Abstract
The complete DNA sequence of the capsular locus 23F of Streptococcus pneumoniae is presented. The 18.6-kb cps23f locus is composed of 18 open reading frames flanked at the 5′ and 3′ ends by the genes dexB and aliA, an arrangement similar to those of some of the other identified cps loci.
Capsular polysaccharides deposited on the outermost surfaces of most clinical strains of Streptococcus pneumoniae are major virulence factors involved with evasion of the host immune system (1, 8). The capsule is the main target of protective antibodies which are specific for the particular polysaccharide (1, 32). Of the 90 different capsular types identified by immunological (17) and chemical (39) techniques, genetic characterization of the capsular biosynthetic genes (cps) is available only for a few capsular loci, namely, 1, 3, 14, 19F, and 19B (4, 9, 10, 14, 15, 19, 25, 27). Understanding the genetic determinants of the capsular polysaccharides should help in unraveling their pathway of biosynthesis which, in turn, could lead to the design of inhibitors capable of blocking the expression of these important pneumococcal virulence factors.
We describe here the complete DNA sequence of the cps locus of a type 23F S. pneumoniae Him18, a recent clinical isolate originating in Mexico. Analysis of a large number of serotype 23F isolates from diverse geographic areas and with a wide variety of isolation dates and chromosomal backgrounds confirmed that all genes identified by sequencing (as well as their structural organization) were conserved in each of the 23F isolates tested.
Bacterial strains, plasmids, and growth conditions.
S. pneumoniae strains were obtained from the American Type Culture Collection (11, 12) or were from the Rockefeller University collection. A semisynthetic medium (C+Y) (20) or tryptic soy agar plates (Difco, Detroit, Mich.) supplemented with 3% sterile sheep blood and incubation at 37°C were used for growth.
Escherichia coli HB101 (16) was obtained from Promega Corp. (Madison, Wis.) and used as the host strain for all recombinant plasmids generated in this study. Plasmid pSP64 (22) was used as the cloning vector. E. coli cells were grown in liquid medium (LB; Difco), SOC (16) with aeration, or solid medium (LA; Difco) incubated at 37°C. Selection of transformants was on LA supplemented with 50 μg of ampicillin/ml.
PFGE.
Chromosomal DNA fragments, generated by SmaI digestion, were separated and analyzed as described previously (36).
Southern blot hybridization.
DNA fragments separated by PFGE or conventional gels were transferred to nylon membranes (Hybond-N+; Amersham, Little Chalfont, Buckinghamshire, United Kingdom) with the Vacuum Gene System (Pharmacia LKB Biotech, Uppsala, Sweden) in accordance with the manufacturer’s instructions. Blots were hybridized to specific DNA probes labelled with the ECL Direct Labelling System (Amersham) in accordance with the manufacturer’s recommendations. Hybridization conditions included a sodium chloride concentration of 0.5 M as recommended by the manufacturer.
Probe preparation.
For the preparation of capsule 19F-specific probes, primers were designed from the published sequence of this capsular type (accession no. U09239) (24, 30) so that the generated probes resembled the ones used by Morona et al. (24). PCR was performed with strain OP5248 (serotype 19F) (13) as template and the GeneAmp system (Perkin-Elmer, Branchburg, N.J.) basically as described previously (28). Amplified products were visualized by agarose gel electrophoresis, and fragments of the expected size were purified by using the Wizard PCR Preps Purification System (Promega) in accordance with the manufacturer’s recommendations. Preparation of serotype 23F-specific probes was based on the sequence determined in this study from the penicillin-resistant serotype 23F clinical isolate Him18 following the same procedure as described above.
Long PCR.
