Abstract
Previous results have indicated that the herpes simplex virus 1 UL31 and UL34 proteins interact and form a complex at the inner nuclear membranes of infected cells, where both play important roles in the envelopment of nucleocapsids at the inner nuclear membrane. In the work described here, mapping studies using glutathione S-transferase pull-down assays indicated that amino acids 137 to 181 of the UL34 protein are sufficient to mediate an interaction with the UL31 protein. A recombinant virus (v3480) lacking UL34 codons 138 to 181 was constructed. Similar to a UL34 null virus, v3480 failed to replicate on Vero cells and grew to a limited extent on rabbit skin cells. A UL34-expressing cell line restored v3480 growth and plaque formation. Similar to the localization of UL31 protein in cells infected with a UL34 null virus, the UL31 protein was present in the nuclei of Hep2 cells infected with v3480. Hep2 cells infected with v3480 contained the UL34 protein in the cytoplasm, the nucleus, and the nuclear membrane, and this was noted to be similar to the appearance of cells infected with a UL31 null virus. In transient expression assays, the interaction between UL34 amino acids 137 to 181 and the UL31 protein was sufficiently robust to target green fluorescent protein and emerin to intranuclear sites that contained the UL31 protein. These data indicate that amino acids 137 to 181 of the UL34 protein are (i) sufficient to mediate interactions with the UL31 protein in vitro and in vivo, (ii) necessary for the colocalization of UL31 and UL34 in infected cells, and (iii) essential for normal viral replication.
Herpes simplex virus (HSV) nucleocapsids bud from the inner nuclear membrane (INM) to form nascent virions in a reaction termed primary envelopment. Recent studies have suggested that HSV type 1 (HSV-1) particles lose this envelope and acquire a secondary envelope in the cytoplasm (10, 13, 15, 22, 23, 28, 29, 35).
Various HSV gene products are required for primary envelopment under different circumstances. For example, the UL31 and UL34 proteins are required for envelopment in Vero cells and Hep2 cells, UL11 facilitates (but is dispensable for) the process in most cell types, and gK plays a major role in envelopment only in quiescent cells (1, 17, 21, 24, 28, 31). Both the gK and UL11 proteins are also required for the efficient movement of viral particles through the cytoplasm (1, 16).
The UL34 protein is likely a type II integral membrane protein with a 22-amino-acid transmembrane domain at the C terminus (27, 31, 33). The UL34 protein localizes to the nuclear rim and the cytoplasm in a pattern reminiscent of the endoplasmic reticulum when expressed transiently in uninfected cells, but it is localized exclusively at the nuclear rim in infected cells (28). The UL31 protein is necessary and sufficient to target the UL34 protein exclusively to the nuclear rim. Both the UL31 and UL34 proteins have been found to associate with newly enveloped virions in the perinuclear space and at the INM and outer nuclear membrane, but not in cytoplasmic virions or extracellular virions, by immunoelectron microscopy of HSV-infected cells (29). The UL34 protein (pUL34) is a substrate of a viral kinase encoded by the US3 protein (26). The absence of US3 delays virion egress but does not preclude primary envelopment and does not obviate phosphorylation of the UL34 protein (26, 27, 29, 32).
The UL31 protein is a hydrophobic, nucleotidylated nuclear phosphoprotein (5, 7). The protein is likely associated with the nuclear matrix, and like many nuclear matrix components such as lamin A/C, it is resistant to extraction with detergent and high salt concentrations (7, 8, 14). It is likely that UL31 plays multiple roles in viral replication. In restrictive cell types such as Vero cells, a UL31 null virus produces 1,000-fold fewer extranuclear particles than the wild-type virus, reminiscent of the phenotype of a UL34 null virus, suggesting that both proteins play important roles in primary envelopment (8, 20). In the absence of UL34, the UL31 protein is more susceptible to degradation by the proteasome and no longer localizes to the nuclear rim, but accumulates almost entirely within the nucleoplasm (28, 29, 42). Transiently expressed UL34 is sufficient to target the UL31 protein to the nuclear rim in uninfected cells (28, 40). In addition, noncomplementing cells infected with the UL31 null mutant generate decreased levels of viral DNA and have a three- to fivefold reduction in the ratio of monomeric to concatemeric viral genomic DNA, suggesting that UL31 has minor roles in both viral DNA synthesis and DNA cleavage and packaging (8).
Its association with the nuclear matrix and presence at the nuclear rim has led to the deduction that the pUL31/pUL34 protein complex is associated with the nuclear lamina of infected cells (7, 28). The observations that both pUL31 and pUL34 interact in vitro with lamin A/C (a major lamina component) and that both are required for the HSV-mediated modification of lamin A/C and chromatin (30, 34) support this hypothesis. pUL31 binds to lamin A domains that normally interact with chromatin (30, 36, 38), suggesting a mechanism for the UL31-mediated chromatin alteration. The overexpression of UL31 is also sufficient to displace lamin A/C from the nuclear rim into the nucleoplasm (30). Disruption of the lamina and associated chromatin may serve to facilitate the passage of nucleocapsids through the lamina barrier to budding sites within the INM. It is also possible that the pUL31/pUL34 complex mediates nucleocapsid budding by interactions with nucleocapsids destined for envelopment at the nuclear membrane. This hypothesis is supported by the observations that (i) pUL34 can interact with ICP5, the major capsid protein, and (ii) both pUL31 and pUL34 become incorporated into nascent virions located within the perinuclear space (29, 41).
To understand the mechanisms by which the UL31 and UL34 proteins mediate nucleocapsid envelopment, we have begun a series of studies to define the structure and function(s) of the pUL31/pUL34 complex. As a first step in this process, we sought to identify domains in the UL34 protein that mediate the interaction with pUL31.
MATERIALS AND METHODS
Cells and viruses.
Hep2, Vero, and rabbit skin cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum (growth medium). Wild-type herpes simplex virus type 1 (strain F), a UL31 deletion virus, and UL34 deletion and repair viruses were described previously (9, 20, 28, 31).
To construct a UL34-expressing cell line, we transfected rabbit skin cells with the plasmid pRR1099 containing full-length UL34 as described previously (31). Cells were selected for the ability to propagate in growth medium containing 200 μg of Geneticin/ml. Several clonal cell lines were derived from the stably transfected population by limiting dilution and were cloned twice before being tested for the ability to support the replication of the UL34 deletion virus. One of the cloned cell lines was chosen for further studies and was designated clone R1310.
Plasmid constructions.
The plasmids used for mapping assays were constructed as described below. All of the plasmids were sequenced to verify the expected genotypes.
Full-length UL31 was cloned into the pCDNA3 vector such that UL31 expression was driven by the human cytomegalovirus immediate early promoter-enhancer. The construct was designated pJB261.
