Abstract
The 3′ noncoding region (3′ NCR) of flaviviruses contains secondary and tertiary structures essential for virus replication. Previous studies of yellow fever virus (YFV) and dengue virus have found that modifications to the 3′ NCR are sometimes associated with attenuation in vertebrate and/or mosquito hosts. The 3′ NCRs of 117 isolates of South American YFV have been examined, and major deletions and/or duplications of conserved RNA structures have been identified in several wild-type isolates. Nineteen isolates (designated YF-XL isolates) from Brazil, Trinidad, and Venezuela, dating from 1973 to 2001, exhibited a 216-nucleotide (nt) duplication, yielding a tandem repeat of conserved hairpin, stem-loop, dumbbell, and pseudoknot structures. YF-XL isolates were found exclusively within one subclade of South American genotype I YFV. One Brazilian isolate exhibited, in addition to the 216-nt duplication, a deletion of a 40-nt repeated hairpin (RYF) motif (YF-XL-ΔRYF). To investigate the biological significance of these 3′ NCR rearrangements, YF-XL-ΔRYF and YF-XL isolates, as well as other South American YFV isolates, were evaluated for three phenotypes: growth kinetics in cell culture, neuroinvasiveness in suckling mice, and ability to replicate and produce disseminated infections in Aedes aegypti mosquitoes. YF-XL-ΔRYF and YF-XL isolates showed growth kinetics and neuroinvasive characteristics comparable to those of typical South American YFV isolates, and mosquito infectivity trials demonstrated that both types of 3′ NCR variants were capable of replication and dissemination in a laboratory-adapted colony of A. aegypti.
The 3′ noncoding region (3′ NCR) of flaviviruses contains cis-acting sequences that are essential for genome replication and translation. Experimental studies indicate that variations in the 3′ NCR are well tolerated but may sometimes lead to altered growth characteristics in cell culture (3, 6, 19), changes in virulence and immunogenicity in vertebrate animals (10, 17, 21, 22), or reduced infectivity in mosquitoes (1, 44). Molecular epidemiological studies of naturally occurring flaviviruses also suggest that 3′ NCR structural differences correlate with patterns of virus distribution and transmission (39, 50). Size heterogeneity has been observed within the 3′ NCRs of several species of mosquito-borne and tick-borne flaviviruses (12, 32, 39, 48), as well as viruses of the no-known-vector group (5). In addition, it has recently been reported that genome fragments representing a major portion of the 3′ NCR accumulate in Japanese encephalitis virus-infected cell cultures (18). Together, these studies suggest that the 3′ NCR may play a regulatory role in RNA synthesis. Evolutionary divergence of the 3′ NCR is thus of considerable interest, as this region is hypothesized to contain molecular determinants of virulence, cell tropism, and/or host specificity.
Previous studies of the 3′ NCR of yellow fever virus (YFV) have examined variability within a limited number (using up to 14 isolates) of African and South American wild-type isolates (8, 13, 29, 49). These studies have shown that YFV 3′ NCRs vary in length from 444 to 524 nucleotides (nt) and contain a series of repeated hairpin motifs (designated RYFs; ∼40 nt) that are genotype specific in number. Sequences examined to date reveal three RYFs among West African isolates of YFV, two RYFs among East African isolates of YFV, and only one RYF among South American isolates of YFV (28, 49). Similar well-conserved and type-specific hairpin repeats are also found among closely related flaviviruses of the YFV genetic group (28). The function of the RYFs remains obscure; indeed, little is known of the biological importance of individual structural components within the 3′ NCR.
The flavivirus 3′ NCR comprises several hairpin-loop structures whose secondary structures are well conserved across virus species, although the primary nucleotide sequences are highly divergent (34). The 3′ NCR has been divided into three regions based on differing levels of sequence conservation (Fig. 1): domain I is the region immediately following the stop codon that is hypervariable and contains insertions-deletions (indels) in most flavivirus species; domain II is a region of moderate conservation comprising several hairpin motifs, including a characteristic dumbell structure and CS2 motif (∼24 nt) present in all mosquito-borne flaviviruses; and domain III is the highly conserved essential region, containing a cyclization domain (∼8 to 14 nt) and the terminal long stable hairpin (∼90 nt), which are essential for viral replication (34, 35). Size heterogeneity among flaviviruses is mostly due to sequence repeats or deletions within domain I. RNA structures within domains I and II are considered dispensable for virus replication (3, 19, 22). However, because deletion mutants typically exhibit some degree of growth restriction or attenuation (3, 22), the structures of domains I and II are believed to serve as replication enhancer elements. The evolutionary conservation of RNA secondary structures across numerous species of mosquito-borne flaviviruses also strongly suggests that they play an important role in virus replication. Based on comparative sequence analysis of 14 wild-type YFV isolates and 7 vaccine isolates, Proutski et al. (33, 34) suggested an association between the predicted folding structure of the 3′ NCR (in particular, the long stable hairpin of domain III and the e3 stem-loop of domain II) and the virulence of YFV.
FIG. 1.
(A) Schematic diagram highlighting conserved features of the prototype YFV 3′ NCR (modified from reference 28). RYF, repeated dual F-G hairpins; CS2, ∼24 nt comprising part of dumbbell B; cyc, conserved cyclization domain; LSH, long stable hairpin. (B) Consensus alignment of YFV 3′ NCR sequences (domains I and II). The numbers of isolates used to build the consensus sequences are provided in parentheses (details in Table 1). Nucleotides identical to the reference Angola71 isolate are indicated by dots. Alignment was optimized by the insertion of gaps (∼). Ambiguity codes are as follows: M = A or C; R = A or G; W = A or T; S = C or G; Y = C or T; K = G or T; V = A, C,or G; H = A, C, or T; D = A, G, or T; B = C, G, or T. Regions involved in basepairing are boxed and labeled to correspond with stem structures as indicated in the schematic in panel A.
In the present study, novel 3′ NCR sequence data for 112 South American and 8 African YFV isolates were generated. Sixteen previously published YFV 3′ NCR sequences were incorporated into the data set for a combined total of 136 sequences. The data were interpreted within the context of predicted RNA folding patterns as determined by Proutski et al. (34) and Olsthoorn and Bol (30). Our examination focused primarily on South American YFV isolates, because preliminary work revealed a novel 3′ NCR conformation (YF-XL) among a subset of South American genotype I viruses. The YF-XL conformation consisted of an imperfect tandem repeat of RNA structures within domain II of the 3′ NCR. In addition, the 3′ NCR of one South American genotype I isolate containing the YF-XL repeat was found to lack all three RYFs (YF-XL-ΔRYF). To investigate the functional significance of the YF-XL 3′ NCR conformation, the growth characteristics of the variant isolates were evaluated in cell culture, suckling mice, and Aedes aegypti mosquitoes. The natural history of duplications and deletions in the 3′ NCR of YFV was examined through phylogenetic reconstructions and interpreted within the context of present models for RNA recombination. The results of this study may have important implications for the molecular evolution of YFV, as they underscore the significant genomic plasticity of the 3′ NCR and suggest a possible role of RNA structural features in promoting template switching during virus replication.
MATERIALS AND METHODS
Virus isolates used in this study.
The wild-type isolates of YFV used in this study were made available through the University of Texas Medical Branch World Arbovirus Reference Center. The majority of South American isolates were low-passage sylvan isolates of virus originally isolated at the Instituto Evandro Chagas in Belém, Brazil, and at the Instituto Nacional de Salud in Lima, Peru. The sources, geographic origins, and passage histories of isolates for which 3′ NCR sequences were obtained, as well as information on isolates with previously published data, are provided in Table 1. The majority of isolates were made via passage in suckling mouse brain, followed by amplification in C6/36 cell culture, and had minimal passages following primary isolation.
TABLE 1.
