Abstract
Oligomeric forms of the HC-Pro protein of the tobacco etch potyvirus (TEV) have been analyzed by analytical ultracentrifugation and single-particle electron microscopy combined with three-dimensional (3D) reconstruction. Highly purified HC-Pro protein was obtained from plants infected with TEV by using a modified version of the virus that incorporates a histidine tag at the HC-Pro N terminus (hisHC-Pro). The purified protein retained a high biological activity in solution when tested for aphid transmission. Sedimentation equilibrium showed that the hisHC-Pro preparations were heterogenous in size. Sedimentation velocity confirmed the previous observation and revealed that the active protein solution contained several sedimenting species compatible with dimers, tetramers, hexamers, and octamers of the protein. Electron microscopy fields of purified protein showed particles of different sizes and shapes. The reconstructed 3D structures suggested that the observed particles could correspond to dimeric, tetrameric, and hexameric forms of the protein. A model of the interactions required for oligomerization of the HC-Pro of potyviruses is proposed.
The HC-Pro protein of plant potyviruses is an important viral product that is involved in many functions during the virus life cycle. HC-Pro (50 to 53 kDa) was initially identified as an accessory helper factor required during the plant-to-plant transmission process of potyviruses by aphid vectors (14), a function that gave the original name to this protein (HC, helper component). Later, it was shown for different members of the genus that HC-Pro also participates in proteolysis of the viral polyprotein (11, 32) and in processes like genome amplification and infectivity (3, 4) and viral movement in the host plant (12, 38). Binding to RNA (48) and interaction with host proteins (16) were also demonstrated for HC-Pro. The multiple roles in many essential processes (reviewed in references 26, 37, and 47) were partially explained by the simultaneous finding in several laboratories that the HC-Pro protein can interfere with plant defense through the suppression of posttranscriptional gene-silencing mechanisms (2, 10, 18). Thus, many of the previously described functions and properties of HC-Pro should be reinterpreted and put in relation with its role in RNA silencing and related plant regulatory mechanisms (19, 20).
In the case of other viral RNA-silencing suppressor proteins such as the p19 of tombusviruses, the resolution of the structure (50, 53) has provided information on the molecular mechanisms of action (21). In spite of being the first characterized suppressor of RNA silencing and the considerable amount of work performed to study HC-Pro functions, very little information is available regarding its structure, and its mode of action in most of the HC-Pro functions remains unknown.
Among other well-established functions of the HC-Pro, the aphid transmission process seems to be highly influenced by the structural properties of the protein. All studied members of the genus Potyvirus are transmitted by aphids in a nonpersistent manner with dependency on the viral-encoded HC-Pro protein in addition to virions. This is a feature shared with other groups of plant viruses (reviewed in references 34 and 13). In the case of potyviruses, successful transmission requires that the aphids have access to HC-Pro before or at the same time as virus particles (36). This fact, along with other experimental data, suggested that HC-Pro might form a “bridge” between the virus particles and the aphid mouthparts, where the virions are likely to be retained and subsequently inoculated to plants (1, 28, 39, 52). Among the findings that supported this hypothesis, two conserved motifs in which modifications were associated with loss of transmission activity were mapped by site-directed mutagenesis in HC-Pro: the first motif, known as KITC, was located in the N-terminal region of the protein and seemed to be critical for virus retention in the stylets of the aphid vector (6), whereas the second motif, PTK, located in the C-terminal half of HC-Pro, was shown to be probably implicated in binding to the coat protein of virions (33). In addition, a HC-Pro binding domain was identified at the N-terminal region of the viral coat protein (7), overlapping with a highly conserved DAG motif that is also essential for the transmission process (24).
Earlier works with different potyviruses showed that the form of HC-Pro biologically active in transmission was probably an oligomer of the protein, with molecular weights ranging between 100 and 150 kDa that could represent dimers or trimers, as determined by high-pressure liquid chromatography (45) and gel filtration (35, 51). Consequently, a self-interaction was proposed to explain the fact that the soluble HC-Pro appeared as oligomers. The yeast two-hybrid system was used to map the expected self-interaction domains, but contradictory results were obtained since in some potyviruses only the N-terminal region was found to be involved in self-interaction (49), while in others the C-terminal region also seemed to be required (15). Cross-linking studies also pointed to the involvement of the central and C-terminal part of the protein in self-interaction and showed the predominant soluble form of HC-Pro to be a dimer (35).
Among other reasons that hindered past structural studies of the protein, it can be mentioned that HC-Pro expressed in bacteria or insect cells resulted in lack of activity when tested for transmission (46). Only its production in plants, either during a potyvirus infection, in transgenic lines (5), or using a heterologous viral vector (42), resulted in functional protein able to support aphid transmission. A recent report from our laboratory demonstrated that active HC-Pro protein of the tobacco etch potyvirus (TEV) can be produced in yeast cells (40), although the transmission rate was reduced compared to that for the protein obtained from plants. Therefore, plant-produced HC-Pro protein seems to be more adequate to perform structural studies that could be correlated with activity. The most relevant recent work on HC-Pro structure was based on the calculation of projection maps from two-dimensional (2D) crystals of purified His-tagged HC-Pro of lettuce mosaic potyvirus (LMV) grown on lipid monolayers (35), although the best-resolution crystals derived from a protein variant with a deletion affecting its N-terminal region (and consequently not active in transmission).
In the present work, we studied the structural properties of purified TEV His-tagged HC-Pro by using both analytical ultracentrifugation and single-particle 3D reconstruction. In an effort to approach the structural knowledge to the functions of HC-Pro, we demonstrated in transmission bioassays that our protein preparations were fully active. Our results give for the first time insights on the 3D structure of the HC-Pro oligomers present in a transmission-active preparation. A model of self-interactions leading to the formation of the identified oligomers of HC-Pro is proposed.
