Abstract
TRIB3, a pseudokinase, was previously studied within only some specific cancer types, leaving its comprehensive functions in pan-cancer contexts largely unexplored. Here, we performed an integrated analysis of TRIB3 expression, prognosis, genetic alterations, functional enrichment and tumor immune-related characteristics in 33 cancer types. Our results showed that TRIB3 exhibits high expression levels across 24 different cancer types and correlates closely with unfavorable prognoses. Meanwhile, TRIB3 shows mutations in a wide spectrum of 22 distinct cancer types, with the predominant mutation types being missense mutations and gene amplifications, and significant changes in DNA methylation levels in 14 types of cancer. We further discovered that TRIB3 expression is significantly associated with cancer immune-related genome mutations, such as tumor mutational burden (TMB), microsatellite instability (MSI) and DNA mismatch repair (MMR), and infiltration of immunosuppressive cells, such as CD4+ Th2 cells and myeloid-derived suppressor cells (MDSCs), into the tumor microenvironment. These results indicated that the expression of TRIB3 might reshape the tumor immune microenvironment (TIME) and lead to immunosuppressive "cold" tumors. In addition, our results confirmed that the loss of function of TRIB3 inhibits cell proliferation, promotes apoptosis, and leads to significant enrichment of "hot" tumor-related immune pathways, at least in breast cancer cells, which further supports the important role of TRIB3 in cancer prognosis and TIME regulation. Together, this pan-cancer investigation provided a comprehensive understanding of the critical role of TRIB3 in human cancers, and suggested that TRIB3 might be a promising prognostic biomarker and a potential target for cancer immunotherapy.
Keywords: TRIB3, Pan-cancer analysis, Tumor immune microenvironment, Prognostic biomarker
Graphical Abstract
1. Introduction
According to data reported by the World Health Organization, cancer is the second leading cause of death globally and poses a serious threat to human health. Unfortunately, the data indicates that cancer cases will continue to increase rapidly until 2040 [1], [2]. Currently, cancer treatment methods primarily include surgery, radiotherapy, chemotherapy, targeted therapy, and immunotherapy [3], [4]. Despite the constant improvement of cancer treatment strategies and the increased understanding of the complex pathogenesis of tumors, issues such as drug resistance, side effects, and differences in the tumor immune microenvironment (TIME) of different patients still result in unsatisfactory patient prognosis and survival rates [5]. Therefore, further research into the potential molecular mechanisms of cancer occurrence and development may reveal new cancer treatment methods. In recent years, immunotherapy has significantly changed the landscape of tumor treatment, particularly immune checkpoint inhibitors (ICIs), which have achieved great success in clinical practice. However, in many patients receiving the same treatment, the difference in the TIME of "cold" and "hot" tumors results in a very low objective response rate in "cold" tumor patients [6], [7]. Hence, it is urgent to discover new immune-related treatment targets and develop new drugs for tumors.
TRIB3 is a member of the pseudokinase family, comprises 358 amino acids, and possesses three distinct functional domains. These domains include the central kinase domain, which lacks ATP-specific binding sites and a catalytic core, rendering it devoid of kinase activity; the N-terminal domain, which is primarily involved in binding to transcription factors; and the C-terminal domain, which interacts mainly with ubiquitin ligases [8]. Although TRIB3 lacks kinase activity, it acts as an adaptor/scaffold protein and directly interacts with various proteins, including ATF4 [9], CHOP [10], COP1 [11], MEK1 [12], MKK7 [13], AKT1/2 [14], PPARγ [15], SIAH1 [16], EGFR [17], β-catenin, and TCF4 [18], and regulates their activity. Therefore, TRIB3 is a key "stress regulator" in cancer. Recent research suggests that TRIB3 may act as both a tumor suppressor gene and an oncogene, and the mechanisms underlying its dual role in cancer are not yet clear [19], [20]. Several studies have found that TRIB3 is upregulated in various types of cancer, including breast cancer [21], liver cancer [22], lung cancer [23], glioblastoma [24], ovarian cancer [25], oral squamous cell carcinoma [26], gastric cancer [27], colorectal cancer [28], and bladder cancer [29], and is closely associated with cancer development and poor prognosis. However, some studies have found that higher levels of TRIB3 protein in breast cancer are associated with better prognosis [30], and knocking down TRIB3 in liver cancer cells increases the survival rate of MHCC02H liver cancer cells [31]. It is worth noting that an increasing number of studies suggest that TRIB3 plays a more prominent role as an oncogene in cancer.
Here, through the implementation of an extensive pan-cancer analysis, this study revealed the abnormal expression, DNA mutations and DNA methylation patterns associated with the TRIB3 gene. Importantly, we also uncovered the pivotal impact of TRIB3 on tumor prognosis and its ability to modulate the immune response within tumors. Our study emphasizes the prognostic significance of TRIB3 in pan-cancer scenarios and its potential as a target for advancing cancer immunotherapy.
2. Materials and methods
2.1. Cancer data collection
We downloaded gene expression data, clinical data, and mutation data for 33 cancers and their corresponding paracancerous tissues in TCGA from the UCSC Xena database (https://xena.ucsc.edu/) [32]. Furthermore, we obtained data from the METABRIC study, which includes both gene expression and clinical information from breast cancer patients, via the cBioPortal database (http://www.cbioportal.org/) [33]. We obtained the GSE116671 dataset from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) [34], which comprises RNA microarray data of control or TRIB3-silenced MCF7 cells samples. The full names and abbreviations of the 33 cancer types we studied can be found in Table 1.
Table 1.
Full names and abbreviations of the 33 cancer types in TCGA.
| Serial number | Cancer Types | Abbreviations |
|---|---|---|
| 1 | Adrenocortical carcinoma | ACC |
| 2 | Bladder Urothelial Carcinoma | BLCA |
| 3 | Breast invasive carcinoma | BRCA |
| 4 | Cervical squamous cell carcinoma and endocervical adenocarcinoma | CESC |
| 5 | Cholangiocarcinoma | CHOL |
| 6 | Colon adenocarcinoma | COAD |
| 7 | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma | DLBC |
| 8 | Esophageal carcinoma | ESCA |
| 9 | Glioblastoma multiforme | GBM |
| 10 | Head and Neck squamous cell carcinoma | HNSC |
| 11 | Kidney Chromophobe | KICH |
| 12 | Kidney renal clear cell carcinoma | KIRC |
| 13 | Kidney renal papillary cell carcinoma | KIRP |
| 14 | Acute Myeloid Leukemia | LAML |
| 15 | Brain Lower Grade Glioma | LGG |
| 16 | Liver hepatocellular carcinoma | LIHC |
| 17 | Lung adenocarcinoma | LUAD |
| 18 | Lung squamous cell carcinoma | LUSC |
| 19 | Mesothelioma | MESO |
| 20 | Ovarian serous cystadenocarcinoma | OV |
| 21 | Pancreatic adenocarcinoma | PAAD |
| 22 | Pheochromocytoma and Paraganglioma | PCPG |
| 23 | Prostate adenocarcinoma | PRAD |
| 24 | Rectum adenocarcinoma | READ |
| 25 | Sarcoma | SARC |
| 26 | Skin Cutaneous Melanoma | SKCM |
| 27 | Stomach adenocarcinoma | STAD |
| 28 | Testicular Germ Cell Tumors | TGCT |
| 29 | Thyroid carcinoma | THCA |
| 30 | Thymoma | THYM |
| 31 | Uterine Corpus Endometrial Carcinoma | UCEC |
| 32 | Uterine Carcinosarcoma | UCS |
| 33 | Uveal Melanoma | UVM |
2.2. Differentially expressed analysis
First, we performed differentially expressed analysis for the TRIB3 gene between tumors and adjacent normal tissues in the TCGA database using the "Exploration-Gene DE" module of the TIMER2.0 network server (http://timer.comp-genomics.org/) [35] and calculated the statistical significance using the Wilcoxon test.
Furthermore, to analyze the expression of TRIB3 in cancers without corresponding normal tissues in the TCGA database (e.g., ACC, DLBC, LAML, LGG, OV, SARC, SKCM, TGCT, THYM, UCS), we utilized the "Expression Analysis" module of the GEPIA 2 network server (http://gepia2.cancer-pku.cn/#analysis) to examine the expression differences between normal tissues in the GTEx database and the corresponding tumor tissues in the TCGA database [36]. We obtained the differentially expressed significance with p value < 0.05 using one-way ANOVA.
Finally, to investigate the differential protein expression of TRIB3 in cancer samples and normal tissues, immunohistochemical images based on the TRIB3 antibody (catalog number: HPA055442; Atlas Antibodies, Sigma—Aldrich) were obtained from the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/) [37].
2.3. Correlation of TRIB3 expression with clinical tumor stage
The correlation between TRIB3 expression and tumor stage for 33 types of cancers was analyzed by the R packages "limma" and "ggpubr". The results were then visualized as box plots.
2.4. Expression and survival prognosis analysis of TRIB3
We utilized three survival prognostic indicators, overall survival (OS), disease-specific survival (DSS), and progression-free period (PFI), to examine the correlation between TRIB3 expression and cancer patient prognosis. Additionally, we also analyzed the prognostic correlation between TRIB3 expression in METABRIC data and OS in breast cancer patients. The median of TRIB3 expression were used as the expression threshold to classify low and high expression subgroups. Kaplan—Meier survival curves were plotted using the R packages "survminer" and "survivor", and statistical significance was determined using the log-rank test. Univariate Cox analysis was performed using the R packages "survival" and "forestplot", and forest plots were created to display the risk ratio (HR), 95% confidence interval, and p value. A risk ratio (HR) less than 1 indicated that TRIB3 was a protective factor for cancer, while an HR greater than 1 indicated that TRIB3 was a risk factor for cancer.