Viruses used in this study
| Isolate | Strain | Passage levela | Source | Stateb | Genotype, cladec | 3′ NCR statusd | GenBank no. | Reference |
|---|---|---|---|---|---|---|---|---|
| 14 FA | Angola71 | SM7 Mosq 1 | Human | ND | Angola | YF-std | AY326413 | 28 |
| OBS 7687 | Bolivia99A | C6/36#2 | Human | Santa Cruz | SAm II, clade 2 | YF-std | AY541326 | |
| OBS 8026 | Bolivia99D | C8/36#2 | Human | La Paz | SAm II, clade 1 | YF-std | AY541327 | |
| JSS | Brazil35 | Mosq1, SM8 | Human | MT | SAm I, Old Pará | YF-std | U52389 | 49 |
| BeH 111 | Brazil54 | C6/36, SM10 | Human | PA | SAm I, Old Pará | ND | AY541335 | 4 |
| BeAr 162 | Brazil56B | SM4, C6/36#1 | Haemagogus janthinomys | PA | SAm I, Old Pará | YF-std | AY541336 | 46a |
| BeAr 189 | Brazil55C | SM1 C6/36#1 | Sabethes sp. | PA | SAm I, Old Pará | YF-std | AY541337 | 4 |
| BeAn 23536 | Brazil60 | SM1 C6/36#1 | Macaco sp. | PA | SAm I, Old Pará | ND | AY541338 | 4 |
| BeAr 46299 | Brazil62A | C6/36#1 | Haemagogus sp. | PA | SAm I, Old Pará | YF-std | AY541339 | 4 |
| BeAr 44824 | Brazil62B | SM1 C6/36#1 | Haemagogus sp. | PA | SAm I, Old Pará | ND | AY541340 | 4 |
| BeAn 142028 | Brazil68A | C6/36#1 | Macaco sp. | PA | SAm I, Old Pará | YF-std | AY541341 | 4 |
| BeAr 142658 | Brazil68C | SM2, C6/36#1 | Haemagogus sp. | PA | SAm I, Old Pará | YF-std | AY541342 | 4 |
| BeAn 142027 | Brazil68D | Original | Saguinus midas | PA | SAm I, Old Pará | YF-std | AY566272 | 4 |
| BeH 171995 | Brazil69A | SM1 C6/36#1 | Human | PA | SAm I, unresolved | ND | AY541343 | 46a |
| BeH 203410 | Brazil71 | Original, C6/36#1 | Human | PA | SAm I, clade 1C | ND | AY541344 | 4 |
| BeAr 233164 | Brazil73A | Mosq 4 | H. janthinomys | GO | SAm I, clade 1A | YF-XL | AY541345 | 4 |
| BeAn 232669 | Brazil73B | Mosq 1, SM2 | Mosquito | GO | SAm I, clade 1A | YF-std | AY541346 | 4 |
| BeAr 233436 | Brazil73C | Original | Haemagogus sp. | GO | SAm I, clade 1A | ND | AY541347 | 4 |
| BeH 233393 | Brazil73D | SM1 C6/36#1 | Human | GO | SAm I, clade 1A | ND | AY541348 | 46a |
| BeAr 301129 | Brazil76A | SM1 C6/36#1 | Haemagogus sp. | PA | SAm I, unresolved | ND | AY541349 | 46a |
| BeH 301035 | Brazil76B | SM1 C6/36#1 | Human | PA | SAm I, unresolved | ND | AY541350 | 46a |
| BeH 324213 | Brazil77 | SM1 C6/36#1 | Human | PA | SAm I, unresolved | ND | AY541351 | 46a |
| BeH 350698 | Brazil78A | SM2, Mosq 1 | Human | PA | SAm I, clade 1C | ND | AY541352 | 4 |
| BeAr 350397 | Brazil78B | SM2, Mosq 1 | Haemagogus sp. | PA | SAm I, unresolved | YF-XL | AY541353 | 46a |
| BeH 340824 | Brazil78C | SM1 C6/36#1 | Human | PA | SAm I, unresolved | ND | AY541354 | 46a |
| BeH 371220 | Brazil79A | SM1 C6/36#1 | Human | PA | SAm I, unresolved | ND | AY541355 | 46a |
| BeAr 378600 | Brazil80A | SM2, C6/36#1 | Haemagogus sp. | GO | SAm I, clade 1C | ND | AY541356 | 46a |
| BeH 385780 | Brazil80B | SM1 C6/36#1 | Human | PA | SAm I, unresolved | YF-XL | AY541357 | 46a |
| BeH 379501 | Brazil80C | SM2 | Human | MA | SAm I, clade 1C | ND | AY541358 | 4 |
| BeH 394880 | Brazil81 | Original, C6/36#1 | Human | PA | SAm I, unresolved | ND | AY541359 | 46a |
| BeH 403366 | Brazil82A | SM1 C6/36#1 | Human | MA | SAm I, clade 1C | ND | AY541360 | 46a |
| BeH 405954 | Brazil82B | SM1 C6/36#1 | Human | PA | SAm I, unresolved | ND | AY541361 | 46a |
| BeH 413820 | Brazil83 | SM1 C6/36#1 | Human | RO | SAm II, clade 1 | YF-std | AY566273 | 46a |
| BeH 425381 | Brazil84A | C6/36#1 | Human | AP | SAm I, unresolved | ND | AY541362 | 4 |
| BeAn 424208 | Brazil84B | Original, C6/36#1 | Chiropotes satanas | PA | SAm I, clade 1C | ND | AY541363 | 46a |
| BeAr 424719 | Brazil84C | SM1 C6/36#1 | Haemagogus sp. | PA | SAm I, clade 1C | ND | AY541364 | 46a |
| BeAr 424083 | Brazil84D | SM1 C6/36#1 | Haemagogus albomaculatus | PA | SAm I, clade 1D | ND | AY541365 | 46a |
| BeAr 424492 | Brazil84E | SM1 C6/36#1 | H. janthinomys | PA | SAm I, clade 1A | ND | AY541366 | 46a |
| BeH 422255 | Brazil84F | SM1 C6/36#1 | Human | PA | SAm 1, clade 1B | ND | AY541367 | 46a |
| BeH 423602 | Brazil84G | SM1 C6/36#1 | Human | PA | SAm I, clade 1C | ND | AY541368 | 46a |
| BeH 422312 | Brazil84H | SM1 C6/36#1 | Human | PA | SAm 1, clade 1B | ND | AY541369 | 46a |
| BeH 436823 | Brazil85A | SM1 C6/36#1 | Human | MT | SAm I, unresolved | ND | AY541370 | 46a |
| BeAr 437159 | Brazil85B | SM1 C6/36#1 | H. janthinomys | MT | SAm I, unresolved | ND | AY541371 | 46a |
| BeH 463676 | Brazil87 | SM1 C6/36#1 | Human | PA | SAm I, clade 1C | ND | AY541372 | 46a |
| BeH 474245 | Brazil88A | SM1 C6/36#1 | Human | GO | SAm I, clade 1C | ND | AY541373 | 46a |
| BeH 474297 | Brazil88B | SM1 C6/36#1 | Human | MG | SAm I, clade 1C | ND | AY541374 | 46a |
| BeH 485717 | Brazil89 | SM1 C6/36#1 | Human | MG | SAm I, clade 1C | ND | AY541375 | 46a |
| BeAr 511437 | Brazil91A | C6/36#1 | H. janthinomys | PA | SAm I, clade 1C | ND | AY541376 | 4 |
| BeH 511843 | Brazil91B | SM1, C6/36#2 | Human | RR | SAm I, clade 1B | YF-XL | AY541377 | 4 |
| BeAn 510266 | Brazil91C | SM1 C6/36#1 | Alouatta sp. | GO | SAm I, clade 1C | YF-ΔRYF-XL | AY541378 | 46a |
| BeAr 512943 | Brazil92A | SM1 C6/36-1 Vero2 | H. janthinomys | MS | SAm 1, clade 1B | YF-XL | AY541379 | 4 |
| BeAr 513008 | Brazil92B | SM1 C6/36#1 | Sabethes sp. | MS | SAm 1, clade 1B | YF-XL | AY541380 | 4 |
| BeH 512772 | Brazil92C | SM2, C6/36#1 | Human | MS | SAm 1, clade 1B | ND | AY541381 | 4 |
| BeAr 513060 | Brazil92D | SM1 C6/36#1 | Haemagogus sp. | MS | SAm 1, clade 1B | YF-XL | AY541382 | 46a |
| BeAr 513292 | Brazil92E | SM1 C6/36-1 Vero1 | Sabethes cloropterus | MS | SAm 1, clade 1B | YF-XL | AY541383 | 4 |
| BeH 520988 | Brazil93A | SM1 C6/36#1 | Human | MA | SAm 1, clade 1B | YF-std | AY541384 | 46a |
| BeH 521244 | Brazil93B | SM1 C6/36#1 | Human | MA | SAm I, clade 1C | YF-XL | AY541385 | 46a |
| BeAr 527785 | Brazil94A | SM1 C6/36#1 | S. cloropterus | MG | SAm I, clade 1C | YF-XL | AY541386 | 4 |
| BeAr 527198 | Brazil94B | SM1 C6/36#1 | Haemagogus sp. | MG | SAm I, clade 1C | YF-XL | AY541387 | 4 |
| BeAr 527547 | Brazil94C | ?, Vero1 | Haemagogus sp. | PA | SAm I, clade 1C | ND | AY541388 | 46a |
| BeH 526722 | Brazil94D | SM2, C6/36#1 | Human | MG | SAm I, clade 1C | YF-std | AY541389 | 46a |
| BeAr 528057 | Brazil94E | Original, C6/36#1 | H. janthinomys | MG | SAm I, unresolved | ND | AY541390 | 46a |
| BeAr 527410 | Brazil94F | Original, C6/36#1 | H. janthinomys | MA | SAm I, clade 1C | ND | AY541391 | 46a |
| BeH 535010 | Brazil95 | SM2, C6/36#1 | Human | MA | SAm I, clade 1C | ND | AY541392 | 46a |
| BeAr 544276 | Brazil96A | SM1 C6/36#1 | H. janthinomys | RO | SAm 1, clade 1B | YF-XL | AY541393 | 4 |
| BeAn 604552 | Brazil98A | Original, C6/36#1 | Alouatta belzebul | PA | SAm 1, clade 1C | ND | AY541394 | 46a |
| BeAr 603401 | Brazil98B | Original, C6/36#1 | H. janthinomys | PA | SAm 1, clade 1D | ND | AY541395 | 46a |
| BeAr 605158 | Brazil98C | SM1 C6/36#1 | H. janthinomys | PA | SAm 1, clade 1D | ND | AY541396 | 46a |
