Abstract
mSur2, a subunit of the Mediator complex, is required for efficient mouse adenovirus type 1 (MAV-1) replication (L. Fang, J. L. Stevens, A. J. Berk, and K. R. Spindler, J. Virol. 78:12888-12900, 2004). We examined the contributions of early-region 1A (E1A) to mSur2 function in MAV-1 replication with E1A mutant viruses. At a multiplicity of infection (MOI) of 1, viruses containing CR3 replicated better in Sur2+/+ mouse embryonic fibroblasts (MEFs) than in Sur2−/− MEFs. In contrast, viruses lacking CR3 replicated no better in Sur2+/+ than in Sur2−/− MEFs. This result supports the hypothesis that the E1A CR3-mSur2 interaction is important for MAV-1 replication. However, at an MOI of 0.05, viruses lacking CR3 showed replication defects in Sur2−/− MEFs compared to Sur2+/+ MEFs, suggesting an E1A CR3 interaction-independent function of mSur2 in MAV-1 replication in cell culture. Paradoxically, CR1Δ, CR2Δ, and CR3Δ mutant viruses replicated slightly more efficiently than wild-type (wt) MAV-1 and E1A null mutant viruses in Sur2−/− MEFs at an MOI of 0.05. Coinfection of Sur2−/− MEFs with wt MAV-1 and CR1Δ, CR2Δ, or CR3Δ mutant viruses rescued the defects of wt MAV-1 replication. This result suggests that an inhibiting effect on wt E1A protein expression and/or E1A function might account for the severe viral replication defect of MAV-1 in Sur2−/− MEFs at an MOI of 0.05. Moreover, titrations of virus yields from infected brains of inbred strains of mice showed that E1A null and CR3Δ mutant viruses had a significant defect in virus replication compared to wt MAV-1. This result supports the hypothesis that the MAV-1 E1A-mSur2 interaction is important in MAV-1 replication in mice.
Study of mouse adenovirus type 1 (MAV-1) infection in its natural host facilitates understanding the pathogenesis of adenoviruses. MAV-1 early-region 1A (E1A) is a virulence factor in virus infection in mice, as demonstrated by 50% lethal dose (LD50) experiments (28). However, the molecular functions of MAV-1 E1A in virus infection have not been fully investigated. Previously, we showed that mSur2, a subunit of Mediator complex, interacts with MAV-1 E1A conserved region 3 (CR3) and that mSur2 is required for efficient MAV-1 replication (8). The Mediator complex connects transcriptional regulators to the basal RNA polymerase II transcriptional machinery and is important for efficient transcription activation (3, 5, 34). Experimental evidence has demonstrated that an interaction between a transcription activation domain and Mediator promotes transcription preinitiation complex assembly on promoter DNA (6). The human adenovirus (hAd) large E1A protein also binds to Sur2 via CR3 (5, 36). The conservation in human and mouse Ads of the ability of E1A to bind to Sur2 indicates the importance of E1A CR3-Sur2 interaction in Ad pathogenesis.
Wild type (wt) MAV-1 exhibits a replication defect in Sur2−/− mouse embryonic fibroblasts (MEFs) in a multiplicity-dependent manner (8). The severe virus replication defect at low-input multiplicities of infection (MOIs) (MOI of 0.05 or 0.1) can be partially overcome by higher-input MOIs (MOI of 1 or 5). The defective viral replication is due at least in part to a defect in virus early gene transcription. This result supports the proposed model that Ad E1A protein transactivates viral early genes by recruiting Mediator complex through E1A CR3-Sur2 interaction (6, 36). In the work reported here, we further test this model with MAV-1 E1A mutant viruses. Our data suggest that mSur2 has broader functions in MAV-1 replication in cell culture than just binding to E1A CR3. In addition, the defective viral replication of E1A null and CR3Δ mutant viruses in inbred stains of mice supports the hypothesis that the MAV-1 E1A CR3-mSur2 interaction is important for virus replication in mice.
MATERIALS AND METHODS
Cells and viruses.
Mouse NIH 3T6 fibroblast cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% heat-inactivated calf serum. MAV-1 E1A-expressing 37.1 cells, derived from NIH 3T6 cells (29), were maintained in DMEM containing 5% heat-inactivated calf serum and 200 μg of G418/ml. Details of the generation of Sur2+/+ and Sur2−/− MEFs were described previously (8). They were maintained in DMEM containing 10% fetal bovine serum. wt MAV-1 was the standard MAV-1 stock originally obtained from S. Larsen (1). pmE109 is an MAV-1 E1A null mutant virus; dlE105, dlE102, and dlE106 viruses are MAV-1 E1A CR1 deletion (CR1Δ), CR2 deletion (CR2Δ), and CR3 deletion (CR3Δ) mutants, respectively (29). The mutant viruses were titrated on 37.1 cells induced by treatment with 1.25 × 10−5 M dexamethasone (Sigma) to express E1A.
Mice.
All animal work complied with all relevant federal guidelines and institutional policies. SJL/JCr (SJL) and BALB/cAnNCr (BALB/c) mice were purchased from the National Cancer Institute. 129S6/SvEvTac (129 Sv/Ev) mice were obtained from the Taconic Company. Mice lacking the alpha/beta interferon (IFN-α/β) receptor (IFNAR−/− mice) (21) are on a 129 Sv/Ev background and were a kind gift from Kate Ryman, who originally obtained them from Barbara Sherry (24). All mice were male and were 3 to 5 weeks old. Mice were maintained in microisolator cages and infected with wt MAV-1, pmE109, or dlE106 at a dose of 100 PFU by intraperitoneal (i.p.) injection in a volume of 0.1 ml of conditioned medium. Infected mice were euthanized by inhalation of CO2 at 8 days postinfection (p.i.), and the brains were harvested and stored at −20°C until use.
Antibodies.
Mouse monoclonal antibody (MAb) (MAb10B10) against MAV-1 E1A and mouse MAb (MAb11H9) against MAV-1 E3gp11k were described previously (8). Anti-MAV-1 E1A (AKO7-147) rabbit polyclonal Abs were as previously described (29). Mouse MAb against β-actin of Arabidopsis thaliana, which cross-reacts with mouse β-actin, was obtained from Richard Meagher at University of Georgia and used at a 1:500 dilution for Western blots.
Viral growth curves.
Sur2+/+ and Sur2−/− MEFs were infected with either wt MAV-1 or mutant viruses at an MOI of 0.05 or 1. Plaque assays were carried out with 37.1 cells as described previously (38). Briefly, cells were harvested at various times p.i. by being scraped in their medium. The cell suspensions were subjected to three cycles of freezing-thawing, and the cell debris was spun out of the supernatant. Tenfold serial dilutions of supernatants were plated in triplicate on 37.1 cells, and plaques were counted 9 days after plating.
Determination of virus loads in mouse brains.
A total weight of 0.1 g of brain tissue from an individual mouse was put into a tube filled with sterile glass beads and homogenized in a volume of 1 ml of phosphate-buffered saline with a Beadbeater (Biospec Products) running at top speed at 1-min intervals for a total of 3 min. The homogenates were aspirated off of the beads and spun in a microcentrifuge at 700 × g for 5 min at room temperature. The supernatants were 10-fold serially diluted and titrated by plaque assays on 37.1 cells (38). wt MAV-1 was used a positive control. Counts of fewer than 20 plaques per 60 mm-diameter plate were considered unreliable. Therefore, 2 × 103 PFU/g of tissue was calculated as the detection limit.