The full-length UL34 sequence (encoding amino acids [aa] 1 to 275) was PCR amplified from HSV-1(F) viral DNA by use of the primers 5′ GTA GTC GAC ATA TGG CGG GAC TGG GCA AG 3′ and 5′ GCG GTC GAC AGG GCT GTG TGG GGC GAA GGC GTC 3′. The PCR product was then removed with SalI and ligated into the pGEX4T-1 SalI restriction site. This plasmid was designated pJB253. The UL34 fragment was then cut from pJB253 with the SalI enzyme and ligated into the XhoI site of pCDNA3, and the construct was designated pJB280a. To include sequences sufficient to promote translation of the gene, we PCR amplified a full-length UL34 fragment from pJB253 by using the primers 5′ GTA CTC GAG ATA TGG CGG GAC TGG GCA AG 3′ and 5′ GTG CTC GAG AGG GCT GTG TGG GGC GAA GGC GTC 3′. The PCR product was digested with XhoI and ligated into pCDNA3, and the construct was named pJB280b.
Codons 1 to 245 of UL34 were PCR amplified from pJB253 by use of the primers 5′ GTA GTC GAC ATA TGG CGG GAC TGG GCA AG 3′ and 5′ GAA GTC GAC GTG CTT AAG ACC CCG CAG 3′. The PCR product was cut with SalI and ligated into the SalI restriction site of pGEX4T-1. The resultant plasmid was designated pJB273. Using the SalI sites, we transferred the UL34 sequences (encoding aa 1 to 245) from pJB273 into the XhoI restriction site of pCDNA3 and named the resultant plasmid pJB274.
Codons 1 to 203 of UL34 were PCR amplified from pJB253 by use of the primers 5′ GTA GTC GAC ATA TGG CGG GAC TGG GCA AG 3′ and 5′ CTA CCC GTA CGC CTC CCG 3′ (UL34-609B-TAG). The PCR product was then ligated into the pCR2.1 vector (Invitrogen). This clone was named pJB336.
Codons 205 to 275 of UL34 were PCR amplified from pJB253 by use of the primers 5′ CGA GGC CGG GCT GGG GGT G 3′ and 5′ GCG GTC GAC AGG GCT GTG TGG GGC GAA GGC GTC 3′. The PCR product was ligated into the pCR2.1 vector, and the construct was named pJB338. The UL34 sequences were then removed by digestion with EcoRI and ligated into the EcoRI site of pGEX4T-1; the resultant plasmid was named pJB339. This EcoRI fragment was also subcloned into the EcoRI site of pEGFPC2 (Clontech) in frame with the gene for green fluorescent protein (GFP), and the resulting plasmid was named pJB354.
UL34 codons 81 to 203 were PCR amplified from pJB253 by use of the primers 5′ CTC GAG ATG GCG TGC AAC CCT TAC CTG 3′ and 5′ CTA CCC GTA CGC CTC CCG 3′, and the resulting amplicon was ligated into pCR2.1. The resulting clone was named pJB337.
UL34 codons 79 to 275 were PCR amplified by use of the primers 5′ CTC GAG ATG GTC CCG TGC AAC CCT TA 3′ and 5′ GTG CTC GAG AGG GCT GTG TGG GGC GAA GGC GTC 3′. The PCR product was ligated into pCR2.1, and the clone was named pJB340. The UL34 sequences were then removed with EcoRI and subcloned into the EcoRI site of pEGFPC3 in frame with the GFP gene. The resulting plasmid was designated pJB341.
UL34 codons 79 to 141 were PCR amplified by use of the primers 5′ TGT CCC GTG CAA CCC TTA CCT G 3′ and 5′ GCC GAG CCG CCC CTT GAT G 3′. The PCR product was ligated into pCR2.1 and subcloned into the pGEX4T-1 EcoRI site. This clone was designated pJB342.
A UL34 gene lacking codons 1 to 80 and 181 to 207 was generated as follows. A UL34 fragment (encoding aa 209 to 275) was PCR amplified by use of the primers 5′ GGA TCC TGG GGG TGG CCG GAA CG 3′ and 5′ GCG GTC GAC AGG GCT GTG TGG GGC GAA GGC GTC 3′, and the amplicon was cloned into pCR2.1. The 210-bp double digestion products created with EcoRI and BamHI were ligated with the EcoRI and BamHI digests from pJB337 into pCR2.1. The clone was named pJB343.
A UL34 gene lacking codons 138 to 181 was generated by a three-step PCR cloning strategy. First, a fragment bearing UL34 codons 182 to 275 was generated by the use of pJB280a as a template with the primers UL34-F-XhoI (5′ CTG GAC ACC ATC AAG TGC CGC GCC GCC GAG CAG 3′) and 5′ GTG CTC GAG AGG GCT GTG TGG GGC GAA GGC GTC 3′. Second, a DNA amplicon bearing UL34 codons 1 to 137 was generated by the use of pJB280a as a template with the PCR primers 5′ GTA CTC GAG ATA TGG CGG GAC TGG GCA AG 3′ and UL34-B-XhoI (5′ CTC GGC GGC GCG GCA CTT GAT GGT GTC CAG GTC 3′). Third, a UL34 DNA fragment lacking the region encoding aa 138 to 181 was generated by the use of gel-purified PCR products from the first two steps as templates with UL34-F-XhoI and UL34-B-XhoI as primers. The final PCR product was cloned into pCR3.1, and the construct was named pJB347. The same fragment was then cut out of pJB347 with XhoI and subcloned into the XhoI site of pGEX4T-2 in frame with the gene encoding glutathione S-transferase (GST). The resultant plasmid was named pJB348.
UL34 codons 137 to 181 were PCR amplified by the use of pJB253 as a template with the primers 5′ CAT GAA GGG GCG GCT CGG C 3′ and 5′ CTA CAG GAT CCG TCT CGT CCG TC 3′, and the product was ligated to the pCR2.1 vector. The UL34 sequences were then subcloned into the EcoRI site of pEGFPC2 in frame with the gene for GFP, and the total fragment was designated pJB345. This fragment was then subcloned into the EcoRI site of pGEX4T-1 and designated pJB346.
Emerin was PCR amplified from the plasmid Emerin-pOTB7 (MGC-2126; American Type Culture Collection [ATCC]) by use of the primers e1F-XhoI (5′ CTA CTC GAG ATG GAC AAC TAC GCA G 3′) and e256B-XhoI (5′ GTT CTC GAG CTA GAA GGG GTT GCC 3′). The PCR product was cloned into pCR2.1, cut with XhoI, and ligated into the XhoI site of pCDNA3. The resulting plasmid was named pJB351.
