdrawal and induction of involucrin expression that accompany suspension of primary keratinocytes in methylcellulose culture (60). If this drug also blocked differentiation-dependent genome amplification, it could be used to test whether template amplification is necessary for p742 activation. The results are shown in Fig. 7.
FIG. 7.
Independence of p742 upregulation from amplification. (a) Southern analysis of CIN-612 9E cells cultured in monolayer or methylcellulose (MC), with or without 5 μM GF109203X. B, digested with BamHI, which does not cut the HPV31b genome; H, digested with HindIII, which digests the HPV31b genome once. (b) RNase protection analysis of CIN-612 9E cells from the same experiment as part a. Total RNA was hybridized with a probe specific to the p742 start site region, subjected to RNase treatment, and analyzed by electrophoresis as described in Materials and Methods. The positions of bands corresponding to the initiation sites of p742 as well as the cyclophilin internal control are indicated. Samples containing yeast RNA hybridized with the probes and treated with RNase or untreated are also indicated.
By Southern analysis (Fig. 7a), HPV31b genomes in CIN-612 9E cells amplified when the cells were differentiated in methylcellulose, in agreement with previous results (51). Treatment of monolayer cells with GF109203X had no effect on copy number, but addition of GF109203X to methylcellulose cultures blocked amplification of the viral genome so that the copy number remained at monolayer levels. When the RNA from these cultures was examined by RNase protection assay with a probe for the p742 region (Fig. 7b), the multiple start sites were clearly visible and there was an increase in these transcripts as a result of methylcellulose culture, as expected (51). Strikingly, aside from minor variations in band intensity at some positions, GF109203X had no effect on p742 activation in response to differentiation in methylcellulose. This result clearly indicates that p742 activation can proceed in the absence of significant genome amplification and that an increase in gene dosage as a result of genome amplification cannot account for the differentiation-dependent increase in p742 transcripts.
Mapping differentiation response elements.
In Fig. 5 and 6, fragment T, the core promoter, had higher activity in monolayer culture than fragment H, whereas in methylcellulose, its activity was equivalent to or less than that of fragment H. This result supports the possibility that fragment H was activated by methylcellulose culture while fragment T was not, so that the activity of H increased relative to that of T. However, this experimental approach is insufficient to test that possibility directly. The monolayer and methylcellulose values shown in Fig. 2 to 6 show patterns of transcriptional activity compared to a reference construct (fragment A or fragment H) under each culture condition. The values for each condition (monolayer or methylcellulose) were obtained in independent experiments, and because the transfection conditions and the treatment of the cells differed between monolayer and methylcellulose cultures, the activities could be compared directly. To determine the locations of differentiation-responsive elements in the DNA with the reporter strategy, it is necessary to directly compare the activity in monolayers to that in methylcellulose for each construct.
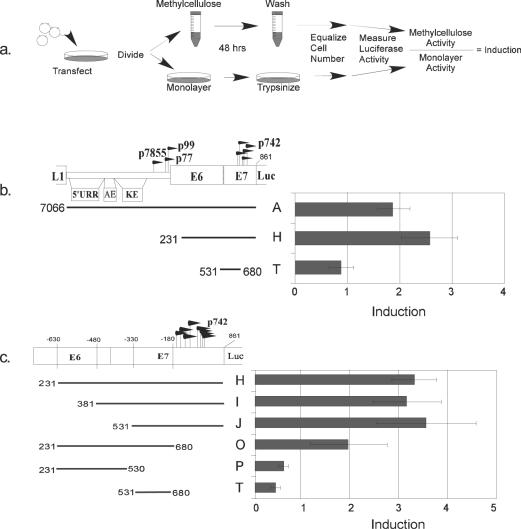
Accordingly, we altered our standard transfection assays according to the scheme diagramed in Fig. 8a. We performed a transfection of each reporter plasmid construct in duplicate and pooled the cells. We split the sample into two and cultured half in methylcellulose and half in monolayer. After harvesting, the number of cells in each sample was equalized for the luciferase reaction. Because the original transfections and the final cell number were the same, any difference in activity must be due to the conditions of the culture, i.e., whether the cells were differentiated in methylcellulose or not. The ratio of activity in methylcellulose to that in monolayer was termed induction, which is the responsiveness of a given reporter to differentiation.
FIG. 8.
