Abstract
The N-proximal region of cowpea chlorotic mottle virus (CCMV) capsid protein (CP) contains an arginine-rich RNA binding motif (ARM) that is also found in the CPs of other members of Bromoviridae and in other RNA binding proteins such as the Tat and Rev proteins of human immunodeficiency virus. To assess the critical role played by this motif during encapsidation, a variant of CCMV RNA3 (C3) precisely lacking the ARM region (C3/Δ919) of its CP gene was constructed. The biology and the competence of the matured CP derived in vivo from C3/Δ919 to assemble and package progeny RNA was examined in whole plants. Image analysis and computer-assisted three-dimensional reconstruction of wild-type and mutant virions revealed that the CP subunits bearing the engineered deletion assembled into polymorphic virions with altered surface topology. Northern blot analysis of virion RNA from mutant progeny demonstrated that the engineered mutation down-regulated packaging of all four viral RNAs; however, the packaging effect was more pronounced on genomic RNA1 and RNA2 than genomic RNA3 and its CP mRNA. In vitro assembly assays with mutant CP subunits and RNA transcripts demonstrated that the mutant CP is inherently not defective in packaging genomic RNA1 (53%) and RNA2 (54%), but their incorporation into virions was competitively inhibited by the presence of other viral RNAs. Northern blot analysis of RNA encapsidation in vivo of two distinct bromovirus RNA3 chimeras, constructed by exchanging CPs having the Δ919 deletion, demonstrated that the role of the conserved N-terminal ARM in recognizing and packaging specific RNA is distinct for each virus.
The first 25 N-terminal amino acids of the capsid proteins (CPs) of the members of genera Bromovirus, brome mosaic virus (BMV) and cowpea chlorotic mottle virus (CCMVs), have a highly conserved arginine-rich motif (ARM) (26, 32). In addition to the bromoviruses, this ARM, which recognizes specific regions in RNA, is also found in other plant virus genera, such as Cucumovirus, Sobemovirus, and Tombusvirus, and in human immunodeficiency virus Tat and Rev proteins, bacterial antiterminators, and ribosomal proteins of nonplant viruses (4, 26, 32).
The genomes of BMV and CCMV are divided among three genomic RNAs. Viral RNA replication is dependent on efficient interaction between two nonstructural proteins, 1a and 2a, encoded by monocistronic RNA1 and RNA2, respectively (16). The two gene products encoded by the dicistronic RNA3 are dispensable for viral replication but are required for infection in plants (10, 20, 25, 26, 30). The genomic RNA3 encodes a nonstructural protein of 32 kDa, designated the movement protein (MP) (20, 25, 26), and a 19-kDa CP that is synthesized from a subgenomic RNA4 (CP mRNA) derived from progeny minus-strand RNA3 by internal initiation (19). The three genomic RNAs and a single subgenomic RNA4 are packaged into three physically and morphologically indistinguishable icosahedral virions (24). These virions having T=3 symmetry are assembled from 180 identical subunits of a single CP (17, 31). The intriguing question of how a single CP discriminates among four RNAs and packages them into three individual particles remains unanswered.
The CPs of BMV and CCMV share 70% identity at the amino acid level (31). Hybrid viruses engineered to express heterologous CPs exhibited neutral effects with respect to the host range (23). The large number of basic residues located in the N-proximal region of BMV CP (seven arginines and one lysine) and CCMV CP (six arginines and three lysines) are envisioned to interact with negative phosphate groups in the RNA during the encapsidation process (34). Previous deletion analysis of the N-terminal ARM region of BMV CP revealed that variants lacking the N-proximal 7, but not 19, amino acids are biologically active and assembled into RNA-containing virions (26). Finer mutational analysis of the N-terminal ARM region further revealed that amino acids located between residues 9 and 19 play crucial roles in RNA packaging and contain determinants specific for directing copackaging of subgenomic RNA4 with genomic RNA3 into a single virion (5, 7). The N-terminal ARMs are highly conserved among the members of the bromoviridae, yet it remains to be verified whether they have evolved to enable conserved mechanism(s) in RNA packaging. Therefore, the working hypothesis for the work described here is to analyze whether the highly conserved N-terminal ARM of CCMV CP exhibits RNA packaging characteristics similar to that of BMV. To address this issue, we have constructed and analyzed the biological activity and packaging competence of several CCMV CP variants. The results indicated that the closely related bromoviruses, BMV and CCMV, with apparently similar N-terminal ARMs, are not functionally analogous in terms of RNA packaging. In addition we also compared the structural features of wild-type (WT) and mutant virion populations by analyzing electron microscopic images of single particles and three-dimensional reconstructions.
MATERIALS AND METHODS
Plasmid constructs.
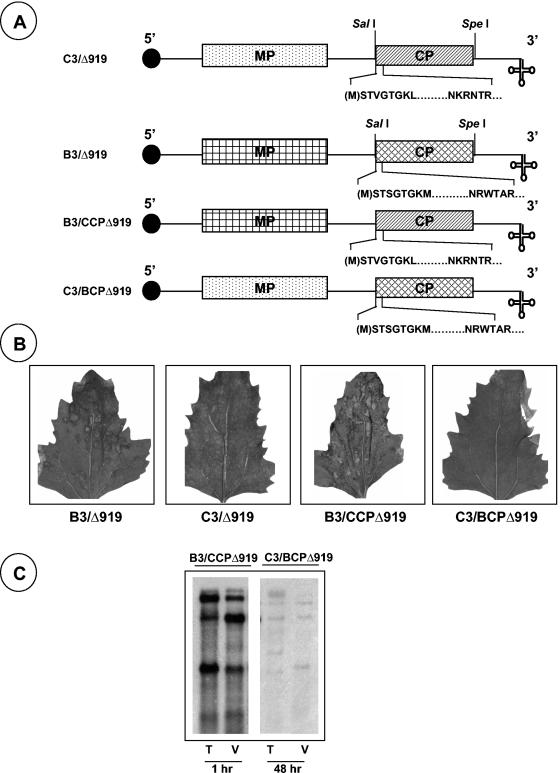
Plasmids pCC1TP1, pCC2TP2, and pCC3TP4 contain full-length cDNA copies of CCMV RNA1, RNA2, and RNA3, respectively, and can be linearized with XbaI prior to transcription with T7 RNA polymerase to yield infectious in vitro transcripts (1). All CCMV RNA3 (C3) variants constructed in this study (Fig. 1) are derived from plasmid pCC3TP4. PCR (5) was used to engineer either a deletion of a desired group of amino acids or substitution of a specific amino acid within the N-terminal ARM region of CCMV CP (CCP) (Fig. 1). Full-length cDNA clones corresponding to the three genomic RNAs of BMV, pT7B1, pT7B2, and pT7B3(−Tth), from which infectious RNAs can be transcribed in vitro, have been described previously (11). To facilitate precise exchange of CP open reading frames (ORFs) between BMV RNA3 (B3) and C3, a SpeI site at the end of BMV CP (BCP) ORF was engineered by site-directed mutagenesis. For constructing a hybrid B3 having C3/Δ919 (Fig. 1A), a combination of a 5′ oligonucleotide primer with SalI (at the start of the CP gene) site and a 3′ oligonucleotide primer with SpeI (at the end of the CP gene) was used amplify the intervening sequence by PCR, the product of which was digested with SalI and SpeI and subcloned into a SalI/SpeI-digested B3 clone. Similarly, a hybrid clone of C3 having the BCP ORF with a Δ919 mutation (7) was constructed by replacing the WT CCP with the entire BCP having the Δ919 mutation as a SalI-SpeI fragment. The presence of the subcloned fragments was confirmed by restriction mapping and DNA sequencing.