A plasmid bearing codons 61 to 100 of emerin replaced with UL34 codons 137 to 181 was constructed by a three-step PCR cloning strategy. First, codons 1 to 60 of emerin were PCR amplified from Emerin-pOTB7 (ATCC) by use of the primers e1F-XhoI (5′ CTA CTC GAG ATG GAC AAC TAC GCA G 3′) and e60-B-new (5′ CGA GCC GCC CGC TAT AAG AGG AGG 3′). UL34 codons 137 to 181 were PCR amplified from pJB253 by use of the primers UL34-138F-ov-e60 (5′ CCT CTT ATA GCG GGC GGCTCG GCC TG 3′) and UL34-181B-ov-e101 (5′ CTG GTG GTC AGG ATCCGT CTC GTC 3′). Emerin codons 101 to 256 were PCR amplified from Emerin-pOTB7 by use of the primers e101F-ov-34-181 (5′ GAC GGA TCC TGA CCA CCA GGA CTT ATG 3′) and e256B-XhoI (5′ GTT CTC GAG CTA GAA GGG GTT GCC 3′). Second, a DNA fragment was generated by a PCR using amplicons containing UL34 codons 137 to 181 and emerin codons 101 to 256 as templates along with the primers UL34-138F-ov-e60 and e256B-XhoI. Third, the final chimeric DNA fragment was generated by using the PCR product for emerin (codons 1 to 60) from step 1 and the PCR product from step 2 as templates. This reaction used the primers e1F-XhoI and e256B-XhoI. The final PCR product was ligated into pCR2.1, removed with XhoI, and ligated into the XhoI site of pCDNA3. The final plasmid was named pJB352.
Construction of a UL34 mutant virus deficient in interaction with UL31.
A DNA fragment containing a UL34 sequence lacking codons 138 to 181 neighboring a kanamycin resistance (Kanr) gene was generated by PCR. These genes were flanked by Flp recombination sites. The UL34-Flp Kanr-Flp construct was, in turn, flanked by UL34 flanking sequences and BamHI restriction sites. This construct was generated as follows. First, a PCR amplicon was generated that contained the UL34 sequence lacking codons 138 to 181. The UL34 sequences were flanked by 50 bp upstream of UL34 and by 12 bp of sequences overlapping Flp-Kanr-Flp. This amplicon was generated by the use of plasmid UL34 (del138-181aa)-pCR3.1 as a template with the primers 50UL34F-Bam (5′ CCT TTG GTG GGT TTA CGC GGG CAC GCA CGC TCC CAT CGC GGG CGC CGG ATC CAT GGC GGG ACT GGG CAA GCC CTA CAC CG 3′) and UL34Bovflip (5′ CGA ACT GCA GGT CGA CTT ATA GGC G 3′). Second, a fragment containing Flp-Kanr-Flp was generated by using Flp-Kanr-Flp as a template (a kind gift of Klaus Osterrieder and Bernd Karsten Tischer, Cornell University) with the primers flpKanF-ov34 (5′ GCG CGC GCC TAT AAG TCG ACC TGC AGT TCG 3′) and flpkanB-Bam-ov34down (5′ GGC GTC CGG AAC GCA CTG GCG ATT AGG GCG GCG GTG CGT CCT TTT GGA TCC GCT TCG AAG TTC CTA TAC TTT CTA GAG 3′). This fragment was flanked by sequences overlapping the 3′ end of UL34 at one end and 50 bp of sequences downstream of UL34 at the other end. Third, the final 2.2-kb fragment was generated by a PCR using the gel-purified PCR products from the amplicons generated in steps 1 and 2 (above) as templates with the primers 50UL34F-Bam and flpkanB-Bam-ov34down.
About 500 ng of the 2.2-kb PCR product was electroporated into a strain of Escherichia coli EL250 harboring (i) the full-length HSV-1(F) genome maintained as a bacterial artificial chromosome (BAC) called Yebac102 (37), (ii) a prophage system encoding the Gam, Exo, and Beta recombinases, and (iii) an arabinose-inducible Flp recombinase to facilitate BAC modification at Flp recombinase recognition target (FRT) sites (19). Electroporation was performed in an ice-cold cuvette at 2.3 kV and 25 μF by use of a Bio-Rad Gene Pulser controller set to 200 Ω. Electroporated bacteria were resuspended in 1 ml of Luria-Bertani medium at room temperature, and the material was transferred to a 1.5-ml microcentrifuge tube and incubated at 30°C for 1 h with shaking. Bacteria were then plated on Luria-Bertani agar plates containing 30 μg of chloramphenicol/ml and 50 μg of kanamycin/ml. Kanr Cmpr colonies were picked, and BAC DNA was extracted by alkaline lysis. The Kanr fragment was deleted by the induction of Flp recombinase with 0.1% l-arabinose overnight at 30°C. BAC DNA was then cotransfected into R1310 cells with a Cre expression plasmid to delete the BAC vector sequences (pCAGGS-nlsCre, a gift from Michael Kotlikoff, Cornell University), and the resulting viral plaques were purified twice. The resulting virus was named v3480.
Purification of GST fusion proteins and GST pull-down assays.
Full-length UL31 fused to GST (encoded by pJB187) and different UL34-GST mutant fusion proteins were purified as described previously (11). The full-length UL34 protein (from plasmid 280b) or UL31 protein (from pJB261) was radiolabeled with [35S]methionine in vitro by use of a rabbit reticulocyte lysate-based transcription and translation expression system (TNT System; Promega). Five microliters of the UL31 radiolabeling reaction was preclarified overnight with Sepharose beads and subsequently incubated for 2 to 5 h at 4°C with 5 μg of GST or a UL34-GST fusion protein conjugated to glutathione-Sepharose beads in cold phosphate-buffered saline supplemented with 1% (vol/vol) Triton X-100. In reciprocal reactions, an [35S]methionine-labeled UL34 mutant protein expressed in rabbit reticulocyte lysates was incubated with 5 μg of a UL31-GST fusion protein or GST. After the incubation periods, the beads and bound proteins were washed six times with cold 1% Triton X-100. The bound proteins were eluted by boiling in 3× sodium dodecyl sulfate (SDS) loading buffer and then electrophoretically separated in an SDS-12% polyacrylamide gel. The gel was soaked for 30 min in 20% sodium salicylate, dried, and either exposed in a PhosphorImager cassette (Molecular Dynamics) followed by quantification with Imagequant software or subjected to fluorography with X-Omat (Kodak) film exposed overnight at −80°C.
Southern blot analysis.