Mapping the major differentiation response element. (a) Schematic diagram of the differentiation response assay. (b) Differentiation response of p742, the core promoter, and the upstream regulatory region (URR). Transfections were performed as described in the text and diagramed in part a. (c) Mapping the differentiation response element (DRE). Bars represent ± 1 standard error of the mean.
When this analysis was performed with our reporters (Fig. 8b), we found that fragment H, which was expected to contain the p742 promoter more or less in its entirety, was inducible by differentiation approximately 2.5-fold. The core promoter, fragment T, was not inducible. Fragment H does not contain the viral origin of replication, adding further evidence that viral genome replication is not necessary for the differentiation responsiveness of p742. Fragment A, which includes the upstream regulatory region and p99, was also inducible by approximately twofold. This level was comparable to that of fragment H, even though the total activity of fragment A is higher than that of H in both monolayer and methylcellulose conditions (Fig. 2a and d). Thus, although the upstream regulatory region contributed to the strength of transcriptional activity, elements surrounding the core promoter control the differentiation response. This result illustrates the distinction between activity and induction.
Using our panel of deletion constructs, we found that a major differentiation response element (DRE) is located downstream of nucleotide 531 (3′ of −330), within fragment J, and that upstream sequences are dispensable for this response (compare fragment J to H and I). We designated this element DRE1. Information important for DRE1 is found between nucleotides 680 and 861 (3′ of −180, compare fragments J and T). Because the region downstream of nucleotide 680 alone (fragment K) had no transcriptional activity (see Fig. 4), it is not by definition inducible. Note that nucleotides 680 to 861 contain the transcriptional start sites and sequences in the 5′ untranslated region of the p742 mRNAs. It also appears that sequences between nucleotides 231 and 530, although not sufficient for a response alone (fragment P), may confer a response when added to the core promoter (fragment T) in the absence of the information downstream of nucleotide 680 (compare fragments O and T). This suggests the possibility of a distinct element in this region that is sufficient for differentiation inducibility but independent the information between nucleotides 680 and 861. The differentiation response element between nucleotides 231 and 680 we tentatively designated DRE2, although further experiments will be required to determine whether it is distinct from DRE1.
DISCUSSION
In this study, we used a reporter strategy to locate enhancer and core promoter elements of p742, the HPV31 late promoter. In many transcription studies, the term core promoter is used to mean the minimal elements needed to drive transcription from a fixed start site (50). In the case of promoters with multiple start sites such as p742, this definition is problematic. We use the term core promoter somewhat more broadly to mean the minimal DNA region necessary and sufficient to drive transcription. We have shown that in CIN-612 9E cells in monolayer culture, mutation of the p99 TATA box in the context of our reporters results in a substantial reduction in transcriptional activity (Fig. 2a). This loss of activity by TATA mutation is mirrored by 5′ deletion of enhancer elements shown by others to be important for p99 activity (30, 35), indicating that total luciferase activity in monolayer has both a p99 and p742 component. On the other hand, mutation of the p99 TATA fails to significantly reduce reporter activity in CIN-612 9E cells in methylcellulose (see Fig. 2d), suggesting strongly that the majority of the reporter activity in methylcellulose originates from p742. The observation that deletion of enhancer elements under differentiating conditions reduces reporter activity (see Fig. 2d and 3) indicates that the enhancers previously identified in the upstream regulatory region have an effect on the activity of p742 even in the absence of a p99. Whether p99 and p742 respond to exactly the same cis elements and same binding factors remains to be determined. Protein factors bound to various DNA elements in the upstream regulatory region change as a function of differentiation (52), so a given element could affect one or both promoters differently under undifferentiated versus differentiated conditions.
We also mapped the major differentiation response element of p742 and demonstrated that the differentiation response is independent of viral genome amplification. Two lines of reasoning illustrate this independence. First, the major differentiation-responsive element does not map to the origin of replication, because the level of induction of the p742 reporter (fragment H) was not augmented upon the addition of the upstream regulatory region (i.e., fragment A), which contains the minimal viral origin (20, 25). Thus, the elements needed for replication are distinct from those that confer differentiation response (Fig. 8b). CIN-612 9E cells contain the E1 and E2 proteins, which are the two viral factors needed for replication, and a transfected origin-containing plasmid such as fragment A could conceivably replicate in these cells. On the other hand, because we have not determined whether such replication actually takes place, the failure of fragment A to be more differentiation responsive than fragment H cannot be taken alone as clear evidence that replication per se is not involved in the differentiation response.