Amplification of a region extending from the gene homologous to cps19fB to the one homologous to cps19fL of strain Him18 (serotype 23F) was accomplished by long PCR (5) with the GeneAmp XL system (Perkin-Elmer). The primers used were XL19Bd1 (AGGGGGTGCGAACCATTGTC) and XL19Lr1 (AGCCAAGCAAAGCCACGTCC). The reaction was performed in a DNA Thermal Cycler 480 (Perkin-Elmer) with a hot start (33), a magnesium acetate concentration of 1.1 mM, and primer and template concentrations of 1 ng/μl. The program used was the following: denaturation at 94°C for 2 min, 16 extension cycles of 94°C for 30 s and 68°C for 11 min, followed by 15 extension cycles of 94°C for 30 s and 68°C for 11 min, with an autoextension of 30 s per cycle.
Cloning of the 23F-specific region.
Long PCR products were purified with the Wizard PCR Preps Purification System in accordance with the manufacturer’s recommendations. The products were digested with either HindIII or PstI (New England Biolabs, Beverly, Mass.), ligated to pSP64, and transformed into E. coli HB101 by standard procedures (34). Colonies resistant to ampicillin were screened for the presence of fragments of correct size by using the procedure described by Corton and Gustafsson (7) with the following modifications. The primers used were pSP64fp (GATAGGGTCTGCTTCAGTAAGC) and pUC/M13F (CGCCAGGGTTTTCCCAGTCACGAC). No glycerol or cresol red was added to the PCR mixture, and LB with ampicillin was used instead of YT plus kanamycin. Clones with inserts of approximately 0.5, 2.4, and 1.3 kb after HindIII digestion and 0.6, 5.7, and 5.1 kb after digestion with PstI were isolated.
Nucleotide sequencing.
Plasmid purified with the Wizard Plus Midiprep DNA Purification System (Promega) was used for automated sequencing. Sequencing of PCR products was performed after purification with the Wizard PCR Preps Purification System (Promega). All sequencing was done by primer walking with the TaqFS fluorescent dye terminator sequencing method run on a Perkin-Elmer Applied Biosystems Division model 377 automated sequencer.
Nucleotide and amino acid sequence analyses were performed with either the DNASTAR (Lasergene, Madison, Wis.) sequence analysis package, the GeneDoc program (29), or servers accessible via the Internet. Homology searches were performed with the Gapped BLAST or PSI-BLAST algorithms (2) or the COG system (37) available at the National Center for Biotechnology Information site, preliminary characterization and analysis of the open reading frames (ORFs) was performed with the GENEQUIZ (35) server, and sequence alignments were performed with the Clustal W (38) algorithm as implemented in DNASTAR or at the Clustal W server at the European Bioinformatics Institute. Promoter analysis was done using the NNPP (31) program available through the Baylor College of Medicine server.
PCR.
Based on the sequences of the clones and the published sequence information for dexB, cps19fA, cps19fB, cps19fL, cps19fM, cps19fN, cps19fO, and aliA genes, primers were designed to obtain a series of overlapping PCR products covering the entire region between the dexB and aliA genes.
Conventional gel electrophoresis.
Total chromosomal DNA was prepared with a modified version of the procedure described by Marmur (21). Restriction was performed with one of the following: EcoRV, HaeIII, HincII, or a mixture of SpeI and HindIII restriction enzymes. All enzymes were obtained from New England Biolabs. Agarose gel electrophoresis was performed in accordance with standard procedures (34).
Nucleotide sequence accession number.
The nucleotide sequences and predicted amino acid translations described in this communication have been submitted to GenBank and are available under accession no. AF057294.
Sequence of the complete type 23F capsular locus.
In a previous study (24), it was shown that a strain of serogroup 23 carried genes with high homology to dexB, cps19fA, cps19fB, cps19fL, cps19fM, cps19fN, cps19fO, and aliA. Using PCR, we established that the organization of the homologous genes in the 23F locus was similar to the one found on serotype 19F, in agreement to what has since been reported by others (6). We therefore designed primers to amplify the region between the cps19fB and cps19fL homologues, which presumably contained sequence information specific for type 23F. A fragment of approximately 14 kb was obtained and subcloned into pSP64. The sequence of the clones was determined by primer walking. In order to confirm the accuracy of the obtained sequence and to close the gaps in the locus due to the cloning strategy, a series of overlapping PCR products covering the entire 23F capsular locus was generated and sequenced.