MATERIALS AND METHODS
Virus, host plants, and aphid vectors.
The pTEV-HCH10 clone, which contains a full-length copy of the TEV genome with a histidine tag fused to the N terminus of HC-Pro (8), was kindly provided by S. Blanc (BGPI, CIRAD-INRA-ENSAM, Montpellier, France). Infectious in vitro transcripts from the pTEV-HCH10 clone were used for virus propagation in Nicotiana tabacum L. cv. Xanthi nc plants. TEV particles were purified as described previously (31) from systemically infected plants 2 weeks postinoculation with pTEV-HCH10 transcripts. Nicotiana benthamiana Domin. seedlings were used as test plants for transmission assays by using as vectors aphids from a clonal population of Myzus persicae Sulzer reared on tobacco plants.
TEV hisHC-Pro protein purification.
A previously described protocol (8) was used, with a modification consisting of a selective precipitation with ammonium sulfate. Briefly, leaves of TEV-infected tobacco plants harvested at 10 and 20 days after inoculation were homogenized in 100 mM Tris-HCl (pH 8.0)-20 mM Mg2SO4-500 mM NaCl-0.5 mM EGTA buffer supplemented with 0.2% Na2SO3, 0.1% polyvinylpyrrolidone, 1 μg of RNase/ml, and 1 μg of DNase/ml. After filtration and centrifugation, two sequential precipitations with 20 and 40% (NH4)2SO4 were carried out, discarding the first pellet and recovering the protein by resuspension of the second pellet. The hisHC-Pro was retained in Ni-nitrilotriacetic acid resin (QIAGEN) packed on a polypropylene column and eluted with 100 mM Tris-HCl (pH 8.0), 20 mM Mg2SO4, 100 mM NaCl, and 400 mM EGTA. The hisHC-Pro protein was concentrated finally using a Centricon 10 concentrator (Pall Gelman) and stored at −70°C until use.
SDS-PAGE and Western blot detection of TEV hisHC-Pro.
Purified TEV hisHC-Pro protein was analyzed in discontinuous sodium dodecyl sulfate (SDS)-4 to 10% polyacrylamide gel electrophoresis (PAGE) stained with Coomassie blue. For Western blot analysis, electrophoretically separated proteins were blotted to polyvinylidene difluoride membranes and incubated with TEV HC-Pro-specific rabbit polyclonal antisera and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G as described previously (40). Bound antibodies were detected using the enhanced chemiluminescence system (Amersham Biosciences).
Aphid transmission assays.
Apterous aphids were collected for 2 h of preacquisition fasting before allowing them to feed through stretched Parafilm membranes as described previously (40). The feeding solution contained a mixture of purified TEV virions (0.2 mg/ml), hisHC-Pro protein purified from infected plants (from 0.1 to 0.4 mg/ml), and 20% sucrose. Volumes were adjusted using 100 mM Tris-HCl (pH 7.2)-20 mM MgCl2 buffer. Following a 10-min acquisition access period, groups of 10 aphids were placed on N. benthamiana test plants for overnight inoculation before spraying with insecticide. Symptoms were recorded 10 days later by visual inspection and confirmed, if required, by reverse transcription-PCR amplification as described previously (40).
Sedimentation equilibrium.
A Beckman Optima XL-A analytical ultracentrifuge equipped with absorbance optics was employed for analytical ultracentrifugation measurements by using an An50Ti rotor. We used standard (12-mm optical path) double-sector center pieces of charcoal-filled Epon. The equilibrium temperature was 4°C. Short-column experiments (70 μl of protein, loading concentrations ranging from 0.4 to 1.3 mg/ml) were done at different speeds (8,000, 10,000, and 13,000 rpm) by taking absorbance scans at the appropriate wavelengths (235, 260, or 280 nm) at sedimentation equilibrium. Samples were judged to be at equilibrium by the absence of systematic deviations in successive scans taken at 2-h intervals. High-speed sedimentation (50,000 rpm) was conducted afterwards for baseline corrections in all cases. Conservation of protein mass in solution was controlled during the experiment. The buoyant molecular masses of the proteins were determined by fitting a sedimentation equilibrium model for a single sedimenting solute to individual datasets with the EQASSOC program (supplied by Beckman Coulter, Inc. [see reference 30]). These values were converted to the corresponding average molecular masses by using 0.731 ml/g as the partial specific volume of TEV hisHC-Pro, calculated from the amino acid composition of the protein (22).
Sedimentation velocity.
Sedimentation velocity experiments were performed at 50,000 rpm and 12°C by using standard double-sector Epon-charcoal center pieces. Absorbance scans were taken at the appropriated wavelength. Differential sedimentation coefficient distributions c(s) were calculated by least-squares boundary modeling of sedimentation velocity data using the program SEDFIT (43). All continuous size distribution calculations were solved on a grid of 100 sedimentation coefficients, and comparison with grids of 200 coefficients produced very similar results. From this analysis, the sedimentation coefficients of all sedimenting species were obtained. The latter were corrected for buffer composition by using the program SEDNTERP (22). The calculated frictional ratios were used to transform the c(s) distribution into the corresponding molar mass distribution c(M) (43, 44).
Negative staining and electron microscopy observations.