Southern blots.
Sur2+/+ and Sur2−/− MEFs were infected at an MOI of 0.05 or 1 and harvested at various times p.i. by scraping the cells off the plates. Viral DNA was isolated by the method of Hirt (12). Equal amounts of DNA samples were digested with HindIII and RNase A and electrophoresed on a 0.7% agarose gel. The DNA was transferred to a positively charged nylon membrane (Boehringer) by capillary transfer. The membrane was prehybridized in 10 ml of PerfectHyb solution (Sigma) at 65°C for 2 h. Then, an MAV-1 genomic DNA-specific γ-32P-labeled probe as previously described (8) was added to 10 ml of PerfectHyb solution to hybridize at 65°C for 5 to 18 h. The membranes were washed and exposed to a phosphorimager (8).
RNase protection assays (RPAs).
Sur2+/+ and Sur2−/− MEFs were infected at an MOI of 0.05. Total RNA was extracted with TRI reagent (Molecular Research Center, Inc.) following the manufacturer's instructions. Equimolar pools of linearized plasmid templates were used to make an [α-32P]UTP-labeled multiplex RPA probe set with T7 or T3 polymerase transcription as described previously (8). One probe set included MAV-1 E1A, E4, and hexon and mouse L32 and was labeled with T7 polymerase. The other probe set included MAV-1 E3gp11K and E2A and mouse β-actin and was labeled with T3 polymerase. The RPAs were carried out as described by Hobbs et al. (13). Briefly, 10 μg of total RNA was hybridized with the probe sets overnight at 56°C. Saccharomyces cerevisiase tRNA was used as a negative control in the RPAs. After digestion with RNase A and RNase T1, samples were ethanol precipitated and electrophoresed on 5% polyacrylamide-8 M urea gels. After the samples were dried, protected mRNA signals were visualized with a phosphorimager, and quantitation was performed by normalizing the mRNA species of interest to L32 or β-actin signals.
Rescue experiment by coinfection.
Sur2−/− MEFs were singly infected with wt MAV-1 (MOI, 0.1), CR1, CR2, or CR3Δ (MOI, 0.1) or coinfected with wt MAV-1 (MOI, 0.05) and CR1Δ (MOI, 0.05), wt MAV-1 (MOI, 0.05) and CR2Δ (MOI, 0.05), or wt MAV-1 (MOI, 0.05) and CR3Δ (MOI, 0.05) on 100-mm-diameter plates. As controls for infection, Sur2+/+ MEFs were singly infected at MOI of 0.1 with wt MAV-1, CR1Δ, CR2Δ, or CR3Δ. Cells were harvested, pelleted, and stored at −20°C until use. The cell pellets were lysed on ice for 30 min in 20 μl of E3 lysis buffer (420 mM NaCl, 50 mM Tris-HCl [pH 7.5], 1% NP-40) containing protease inhibitor cocktail (1:50) (Sigma) and 1 mM phenylmethylsulfonyl fluoride. Twenty microliters of 50 mM Tris-HCl (pH 7.5) was added to the lysates. The lysates were centrifuged at 12,000 × g for 15 min at 4°C, and 13 μl of 4× sample loading buffer (17) was added to the 40 μl of supernatants and boiled for 10 min. Fifteen microliters of each sample was analyzed by Western blotting.
Western blots.
Protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a 6 to 18% polyacrylamide gradient gel and electrotransferred onto polyvinylidene difluoride membranes at 120 V for 3 h. The membranes were blocked by incubation at room temperature for 1 h in Tris-buffered saline-Tween (150 mM NaCl, 10 mM Tris [pH 7.4], 0.1% Tween 20) containing 5% nonfat dry milk. Anti-rabbit (1:12,000 from Amersham Biosciences) or anti-mouse (1:12,000 from Amersham Biosciences) immunoglobulin G-horseradish peroxidase-conjugated antibody was used as the secondary antibody. Immunoblots were developed with SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology, Inc.).
RESULTS
Functions of mSur2 in MAV-1 E1A mutant virus replication.
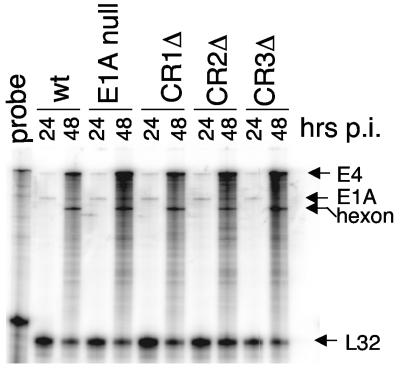
Infection of Sur2+/+ and Sur2−/− MEFs with wt MAV-1 showed that mSur2 is required for efficient viral replication (8). Using E1A mutant viruses, we also showed that MAV-1 E1A is dispensable for viral replication in 3T6 cells infected at an MOI of 5 (38). Therefore, it was of interest to test whether mSur2 was required for E1A mutant viruses to replicate. Since there is a multiplicity-dependent effect in Sur2−/− MEFs upon wt MAV-1 infection (8), it was important to ensure that equivalent input amounts of all the viruses were used. Accordingly, wt MAV-1, E1A null, CR1Δ, CR2Δ, and CR3Δ mutants were titrated on 37.1 cells (a complementing cell line for E1A mutants) in the same experiment. To confirm equivalent input virus in subsequent infections based on these plaque assays, we analyzed viral gene expression by RPAs. We infected 37.1 cells at an MOI of 0.05 with these viruses and extracted total RNAs at 24 and 48 h p.i. As shown in Fig. 1, viral E4 and hexon gene expression appeared similar in the wt and E1A mutant virus infections, indicating equivalent amounts of input virus. We confirmed this by quantitation with a phosphorimager, normalizing viral RNA levels to the mouse L32 gene RNA levels (data not shown).
FIG. 1.
RPA analysis of mRNA levels from equal MOI E1A mutant infections. 37.1 cells were infected with wt MAV-1 (wt), E1A null mutant, CR1Δ, CR2Δ, or CR3Δ at an MOI of 0.05. Total RNA was isolated at the indicated time points. The probe lane shows the full-length of the probe set, including MAV-1 E4, E1A, and hexon and mouse L32. The protected probe sizes are shown by arrows for each gene. L32 was used as an internal loading control. The wt E1A bands detected in all samples were from 37.1 cells, which are NIH 3T6 cell derivatives stably transfected with MAV-1 E1A (29). Quantitation with a phosphorimager with normalization to mouse L32 confirmed that viral gene expression levels were similar among the viruses (data not shown). A band appears just below the hexon band, between the wt 48-h lane and the E1A null 24-h lane. Because this band spans the space between the two lanes and was found in both the phosphorimager and X-ray film exposures, it is likely an artifact that occurred when the gel was dried.