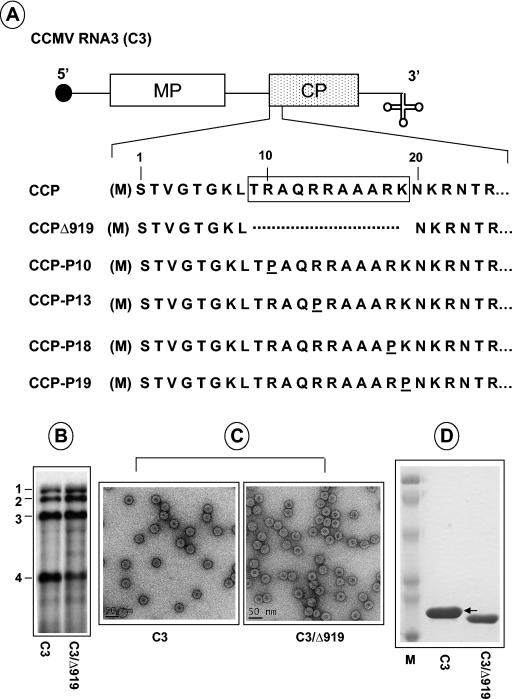
FIG. 1.
Characteristics of CCMV CP variants. (A) The structure of C3 is shown, with noncoding sequences represented as single lines and MP and CP genes shown as open and stippled boxes, respectively. A filled circle at the 5′ end and a cloverleaf at the 3′ end represent cap and tRNA-like structures, respectively. The first 25 N-proximal amino acids are shown, and the boxed region represents the N-terminal ARM conserved among plant and nonplant viruses. In CCMV, the initiating methionine (enclosed in parenthesis) is removed, and the resultant N-terminal serine is acetylated in the mature CP (21). In variant C3/Δ919, the deletion of the ARM region, located between amino acids 8 and 20, is indicated by a broken line. In variants C3/P10, C3/P13, CP/P18, and C3/CP19, arginine residues located at positions 10, 13, 18, and 19, respectively, are replaced by a proline residue. (B) Replication competence of C3/Δ919 in protoplasts. Protoplasts were transfected with WT C1 and C2 and either WT C3 or C3/Δ919. After a 24-h incubation, total RNA was isolated and subjected to Northern blot analysis. The positions of four WT CCMV RNAs are shown to the left. (C) Electron microscopy of purified virions. Virions were purified from symptomatic leaves, applied to glow-discharged carbon-coated copper grids and negatively stained with 1% uranyl acetate prior to viewing under an electron microscope. Prior to application of the sample onto the grids, WT samples were diluted 1:10 while the Δ919 samples remain undiluted. (D) Analysis of viral capsid protein. Purified virions resulting from infections of either WT C3 or C3/Δ919 were suspended in sample buffer, boiled for 5 min, and subjected to SDS-10% PAGE. The gel was stained with Coomassie brilliant blue prior to photography. An arrow indicates the position of WT CP. Lane M, molecular weight marker proteins.
In vitro transcription and whole-plant inoculations.
Prior to in vitro transcription, all WT and variant clones of CCMV and BMV were linearized with XbaI and BamHI, respectively. Capped full-length transcripts were synthesized in vitro by using a MEGAscript T7 kit (Ambion Inc., Austin, Tex.). Unless specified otherwise, each RNA3 variant was always coinoculated with RNA1 and RNA2. Control inoculations contained in vitro transcripts of all three WT RNAs. For inoculating Chenopodium quinoa or cowpea plants, a mixture containing all three transcripts at a concentration of 150 μg/ml was used. The purification of virions from symptomatic leaves and RNA isolation were performed as described previously (5).
Progeny analysis.
For Northern blot analysis virion RNA (0.5 μg) or plant total RNAs (5 μg) were dried in a microcentrifuge tube and suspended in 10 μl of sample buffer (10× MOPS [morpholinepropanesulfonic acid] buffer-formamide-formaldehyde-H2O in a ratio of 1:1.8:5:2.2, respectively), heated at 65oC for 10 min, and electrophoresed in 1.2% agarose-formaldehyde gel (29). Following a 3-h electrophoresis, fractionated RNA was transferred to a nylon membrane with a VacuGene XL blotting unit (Pharmacia Biotech). The blot was then processed for prehybridization and hybridization by using riboprobes corresponding to the 3′ conserved region as described previously (27). CP samples were analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) according to Osman et al. (22).
Capsid protein preparation and in vitro assembly assays.
Purified virions of either WT or variants were dissociated into CP by dialyzing at 4°C for 24 h against a buffer containing 500 mM CaCl2, 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride. Following centrifugation at 12,000 × g for 30 min, any traces of viral RNA contaminating the supernatant were removed by dialyzing against RNA assembly buffer (see below). The dialyzed mixture was then centrifuged for 90 min at 220,000 × g in a Beckman TL 100 centrifuge to pellet the assembled virions. The concentration of the dissociated CP subunits present in the supernatant was determined by a spectrophotometer. For in vitro assembly of RNA containing virions, CP and desired RNA transcripts were mixed in a ratio of 1:5 (wt/wt) for RNA1 and RNA2 and 1:2 (wt/wt) for RNA3 and dialyzed at 4°C for 24 h against RNA assembly buffer (50 mM NaCl, 50 mM Tris-HCl [pH 7.2], 10 mM KCl, 5 mM MgCl2, and 1 mM dithiothreitol). The assembled virions were concentrated by using Centricon-100 microconcentrators (Amicon, Beverly, Mass.).
Gel retardation assays.
The interaction between various molar ratios of either WT or mutant CP dimers and the desired genomic RNA component leading to virion assembly following a short (20 min) or long (24 h) incubation time was analyzed (15). Approximately 0 to 150 CP dimers were titrated against a constant RNA concentration in a typical 20-μl reaction mixture containing 50 mM MOPS (pH 7.2), 150 mM NaCl, and 2 mM MgCl2. The samples were then subjected to 1% agarose gel electrophoresis in Tris-acetate-EDTA buffer (29), stained with ethidium bromide, and photographed with a Bio-Rad gel documentation system. In these assays, a sample of native WT CCMV virions purified from symptomatic leaves was always coelectrophoresed as a control.
Electron microscopy.