2.5. TRIB3 mutation characterization
To examine the mutational signature of TRIB3 in various cancers, we utilized the cBioPortal tool. First, we selected the TCGA Pan-cancer Atlas Studies cohort. Next, we entered "TRIB3" in the "Query" module, and the tool provided information on TRIB3 change sites, types, and numbers in both the "Cancer Types Summary" and "Mutations" modules.
2.6. DNA methylation analysis for TRIB3
To assess the relationship between TRIB3 expression and DNA methylation in 33 different types of cancers found within the TCGA database, we utilized the SMART platform (http://www.bioinfo-zs.com/smartapp/), which is an interactive web server that provides a comprehensive analysis of DNA methylation in the TCGA project [38]. Specifically, we entered "TRIB3" into the "Quick Start" module. Then, we used the "CpG-aggregated methylation" module to calculate the pan-cancer TRIB3 methylation level and displayed the resulting data in a box plot.
2.7. Functional enrichment analysis of TRIB3
To explore the biological function of TRIB3, we conducted functional enrichment analyses of TRIB3 using the LinkedOmics (www.linkedomics.org/login.php), which is a powerful platform designed for acquiring, analyzing, and comparing multi-omics cancer data across various tumor types, providing a unique resource for cancer research [39]. To be more precise, we employed the Pearson correlation test to establish the correlation between TRIB3 and co-expressed genes. Subsequently, we generated an enriched result bar graph by selecting the "GSEA Enrichment Analysis" and "KEGG pathway" options using the "LinkInterpreter" module.
2.8. Correlation analysis of TRIB3 expression with the tumor microenvironment (TME)
ESTIMATE (Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data) is a sophisticated scoring system designed to evaluate the tumor purity, stromal cell presence, and immune cell infiltration in tumor tissue by analyzing expression data [40]. Hence, this scoring system has been used to calculate the ImmuneScore, StromalScore, and ESTIMATEScore for 33 different types of cancer in TCGA. The scores indicate the proportion of corresponding components present in the TME. Higher scores correspond to a greater proportion of the corresponding component in the TME. The ESTIMATEScore is the sum of the ImmuneScore and StromalScore, which reflects the combined proportion of the two components in the TME. The "estimate" R package and Spearman correlation test were used to estimate the ImmuneScore and StromalScore as well as tumor purity for each tumor. The resulting data were then visualized using the "ggplot2", "ggpubr", "heatmap" and "ggExtra" R packages.
2.9. Correlation analysis of TRIB3 expression with immune infiltration
We utilized the TIMER 2.0 database (http://timer.comp-genomics.org/) [35] "immune-gene" module, with purity adjustment applied, to investigate the association between TRIB3 expression and tumor-infiltrating immune cells (TIICs) across various cancer types. The analysis incorporated TIMER, EPIC, MCPCOUNTER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, XCELL, and TIDE algorithms. The Spearman correlation test was utilized to determine p values and partial correlation values, with the results presented as a heatmap. Additionally, the R package "limma" was utilized to identify the correlation between TRIB3 expression and immune-related genes, including immunostimulator genes, immunoinhibitor genes, chemokine receptors genes, chemokines genes, and major histocompatibility complex (MHC) genes, which were also visualized as a heatmap using the R package "heatmap".
2.10. Correlation analysis of TRIB3 expression with tumor mutational burden (TMB), microsatellite instability (MSI), and mismatch repair (MMR) genes
Previous studies have demonstrated that tumor mutational burden-high (TMB-H), mismatch repair deficiency (dMMR), and high microsatellite instability (MSI-H) are predictive biomarkers that have gained significant attention in recent years for assessing the response to immune checkpoint blockade (ICB) therapy [41], [42]. To explore the correlation between TRIB3 expression and TMB or MSI, Spearman's rank correlation coefficient was employed, and the results were presented using a radar chart generated by the "fmsb" package in R. The correlation between TRIB3 and MMR gene expression was determined based on gene expression profiling data from the TCGA cohort using the "limma" package in R, and the results were visualized as a heatmap using the "heatmap" package in R.
2.11. RNA microarray data analysis upon TRIB3 silencing in MCF-7 breast cancer cells
Differentially expressed analysis for the GSE116671 expression matrix was conducted using the "limma" R package. The result was visualized as a volcano plot using the "ggplot" R package. Subsequently, the "clusterProfiler" R package was employed to conduct KEGG and GSEA enrichment analyses. The enriched KEGG pathways were visualized as bar plots using the "ggplot" R package, while the GSEA enrichment results were visualized using the "GseaVis" R package.
2.12. Cell lines and cultures
Triple-negative breast cancer (TNBC) MDA-MB-231 cells were obtained from ATCC (Manassas, USA). Additionally, TNBC HCC1806 cells were acquired from Zhejiang Mason Cell Technology Co. (Zhejiang, China). The MDA-MB-231 and HCC1806 cell lines were maintained in RPMI 1640 medium (Gibco, Thermo Fisher, China) supplemented with 10% fetal bovine serum (FBS, ExCell, China) and 100 U/ml penicillin-streptomycin (HyClone™, Utah, USA), at 37 ℃ in a 5% CO2 incubator.
2.13. siRNA reverse transfection assay
Following the transfection reagent kit protocol, we used 40 pmol of siRNA per well for the 12-well plate containing 1 ml of culture medium and 100 pmol of siRNA per well for the 6-well plate containing 2 ml of cell culture medium. Opti-MEM™(Gibco, Thermo Fisher, China), TRIB3-siRNA (siTRIB3), and NC-siRNA (siNC) were mixed with the transfection reagent (Lipo8000, Beyotime, China) and incubated. Subsequently, the resulting mixture was evenly distributed at the bottom of the plates: 7 × 104 cells were seeded in each well of the 12-well plate, and 2 × 105 cells were seeded in each well of the 6-well plate. After 48 h of transfection, we extracted total mRNA or protein to assess the transfection efficiency of the siRNA. The sequence of TRIB3 siRNA is as follows:
TRIB3 siRNA-1: GGUGUACCCCGUCCAGGAATT;
TRIB3 siRNA-2: GGACCUGAGAUACUCAGCUTT;
TRIB3 siRNA-3: GAUGAUUCCCUGUGGGACA;
TRIB3 siRNA-4: UCGCUGACCGUGAGAGGAA;
TRIB3 siRNA-5: ACAGAGAAGUGGUUCUGUA;
Negative control siRNA: UUCUCCGAACGUGUCACGUTT. These siRNAs were purchased from GenePharma (Shanghai, China).
2.14. Cloning formation experiment
Based on the transfection reagent kit protocol, Opti-MEM™ (Gibco, Thermo Fisher, China), siTRIB3/siNC and the transfection reagent (Lipo8000, Beyotime, China) were mixed and left to stand. The resulting mixture was then evenly added to the bottom of a 12-well plate, and cells were seeded into each well. Once cell colonies formed, they were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Finally, the cell colonies were imaged and counted for statistical analysis.
2.15. RNA extraction and quantitative real-time fluorescence quantitative PCR
Total mRNA was extracted using the RNA isolater kit (Vazyme Biotech Co., Ltd, China) following the manufacturer's protocol. The extracted total mRNA was then reversely transcribed into cDNA using the HiScript II Q RT SuperMix for qPCR kit (Vazyme, China), followed by RT-PCR amplification using ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) with GAPDH serving as the reference gene. Data analysis was performed using the 2−ΔΔCT method. The forward and reverse primers used are as follows:
TRIB3-F: TGCGTGATCTCAAGCTGTG;
TRIB3-R: CTTGTCCCACAGGGAATCA.
GAPDH-F: GTCTCCTCTGACTTCAACAGCG;
GAPDH-R: ACCACCCTGTTGCTGTAGCCAA.
2.16. Western blotting
After 48 h of cell transfection in a 6-well plate, the cells were lysed on ice for 30 min using RIPA buffer (Beyotime, Shanghai, China), which included protease inhibitors and phosphatase inhibitors (BOSTER, Wuhan, China). The lysate was then centrifuged to obtain the supernatant. Subsequently, the protein concentration in the samples was determined using a BCA protein assay kit (CWBIO, Jiangsu, China). The protein samples were mixed with loading buffer (Epizyme, Shanghai, China), heated at 95 °C for 10 min to denature the proteins, and then loaded onto a 10% SDS-polyacrylamide gel for electrophoresis. The proteins were subsequently transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, USA). After a 20 min incubation in a QuickBlock™ Western blocking solution (Beyotime, Shanghai, China), primary antibodies and HRP-conjugated secondary antibodies were added sequentially to incubate the membrane. Finally, the target proteins were visualized using the ECL High Sensitivity Chemiluminescent Substrate (4 A BIOTECH, Suzhou, China). GAPDH (1:20,000, 60004–1-Ig), TRIB3 (1:500, 13300–1-AP), HRP-conjugated anti mouse IgG (1:5000, SA00001–1) and anti-rabbit IgG (1:5000, SA00001–2) were obtained from Proteintech (Wuhan, Hubei, China).