For negative staining, purified TEV hisHC-Pro protein was diluted to an approximate concentration of 0.05 mg/ml. Samples were applied to glow-discharged copper-rhodio grids supporting a carbon film. The grids were negatively stained with 2% (wt/vol) uranyl acetate, blotted, and air dried. Observations were conducted at 80 kV in a JEOL 1230 electron microscope. Areas covered with individual molecules were recorded at 0 and 15 degrees tilt under low-dose conditions at a nominal magnification of ×40,000. Micrographs were digitalized in a Dimage Scan Multi PRO scanner (Minolta) with a sampling window corresponding to 4 Å/pixel.
Image processing and reconstruction.
Particles observed on purified TEV hisHC-Pro protein preparations were selected from the micrographs, centered, and averaged in two dimensions by using XMIPP (27). Image classification and 3D refinement were carried out without assuming any starting model by using EMAN (25). Resolution was estimated by Fourier shell correlation between two independent volumes by using the criteria of a 0.5 correlation coefficient.
RESULTS
Purification of TEV hisHC-Pro.
Although different purification methods have been described for the HC-Pro protein of potyviruses (17, 41, 45), we chose a purification system based on the use of a histidine tag fused to the N terminus of the protein (8) to obtain a highly active HC-Pro of TEV. We selected this virus because it has been used before as a model system in aphid transmission studies (23, 24, 52).
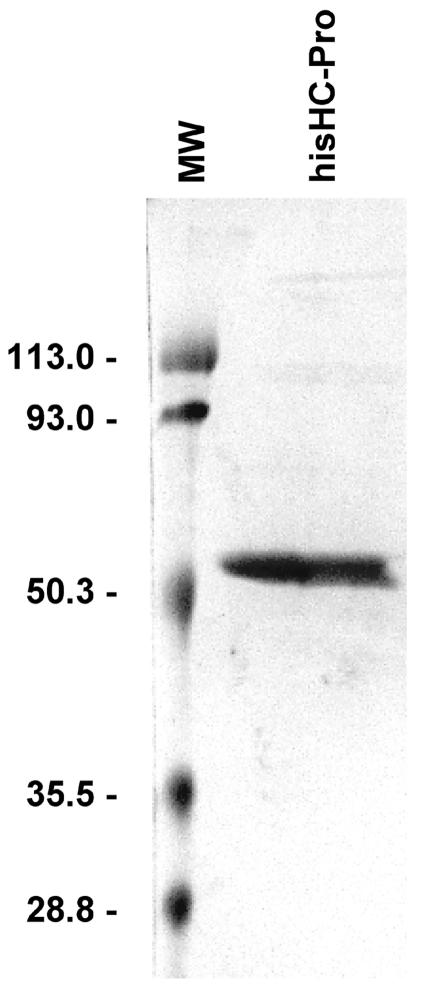
As previously described (8), transcripts derived from the TEV HCH10 construct were infectious when inoculated to tobacco plants, displaying typical symptoms. The hisHC-Pro protein was purified from infected plants with a yield similar to the one reported earlier (8). In our preparations, the performance of sequential selective ammonium sulfate precipitations yielded a final product with the unique expected 53-kDa band when analyzed by SDS-PAGE (Fig. 1). Other contaminants that usually copurify with TEV HC-Pro, in particular, a 12-kDa peptide (8), were completely absent when the described precipitation steps were performed.
FIG. 1.
Electrophoretic analysis of purified TEV hisHC-Pro protein. Coomassie blue staining of SDS-PAGE analysis of the TEV hisHC-Pro eluted from Ni-nitrilotriacetic acid resin and concentrated using a Centricon 10,000-Da membrane. Molecular weights (MW) of standard marker proteins (Bio-Rad) are indicated to the left of the panel.
Aphid transmission experiments.
To determine the activity of the purified TEV hisHC-Pro protein, aphid transmission experiments were performed using increasing concentrations of the protein (from 0.1 to 0.4 mg/ml). The purified protein was active in transmission bioassays when mixed with purified virus particles, and the efficiency of aphid transmission was directly correlated with the hisHC-Pro concentration, reaching 100% of transmission for the highest concentration (Table 1). As expected, the negative control (purified virions mixed with buffer) was not transmitted in parallel experiments.
TABLE 1.
Aphid transmission of purified TEV virions supplemented with purified hisHC-Pro
| Concn (mg/ml) in aphid feeding solutiona of:
|
Transmission efficiencyb
|
||
|---|---|---|---|
| TEV virions | TEV hisHC-Pro protein | Results | % |
| 0.2 | 0/45 | 0% | |
| 0.2 | 0.1 | 20/50 | 40% |
| 0.2 | 0.2 | 51/65 | 79% |
| 0.2 | 0.3 | 41/45 | 91% |
| 0.2 | 0.4 | 45/45 | 100% |
Aphid feeding solutions contained 20% sucrose and plant-purified TEV virus particles at the indicated concentration, mixed with different amounts of purified TEV hisHC-Pro protein. Volumes were adjusted with buffer. For the negative control (first row), only virus and buffer were added.
Several independent experiments were performed for each treatment, and the pooled values are indicated as well as the final percentages. The number of infected N. benthamiana plants over total number of test plants, using 10 aphids (M. persicae) per plant, is shown in the Results column. Significant differences (P < 0.05) were found according to a χ2 test between the negative control and each one of the treatments.
Analysis of functional TEV hisHC-Pro oligomers by analytical ultracentrifugation.