For negative staining, purified virus preparation at a concentration of between 20 and 50 μg/ml was applied to glow-discharged carbon-coated copper grids. Grids were washed once with water, stained with 1% uranyl acetate, and air dried. Grids were examined with an FEI Tecnai12 transmission electron microscope operating at 100 kV. Images were recorded in low-dose mode with a 2,048-by-2,048 pixel charge-coupled device (Gatan Inc.) in single frame or 2-by-2 montage mode. Images of stained samples were recorded with an underfocus of between 800 and 1,000 nm and an electron optical magnification of ×30,000, placing the first zero of the contrast transfer function at around 1/20 Å−1.
Image analysis.
Electron microscopic images were analyzed with the EMAN (18) and IMAGIC 5 (33) software packages running on SGI workstations (O2 and Octane) essentially as described (35, 36). Briefly, WT and mutant (Δ919) CCMV particles were selected from 15 and 25 electron micrographs, respectively, and excised as 140-by-140 pixel images. Data sets of 860 (WT) and 2,200 (mutant) single images were normalized and band-pass filtered to remove low (<0.005 Å−1) and high (>0.1 Å−1) spatial frequencies. The data sets were subsequently self-centered and analyzed by the procedure of alignment by classification (12). At this stage, two sizes of mutant virions with about equal abundances could be identified. The data set of mutant virions was divided into a class of particles with a diameter similar to that of WT virions (∼28 nm) and a class of smaller particles (∼26.5 nm in diameter). The resulting data sets for the mutant particles were analyzed separately. The data sets (the WT and the two mutant data sets) were aligned to their total averages as references (two iterations), and the aligned data sets were sorted into 24 classes by multivariate statistical analysis and hierarchical ascendant classification. Averages of the classes served as input images for the three-dimensional reconstruction, which was done assuming icosahedral symmetry. The image size was reduced to 128 by 128 pixels, and initial angles were assigned with the angular reconstitution method as implemented in IMAGIC 5. Initial angles were refined by using increasing numbers of forward projections as anchor sets. Reprojection errors for the WT reconstruction stabilized after four iterations (going from 8.1 to 5.3% after three iterations and to 4.8% after four iterations), whereas for the WT size Δ919 reconstruction, reprojection errors did not improve significantly (going from an initial 8.2 to 7.6% after three iterations). At this stage, the reconstructions were forward projected along 22 directions uniformly distributed on the asymmetric triangle of the Euler sphere, and the projections were used as references in a multireference alignment (MRA) step. Averages were obtained by multivariate statistical analysis classification and based on the cross-correlation coefficients obtained during the MRA. The resolution of the final reconstruction was estimated by calculating the Fourier shell correlation between two three-dimensional models, each calculated from half the input projections (3).
RESULTS
Physical and biological characteristics of C3/Δ919.
In this study, deletion of the N-terminal ARM region (sequences encompassing amino acids 9 to 19) was engineered in the cDNA clone of C3, yielding C3/Δ919 (Fig. 1A). Since sequences encoding the CP gene do not contribute to bromovirus replication (14, 26), C3/Δ919 replicated to near WT levels in protoplasts (Fig. 1B), and the levels of mutant CP synthesized is similar to the level of WT (data not shown).
The requirements of CP and MP in promoting cell-to-cell and long-distance spread in a given susceptible host are distinct for BMV and CCMV (20, 30). For example, BMV requires both MP and CP for efficient cell-to-cell movement (30), whereas CCMV can be transported between cells independent of CP (25). However, both viruses require encapsidation-competent CP for efficient long-distance movement (26, 28). In order to examine whether the truncated CP expressed from the subgenomic RNA4 of C3/Δ919 (hereafter, the CCMV CP harboring the Δ919 deletion will be referred to as CCPΔ919) is competent for virion formation and subsequently promotes systemic spread of infection in whole plants, a mixture containing in vitro synthesized RNA transcripts of WT CCMV RNA1 (C1), CCMV RNA2 (C2), and C3/Δ919 was mechanically inoculated to cowpea plants. Plants inoculated with all three WT genomic CCMV transcripts served as controls. Reddish necrotic local lesions, characteristic of CCMV infection, appeared approximately 4 to 5 days postinoculation on leaves inoculated with WT control as well as with C3/Δ919 (Table 1). However, at 12 to 15 days postinoculation, control plants displayed visible chlorotic mottling symptoms on uninoculated upper leaves, whereas plants inoculated with transcripts of C3/Δ919 did not develop any visible systemic symptoms even at 3 weeks postinoculation (Table 1). Northern blot analysis of total RNA isolated from uninoculated systemic leaves failed to reveal the presence of any viral RNA (data not shown), confirming that C3/Δ919 is incompetent to support systemic infection. To characterize the packaging profiles of C3/Δ919, virus was purified from symptomatic inoculated leaves and examined by electron microscope. Although purified virions of C3/Δ919 displayed icosahedral morphology similar to those of WT CCMV (Fig. 1C), the majority of them, unlike WT CCMV, appeared to have larger electron-dense centers. A close examination further revealed that the mutant particles were polymorphic (see below). The particles of one of the populations were about the same size as those of the WT, while the other contained virions approximately 1.5 nm smaller in diameter than those of the WT. However, on sucrose gradients these two virion populations sedimented as a single peak (data not shown). Protein analysis by SDS-PAGE (Fig. 1D) confirmed that these virions are assembled from truncated CP subunits.
TABLE 1.
Biological properties of WT CCMV and BMV and their variants used in this study
| Inoculuma | Symptomsb
|
|||
|---|---|---|---|---|
|
C. quinoa
|
Cowpea
|
|||
| L | S | L | S | |
| CCMV (WT) | NL | NI | RLL | SM |
| C3/Δ919 | NL | NI | RLL | NI |
| C3/P10 | NT | NT | RLL | SM |
| C3/P13 | NT | NT | RLL | SM |
| C3/P18 | NT | NT | RLL | SM |
| C3/P19 | NT | NT | RLL | SM |
| BMV (WT) | CLL | SM | NI | NI |
| B3/CCPΔ919 | NL | NI | NI | NI |
| C3/BCPΔ919 | NLc | NI | RLLc | NI |
Each inoculum contained a mixture of WT RNA1 and RNA2 transcripts and desired WT or mutant RNA3 transcript adjusted to a concentration of 150 μg/ml.
L, local; S, systemic; NL, necrotic local lesions; CLL, chlorotic local lesions; RLL, reddish necrotic local lesions; SM, systemic mottling; NI, not infected, NT, not tested.
In each case, few local lesions that are hard to visualize appeared 3 to 4 weeks post-inoculation.
Comparative structural analysis of WT and mutant virions by three-dimensional reconstruction.