More direct evidence that p742 is independent of amplification is that inhibition of protein kinase C activity in methylcellulose culture can separate amplification of genomes from activation of p742 (Fig. 7). Since p742 was upregulated in the same cultures at the same time that amplification was inhibited clearly demonstrates that increased genome copy number was not necessary for increased transcripts from p742 in methylcellulose. The results of Flores et al. (18) demonstrate that late replication involves not only a quantitative change in genome copy number (i.e., amplification) but also a qualitative change in replication mechanism. This change in mechanism undoubtedly involves changes in template structure in addition to template number. We have shown that inhibition of protein kinase C can block amplification, but we have not shown that GF109203X can block any qualitative changes in the template associated with late replication. Although our results indicate that the quantitative phenomenon of amplification is not necessary for p742 activation, it is possible that as yet unidentified structural changes associated with late replication may still be needed. A more thorough understanding of the structural and mechanistic basis of amplification will help resolve this question. The observation that amplification was effectively blocked by GF109203X indicates that protein kinase C activity is important for differentiation-dependent viral replication, and experiments are under way to explore this observation.
Although transcripts from p742 become abundant upon cellular differentiation, it has not been shown definitively that this increase in transcripts is mediated at the transcriptional level, that is, by direct promoter activation, as opposed to stabilization of transcripts during differentiation. The experiments reported here also did not distinguish between these possibilities. Posttranscriptional modification of papillomavirus transcripts can change with differentiation (5, 27, 59), and these changes could result in stabilization of transcripts from p742, although this has not been demonstrated.
A noteworthy finding in this study is that information needed for the function of DRE1 lies downstream of the transcription start sites. Because of this location, information contained in part of DRE1 would be present in the 5′ untranslated region of all p742 transcripts, leaving open the possibility that this element is in fact an RNA stability element. Although many elements controlling RNA stability are located at the 3′ ends of mRNAs, some elements in 5′ untranslated regions also have been described (23). p742-derived transcripts have either of two 3′ ends, corresponding to either the early or late polyadenylation signal (44, 59). Conservation of a stability element at the 5′ end of p742 transcripts would allow both classes of transcripts to be upregulated by the same mechanism. However, the location of essential DRE1 elements downstream of the core promoter does not exclude the possibility that the relevant elements are DNA elements, as transcription factor binding sites downstream of initiation sties are hardly rare, especially in multiple-start promoters (8, 9, 13, 14, 17, 24, 29, 33, 38, 63).
It is possible that both mechanisms contribute to the final activation. Perhaps transactivation versus stabilization may turn out to distinguish DRE2 versus DRE1. The resolution of our mapping studies was insufficient to unambiguously separate DRE1 from DRE2, so they may actually share important information (e.g., within fragment T). We note that the level of induction as measured by luciferase reporter assays is less than that observed at the RNA level in infected cells (compare Fig. 7 and 8). Assuming that the luciferase assays predominantly measure transcriptional effects, the remaining induction observed at the RNA level may be due to additional posttranscriptional effects. A similar phenomenon was observed in the simian virus 40 late promoter, in which both transcriptional and nontranscriptional mechanisms contribute to the final transcript level (31).
We favor the hypothesis that p742 is transcriptionally activated at least to some degree as a function of differentiation because of several circumstantial considerations. First, a marked increase in DNase I hypersensitivity occurs around the p742 region as a function of differentiation (16), suggesting that the DNA template may become more competent for transcription under these conditions. Second, if transcription were to be upregulated by differentiation, two nonexclusive mechanisms could be responsible: gain of a differentiation-specific activator or loss of a repressor. Binding of human Skn-1a, an Oct family transcriptional activator specifically expressed in differentiating epithelia (2), has been shown for the analogous promoter (p670) of HPV16 (34). Binding of CCAAT displacement protein (CDP/cut), a transcriptional repressor not found in differentiating cells, has been demonstrated for both HPV6 and HPV31 and may also result in alleviation of repression as a function of differentiation (1).