| BeH 603325 | Brazil98D | Original, C6/36#1 | Human | PA | SAm 1, clade 1D | ND | AY541397 | 46a |
| BeH 605427 | Brazil98E | SM1 C6/36#1 | Human | MT | SAm 1, clade 1D | ND | AY541398 | 46a |
| BeAr 614320 | Brazil99B | SM1 C6/36#1 | Haemagogus sp. | PA | SAm 1, clade 1D | ND | AY541399 | 46a |
| BeAr 617127 | Brazil99D | SM1 C6/36#1 | H. janthinomys | TO | SAm 1, clade 1D | ND | AY541400 | 46a |
| BeH 613582 | Brazil99E | SM1 C6/36#1 | Human | PA | SAm 1, clade 1D | ND | AY541401 | 46a |
| BeAr 628124 | Brazil2000A | SM1 C6/36#1 | H. janthinomys | TO | SAm 1, clade 1D | YF-std | AY541328 | 4 |
| BeH 622491 | Brazil2000B | ?, C6/36#1 | Human | DF | SAm 1, clade 1D | ND | AY541329 | 46a |
| BeAn 625923 | Brazil2000C | Original, C6/36#1 | Alouatta sp. | GO | SAm 1, clade 1D | ND | AY541330 | 46a |
| BeH 622205 | Brazil2000D | SM1 C6/36#1 | Human | GO | SAm 1, clade 1D | ND | AY541331 | 46a |
| BeAr 630768 | Brazil2001A | ?, C6/36#1 | H. janthinomys | GO | SAm 1, clade 1D | ND | AY541332 | 46a |
| BeAr 631464 | Brazil2001B | SM1 C6/36#1 | S. cloropterus | BA | SAm 1, clade 1D | YF-XL | AY541333 | 46a |
| BeAr 645693 | Brazil2001D | SM1 C6/36#1 | Haemagogus sp. | MG | SAm 1, clade 1D | ND | AY541334 | 46a |
| Ar B 8883 | CAR77A | SM5, C6/36#1 | Mosquito | Boyo | East Central Africa | ND | U52392 | 4a |
| Ar B 9005 | CAR77B | SM5 Mosq1 | Aedes africanus | Boyo | East Central Africa | ND | U52395 | 4a |
| DAK Ar B 17239 | CAR80 | AP61#1, C6/36#1 | Aedes sp. | Bozo | East Central Africa | ND | AY541402 | |
| V528A | Colombia79 | Monkey 1 Mosq2 | Human | ND | SAm I, unresolved | ND | AY541403 | |
| INS347613 | Colombia85 | C6/36#3 | Human | ND | SAm I, unresolved | ND | AY541404 | |
| 1337 | Ecuador79 | SM1, Mosq2 | Human | ND | SAm I, unresolved | YF-std | U52399 | 4a |
| 1345 | Ecuador81 | SM1, Vero1, c6/36#2 | Human | ND | SAm I | ND | AY541405 | |
| OBD 5041 | Ecuador97 | C6/36#1 | Human | Pastaza | SAm II, clade 1 | YF-std | AY541406 | |
| Serie 227 | Ethiopia61A | Mosq1, SM7 | Human | ND | East Africa | ND | AY541407 | |
| Asibi | Ghana27 | C6/36#1 | Human | ND | West Africa II | ND | Not deposited | 37a |
| 85-82H | IvoryCoast85 | ?, C6/36#1 | Human | ND | West Africa II | ND | O54798 | 49 |
| BC 7914 | Kenya93a | SM1, c636#1 | Human | Baringo | East Africa | ND | AY541408 | |
| 69056 | Nigeria46 | ?, C6/36#2 | Human | ND | West Africa I | ND | U52403 | 29 |
| IB AR 45244 | Nigeria69 | SM4 | Mosquito | ND | West Africa I | ND | AY541409 | |
| H117505 | Nigeria87C | SM4 | Human | ND | West Africa I | ND | AY541410 | |
| 56205 | Nigeria91 | ?, C6/36#1 | Human | ND | West Africa I | ND | AY541411 | |
| 614819 | Panama74A | ?, C6/36#1 | Human | ND | SAm I | YF-XL | AY541412 | |
| 1362/77 | Peru77A | C6/36#2 | Human | Ayacucho | SAm II, clade 2 | YF-std | AY541413 | 4 |
| 1368 | Peru77B | SM1, Vero1, C6/36#2 | Human | Ayacucho | SAm II, clade 1 | ND | AY541414 | 4 |
| 1371 | Peru77C | SM1, Vero1, C6/36#2 | Human | Ayacucho | SAm II, clade 2 | ND | AY541415 | 4 |
| 287/78 | Peru78 | SM2, Mosq 2 | Human | Ayacucho | SAm II, clade 2 | ND | AY541416 | 4 |
| R 35740 | Peru79 | SM1, Mosq 2 | Human | Ayacucho | SAm II, clade 2 | ND | AY541417 | 4 |
| B4.1 | Peru81A | Plaque pick | Plaque pick | Cusco | SAm II, clade 2 | YF-std | U52411 | 4a |
| 1914 | Peru81B | MILLC 1, LLCMK2 | Sentinel mice | Cusco | SAm II, clade 2 | ND | AY541418 | 4 |
| ARV 0544 | Peru95A | SM1, Vero1, c6/36#1 | Human | S. Martin | SAm II, clade 1 | YF-std | AY541419 | 4 |
| HEB4224 | Peru95B | SM1, C6/36#1 | Human | S. Martin | SAm II, clade 1 | ND | AY541420 | 4 |
| HEB4236 (153) | Peru95C | C6/36#1 | Human | Pasco | SAm II, clade 2 | YF-std | AY541421 | 4a |
| 149 | Peru95D | SM1, C6/36#1 | Human | Pasco | SAm II, clade 2 | YF-std | AY541422 | 4a |
| cepa #2 | Peru95E | SM1, C6/36#1 | Human | Puno | SAm II, clade 2 | YF-std | AY541423 | 4 |
| Cepa#1 | Peru 95F | C6/36#2 | Human | Puno | SAm II, clade 1 | ND | AY541424 | 4 |
| OBS 2240 | Peru95G | C6/36#2 | Human | Huanuco | SAm II, clade 1 | YF-std | AY541425 | 4 |
| OBS 2250 | Peru95H | SM1, C6/36#1 | Human | Huanuco | SAm II, clade 2 | ND | AY541426 | 4 |
| HEB 4240 | Peru95I | C6/36#1, SM1 | Human | Junin | SAm II, clade 2 | ND | AY541427 | 4 |
| HEB 4245 | Peru95J | SM1, C6/36#1 | Human | Junin | SAm II, clade 2 | ND | AY541428 | 4 |
| HEB 4246 | Peru95K | SM1, C6/36#1 | Human | Junin | SAm II, clade 2 | YF-std | AY541429 | 4 |
| ARV 0548 | Peru95M | SM1, C6/36#1 | Human | S. Martin | SAm II, clade 1 | ND | AY541430 | 4 |
| OBS 6530 | Peru98A | ?, C6/36#1 | Human | Cusco | SAm II, clade 2 | ND | AY541431 | 4 |
| 03-5350-98 | Peru98B | C6/36#2 | Human | Cusco | SAm II, clade 2 | ND | AY541432 | 4 |
| OBS 6745 | Peru98C | C6/36#2 | Human | Cusco | SAm II, clade 2 | YF-std | AY541433 | 4 |
| IQT 5591 | Peru98D | C6/36#2 | Human | Loreto | SAm II, clade 1 | ND | AY541434 | 4 |
| OBS 7904 | Peru99 | Vero1, C6/36 3 | Human | S. Martin | SAm II, clade 1 | YF-std | AY541435 | 4 |
| FVV | Senegal27 | Monkey, c6/36#1 | Human | ND | West African II | ND | GI694115 | 49a |
| Dak 1279 | Senegal65C | SM7 | Human | ND | West African II | ND | U52413 | 49 |
| M 90-5 | Sudan40B | C6/36#1, SM4 | Human | ND | East Central Africa | ND | AY326414 | 28 |
| GML902621 | Trinidad54 | Monk1, C6/36#1 | Alouatta sp. | ND | SAm I, unresolved | ND | AY541436 | |
| CAREC 889920 | Trinidad88 | AP61#2, C6/36#1 | Haemagogus sp. | S.E. Trin | SAm I | YF-XL | AY541437 | |
| CAREC 890692 | Trinidad89A | ?, C6/36#1 | S. cloropterus | S.E. Trin | SAm I | YF-XL | AY541438 | |
| CAREC 891957 | Trinidad89C | AP61, C6/36#1 | Alouatta sp. | S.E. Trin | SAm I | ND | AY541439 | |
| A 709-4-A2 | Uganda48A1 | C6/36#1 | Human | ND | East Africa | ND | U52424 | |
| MR 896 | Uganda48B1 | C6/36#1 | Human | ND | East Africa | ND | U52422 | |
| Z 19039 | Uganda72 | C6/36#1, SM3 | Monkey | ND | East Central Africa | ND | AY541440 | |
| P128MC | Venezuela59 | SM3, Mosq 1 | Monkey | Cojedes | SAm 1 | ND | AY541441 | |
| PHO 42H | Venezuela61 | C6/36#2, SM2 | Human | Tachira | SAm 1 | YF-std | AY541442 | |
| 35720 | Venezuela98A | Vero#1, C6/36#1 | Human | Amazonas | SAm I | YF-XL | AY541443 | |
| 35708 | Venezuela98B | SM1, Vero1 | Human | Amazonas | SAm I | YF-XL | AY541444 | |
| STA-LSF-4-4143 | Zaire58 | SM2, Mosq 1, C6/36#1 | Human | ND | East Central Africa | ND | AY541445 |
Passage of seed stock virus, cell line, or animal model followed by passage number. SM, suckling mouse; Monk, monkey; Mosq. mosquito. Question mark indicates unknown passage level prior to collection catalogue.
AP, Amapa; BA, Bahia; DF, Federal District; GO, Goiás; MA, Maranhão; MG, Minas Gerais; MS, Mato Grosso do Sul; MT, Mato Grosso; PA, Para; RO, Rondônia; RR, Roraima; TO, Tocantins; ND, no data.
SAm I, South American genotype I; SAm II, South American genotype II.
YF-XL, viruses containing the 216-nt duplication; YF-std, viruses with prototype conformation of 3′ NCR; ND, no data, viruses for which the genome terminus has not been sequenced.
RT-PCR and sequencing.