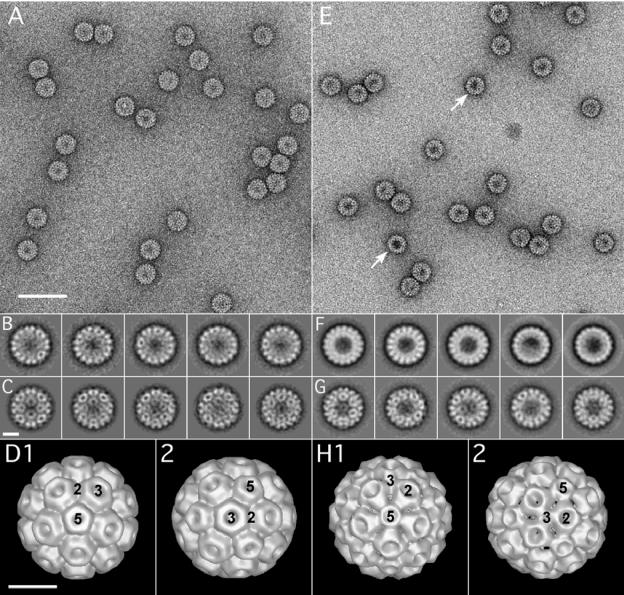
To compare the structural features of the WT and Δ919 virions, we used electron microscopy of negatively stained virions to generate projection images for three-dimensional reconstruction. These results are summarized in Fig. 2. In the case of the Δ919 mutant, many of the virions seemed to be darker in the central region (Fig. 2E, arrows), indicating that a larger amount of the negative stain had entered the mutant particles compared to WT. This can be explained by the fact that deletion of the N terminus of the capsid protein will remove 1,080 (180 × 6) positively charged residues from the interior of the virion, thus allowing more of the positively charged uranyl cation to enter the inside of the mutant particles. Data sets of 860 and 2,200 particles were collected for WT and Δ919 virions, respectively, and analyzed by alignment by classification. Some of the resulting class sums are shown in Fig. 2B and F. At this stage, a number of averages show some degree of substructure typical for icosahedral virions. Further inspection of the class averages of the sorted data sets revealed that the mutant particles were polymorphic with approximately equal size populations. The two mutant populations were separated into two classes, one class of virions of WT size (28 nm) and one class of smaller particles (26.5 nm), and the two resulting data sets were then analyzed separately. Figures 2C and G show selections of projection averages after the final MRA. The averages obtained after the final alignment, including the ones shown in Fig. 2C and G, served as input images to calculate the final three-dimensional reconstructions of WT and mutant virions, which are shown in Fig. 2D and H, respectively. As Fig. 2 shows, the WT reconstruction, oriented along the five- and threefold axes (images D1 and D2, respectively), appears very similar to reconstructions of CCMV obtained by cryoelectron microscopy and X-ray crystallography (31). The structure is different for the mutant particles. While the fivefold axes coincide with the small pores (image H1) as in WT (image D1), the twofold axes seem to be positioned in the large pores (image H2), which is where the threefold axes are positioned in the WT (image D2). Although the significance of this structural difference is not immediately obvious, it might reflect a change in the biology of the virus (defective in long-distance movement in the natural host) (Table 1) as well as the efficiency of RNA packaging (see below). Alternatively, it is possible that deletion of the N-terminal residues could have resulted in a different subunit assembly; however, it cannot be excluded that the mutant particles exhibit a structural heterogeneity leading to a somewhat artificial icosahedral arrangement of the capsid proteins in the reconstruction.
FIG. 2.
Electron microscopy and image analysis of WT CCMV and C3/Δ919 virions. Electron micrographic images of purified virions of WT CCMV (A to D) and Δ919 (E to F). Images shown in panels A and E represent typical regions of electron micrographs of WT and Δ919 mutant virus particles, respectively, negatively stained with 1% uranyl acetate. Images shown in panels B and F represent class sums after the alignment by classification step. Uniform size was observed for the WT (B), whereas for the Δ919 mutant, two particle sizes were observed In panel F, particles shown in the first three images from the left are WT size, while particles shown in the remaining two images are approximately 1.5 nm smaller in diameter than those of the WT. Images C and G show averages after the final MRA for WT and the larger of the two Δ919 populations, respectively. Images D and H represent the final three-dimensional reconstructions for the WT and the larger Δ919 particles, respectively. The positions of the five-, three-, and twofold symmetry axes are indicated. Scale bars are 60 nm (A and E) and 10 nm (B through H). The resolution of the final models was estimated to 25 Å by the Fourier shell criterion with a cutoff of 0.5 for both WT and mutant virions.
Effect of N-terminal ARM deletion on RNA packaging.
Bromoviruses exhibit specificity in RNA packaging (9, 23). Furthermore, bromovirus infections are not known to result in the assembly of empty virions, since they are stabilized by RNA-protein interactions (13). Therefore, it was surmised that virions assembled with CCPΔ919 possibly have packaged the RNA progeny resulting from replication of genomic C3/Δ919 and its WT counter parts. To substantiate this assumption, RNA was extracted from purified virions of C3/Δ919 and analyzed by native agarose gel electrophoresis. Interestingly, the RNA profile for variant C3/Δ919 is distinct from that of the WT control (Fig. 3A). In contrast to the characteristic profile of four CCMV RNAs found in the WT control (Fig. 3A), RNA recovered from virions of C3/Δ919 contained only two major RNAs coelectrophoresing with genomic RNA3 and subgenomic RNA4 (Fig. 3A). This RNA profile was reproducible with several independently isolated virus preparations.
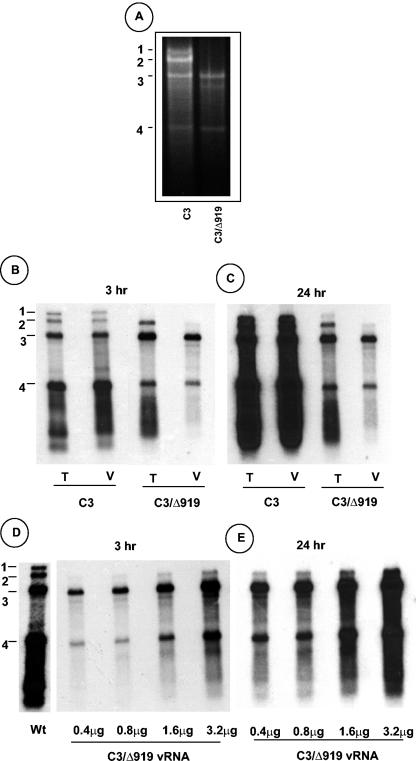
FIG. 3.
Analysis of virion RNA content of C3/Δ919 virions. (A) Native agarose gel analysis of viral RNA. RNA isolated from purified virion preparations of WT C3 or C3/Δ919 was subjected to electrophoresis in 1% agarose and stained with ethidium bromide. (B) Northern blot analysis of total nucleic acid (T) and virion RNA (V) preparations recovered from symptomatic cowpea leaves inoculated with WT C1 and C2 and either C3 or C3/Δ919. Approximately 5 μg of total nucleic acid and 200 ng of virion RNA were denatured with formamide-formaldehyde and subjected to 1.5% agarose gel electrophoresis prior to vacuum blotting to a nylon membrane. The blot was hybridized with a 32P-labeled riboprobe complementary to the commonly shared 3′ noncoding region of CCMV RNAs. The autoradiograph shown in panel C represents a longer exposure image of panel B. (D) Concentrations of C3/Δ919 virion RNA ranging between 0.4 to 3.2 μg per ml were subjected to Northern hybridization as described above. The autoradiograph shown in panel E represents a longer exposure image of panel D. The positions of four CCMV RNAs are shown to the left.