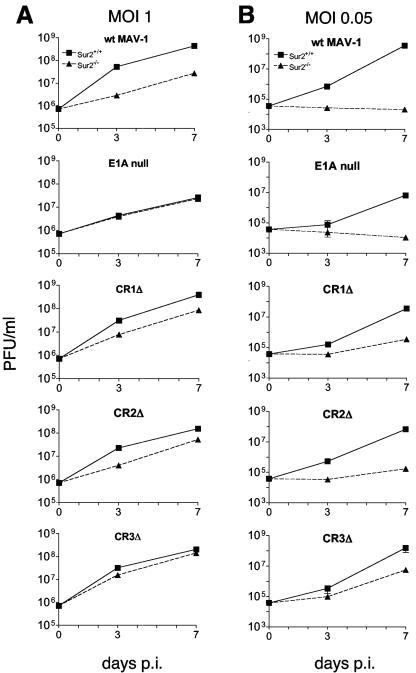
We performed viral growth assays to compare growth of wt and E1A mutants on Sur2+/+ and Sur2−/− MEFs. wt virus yields were similar on NIH 3T6, 37.1, and Sur2+/+ cells (L. Fang and K. R. Spindler, data not shown); all are immortalized MEF lines. This result suggests that infectivity of the three cell types by MAV-1 was equivalent. We infected Sur2+/+ and Sur2−/− MEFs with wt, E1A null mutant, CR1Δ mutant, CR2Δ mutant, or CR3Δ mutant viruses at an MOI of 1. We titrated viral yields by plaque assays on 37.1 cells, and the results are shown in Fig. 2A. There was about a 10-fold difference in wt virus yield between Sur2+/+ and Sur2−/− MEFs. This is consistent with previous results (8). There were about fivefold-lower yields of CR1Δ and CR2Δ viruses in Sur2−/− than in Sur2+/+ MEFs. MAV-1 E1A CR3 is required for interacting with mSur2 (8). These data, taken together with the wt MAV-1 result, indicate that mSur2 is important for these viruses, which all contain CR3, to replicate. No significant difference was detected in the yields of E1A null mutant or CR3Δ viruses between Sur2+/+ and Sur2−/− MEFs, indicating that mSur2 is not required for them to replicate, at least at an input MOI of 1. Together, the viral replication data of viruses containing CR3 (wt, CR1Δ, and CR2Δ) and viruses lacking CR3 (E1A null and CR3Δ) support the hypothesis that E1A CR3-mSur2 interaction was important for MAV-1 replication.
FIG. 2.
Growth of MAV-1 E1A mutant viruses in Sur2+/+ and Sur2−/− MEFs. The indicated cell types were infected with wt MAV-1, E1A null mutant, CR1Δ, CR2Δ, or CR3Δ at an MOI of 1 (A) or 0.05 (B) and harvested at the indicated times. The viral yields were determined by plaque assays of 37.1 cells. The legend for Sur2+/+ MEFs and Sur2−/− MEFs is shown at the top and is the same for all panels. The experiments were reproduced two additional times. Error bars are only distinguishable in a few samples (e.g., CR3Δ, MOI 0.05); other error bars were too small to be visualized.
We tested whether there was a multiplicity-dependent effect in viral replication of E1A mutant viruses, as was shown previously for wt virus (8). We infected Sur2+/+ and Sur2−/− MEFs at an MOI of 0.05 and analyzed virus yields by plaque assays of 37.1 cells. There were virus replication defects for all strains of viruses in Sur2−/− MEFs compared to Sur2+/+ MEFs at an MOI of 0.05 (Fig. 2B). The wt MAV-1 replication was severely restricted in Sur2−/− MEFs, since the virus yields were the same as the level of input viruses. This was consistent with previous results for wt MAV-1 growth in Sur2−/− MEFs titrated on 3T6 cells (8). The E1A null mutant showed the same severe virus replication restriction in Sur2−/− MEFs as wt MAV-1. Yields were 1 to 2 log units lower for CR1Δ, CR2Δ, and CR3Δ mutant viruses in Sur2−/− MEFs than in Sur2+/+ MEFs. These data indicate that mSur2 was important for efficient viral replication for all the E1A mutant viruses at an MOI of 0.05. For each virus (wt and E1A mutants), the severity of the virus replication defect in Sur2−/− MEFs at an MOI of 0.05 was partially overcome at an MOI of 1. Thus, there is a multiplicity-dependent effect for all these viruses. However, E1A null and CR3Δ mutants, which lack CR3 and therefore lack the ability to interact with mSur2, also showed virus replication defects in Sur2−/− MEFs at an MOI of 0.05. Therefore, E1A null and CR3Δ mutant viruses are dependent on mSur2 for replication at an MOI of 0.05. But this was independent of an E1A CR3-mSur2 interaction, since these viruses lack CR3. Surprisingly, CR1Δ, CR2Δ, and CR3Δ mutant viruses replicated slightly better than wt MAV-1 and E1A null mutant in Sur2−/− MEFs at an MOI of 0.05.
Multiplicity-dependent defects in viral DNA replication in E1A mutant-infected Sur2−/− MEFs.
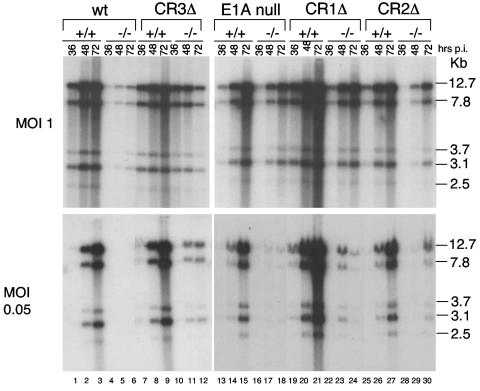
wt MAV-1 infection of Sur2−/− MEFs shows reduced accumulation of viral DNA compared to infection of Sur2+/+ MEFs (8), indicating that mSur2 is important for viral DNA replication. To determine whether the accumulated viral DNA levels correlated with reduced virus yields in Sur2−/− MEFs compared to Sur2+/+ MEFs for E1A mutant infections, we infected Sur2−/− and Sur2+/+ MEFs at an MOI of 0.05 or 1 with wt MAV-1 and E1A mutant viruses. Viral DNAs were isolated by the method of Hirt (12), digested with HindIII, and analyzed by Southern blotting (Fig. 3). Consistent with previous results (8), we did not detect viral DNA in wt MAV-1-infected Sur2−/− MEFs at an MOI of 0.05 (Fig. 3, bottom, lanes 4 to 6), and we detected a reduced amount of viral DNA at an MOI of 1 compared to Sur2+/+ MEFs (top, lanes 4 to 6 and lanes 1 to 3). Little viral DNA was detected in Sur2−/− MEFs infected with the E1A null mutant at an MOI of 0.05 (bottom, lanes 16 to 18), but the viral DNA levels were comparable between Sur2+/+ and Sur2−/− MEFs at an MOI of 1 (top, lanes 16 to 18 and 13 to 15). The defects of viral DNA accumulation in Sur2−/− MEFs compared to Sur2+/+ MEFs were more severe at an MOI of 0.05 than at an MOI of 1 for CR1Δ, CR2Δ, and CR3Δ mutant viruses, respectively, although we detected low levels of viral DNA at an MOI of 0.05. Therefore, the multiplicity-dependent defects in viral DNA replication in Sur2−/− MEFs seen with wt virus were also observed with E1A mutant viruses. At an MOI of 1, we noted that viral DNA levels in Sur2−/− MEFs were comparable to those in Sur2+/+ MEFs for E1A null and CR3Δ virus infection (Fig. 3, top, lanes 7 to 18). This result was consistent with plaque assay results (Fig. 2A). The low levels of viral DNA in Sur2−/− MEFs infected with CR1Δ, CR2Δ, or CR3Δ mutant viruses at an MOI of 0.05, taken together with the lack of viral DNA detected in Sur2−/− MEFs infected with the wt MAV-1 or E1A null mutant, were also consistent with virus replication data (Fig. 2).