2.17. Apoptosis assay
After 48 h of cell transfection in a 6-well plate, following the protocol of the Annexin V-FITC Cell Apoptosis Detection Kit (Beyotime, Shanghai, China), the cell culture medium was removed, and the cells were washed once with PBS. Subsequently, Annexin V-FITC binding solution was added, followed by the addition of Annexin V-FITC and propidium iodide staining solution. The mixture was gently mixed and then incubated in a light-avoiding environment at room temperature for 15 min. This was followed by capturing images using a fluorescence microscope (Olympus, IX73, Japan) and subsequent statistical analysis.
2.18. Statistical analysis
For this study, all data were calculated, graphed, and statistically analyzed using GraphPad Prism 8.3.0 and R software (version 4.1.1). The differentially expressed analysis was performed using either the Wilcoxon test or one-way ANOVA, while the log-rank test was used to determine statistically significance in prognosis analysis. The correlation between the two groups was calculated using either Spearman or Pearson correlation analysis. Statistical analysis of qPCR and cloning formation data was conducted using Student's t-test or one-way ANOVA. Significance levels are defined as P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****).
3. Results
3.1. TRIB3 is significantly highly expressed in 24 types of cancers
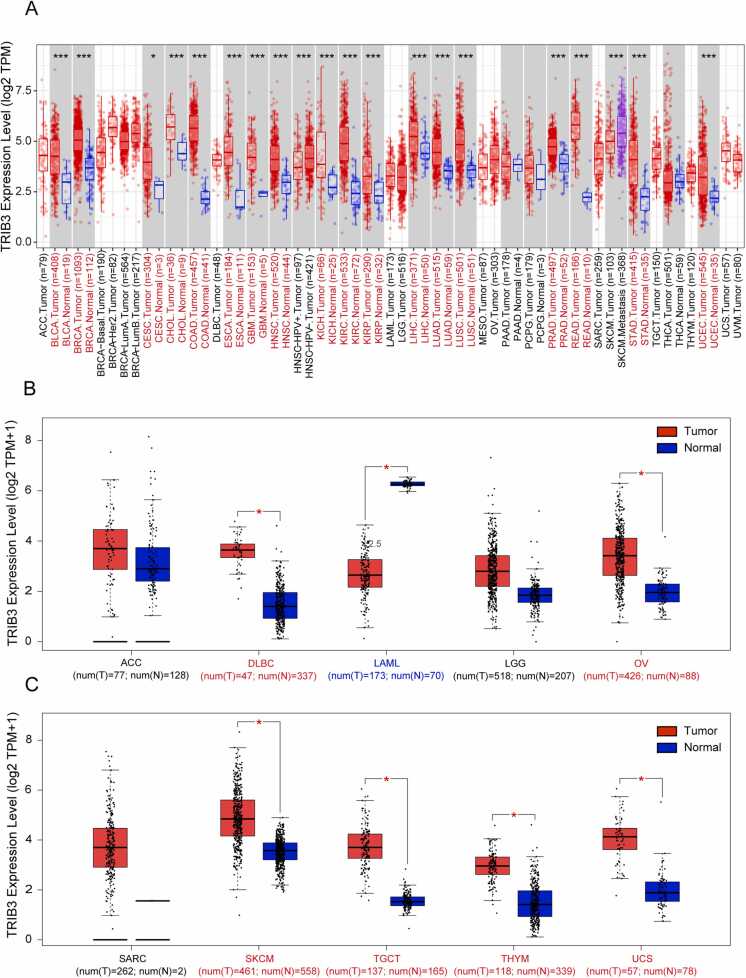
To investigate the differential expression of TRIB3 in various human cancers and their corresponding adjacent tissues, we utilized the TIMER2.0 platform to analyze RNA sequencing data of 33 types of cancer in TCGA (Fig. 1A). Simultaneously, we used the GEPIA2 platform to analyze various cancers in TCGA without normal samples by combining the GTEx and TCGA databases (Fig. 1B-C). Our findings indicated that TRIB3 expression was statistically significant in 25 types of cancer, except for tumors without available normal tissue data (MESO and UVM). Among these, TRIB3 was highly expressed in 24 types of cancer, including BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, OV, PRAD, READ, SKCM, STAD, TGCT, THYM, UCEC, and UCS. Notably, READ and COAD exhibited the highest upregulation of TRIB3 expression compared to normal tissue, while TRIB3 expression was significantly decreased in LAML.
Fig. 1.
TRIB3 exhibits high expression levels in 24 types of tumors, but demonstrates low expression levels specifically in LAML. (A) Differences in TRIB3 expression between 33 normal and tumor tissues in TCGA from the TIMER2.0 database (*P < 0.05, **P < 0.01, ***P < 0.001). (B-C) Differences in TRIB3 expression between cancers from the TCGA database and normal samples from the GTEx database (*P < 0.05). Red indicates high expression, and blue indicates low expression.
We carried out further investigation into the protein expression levels of TRIB3 in both normal tissues and tumors. Immunohistochemical images were obtained from the HPA database. Our findings indicate that TRIB3 expression may be higher in BRCA, COAD, HNSC, LIHC, and SKCM than that in the corresponding normal tissues (Fig. S1). These results seemed to align with the mRNA expression results obtained from our analysis.
3.2. TRIB3 is significantly highly expressed in the late stages of 8 cancer types
As pathological staging is a critical prognostic indicator for cancer patients, the study evaluated the mRNA expression of TRIB3 in stages I, II, III, IV, and X of cancer patients. The findings demonstrated a significant correlation between TRIB3 expression and tumor stage across eight types of cancer (Fig. S2), namely, ACC, BLCA, BRCA, COAD, HNSC, KIRC, KIRP, and LIHC. Additionally, TRIB3 expression was found to be higher in the later stages of most cancers. While TRIB3 expression in ESCA, LUAD, MESO, and UVM did not show a significant association with tumor staging, its expression increased along with higher stages.
3.3. High TRIB3 expression indicates poor prognosis in multiple cancer types
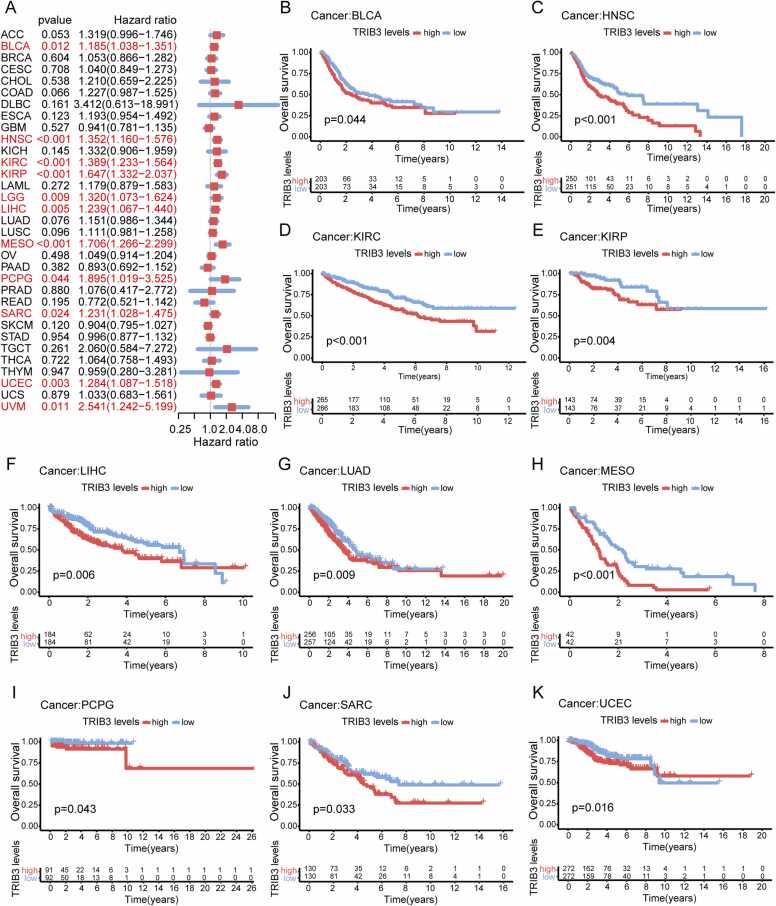
After conducting research previously on the expression level of TRIB3 and tumor pathological staging, we performed further analysis using single-factor Cox regression analysis and Kaplan—Meier survival analysis to determine the correlation between the TRIB3 expression and the prognosis of various cancer patients. Notably, this study investigated three survival indicators: OS, DSS, and PFI.
Initially, the relationship between TRIB3 expression and cancer patient OS was studied, and single-factor Cox regression analysis indicated that TRIB3 is a hazard factor for BLCA, HNSC, KIRC, KIRP, LGG, LIHC, MESO, PCPG, SARC, UCEC, and UVM patients (Fig. 2A). Kaplan—Meier survival analysis also established that high TRIB3 expression is significantly correlated with poor prognosis in BLCA, HNSC, KIRC, KIRP, LIHC, LUAD, MESO, PCPG, SARC, and UCEC patients (Fig. 2B-K).
Fig. 2.