Computer analysis of the region located between the dexB and aliA homologues.
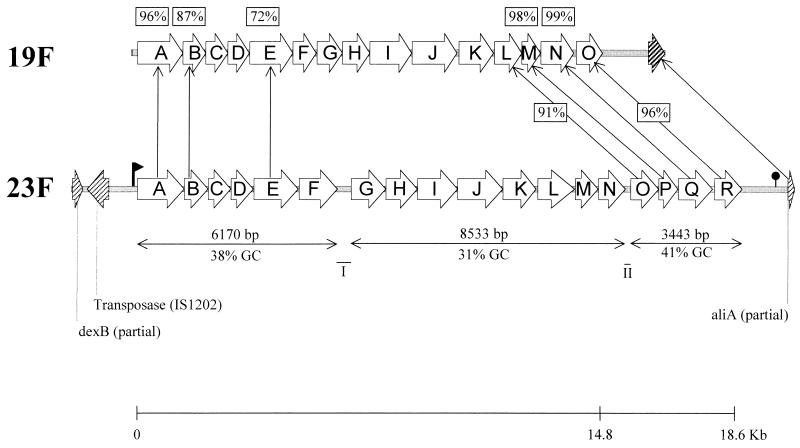
Computer analysis of the region located between the dexB and aliA homologues revealed the existence of 19 ORFs (Fig. 1).
FIG. 1.
Structures of capsular determinants 19F and 23F in S. pneumoniae. The structural organizations of the genes encoding the enzymes necessary for capsule 19F and 23F synthesis are compared. The designation of the genes is simplified, omitting the cps designation. Arrows connect genes that had positive signals in cross-hybridization experiments. Boxed numbers indicate DNA sequence identity. Two intergenic regions larger than 100 nucleotides are identified as I and II. A putative promoter ( ) and terminator ( ) are depicted. The scale at the bottom compares the size of type 19F and 23F coding regions. Hatched genes are not part of the cps locus.
The first ORF is transcribed in the orientation opposite that of dexB, and its product has a high degree of similarity to the C-terminal portion of IS1202 transposase (23) up to a deletion of one base in a stretch of T residues at position 630 (accession no. AF057294) that causes a frameshift. Moreover, a detailed examination of the DNA sequence in this region revealed a high degree of identity (95%) at the DNA level between the region encompassing nucleotides 495 to 1892 (accession no. AF057294) and the previously reported coding sequence for IS1202 transposase (23). The insertion of a C residue at position 135 relative to the IS1202 transposase coding sequence (accession no. U04047) creates a frameshift that generates a premature stop codon 53 bp downstream, preventing the synthesis of a correct polypeptide.
Although regions with homology to the repeats previously recognized to be associated with IS1202 (23) could be found immediately upstream of cps23fA, these did not present the same organization described before, and the corresponding regions downstream of the dexB homologue could not be identified. Taken together, these findings suggest that the insertion sequence associated with the capsule 23F genes is no longer functional but is an inactivated form of IS1202.
The location, properties, significant similarities with known proteins, and proposed functions for each of the remaining ORFs associated with serotype 23F are summarized in Table 1.
TABLE 1.