FIG. 3.
Multiplicity-dependent defects in viral DNA replication of wt MAV-1 and E1A mutant viruses in Sur2−/− MEFs. Sur2+/+ (+/+) and Sur2−/− (−/−) MEFs were infected with wt MAV-1, E1A null mutant, CR1Δ, CR2Δ, or CR3Δ at an MOI of 0.05 or 1. Viral DNA was isolated at the indicated times by the Hirt method (12). Equal amounts of DNA were digested with HindIII and Southern blotted with MAV-1-specific DNA probes. DNA fragment sizes are indicated in kilobases on the right.
Importance of mSur2 for viral mRNA expression in E1A mutant infections.
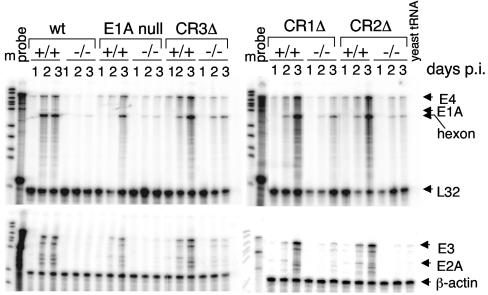
MSur2 is important for efficient wt MAV-1 replication, and the defect in viral replication is due at least in part to a defect in virus early gene transcription (8). We tested whether there was a defect in viral mRNA expression in E1A mutant virus infections of Sur2−/− MEFs that correlated with the virus replication and DNA accumulation defects. We infected Sur2+/+ and Sur2−/− MEFs with wt or E1A mutant MAV-1 at an MOI of 0.05 and analyzed virus early genes (E1A, E2A, E3, and E4) and one virus late gene, hexon, by RPA (Fig. 4). We detected wt E1A only in wt MAV-1-infected Sur2+/+ MEFs at 2 and 3 days p.i. The CR1, CR2, and CR3 deletion E1A mRNAs expressed from the E1A mutant-infected cells cannot be detected by the MAV-1 E1A probe in these RPAs. The mRNA levels of all the assayed viral genes were markedly reduced in Sur2−/− MEFs compared to Sur2+/+ MEFs for wt virus and each mutant virus, indicating that mSur2 is important for virus gene transcription in both wt MAV-1 and E1A mutant infections. The viral mRNA levels were severely diminished or not detectable in Sur2−/− MEFs upon wt MAV-1 infection, results which were consistent with previous data (8). Similar to wt virus, there were severely diminished or nondetectable levels of viral mRNAs in Sur2−/− MEFs infected with the E1A null mutant. However, we detected low levels of viral mRNAs, particularly E4 and hexon, in Sur2−/− MEFs infected with CR1Δ, CR2Δ, or CR3Δ mutant viruses. These data were consistent with viral replication and viral DNA accumulation defects observed in Fig. 2 and 3.
FIG. 4.
Importance of mSur2 for viral mRNA expression in E1A mutant infections. Sur2+/+ (+/+) and Sur2−/− (−/−) MEFs were infected with wt MAV-1, E1A null mutant, CR1Δ, CR2Δ, or CR3Δ at an MOI of 0.05. Total RNA was isolated at the indicated times. The sizes of probes protected from RNase treatment are shown by the arrows for each gene at the right side. Even with longer exposures, we did not observe wt E1A expression in the E1A mutant-infected samples (data not shown). L32 and β-actin were used as internal loading controls. Yeast tRNA was used as a negative control. m, DNA size marker; probe, full-length probe set (no RNase).
CR1Δ, CR2Δ, and CR3Δ mutant viruses are able to rescue wt MAV-1 in Sur2−/− MEFs.
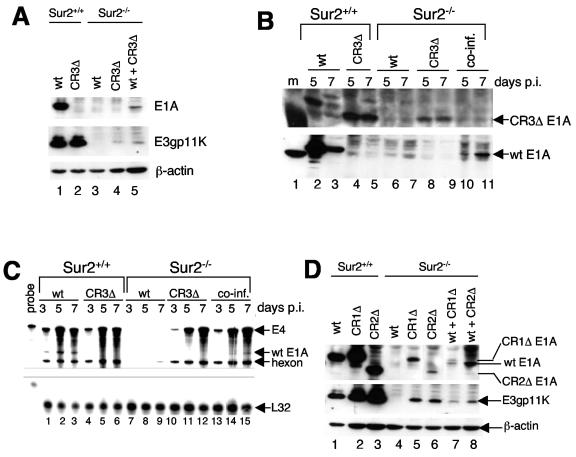
wt MAV-1 replication is severely restricted and there are no detectable levels of viral DNA, viral mRNAs, or viral proteins (E1A or E3gp11K) in Sur2−/− MEFs at an MOI of 0.05 (8). However, CR1Δ, CR2Δ, and CR3Δ mutants did replicate in Sur2−/− MEFs, although not as efficiently as in Sur2+/+ MEFs (Fig. 2). We also detected low levels of virus DNA and virus mRNAs in Sur2−/− MEFs infected with CR1Δ, CR2Δ, or CR3Δ mutant viruses (Fig. 3 and 4). We speculated that there might be a negative regulatory factor that inhibited the replication of wt MAV-1, but not CR1Δ, CR2Δ, or CR3Δ mutant viruses in Sur2−/− MEFs. To test this hypothesis, we examined whether the CR3Δ mutant could rescue virus replication of wt MAV-1 in Sur2−/− MEFs in a coinfection experiment. Although we could not distinguish plaques between wt MAV-1 and CR3Δ infection, with Western blots we could distinguish wt and CR3Δ E1A protein expression with a MAb that only recognized the wt E1A protein. A mouse MAb for E3gp11K recognized E3 proteins from both wt- and CR3Δ mutant-infected cells. We infected Sur2−/− MEFs with wt MAV-1 or CR3Δ at an MOI of 0.1 or coinfected them with wt MAV-1 and CR3Δ (MOI of 0.05 each). We singly infected Sur2+/+ MEFs as controls. As shown in Fig. 5A, lanes 1 and 2, we detected E3gp11K protein at 5 days p.i. in Sur2+/+ MEFs infected with wt MAV-1 or CR3Δ. These data demonstrated that the cells were successfully infected. We also detected wt E1A protein recognized by the MAb (Fig. 5A, lane 1). As expected, we did not detect any CR3Δ mutant E1A protein in Sur2+/+ MEFs infected with CR3Δ (Fig. 5A, lane 2, and Fig. 5B, bottom, lanes 4 and 5) even after a long exposure (data not shown), demonstrating that the MAb only recognized wt E1A protein, not the CR3Δ mutant E1A protein. We further showed that CR3Δ E1A protein was expressed from CR3Δ-infected Sur2+/+ MEFs (Fig. 5B, top, lanes 4 and 5) by using a rabbit polyclonal antibody (AKO7-147) against MAV-1 E1A that can recognize both wt and CR3Δ E1A. The reduced amount of E1A protein at 7 days p.i. in Sur2+/+ MEFs infected with wt MAV-1 (Fig. 5B, lane 3) is due to the cytopathic effects at late times in infections, as observed previously (8). There were no detectable levels of E1A or E3gp11K protein in Sur2−/− MEFs infected with wt MAV-1 (Fig. 5A, lane 3, and Fig. 5B, lanes 6 and 7), consistent with previous data (8). In contrast, there were low levels of E3gp11K protein at 7 days p.i. in Sur2−/− MEFs infected with CR3Δ (Fig. 5A, lane 4), correlating with better replication of CR3Δ compared to wt MAV-1 in Sur2−/− MEFs (Fig. 2B). In Sur2−/− MEFs coinfected with wt MAV-1 and CR3Δ, we detected wt E1A protein (Fig. 5A, lane 5, and Fig. 5B, bottom, lanes 10 to 11). This result demonstrated that coinfection with CR3Δ rescued the E1A protein expression in wt virus-infected Sur2−/− MEFs.