TRIB3 expression is correlated with an unfavorable prognosis of OS in 10 types of cancer patients. (A) Forest plot depicting the results of survival analysis of TRIB3 expression on OS in pan-cancer; cancers marked in red are significant (P < 0.05). (B-K) Results of Kaplan—Meier analysis of significance between TRIB3 expression and OS (P < 0.05).
Subsequently, a single-factor Cox regression analysis of DSS data found a significant positive correlation between high TRIB3 expression and poor prognosis in ACC, HNSC, KIRC, KIRP, LGG, LIHC, MESO, PCPG, UCEC, and UVM patients (Fig. S3A). Additionally, Kaplan—Meier survival analysis of DSS data showed that high TRIB3 expression is significantly correlated with poor prognosis in ESCA, HNSC, KIRC, KIRP, MESO, PCPG, UCEC, and UVM patients (Fig. S3B-I).
Finally, investigating the association between TRIB3 expression and cancer patient PFI, the study discovered that TRIB3 expression is a hazard factor in CESC, HNSC, KICH, KIRC, KIRP, MESO, PCPG, PRAD, UCEC, and UVM patients determined via a single-factor Cox regression analysis (Fig. S4A). Kaplan—Meier survival curve analysis also showed a significant correlation between high TRIB3 expression and poor prognosis in ESCA, HNSC, KIRC, KIRP, LIHC, LUAD, MESO, PCPG, UCEC, and UVM patients (Fig. S4B-K). The results showed that high TRIB3 expression is closely related to poor prognosis of various cancers, revealing the potential of TRIB3 as a pan-cancer prognostic biomarker.
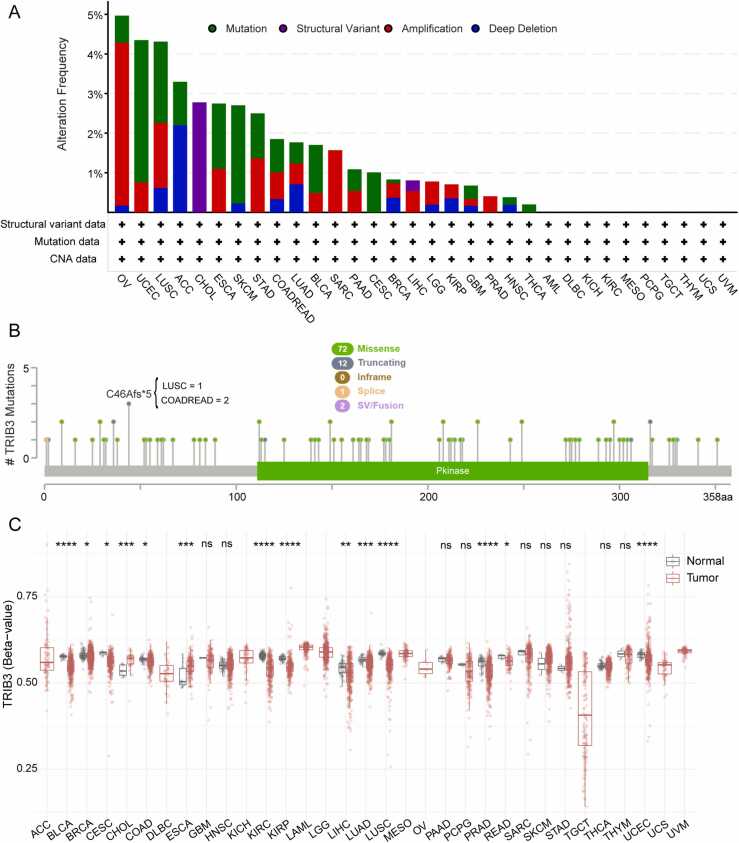
3.4. TRIB3 exhibits mutations in a wide spectrum of 22 distinct cancer types, with the predominant mutation types being missense mutations and gene amplifications
Due to the significant role of gene mutations in tumor development, we utilized the CBioPortal platform to conduct a comprehensive analysis of TRIB3 mutations in pan-cancer using data from 10,953 patients in the TCGA database. The findings revealed that missense mutations and amplifications were the primary types of TRIB3 mutations, with OV (4.97%), UCEC (4.35%), and LUSC (4.31%) being the top three mutated cancer types (Fig. 3A). Additionally, we identified 87 mutation points within amino acids 0–358, with 72 missense mutations, 12 truncating mutations, 1 splice site mutation, and 2 SV/fusion mutations mainly located in TRIB3 protein structural domains. The C46Afs* 5 mutation is the most frequent mutation site and has been found in LUSC and COADREAD cases (Fig. 3B).
Fig. 3.
Genetic alterations of TRIB3 in pan-cancer. (A) Frequency and mutation types of TRIB3 gene alterations in pan-cancer. COADREAD refers to the COAD and READ. (B) TRIB3 mutations across protein structural domains. (C) DNA methylation levels of TRIB3 in pan-cancer; *P < 0.05, * *P < 0.01, * **P < 0.001, * ** *P < 0.0001.
3.5. TRIB3 shows significant changes in DNA methylation levels in 14 types of cancer
There is mounting evidence that DNA methylation could be a promising diagnostic, prognostic, and predictive biomarker in cancer [43]. Therefore, we employed the SMART platform to examine the DNA methylation levels of TRIB3 in tumor tissues and their corresponding normal tissues in the TCGA database (Fig. 3C). The results showed significant variations in TRIB3 DNA methylation across 14 types of cancer. Notably, TRIB3 methylation levels were significantly decreased in BLCA, BRCA, CESC, COAD, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, READ, and UCEC patient tissues, while the opposite was true in CHOL and ESCA. Furthermore, the findings indicated that DNA methylation levels of TRIB3 were negatively correlated with its mRNA expression in most cancers.
3.6. The expression of TRIB3 is significantly associated with immune-related pathways in 11 types of cancers
Following above results, we conducted a deeper investigation into the biological functions of TRIB3 across different types of cancers. To this end, we utilized the LinkedOmics database to conduct gene set enrichment analysis (GSEA) on TRIB3. The results of the analysis revealed the top 10 ranked KEGG pathways in GSEA. Remarkably, we found that immune-related pathways were enriched in 11 types of cancers, including BRCA, CHOL, COADREAD, DLBC, ESCA, HNSC, LUAD, LUSC, PAAD, PRAD, and STAD (Fig. 4). Furthermore, we observed that the high expression of TRIB3 was negatively associated with immune-related pathways, such as cytokine—cytokine receptor interaction, chemokine signaling pathway, T cell receptor signaling pathway, and Th17 cell differentiation pathway. Conversely, we found that the high expression of TRIB3 was the positively enriched KEGG pathways primarily included ribosome, proteasome, cell cycle, RNA transport, and carbon metabolism. Taken together, these findings suggested potential correlation between high TRIB3 expression and immune suppression in the TIME.
Fig. 4.
TRIB3 expression in 11 cancers with GSEA-KEGG functional enrichment results significantly correlated with immune-related pathways. Top 10 GSEA functionally enriched pathways of TRIB3 in (A) BRCA, (B) CHOL, (C) COADREAD, (D) DLBC, (E) ESCA, (F) HNSC, (G) LUAD, (H) LUSC, (I) PAAD, (J) PRAD, and (K) STAD. Those marked in red are immune-related pathways (FDR ≤ 0.05). Blue represents positive enrichment, and yellow represents negative enrichment.
3.7. High TRIB3 expression indicates lower abundance of stromal and immune cells in the TME across 14 cancer types
The TME plays a critical role in promoting tumor occurrence, development, metastasis, recurrence, drug resistance, and immune suppression through complex cell-to-cell signaling [44]. Our functional enrichment analysis of TRIB3 revealed its association with immune-related signaling pathways. Therefore, it is of great significance to further investigate the pan-cancer relationship between TRIB3 expression and the TME. To do so, we used the ESTIMATE algorithm to calculate StromalScore, ImmuneScore, ESTIMATEScore, and tumor purity of TRIB3 in 33 types of cancers. According to our research findings, we observed a significant negative correlation between the expression of TRIB3 and the StromalScore, ImmuneScore, and ESTIMATEScore in 14 different types of cancers, namely, BRCA, CHOL, COAD, DLBC, ESCA, HNSC, LAML, LUAD, LUSC, PAAD, PRAD, SKCM, STAD, and THCA. However, the opposite result was observed in BLCA. Interestingly, among 15 types of cancers, including BRCA, CHOL, COAD, DLBC, ESCA, HNSC, LAML, LUAD, LUSC, PAAD, PRAD, SKCM, STAD, THCA, and UCEC, the expression of TRIB3 was significantly positively correlated with tumor purity, while the opposite result was observed in BLCA, KIRC, and TGCT (Fig. 5A). Moreover, the scatterplot provides a more detailed illustration of the correlation between TRIB3 expression and StromalScore, ImmuneScore, and tumor purity in BRCA, DLBC, and LUAD (Fig. 5B-J). Specifically, our results suggested that the proportions of immune cells and stromal cells are lower in most tumors with high TRIB3 expression.
Fig. 5.
Elevated TRIB3 expression is linked to reduced levels of stromal and immune cells in the TME across 14 cancer types. (A) Heatmap showing the correlation between TRIB3 in pan-cancer and StromalScore, ImmuneScore, ESTIMATEScore and tumor purity; red represents positive correlation, and blue represents negative correlation. (B-J) Specific correlation plots between TRIB3 expression in BRCA, DLBC, LUAD and StromalScore, ImmuneScore, tumor purity; * P < 0.05, * * P < 0.01, * ** P < 0.001.