Vero cells in 25-cm2 dishes were infected with a wild-type virus derived from Yebac102 or with the v3480, v3480-Kan, or v3480-Kan-BAC virus until a cytopathic effect was visible in 80% of the cells. Viral DNAs were then purified as described previously (20). The viral DNAs were digested with BamHI. The viral DNA digests were then separated in 1% agarose gels, transferred to nitrocellulose membranes, probed with [α-32P]dCTP (Amersham)-labeled DNA, and exposed to radiographic film as described previously (20). The DNA probes used for the experiments were derived from the purified full-length UL34 PCR product, a fragment bearing the Kanr gene generated by PCR, and the True Blue BAC II plasmid (Genomics One Corporation) (BAC probe).
Confocal microscopy.
Hep2 cells were seeded on glass coverslips in a six-well plate to reach 80% confluence. The cells were transfected with the plasmid pJB345, bearing UL34 codons 137 to 181 fused to the GFP gene or UL34 codons 205 to 275 fused to the GFP gene, either alone or together with pJB261 containing full-length UL31. The cells were fixed in 3% paraformaldehyde for 15 min at room temperature and then permeabilized with 0.1% Triton X-100 for 3 min. The cells were blocked in 10% human serum and stained with a pUL31-specific rabbit antibody diluted 1:50 (28), followed by extensive washing and incubation with a Texas Red-conjugated donkey anti-rabbit antibody (Jackson Immunoresearch). Fluorescence values attributable to GFP and Texas Red were recorded as described previously (28).
In a separate experiment, Hep2 cells were transfected with emerin (pJB351) or emerin-UL34 (pJB352) alone or together with full-length UL31. The cells were fixed and permeabilized with ice-cold methanol for 20 min at −20°C. Nonspecific immunoreactivity was blocked with 10% human serum for 1 h, and the cells were stained with the pUL31-specific rabbit antibody diluted 1:50 and with a mouse monoclonal anti-emerin antibody (NCL-Emerin; Novocastra) diluted 1:40. The cells were then incubated with fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit and Texas Red-conjugated donkey anti-mouse antibodies.
In a final experiment, Hep2 cells were infected with the UL31 deletion virus, the UL34 deletion virus, the UL34 mutant virus v3480, or the UL34 repair virus. Sixteen hours after infection, the cells were fixed with ice-cold methanol for 20 min at −20°C. The cells were then stained with pUL31- and pUL34-specific antibodies as described above.
In all cases, glass coverslips were examined with an Olympus confocal laser scanning microscope using 10× ocular and 40× stage objectives and excitation wavelengths of 488 and 568 nm. The cells were also examined by Nomarski differential interference contrast imaging.
RESULTS
Mapping of UL34 protein domains that interact with UL31 protein.
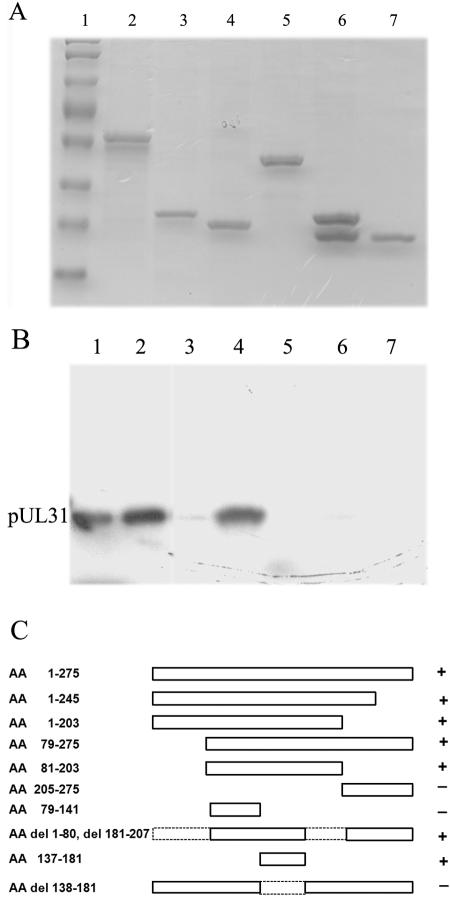
Different portions of UL34 were cloned into plasmid vectors that placed fragments of UL34 in frame with the gene encoding GST, as described in Materials and Methods. The resultant GST fusion proteins were affinity purified as detailed in Materials and Methods. Equal amounts of the fusion proteins bound to glutathione-Sepharose beads were incubated with 35S-labeled UL31 protein, followed by extensive washing, elution in an SDS-containing buffer, separation by electrophoresis, fluorography, and analysis on a Molecular Dynamics PhosphorImager. Some of the results are shown in Fig. 1A and B. The results of all experiments are summarized in Fig. 1C.
FIG. 1.
Mapping of regions of UL34 that interact with pUL31. (A) GST fusion proteins bearing portions of pUL34 were purified by the use of Sepharose beads, electrophoretically separated, and stained with Coomassie blue. Lane 1, protein standard; lane 2, full-length pUL34-GST. The other lanes contain portions of pUL34 fused to GST as follows: lane 3, amino acids 79 to 141; lane 4, amino acids 137 to 181; lane 5, full-length pUL34 lacking amino acids 138 to 181; lane 6, amino acids 205 to 275; lane 7, GST alone. (B) Equal amounts of the bound proteins were mixed with [35S]methionine-labeled pUL31 produced in a rabbit reticulocyte lysate. After extensive washing, the proteins bound to the GST fusion protein were eluted and separated in a denaturing polyacrylamide gel, followed by fluorography and quantification by a Molecular Dynamics PhosphorImager. Lane 1, in vitro-translated pUL31; lane 2, GST pull-down reaction containing full-length pUL34-GST. The other lanes show the results of similar pull-down reactions containing the same segments of pUL34 fused to GST as those shown in panel A. (C) Summary of mapping studies. The schematic diagram shows the pUL34 sequences that were fused to GST and tested in GST pull-down assays. Dashed lines in the diagram indicate portions of pUL34 that are missing from some of the constructs. +, the fusion protein pulled down levels of pUL31 above the background levels obtained with GST alone.
We determined that full-length pUL34 pulled down 43 times more radiolabeled pUL31 than did GST alone, confirming previous results indicating that the UL31 and UL34 proteins interact in this assay (28). In contrast, codons 1 to 137 (codon 1 encodes methionine) and 182 to 275 of the 275-codon UL34 open reading frame were dispensable for the interaction inasmuch as fusion proteins bearing these sequences did not pull down the radiolabeled UL31 protein significantly more than did GST alone. Importantly, a GST fusion protein containing only UL34 amino acids 137 to 181 was sufficient to pull down 20 times more radiolabeled UL31 protein than was GST alone. The deletion of this region from full-length UL34 produced a protein that pulled down only five times more pUL31 than did GST alone. This was significantly less than the amount that was pulled down with full-length pUL34-GST. We therefore concluded that amino acids 137 to 181 were necessary and sufficient to mediate an interaction with pUL31 in vitro.