FIG. 5.
The defect in wt E1A expression in Sur2−/− MEFs was rescued by coinfection. (A) Sur2+/+ and Sur2−/− MEFs were infected with wt MAV-1 or CR3Δ at an MOI of 0.1 or coinfected with wt MAV-1 and CR3Δ (both at an MOI of 0.05). Cells were harvested at 7 days p.i. and lysed in E3 lysis buffer. Samples were analyzed by sodium dodecyl sulfate-PAGE on an 8 to 15% gradient PAGE gel and Western blotting with antibodies as indicated at the right: MAb10B10 was the E1A antibody and MAb11H9 was the E3gp11K antibody. β-Actin was assayed as a loading control. (B) Sur2+/+ and Sur2−/− MEFs were infected and processed as described for the results shown in panel A. Rabbit polyclonal antibody against MAV-1 E1A (AKO7-147) was used to detect the CR3Δ mutant E1A (top), and MAb (mAb10B10) against MAV-1 E1A was used panel to detect wt E1A (bottom). (C) Total RNAs were isolated at the times indicated from cells infected as described for panel A, and RPAs with the probe set that includes viral gene E1A, E4, and hexon and mouse L32 were carried out. The sizes of protected probes are shown by the arrows on the right. L32 was used as an internal loading control. (D) Sur2+/+ and Sur2−/− MEFs were singly infected with wt MAV-1 (lanes 1 and 4), CR1Δ (lanes 2 and 5), or CR2Δ (lanes 3 and 6) at an MOI of 0.1 or coinfected with wt MAV-1 and CR1Δ (both at an MOI of 0.05) (lane 7) or wt MAV-1 and CR2Δ (both at an MOI of 0.05) (lane 8). Cells were harvested at 7 days p.i. and processed as described for the results shown in panel A with MAb10B10 for E1A and MAb11H9 for E3gp11K.
In addition, we tested whether coinfection could rescue the transcription defects of wt E1A in Sur2−/− MEFs. We singly infected or coinfected Sur2+/+ and Sur2−/− MEFs with wt MAV-1 and CR3Δ as in Fig. 5A and analyzed the wt E1A mRNA by RPA. Wt E1A mRNA was detected in Sur2+/+ infected cells (Fig. 5C, lanes 1 to 3). As expected, there was no wt E1A mRNA in CR3Δ-infected cells (Fig. 5C, lanes 4 to 6). We detected wt E1A mRNA expression in coinfected Sur2−/− MEFs (Fig. 5C, lanes 14 and 15) but not in wt virus-infected Sur2−/− MEFs (Fig. 5C, lanes 8 and 9), demonstrating that coinfection rescued the defects of wt E1A expression at the transcription level. In addition, the wt viral DNA replication defect in Sur2−/− MEFs (Fig. 3) (8) and was rescued by coinfection with the CR3Δ mutant virus (data not shown). These data indicated that the CR3Δ mutant virus was able to rescue wt MAV-1 replication in Sur2−/− MEFs.
We also tested whether CR1Δ and CR2Δ mutants could rescue wt MAV-1 viral replication in Sur2−/− MEFs, as did the CR3 mutant. We singly infected or coinfected Sur2+/+ MEFs and Sur2−/− MEFs, as indicated in Fig. 5D. E3gp11K virus proteins were expressed from virus-infected Sur2+/+ MEFs, demonstrating successful virus infections (Fig. 5D, lanes 1 to 3). The MAb against E1A recognized wt E1A, CR1Δ, and CR2Δ mutant E1A with distinguishable positions in the membrane, as shown in Fig. 5D, lanes 1 to 3. The levels of CR1Δ E1A protein ran slightly higher than wt E1A protein, as previously reported (29). We detected E3gp11K proteins and CR1Δ E1A and CR2Δ E1A protein in Sur2−/− MEFs (Fig. 5D, lanes 5 to 6), correlating with better replication of CR1Δ and CR2Δ mutants than of wt MAV-1 in Sur2−/− MEFs (Fig. 2B). There was no detectable wt E1A protein in wt MAV-1-infected Sur2−/− MEFs (Fig. 5D, lane 4). In contrast, we detected wt E1A proteins in coinfected Sur2−/− MEFs (Fig. 5D, lanes 7 and 8). The data indicated that, like the CR3Δ mutant, the CR1Δ and CR2Δ mutants could rescue wt MAV-1 viral replication in Sur2−/− MEFs.
Replication defects of E1A null mutant and CR3Δ mutant viruses in mice.
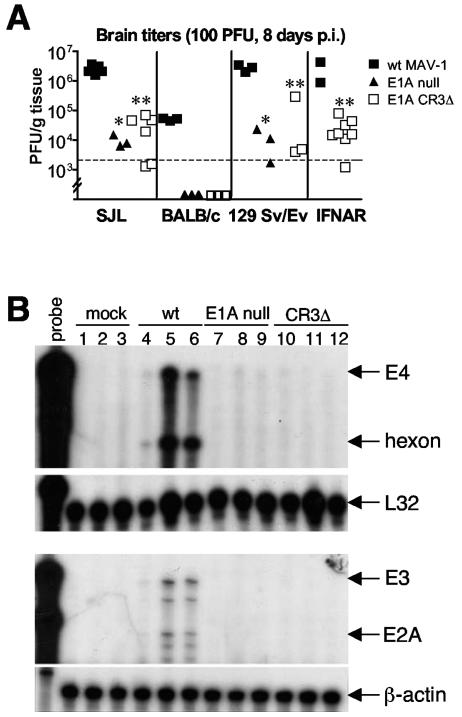
The E1A gene product is a virulence factor in MAV-1 infection in Swiss outbred mice (28) and inbred SJL mice (31), as demonstrated by LD50 experiments. Since the mSur2 knockout is embryonically lethal in mice (J. L. Stevens and A. J. Berk, personal communication), we cannot directly address the function of mSur2 in MAV-1 replication in vivo. Therefore, we indirectly tested whether E1A-mSur2 interaction is one factor that may contribute to virulence by infecting mice with the E1A null mutant and CR3Δ mutant viruses. Because E1A null mutant and CR3Δ mutant viruses lack E1A CR3-mSur2 interactions (8), we tested whether there were virus replication defects of these virus mutants in inbred mice. We infected four different inbred strains of mice (SJL, BALB/c, 129 Sv/Ev, and IFNAR−/−) i.p. with wt MAV-1, E1A null mutant, or CR3Δ at 100 PFU. We harvested mouse brains at 8 days p.i. and titrated them for viral yields on 37.1 cells. As shown in Fig. 6A, the virus yields of both E1A mutants were reduced compared to wt MAV-1 in all four strains of mice. These results are consistent with higher LD50s of E1A null mutant and CR3Δ mutant viruses in outbred mice (28). Viral loads in the brains of SJL mice were about 2 log units lower for E1A null and CR3Δ mutant viruses than for wt MAV-1 (P < 0.0001 and P = 0.0001, respectively). Virus yields from wt MAV-1-infected BALB/c mice were 2 log units lower than for SJL mice, consistent with BALB/c mice being resistant to MAV-1 infection (10, 31). We did not detect any infectious viruses in brains of E1A null or CR3Δ-infected BALB/c mice. 129 Sv/Ev mice are the parental strain of IFNAR−/− mice, which are deficient for the IFN-α/β receptor. The virus replication patterns with 129 Sv/Ev mice were similar to those with SJL mice, with about 2 log units lower virus yields from E1A null and CR3Δ mutant viruses than from wt MAV-1 infection. There was also about a 2 log unit difference between virus yields of CR3Δ and wt MAV-1 infection in IFNAR−/− mice. This result of CR3Δ mutant infection of IFNAR−/− mice was consistent with results we have seen with E1A null mutant infection of IFNAR−/− mice, albeit under slightly different dose and time conditions. Those results were that E1A null mutant virus titers in brains were lower than wt virus titers (M. Moore and K. R. Spindler, unpublished data).