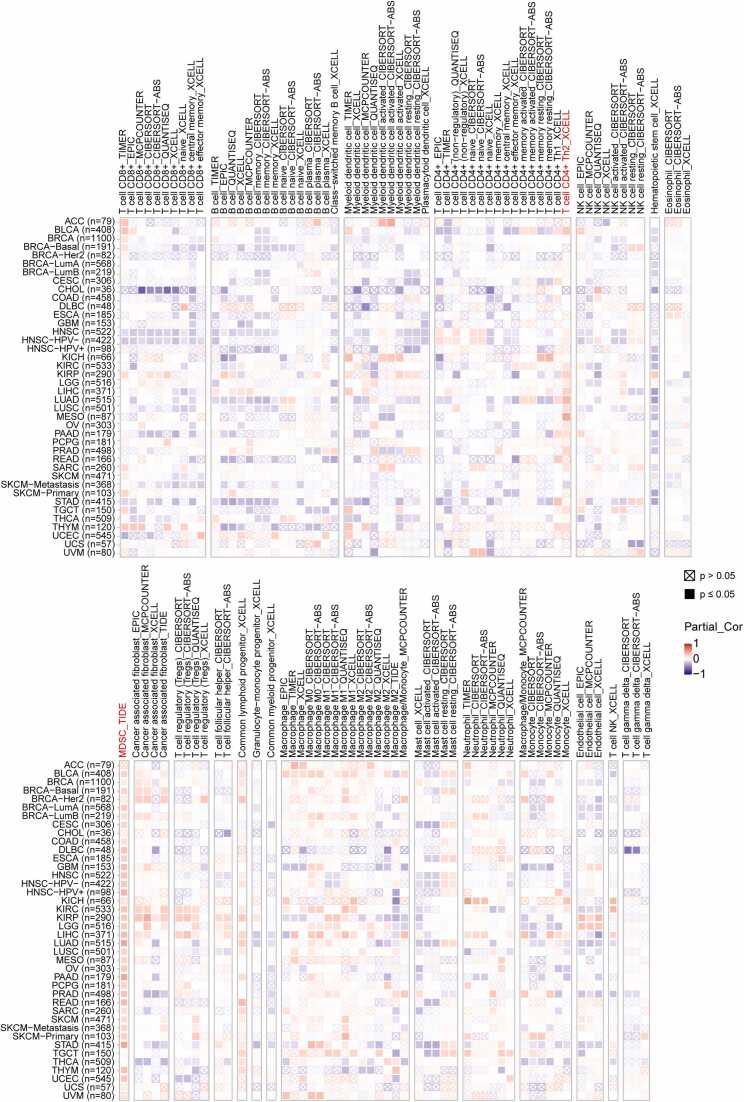
3.8. TRIB3 expression is positively associated with the infiltration of immunosuppressive cells, such as CD4+ Th2 cells and myeloid-derived suppressor cells (MDSCs), in the majority of cancer types
The TIME refers to all the immune components within the TME that have been shown to play crucial roles in tumor immune suppression, metastasis, recurrence, and resistance [45]. Previous research has indicated that the expression of TRIB3 is closely related to the TME. To investigate this further, we used the TIMER 2.0 database, which employs multiple immune prediction methods, to evaluate the correlation between TRIB3 expression and immune infiltrating cells. Our results demonstrate that TRIB3 expression was positively correlated with most MDSCs, CD4+ Th2 cells, M0 macrophages, resting mast cells, and common lymphoid progenitors in most tumors, whereas TRIB3 expression was negatively correlated with CD8+ T cells, B cells, and hematopoietic stem cells in most cancers (Fig. 6). The above results provided further evidence that high expression of TRIB3 promotes the formation of an immunosuppressive TME.
Fig. 6.
TRIB3 expression positively correlates with recruitment of immunosuppressive cells, such as CD4+ Th2 cells and MDSCs, in most cancer types, while showing a negative correlation with CD8+ T cell infiltration in certain cancers. Orange color indicates significant positive correlation and purple color indicates significant negative correlation.
3.9. The expression of TRIB3 is significantly correlated with immune-related genes in multiple types of cancer
We conducted gene co-expression analysis to explore the relationship between TRIB3 expression and the expression of genes encoding MHC, immunostimulator, immunoinhibitor, chemokines, and chemokine receptor proteins. Our findings indicate that TRIB3 is significantly correlated with most immunostimulator genes in various cancers. Specifically, TRIB3 expression was negatively correlated in BRCA, CHOL, COAD, ESCA, HNSC, LUAD, LUSC, PAAD, PRAD, SKCM, STAD, and THCA, while it was significantly positively correlated in BLCA, KIRC, and KIRP. Interestingly, TRIB3 expression was significantly positively correlated with the expression of the ULBP1, PVR, and CD276 genes in almost all cancers (Fig. S5A). Additionally, TRIB3 expression was negatively correlated with immunoinhibitor genes, particularly in BRCA, COAD, ESCA, HNSC, LUSC, PAAD, PRAD, SKCM, STAD, and THCA. In contrast, TRIB3 expression was positively correlated with immunoinhibitor in BLCA, KIRC, KIRP, and TGCT (Fig. S5B). With respect to chemokine receptors, TRIB3 expression was negatively correlated in BRCA, CHOL, COAD, HNSC, LUAD, LUSC, PAAD, PRAD, SKCM, STAD, and THCA (Fig. S5C) and positively correlated with most chemokines in most cancers, except for CHOL, PAAD, and PRAD (Fig. S5D). Finally, TRIB3 expression was positively correlated with almost all MHC genes in BLCA and TGCT, while it was negatively correlated with most MHC genes in BRCA, COAD, ESCA, LUAD, LUSC, PAAD, PRAD, SKCM and STAD (Fig. S5E). Taken together, these findings suggested that TRIB3 might be involved in regulating the biological functions of immune cell infiltration and various immune-related genes.
3.10. The expression of TRIB3 is significantly correlated with cancer immune-related TMB, MSI, and MMR in multiple types of tumors
Many studies have shown that patients with MSI-H and TMB-H tumors have a higher response rate to immunotherapy [41], [42]. Thus, we investigated the relationship between TMB, MSI, and TRIB3 expression across cancers. TRIB3 expression was significantly correlated with TMB in 18 types of cancer, with positive correlations observed in BLCA, BRCA, HNSC, KICH, KIRC, LGG, LUAD, LUSC, OV, PAAD, PCPG, PRAD, SARC, SKCM, STAD, and UCEC and negative correlations in COAD and KIRP (Fig. 7A). Additionally, we found that TRIB3 expression was significantly positively correlated with MSI in 6 types of cancer, including BLCA, HNSC, KICH, PRAD, STAD, and UCEC, but significantly negatively correlated with MSI in COAD, LAML, and TGCT (Fig. 7B).
Fig. 7.
The expression of TRIB3 is significantly associated with TMB, MSI, and MMR in multiple types of cancer. (A) Significant positive correlation between TRIB3 expression and TMB in 16 cancer types (marked in red) and significant negative correlation in 2 cancer types (marked in blue). (B) Significant positive correlation between TRIB3 expression and MSI in 6 cancer types (marked in red) and significant negative correlation in 3 cancer types (marked in blue). (C) Heat map of the correlation between TRIB3 expression and MMR genes; red represents positive correlation, and blue represents negative correlation; *P < 0.05, * *P < 0.01, * **P < 0.001.
Moreover, MMR can repair base mismatches during DNA replication to maintain genomic stability, and MMR deficiency can generate new antigens, increasing the sensitivity of tumor patients to immunotherapy [42]. Therefore, we evaluated the correlation between TRIB3 and MMR genes, including MLH1, MSH2, MSH6, PMS2, and EPCAM. The results showed that TRIB3 expression was closely related to MMR genes in 12 types of cancer, including BLCA, BRCA, COAD, ESCA, KIRC, KIRP, LGG, LUAD, LUSC, SARC, STAD, and UCEC (Fig. 7C). Overall, these findings suggested that TRIB3 is closely related to various tumors and may be a potential immune biomarker for indicating the immune therapy response of these tumor types.
3.11. Loss-of-function TRIB3 inhibits the proliferation and promotes the apoptosis of breast cancer cells
Due to the limited research on the TRIB3 gene in breast cancer, some studies have yielded conflicting results. For instance, while increase in TRIB3 mRNA has been linked to a poorer prognosis [46], other studies have found that high TRIB3 protein expression is associated with a better prognosis in luminal breast cancer [47]. Although, the expression of TRIB3 was not significantly correlated with the prognosis of BRCA in TCGA, we found that high TRIB3 expression was significantly associated with a poor OS prognosis in breast cancer patients in METABRIC database (Fig. 8A). Furthermore, several studies have indicated that TRIB3 contributes to the advancement of TNBC [48], [49]. So, we speculated that TRIB3 could be a potential target for TNBC. To this end, we delved deeper into the influence of TRIB3 on the proliferation and apoptosis of TNBC cells. To accomplish this, we transfected HCC1806 and MDA-MB-231 cells with five distinct siRNAs targeting TRIB3. The results indicate that siTRIB3-2, siTRIB3-3, siTRIB3-4, and siTRIB3-5 achieved a higher gene knockout efficiency (Fig. 8B-E, Fig. S6A-B). Subsequently, we chose these four siRNAs for cloning formation experiments and apoptosis experiments. The findings demonstrated a significant reduction in the proliferation capacity of HCC1806 and MDA-MB-231 cells as a result of TRIB3 function loss (Fig. 8F-I, Fig. S6C-F). Additionally, this loss-of-function of TRIB3 induces apoptosis in HCC1806 cells (Fig. 8J-K). These observations furnished compelling evidence highlighting the oncogenic role of TRIB3 in breast cancer.