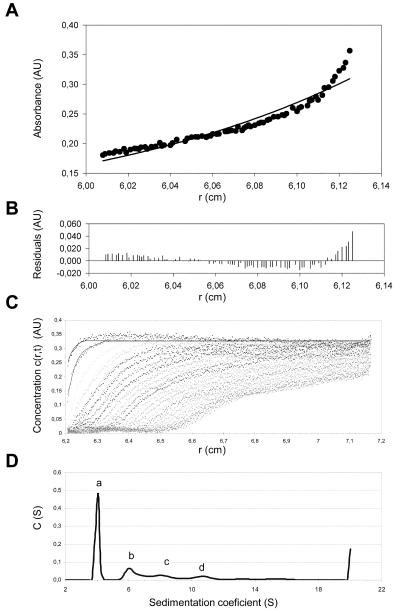
Sedimentation equilibrium was performed to analyze the association state of TEV hisHC-Pro in solution. Figure 2A shows a representative sedimentation equilibrium gradient of the protein (dots) together with the best fits to a single sedimenting species model (solid line). It is clear from the poor residual distribution (Fig. 2B) that the aphid-transmissible TEV hisHC-Pro was not behaving like a protein homogeneous in size but as a mixture of different species in solution. This result was consistently observed at different speeds and protein concentrations. The two-species model was also not fully compatible with the experimental data (not shown), suggesting that more than two oligomeric species were present in the biological active protein preparation. The latter fact precluded further analysis using this method. Therefore, we used sedimentation velocity to characterize the oligomerization pattern of TEV hisHC-Pro. Figure 2C shows a typical sedimentation velocity profile of the protein together with the corresponding best-fit boundaries from the c(s) sedimentation coefficient distribution (Fig. 2D). This analysis consistently revealed the presence of four species (Fig. 2D, peaks a through d) with sedimentation coefficients of 4.0, 6.3, 8.2, and 10.6S. Taking into account the calculated frictional coefficient ratio, these sedimentation coefficients were consistent with protein species of molar masses 115, 222, 331, and 494 kDa, respectively. These peaks amount to relative concentrations of 54, 23, 13, and 10%, respectively. Considering that the expected molecular mass of the HC-Pro monomer is 53 kDa, the major species found in this analysis was compatible with a nonspherical dimer and the rest of the oligomers could be assigned to tetramers, hexamers, and octamers of the protein, respectively.
FIG. 2.
Association states of TEV hisHC-Pro in solution. Sedimentation equilibrium at 10,000 rpm (Beckman Optima XL-A; An50Ti rotor) and 4°C of a sample containing TEV hisHC-Pro at a 1.3-mg/ml concentration (panels A and B) shows that the purified protein in solution is present in more than one form. (A) Sedimentation equilibrium data (dots) and fit corresponding to one homogeneous component (solid line). (B) Residuals between estimated values and experimental data for the one-component fit. Sedimentation velocity data (panels C and D) obtained with a 0.3-mg/ml sample of TEV hisHC-Pro subjected to a 50,000 rpm run at 12°C are shown. (C) Concentration profiles where dotted lines represent the experimental data and solid lines represent the fittings performed by application of the SEDFIT program. (D) Concentration distribution versus sedimentation coefficient showing four different association states (peaks a, b, c, and d), compatible with the dimers, tetramers, hexamers, and octamers of the protein.
Electron microscopy and 3D reconstruction of TEV hisHC-Pro.
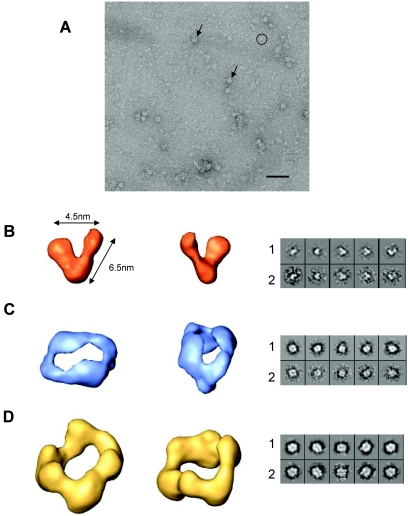
Purified and transmission-active TEV hisHC-Pro protein was applied to carbon-coated grids and negatively stained for observation under the electron microscope. Particles with different sizes and shapes could be observed on electron micrographs (Fig. 3A). As one type of molecule can give several projection images of variable size and shape, it could not be concluded that each type of view corresponded to a different molecule. However, size differences were sufficiently large to suggest that, most likely, different oligomerization states of the protein were present. To analyze the structures of putative oligomers, the particles extracted from the micrographs were classified in two groups, a first group of small particles (Fig. 3A) and a second group of large ones (Fig. 3A), and processed separately.
FIG. 3.
The 3D structure of TEV hisHC-Pro protein oligomers. (A) Electron microscopy field of a negatively stained sample of the purified TEV hisHC-Pro protein showing the presence of oligomeric structures of different sizes and shapes, indicated by a circle (smaller particles) and arrows (larger particles). Scale bar, 50 nm. The 3D structure of each one of the volumes reconstructed (B through D) is shown as two views corresponding to the putative dimer (panel B), tetramer (panel C), and hexamer (panel D). Each pair of images shown in the right panels represents a projection of the final volume (row 1) and the corresponding average of selected images (row 2). Videos of the reconstructed volumes are provided (see supplemental material).
In the case of small particles, 1,463 individual images were used for image classification and 3D refinement (25). Particles were classified into classes that corresponded to similar projections and aligned to form class averages. An initial volume was generated after model-free average classification using common lines. This model was used to generate projections with uniform distribution of orientations. Images were then grouped into classes by using these projections as references and aligned to create new averages. In the last refinement steps, the projections of the volume correlated well with the corresponding class averages (Fig. 3B, rows 1 and 2). The resulting volume showed a V-shaped molecule representing probably a dimeric form of the protein. This putative dimer had two arms of about 6.5 nm in length, and the two arms had structurally different ends that might correspond to different extremes of the monomers, suggesting that the monomers interacted in antiparallel orientation to form a dimer.