It is possible that the CCPΔ919 subunits could have packaged genomic RNA1 and RNA2 with much lower efficiency and, hence, remain undetectable by the less sensitive native agarose gel electrophoresis. Therefore, a Northern blot containing total and virion RNA progeny preparations from cowpea leaves infected with WT control and C3/Δ919 variant was hybridized with riboprobes complementary to the 3′ noncoding region conserved among all four CCMV RNAs (Fig. 3B). In contrast to the WT control, total RNA preparations of C3/Δ919 contained detectable levels of genomic RNA2 and RNA3, and subgenomic RNA4 (Fig. 3B) and RNA1 could be detected only when the blot was subjected to a prolonged exposure time (Fig. 3B). Virion RNA preparations of C3/Δ919 contained detectable amounts of genomic RNA3 and its subgenomic RNA4 and comparatively lower but detectable amounts of RNA2 (Fig. 3B). The fact that prolonged exposure of the blot failed to detect any genomic RNA1 (Fig. 3C) suggests that subunits of CCPΔ919 are incompetent in packaging genomic RNA1. To further substantiate this assumption, two additional experiments were performed. In the first experiment, increasing concentrations of C3/Δ919 virion RNA was subjected to Northern hybridization, and the data are shown in Fig. 3D and E. Although packaging of genomic RNA2 into the virions assembled with CCPΔ919 is obvious, the presence of genomic RNA1 could not be established even when RNA concentrations as high as 3.2 μg/ml were allowed to hybridize (Fig. 3D and E). A second experiment was performed by inoculating C. quinoa (a local lesion host for CCMV) (Table 1) and cowpea plants with RNA preparations recovered from purified virions of variant C3/Δ919. The rationale for this experiment was that any amount of RNA1 undetectable by the Northern hybridization is likely to be amplified in vivo, resulting in the induction of infection in these host plants. However, none of the inoculated plants displayed symptoms even at 4 weeks postinoculation, confirming that the subunits of CCPΔ919 are incompetent to package genomic RNA1.
Polymerization kinetics of WT and mutant CP leading to virion assembly.
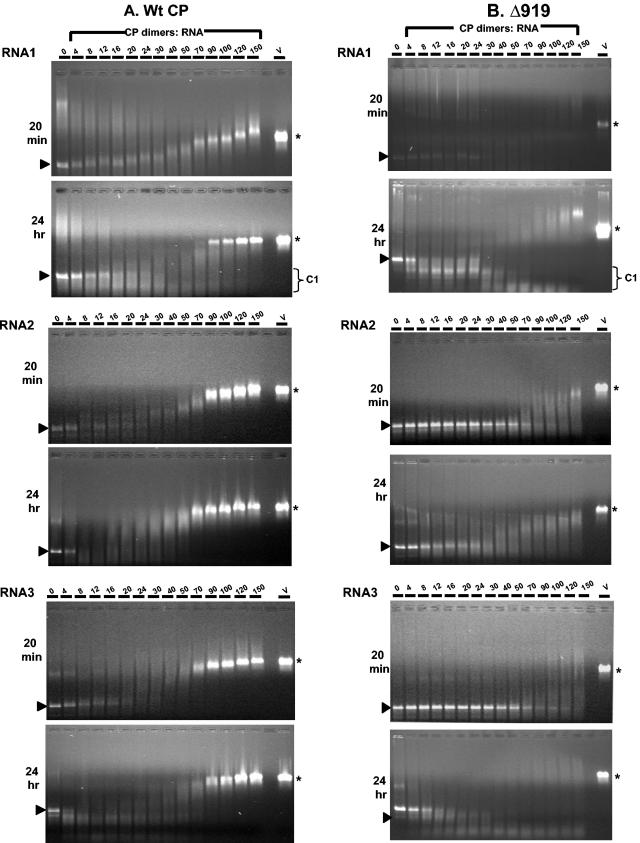
Recently, Johnson et al. (15) demonstrated that CCP dimers bind RNA1 cooperatively, leading to virus assembly. Our inability to isolate RNA1 containing virions from symptomatic leaves (Fig. 3) prompted us to examine the binding of mutant CP subunits to RNA1. Thus, stoichiometric ratios of WT and mutant CP subunits were titrated against a constant concentration (30 nM) of RNA1 and incubated for 20 min (short incubation) and 24 h (long incubation). As observed by Johnson et al. (15), increasing concentrations of WT CP dimers progressively retarded the migration of RNA1 (Fig. 4A). Agarose gel analysis of 20-min reaction products revealed that the migration of CP-RNA1 complex at a stoichiometric ratio of 90 or more dimers per RNA1 molecule paralleled that of native virions (Fig. 4A) (15). The formation of a faster migrating C1 complex, postulated to be an important intermediate in the assembly pathway of CCMV RNA1 virions (15), was evident in 24-h incubation products (Fig. 4A). Electron microscopic examination of samples assembled with 70 CP dimers showed partially assembled virus-like particles, and those assembled with 90 dimers or more displayed virus-like particles indistinguishable from virions recovered from natural infections (data not shown).
FIG. 4.
Gel retardation analysis of RNAs by WT and Δ919 CP subunits. The desired CCMV RNA transcript (approximately 30 nM) was titrated with the indicated amounts of CP dimers of either WT or Δ919 for 20 min or 24 h at 20°C. After incubation the samples were loaded onto prepared 1% agarose gels and electrophoresed in Tris-acetate-EDTA buffer. A sample of native CCMV virions, assembled with 90 CP dimers, of purified symptomatic cowpea leaves is shown on the right. The arrowhead shown to the left and the asterisk shown to the right of each panel, respectively, indicate the relative mobility of free viral RNA and purified WT virions (v). The position of the C1 complex formed in a 24-h incubation with RNA1 samples is indicated by a bracket.
Results of similar in vitro assembly assays performed with CCPΔ919 and RNA1 are shown in Fig. 4B. Unlike WT CP subunits, a short incubation period (20 min) of various concentrations of CP dimers resulted in faint but not discrete complex formation (Fig. 4B). Although the 24-h incubation period revealed a bimodal distribution similar to WT CP subunits, two major differences are clearly evident. First, the C1 complex with 30 or more dimers migrated faster than similar products assembled with WT CP subunits (Fig. 4B). It is likely that RNA1 was folded imperfectly due to the interaction with mutant CP subunits, resulting in faster migration. These observations suggest that the engineered mutation affected the interaction between CP and RNA1, resulting in an altered structure and thereby interfering with proper virion assembly (15). Second, unlike WT, the RNA complexes formed with 70 or more dimers migrated more slowly than native virions (Fig. 4B). This slower migration can perhaps be attributed to changes in virion topology observed in the three-dimensional reconstruction experiments described above (Fig. 2).