The methods for genomic RNA extraction and amplification of viral sequences by reverse transcriptase (RT) PCR were as previously described (49) with the following modifications. RNA extraction was achieved using the QIAamp Viral RNA Mini kit (QIAGEN) according to the manufacturer's instructions, and the reverse transcription protocol employed Superscript II RNase H− RT (Invitrogen). Sequences of the genome terminus were obtained using either the sNS5i-aNS5i or the emf-vd8 primer pairs. The sNS5i primer amplifies from genomic positions 10167 to 10188 (5′-CCAAGAGACAAGACAAGCTGTG-3′); aNS5i binds to the genome terminus positions 10838 to 10862 (5′-AGTGGTTTTGTGTTTTTCATCCAAAG-3′); emf corresponds to positions 10055 to 10075 (5′-TGGATGACSACHGARGAYA-3′), and vd8 binds to 10709 to 10728 (5′-TAGAGGTTAGAGGAGACCC-3′). Amplicons were visualized by gel electrophoresis, gel extracted (QIAGEN gel extraction kit), and sequenced by direct sequencing using a Perkin-Elmer 373XL. Sequences were obtained from both strands of each RT-PCR amplicon for sequence confirmation (using the same primers for amplification reactions). Amplicons of isolates that did not produce sufficient quantities for direct sequencing were cloned into the pGEM-EZ cloning vector (Promega); three clones were sequenced in both directions to provide a representative consensus sequence.
Sequence analysis.
Initial editing of 3′ NCR sequence fragments was performed using Vector NTI (Informax). Sequences were aligned and manually edited using the Genomics Computer Group Wisconsin Package version 10.3 (Accelrys, Inc., San Diego, Calif.). The PAUP* program (41) was used to infer maximum-likelihood trees of the multiple sequence alignment.
Growth characteristics in cell culture.
Twelve-day growth curves for YF-XL and YF-XL-ΔRYF, as well as representative South American genotype I and II isolates with “standard” 3′ NCR conformations (YF-std), were determined in C6/36 and Vero cells. Cell monolayers at 75 to 80% confluency were incubated with virus stock at an approximate multiplicity of infection (MOI) of 0.03 for 1 h at 37 (Vero) or 28°C (C6/36). After the 1-h incubation, the monolayers were washed twice with phosphate-buffered saline and then incubated in 2% minimal essential medium. Aliquots of the supernatant were harvested with replacement for the subsequent 12 days, and titrations were performed by endpoint infectivity assay in Vero cells.
Mouse neuroinvasiveness studies.
The ability of YFV isolates to be neuroinvasive in 8-day-old mice was evaluated following intraperitoneal inoculation. Serial 10-fold dilutions of virus in phosphate-buffered saline were inoculated intraperitoneally into litters of outbred NIH Swiss mice and observed over a period of 15 days. Fifty percent lethal dose (LD50) calculations were performed as described by Reed and Muench (37).
Mosquito infectivity trials.
The abilities of YF-XL and YF-XL-ΔRYF isolates to replicate and produce disseminated infections in A. aegypti mosquitoes were tested using a white-eyed strain that is known to be highly susceptible to YFV infection (24). Four groups of 50 female A. aegypti mosquitoes were orally infected with a YF-XL isolate (Brazil92e), YF-XL-ΔRYF (Brazil91c), Asibi (Ghana27), or vaccine isolate 17D. The Asibi and 17D isolates served as positive and negative controls, respectively; the 17D vaccine virus is incapable of disseminating from the midgut to peripheral body tissues (23). Mosquitoes were exposed to YFV by feeding on infected blood meals consisting of equal volumes of virus and medium containing defibrinated sheep blood and 2 mM ATP as a phagostimulant. Virus inocula were in the form of supernatants from infected Vero cell cultures harvested at 4 (for Asibi, 17D, and Brazil91c) and 9 (for Brazil92e) days postinfection. Ten-day-old female mosquitoes were starved for 24 h prior to the infectious meal and then allowed to feed for 45 min through a chicken skin membane covering a Hemotek feeding apparatus (Discovery Workshops, Accrington, Lancashire, United Kingdom) containing the feeding mixture maintained at 37°C. The fully engorged mosquitoes were sorted and maintained for 14 days at 28°C and 80% humidity. The titer of the infectious meal was determined by direct sampling of the fresh blood meal, as well as by sampling one engorged mosquito (day zero) from each treatment group; titrations were performed by endpoint infectivity assay in Vero cells. All mosquitoes were processed as follows. Heads and bodies were separated and triturated in 140 μl and 1 ml of 2% minimal essential medium, respectively. The body and head homogenates were filtered using a 0.22-μm-pore-size filter (Millipore, Molsheim, France), and virus infection of mosquitoes was determined by RT-PCR. Midgut infection rates (MIR) and disseminated infection rates (DIR) were calculated as the ratio of percent infected over total mosquitoes tested.
RESULTS
Genetic variability of the YFV 3′ NCR.
Alignment of all available 3′ NCR sequences for wild-type YFV isolates included 136 sequences (117 South American and 19 African) (Table 1) and required extensive manual editing for indels ranging from 4 to 216 nt in length. As it was not feasible to show an alignment of all 136 sequences, we have combined groups of related viruses in Fig. 1B for clarity and simplicity. The overall conformation of 3′ NCR RNA secondary structures as previously defined (30, 33) appeared to be well conserved among both South American and African isolates of YFV (Fig. 1B). Substitutions that distinguished South American isolates from African isolates are indicated in Fig. 2A. Variable sites were found more frequently in single-stranded regions than in regions with predicted secondary structures (36) (Fig. 2B). The greatest divergence between South American and African isolates occurred within the 5′ side loop (e3) of structure E (Fig. 2A). The e3 region is composed of 22 sites (among African YFVs) and 21 sites (among South American YFVs). Of these sites, 13 were conserved and 9 were variable, making this the region with the highest density of substitutions within the stem-loop structures of the 3′ NCR.
FIG. 2.
(A) Nucleotide substitutions within the 3′ NCR of South American YFVs compared to the prototype Asibi isolate. Substitutions shared among all South American isolates are indicated in red; sites that are partially conserved within genotypes I and II are indicated in black. (B) Schematic diagram showing the positions of informative sites within multiple sequence alignment of partial 3′ NCR sequences. Note that the alignment ends after the b4 loop.
As predicted, domain I of the 3′ NCR was hypervariable, with numerous single-nucleotide polymorphisms in the single-stranded region upstream of the F-G dual-hairpin RYF motifs and size variation resulting from deletions of 40 to 80 nt between the different genotypes (49). The 117 South American isolates contained a single RYF motif, whereas East African isolates contained two RYFs and West African isolates contained three RYFs (28, 49). However, one South American genotype I isolate (Brazil91c; YF-XL-ΔRYF) had a deletion of 45 nt from positions 61 to 106 of the 3′ NCR, thus removing the RYF (F-G dual hairpin) plus an additional 10 nt of downstream single-stranded RNA (Fig. 1B). This isolate was obtained from a fatally infected Alouatta monkey in 1991 in Goias State, Brazil. Another notable polymorphism was a 4-nt deletion (3′ NCR positions 178 to 181) in the single-stranded region immediately upstream of the RYF dual hairpin, found in each of the 20 isolates of the Brazil clade 1C (represented as a single sequence in Fig. 1B), as well as in Brazil91C. Two isolates, Trinidad79b and Brazil60, exhibited substitutions in the pk2′ binding site of the C loop that are potentially disruptive to the formation of this pseudoknot structure (data not shown). In these isolates, the corresponding pk2 that binds to pk2′ lacked a covariant change. Similarly, 15 South American isolates exhibited a G→U substitution in stem D, but only one isolate (Brazil91b) showed a compensatory change to preserve the D stem without introducing a bulge (data not shown). The cumulative effect of these substitutions on the RNA folding patterns and secondary structure of the 3′ NCR is unclear.
Structural elements of domain II are duplicated in selected genotype I isolates.