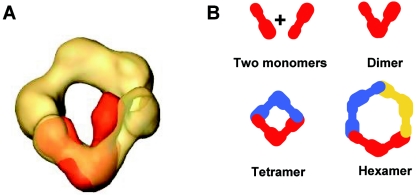
In the group of large particles, 2,828 images were selected and used in model-free refinement to obtain a 3D structure (25). To generate the volume, only the more similar particles were used to create averages, and the volume was made with class averages that best correlated with a volume. The resulting 3D structure (Fig. 3D) had a circular shape where three V-shaped domains could be distinguished. Correlation between volume projections and corresponding class averages is shown in Fig. 3D, rows 1 and 2. To perfectly fit a 3D structure of the putative dimer of hisHC-Pro to this volume, only a minor angle adjustment at the hinge would be required (Fig. 4A). Therefore, we can conclude that the reconstructed volume corresponds to the 3D structure of a hexameric form of hisHC-Pro.
FIG. 4.
Interpretation of the structure of the TEV hisHC-Pro oligomers. (A) Docking of the putative dimer (orange) into the hexamer (yellow). (B) Schematic representation of a model of the interactions required between two putative monomers (represented by an elongated shape with two structural domains at each end, connected by a constriction) in antiparallel orientation to form the dimer state of the protein and the proposed model of formation of tetramers and hexamers by the interaction of two or three dimers.
Using the above-mentioned selection criteria to reconstruct the hexameric form, a number of images were always excluded from the 3D structure of the volume. These excluded images were smaller in size than those used in the reconstruction, and they were selected and used to generate a new reference-free volume by using a common-lines procedure. This new volume and the volume corresponding to the hexamer were then used as two simultaneous reference models during refinement (25). As a first step of refinement, both volumes were used to create two independent sets of projections. Each single image was classified using all projections as a reference, and two sets of class averages were created and used to generate two independent volumes. These new models were projected in the following refinements. In these processes, a 3D model corresponding to a hexamer did not change and a new volume was stabilized (Fig. 3C) with good correlation of projections and averages (Fig. 3C, rows 1 and 2). This new 3D structure had two V-shaped domains connected by the tips of their arms. The volume had clear twofold symmetry imposed just in the two last rounds of refinement. A 3D structure representing a dimer could easily be fitted in each side of this new volume (data not shown), indicating that we had elucidated a 3D structure of the putative tetramer of hisHC-Pro.
Using the criteria of a 0.5 correlation coefficient to estimate the resolution achieved, the 3D reconstructions were estimated to have a resolution of 36 Å for the volumes of dimer and hexamer, and 30 Å in the case of the tetramer. Videos showing the reconstructed volumes are supplied (see supplemental material).
Interpretation of the structure of TEV hisHC-Pro oligomers.
The structure of the putative dimer showed an open compass shape with two arms of about 6.5 nm in length and with a maximal separation of 4.5 nm, which corresponds roughly to a 40° angle (Fig. 3B and supplemental material). Apparently, the two arms had different sizes, one being larger than the other. This dimeric state of the protein could represent a basic unit to form higher oligomerization states like tetramers (Fig. 3C and supplemental material) and hexamers (Fig. 3D and supplemental material). The size of the resolved structures resulting was compatible with this interpretation, as demonstrated by superimposing images with the same scale (Fig. 4A). Figure 4B shows an interpretation of how two hypothetical elongated monomers which have structurally different ends could interact in antiparallel orientation to form a dimer, and from this structural unit, similar interactions involving large and small arms of two or three dimers could be arranged to create tetrameric and hexameric structures, respectively.
DISCUSSION
A better knowledge of the HC-Pro structure is clearly required to help us to understand its multiple functions. Several important roles of the protein await structural information to gain insights into its modes of action. Among other functions, assistance during aphid transmission is supposed to be highly influenced by the structural and oligomerization properties of HC-Pro.
Preliminary attempts to obtain structural information of HC-Pro of potyviruses by using in silico approaches yielded unsuccessful results (not shown). As several functions are present in the same protein, it was tempting to assume that they might be distributed along the folded structure in a modular fashion. However, even the well-characterized protease domain, supposed to be restricted to the C-terminal part of the protein (11, 32), does not allow a convincing prediction. Another documented fact is the capacity of HC-Pro to form oligomers (15, 35, 45, 49, 51), and again, attempts to use in silico interaction approaches to predict contacts between HC-Pro domains were unsuccessful.
The failure of these prediction methods prompted us to obtain more experimental data about the structural nature of HC-Pro and the oligomers that it might form. Therefore, purified hisHC-Pro protein of TEV was used for structural studies. Since we were interested in the activity of HC-Pro, particularly in transmission, we demonstrated first that the protein preparations used for structural determinations remained fully active in transmission bioassays.
Sedimentation equilibrium unambiguously showed that the protein in solution was present as a mixture of molecular species. This result confirmed for TEV HC-Pro the heterogeneity previously found and with different methods for the HC-Pro protein of other potyviruses (35, 45, 51). The polydispersity of the protein prompted us to use sedimentation velocity to determine the nature of the main protein species present in the solution. Using this method, the presence of at least four oligomeric species of the hisHC-Pro of TEV was detected. The calculated molar masses of the more abundant species were compatible with dimers, tetramers, hexamers, and octamers of the protein. This result strongly suggested that the dimer is the minimal species required for oligomerization. The presence of dimers in HC-Pro preparations had been proposed by other workers (45), although the molecular size determined by gel filtration indicated an intermediate value between dimer and tetramer (35) that might also be interpreted as trimers. With LMV HC-Pro, a similar result was found, but the authors excluded the existence of trimers by using cross-linking experiments (35). Our results with TEV HC-Pro also pointed to the existence of abundant nonspherical dimers in a fully active solution, stressing its possible role as the functional molecule that was proposed earlier (45). In addition to dimers, we also identifyed tetramers, hexamers, and possibly octamers, and at this point, it can not be ruled out that they might contribute to the activity or that different oligomerization states of HC-Pro might fulfill different functions.