Fig. 8.
Loss-of-function TRIB3 in breast cancer cells significantly suppresses cell proliferation and induces apoptosis. (A) METABRIC data analysis showed that patients with high TRIB3 expression had a worse prognosis for OS. (B-C) The knockdown efficiency of TRIB3 mRNA in HCC1806 and MDA-MB-231 cells. (D-E) The knockdown efficiency of TRIB3 protein levels in HCC1806 and MDA-MB-231 cells. (F-I) Silencing of TRIB3 significantly inhibited the proliferation of HCC1806 and MDA-MB-231 cells. (J-K) Silencing TRIB3 can significantly induce apoptosis in HCC1806 cells. * P < 0.05, * * P < 0.01, * ** P < 0.001, * ** * P < 0.0001.
3.12. The functional deficiency of TRIB3 in breast cancer cells might reverse the immunosuppressive TME
Differential expression analysis was performed on the mRNA expression profile derived from TRIB3 silencing in MCF-7 cells, confirming the significant downregulation of TRIB3 and indicating the high quality of the GSE116671 dataset (Fig. 9A). The KEGG enrichment analysis demonstrated significant enrichment of "hot" tumor-related pathways, such as the chemokine signaling pathway and the T-cell receptor signaling pathway (Fig. 9B). Notably, the GSEA results also exhibited a similar trend with positive enrichments in the T cell receptor signaling pathway (Fig. 9C-F) and cytokine signaling in immune system (Fig. S6G-J). Together, these results further supported the role of TRIB3 in fostering the formation of an immunosuppressive tumor microenvironment.
Fig. 9.
Loss-of-function TRIB3 in breast cancer cells is associated with a significant enrichment of multiple immune-related pathways. (A) The volcano plot depicts the differential expression of genes (DEGs) in TRIB3-silenced MCF-7 breast cancer cells, where upregulated genes are denoted in red and downregulated genes in blue. (B) The DEGs were enriched in immune-related pathways as identified by KEGG analysis. (C-F) The GSEA-KEGG enrichment analysis of TRIB3-silenced MCF-7 cells demonstrated a positive enrichment of immune-related pathways.
4. Discussion
Through comprehensive pan-cancer analysis, our study revealed that TRIB3 exhibited elevated mRNA expression levels in as many as 24 different cancers. This abnormal expression pattern was influenced by gene mutations and DNA methylation and showed a significant correlation with poor prognosis across multiple cancers. Additionally, we performed functional enrichment analysis of TRIB3 and found a significant enrichment of immune-related pathways in 11 cancers. Notably, we observed associations between the expression of TRIB3 and the recruitment of immunosuppressive cells such as CD4+ Th2 cells and MDSCs. Of particular interest, in breast cancer, our study revealed that loss-of-function of TRIB3 inhibits cell proliferation, promotes apoptosis, and leads to the significant enrichment of signaling pathways associated with "hot" tumors. These findings suggested that high TRIB3 expression might reshape the TIME, thereby impeding the transition of "cold" tumors to "hot" tumors. Hence, our research revealed the latent potential of TRIB3 as a prognostic biomarker, and a target for pharmaceutical development and immunotherapeutic interventions.
Genetic mutations and aberrant DNA methylation are known to contribute to dysregulated gene expression in cancer, thereby impacting the onset and progression of cancer [50], [51]. The utilization of gene expression profiling data offers a powerful means to investigate the clinical prognosis of genes in cancer patients. Our study reveals consistent and significant overexpression of TRIB3 in almost all types of cancer, corroborating previous findings. This widespread upregulation of TRIB3 suggests its potential significance across multiple cancer types, prompting us to further investigate its clinical prognostic implications. Through rigorous analysis of clinical prognosis data, we identified TRIB3 as an unfavorable prognostic factor in diverse cancers.
Currently, the molecular mechanisms of TRIB3 have primarily been investigated in specific cancer types, and its role in pan-cancer remains unclear. Previous studies have suggested that TRIB3 functions as an adapter/scaffold protein for various proteins, mediating interactions with transcription factors, ubiquitin ligases, and kinase-dependent proteins to regulate diverse signaling pathways, including MAPK, PI3K/AKT, NF-κB, and TGF-β [19]. However, the precise involvement of TRIB3 in pan-cancer contexts remains incompletely understood. Notably, pathway enrichment analyses based on GSEA-KEGG in pan-cancer have demonstrated a negative enrichment of immune-related pathways, such as cytokine—cytokine receptor interaction and chemokine signaling pathways. Consistently, functional enrichment analysis of mRNA expression profile from TRIB3 silencing experiments in MCF-7 cells revealed enrichment of immune-related pathways, including the chemokine signaling pathway and T cell receptor signaling pathway, thereby further confirming the immunomodulatory properties of TRIB3. Remarkably, it has been reported that TRIB3 inhibits the release of macrophage cytokines [52] and can also suppress CXCL10 expression by inhibiting STAT1 activation [53]. Consistent with previous findings, our study's results corroborate the notion that TRIB3 exerts immunoregulatory effects by modulating the release of cytokines and chemokines.
Tumor immunotherapy has shown significant efficacy in various solid tumors and is considered a groundbreaking advancement. However, its clinical response rates remain relatively low [54]. Therefore, the need to identify more effective targets for immune-based therapies is urgent. Numerous studies have indicated that immune cells present in the TME can either promote or inhibit tumor immune responses, thus representing a dual-edged sword in tumor development [55], [56]. The composition and activity of immune cells within the TME play a critical role in determining the response to immunotherapy [57]. Prior investigations have demonstrated that cytotoxic CD8+ T lymphocytes (CTLs), CD4+ T helper 1 cells (Th1 cells), and natural killer (NK) cells predominantly exert antitumor effects, while CD4+ Th2 cells and MDSCs are often associated with immune-suppressive microenvironment formation [58], [59].
Our study exploring the relationship between TRIB3 expression and immune cell infiltration reveals a positive correlation between TRIB3 expression and MDSCs, CD4+ Th2 cells, macrophages of the M0 phenotype, and resting mast cells. Conversely, TRIB3 expression exhibits a negative correlation with CD8+ T cells and B cells in multiple cancer types. Notably, existing research has demonstrated that overexpression of TRIB3 inhibits cytokine release from macrophages [52] and contributes to the survival and vitality of bone marrow-derived mast cells [60]. Furthermore, single-cell transcriptome analysis has indicated elevated expression of TRIB3 in CD4+ Th2 cells, highlighting its high specificity [61]. Another study confirms that TRIB3 impedes CD8+ T-cell infiltration and induces immune evasion by suppressing the release of the chemokine CXCL1053. Consistent with previous reports, our results indicate that TRIB3, through its regulation of chemokine and cytokine release, not only hinders the infiltration of CD8+ T cells but also potentially promotes the recruitment of MDSCs and CD4+ Th2 cells into the TME, thereby contributing to the establishment of an immunosuppressive microenvironment.
Due to the limited research on TRIB3 in breast cancer and the contradictory conclusions, the role of TRIB3 in breast cancer remains complex and poorly understood. While some studies have associated high TRIB3 mRNA expression with poor prognosis in breast cancer patients [46], others have found that elevated TRIB3 protein levels are linked to a favorable prognosis [30]. Additionally, the effects of TRIB3 in breast cancer are further complicated by conflicting findings, such as enhanced cloning formation ability in breast cancer cells (BT474) following TRIB3 knockdown [62], while TRIB3 gene silencing suppresses the growth of MDA-MB-231 xenograft tumors [48] and inhibits proliferation in radiation-resistant MDA-MB-231 cells [49]. Consequently, we conducted cloning formation assays and apoptosis assays, which further confirmed that TRIB3 silencing strongly inhibits cell proliferation in the TNBC cell lines, HCC1806 and MDA-MB-231. Additionally, this loss of function induces apoptosis in HCC1806 cells. Our research results align with the conclusions from previous studies on TRIB3 in TNBC, which leads us to believe that TRIB3 plays an oncogenic role in breast cancer, particularly in TNBC.
Encouragingly, recent research has demonstrated that TRIB3 expression is suppressed by knocking down P300 or treating 4T1 murine breast cancer cells with C646, a P300 inhibitor. Moreover, the combined treatment of C646 and PD-L1 antibody in situ breast cancer models (4T1) showed partial inhibition of tumor progression, with significantly enhanced antitumor efficacy compared to using either treatment alone. Furthermore, treatment with C646, either alone or in combination, significantly increased the infiltration of CD8+ T cells [53]. These findings suggest that targeting TRIB3 is a crucial strategy for converting "cold tumors" into "hot tumors", particularly in the case of TNBC, where effective therapeutic targets are lacking. These findings underscore the potential of TRIB3 as a promising immunotherapeutic target in TNBC. Notably, studies have shown that TRIB3 gene knockout mice do not exhibit significant phenotypic differences compared to wild-type mice, indicating that the gene may not be essential [63]. However, this observation also suggests that TRIB3 represents a highly promising target that can be selectively modulated for therapeutic purposes.