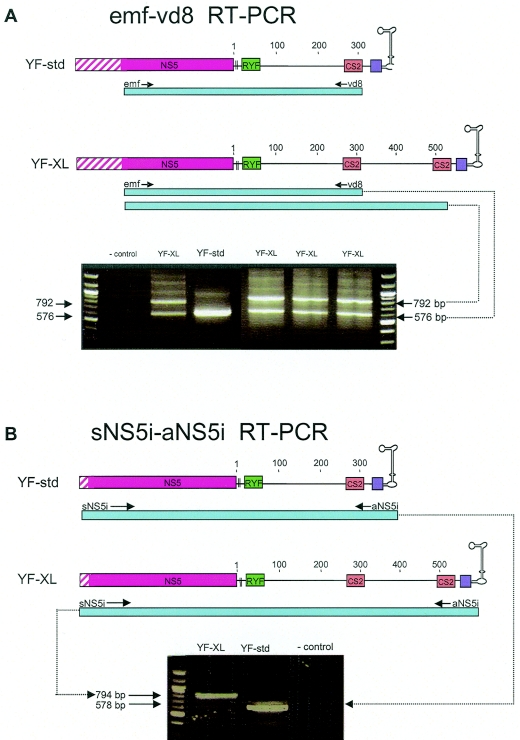
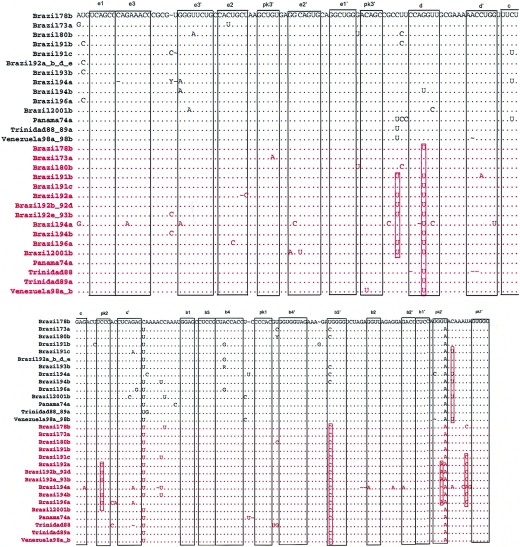
Nineteen of the 89 South American genotype I isolates of YFV (21%) were found to contain a duplication of 216 nt from positions 182 to 400 of the 3′ NCR (designated YF-XL). The presence of the YF-XL conformation was initially detected as a double banding pattern observed upon RT-PCR using the emf-vd8 primer pair (Fig. 3A). Sequence analysis of subcloned emf-vd8 band size variants revealed the presence of a duplication; subsequently, confirmation of the duplication was achieved by sequencing sNS5i-aNS5i amplicons (Fig. 3B). The sNS5i forward primer binds to genomic positions in the NS5 coding region, and the aNS5i reverse primer binds to the extreme 3′ genomic terminus; thus, sNS5i-aNS5i amplicons span the entire 3′ NCR and allow analysis of the junction sites at either end. The YF-XL duplication comprised an imperfect repeat of the RNA structures within domain II of the 3′ NCR, including four hairpins (E, D, C, and B) and binding sites for two pseudoknots (pk2 and pk1). The repeat region commenced 3 nt upstream of the E1 stem and ended immediately after the pk2 motif located downstream of dumbbell B. Insertion breakpoints were 100% conserved among the 19 YF-XL isolates at both the 3′ and 5′ ends of the duplication. Alignment of the upstream and downstream copies of the duplicated region revealed variation at a total of 63 sites (29%) over the length of the duplicated region (216 nt; genomic positions 10533 to 10749; 3′ NCR 182 to 400 [Asibi reference numbering]) (Fig. 4). Thirteen variable sites were due to indels. The duplicates within each YF-XL isolate varied at 8.2 ± 4.7 sites. Notable substitutions included a G→U transversion at 3′ NCR position 197 in downstream copies of 16 of the YF-XL isolates; for this position, there was no covariant substitution to help preserve the predicted stem of structure D. In addition, pseudoknot binding sites (pk2 and pk2′) of the upstream sequence differed from the downstream copy in six YF-XL isolates (Brazil92a, -b, -d, and -e; Brazil93b; and Brazil96a) (Fig. 4; also see Fig. 6). The typical pk2-pk2′ interaction of YF-std isolates involved predicted complementary binding between a UCCC motif in the loop of structure C and GGGA sequence on the 3′ side of dumbbell B. In these six YF-XL isolates (Brazil92a, -b, -d, and -e; Brazil93b; and Brazil96a), which were closely related variants of the IB clade (see Fig. 6), covariant substitutions in the pk2-pk2′ sites of the duplicated region were evident: C→U at position 226 of the 3′ NCR in the loop of structure C and a G→A substitution at position 323 in pk2′ (Fig. 4). As a result, an altered pseudoknot interaction was possible, involving 5 rather than 4 bp (UUUCC to GGAAA) (Fig. 2A).
FIG. 3.
RT-PCR amplification of YF-XL isolates using the emf-vd8 primer pair results in double bands (A), whereas amplification using the sNS5i-aNS5i primer pair results in a larger fragment size (B).
FIG. 4.
Alignment of the duplicated region of 19 YF-XL isolates, genomic positions 10533 to 10749 (3′ NCR 182 to 398; Asibi reference numbering). Sequences shown in red are the downstream duplicates of the sequences shown in black. Multiple isolates with 100% identity are represented by a single sequence. Regions involved in basepairing are boxed and labeled to correspond to stem structures as indicated in Fig. 1A and 2A. Hatched boxes indicate pseudoknot binding sites; red boxes indicate conserved substitutions among duplicated structures.
FIG. 6.
Neighbor-joining phylogenetic tree based on partial NS5-3′ NCR sequences (576 nt) of 117 South American isolates. To obtain sequences of identical lengths, the alignment was trimmed at the 3′ side of pk1′; thus, the alignment did not incorporate the duplicate copy of domain II structures for the YF-XL isolates. Sequences with 100% identity are represented by single branches. Isolates with no color highlighting have undetermined 3′ NCR status, because sequence for the genome terminus was not obtained.
Growth characteristics in cell culture.
To determine the growth characteristics for representatives of the different YF 3′ NCR variants, we infected mosquito C6/36 cells and mammalian Vero cells. The cells were infected at an MOI of 0.03, and infectivity titers in the media were followed for 12 days. Four YF-std and four YF-XL strains were evaluated in duplicate. YF-std isolates grew faster and reached higher peak titers than YF-XL isolates in mosquito cells but not in mammalian cells (Fig. 5). On 10 of the 12 sampling days, titers for the YF-std isolates grown in the C6/36 cultures exceeded those of YF-XL isolates by an average of 1.2 ± 0.5 log10 50% tissue culture infective dose (TCID50) units/ml; however, differences in daily titers were statistically significant only on day 3 (P = 0.04), reflecting an ∼24-h delay in growth kinetics. There was no evident trend in different growth rates of YF-std and YF-XL in Vero cells (Fig. 5E). The YF-XL-ΔRYF isolate (Brazil91c) grew better than other YF-XL isolates in C6/36 cells, but not in Vero cells. Average peak titers for YF-XL-ΔRYF (5.6 and 5.2 log10 TCID50 units/ml in C6/36 and Vero cells, respectively) were equivalent to those observed for YF-std isolates.
FIG. 5.
Comparison of growth characteristics of YF-XL, YF-XL-ΔRYF, and YF-std isolates in Vero and C6/36 cells. Cultures were infected with equal volumes of virus stocks at an approximate MOI of 0.03; 0.5-ml aliquots of supernatant were sampled with replacement for 12 consecutive days. The values represent the mean infectivity titers by endpoint assay; error bars indicate the standard deviations of the results for triplicate samples. (A) YF-std in C6/36 cells; (B) YF-XL in C6/36 cells; (C) YF-std in Vero cells; (D) YF-XL in Vero cells; (E) YF-XL-ΔRYF compared to mean of four YF-std isolates, each evaluated in duplicate.
Mouse neuroinvasiveness.
The abilities of YFV strains to be neuroinvasive in suckling outbred mice were evaluated following intraperitoneal inoculation of 8-day-old litters, as that is the age previously determined to allow optimal discrimination of neuroinvasiveness among strains (11). LD50 values were obtained for a total of 10 YF-XL strains (including YF-XL-ΔRYF), 5 YF-std South American genotype I strains, and 7 YF-std South American genotype II strains. LD50 values and average survival times for the 22 isolates of South American wild-type YFV revealed significant heterogeneity in the mouse neuroinvasiveness phenotype (Table 2). In general, the neuroinvasiveness phenotype did not appear to be correlated with the genotype or 3′ NCR conformation; there was no significant difference between the lethality of YF-XL (LD50 = 3.3 ± 1.3 log10 TCID50) and YF-std (LD50 = 2.5 ± 1.9 log10 TCID50) isolates or between South American genotype I (LD50 = 2.6 ± 2.6 log10 TCID50) and genotype II (LD50 = 2.5 ± 1.3 log10 TCID50). Brazil91c (YF-XL-ΔRYF) yielded an LD50 of 3.8 log10 TCID50. The average survival times were remarkably consistent across all isolates tested (average, 10.2 ± 1.1 days).
TABLE 2.
Mouse neuroinvasive phenotypes of selected wild-type strains of YFV following intraperitoneal inoculation into litters of 8-day-old mice
| Sequence | Strain | Source | Passage historya | Log10 TCID50 | LD50 (log10 TCID50) | Avg survival time (days) |
|---|---|---|---|---|---|---|
| South American genotype I | ||||||
| Brazil55B | BeAR 162 | Haemagogus janthinomys | SM4, C6/36#1 | 4.8 | 0.1 | 9.2 |
| Brazil60 | BeAN 23536 | Monkey | SM1, C6/36#1 | 6.8 | 5.5 | 1 |
| Brazil68C | BeAR 142658 | Haemagogus sp. | ?, C6/36#1 | 6.8 | 5.5 | 8.8 |
| Brazil73B | BeAN 232869 | Mosquito | Mosq 1, SM2 | 2.2 | 0.9 | 10.2 |
| Venezuela61 | PHO 424 | Human | C6/36#2, SM2 | 6.4 | 0.9 | 9 |
| South American genotype I-XL | ||||||
| Brazil73A | BeAN 233164 | Haemagogus | Mosq 4 | 4.2 | 3.7 | 10.4 |
| Panama74B | Jimenez | Human | Mk1 | 5.3 | 0.9 | 10.2 |
| Brazil73D | BeH 233393 | Human | SM1 c6/36#1 | 4.8 | 2.6 | 12 |
| Brazil91C | BeAN 510268 | Alouatta | SM1 c6/36#1 | 4.5 | 3.8 | 11 |
| Brazil92A | BeAR 512943 | Haemagogus | C6/36#1 | 5.0 | 3.9 | 10.4 |
| Brazil92B | BeAR 513008 | Sabethes | SM1, C6/36#1 | 4.5 | 4.3 | 6 |
| Brazil92D | BeAR 513060 | Haemagogus | SM1, C6/36#1 | 4.0 | 1.7 | 10.2 |
| Brazil92E | BeAR 513292 | Sabethes | SM1, C6/36#1 | 6.8 | 5.3 | 9.1 |
| Brazil94A | BeAN 527785 | Sabethes | SM1, C6/36#1 | 5.0 | 4.1 | 9.5 |
| Trinidad89A | CAREC 890692 | Sabethes | (NH) C6/36#1 | 4.4 | 2.8 | 10.7 |
| South American genotype II | ||||||
| Bolivia99A | OBS 7687 (JR 035) | Human | C6/36#2 | 5.6 | 1.9 | 9.3 |
| Peru77A | 1362/77 | Human | C6/36#2 | 4.6 | 2.7 | 9.3 |
| Peru78 | 287/78 | Human | SM1: Mosq 2 | 4.8 | 1.8 | 8.5 |
| Peru81A | B4.1 | Human | plaque pick | 5.8 | 2.8 | 9.1 |
| Peru95a | ARV 0544 | Human | SM1, Vero1, C6/36#1 | 3.8 | 1.2 | 10.8 |
| Peru95C | 153 (HEB4236) | Human | C6/36#1 | 7.3 | 1.9 | 8.4 |
| Peru95J | HEB 4245 | Human | SM1, C6/36#1 | 5.8 | 5.3 | 10.8 |
Passage history of seed strain in collection. SM, suckling mouse; Mosq, mosquito; Mk, monkey; ham, hamster.