The existence of multimers of dimers was also suggested for HC-Pro proteins from another potyvirus (35). Working with the same TEV hisHC-Pro protein, we found in a previous work that several forms with higher molecular masses were detectable in Western blot analysis (40). In the present work, we were able to quantify the different forms in solution, finding that the majority (above 50%) of HC-Pro is in the dimeric state, with significant amounts of tetramers (above 20%) and also hexamers and octamers. Higher-than-octamer forms were not detected in our centrifugation experiments. In collaboration with S. Blanc and M. Drucker, we performed preliminary analytical ultracentrifugation experiments using purified LMV hisHC-Pro and found a similar oligomerization profile, which suggests that the presence of oligomers in solution might be a general characteristic of most potyviral HC-Pro proteins (unpublished results).
Monomeric hisHC-Pro protein is probably too small (53 kDa) to be distinguished from the background in electron microscopy field. Indeed, our electron microscope observations pointed out that several oligomeric forms were present in the active solution. Without any previous assumption, we were able to reconstruct three of the visualized structures, finding that they had a compass-like V shape with different ends in the case of the smaller form and that the larger ones resembled two or three V forms next to each other. If we assume that the V-shaped structure was dimeric, a tetramer could be obtained with two dimers together and piling up three dimers would result in the hexamer. These facts, taken together with our analytical ultracentrifugation results, strongly suggest that the observed structures were indeed dimers, tetramers, and hexamers of hisHC-Pro in which the dimer was acting as a structural unit to build up larger oligomers. In addition, the hisHC-Pro dimer is among the smallest structures solved by single-particle methodology (9, 29).
There is structural information available on hisHC-Pro of another potyvirus obtained by projection maps of negatively stained 2D crystals (35). The LMV hisHC-Pro protein formed 2D crystals with a tetrameric organization, in which the monomer was proposed to have an elongated shape composed of two structural domains (1 and 2) with different densities, separated by a flexible constriction. The authors suggested that a dimer of hisHC-Pro is the structural unit of the tetramer. Projections of 2D crystals of this tetramer showed interactions between type 1 domains, although intermolecular contacts between domains 1 and 2 were also observed (35). Our results from single-particle microscopy and 3D reconstruction showed a volume where the shape of the dimer could easily be interpreted as a junction of two elongated monomers. Certainly, the shape of the monomer required to form our TEV hisHC-Pro dimeric structure resembles the elongated dumbbell-like shape proposed for the LMV hisHC-Pro in the 2D crystal projection (35). As the ends of the two arms in the V shape were not equivalent, we consider that the two monomers could bind in antiparallel orientation to form a dimer. This would imply interaction between different domains of two HC-Pro monomers, in contrast with the 2D projection map of LMV HC-Pro that showed tetramers where the equivalent domains were in close contact. This apparent discrepancy is probably due to the different experimental conditions used. Whereas our protein was studied in solution, the LMV data were obtained with crystals grown on Ni2+-chelating lipid monolayers (35) where reorientation of the monomers might have occurred.
If our proposed model of interactions shown in Fig. 4B is correct, and assuming that the putative elongated monomer of TEV hisHC-Pro follows the predicted secondary structure as in the case of LMV hisHC-Pro (35), then the KITC and PTK motifs should be rather separated in the monomer, each one being close to a different end the molecule. Furthermore, since the dimer seemed to be formed by two antiparallel monomers, the two motifs might be placed near the opposite arms of the final structure, and this geometrical arrangement could explain how the HC-Pro dimer could interact with virions on one side and with the aphid vector inner stylet structures on the other during the transmission process, as suggested by the bridge hypothesis. Of course, at this point, it is not possible to allocate either the N- and C-terminal regions or particular motifs in the structure, and further analysis will be required to clarify this.
The present work provides the first experimental information on the structural organization of the soluble oligomers formed by the HC-Pro protein of potyviruses. The structural data have been obtained with preparations that maintained a high biological activity, as demonstrated using aphid transmission bioassays. The mode of action of the protein in this and other important functions of the HC-Pro, including its ability to suppress RNA silencing in plants, could benefit from the structural knowledge generated. Further structural analysis could serve for purposes like the design of HC-Pro variants capable of interfering with any of the functions and processes in which the protein is involved.
Supplementary Material
Acknowledgments
We thank S. Blanc and M. Drucker for the LMV hisHC-Pro protein preparation and critical reading of the manuscript. Advice by Jose Luis Garcia is greatly acknowledged. We also thank Ramon Roca and Antonio Ruiz Mateo for their valuable help and assistance.