We note (Fig. 9) that DRE1 contains potential binding sites for both of these factors as well as others, such as Tst-1 and C/EBP, that have also been implicated in epithelial or HPV differentiation-dependent transcription (3, 52, 64). That fragment T is more active than fragment H in monolayer but approximately equal in methylcellulose (Fig. 5 and 6) supports the idea that H may contain an element that is repressive in undifferentiated conditions and would result in activation of p742 as the cells differentiate. Further studies are under way to determine whether transactivation is in fact a mechanism by which p742 transcripts are upregulated and what sites and mechanisms are responsible for the activity of the differentiation response elements in vivo.
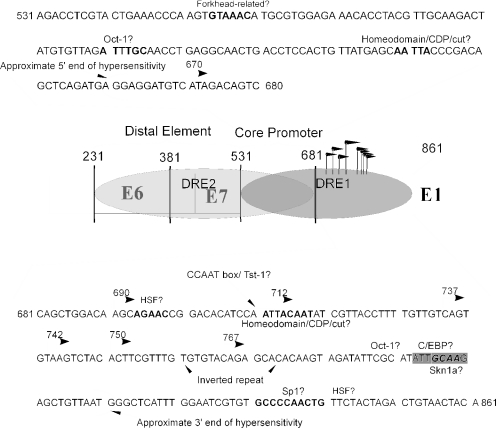
FIG.9.
Diagram of elements identified in this report. A schematic of the HPV genome with the nucleotide numbers, open reading frames, and other features shown. Bent arrows represent selected transcriptional start sites. At the top of the figure is the sequence of the core promoter (fragment T) with potential transcription factor binding sites in bold, as predicted by the Match program available at http://www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi. At the bottom is the sequence of fragment K, which contains information necessary for DRE1, with predicted factor binding sites in bold, shadowed, or boxed. Also indicated are the approximate boundaries of the differentiation-dependent DNase-hypersensitive region as described earlier (16).
The use of genetic reporters to analyze promoter activity is a widely used strategy but can lead to artifactual results, especially in the case of HPV, in which the small viral genome is densely packed with cis- and trans-acting control elements (E. Sen, J. M. Bodily, S. Alam, and C. Meyers, unpublished data). The fact that the pattern of transcriptional start sites used by the reporter resembles that seen from native genomes (Fig. 1) lends some confidence to the accuracy of the mapping results reported here. Certainly the definitive test of the function of a transcriptional control element would be mutation in the context of the whole genome followed by analysis of the functionality of the mutant virus throughout the life cycle in raft culture. Because this is the first study using genetic strategies to examine the activity and regulation of p742, elements at the resolution of individual transcription factor binding sites have yet to be identified. We anticipate that the studies in this report will serve as a guide to identify such sequence elements, leading to their analysis in the context of the whole virus genome.
HPV31 has evolved a late promoter with multiple start sites. Neither the mechanistic basis nor any adaptive significance of such a promoter strategy is understood. It is intriguing that in addition to the mucosal papillomaviruses, simian virus 40 (21, 56), and polyomavirus (8), which are also small, circular, oncogenic DNA viruses, have multiple-start late promoters. The common occurrence of multiple-start promoters in both cellular and viral systems raises an important question in the field of transcription that remains unanswered: why are there so many different kinds of promoters in the first place? Is it biologically significant that some promoters have TATA boxes, some have initiator elements, some have both, some have neither, some have a single start, and some have dozens? Despite some suggestions that different classes of promoters may have different regulatory properties (37, 54, 55), real understanding of this basic question remains elusive.
As a model multiple-start promoter, p742 has many important features, including responsiveness to experimentally controllable conditions (16, 26, 44) and its location on a small, genetically self-contained virus for which the technology exists to study its regulation at any point in its natural history (41-43). Because of these considerations, we expect that a full understanding of the components, regulation, and biological function of p742 will deepen not only our understanding HPV biology but also the function of multiple-start promoters in general and how they uniquely contribute to gene expression.
Acknowledgments
We thank David Spector and the members of the Meyers laboratory for many helpful discussions and the laboratory of Robert Bonneau for use of their Coulter counter.
This work was supported by a National Science Foundation Graduate Research Fellowship (J.M.B.) and Public Health Service grant CA79006.