Growth characteristics in mosquitoes.
Experiments were performed to determine the abilities of YF-XL and YF-XL-ΔRYF strains to produce disseminated infections in Rexville D A. aegypti. (Rexville D is a laboratory strain of A. aegypti originally established using mosquitoes from Rexville, Puerto Rico.) Four treatment groups of mosquitoes were orally infected with a representative YF-XL strain (Brazil92e); YF-XL-ΔRYF (Brazil91c); Asibi (prototype West African strain; positive control); and vaccine strain 17D-204 (negative control). Midgut infection rates for YF-XL-ΔRYF and YF-XL were 81 and 89% (Table 3), respectively, and were similar to the infection rate observed for the positive control (Asibi; 80% MIR). There was no evidence of infection in mosquitoes that fed on 17D-204 vaccine (0% MIR; 0% DIR). Dissemination to head tissues was highest in mosquitoes infected with Asibi (61% DIR) and lower in mosquitoes infected with the YF-XL-ΔRYF and YF-XL isolates (35 and 28% DIR, respectively).
TABLE 3.
Oral infection rates of A. aegypti (Rexville D) with four strains of YFVa
| Sequence | Strain | Blood meal titerb | Engorged day 0 midgut titerb | MIR (% infected midguts) (n)c | DIR (% infected heads) (n)c |
|---|---|---|---|---|---|
| Ghana27 | Asibi | 4.2 | 4.1 | 80 (5) | 61.1 (18) |
| 17D | Vaccine strain | 5.6 | 5.2 | 0 (14) | 0 (13) |
| Brazil91c (YF-XL-ΔRYF) | BeAN 510268 | 3.3 | 3.8 | 80.7 (26) | 35.3 (17) |
| Brazil92e (YF-XL) | BeAR 513292 | 2.8 | 3.9 | 89.2 (28) | 28.5 (21) |
The titer for 17D on day zero indicates that mosquitoes ingested live virus at the time of taking the blood meal, but it does not imply or suggest that infections were established within the midgut at this time. In order to demonstrate infection of the midgut, mosquitoes must test positive for viruses after a 2-week incubation period.
log10 TCID50/ml determined by endpoint assay in Vero cells.
By RT-PCR. n, number of females.
Evolutionary relationships among YF-XL isolates.
Partial 3′ NCR fragments (∼570 nt) were obtained for 73 South American isolates, and complete 3′ NCR sequences (440 to 660 nt) were obtained for 46 South American isolates. Among those isolates for which complete 3′ NCR sequences were obtained, 19 viruses (41%) were identified as YF-XL. The YF-XL isolates originated from the eastern and western states of Brazil (Goiás, Pará, Roraima, Mato Grosso do Sul, Maranhao, Minas Gerais, Rondônia, and Bahia), as well as Trinidad, Venezuela, and Panama (Table 1). YF-XL isolates were found exclusively among the dominant lineage of presently circulating South American genotype I viruses and were not present among the older Brazilian isolates from Pará state dating from 1954 to 1968 (9 isolates) or among South American genotype II isolates (29 isolates) (Fig. 6). The oldest isolate with the YF-XL conformation dated from 1973. There was no apparent correlation between the presence of the YF-XL repeat and the source of the isolate (i.e., mosquito or primate species) or any correlation with passage history. Four of the seven human isolates identified as YF-XL were obtained from fatal human cases (Table 1).
Although YF-XL isolates were confined to one subclade of genotype I, they did not appear to be monophyletic (Fig. 6), and closely related isolates did not appear to share the same 3′ NCR status. There was one instance in which a pair of isolates (Brazil73a and -b) were 100% identical over the partial NS5-3′ NCR fragment (576 nt); however, Brazil73a exhibited the YF-XL conformation and Brazil73b was YF-std (Fig. 6). The prM/E sequences of Brazil73a and Brazil73b were shown to differ by as much as 3.5% (24 nt) (4). Similarly, a group of mosquito isolates from Mato Grosso do Sul were each confirmed as YF-XL (Brazil92a, -b, -d, and -e), but the sequence of the closely related Brazil93a isolate did not show evidence of the duplication.
DISCUSSION
Data presented here provide the first evidence of variation (216 nt) in the length of domain II of the YFV 3′ NCR (YF-XL) and the first report of a naturally occurring wild-type YFV strain lacking all three copies of RYF (YF-XL-ΔRYF). The breakpoints of the duplicated regions of the YF-XL strains coincided precisely with predicted structural features. Specifically, the 5′ junction occurred at the base of stem-loop E, and the 3′ junction was immediately upstream of the cyclization motif at the beginning of zone III. There are several critical questions to be addressed regarding the finding of novel 3′ NCR duplications. Most important is whether YF-XL isolates represent naturally occurring virus populations or whether these findings reflect artifactual changes during cell growth or viral amplification by RT-PCR.
Three observations from the sequence data argue against the possibility that YF-XL sequences were generated during RT-PCR amplification. First, the YF-XL sequences were obtained using two different primer pairs (emf-vd8 and sNS5i-aNS5i) (Fig. 3). The sNS5i-aNS5i primer pair generated intact full-length 3′ NCR fragments containing the duplicated regions, whereas the emf-vd8 primer pair generated two fragments, corresponding to fragments both with and without the repeat. Second, an in vitro enzymatic process cannot easily explain the accumulation of conserved point mutations in the duplicated region. Third, the presence of precise breakpoints among YF-XL isolates suggests an in vivo replication process in which selection has operated to preserve the integrity of specific secondary structures. The presence of precise breakpoints provides indirect evidence in support of the presently proposed model for YFV RNA folding patterns (30, 33).
More difficult to discern is whether the YF-XL duplication may have occurred as a cell culture adaptation during laboratory passage. The YF-XL isolates identified in this study originated from at least four different regional reference laboratories (Instituto Evandro Chagas in Brazil, Gorgas Memorial Laboratory in Panama, Trinidad Regional Virus Laboratory, and the Instituto Nacional de Salud in Venezuela). Isolation procedures for the majority of YFV isolates were uniform (1 passage in suckling mouse brain, followed by growth in C6/36 cells), and most were low passage (<5 passages). Viruses with the XL duplication were found exclusively among isolates from one subclade of South American genotype I, suggesting that the duplication was a unique characteristic of one lineage rather than the result of shared passage histories. It is worth noting that spontaneous deletion of two RYF elements has been reported during passage of a YF vaccine virus (French neurotropic vaccine) (28), and deletions in the 3′ NCR have also been reported following repeated passage of tick-borne flaviviruses (12). Although passage-induced sequence deletions may be a relatively frequent occurrence, the generation of novel duplications is likely to be a rarer event (31). To date, passage-induced duplications have not been reported for any viruses in the genus Flavivirus. Definitive proof of the existence of YF-XL in nature will require direct amplification from field-collected material (sera or tissue of infected humans, wild-caught vertebrates, or mosquitoes).
The two copies of domain II RNA structures in YF-XL strains contained an average of 7.6 (3.5%) nucleotide differences. Importantly, a number of substitutions within the duplicated region appeared to be partially conserved (Fig. 4). A pair of covariant substitutions observed in six YF-XL isolates was predicted to increase the complementary interactions of pseudoknot binding (pk2-pk2′) and may serve to differentiate the two pseudoknot binding sites of the duplicated region (Fig. 4). The upstream copy showed higher identity to the homologous region of prototype South American isolates, suggesting that this copy may represent the progenitor sequence. It is particularly interesting that the YF-XL conformation contained two dumbbell structures, since all mosquito-borne flaviviruses studied to date—with the exception of YFV—also contain two dumbbells in domain II of the 3′ NCR (20, 30). Thus, identification of YF-XL isolates suggests the possibility of convergent evolution toward a conformation commonly held by most other mosquito-borne flaviviruses. In viruses of the Japanese encephalitis and dengue serogroups, the two dumbbells are adjacent to one another but separated by flanking A-rich single-stranded regions. In contrast, the spacing between the two dumbbells of YF-XL isolates is significantly greater, due to the intervening duplicated C, D, and E hairpins.
Interestingly, there are two published reports in the clinical and diagnostic literature of aberrant large-size amplicons obtained during analysis of the 3′ NCR of South American YFV (7, 9). These reports involved fatal cases of YF in an unvaccinated tourist who became infected while visiting Manaus, Brazil, in 1997 (9) and within a YF outbreak in Minas Gerais, Brazil, in 2001 (7). Both reports used RT-PCR emf-vd8 amplification to identify South American genotype I isolates, and both reported unexpected double bands from sample viruses but not from controls (wild-type and attenuated West African isolates). The large bands were dismissed as PCR artifacts; however, no apparent attempts were made to sequence the fragments. These findings indirectly support the hypothesis that the extra bands (YF-XL) are characteristic of South American genotype I YFV. To date, complete 3′ NCR sequences have been obtained for a total of 46 South American YFV isolates, and 19 (41%) have shown evidence of the YF-XL duplication, as confirmed by multiple sequencing reactions. As mentioned previously, the YF-XL isolates have been identified exclusively from the dominant lineage of contemporary South American genotype I isolates (Fig. 6). Sequence determinations for the remaining South American genotype I isolates will likely reveal more examples of the YF-XL 3′ NCR conformation.