Amino acids 137 to 181 of UL34 can interact with full-length UL31 in eukaryotic cells.
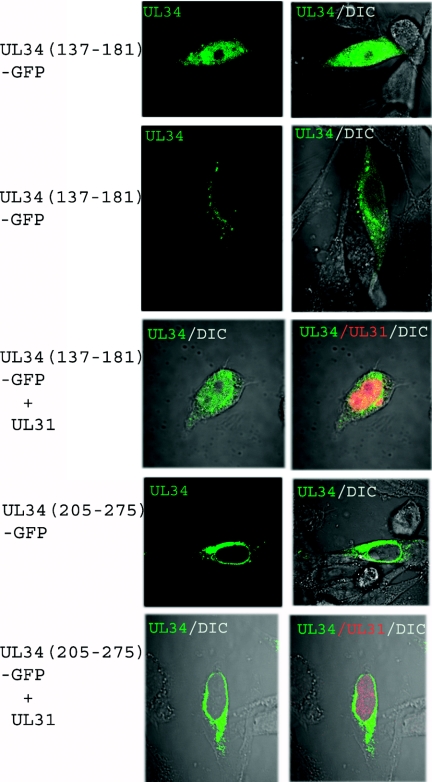
Because GST pull-down assays do not necessarily indicate that a given region mediates the interaction in more relevant contexts, a series of experiments were undertaken to investigate the role of UL34 amino acids 137 to 181 in mediating the interaction in both HSV-infected and uninfected eukaryotic cells. Specifically, the putative interacting region (UL34 codons 137 to 181) and pUL34 codons 205 to 275 (a region shown not to interact with pUL34 in the GST pull-down assays) were fused to the gene encoding GFP, and the fluorescence levels in cells that transiently expressed the construct in the presence or absence of pUL34 were compared. The cells were also immunostained with antibodies specific for the pUL31 protein.
When expressed alone, pUL34 137-181/GFP localized in a smooth pattern either within the cytoplasm or in the nucleus and cytoplasm (Fig. 2, top two rows). When coexpressed with the UL31 protein, however, the majority of pUL34 137-181/GFP localized to the nucleus, where it partially colocalized with the UL31 protein (Fig. 2). In contrast, the construct encoding pUL34 (aa 205 to 275) fused to GFP did not colocalize with coexpressed UL31.
FIG. 2.
Confocal microscopic images of chimeric pUL34-containing proteins in the presence and absence of wild-type pUL31. Hep2 cells were transfected with a plasmid encoding pUL34 (137-181) or pUL34 (205-275) fused to GFP alone or together with the wild-type UL31 protein. The cells were fixed, permeabilized, and stained with a pUL31-specific rabbit polyclonal antibody, and bound IgG was detected with a Texas Red-conjugated donkey anti-rabbit secondary antibody.
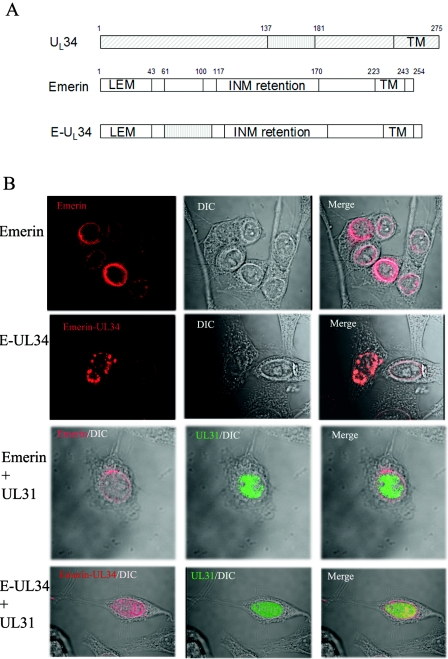
To further show that pUL34 aa 137 to 181 were sufficient to interact with pUL31 in eukaryotic cells, UL34 codons 137 to 181 were used to replace codons 61 to 100 of emerin, a type II integral membrane protein that localizes within the inner nuclear membrane (2, 3, 25, 39). The construct bearing the emerin chimera was designated pJB352. The localization of the pJB352-encoded protein was similar to the localization of emerin cloned into the same vector, pJB351 (Fig. 3B), inasmuch as both proteins localized to the nuclear rim. However, the appearance of the emerin chimera-specific immunofluorescence at the nuclear rim was slightly altered from that of native emerin and often exhibited a punctate distribution, especially in cells containing large amounts of chimeric protein. It should also be noted that under these conditions, the staining of native emerin was readily distinguished from that of the expressed transgenes, presumably as a consequence of the relatively low expression level of the former. In control experiments, the localization of transiently expressed emerin did not change, regardless of whether the UL31 protein was coexpressed (Fig. 3B, third row). In contrast, the localization of the emerin chimera bearing UL34 amino acids 137 to 181 changed significantly upon coexpression of the UL31 protein such that the chimera largely colocalized with pUL31 in the nucleoplasm, with some emerin-specific staining remaining at the nuclear rim. We suspect but cannot prove that the residual fluorescence at the nuclear rim represents native emerin as opposed to that derived from the transiently expressed chimera. Irrespective of this possibility, these experiments indicate that UL34 amino acids 137 to 181 are sufficient to cause a partial colocalization of two different proteins (GFP and emerin) with the UL31 protein in the context of a eukaryotic cell.
FIG. 3.
(A) Schematic diagram of emerin, pUL34, and chimeric pUL34. Most pUL34 sequences are indicated by diagonally hatched rectangles, whereas the putative pUL31 interaction region is indicated by a vertically hatched rectangle. Emerin sequences are indicated by open rectangles. The final chimera (E-UL34) contains the putative pUL31 interaction region of pUL34 in place of codons 61 to 100 of emerin. Numbers above the diagrams indicate the amino acids delimiting the domains. TM, transmembrane domain; INM, region shown to be necessary for targeting emerin to the inner nuclear membrane; LEM, domain common to many INM-targeted proteins. (B) Hep2 cells were transfected with a plasmid encoding emerin or the emerin-UL34 chimeric construct (E-UL34) alone or together with a plasmid encoding the UL31 protein. The cells were stained with an emerin-specific mouse monoclonal antibody and a pUL31-specific rabbit polyclonal antibody. Bound antibodies were detected with Texas Red-conjugated donkey anti-mouse and FITC-conjugated donkey anti-rabbit antibodies.
Construction and characterization of UL34 (del 138-181) virus.
The next set of experiments was designed to determine if amino acids 138 to 181 of UL34 were essential for colocalization of the UL31 and UL34 proteins in infected cells and to determine if this region was also essential for the contribution of UL34 to viral replication.