Although this study systematically revealed the relationship between TRIB3 and various types of cancer, it still has some limitations. The research primarily relies on bioinformatics analysis, while TRIB3 is closely associated with multiple cellular signaling pathways. Although there are reports suggesting that TRIB3 inhibits immune infiltration of CD8+ T cells and exhibits high expression and specificity in CD4+ Th2 cells, the precise biological functions of TRIB3 in T cells and its specific relationship with CD4+ Th2 cells have not been fully elucidated. Additionally, there is a lack of research linking TRIB3 to MDSCs. Therefore, further investigations using basic experimental approaches are needed to clarify the biological functions and molecular mechanisms of TRIB3 in tumor immunity. Moreover, due to the absence of a crystalline protein structure for TRIB3, we plan to employ gene expression profiling after drug perturbation to identify small molecule compounds that can effectively inhibit the functional activity of TRIB3.
5. Conclusion
Our research reveals that TRIB3 is significantly overexpressed in 24 different cancer types and is closely associated with adverse prognosis. Additionally, TRIB3 appears to play a critical role in orchestrating the infiltration of immune cells into the tumor microenvironment. It promotes the recruitment of MDSCs and CD4+ Th2 cells by regulating chemokines and cytokines, while concurrently suppressing the infiltration of CD8+ T cells. This intricate modulation might ultimately contribute to the formation of an immunologically "cold" tumor phenotype, facilitating tumor immune escape. These findings provide valuable insights into the potential of TRIB3 as a target for drug development and immunotherapy, as well as its promising prospects as a prognostic biomarker.
Author Statement
Chao Hu, Yumei Wang, and Dong Wang contributed to the conception of the study. Data collection, analysis, and figure generation were performed by Chao Hu, Lei Xiang, Shengrong Li, Xiankuo Yu, and Guochen Zhang. Experiments were conducted by Qingzhou Li, Jun An, Yan Luo, Yuhui Chen. The manuscript was drafted by Chao Hu. Chao Hu, Yumei Wang, and Dong Wang revised the manuscript. All authors made contributions to the article and have approved the final submitted version.
Declaration of Competing Interest
The authors confirm that they have no known conflicting financial interests or personal associations that might have influenced the findings presented in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 82172723), Science and Technology Department of Sichuan Province (No. 2021ZYD0079 and No. 2023NSFSC1828), and Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (No. ZYYCXTD-D-202209).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.11.043.
Contributor Information
Yumei Wang, Email: yumeiwang@cdutcm.edu.cn.
Dong Wang, Email: dwang@cdutcm.edu.cn.
Appendix A. Supplementary material
Figure S1. The immunohistochemistry (IHC) results obtained from the HPA database indicate that the expression of TRIB3 protein is higher in tumors (right) than in normal tissues (left). (A) BRCA, (B) COAD, (C) HNSC, (D) LIHC, (E) SKCM. The numbers in the picture are patient ID numbers.
.
Figure S2. The expression of TRIB3 is higher in the late clinical stages of 8 cancers. (A) ACC, (B) BLCA, (C) BRCA, (D) COAD, (E) ESCA, (F) HNSC, (G) KIRC, (H) KIRP, (I) LIHC, (J) LUAD, (K) MESO, (L) UVM.
.
Figure S3.TRIB3 expression is significantly correlated with an unfavorable prognosis of DSS in 8 types of cancer patients. (A) Forest plot depicting the results of survival analysis for TRIB3 expression on DSS in pan-cancer; cancers marked in red are significant (P < 0.05). (B-I) Results of Kaplan–Meier analysis of significance between TRIB3 expression and DSS (P < 0.05).
.
Figure S4. The expression of TRIB3 is significantly associated with poor prognosis of PFI in 10 types of cancer patients. (A) Forest plot depicting the results of survival analysis of TRIB3 expression on PFI in pan-cancer; cancers marked in red are significant (P < 0.05). (B-K) Results of Kaplan–Meier analysis of significance between TRIB3 expression and PFI (P < 0.05).
.
Figure S5.TRIB3 expression is significantly correlated with the majority of immune-related genes in various types of tumors. The co-expression relationship between TRIB3 and (A) Immunostimulator genes, (B) Immunoinhibitor genes, (C) Chemokine receptors, (D) Chemokines, and (E) MHC genes in pan-cancer; *P < 0.05, * *P < 0.01, * **P < 0.001. Red represents positive correlation, and blue represents negative correlation.
.
Figure S6. The silencing of TRIB3 in breast cancer cells leads to a substantial reduction in cell proliferation and a significant positive enrichment of immune-related signaling pathways. (A-B) The knockdown efficiency of TRIB3 mRNA in MDA-MB-231 and HCC1806 cells. (C-F) Silencing of TRIB3 significantly inhibited the proliferation of HCC1806 and MDA-MB-231 cells cloning formation ability. * P < 0.05, * * P < 0.01, * ** P < 0.001, * ** * P < 0.0001. (G-J) The GSEA-Reactome enrichment analysis indicates a significant enrichment of immune-related signaling pathways in TRIB3-silenced MCF-7 breast cancer cells; NES, normalized enrichment score.
.
References
- 1.Sung H., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Miller K.D., et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409–436. doi: 10.3322/caac.21731. [DOI] [PubMed] [Google Scholar]
- 3.DeVita V.T., Chu E. A history of cancer chemotherapy. Cancer Res. 2008;68:8643–8653. doi: 10.1158/0008-5472.CAN-07-6611. [DOI] [PubMed] [Google Scholar]
- 4.Zhu S., et al. Combination strategies to maximize the benefits of cancer immunotherapy. J Hematol Oncol. 2021;14 doi: 10.1186/s13045-021-01164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meric-Bernstam F., Larkin J., Tabernero J., Bonini C. Enhancing anti-tumour efficacy with immunotherapy combinations. Lancet. 2021;397:1010–1022. doi: 10.1016/S0140-6736(20)32598-8. [DOI] [PubMed] [Google Scholar]
- 6.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Huang D., Saw P.E., Song E. Turning cold tumors hot: from molecular mechanisms to clinical applications. Trends Immunol. 2022;43:523–545. doi: 10.1016/j.it.2022.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Stefanovska B., André F., Fromigué O. Tribbles pseudokinase 3 regulation and contribution to cancer. Cancers (Basel) 2021;13:1822. doi: 10.3390/cancers13081822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liew C.W., et al. The pseudokinase tribbles homolog 3 interacts with ATF4 to negatively regulate insulin exocytosis in human and mouse beta cells. J Clin Invest. 2010;120:2876–2888. doi: 10.1172/JCI36849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohoka N., Hattori T., Kitagawa M., Onozaki K., Hayashi H. Critical and functional regulation of CHOP (C/EBP homologous protein) through the N-terminal portion. J Biol Chem. 2007;282:35687–35694. doi: 10.1074/jbc.M703735200. [DOI] [PubMed] [Google Scholar]
- 11.Qi L., et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312:1763–1766. doi: 10.1126/science.1123374. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama T., et al. Trib1 links the MEK1/ERK pathway in myeloid leukemogenesis. Blood. 2010;116:2768–2775. doi: 10.1182/blood-2009-10-246264. [DOI] [PubMed] [Google Scholar]
- 13.Kiss-Toth E., et al. Human Tribbles, a Protein Family Controlling Mitogen-activated Protein Kinase Cascades. J Biol Chem. 2004;279:42703–42708. doi: 10.1074/jbc.M407732200. [DOI] [PubMed] [Google Scholar]
- 14.Du K., Herzig S., Kulkarni R.N., Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi Y., Ohoka N., Hayashi H., Sato R. TRB3 suppresses adipocyte differentiation by negatively regulating PPARgamma transcriptional activity. J Lipid Res. 2008;49:880–892. doi: 10.1194/jlr.M700545-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y., et al. E3 ubiquitin ligase SIAH1 mediates ubiquitination and degradation of TRB3. Cell Signal. 2008;20:942–948. doi: 10.1016/j.cellsig.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Yu J.-J., et al. TRIB3-EGFR interaction promotes lung cancer progression and defines a therapeutic target. Nat Commun. 2020;11 doi: 10.1038/s41467-020-17385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua F., et al. TRIB3 interacts with β-catenin and TCF4 to increase stem cell features of colorectal cancer stem cells and tumorigenesis. Gastroenterology. 2019;156:708–721.e15. doi: 10.1053/j.gastro.2018.10.031. [DOI] [PubMed] [Google Scholar]
- 19.Arif A., et al. The functions and molecular mechanisms of Tribbles homolog 3 (TRIB3) implicated in the pathophysiology of cancer. Int Immunopharmacol. 2023;114 doi: 10.1016/j.intimp.2022.109581. [DOI] [PubMed] [Google Scholar]
- 20.Salazar M., et al. Oncosuppressive functions of tribbles pseudokinase 3. Biochem Soc Trans. 2015;43:1122–1126. doi: 10.1042/BST20150124. [DOI] [PubMed] [Google Scholar]
- 21.Yu J., et al. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat Commun. 2019;10 doi: 10.1038/s41467-019-13700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R.-Q., et al. Tribbles pseudokinase 3 (TRIB3) contributes to the progression of hepatocellular carcinoma by activating the mitogen-activated protein kinase pathway. Ann Transl Med. 2021;9:1253. doi: 10.21037/atm-21-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W., Ma J., Meng L., Liu D., Chen J. Deletion of TRIB3 disrupts the tumor progression induced by integrin αvβ3 in lung cancer. BMC Cancer. 2022;22:459. doi: 10.1186/s12885-022-09593-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z., et al. TRIB3 facilitates glioblastoma progression via restraining autophagy. Aging (Albany NY) 2020;12:25020–25034. doi: 10.18632/aging.103969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S., et al. Down-regulation of TRIB3 inhibits the progression of ovarian cancer via MEK/ERK signaling pathway. Cancer Cell Int. 2020;20 doi: 10.1186/s12935-020-01509-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen P., Zhang T.-Y., Wang S.-Y. TRIB3 promotes oral squamous cell carcinoma cell proliferation by activating the AKT signaling pathway. Exp Ther Med. 2021;21:313. doi: 10.3892/etm.2021.9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong S., et al. Overexpression of TRIB3 promotes angiogenesis in human gastric cancer. Oncol Rep. 2016;36:2339–2348. doi: 10.3892/or.2016.5017. [DOI] [PubMed] [Google Scholar]