Growth characteristics of YF-XL isolates.
Because RNA secondary structures of the 3′ NCR are involved in virus replication and are believed to serve as replication enhancer elements (22, 30, 33, 47), it was hypothesized that YF-XL isolates would exhibit a phenotype measurably different from those of YF-std isolates in one or more model systems (i.e., cell culture, suckling mice, or mosquitoes). Evaluation of South American YFV isolates in the mouse model showed no significant difference among the neuroinvasive properties of YF-XL, YF-XL-ΔRYF, and YF-std isolates (Table 2) or differences in lethality between South American genotype I YFV (n = 15) and South American genotype II (n = 7). The YF-XL-ΔRYF isolate showed cell culture growth characteristics similar to those of YF-std isolates in both C6/36 cells and monkey kidney Vero cells. In contrast, three representative YF-XL isolates grew more slowly than YF-std isolates in Aedes albopictus mosquito larval C6/36 cells, but not in monkey kidney Vero cells. The observation that YF-XL isolates exhibited delayed replication in C6/36 cells suggests the possibility that the duplicated domain II RNA secondary structures either directly or indirectly influence replication efficiency within cell lines of arthropod origin. Nevertheless, the YF-XL isolate Brazil92e and YF-XL-ΔRYF Brazil91c successfully established midgut and disseminated infections in A. aegypti mosquitoes following oral feeding on infectious blood meals (Table 3). MIRs for YF-XL and YF-XL-ΔRYF were 89 and 81%, respectively, and DIRs were 28 and 35%, respectively (Table 3). The infectivity of these strains was particularly remarkable given the relatively low titer of the infectious blood meal, which was 1 log unit lower than for 17D-204. Previously published studies using Rexville D A. aegypti mosquitoes have estimated MIRs for South American YFV in the range of 42 to 100% and DIRs from 46 to 79% (14, 24-26). Thus, the infectivity rates of YF-XL and YF-XL-ΔRYF fell within the expected variability for isolates with standard 3′ NCR conformations.
The biological function of the domain I RYFs has been the source of much speculation. Because the RYF copy numbers of the major lineages of YFV (49) that are vectored by different mosquito genera differ, vector coadaptation has been postulated. Recent mutagenesis experiments using the 17D infectious clone demonstrated that all three copies of the RYF motif could be deleted without eliminating virus replication in vertebrate cell culture (3). The present study extends those observations considerably, providing mouse neuroinvasiveness and mosquito infectivity data indicating that RYF motifs are not required for in vivo replication in either vertebrate or arthropod hosts and that phenotypic differences from YF-std isolates, if any exist, are subtle (they were not detected in our experiments).
The failure to identify pronounced phenotypic differences among YF-XL, YF-XL-ΔRYF, and YF-std in these model systems does not negate the possibility that there are as-yet-undiscovered phenotypic differences in the pathogeneses, virulences, or transmissibilities of these isolates. It seems reasonable to hypothesize that the variant 3′ NCR conformations of YF-XL-ΔRYF and YF-XL viruses must confer a fitness advantage during in vivo replication under some circumstances, as otherwise they would not be retained within the lineage of wild-type circulating viruses. Although it is tempting to speculate that differing 3′ NCR conformations reflect adaptation to novel arthropod vectors or to altered transmission cycles, there is no evidence at this point to support such claims. Future studies using reverse-genetics techniques and alternative measures of replication efficiency and/or infectivity may provide more refined assessments of the phenotypic effects conferred by YF-XL and YF-XL-ΔRYF 3′ NCR conformations.
The identification of YF-XL strains may have important evolutionary implications for the virus, insofar as it provides evidence that intra- or intermolecular reactions akin to recombination have occurred at least once in the recent history of YFV. Recombinational insertions similar to the YF-XL duplication have been described for a large number of positive-sense RNA viruses; several studies have identified recombination hot spots in the 3′ NCR that correspond to direct imperfect sequence repeats, stem-loop structures, and cryptic promoter-like elements recognized by the RNA-dependent RNA polymerase (15, 40, 51). It is worth noting that until 1999, there was no evidence for recombination among flaviviruses. Since the first report of mosaic genomes among selected strains of dengue type 2 virus in 1999, examples of homologous recombination have been proposed for all four dengue viruses (42, 43, 46, 53), Japanese encephalitis virus, and St. Louis encephalitis virus (45), as well as for other viruses in the family Flaviviridae, including pestiviruses (bovine viral diarrhea virus) (2), hepaciviruses (hepatitis C virus) (16), and GB virus C/hepatitis C (52). Identification of recombinant (mosaic) forms of flaviviruses has been controversial, however, due to issues of strain identification and potential contamination. Furthermore, the evolutionary importance of recombination continues to be debated, as these events are believed to be extremely rare in nature and most frequently have a deleterious effect on virus fitness (27, 38). Key areas for future research include determining the mechanism by which 3′ NCR duplications are generated and maintained within virus populations, continued investigations of the phenotypic importance of 3′ NCR variations, and elucidation of the precise regulatory role of RNA secondary structures.
Acknowledgments
We thank Robert Tesh and John Roehrig, who provided virus isolates from collections maintained at the World Arbovirus Reference Collection at the University of Texas Medical Branch, Galveston, Tex.; the Centers for Disease Control and Prevention, Fort Collins, Colo.; and the Instituto Evandro Chagas in Belém, Brazil, respectively. We thank the University of Texas Medical Branch Protein Core Facility for conducting sequencing reactions.
This research was supported by the CDC Fellowship Training Program in Vector-Borne Infectious Diseases no. T01/CCT622892 and by the Zelda Zinn Casper fund for graduate research. The participation of P. F. C. Vasconcelos had the financial support of a 2002 Lancet International Fellowship Award and CNPq (process 302770/02-0).
REFERENCES
- 1.Armstrong, P. M., and R. Rico-Hesse. 2001. Differential susceptibility of Aedes aegypti to infection by the American and Southeast Asian genotypes of dengue type 2 virus. Vector Borne Zoonotic Dis. 1:159-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becher, P., M. Orlich, and H. J. Thiel. 2001. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J. Virol. 75:6256-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bredenbeek, P. J., E. A. Kooi, and B. D. Lindenbach. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261-1268. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, J. E., and A. D. T. Barrett. 2003. Comparative phylogenies of yellow fever isolates from Peru and Brazil. FEMS Immunol. Med. Microbiol. 39:103-118. [DOI] [PubMed] [Google Scholar]
- 5.Charlier, N., P. Leyssen, C. W. Pleij, P. Lemey, F. Billoir, K. Van Laethem, A. M. Vandamme, E. De Clercq, X. de Lamballerie, and J. Neyts. 2002. Complete genome sequence of Montana Myotis leukoencephalitis virus, phylogenetic analysis and comparative study of the 3′ untranslated region of flaviviruses with no known vector. J. Gen. Virol. 83:1875-1885. [DOI] [PubMed] [Google Scholar]
- 6.Cologna, R., and R. Rico-Hesse. 2003. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 77:3929-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Filippis, A. M., R. M. Nogueira, H. G. Schatzmayr, D. S. Tavares, A. V. Jabor, S. C. Diniz, J. C. Oliviera, E. Moreira, M. P. Miagostovich, E. V. Costa, and R. Galler. 2002. Outbreak of jaundice and hemorrhagic fever in the Southeast of Brazil in 2001: detection and molecular characterization of yellow fever virus. J. Med. Virol. 68:620-627. [DOI] [PubMed] [Google Scholar]
- 8.Deubel, V., and M.-T. Drouet. 1997. Biological and molecular variations of yellow fever virus strains, p. 157-164. In J. F. Saluzzo and B. Dodet (ed.), Factors in the emergence of arbovirus diseases. Elsevier, Paris, France.
- 9.Deubel, V., M. Huerre, G. Cathomas, M. T. Drouet, N. Wuscher, B. Le Guenno, and A. F. Widmer. 1997. Molecular detection and characterization of yellow fever virus in blood and liver specimens of a non-vaccinated fatal human case. J. Med. Virol. 53:212-217. [PubMed] [Google Scholar]
- 10.Durbin, A. P., R. A. Karron, W. Sun, D. W. Vaughn, M. J. Reynolds, J. R. Perreault, B. Thumar, R. Men, C. J. Lai, W. R. Elkins, R. M. Chanock, B. R. Murphy, and S. S. Whitehead. 2001. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3′-untranslated region. Am. J. Trop. Med. Hyg. 65:405-413. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgeorge, R., and C. J. Bradish. 1980. The in vitro differentiation of strains of yellow fever virus in mice. J. Gen. Virol. 46:1-13. [DOI] [PubMed] [Google Scholar]
- 12.Gritsun, T. S., K. Venugopal, P. M. Zanotto, M. V. Mikhailov, A. A. Sall, E. C. Holmes, I. Polkinghorne, T. V. Frolova, V. V. Pogodina, V. A. Lashkevich, and E. A. Gould. 1997. Complete sequence of two tick-borne flaviviruses isolated from Siberia and the UK: analysis and significance of the 5′ and 3′-UTRs. Virus Res. 49:27-39. [DOI] [PubMed] [Google Scholar]
- 13.Heraud, J. M., D. Hommel, A. Hulin, V. Deubel, J. D. Poveda, J. L. Sarthou, and A. Talarmin. 1999. First case of yellow fever in French Guiana since 1902. Emerg. Infect. Dis. 5:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgs, S. T., J. O. Rayner, K. E. Olson, B. S. Davis, B. J. Beaty, and C. Blair. 1998. Engineered resistance in Aedes aegypti to a West African and a South American strain of yellow fever virus. Am. J. Trop. Med. Hyg. 58:663-670. [DOI] [PubMed] [Google Scholar]
- 15.Hill, K. R., M. Hajjou, J. Y. Hu, and R. Raju. 1997. RNA-RNA recombination in Sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching. J. Virol. 71:2693-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leitmeyer, K. C., D. W. Vaughn, D. M. Watts, R. Salas, I. Villalobos de Chacon, C. Ramos, and R. Rico-Hesse. 1999. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 73:4738-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, K.-C., H. Chang, and R.-Y. Chang. 2004. Accumulation of a 3′-terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells. J. Virol. 78:5133-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo, M. K., M. Tilgner, K. A. Bernard, and P. Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 77:10004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markoff, L. 2004. 5′ and 3′-noncoding regions in flavivirus RNA, p. 177-229. In T. J. Chambers and T. P. Monath (ed.), The flaviviruses: structure, replication, and evolution. Elsevier Press, Amsterdam, The Netherlands.