A recombinant virus bearing a mutant UL34 protein lacking codons 138 to 181 was used to replace native UL34 within a BAC containing the HSV-1(F) genome as detailed in Materials and Methods. Briefly, the recombinant virus was constructed as follows. (i) A DNA fragment that contained a mutant UL34 gene lacking codons 137 to 181 and a kanamycin resistance gene that was flanked by Flp recombination sites and by 50 bp of UL34 sequence and BamHI restriction sites was generated by PCR. (ii) The PCR amplicon was electroporated into bacteria harboring the HSV-1(F) genome within the BAC pYEC102 (37), and kanamycin-resistant colonies were selected at 30°C. (iii) One colony was picked, genotypically verified (see below), and designated v3480-Kan-BAC. (iv) Flp recombinase was induced in E. coli cells harboring v3480-BAC-Kn to remove the kanamycin resistance gene. The resulting recombinant BAC was designated v3480-BAC. (v) The BAC DNA was purified from bacteria and cotransfected with a Cre expression plasmid into R1310 cells (a UL34-expressing cell line derived from rabbit skin cells), viral plaques were purified, and the virus from one of the plaques was designated v3480. The Cre recombinase-mediated removal of the BAC vector sequences inserted between UL3 and UL4 was expected inasmuch as the BAC sequences were flanked by Lox recombination sites (37).
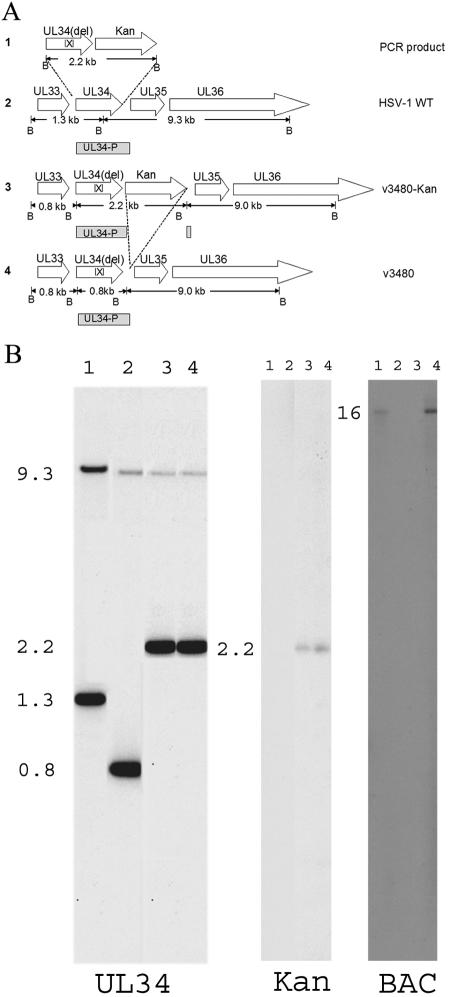
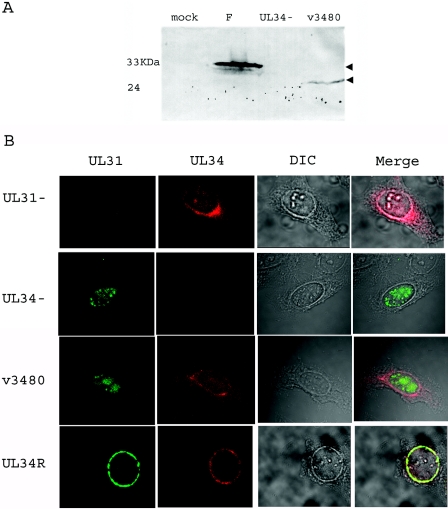
For verification of the expected genotypes of the recombinant BACs, HSV-1(F) BAC, v3480, v3480-Kan, and v3480-Kan-BAC DNAs were purified, digested with BamHI, and electrophoretically separated in a 1% agarose gel. The digested DNAs were transferred to nitrocellulose and probed with [α-32P]dCTP-labeled DNA from either UL34, the gene encoding kanamycin resistance, or the BAC vector sequence. As shown in Fig. 4B, the UL34 probe recognized 9.3- and 1.3-kb bands from the HSV-1(F) BAC (lane 1, left panel). However, both bands were missing from the v3480, v3480-Kan, and v3480-Kan-BAC DNAs; instead, a 0.8- or 2.2-kb band was recognized due to the presence or absence of the kanamycin resistance gene, respectively (lanes 2 to 4). A band of approximately 9.0 kb was also recognized for all three recombinant viruses due to sequences within the probe that hybridized to UL34 (Fig. 4A shows a diagrammatic representation of these sequences). As expected, the kanamycin probe only recognized bands from the v3480-Kan and v3480-Kan-BAC DNAs (lanes 3 and 4, middle panel), and the BAC probe only recognized bands from the HSV-BAC and v3480-Kan-BAC DNAs (lanes 1 and 4, right panel). In addition, the UL34 gene of v3480 was amplified by PCR and sequenced to ensure that the viral genotype was as designed (not shown). As confirmation of the truncation of pUL34, as shown in Fig. 5A, the pUL34-specific antibody recognized a protein with an apparent relative molecular weight of 34,000 in lysates of cells infected with the wild-type virus, whereas cells infected with v3480 contained a protein with an apparent relative molecular weight of 28,000. As expected, no UL34 protein was detected in lysates of mock-infected cells or cells infected with the UL34 deletion virus.
FIG. 4.
(A) Schematic diagram of steps involved in the construction of a virus bearing a UL34 gene lacking codons 137 to 181. Line 1, sequences within a 2.2-kb PCR product containing the UL34 gene lacking codons 137 to 181 and a gene encoding kanamycin resistance (Kan) flanked by Frt recombination sites (not shown), with 50 bp of sequence homologous to regions flanking UL34 in wild-type viral genomes and BamHI restriction sites. Line 2, wild-type HSV-1 sequences containing UL34 and schematic diagram of the UL34 probe (UL34-P, gray rectangle) used to hybridize to DNA sequences, as shown in panel B. The lengths of the expected DNA fragments are indicated in kilobase pairs and are delimited by BamHI (B) sites. Line 3, genome of the UL34 mutant virus v3480-Kan after homologous recombination within the 50-bp sequences flanking the Kanr gene and UL34. The sizes of the expected BamHI fragments are indicated in kilobase pairs below the rectangles. Line 4, genome of the UL34 mutant virus v3480 after deletion of the kanamycin resistance gene at the Frt sites by the induction of Flp recombinase. (B) Digitally scanned image of a fluorograph of viral DNAs probed with radiolabeled DNAs. Viral DNAs were purified, digested with BamHI, and electrophoretically separatedin a 1% agarose gel. The DNAs were transferred to nitrocellulose and probed with [α-32P]dCTP-labeled UL34 (schematically diagramed as UL34-P in panel A), the gene encoding kanamycin resistance, or the BAC vector sequence. Lanes 1, HSV-BAC; lanes 2, v3480 DNA; lanes 3, v3480-Kan DNA; lanes 4, v3480-Kan-BAC DNA. The numbers indicate the sizes of the DNA fragments in kilobase pairs.