REFERENCES
- 1.Ai, W., J. Narahari, and A. Roman. 2000. Yin yang 1 negatively regulates the differentiation-specific E1 promoter of human papillomavirus type 6. J. Virol. 74:5198-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, B., M. D. Schonemann, S. E. Flynn, R. V. Pearse, 2nd, H. Singh, and M. G. Rosenfeld. 1993. Skn-1a and Skn-1i: two functionally distinct Oct-2-related factors expressed in epidermis. Science 260:78-82. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, B., W. C. Weinberg, O. Rennekampff, R. J. McEvilly, J. R. Bermingham, Jr., F. Hooshmand, V. Vasilyev, J. F. Hansbrough, M. R. Pittelkow, S. H. Yuspa, and M. G. Rosenfeld. 1997. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev. 11:1873-1884. [DOI] [PubMed] [Google Scholar]
- 4.Ayer, D. E., and W. S. Dynan. 1988. Simian virus 40 major late promoter: a novel tripartite structure that includes intragenic sequences. Mol. Cell. Biol. 8:2021-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barksdale, S., and C. C. Baker. 1995. Differentiation-specific alternative splicing of bovine papillomavirus late mRNAs. J. Virol. 69:6553-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedell, M. A., J. B. Hudson, T. R. Golub, M. E. Turyk, M. Hosken, G. D. Wilbanks, and L. A. Laimins. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J. Virol. 65:2254-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benoist, C., and P. Chambon. 1981. In vivo sequence requirements of the SV40 early promotor region. Nature 290:304-310. [DOI] [PubMed] [Google Scholar]
- 8.Bourachot, B., M. Yaniv, and P. Herbomel. 1989. Control elements situated downstream of the major transcriptional start site are sufficient for highly efficient polyomavirus late transcription. J. Virol. 63:2567-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bream, G. L., P. Vaillancourt, and M. R. Botchan. 1992. A constitutive enhancer in the bovine papillomavirus upstream regulatory region shares genetic elements with the viral P1 promoter. J. Virol. 66:7319-7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bromberg-White, J. L., and C. Meyers. 2003. Comparison of the basal and glucocorticoid-inducible activities of the upstream regulatory regions of HPV18 and HPV31 in multiple epithelial cell lines. Virology 306:197-202. [DOI] [PubMed] [Google Scholar]
- 11.Bromberg-White, J. L., and C. Meyers. 2002. The upstream regulatory region of human papillomavirus type 31 is insensitive to glucocorticoid induction. J. Virol. 76:9702-9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, W., and K. Struhl. 1985. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 4:3273-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, R. B., L. Yang, J. A. Thompson, and B. Safer. 1988. Identification of a downstream sequence and binding protein that regulate adenovirus major late promoter transcription in vitro. J. Biol. Chem. 263:10377-10385. [PubMed] [Google Scholar]
- 14.Cornwell, M. M. 1990. The human multidrug resistance gene: sequences upstream and downstream of the initiation site influence transcription. Cell Growth Differ. 1:607-615. [PubMed] [Google Scholar]
- 15.Crook, T., D. Wrede, and K. H. Vousden. 1991. p53 point mutation in HPV negative human cervical carcinoma cell lines. Oncogene 6:873-875. [PubMed] [Google Scholar]
- 16.del Mar Pena, L. M., and L. A. Laimins. 2001. Differentiation-dependent chromatin rearrangement coincides with activation of human papillomavirus type 31 late gene expression. J. Virol. 75:10005-10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farnham, P. J., and A. L. Means. 1990. Sequences downstream of the transcription initiation site modulate the activity of the murine dihydrofolate reductase promoter. Mol. Cell. Biol. 10:1390-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores, E. R., and P. F. Lambert. 1997. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 71:7167-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frattini, M. G., and L. A. Laimins. 1994. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology 204:799-804. [DOI] [PubMed] [Google Scholar]
- 21.Gong, S. S., and K. N. Subramanian. 1988. Functional anatomy of the simian virus 40 late promoter. Virology 163:481-493. [DOI] [PubMed] [Google Scholar]
- 22.Grassmann, K., B. Rapp, H. Maschek, K. U. Petry, and T. Iftner. 1996. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol. 70:2339-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guhaniyogi, J., and G. Brewer. 2001. Regulation of mRNA stability in mammalian cells. Gene 265:11-23. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto-Gotoh, T., R. Kikuno, M. Takahashi, and H. Honkawa. 1988. Possible role of the first intron of c-H-ras in gene expression: anti-cancer elements in oncogenes. Anticancer Res. 8:851-859. [PubMed] [Google Scholar]
- 25.Hubert, W. G., T. Kanaya, and L. A. Laimins. 1999. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J. Virol. 73:1835-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hummel, M., H. B. Lim, and L. A. Laimins. 1995. human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J. Virol. 69:3381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenson, B., and W. Lancaster. 1990. Association of human papillomaviruses with benign, premalignant, and malignant anogenital lesions, p. 11-43. In H. Pfister (ed.), Papillomaviruses and human cancer. CRC Press, Boca Raton, Fla.