FIG. 6.
Virus replication defects of E1A null and CR3Δ mutants in mice. (A) Four strains of mice (SJL/J, BALB/c, 129 Sv/Ev, and IFNAR−/−) were infected by the i.p. route with wt MAV-1, E1A null mutant, or CR3Δ at 100 PFU. Brains were harvested at 8 days p.i., and homogenates were titrated for viruses by plaque assays of 37.1 cells. Each symbol represents an individual mouse. The dotted line at 2 × 103 represents the lower limit of detection of the assay. Data points below the limit of detection were excluded from statistical calculations. *, P < 0.0001 and P = 0.002 for E1A null mutant titers compared to wt MAV-1 titers in SJL/J and 129 Sv/Ev mice, respectively; **, P = 0.0001, P = 0.02, and P = 0.003 for CR3Δ titers compared to wt MAV-1 titers in SJL/J, 129 Sv/Ev, and IFNAR−/− mice, respectively. (B) SJL/J mice were infected by the i.p. route with wt MAV-1, E1A null mutant, or CR3Δ at 100 PFU. Brains were harvested at 8 days p.i. Total RNA was isolated, and RPAs were carried out with probe sets as described for the results shown in Fig. 4.
Previously we showed that the defective virus replication of MAV-1 in Sur2−/− MEFs compared to Sur2+/+ MEFs is due at least in part to reduced viral mRNA expression, indicating that mSur2 is important for virus gene transcription in vitro (8). We tested whether there was a defect in virus mRNA expression of E1A null and CR3Δ mutant viruses compared to wt MAV-1 in mice. We infected SJL mice i.p. with wt MAV-1, E1A null, or CR3Δ at 100 PFU. We harvested mouse brains at 8 days p.i. and isolated total RNA for RPA of the virus mRNA levels. We detected virus mRNAs (E4, hexon, E3, and E2A) from wt MAV-1-infected mouse brains, but not from E1A null or CR3Δ- infected mouse brains (Fig. 6B). Therefore, the defective viral mRNA expression of E1A null and CR3Δ may account at least in part for the reduced viral replication of E1A null and CR3Δ in mice.
DISCUSSION
mSur2 is required for efficient wt MAV-1 replication in cell culture (8). Here, we expanded the study of mSur2 function with E1A mutant viruses. We found that similar to wt virus (8), the E1A mutants exhibited a multiplicity-dependent effect, that is, they were more defective in Sur2−/− MEFs at low-input MOIs than at higher MOIs (Fig. 2 and 3).
Our data showed that mSur2 may function in MAV-1 replication via E1A-CR3 interaction-dependent and -independent pathways. MAV-1 E1A CR3 is required for interaction with mSur2 (8). Since wt MAV-1, CR1Δ, and CR2Δ mutant viruses contain an intact CR3 domain, presumably there is E1A CR3-mSur2 interaction in Sur2+/+ MEFs upon infection. In contrast, E1A null and CR3Δ lack the CR3 domain. As expected, we observed a replication defect in Sur2−/− MEFs infected at an MOI of 1 with the CR3-containing viruses, wt, CR1Δ, and CR2Δ, compared to Sur2+/+ MEFs. Similarly, as expected, in E1A null or CR3Δ infection we observed no replication difference between Sur2+/+ and Sur2−/− MEFs. Although there may be CR3 domain functions that are independent of mSur2 interaction, our data with E1A mutant viruses indicated that the E1A CR3-mSur2 interaction was important for MAV-1 replication.
At an MOI of 0.05, we observed an E1A CR3 interaction-independent function of mSur2. E1A null and CR3Δ mutant viruses showed virus replication defects in Sur2−/− MEFs compared to Sur2+/+ MEFs (Fig. 2) that were paralleled by virus DNA and virus mRNA defects (Fig. 3 and 4). Since MAV-1 replication apparently was not exclusively dependent on E1A CR3-mSur2 interaction (Fig. 2B), these results suggested that mSur2 must have a broader function in virus replication in cell culture at an MOI of 0.05 than just the consequences of its binding to E1A CR3. The defects of virus replication of E1A null and CR3Δ in Sur2−/− MEFs (Fig. 2) were due at least in part to a defect in transcription of virus early genes (Fig. 4). The physiological functions of Sur2 might have direct and/or indirect effects on MAV-1 replication. Further study of Sur2 functions may help to understand the E1A CR3-mSur2 interaction-independent mechanisms.
The fact that the E1A null mutant was able to replicate in Sur2−/− MEFs at an MOI of 1 (Fig. 2A) demonstrated that there was an E1A-independent, mSur2-independent viral replication pathway. hAd E1A CR3 is primarily responsible for transactivating transcription of viral early genes (5, 6, 36). Berk and colleagues have proposed that recruiting Mediator complex to enhance the efficiency of transcription via E1A CR3-Sur2 interaction is the primary molecular mechanism of function of the interaction (5, 6, 36). Our previous (8) and present studies of MAV-1 infection using Sur2−/− MEFs also support this proposed model. However, neither hAd E1A nor MAV-1 E1A is absolutely required for viral replication at high-input MOIs (8, 9, 22, 27). hAd E4 open reading frame 6/7 (designated ORF6/7) can functionally compensate for the loss of function of E1A (23, 26). Cellular E1A-like molecules (14, 18, 19) in the host cells may also support E1A null mutant viral replication. Human CCAAT/enhancer binding protein β (C/EBPβ) has been shown to have E1A-like activity (30). It interacts with human Sur2 (20), and hAd E1A protein can totally block C/EBPβ-human Sur2 interaction (20). It is possible that MAV-1 E4 ORFd (1), which has 17% identity and 43% similarity to hAd E4 ORF6/7 protein (Fang and Spindler, unpublished), and/or mouse C/EBPβ could compensate for the loss of function of MAV-1 E1A in virus replication in E1A mutants. Moreover, our data presented in this paper suggest mSur2 has an E1A CR3 interaction-independent function. Further study of the functions of MAV-1 E4, mouse C/EBPβ, and mSur2 may help to elucidate the molecular mechanisms of multiple replication pathways of MAV-1.