Characteristics of ORFs associated with S. pneumoniae Him18 serotype 23F
| Protein | Gene size (bp) | % G+C | Predicted protein size
|
Localization of protein on membranea | Highest scoring homologue(s) | Amino acid identity (%)c | Organism(s) | Proposed function | |
|---|---|---|---|---|---|---|---|---|---|
| Amino acids | kDa | ||||||||
| Cps23fA | 1,443 | 38 | 481 | 53.4 | Yes | Cps14A | 99 | S. pneumoniae | Regulatory protein |
| Cps23fB | 729 | 41 | 243 | 28.2 | No | Cps19fB, Cap1B | 88 | S. pneumoniae | Unknown |
| Cps23fC | 693 | 42 | 230 | 25.4 | Yesd | Cap1C | 72 | S. pneumoniae | Chain length regulation and/or export |
| Cps23fD | 687 | 38 | 229 | 25.0 | No | Cps19fD, Cps14D | 78 | S. pneumoniae | Chain length regulation and/or export |
| Cps23fE | 1,365 | 38 | 455 | 52.2 | No | Cps19fEb | 71 | S. pneumoniae | Glucosyl transferase |
| Cps23fF | 1,176 | 36 | 392 | 45.1 | No | MJ1069 | 4 | Methanococcus jannaschii | Rhamnosyl transferase |
| Cps23fG | 1,191 | 27 | 397 | 45.3 | Yes | BLAST score of <40 | Unknown | ||
| Cps23fH | 969 | 32 | 323 | 37.4 | No | EpsI, Cps14Jb | 32 | Streptococcus thermophilus, S. pneumoniae | β-1,4-Galactosyl transferase |
| Cps23fI | 1,230 | 31 | 410 | 47.8 | No | BLAST score of <40 | Unknown | ||
| Cps23fJ | 1,383 | 29 | 461 | 51.9 | Yes | EpsM | 31 | S. thermophilus | Repeating unit transporter |
| Cps23fK | 1,146 | 31 | 382 | 44.9 | No | TasA | 34 | S. pneumoniae | Glycerophosphotransferase |
| Cps23fL | 822 | 37 | 274 | 30.2 | Yes | GldA | 27 | Archaeoglobus fulgidus | Synthesis of glycerol-2-phosphate |
| Cps23fM | 702 | 33 | 234 | 26.1 | No | RfbF | 18 | Archaeoglobus fulgidus | Nucleotidyl transferase |
| Cps23fN | 831 | 33 | 277 | 31.1 | No | YutF | 27 | Bacillus subtilis | Unknown |
| Cps23fO | 867 | 41 | 289 | 32.2 | No | Cps19fLb | 95 | S. pneumoniae | Glucose-1-phosphate thymidyl transferase |
| Cps23fP | 591 | 42 | 197 | 22.2 | No | Cps19fMb | 99 | S. pneumoniae | dTDP-4-keto-6 deoxyglucose-3,5-epimerase |
| Cps23fQ | 1,047 | 42 | 349 | 39 | No | Cps19fNb | 99 | S. pneumoniae | dTDP-glucose-4,6 dehydratase |
| Cps23fR | 849 | 41 | 283 | 32.3 | No | Cps19fOb | 98 | S. pneumoniae | dTDP-l-rhamnose synthase |
Predicted localization of the protein based on the PHDacc program output. If no transmembrane segment was identified, the protein was considered not to be membrane associated. If several transmembrane regions were predicted, the protein was considered to target to the membrane.
Protein for which prediction of function was based on genetic or biochemical evidence.
Identity at the amino acid level between the two complete proteins.
In the case of CpsC, a single transmembrane segment was predicted.
Analysis of other isolates expressing serotype 23F.
In order to assess possible variations at the 23F locus, 11 strains from diverse genetic backgrounds (as defined by SmaI PFGE restriction profile) and different geographic origins (Brazil, United States, South Korea, and Bulgaria) were analyzed for their hybridization pattern to probes specific for the cps23f genes after enzymatic digestion. All ORFs identified in strain Him18, i.e., the strain used for the molecular characterization of the cps23f locus, were also present in all serotype 23F strains examined, and their organization was similar. Strain-to-strain differences were localized to the ends of the locus and were attributable to two factors: sequence variation in the genes (e.g., cps23fA, cps23fM, and cps23fR) and/or variation in the intergenic regions between cps23fR and aliA or dexB and cps23fA (data not shown). These results agree with findings recently reported by other authors (6), who found substantial variation in the regions flanking the cps locus among different pneumococcal isolates expressing serogroup 19 polysaccharides.
Structure of 23F capsular determinant of the Spanish/USA clone.