V.R.-F. is a recipient of a Plan nacional de formación de personal investigador (PNFPI) fellowship from MCyT (Spain). This work was supported by Ministerio de ciencia y tecnología (MCyT, Spain) projects AGL2001-2141 to J.J.L.-M. and SAF2002-01715 to O.L., and by Comunidad de Madrid (Spain) grant 07 M-0072-2002 to J.J.L.-M.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ammar, E. D., U. Jarlfors, and T. P. Pirone. 1994. Association of potyvirus helper component protein with virions and the cuticle lining the maxillary food canal and foregut of an aphid vector. Phytopathology 84:1054-1060. [Google Scholar]
- 2.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atreya, C. D., and T. P. Pirone. 1993. Mutational analysis of the helper component-proteinase gene of a potyvirus: effects of amino acid substitutions, deletions, and gene replacement on virulence and aphid transmissibility. Proc. Natl. Acad. Sci. USA 90:11919-11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atreya, C. D., P. L. Atreya, D. W. Thornbury, and T. P. Pirone. 1992. Site-directed mutations in the potyvirus HC-Pro gene affect helper component activity, virus accumulation, and symptom expression in infected tobacco plants. Virology 191:106-111. [DOI] [PubMed] [Google Scholar]
- 5.Berger, P. H., A. G. Hunt, L. L. Domier, G. M. Hellman, Y. Stram, D. W. Thornbury, and T. P. Pirone. 1989. Expression in transgenic plants of a viral gene product that mediates insect transmission of potyviruses. Proc. Natl. Acad. Sci. USA 86:8402-8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc, S., E. D. Ammar, S. García-Lampasona, V. Dolja, C. Llave, J. Baker, and T. P. Pirone. 1998. Mutations in the potyvirus helper component protein: effects on interactions with virions and aphid stylets. J. Gen. Virol. 79:3119-3122. [DOI] [PubMed] [Google Scholar]
- 7.Blanc, S., J. J. Lopez-Moya, R. Y. Wang, S. Garcia-Lampasona, D. W. Thornbury, and T. P. Pirone. 1997. A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology 231:141-147. [DOI] [PubMed] [Google Scholar]
- 8.Blanc, S., V. Dolja, C. Llave, and T. P. Pirone. 1999. Histidine-tagging and purification of tobacco etch potyvirus helper component protein. J. Virol. Methods 77:11-15. [DOI] [PubMed] [Google Scholar]
- 9.Boskovic, J., A. Rivera-Calzada, J. D. Maman, P. Chacón, K. R. Willison, L. H. Pearl, and O. Llorca. 2003. Visualization of DNA-induced conformational changes in the DNA repair kinase DNA-PKcs. EMBO J. 22:5875-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Carrington, J. C., S. M. Cary, T. D. Parks, and W. G. Dougherty. 1989. A second proteinase encoded by a plant potyvirus genome. EMBO J. 8:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronin, S., J. Verchot, R. Haldeman-Cahill, M. Schaad, and J. Carrington. 1995. Long-distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell 7:549-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Froissart, R., Y. Michalakis, and S. Blanc. 2002. Helper component-transcomplementation in the vector transmission of plant viruses. Phytopathology 92:576-579. [DOI] [PubMed] [Google Scholar]
- 14.Govier, D. A., and B. Kassanis. 1974. A virus-induced component of plant sap needed when aphids acquire potato virus Y from purified preparations. Virology 61:420-426. [DOI] [PubMed] [Google Scholar]
- 15.Guo, D., A. Merits, and M. Saarma. 1999. Self-association and mapping of interaction domains of helper component-proteinase of potato A potyvirus. J. Gen. Virol. 80:1127-1131. [DOI] [PubMed] [Google Scholar]
- 16.Guo, D., C. Spetz, M. Saarma, and J. P. Valkonen. 2003. Two potato proteins, including a novel RING finger protein (HIP1), interact with the potyviral multifunctional protein HCpro. Mol. Plant Microbe Interact. 16:405-410. [DOI] [PubMed] [Google Scholar]
- 17.Kadouri, D., Y. Peng, Y. Wang, S. Singer, H. Huet, B. Raccah, and A. Gal-On. 1998. Affinity purification of HC-Pro of potyviruses with Ni2+-NTA resin. J. Virol. Methods 76:19-29. [DOI] [PubMed] [Google Scholar]
- 18.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 19.Kasschau, K. D., and J. C. Carrington. 2001. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285:71-81. [DOI] [PubMed] [Google Scholar]
- 20.Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 21.Lakatos, L., G. Szittya, D. Silhavy, and J. Burgyan. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, United Kingdom.
- 23.Llave, C., B. Martinez, J. R. Diaz-Ruiz, and D. Lopez-Abella. 2002. Amino acid substitutions within the Cys-rich domain of the tobacco etch potyvirus HC-Pro result in loss of transmissibility by aphids. Arch. Virol. 147:2365-2375. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Moya, J. J., R. Y. Wang, and T. P. Pirone. 1999. Context of the coat protein DAG motif affects potyvirus transmissibility by aphids. J. Gen. Virol. 80:3281-3288. [DOI] [PubMed] [Google Scholar]
- 25.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 26.Maia, I. G., A. Haenni, and F. Bernardi. 1996. Potyviral HC-Pro: a multifunctional protein. J. Gen. Virol. 77:1335-1341. [DOI] [PubMed] [Google Scholar]
- 27.Marabini, R., I. M. Masegosa, M. C. San Martin, S. Marco, J. J. Fernandez, L. G. de la Fraga, C. Vaquerizo, and J. M. Carazo. 1996. Xmipp: an image processing package for electron microscopy. J. Struct. Biol. 116:237-240. [DOI] [PubMed] [Google Scholar]
- 28.Martín, B., J. L. Collar, W. F. Tjallingii, and A. Fereres. 1997. Intracellular ingestion and salivation by aphids may cause the acquisition and inoculation of non-persistently transmitted plant viruses. J. Gen. Virol. 78:2701-2705. [DOI] [PubMed] [Google Scholar]
- 29.Martin-Benito, J., J. Boskovic, P. Gomez-Puertas, J. L. Carrascosa, C. T. Simons, S. A. Lewis, F. Bartolini, N. J. Cowan, and J. M. Valpuesta. 2002. Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 21:6377-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minton, A. P. 1994. Conservation of signal: a new algorithm for the elimination of the reference concentration as an independently variable parameter in the analysis of sedimentation equilibrium, p. 81-93. In T. M. Schuster and T. M. Laue (ed.), Modern analytical ultracentrifugation. Birkhäuser, Boston, Mass.