- 29.Kadonaga, J. T. 2002. The DPE, a core promoter element for transcription by RNA polymerase II. Exp. Mol. Med. 34:259-264. [DOI] [PubMed] [Google Scholar]
- 30.Kanaya, T., S. Kyo, and L. A. Laimins. 1997. The 5′ region of the human papillomavirus type 31 upstream regulatory region acts as an enhancer which augments viral early expression through the action of YY1. Virology 237:159-169. [DOI] [PubMed] [Google Scholar]
- 31.Keller, J. M., and J. C. Alwine. 1984. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell 36:381-389. [DOI] [PubMed] [Google Scholar]
- 32.Klumpp, D. J., and L. A. Laimins. 1999. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology 257:239-246. [DOI] [PubMed] [Google Scholar]
- 33.Kotova, I., A. L. Chabes, S. Lobov, L. Thelander, and S. Bjorklund. 2003. Sequences downstream of the transcription initiation site are important for proper initiation and regulation of mouse ribonucleotide reductase R2 gene transcription. Eur. J. Biochem. 270:1791-1801. [DOI] [PubMed] [Google Scholar]
- 34.Kukimoto, I., and T. Kanda. 2001. Displacement of YY1 by differentiation-specific transcription factor hSkn-1a activates the P(670) promoter of human papillomavirus type 16. J. Virol. 75:9302-9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kyo, S., A. Tam, and L. A. Laimins. 1995. Transcriptional activity of human papillomavirus type 31b enhancer is regulated through synergistic interaction of AP1 with two novel cellular factors. Virology 211:184-197. [DOI] [PubMed] [Google Scholar]
- 36.Lu, J., W. Lee, C. Jiang, and E. B. Keller. 1994. Start site selection by Sp1 in the TATA-less human Ha-ras promoter. J. Biol. Chem. 269:5391-5402. [PubMed] [Google Scholar]
- 37.Mack, D. H., J. Vartikar, J. M. Pipas, and L. A. Laimins. 1993. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature 363:281-283. [DOI] [PubMed] [Google Scholar]
- 38.McCready, P. M., R. K. Hansen, S. L. Burke, and J. F. Sands. 1997. Multiple negative and positive cis-acting elements control the expression of the murine CD4 gene. Biochim. Biophys. Acta 1351:181-191. [DOI] [PubMed] [Google Scholar]
- 39.McLaughlin-Drubin, M. E., and C. Meyers. 2004. Evidence for the coexistence of two genital HPV types within the same host cell in vitro. Virology 321:173-180. [DOI] [PubMed] [Google Scholar]
- 40.McLaughlin-Drubin, M. E., S. Wilson, B. Mullikin, J. Suzich, and C. Meyers. 2003. Human papillomavirus type 45 propagation, infection, and neutralization. Virology 312:1-7. [DOI] [PubMed] [Google Scholar]
- 41.Meyers, C., J. L. Bromberg-White, J. Zhang, M. E. Kaupas, J. T. Bryan, R. S. Lowe, and K. U. Jansen. 2002. Infectious virions produced from a human papillomavirus type 18/16 genomic DNA chimera. J. Virol. 76:4723-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyers, C., M. G. Frattini, J. B. Hudson, and L. A. Laimins. 1992. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971-973. [DOI] [PubMed] [Google Scholar]
- 43.Meyers, C., T. J. Mayer, and M. A. Ozbun. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J. Virol. 71:7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozbun, M. A., and C. Meyers. 1997. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 71:5161-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozbun, M. A., and C. Meyers. 1998. human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology 248:218-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozbun, M. A., and C. Meyers. 1998. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J. Virol. 72:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfister, H. 1990. General introduction to Papillomaviruses, p. 1-9. In H. Pfister (ed.), Papillomaviruses and human cancer. CRC Press, Boca Raton, Fla.