The structure of the 23F capsular determinant of the widespread multiresistant Spanish/USA clone (26) was analyzed by using representatives of this clone recovered from different geographic sites but sharing a similar SmaI PFGE restriction pattern. We found that these isolates constituted a very homogeneous group (data not shown). Differences detected between the capsular determinant of this group and strain Him18 (whose sequence was determined in this study) could be explained solely by differences in the region between dexB and cps23fA.
Diversity of pneumococcal capsular loci.
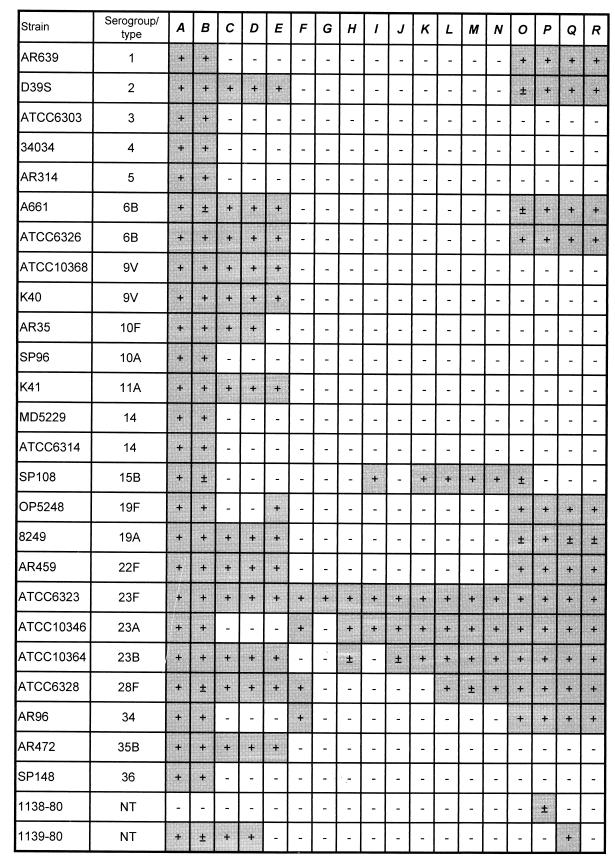
To examine the relationship between the genes found in the capsule 23F locus and other pneumococcal capsular determinants, individual genes of strain Him18 were used to probe Southern blots of restricted chromosomal DNA from strains belonging to 21 different serotypes and two nontypeable strains. The results obtained are summarized in Fig. 2.
FIG. 2.
Cross-reactivity of 23F genes with S. pneumoniae isolates with different capsular types. A strong signal, a weak signal, and no signal are indicated by +, ±, and −, respectively. All positive signals are shaded.
The properties and proposed functions of the 18 ORFs of the 23F capsular locus are summarized in Table 1. In most cases, the predicted functions of the homologous genes were themselves based on DNA or amino acid sequence homology. In some of the cases, proposed functions were based on genetic or biochemical evidence.
Upstream of cps23fA, a putative promoter could be identified. Although promoters could also be detected in intergenic region I and II by using the NNPP software (31) (Fig. 1 and results not shown), the fact that these transcripts lack a convincing Rho independent terminator makes them less biologically plausible. Downstream of cps23fR, a putative Rho independent terminator could be identified (−23.4 Kcal). Taken together, these findings suggest that the entire locus could be transcribed in a single 18.6-kb transcript.
The variable G+C composition of the locus and the location of intergenic spaces suggest a modular organization in which different parts of the locus could have been acquired independently (Fig. 1). It is noteworthy that the four genes responsible for the synthesis and activation of rhamnose have the same organization and have a very high sequence identity to genes identified in the serotypes 19F and 1 loci (24, 27).
The 115-nucleotide sequence found upstream of cap1A and conserved in all pneumococcal capsular loci (27) was also found associated with the 23F locus (89.6% identity).
Analysis of isolates expressing the 23F serotype.