The polymerization kinetics for CCMV RNA2 and RNA3 are not known. Data presented in Fig. 4 indicate that the polymerization kinetics for CCMV RNA2 and RNA3 appears to be same as that of RNA1 since the migration of a CP-RNA complex at a stoichiometric ratio of 90 or more dimers paralleled that of native virions (Fig. 4). However, unlike RNA1, no C1 complex formation was observed either for RNA2 or RNA3. Incubation of CCPΔ919 subunits with RNA2 and RNA3, like RNA1, resulted in weaker interactions in 20-min and 24-h incubation reactions. Collectively, these observations demonstrate that the interaction between CCPΔ919 subunits and viral RNAs is severely debilitated, resulting in inefficient virion formation.
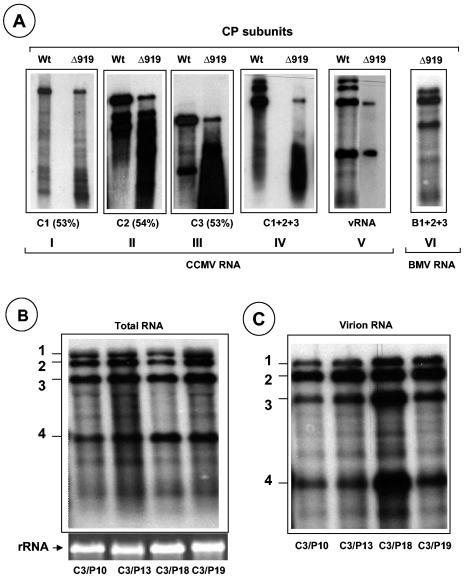
In vitro assembly assays demonstrate that subunits of CCPΔ919 display differential packaging competence with CCMV genomic RNAs. Bromovirus CP and RNA are amenable for assembly in vitro (2, 5, 8, 37). Previous studies with BCP demonstrated that in vitro assembly is ideal for confirming the packaging defects observed in vivo (5). Therefore, in vitro assembly assays were performed to quantitatively analyze the relative competence of WT and CCPΔ919 subunits to interact with transcripts of WT CCMV genomic RNAs (independently or together) during packaging. The results of these in vitro assembly assays are shown in Fig. 5A. It is interesting that, although RNA1 was not detected in virions of C3/Δ919 purified from symptomatic leaves (Fig. 3A), in the absence of other viral RNAs, RNA1 was assembled by Δ919 into virions at 53% of the level assembled by WT (Fig. 5A, panel I). Similarly the packaging efficiency of genomic RNA2 and RNA3 was also reduced to approximately 54 and 53%, respectively, when individual transcripts are provided as substrates (Fig. 5A, panels II and III). However, when mixtures of all three genomic RNA transcripts or virion RNAs (containing all four RNAs) were provided as substrates to CCPΔ919 subunits, packaging of RNA1 and RNA2 was inhibited beyond detection (Fig. 5A, panels IV and V), a scenario reminiscent of in vivo results (Fig. 3). Collectively, these results suggest that the packaging of genomic RNA1 and RNA2 was inhibited competitively by the genomic RNA3 and its subgenomic RNA4. Surprisingly, when the three WT genomic BMV RNA transcripts were allowed to assemble with the subunits of CCPΔ919, efficient packaging of all three RNA transcripts was observed (Fig. 5A, panel VI).
FIG. 5.
Encapsidation competence of CCMV RNAs with N-terminal ARM mutants. (A) In vitro assembly assays. Northern blot analysis of RNA isolated from virions assembled in vitro with Δ919 CP subunits and each of the three genomic RNA transcripts (panels I to III), their mixture (panel IV), virion RNA (panel V), and a mixture containing all three BMV RNA transcripts (panel VI). Purified CP subunits and the indicated RNAs were allowed to assemble in vitro as described in Materials and Methods. Conditions for denaturizing RNA, electrophoresis, and hybridization with riboprobes are as described in the legend of Fig. 3. RNA samples shown in panels I to V were hybridized with a probe complementary to the 3′ end of CCMV RNA, whereas the RNA sample shown in panel VI was hybridized with a probe complementary to the 3′ end of BMV RNA. The numbers shown in parentheses below panels I to III represent the percentages of assembly efficiency of Δ919 CP subunits for each WT RNA transcript with respect to WT CP subunits. (B and C) Packaging profiles of CCMV CP bearing N-terminal proline mutations (Fig. 1A). Shown are Northern blots of total nucleic acids or virion RNA recovered from symptomatic leaves of cowpea infected with the indicated C3 mutant. The blots were hybridized with 32P-labeled RNA probes complementary to the homologous 3′ region present on each of the four CCMV RNAs. The positions of CCMV RNAs are shown to the left of each blot.
Mutation of positively charged amino acids of the N-terminal ARM region of CCP did not display packaging defects.
Previous nuclear magnetic resonance studies (34) with CCP suggested that, upon interaction of RNA with the positively charged arginine and lysine residues, the N-terminal basic arm rolls up into a rigid α-helix. Additional circular dichroism experiments showed that peptides of CCP encompassing the ARM region exhibit α-helical conformation and that such an α-helical region is situated between residues 9 and 19 (32). Furthermore, for proteins with ARMs, maintenance of the α-helical region is critical for RNA interaction (32). Therefore, one likely explanation for the defective packaging exhibited by the subunits of CCPΔ919 is that CP lacking an α-helical region failed to interact with RNAs efficiently, resulting in defective encapsidation. To examine this possibility directly in vivo, proline residues (expected to break the α-helix) were substituted for each of the three arginine residues and one lysine residue located at positions 10, 13, 18, and 19, respectively (Fig. 1), into the biologically active clone of C3. Cowpea plants were inoculated with a mixture containing WT transcripts of C1, C2, and either C3/P10, C3/P13, C3/P18, or C3/P19. All four mutant inocula induced local and systemic chlorotic mottling symptoms on a time scale similar to that for control plants inoculated with all three WT transcripts (Table 1).
In CCMV successful systemic movement requires efficient encapsidation (25). Therefore, each mutant CP having the proline mutation must have been competent to assemble into virions. To substantiate this assumption, virions were purified from symptomatic leaves for electron microscopic examination. All preparations contained virions similar to those of WT CCMV (Fig. 3A). Northern blot analysis of virion RNA revealed no defects in RNA packaging, and the virion profile was indistinguishable from that of WT CCMV (Fig. 5B and C). For each case, reverse transcription-PCR analysis of progeny RNA3 for each variant confirmed the maintenance of the engineered mutation (data not shown). These observations clearly show that CCP with disrupted N-proximal α-helical conformation has no influence on RNA packaging. However, unlike in BCP (5, 7), the N-terminal ARM region did not contain determinants to dictate packaging of any given CCMV RNA.
Packaging competence of a bromovirus RNA3 chimera expressing heterologous CPs with Δ919 mutation.