- 28.Miyoshi N., et al. Abnormal expression of TRIB3 in colorectal cancer: a novel marker for prognosis. Br J Cancer. 2009;101:1664–1670. doi: 10.1038/sj.bjc.6605361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J., et al. TRIB3 promotes the malignant progression of bladder cancer: an integrated analysis of bioinformatics and in vitro experiments. Front Genet. 2021;12 doi: 10.3389/fgene.2021.649208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wennemers M., Bussink J., Grebenchtchikov N., Sweep F.C.G.J., Span P.N. TRIB3 protein denotes a good prognosis in breast cancer patients and is associated with hypoxia sensitivity. Radio Oncol. 2011;101:198–202. doi: 10.1016/j.radonc.2011.05.057. [DOI] [PubMed] [Google Scholar]
- 31.Li Y., et al. TRB3 reverses chemotherapy resistance and mediates crosstalk between endoplasmic reticulum stress and AKT signaling pathways in MHCC97H human hepatocellular carcinoma cells. Oncol Lett. 2018;15:1343–1349. doi: 10.3892/ol.2017.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldman M.J., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerami E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett T., et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.T L., et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48 doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Z T., B K., C L., T C., Z Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;(47) doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlen M., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Ge D., Lu C. The SMART App: an interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin. 2019;12:71. doi: 10.1186/s13072-019-0316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasaikar S.V., Straub P., Wang J., Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshihara K., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4 doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan T.A., et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi M., et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17 doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch A., et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15:459–466. doi: 10.1038/s41571-018-0004-4. [DOI] [PubMed] [Google Scholar]
- 44.Anderson N.M., Simon M.C. The tumor microenvironment. Curr Biol. 2020;30:R921–R925. doi: 10.1016/j.cub.2020.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu T., et al. Spatial architecture of the immune microenvironment orchestrates tumor immunity and therapeutic response. J Hematol Oncol. 2021;14 doi: 10.1186/s13045-021-01103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wennemers M., et al. Tribbles homolog 3 denotes a poor prognosis in breast cancer and is involved in hypoxia response. Breast Cancer Res. 2011;13 doi: 10.1186/bcr2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orea-Soufi A., et al. The Pseudokinase TRIB3 negatively regulates the HER2 receptor pathway and is a biomarker of good prognosis in luminal breast cancer. Cancers (Basel) 2021;13:5307. doi: 10.3390/cancers13215307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Izrailit J., Berman H.K., Datti A., Wrana J.L., Reedijk M. High throughput kinase inhibitor screens reveal TRB3 and MAPK-ERK/TGFβ pathways as fundamental Notch regulators in breast cancer. Proc Natl Acad Sci USA. 2013;110:1714–1719. doi: 10.1073/pnas.1214014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee Y.-C., et al. Tribbles homolog 3 involved in radiation response of triple negative breast cancer cells by regulating notch1 activation. Cancers (Basel) 2019;11:127. doi: 10.3390/cancers11020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endicott J.L., Nolte P.A., Shen H., Laird P.W. Cell division drives DNA methylation loss in late-replicating domains in primary human cells. Nat Commun. 2022;13:6659. doi: 10.1038/s41467-022-34268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishiyama A., Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021;37:1012–1027. doi: 10.1016/j.tig.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Steverson D., et al. Tribbles homolog 3 promotes foam cell formation associated with decreased proinflammatory cytokine production in macrophages: evidence for reciprocal regulation of cholesterol uptake and inflammation. Metab Syndr Relat Disord. 2016;14:7–15. doi: 10.1089/met.2015.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shang S., et al. TRIB3 reduces CD8+ T cell infiltration and induces immune evasion by repressing the STAT1-CXCL10 axis in colorectal cancer. Sci Transl Med. 2022;14 doi: 10.1126/scitranslmed.abf0992. [DOI] [PubMed] [Google Scholar]
- 54.Hiam-Galvez K.J., Allen B.M., Spitzer M.H. Systemic immunity in cancer. Nat Rev Cancer. 2021;21:345–359. doi: 10.1038/s41568-021-00347-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Locy H., et al. Immunomodulation of the tumor microenvironment: turn foe into friend. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.02909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LV B., et al. Immunotherapy: reshape the tumor immune microenvironment. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.844142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petitprez F., Meylan M., de Reyniès A., Sautès-Fridman C., Fridman W.H. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front Immunol. 2020;11:784. doi: 10.3389/fimmu.2020.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Basu A., et al. Differentiation and regulation of TH cells: a balancing act for cancer immunotherapy. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.669474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Q., et al. CCL5-mediated Th2 immune polarization promotes metastasis in luminal breast cancer. Cancer Res. 2015;75:4312–4321. doi: 10.1158/0008-5472.CAN-14-3590. [DOI] [PubMed] [Google Scholar]
- 60.Ord T., Ord D., Kuuse S., Plaas M., Ord T. Trib3 is regulated by IL-3 and affects bone marrow-derived mast cell survival and function. Cell Immunol. 2012;280:68–75. doi: 10.1016/j.cellimm.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Jaroušek R., et al. Single-cell RNA sequencing analysis of T helper cell differentiation and heterogeneity. Biochim Biophys Acta Mol Cell Res. 2022;1869 doi: 10.1016/j.bbamcr.2022.119321. [DOI] [PubMed] [Google Scholar]
- 62.Salazar M., et al. Loss of tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ. 2015;22:131–144. doi: 10.1038/cdd.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okamoto H., et al. Genetic deletion of Trb3, the mammalian Drosophila tribbles homolog, displays normal hepatic insulin signaling and glucose homeostasis. Diabetes. 2007;56:1350–1356. doi: 10.2337/db06-1448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The immunohistochemistry (IHC) results obtained from the HPA database indicate that the expression of TRIB3 protein is higher in tumors (right) than in normal tissues (left). (A) BRCA, (B) COAD, (C) HNSC, (D) LIHC, (E) SKCM. The numbers in the picture are patient ID numbers.
Figure S2. The expression of TRIB3 is higher in the late clinical stages of 8 cancers. (A) ACC, (B) BLCA, (C) BRCA, (D) COAD, (E) ESCA, (F) HNSC, (G) KIRC, (H) KIRP, (I) LIHC, (J) LUAD, (K) MESO, (L) UVM.
Figure S3.TRIB3 expression is significantly correlated with an unfavorable prognosis of DSS in 8 types of cancer patients. (A) Forest plot depicting the results of survival analysis for TRIB3 expression on DSS in pan-cancer; cancers marked in red are significant (P < 0.05). (B-I) Results of Kaplan–Meier analysis of significance between TRIB3 expression and DSS (P < 0.05).
Figure S4. The expression of TRIB3 is significantly associated with poor prognosis of PFI in 10 types of cancer patients. (A) Forest plot depicting the results of survival analysis of TRIB3 expression on PFI in pan-cancer; cancers marked in red are significant (P < 0.05). (B-K) Results of Kaplan–Meier analysis of significance between TRIB3 expression and PFI (P < 0.05).
Figure S5.TRIB3 expression is significantly correlated with the majority of immune-related genes in various types of tumors. The co-expression relationship between TRIB3 and (A) Immunostimulator genes, (B) Immunoinhibitor genes, (C) Chemokine receptors, (D) Chemokines, and (E) MHC genes in pan-cancer; *P < 0.05, * *P < 0.01, * **P < 0.001. Red represents positive correlation, and blue represents negative correlation.
Figure S6. The silencing of TRIB3 in breast cancer cells leads to a substantial reduction in cell proliferation and a significant positive enrichment of immune-related signaling pathways. (A-B) The knockdown efficiency of TRIB3 mRNA in MDA-MB-231 and HCC1806 cells. (C-F) Silencing of TRIB3 significantly inhibited the proliferation of HCC1806 and MDA-MB-231 cells cloning formation ability. * P < 0.05, * * P < 0.01, * ** P < 0.001, * ** * P < 0.0001. (G-J) The GSEA-Reactome enrichment analysis indicates a significant enrichment of immune-related signaling pathways in TRIB3-silenced MCF-7 breast cancer cells; NES, normalized enrichment score.