FIG. 5.
(A) Scanned digital image of immunoblot of cell lysates probed with an antibody to pUL34. Hep2 cells were either mock infected or infected with wild-type HSV-1(F), the UL34 deletion virus, or the mutant virus v3480. The cells were lysed, and associated proteins were denatured, electrophoretically separated, transferred to nitrocellulose, and probed with a pUL34-specific antibody. (B) Confocal images of the localization of pUL31 and pUL34 in infected cells. Hep2 cells were infected with the UL31 deletion virus (UL31−), the UL34 deletion virus (UL34−), the mutant virus v3480, or the UL34 repair virus (UL34R) for 16 h and then fixed with ice-cold methanol at −20°C. The cells were then stained with pUL31- and pUL34-specific rabbit and pUL34-specific chicken antibodies, and bound antibodies were detected with a FITC-conjugated donkey anti-rabbit antibody and a Texas Red-conjugated donkey anti-chicken antibody. Differential interference contrast (DIC) images were collected separately. The separately collected channels of fluorescent and DIC images were coprojected in the rightmost column. The intensity of the red and green channels was increased in the merged image to allow comparison with light-refracting features.
To determine whether precluding an interaction with UL31 played a role in viral replication, we infected Vero cells, rabbit skin cells, and R1310 cells with 1.0 PFU of virus per cell. Twenty-four hours after infection, the cells were lysed by freezing and thawing, and infectious titers were determined by titration and a plaque assay on R1310 cells; the data are shown in Table 1. Like the UL34 null virus (31), v3480 failed to replicate on Vero cells to an appreciable extent and replicated up to 6.9 × 104 PFU/ml on rabbit skin cells. Titers produced in rabbit skin cells containing UL34 were increased >10- to 100-fold compared to titers obtained from normal rabbit skin cells. Experiments to characterize the ability of v3480 to form viral plaques were also performed (data not shown). Neither the UL34 deletion mutant nor v3480 formed plaques on Vero cells. However, both the UL34 deletion mutant and v3480 formed small plaques on the more permissive rabbit skin cells. In summary, these data indicate that v3480 replication in Vero and rabbit skin cells resembles that of the UL34 deletion virus, suggesting that amino acids 137 to 181 are essential for normal pUL34 function.
TABLE 1.
Peak titers of viruses on different cell lines
| Virus | Peak virus titer on cell line
|
||
|---|---|---|---|
| Vero | R1310 | RSC | |
| F | 4.3 × 107 | 2.2 × 107 | 2.0 × 107 |
| UL34 Rep | 3.0 × 107 | 3.3 × 107 | 1.7 × 107 |
| ΔUL34 | 1.8 × 103 | 2.4 × 106 | 1.5 × 104 |
| v3480 | 3.1 × 103 | 1.0 × 106 | 6.9 × 104 |
To determine if the inability of mutant UL34 to support viral replication was reflected in an inability to properly localize UL31 to the nuclear rim of infected cells, we infected Hep2 cells with the UL31 deletion virus, the UL34 deletion virus, the UL34 restored virus, or v3480. The cells were then fixed 16 h after infection and stained with antibodies directed against the UL31 and UL34 proteins. The results are shown in Fig. 5. As previously described (28), cells infected with the virus bearing a restored UL34 gene contained both UL31 and UL34 proteins almost exclusively at the nuclear rim. In contrast, cells infected with the UL34 deletion mutant contained UL31 exclusively within the nucleoplasm, and cells infected with the UL31 deletion virus contained the UL34 protein at the nuclear rim and within the nucleus and the cytoplasm. The latter was present in a pattern emanating from the nuclear membrane that strongly resembled the distribution of the endoplasmic reticulum.
In cells infected with v3480, the UL31 protein localized exclusively in the nucleoplasm in a pattern that closely resembled that of the distribution of the UL31 protein in cells infected with the UL34 null mutant. We also noted that some of the nucleoplasmic aggregates containing the UL31 protein also contained lamin A/C (not shown). The colocalization of the UL31 and UL34 proteins at the nuclear membrane was restored in the complementing cell line R1310 infected with v3480 (not shown). We therefore concluded that the UL34 protein of v3480 lacks the capacity to correctly target the UL31 protein to the nuclear membrane. Although we cannot exclude the possibility that amino acids 137 to 181 mediate other functions that contribute to viral replication, these data suggest that these amino acids constitute a domain that is necessary for interacting with the UL31 protein in vitro and in vivo.
DISCUSSION
Previous studies have shown that the UL31 and UL34 proteins form a complex at the nuclear membrane and act together to play important roles in the primary envelopment of both HSV-1 and pseudorabies virus (8, 12, 18, 28, 31). The deletion of either protein from HSV-1 abolishes its replication on some cell lines and causes mislocalization of the other protein (28, 29, 31).
The goal of this study was to identify the domain in pUL34 that mediates the interaction with pUL31 and to test the significance of the pUL31-pUL34 interaction for viral replication. Previous studies showed that charged cluster mutations did not disrupt the interaction with pUL31, but some of these mutations were lethal for virus replication and mislocalized the pUL31/pUL34 complex elsewhere in the cell (4). For the present study, it was determined that amino acids 137 to 181 of pUL34 were sufficient to mediate the interaction with pUL31 in three different contexts, including GST pull-down assays, targeting of GFP to sites containing pUL31, and targeting of chimeric emerin to intracellular sites that contained pUL31. Moreover, pUL34 amino acids 137 to 181 were necessary for viral replication and were shown to be essential for colocalization of the pUL31 and pUL34 proteins in infected cells.
The peptide comprising amino acids 137 to 181 is less polar (9.7% by weight) and more basic (26.4% by weight) than pUL34 as a whole (18.3 and 17.0% by weight, respectively [not shown]). A comparison of the sequences of the UL34 homologs of HSV-1, pseudorabies virus, varicella zoster virus, and equine herpesvirus 1 has shown that the identified interaction region is highly conserved (not shown). In particular, the consensus for residues 155 to 173 is 155CF(V/T)RMP(X)VQL(A/S)FRFMGP(E/D)D173, with 79% identity among the various homologs. Two HSV-1 mutations that were assessed for their effects on the pUL31-pUL34 interaction are particular interesting inasmuch as they are located near or in the pUL31 interaction region detected in the present study. Thus, while changing 136IKGR139 (a mutant designated CL10) or 158RMPR161(designated CL13) to alanines did not disrupt the interaction with pUL31 or the localization of the complex at the nuclear membrane, it precluded complementation of a UL34 viral deletion mutant when the proteins were expressed transiently (4). These data indicate that the alanine clusters, while insufficient to disrupt the pUL31-pUL34 interaction, may have functionally altered the complex such that it was unable to perform its role(s) in viral egress.