In contrast to the effect of a Δ919 deletion on CCMV RNA packaging (Fig. 3A), BCP having an identical deletion (BCPΔ919) is exclusively defective in packaging RNA4 (5). These observations, together with the fact that B3 and C3 expressing heterologous WT CPs efficiently package all four progeny RNAs into virions and maintain a host range similar to the parental viruses donating genomic RNA1 and RNA2 (22), led us to examine what packaging defects will be manifested by each CP with a Δ919 deletion when incorporated into the heterologous genetic background. Therefore, B3 and C3 chimeras were constructed by precisely substituting the WT CP ORF with each heterologous CP ORF having the Δ919 deletion (Fig. 6A). Unlike WT CCMV, which induces only local necrotic lesions in C. quinoa, WT BMV induces chlorotic local lesions followed by a systemic mottling phenotype (Table 1). Inoculation of C. quinoa plants with a mixture of inoculum containing WT BMV RNA1, BMV RNA2, and B3/CCPΔ919 resulted in the induction of numerous necrotic local lesions that coalesced with time (Fig. 6B) but failed to move systemically even at 4 weeks postinoculation (Table 1). By contrast, inoculum containing C1, C2, and C3/BCPΔ919 is poorly infectious, resulting in the induction of inconspicuous necrotic lesions that are hard to visualize even at 3 to 4 weeks postinoculation (Fig. 6B and Table 1). As demonstrated by in vitro assembly assays (Fig. 5A, panel VI), Northern blot analysis of virion RNA confirmed the efficient packaging of all four BMV RNAs by CCPΔ919 (Fig. 6C). In contrast, total and virion RNA preparations obtained from leaves inoculated with a combination of C1, C2, and C3/BCPΔ919 contained only trace amounts of progeny RNA, and their detection required prolonged exposure of the blot (Fig. 6C).
FIG. 6.
Characteristic features and biological activity of RNA3 chimeras of CCMV and BMV. (A) Schematic representation of B3 and C3 chimeras bearing heterologous CP with Δ919 mutation. The structures of C3/Δ919 and B3/Δ919 are shown, with noncoding sequences represented as single lines and the MP and CP as rectangle boxes. A filled circle at the 5′ end and a cloverleaf at the 3′ end represent cap and tRNA-like structures, respectively. In each case, the sequence of the N-proximal amino acid region lacking amino acids 9 to 19 (indicated by a dashed line) is shown. (B) Symptom phenotypes induced by WT and chimeras in C. quinoa. (C) Northern hybridization of total nucleic acid (T) and virion RNA (V) preparations recovered from C. quinoa plants inoculated with a mixture containing WTB1+WTB2+B3/CCPΔ919 (B1, BMV RNA1; B2, BMV RNA2) and WTC1+WTC2+C3/BCPΔ919. The conditions for Northern hybridization are as described in the legend of Fig. 3.
DISCUSSION
Results of this study provide new directions in understanding the role of the N-terminal ARM region conserved among bromoviruses. A significant outcome of this study is the evidence that, although the N-terminal ARM is highly conserved between BMV and CCMV, its interaction with respective genomic RNAs during packaging is distinct (references 5 and 7 and this study). Likewise, as discussed below, the interaction between RNA and CP leading to encapsidation also appears to be distinct for these two viruses.
Role of N-terminal ARM in the assembly pathway of BMV and CCMV capsids.
For the bromoviruses, the conserved N-terminal ARMs have been proposed to interact with the RNA (31). Consistent with this role, mutational analysis of CCP showed that deletion of the N-terminal 25 amino acid residues eliminated the assembly of RNA containing virions but not empty particles (37). However, the N-terminal residues were not visible in currently available crystal structures (31), making it difficult to generate more precise hypotheses concerning the mechanism of action. Our in vivo analysis of several CP variants of BMV revealed that the first 18, but not 19 or more, residues could be deleted without affecting virion formation (26, 28). The results in the present work are consistent with our previous observations and indicate that the general capsid assembly pathway features are conserved between BMV and CCMV and that an intact N-terminal basic region may not be required for the assembly of empty particles.
Then what is the role of the N terminus in virus assembly? Based on the mutational analysis of the N terminus, we speculate that an intact N terminus is essential to maintain the optimal structural conformation of the CP dimers that subsequently yield pentamers or dimers, the predicted backbones of icosahedral virus assembly (38). Support for this conjecture comes from our preliminary observations that, unlike the assembly-incompetent nature of a BMV CP lacking the first 25 amino acids (26, 28), a BCP chimera having nonbasic heterologous N-terminal 25 amino acids derived from the tobacco mosaic virus CP is competent for efficient RNA packaging (Y. G. Choi and A. L. N. Rao, unpublished data). These results suggest that in the absence of basic N-terminal residues, other regions of the CP are likely to interact with RNA during packaging. Alternatively, the N-terminal basic ARM might be functioning as high affinity binding sites to distinguish viral RNA from host RNAs. Thus, examination of the biological activity and packaging competence of progeny RNA from full-length CPs with a mosaic of residues from BMV and CCMV may generate insight into this process.
Role of N-terminal ARM in RNA packaging.
The basic residues of the first 25 N-terminal amino acids of BCP (seven arginines and one lysine) and CCP (six arginines and three lysines) are predicted to interact with the negative phosphate groups in the RNA during the encapsidation process (31). Although the N-terminal ARM of BCP has been shown to harbor determinants specific for selected viral RNAs (5), no such determinants were identified for CCP (this study). Experimental evidence suggests that RNA packaging in BMV is a highly specific process (5, 8, 9, 23) resulting from the interaction between determinants localized within the N-terminal ARM and a specific packaging signal(s) encoded within a given genomic RNA (6). By contrast, the interaction between CCP and a given RNA species is nonspecific since a variety of heterologous RNAs have been shown to be efficient substrates for packaging (2; P. Annamalai and A. L. N. Rao, unpublished data). Despite this nonspecific interaction between CCP and nucleic acid species, evidence for the existence of cellular RNAs in the mature virions of CCMV is meager. Consequently a mechanism inherent to CCMV, distinct from that of BMV, functioning as a selective filter during the encapsidation process exists so that mature virions exclusively package viral RNAs. One such mechanism, independent of RNA sequence, has been proposed for packaging C1 by Johnson et al. (15). According to these authors, binding of CP slowly folds RNA into a compact structure (C1 complex), and when CP concentration peaks during infection, it preferentially binds the C1 complex cooperatively, leading to encapsidation. Using this model as a basis, we propose that, unlike WT, binding of CP subunits of Δ919 to RNA1 did not result in the optimally structured C1 complex (Fig. 4, compare migration patterns of C1 complex formed between CP subunits of WT and Δ919 with RNA1) that is amenable for subsequent addition of CP dimers leading to virion formation. Since no such C1 complexes appear to form with genomic RNA2 or RNA3 (Fig. 4), it remains to be seen whether these genomic RNAs are encapsidated.