The fact that isolates from unrelated genetic backgrounds (as defined by SmaI PFGE restriction profile) had homologues to all the genes identified in strain Him18 is in agreement with the hypothesis that these genes are essential for capsular biosynthesis. The observed differences could be explained by two phenomena: the loss in some strains of a HaeIII site near the 5′ end of the cps23fO coding region and the variability in the regions flanking the cps genes. The highly variable nature of these regions is apparent from the cps loci sequenced to date (3, 6, 19, 23).
Relatedness of cps loci of S. pneumoniae.
In agreement with other authors (26, 35), we found that sequences homologous to cps23fA were present in all strains with the exception of one of the nontypable isolates (Fig. 2). Sequences homologous to cps23fB were also detected in all isolates hybridizing to cps23fA. This observation is in apparent contradiction to results of a previous study (18), which found that strains expressing serotypes 9V, 11A, and 11B did not hybridize to the cps14B probe. However, the discrepancies may be explained by the nature of the probes used in the two studies. The probe used in the study described here spans nucleotides 99 to 389 of the coding region of cps23fB while the probe used in the previous analysis (18) covered the region from nucleotide 260 of the cps14B coding region to nucleotide 26 of the cps14C coding region. The 5′ regions of the cpsB genes sequenced to date show a high sequence identity that rapidly degenerates as one passes the middle of the gene. Therefore, if the structure of the cpsB genes of these serotypes follows the same principles, but with an even higher divergence, the construction of the probes would explain the differences in results.
Genes homologous to at least some of the cps23fC to -E genes were also found in 11 other serotypes in agreement with their proposed functions (Table 1).
Of the 21 different serotypes analyzed, 7 contained rhamnose. All strains with rhamnose containing polysaccharides had genes homologous to cps23fO to -Q similar to what was previously reported (24). It is interesting that isolates expressing serotypes 1 and 34, although having no rhamnose constituent, nevertheless carried a set of genes homologous to cps23fO to -Q. Dispensability of these genes for capsule 1 expression and their presence in a set of 19 different isolates of type 1 pneumococci were interpreted as evidence of a clonal origin of type 1 strains through recombination with DNA from an unknown origin (27). A similar phenomenon could explain the presence of these genes in serotype 34, which also does not have rhamnose as a capsule constituent.
It is interesting that 23A, 23B, and 28F, which are serologically related to 23F (17), had the highest percentage of genes homologous to the 23F locus. The structure of type 28F is known to include rhamnose, glucose, and phosphoglycerol in common with type 23F. The chemical compositions of types 23A and 23B are unknown (39). Our results show that type 15B also has a large set of homologues with type 23F. Elucidation of the genetic determinants of capsules 23A, 23B, and 28F should provide insights into the structure of these polysaccharides.
Acknowledgments
These investigations received partial support from a grant from the National Institutes of Health (RO1AI37275) and the Aaron Diamond Fund.
REFERENCES
- 1.Alonsodevelasco E, Verheul A F M, Verhoef J, Snippe H. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol Rev. 1995;59:591–603. doi: 10.1128/mr.59.4.591-603.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrecubieta C, Garcia E, Lopez R. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniaetype 3. Gene. 1995;167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 4.Arrecubieta C, Lopez R, Garcia E. Molecular characterization of cap3A, a gene from the operon required for the synthesis of the capsule of Streptococcus pneumoniaetype 3: sequencing of mutations responsible for the unencapsulated phenotype and localization of the capsular cluster on the pneumococcal chromosome. J Bacteriol. 1994;176:6375–6383. doi: 10.1128/jb.176.20.6375-6383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng S, Fockler C, Barnes W M, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 7.Corton J C, Gustafsson J A. Increased efficiency in screening large numbers of cDNA fragments generated by differential display. BioTechniques. 1997;22:802–804. doi: 10.2144/97225bm03. [DOI] [PubMed] [Google Scholar]