- 48.Piatak, M., K. N. Subramanian, P. Roy, and S. M. Weissman. 1981. Late messenger RNA production by viable simian virus 40 mutants with deletions in the leader region. J. Mol. Biol. 153:589-618. [DOI] [PubMed] [Google Scholar]
- 49.Rapp, B., A. Pawellek, F. Kraetzer, M. Schaefer, C. May, K. Purdie, K. Grassmann, and T. Iftner. 1997. Cell-type-specific separate regulation of the E6 and E7 promoters of human papillomavirus type 6a by the viral transcription factor E2. J. Virol. 71:6956-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 51.Ruesch, M., F. Stubenrauch, and L. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin 10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sen, E., S. Alam, and C. Meyers. 2004. Genetic and biochemical analysis of cis regulatory elements within the keratinocyte enhancer region of the human papillomavirus type 31 upstream regulatory region during different stages of the viral life cycle. J. Virol. 78:612-629. [DOI] [PMC free article] [PubMed]
- 53.Sen, E., J. L. Bromberg-White, and C. Meyers. 2002. Genetic analysis of cis regulatory elements within the 5′ region of the human papillomavirus type 31 upstream regulatory region during different stages of the viral life cycle. J. Virol. 76:4798-4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smale, S. T. 2001. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 15:2503-2508. [DOI] [PubMed] [Google Scholar]
- 55.Smale, S. T., A. Jain, J. Kaufmann, K. H. Emami, K. Lo, and I. P. Garraway. 1998. The initiator element: a paradigm for core promoter heterogeneity within metazoan protein-coding genes. Cold Spring Harb. Symp. Quant. Biol. 63:21-31. [DOI] [PubMed] [Google Scholar]
- 56.Somasekhar, M. B., and J. E. Mertz. 1985. Sequences involved in determining the locations of the 5′ ends of the late RNAs of simian virus 40. J. Virol. 56:1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steele, B. K., C. Meyers, and M. A. Ozbun. 2002. Variable expression of some “housekeeping” genes during human keratinocyte differentiation. Anal. Biochem. 307:341-347. [DOI] [PubMed] [Google Scholar]
- 58.Stoler, M. H., S. M. Wolinsky, A. Whitbeck, T. R. Broker, and L. T. Chow. 1989. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology 172:331-340. [DOI] [PubMed] [Google Scholar]
- 59.Terhune, S. S., C. Milcarek, and L. A. Laimins. 1999. Regulation of human papillomavirus type 31 polyadenylation during the differentiation-dependent life cycle. J. Virol. 73:7185-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tibudan, S. S., Y. Wang, and M. F. Denning. 2002. Activation of protein kinase C triggers irreversible cell cycle withdrawal in human keratinocytes. J. Investig. Dermatol. 119:1282-1289. [DOI] [PubMed] [Google Scholar]
- 61.Tommasino, M., and P. Jansen-Durr. 1997. E7 protein, p. 103-136. In M. Tommasino (ed.), Papillomaviruses in human cancer: the role of E6 and E7 oncoproteins. Landes Bioscience, Austin, Tex.
- 62.Toullec, D., P. Pianetti, H. Coste, P. Bellevergue, T. Grand-Perret, M. Ajakane, V. Baudet, P. Boissin, E. Boursier, F. Loriolle, et al. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266:15771-15781. [PubMed] [Google Scholar]
- 63.Yoshida, T., and T. Sakai. 2004. Promoter of TRAIL-R2 gene. Vitam. Horm. 67:35-49. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, W., L. T. Chow, and T. R. Broker. 1999. A distal element in the HPV-11 upstream regulatory region contributes to promoter repression in basal keratinocytes in squamous epithelium. Virology 253:219-229. [DOI] [PubMed] [Google Scholar]
- 65.zur Hausen, H. 1991. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology 184:9-13. [DOI] [PubMed] [Google Scholar]
- 66.zur Hausen, H. 1996. Papillomavirus infections-a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-78. [DOI] [PubMed] [Google Scholar]
- 67.zur Hausen, H. 1999. Papillomaviruses in human cancers. Proc. Assoc. Am Physicians 111:581-587. [DOI] [PubMed] [Google Scholar]