Acknowledgments
We thank George Grantham for excellent technical assistance and Paul Ahlquist for providing CCMV RNA cDNA clones.
Research in this laboratory was supported by a grant from the National Institutes of Health (GM064465-01A2).
REFERENCES
- 1.Allison, R. F., M. Janda, and P. Ahlquist. 1988. Infectious in vitro transcripts from cowpea chlorotic mottle virus cDNA clones and exchange of individual RNA components with brome mosaic virus. J. Virol. 62:3581-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bancroft, J. B. 1970. The self-assembly of spherical plant viruses. Adv. Virus Res. 16:99-134. [DOI] [PubMed] [Google Scholar]
- 3.Böttcher, B., S. A. Wynne, and R. A. Crowther. 1997. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386:88-91. [DOI] [PubMed] [Google Scholar]
- 4.Burd, C. G., and G. Dreyfuss. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615-621. [DOI] [PubMed] [Google Scholar]
- 5.Choi, Y. G., and A. L. N. Rao. 2000. Molecular studies on bromovirus capsid protein. VII. Selective packaging of BMV RNA4 by specific N-terminal arginine residues. Virology 275:207-217. [DOI] [PubMed] [Google Scholar]
- 6.Choi, Y. G., and A. L. N. Rao. 2003. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 77:9750-9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, Y. G., G. L. Grantham, and A. L. N. Rao. 2000. Molecular studies on bromovirus capsid protein. VI. Contributions of the N-terminal arginine-rich motif of BMV capsid protein to virion stability and RNA packaging. Virology 270:377-385. [DOI] [PubMed] [Google Scholar]
- 8.Choi, Y. G., T. W. Dreher, and A. L. N. Rao. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 99:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullel, M., M. Herzog, and L. Hirth. 1979. Specificity in vitro reconstitution of bromegrass mosaic virus. Virology 95:146-153. [DOI] [PubMed] [Google Scholar]
- 10.De Jong, W., and P. Ahlquist. 1991. Bromovirus host specificity and systemic infection. Semin. Virol. 2:97-105. [Google Scholar]
- 11.Dreher, T. W., A. L. N. Rao, and T. C. Hall. 1989. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 206:425-438. [DOI] [PubMed] [Google Scholar]
- 12.Dube, P., P. Tavares, R. Lurz, and M. van Heel. 1993. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 12:1303-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, J. M., J. E. Johnson, and M. J. Young. 1994. RNA/protein interactions in icosahedral virus assembly. Semin. Virol. 5:51-60. [Google Scholar]
- 14.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, J. M., D. A. Willits, M. J. Young, and A. Zlotnick. 2003. Interaction with capsid protein alters RNA structure and the pathway for in vitro assembly of cowpea chlorotic mottle virus. J. Mol. Biol. 335:455-464. [DOI] [PubMed] [Google Scholar]
- 16.Kao, C. C., and K. Sivakumaran. 2000. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 1:91-98. [DOI] [PubMed] [Google Scholar]
- 17.Lucas, R. W., S. B. Larson, and A. McPherson. 2002. The crystallographic structure of brome mosaic virus. J. Mol. Biol. 317:95-108. [DOI] [PubMed] [Google Scholar]
- 18.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstruction. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 19.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by initiation on (−)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 20.Mise, K., R. F. Allison, M. Janda, and P. Ahlquist. 1993. Bromovirus movement protein genes play a crucial role in host specificity. J. Virol. 67:2815-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moosic, J. P., D. J. McKean, D. S. Sish, and P. Kaesberg. 1983. Primary structure of brome mosaic virus subgenomic virus coat protein. Virology 129:517-520. [DOI] [PubMed] [Google Scholar]
- 22.Osman, F., G. L. Grantham, and A. L. N. Rao. 1997. Molecular studies on bromovirus capsid protein. IV. Coat protein exchanges between brome mosaic and cowpea chlorotic mottle viruses exhibit neutral effects in heterologous hosts. Virology 238:452-459. [DOI] [PubMed] [Google Scholar]
- 23.Osman, F., Y. G. Choi, G. L. Grantham, and A. L. N. Rao. 1998. Molecular studies on bromovirus capsid protein. V. Evidence for the specificity of brome mosaic virus encapsidation using chimera of brome mosaic and cucumber mosaic viruses expressing heterologous coat proteins. Virology 251:438-448. [DOI] [PubMed] [Google Scholar]
- 24.Rao, A. L. N. 2001. Bromoviruses, p. 155-158. In O. C Maloy and T. D. Murray (ed.), Encyclopedia of plant pathology. John Wiley & Sons, Mississauga, Ontario, Canada.
- 25.Rao, A. L. N. 2000. Molecular studies on bromovirus capsid protein. III. Analysis of cell-to-cell movement competence of coat protein defective variants of cowpea chlorotic mottle virus. Virology 232:385-395. [DOI] [PubMed] [Google Scholar]
- 26.Rao, A. L. N., and G. L. Grantham. 1996. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino terminal arginine rich motif and its role in encapsidation, movement and pathology. Virology 226:294-305. [DOI] [PubMed] [Google Scholar]
- 27.Rao, A. L. N., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 86:5335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sacher, R., and P. Ahlquist. 1989. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packing and systemic infection. J. Virol. 63:4545-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schmitz, I., and A. L. N. Rao. 1996. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell defective RNA3 variants of brome mosaic virus. Virology 226:281-293. [DOI] [PubMed] [Google Scholar]
- 31.Speir, J. A., S. Munshi, S. Wang, T. S. Baker, and J. E. Johnson. 1995. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 3:63-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan, R., and A. D. Frankel. 1995. Structural variety of arginine-rich RNA-binding peptides. Proc. Natl. Acad. Sci. USA 92:5282-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 34.Vriend, G., B. J. Verduin, and M. A. Hemminga. 1986. Role of the N-terminal part of the coat protein in the assembly of cowpea chlorotic mottle virus. A 500 MHz proton nuclear magnetic resonance study and structural calculations. J. Mol. Biol. 191:453-460. [DOI] [PubMed] [Google Scholar]
- 35.Wilkens, S., and M. Forgac. 2001. Three-dimensional structure of the vacuolar ATPase proton channel by electron microscopy. J. Biol. Chem. 276:44064-44068. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Z., C. Charsky, P. M. Kane, and S. Wilkens. 2003. Yeast V1-ATPase: affinity purification and structural features by electron microscopy. J. Biol. Chem. 278:47299-47306. [DOI] [PubMed] [Google Scholar]
- 37.Zhao, X., J. Fox, N. Olson, T. S. Baker, and M. J. Young. 1995. In vitro assembly of cowpea chlorotic mottle virus from coat protein expressed in Escherichia coli and in vitro transcribed viral cDNA. Virology 207:486-494. [DOI] [PubMed] [Google Scholar]
- 38.Zlotnick, A., R. Aldrich, J. M. Johnson, P. Ceres, and M. J. Young. 2000. Mechanism of capsid assembly for an icosahedral plant virus. Virology 277:450-456. [DOI] [PubMed] [Google Scholar]