- 8.Cross A S. The biologic significance of bacterial encapsulation. Curr Top Microbiol Immunol. 1990;150:87–95. doi: 10.1007/978-3-642-74694-9_5. [DOI] [PubMed] [Google Scholar]
- 9.Dillard J P, Vandersea M W, Yother J. Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med. 1995;181:973–983. doi: 10.1084/jem.181.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillard J P, Yother J. Genetic and molecular characterization of capsular polysaccharide biosynthesis in Streptococcus pneumoniaetype 3. Mol Microbiol. 1994;12:959–972. doi: 10.1111/j.1365-2958.1994.tb01084.x. [DOI] [PubMed] [Google Scholar]
- 11.Eddy B E. Cross reactions between the several pneumococcic types and their significance in the preparation of polyvalent antiserum. Public Health Rep. 1944;59:485–499. [Google Scholar]
- 12.Eddy B E. A study of cross reactions among the pneumococcic types and their application to the identification of the types. Public Health Rep. 1944;59:451–468. [Google Scholar]
- 13.Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniaein the Czech Republic and in Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 14.Garcia E, Garcia P, Lopez R. Cloning and sequencing of a gene involved in the synthesis of the capsular polysaccharide of Streptococcus pneumoniaetype 3. Mol Gen Genet. 1993;239:188–195. doi: 10.1007/BF00281617. [DOI] [PubMed] [Google Scholar]
- 15.Guidolin A, Morona J K, Morona R, Hansman D, Paton J C. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniaetype 19F. Infect Immun. 1994;62:5384–5396. doi: 10.1128/iai.62.12.5384-5396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolkman M A. Ph.D. thesis. Utrecht, The Netherlands: University of Utrecht; 1997. [Google Scholar]
- 19.Kolkman M A, Wakarchuk W, Nuijten P J, van der Zeijst B A. Capsular polysaccharide synthesis in Streptococcus pneumoniaeserotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 20.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in penumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 21.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 22.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morona J K, Guidolin A, Morona R, Hansman D, Paton J C. Isolation, characterization, and nucleotide sequence of IS1202, an insertion sequence of Streptococcus pneumoniae. J Bacteriol. 1994;176:4437–4443. doi: 10.1128/jb.176.14.4437-4443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniaetype 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 25.Morona J K, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniaetype 19B. J Bacteriol. 1997;179:4953–4958. doi: 10.1128/jb.179.15.4953-4958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz R, Coffey T J, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, et al. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 27.Munoz R, Mollerach M, Lopez R, Garcia E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 28.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 29.Nicholas K B, Nicholas H B, Jr, Deerfield D W I. GeneDoc: analysis and visualization of genetic variation. EMBNET News. 1997;4:14. [Google Scholar]
- 30.Paton J C, Morona J K, Morona R. Characterization of the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniaetype 19F. Microb Drug Resist. 1997;3:89–99. doi: 10.1089/mdr.1997.3.89. [DOI] [PubMed] [Google Scholar]
- 31.Reese M G, Harris N L, Eeckman F H. Presented at the Pacific Symposium on Biocomputing, Kona, Hawaii. 1996. [Google Scholar]
- 32.Robbins J B, Austrian R, Lee C J, Rastogi S C, Schiffman G, Henrichsen J, Makela P H, Broome C V, Facklam R R, Tiesjema R H, et al. Considerations for formulating the second-generation pneumococcal capsular polysaccharide vaccine with emphasis on the cross-reactive types within groups. J Infect Dis. 1983;148:1136–1159. doi: 10.1093/infdis/148.6.1136. [DOI] [PubMed] [Google Scholar]
- 33.Roux K H. Optimization and troubleshooting in PCR. PCR Methods Appl. 1995;4:S185–S194. doi: 10.1101/gr.4.5.s185. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Scharf M, Schneider R, Casari G, Bork P, Valencia A, Ouzounis C, Sander C. Presented at the Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. 1994. [PubMed] [Google Scholar]
- 36.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniaefrom Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 37.Tatusov R L, Koonin E V, Lipman D J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Dam J E, Fleer A, Snippe H. Immunogenicity and immunochemistry of Streptococcus pneumoniaecapsular polysaccharides. Antonie Leeuwenhoek. 1990;58:1–47. doi: 10.1007/BF02388078. [DOI] [PubMed] [Google Scholar]