- 21.Markoff, L., X. Pang, H. S. Houng, B. Falgout, R. Olsen, E. Jones, and S. Polo. 2002. Derivation and characterization of a dengue type 1 host range-restricted mutant virus that is attenuated and highly immunogenic in monkeys. J. Virol. 76:3318-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Men, R., M. Bray, D. Clark, R. M. Chanock, and C. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, B. R., and D. Adkins. 1988. Biological characterization of plaque-size variants of yellow fever virus in mosquitoes and mice. Acta Virol. 32:227-234. [PubMed] [Google Scholar]
- 24.Miller, B. R., and M. E. Ballinger. 1988. Aedes albopictus mosquitoes introduced into Brazil: vector competence for yellow fever and dengue viruses. Trans. R. Soc. Trop. Med. Hyg. 82:476-477. [DOI] [PubMed] [Google Scholar]
- 25.Miller, B. R., and C. J. Mitchell. 1986. Passage of yellow fever virus: its effect on infection and transmission rates in Aedes aegypti. Am. J. Trop. Med. Hyg. 35:1302-1309. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell, C. J., B. R. Miller, and D. J. Gubler. 1987. Vector competence of Aedes albopictus from Houston, Texas, for dengue serotypes 1 to 4, yellow fever and Ross River viruses. J. Am. Mosquito Control Assoc. 3:460-465. [PubMed] [Google Scholar]
- 27.Murhpy, B. R., J. E. Blaney, Jr., and S. S. Whitehead. 2004. Arguments for live flavivirus vaccines. Lancet 364:499-500. [DOI] [PubMed] [Google Scholar]
- 28.Mutebi, J. P., R. C. A. Rijnbrand, H. Wang, K. Ryman, E. Wang, L. D. Fulop, R. Titball, and A. D. T. Barrett. 2004. Genetic relationships and evolution of genotypes of yellow fever virus and other members of the yellow fever virus group within the Flavivirus genus based on the 3′ noncoding region. J. Virol. 78:9652-9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mutebi, J. P., H. Wang, L. Li, J. E. Bryant, and A. D. Barrett. 2001. Phylogenetic and evolutionary relationships among yellow fever virus isolates in Africa. J. Virol. 75:6999-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsthoorn, R. C., and J. F. Bol. 2001. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA 7:1370-1377. [PMC free article] [PubMed] [Google Scholar]
- 31.Peerenboom, E., V. Jacobi, E. J. Cartwright, M. Adams, H.-H. Steinbiss, and J. F. Antoiw. 1997. A large duplication in the 3′-untranslated region of a subpopulation of RNA2 of the UK-M isolate of barley mild mosaic bymovirus. Virus Res. 47:1-6. [DOI] [PubMed] [Google Scholar]
- 32.Poidinger, M., R. A. Hall, and J. S. Mackenzie. 1996. Molecular characterization of the Japanese encephalitis serocomplex of the Flavivirus genus. Virology 218:417-421. [DOI] [PubMed] [Google Scholar]
- 33.Proutski, V., M. W. Gaunt, E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′-untranslated region of yellow fever virus: implications for virulence, attenuation and vaccine development. J. Gen. Virol. 78:1543-1549. [DOI] [PubMed] [Google Scholar]
- 34.Proutski, V., E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Proutski, V., T. S. Gritsun, E. A. Gould, and E. C. Holmes. 1999. Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res. 64:107-123. [DOI] [PubMed] [Google Scholar]
- 36.Rauscher, S., C. Flamm, C. W. Mandl, F. X. Heinz, and P. F. Stadler. 1997. Secondary structure of the 3′-noncoding region of flavivirus genomes: comparative analysis of base pairing probabilities. RNA 3:779-791. [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 37a.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 38.Seligman, S. J., and E. A. Gould. 2004. Live flavivirus vaccines: reasons for caution. Lancet 363:2073-2075. [DOI] [PubMed] [Google Scholar]
- 39.Shurtleff, A. C., D. W. Beasley, J. J. Chen, H. Ni, M. T. Suderman, H. Wang, R. Xu, E. Wang, S. C. Weaver, D. M. Watts, K. L. Russell, and A. D. Barrett. 2001. Genetic variation in the 3′ non-coding region of dengue viruses. Virology 281:75-87. [DOI] [PubMed] [Google Scholar]
- 40.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254:6-14. [DOI] [PubMed] [Google Scholar]
- 41.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland, Mass.
- 42.Tolou, H., P. Couissinier-Paris, V. Mercier, M. R. Pisano, X. de Lamballerie, P. de Micco, and J. P. Durand. 2000. Complete genomic sequence of a dengue type 2 virus from the French West Indies. Biochem. Biophys. Res. Commun. 277:89-92. [DOI] [PubMed] [Google Scholar]
- 43.Tolou, H., J. Nicoli, and C. Chastel. 2002. Viral evolution and emerging viral infections: what future for the viruses? A theoretical evaluation based on informational spaces and quasispecies. Virus Genes 24:267-274. [DOI] [PubMed] [Google Scholar]
- 44.Troyer, J. M., K. A. Hanley, S. S. Whitehead, D. Strickman, R. A. Karron, A. P. Durbin, and B. R. Murphy. 2001. A live attenuated recombinant dengue-4 virus vaccine candidate with restricted capacity for dissemination in mosquitoes and lack of transmission from vaccinees to mosquitoes. Am. J. Trop. Med. Hyg. 65:414-419. [DOI] [PubMed] [Google Scholar]
- 45.Twiddy, S. S., and E. C. Holmes. 2003. The extent of homologous recombination in members of the genus Flavivirus. J. Gen. Virol. 84:429-440. [DOI] [PubMed] [Google Scholar]
- 46.Uzcategui, N. Y. C. 2001. Molecular epidemiology of dengue type 2 virus in Venezuela: evidence for in situ virus evolution and recombination. J. Gen. Virol. 82:2945-2953. [DOI] [PubMed] [Google Scholar]
- 46a.Vasconcelos, P. F. C., J. E. Bryant, A. P. A. T. Rosa, R. B. Tesh, S. G. Rodrigues, and A. D. T. Barrett. 2004. Genetic divergence and dispersal of yellow fever virus in Brazil: periodic expansions of the enzootic zone. Emerg. Infect. Dis. 10:1578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallner, G., C. W. Mandl, M. Ecker, H. Holzmann, K. Stiasny, C. Kunz, and F. X. Heinz. 1996. Characterization and complete genome sequences of high- and low-virulence variants of tick-borne encephalitis virus. J. Gen. Virol. 77:1035-1042. [DOI] [PubMed] [Google Scholar]
- 48.Wallner, G., C. W. Mandl, C. Kunz, and F. X. Hienz. 1995. The flavivirus 3′-non coding region: extensive size heterogeneity independent of evolutionary relationships among strains of tick-borne encephalitis virus. Virology 213:169-178. [DOI] [PubMed] [Google Scholar]
- 49.Wang, E., S. C. Weaver, R. E. Shope, R. B. Tesh, D. M. Watts, and A. D. Barrett. 1996. Genetic variation in yellow fever virus: duplication in the 3′ noncoding region of strains from Africa. Virology 225:274-281. [DOI] [PubMed] [Google Scholar]
- 49a.Wang, E., K. D. Ryman, A. D. Jennings, D. J. Wood, F. Taffs, P. D. Minor, P. G. Sanders, and A. D. Barrett. 1995. Comparison of the genomes of the wild-type French viscerotropic strain of yellow fever virus with its vaccine derivative French neurotropic vaccine. J. Gen. Virol. 76:2749-2755. [DOI] [PubMed] [Google Scholar]
- 50.Watts, D., G. Ramirez, C. Cabezas, M. Wooster, C. Carrillo, C. Chuy, E. J. Gentrau, and C. G. Hayes. 1998. Arthropod-borne viral diseases in Peru, p. 193-218. In A. P. A. Travassos da Rosa, P. F. C. Vasconcelos, and J. F. S. Travassos da Rosa (ed.), An overview of arbovirology in Brazil and neighboring countries. Instituto Evandro Chagas, Belem, Brazil.
- 51.White, K. A., and T. J. Morris. 1995. RNA determinants of junction site selection in RNA virus recombinants and defective interfering RNAs. RNA 1:1029-1040. [PMC free article] [PubMed] [Google Scholar]
- 52.Worobey, M., and E. C. Holmes. 2001. Homologous recombination in GB virus C/hepatitis G virus. Mol. Biol. Evol. 18:254-261. [DOI] [PubMed] [Google Scholar]
- 53.Worobey, M., A. Rambaut, and E. C. Holmes. 1999. Widespread intra-serotype recombination in natural populations of dengue virus. Proc. Natl. Acad. Sci. USA 96:7352-7357. [DOI] [PMC free article] [PubMed] [Google Scholar]