A comprehensive genetic analysis of the more distantly related murine cytomegalovirus homolog of UL34 (M50) has also been conducted in the context of viral infection (6). In previous studies, residues 54YPVEY57 of M50 were shown to be essential for coimmunoprecipitation with M53, the HSV-1 UL31 homolog. Although this region is conserved in the alphaherpesviruses equine herpesvirus 1, varicella zoster virus, and HSV [consensus sequence, (F/Y)P(I/L/V)EY], the N-terminal 80 amino acids of pUL34 were dispensable for the interaction with pUL31 in GST pull-downs (Fig. 1C), and a UL34 protein lacking the first 78 residues maintained the ability to colocalize with UL31 at the nuclear membrane (not shown). Similarly, a cluster mutation near this region (67EYVLR71) did not prevent colocalization of this UL34 mutant protein with the UL31 protein at the nuclear membrane in cells infected with a UL34 null virus (4). HSV-1 pUL34 amino acids 137 to 181 are mostly conserved in murine cytomegalovirus M50, but this region of M50 also contains insertions of 12, 12, 8, and 9 amino acids that are not conserved in alphaherpesviruses (not shown). The insertion of 5 amino acids into the 12-amino-acid nonconserved regions precluded rescue of the murine cytomegalovirus M50 null mutant (6). Other lethal mutations in M50 mapped within regions that are conserved in HSV-1 pUL34 137-181. The data therefore suggest that the interactions of the HSV and cytomegalovirus homologs and the structures of the resulting complexes may differ significantly. It is also possible that mutations at the N terminus of M50 cause it to aberrantly interact with other regions of M50 (or other proteins) and thus preclude an interaction with M53. In our studies with pUL34, such effects would not be expected because the N-terminal region was deleted from the constructs tested in vitro.
In cells infected with the UL34 null virus or v3480, the UL31 protein localized to the nucleus and within intranuclear aggregates (Fig. 5). Recent analyses indicated that these aggregates also contained lamin A/C (not shown). This is consistent with the observation that overexpression of the UL31 protein is sufficient to relocalize lamin A/C to the nucleoplasm and may reflect the possibility that the interaction between UL31 and lamin A/C is sufficient to mediate a partial dissolution of the lamina (30). Such relocalization may not be readily apparent in infected cells expressing wild-type pUL34 because pUL31 is retained at the nuclear rim, as is the lamin A/C to which the pUL31/pUL34 complex binds.
Acknowledgments
This study was supported by National Institutes of Health grant R01 AI52341.
We thank Jarek Okulicz Kozaryn for technical support and Yasushi Kawaguchi, Richard Roller, Michael Kotlikoff, Klaus Osterrieder, Bernd Karsten Tischer, and Carol Duffy for reagents and helpful discussions.
REFERENCES
- 1.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bengtsson, L., and K. L. Wilson. 2004. Multiple and surprising new functions for emerin, a nuclear membrane protein. Curr. Opin. Cell Biol. 16:73-79. [DOI] [PubMed] [Google Scholar]
- 3.Bione, S., E. Maestrini, S. Rivella, M. Mancini, S. Regis, G. Romeo, and D. Toniolo. 1994. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 8:323-327. [DOI] [PubMed] [Google Scholar]
- 4.Bjerke, S. L., J. M. Cowan, J. K. Kerr, A. E. Reynolds, J. D. Baines, and R. J. Roller. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601-7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J. Biol. Chem. 269:17401-17410. [PubMed] [Google Scholar]
- 6.Bubeck, A., M. Wagner, Z. Ruzsics, M. Lötzerich, M. Iglesias, I. R. Singh, and U. H. Koszinowski. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J. Virol. 78:8026-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. E., and B. Roizman. 1993. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J. Virol. 67:6348-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype of infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frangioni, J. V., and B. J. Neel. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210:179-187. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3673-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, D. C., J. A. Nickerson, and S. Penman. 1990. Core filaments of the nuclear matrix. J. Cell Biol. 110:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holland, D. J., M. Miranda-Saksena, R. A. Boadle, P. Armati, and A. L. Cunningham. 1999. Anterograde transport of herpes simplex virus proteins in axons of peripheral human fetal neurons: an immunoelectron microscopy study. J. Virol. 73:8503-8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchinson, L., and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 69:5401-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayachandra, S., A. Baghian, and K. G. Kousoulas. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 71:5012-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, E.-C., D. Yu, J. M. de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 20.Liang, L., M. Tanaka, M. Kawaguchi, and J. D. Baines. 2004. Cell lines that support replication of a novel herpes simplex virus 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 329:68-76. [DOI] [PubMed] [Google Scholar]
- 21.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miranda-Saksena, M., P. Armati, R. A. Boadle, D. J. Holland, and A. L. Cunningham. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neubauer, A., J. Rudolph, C. Brandmüller, F. Just, and N. Osterrieder. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189-204. [DOI] [PubMed] [Google Scholar]
- 25.Ostlund, C., J. Ellenberg, E. Hallberg, J. Lippincott-Schwartz, and H. J. Worman. 1999. Intracellular trafficking of emerin, the Emery-Dreifuss muscular dystrophy protein. J. Cell Sci. 112:1709-1719. [DOI] [PubMed] [Google Scholar]
- 26.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purves, F. C., D. Spector, and B. Roizman. 1992. UL34, the target gene of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J. Virol. 66:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78:399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiba, C., T. Daikoku, F. Goshima, H. Takakuwa, Y. Yamauchi, O. Koiwai, and Y. Nishiyama. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 81:2397-2405. [DOI] [PubMed] [Google Scholar]
- 34.Simpson-Holley, M., J. D. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment→deenvelopment→reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stierle, V., J. Couprie, C. Ostlund, I. Krimm, S. Zinn-Justin, P. Hossenlopp, H. J. Worman, J. C. Courvalin, and I. Duband-Goulet. 2003. The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry 42:4819-4829. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniura, H., C. Glass, and L. Gerace. 1995. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol. 131:33-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tews, D. S. 1999. Emerin. Int. J. Biochem. Cell Biol. 31:891-894. [DOI] [PubMed] [Google Scholar]
- 40.Yamauchi, Y., C. Shiba, F. Goshima, A. Nawa, T. Murata, and Y. Nishiyama. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 82:1428. [DOI] [PubMed] [Google Scholar]
- 41.Ye, G.-J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye, G.-J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. USA 97:11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]