- 31.Murphy, J. F., R. E. Rhoads, A. G. Hunt, and J. G. Shaw. 1990. The VPg of tobacco etch virus RNA is the 49-kDa proteinase or the N-terminal 24-kDa part of the proteinase. Virology 178:285-288. [DOI] [PubMed] [Google Scholar]
- 32.Oh, C. S., and J. C. Carrington. 1989. Identification of essential residues in potyvirus proteinase HC-Pro by site-directed mutagenesis. Virology 173:692-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng, Y. H., D. Kadoury, A. Gal-On, H. Huet, Y. Wang, and B. Raccah. 1998. Mutations in the HC-Pro gene of zucchini yellow mosaic potyvirus: effects on aphid transmission and binding to purified virions. J. Gen. Virol. 79:897-904. [DOI] [PubMed] [Google Scholar]
- 34.Pirone, T. P., and S. Blanc. 1996. Helper-dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34:227-247. [DOI] [PubMed] [Google Scholar]
- 35.Plisson, C., M. Drucker, S. Blanc, S. German-Retana, O. Le Gall, D. Thomas, and P. Bron. 2003. Structural characterization of HC-Pro, a plant virus multifunctional protein. J. Biol. Chem. 278:23753-23761. [DOI] [PubMed] [Google Scholar]
- 36.Raccah, B., H. Huet, and S. Blanc. 2001. Potyviruses, p. 181-206. In K. Harris, J. E. Duffus, and O. P. Smith (ed.), Virus-insect-plant interactions. Academic Press, San Diego, Calif.
- 37.Revers, F., O. LeGall, T. Candresse, and A. J. Maule. 1999. New advances in understanding the molecular biology of plant/potyvirus interactions. Mol. Plant-Microbe Interact. 12:367-376. [Google Scholar]
- 38.Rojas, M. R., F. M. Zerbini, R. F. Allison, R. L. Gilbertson, and W. J. Lucas. 1997. Capsid protein and helper component-proteinase function as potyvirus cell-to-cell movement proteins. Virology 237:283-295. [DOI] [PubMed] [Google Scholar]
- 39.Roudet-Tavert, G., S. German-Retana, T. Delaunay, B. Delecolle, T. Candresse, and O. Le Gall. 2002. Interaction between potyvirus helper component-proteinase and capsid protein in infected plants. J. Gen. Virol. 83:1765-1770. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz-Ferrer, V., E. Goytia, B. Martinez-Garcia, D. Lopez-Abella, and J. J. Lopez-Moya. 2004. Expression of functionally active helper component protein of Tobacco etch potyvirus in the yeast Pichia pastoris. J. Gen. Virol. 85:241-249. [DOI] [PubMed] [Google Scholar]
- 41.Sako, N., and K. Ogata. 1981. Different helper factors associated with aphid transmission of some potyviruses. Virology 112:762-765. [DOI] [PubMed] [Google Scholar]
- 42.Sasaya, T., L. Torrance, G. Cowan, and A. Ziegler. 2000. Aphid transmission studies using helper component proteins of potato virus Y expressed from a vector derived from potato virus X. J. Gen. Virol. 81:1115-1119. [DOI] [PubMed] [Google Scholar]
- 43.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78:1606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuck, P., M. A. Perugini, N. R. Gonzales, G. J. Howlett, and D. Schubert. 2002. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 82:1096-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thornbury, D. W., G. M. Hellmann, R.E. Rhoads, and T. P. Pirone. 1985. Purification and characterization of potyvirus helper component. Virology 144:260-267. [DOI] [PubMed] [Google Scholar]
- 46.Thornbury, D. W., J. F. van den Heuvel, J. A. Lesnaw, and T. P. Pirone. 1993. Expression of potyvirus proteins in insect cells infected with a recombinant baculovirus. J. Gen. Virol. 74:2731-2735. [DOI] [PubMed] [Google Scholar]
- 47.Urcuqui-Inchima, S., A. L. Haenni, and F. Bernardi. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74:157-175. [DOI] [PubMed] [Google Scholar]
- 48.Urcuqui-Inchima, S., I. G. Maia, P. Arruda, A. L. Haenni, and F. Bernardi. 2000. Deletion mapping of the potyviral helper component-proteinase reveals two regions involved in RNA binding. Virology 268:104-111. [DOI] [PubMed] [Google Scholar]
- 49.Urcuqui-Inchima, S., J. Walter, G. Drugeon, S. German-Retana, A. L. Haenni, T. Candresse, F. Bernardi, and O. Le Gall. 1999. Potyvirus helper component-proteinase self-interaction in the yeast two-hybrid system and delineation of the interaction domain involved. Virology 258:95-99. [DOI] [PubMed] [Google Scholar]
- 50.Vargason, J. M., G. Szittya, J. Burgyan, and T. M. Tanaka Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799-811. [DOI] [PubMed] [Google Scholar]
- 51.Wang, R. Y., and T. P. Pirone. 1999. Purification and characterization of turnip mosaic virus helper component protein. Phytophatology 89:564-567. [DOI] [PubMed] [Google Scholar]
- 52.Wang, R. Y., E. D. Ammar, D. W. Thornbury, J. J. Lopez-Moya, and T. P. Pirone. 1996. Loss of potyvirus transmissibility and helper-component activity correlate with non-retention of virions in aphid stylets. J. Gen. Virol. 77:861-867. [DOI] [PubMed] [Google Scholar]
- 53.Ye, K., L. Malinina, and D. J. Patel. 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.