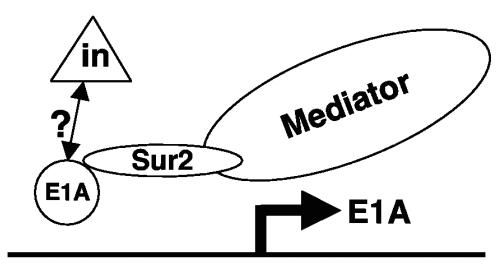
We also noted that CR1Δ, CR2Δ, and CR3Δ mutant viruses replicated slightly better than wt MAV-1 and the E1A null mutant in Sur2−/− MEFs at an MOI of 0.05 (Fig. 2B), paralleled by virus DNA (Fig. 3), mRNA (Fig. 4), and protein levels (Fig. 5; single virus-infected samples). However, it is paradoxical that wt MAV-1 had more severely restricted replication than CR1Δ, CR2Δ, or CR3Δ mutant viruses. One model to explain this is pictured in Fig. 7. There may be a negative regulator that inhibits wt E1A functions during wt MAV-1 infection in Sur2−/− MEFs. Consistent with this are previous data showing that the steady-state levels of virus early mRNAs (E4, E1B, and E1A) in CR1Δ, CR2Δ, or CR3Δ mutant-infected 3T6 cells at an MOI of 5 are higher than in wt MAV-1-infected cells (38). wt MAV-1 E1A may associate directly or indirectly with an inhibitor activity to repress viral gene expression, including its own. hAd wt E1A represses transcription from viral (2, 35, 37) and cellular (11, 15, 32, 33) enhancers. We postulate that the inhibiting effect could be relieved by CR1Δ, CR2Δ, and CR3Δ mutant E1A proteins, for example, if the mutant E1A proteins had conformational changes such that they could not be influenced by the negative regulator. In this case in the absence of mSur2, wt MAV-1 E1A would directly or indirectly repress the transcription of virus early genes, including E1A itself, by recruiting some repressor-like activity. However, due to conformational changes of CR1Δ, CR2Δ, and CR3Δ mutant E1A proteins, the repressor-like activity would not be associated, and virus early genes could be transcribed in the mutant virus infections. Interestingly, our coinfection data indicated that CR1Δ, CR2Δ, and CR3Δ mutant viruses could rescue the viral replication of wt MAV-1 in Sur2−/− MEFs. The rescued wt E1A protein expression (Fig. 5A and D) and mRNA expression (Fig. 5C) are consistent with a model where there is an inhibiting effect (at least in part) on wt E1A transcription and translation. hAd E1A protein binds C-terminal binding protein (CtBP), a transcriptional corepressor (4, 25). Even though the C terminus of MAV-1 E1A lacks the conserved motif PXDLS that is required for interaction with CtBP and thus is unlikely to bind to a mouse homolog of CtBP, it is possible that MAV-1 E1A could bind to a cellular protein that contains CtBP-like activity. We are currently investigating a cellular protein that potentially interacts with the C terminus of MAV-1 E1A.
FIG. 7.
Model of restricted viral replication of wt MAV-1 in Sur2−/− MEFs. We propose that there is a negative regulator (triangle) that inhibits wt E1A function during wt MAV-1 infection. This negative regulator might directly or indirectly associate with E1A protein. In the presence of Sur2 (Sur2+/+ MEFs), the interaction between MAV-1 E1A and Sur2 is the predominant mechanism to transactivate viral genes and is important for efficient viral replication, and the inhibitor has little influence. In contrast, in the absence of Sur2 (Sur2−/− MEFs) due to the lack of E1A-Sur2 interaction, the negative regulator inhibits E1A functions, resulting in the restricted viral replication of wt MAV-1 at low MOIs. We propose that the negative regulator would not inhibit the function of mutant E1As (CR1Δ, CR2Δ, and CR3Δ) due to conformational changes resulting from the deletions. Therefore, these mutant viruses would replicate in Sur2−/− MEFs. The presence of a negative regulator is consistent with coinfection experiments in which wt E1A expression was rescued by CR1Δ, CR2Δ, and CR3Δ mutant viruses.
It should be noted that in the coinfection experiments at an MOI of 0.05, only 0.25% of the cells (5 × 5%) would initially have been productively infected with both viruses. Thus, it might appear surprising that we were able to detect the coinfection (as indicated by the presence of wt E1A mRNA and protein) (Fig. 5). One possibility is that coinfection was indeed detectable from that very small fraction of initially coinfected cells. It is also possible that multiple rounds of infection and coinfection occurred during the 7-day time course. However, we favor the explanation that because the particle:PFU ratio for MAV-1 is 1,000:1 (31), it is likely that >0.25% of the cells were coinfected with MAV-1 particles. In any case, we found clear evidence of coinfection, since wt E1A expression was rescued by E1A mutant viruses in multiple experiments.
MAV-1 E1A is a virulence factor in mice (28), but the mechanism of E1A virulence has not been elucidated. Due to the unavailability of Sur2 knockout mice, we tested indirectly whether the E1A-mSur2 interaction is one factor that may contribute to virulence by infecting mice with E1A mutants. SJL mice are susceptible and BALB/c mice are resistant to MAV-1 infection (31). 129 Sv/Ev mice are the immunocompetent parent strain of IFNAR−/− mice, which are immunodeficient, lacking the interferon-α/β receptor. In all four strains of mice, the E1A null mutant and CR3 mutant viruses replicated at significantly reduced levels than that of wt MAV-1 (Fig. 6A). This suggests that the reduced level of virus replication was due to a defect in the mutant viruses themselves, rather than to host-specific factors. There is no E1A-mSur2 interaction in E1A null mutant- or CR3 mutant-infected mouse brain microvascular endothelial cells (8). Endothelial cells are a target cell type in MAV-1-infected mice (7, 16). We further showed that there is defective viral mRNA expression of E1A null mutant and CR3Δ viruses in mouse brains (Fig. 6B), which correlated with the virus replication defects (Fig. 6A). This defect in viral mRNA expression might be due to the lack of E1A CR3-mSur2 interaction, since Sur2 is primarily responsible for transactivating transcription of viral genes through interaction with E1A CR3 (34). Although we cannot rule out the possibility that other functions of E1A CR3 besides its interaction with mSur2 also cause the virus replication defects in mice, the results support a hypothesis that E1A CR3-mSur2 interaction is critical for virus replication in mice, thereby contributing to E1A being a virulence factor in mice. An MAV-1 mutant that is only defective in binding to mSur2 may address this question directly.
Acknowledgments
We thank Amanda Welton for excellent technical assistance and Jason Weinberg for assistance with RPAs. We thank Arnie Berk for helpful discussions. We thank Mike Imperiale for suggesting the coinfection experiments and critical review of the manuscript. We thank Sidney Kushner for helpful comments on the manuscript.
This work was supported by NIH grant R01 AI023762 to K.R.S.
REFERENCES
- 1.Ball, A. O., C. W. Beard, P. Villegas, and K. R. Spindler. 1991. Early region 4 sequence and biological comparison of two isolates of mouse adenovirus type 1. Virology 180:257-265. [DOI] [PubMed] [Google Scholar]
- 2.Borelli, E., R. Hen, and P. Chambon. 1984. Adenovirus-2 E1A products repress enhancer-induced stimulation of transcription. Nature 312:608-612. [DOI] [PubMed] [Google Scholar]
- 3.Boube, M., L. Joulia, D. L. Cribbs, and H.-M. Bourbon. 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, J. M., T. Subramanian, U. Schaeper, M. La Regina, S. Bayley, and G. Chinnadurai. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis, and metastasis. EMBO J. 12:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, T. G., M. E. D. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 6.Cantin, G. T., J. L. Stevens, and A. J. Berk. 2003. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl. Acad. Sci. USA 100:12003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charles, P. C., J. D. Guida, C. F. Brosnan, and M. S. Horwitz. 1998. Mouse adenovirus type-1 replication is restricted to vascular endothelium in the CNS of susceptible strains of mice. Virology 245:216-228. [DOI] [PubMed] [Google Scholar]
- 8.Fang, L., J. L. Stevens, A. J. Berk, and K. R. Spindler. 2004. Requirement of Sur2 for efficient mouse adenovirus type 1 replication. J. Virol. 78:12888-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaynor, R. B., and A. J. Berk. 1983. cis-Acting induction of adenovirus transcription. Cell 33:683-693. [DOI] [PubMed] [Google Scholar]
- 10.Guida, J. D., G. Fejer, L.-A. Pirofski, C. F. Brosnan, and M. S. Horwitz. 1995. Mouse adenovirus type 1 causes a fatal hemorrhagic encephalomyelitis in adult C57BL/6 but not BALB/c mice. J. Virol. 69:7674-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hen, R., E. Borelli, and P. Chambon. 1985. Repression of the immunoglobulin heavy chain enhancer by the adenovirus-2 E1A products. Science 230:1391-1394. [DOI] [PubMed] [Google Scholar]
- 12.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 13.Hobbs, M. V., W. O. Weigle, D. J. Noonan, B. E. Torbett, R. J. McEvilly, R. J. Koch, G. J. Cardenas, and D. N. Ernst. 1993. Patterns of cytokine gene expression by CD4+ T cells from young and old mice. J. Immunol. 150:3602-3614. [PubMed] [Google Scholar]
- 14.Imperiale, M. J., H.-T. Kao, L. T. Feldman, J. R. Nevins, and S. Strickland. 1984. Common control of the heat shock gene and early adenovirus genes: evidence for a cellular E1A-like activity. Mol. Cell. Biol. 4:867-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janaswami, P. M., D. V. R. Kalvakolanu, Y. Zhang, and G. C. Sen. 1992. Transcriptional repression of interleukin-6 gene by adenoviral E1A proteins. J. Biol. Chem. 267:24886-24891. [PubMed] [Google Scholar]
- 16.Kajon, A. E., C. C. Brown, and K. R. Spindler. 1998. Distribution of mouse adenovirus type 1 in intraperitoneally and intranasally infected adult outbred mice. J. Virol. 72:1219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lee, H. J., J. K. Lee, S. Miyake, and S. J. Kim. 2004. A novel E1A-like inhibitor of differentiation (EID) family member, EID-2, suppresses transforming growth factor (TGF)-β signaling by blocking TGF-β-induced formation of Smad3-Smad4 complexes. J. Biol. Chem. 279:2666-2672. [DOI] [PubMed] [Google Scholar]
- 19.MacLellan, W. R., G. Xiao, M. Abdellatif, and M. D. Schneider. 2000. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol. Cell Biol. 20:8903-8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mo, X., E. Kowenz-Leutz, H. Xu, and A. Leutz. 2004. Ras induces mediator complex exchange on C/EBP beta. Mol. Cell 13:241-250. [DOI] [PubMed] [Google Scholar]
- 21.Müller, U., U. Steinhoff, L. F. L. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 22.Nevins, J. R. 1981. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell 26:213-220. [DOI] [PubMed] [Google Scholar]
- 23.O'Connor, R. J., and P. Hearing. 2000. The E4-6/7 protein functionally compensates for the loss of E1A expression in adenovirus infection. J. Virol. 74:5819-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryman, K. D., W. B. Klimstra, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 2000. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J. Virol. 74:3366-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaley, J., R. J. O'Connor, L. J. Taylor, D. Bar-Sagi, and P. Hearing. 2000. Induction of the cellular E2F-1 promoter by the adenovirus E4-6/7 protein. J. Virol. 74:2084-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenk, T., N. Jones, W. Colby, and D. Fowlkes. 1979. Functional analysis of adenovirus-5 host-range deletion mutants defective for transformation of rat embryo cells. Cold Spring Harbor Symp. Quant. Biol. 44:367-375. [DOI] [PubMed] [Google Scholar]
- 28.Smith, K., C. C. Brown, and K. R. Spindler. 1998. The role of mouse adenovirus type 1 early region 1A in acute and persistent infections in mice. J. Virol. 72:5699-5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, K., B. Ying, A. O. Ball, C. W. Beard, and K. R. Spindler. 1996. Interaction of mouse adenovirus type 1 early region 1A protein with cellular proteins pRb and p107. Virology 224:184-197. [DOI] [PubMed] [Google Scholar]
- 30.Spergel, J. M., W. Hsu, S. Akira, B. Thimmappaya, T. Kishimoto, and S. Chen-Kiang. 1992. NF-IL6, a member of the C/EBP family, regulates E1A-responsive promoters in the absence of E1A. J. Virol. 66:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spindler, K. R., L. Fang, M. L. Moore, G. N. Hirsch, C. C. Brown, and A. Kajon. 2001. SJL/J mice are highly susceptible to infection by mouse adenovirus type 1. J. Virol. 75:12039-12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stein, R. W., M. Corrigan, P. Yaciuk, J. Whelan, and E. Moran. 1990. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J. Virol. 64:4421-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein, R. W., and E. B. Ziff. 1987. Repression of insulin gene expression by adenovirus type 5 E1A proteins. Mol. Cell. Biol. 7:1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, A. Shevchenko, and A. J. Berk. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296:755-758. [DOI] [PubMed] [Google Scholar]
- 35.Velcich, A., and E. Ziff. 1985. Adenovirus E1A proteins repress transcription from the SV40 early promoter. Cell 40:705-716. [DOI] [PubMed] [Google Scholar]
- 36.Wang, G., and A. J. Berk. 2002. In vivo association of adenovirus large E1A protein with the human mediator complex in adenovirus-infected and -transformed cells. J. Virol. 76:9186-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, H. G., Y. Rikitake, M. C. Carter, P. Yaciuk, S. E. Abraham, B. Zerler, and E. Moran. 1993. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J. Virol. 67:476-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying, B., K. Smith, and K. R. Spindler. 1998. Mouse adenovirus type 1 early region 1A is dispensable for growth in cultured fibroblasts. J. Virol. 72:6325-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]