Abstract
The avian sarcoma/leukosis virus (ASLV) is activated for fusion by a two-step mechanism. For ASLV subgroup A (ASLV-A), association with its receptor (Tva) at neutral pH converts virions to a form that can bind target membranes and, in some assays, induce the lipid-mixing stage of fusion. Low pH is necessary to complete the fusion reaction. ASLV-A env (EnvA) exists on the viral surface as a trimer of heterodimers consisting of receptor binding (SU-A) and fusion-mediating (TM-A) subunits. As the receptor binding and fusion-mediating functions reside in separate subunits, we hypothesize that SU-A and TM-A are conformationally coupled. To begin to understand the effect of the binding of a soluble 47-residue domain of the receptor (sTva) on this coupling and the subsequent function of low pH, we prepared recombinant proteins representing full-length SU-A and a nested set of deletion mutant proteins. Full-length SU-A binds sTva with high affinity, but even small deletions at either the N or the C terminus severely impair sTva binding. We have purified the full-length SU-A subunit and characterized its interactions with sTva and the subsequent effect of low pH on the complex. sTva binds SU-A with an apparent KD of 3 pM. Complex formation occludes hydrophobic surfaces and tryptophan residues and leads to a partial loss of α-helical structure in SU-A. Low pH does not alter the off rate for the complex, further alter the secondary structure of SU-A, or induce measurable changes in tryptophan environment. The implications of these findings for fusion are discussed.
Enveloped viruses initiate infection by fusing their membranes with those of target cells. Virus-encoded fusion proteins mediate this process. Fusion proteins exist on the virion surface in metastable states that are created by posttranslational processing during assembly and/or budding of the virus particle. The metastable viral surface proteins must first bind target cell receptors and then unleash the fusion process. Two primary modes of triggering the fusion process have been established: exposure to low pH and receptor binding. Low-pH-triggered fusion is activated by the decreasing pH of the endosome following endocytosis of the receptor-bound virion. Receptor-triggered fusion can occur at the plasma membrane and, as its name implies, is triggered by interaction with the receptor. Recently a hybrid two-step mechanism has been identified in which receptor binding initiates the fusion process but low pH is required to complete it (reviewed in reference 20).
Class I fusion proteins are type I membrane proteins that extend their ectodomains from the virion surface. Many can be considered to have a “ball-and-stick” morphology in which the ball (also known as the “head” group) contains the receptor binding function and also serves as a clamp to hold the stick-like fusion subunit in an inactive conformation. The triggering process releases this clamp. The fusion subunit contains a hydrophobic sequence at or near its N terminus that serves as a fusion peptide, two heptad repeat regions, a transmembrane domain, and a cytoplasmic tail. For retroviruses, the receptor binding (ball) and fusion-mediating (stick) proteins are two subunits generated from a single precursor by posttranslational proteolytic processing. They are referred to as SU (for surface subunit) and TM (for transmembrane subunit), respectively. The functional fusion protein is a trimer of SU-TM heterodimers.
To date, two structural motifs have been identified for retroviral SUs. In one, exemplified by the murine leukemia virus SU, the receptor binding domain (RBD) occurs in the N-terminal third of the subunit, followed by a proline-rich hinge region and a C-terminal domain (44). The RBD can be prepared in the absence of the other domains (24). An interaction between the RBD and the C-terminal domain is required to trigger fusion. Interestingly, the RBD can be supplied in trans as a soluble protein (3, 6, 44). The other type of structure is exemplified by the human immunodeficiency virus (HIV) SU, gp120. gp120 has multiple variable regions interspersed with conserved regions (46). An independent RBD cannot be isolated from gp120 because sequences throughout the SU contribute to its structure (43). The N- and C-terminal conserved sequences appear to interact with the TM subunit (8, 45, 54). Interestingly, the receptor binding subunit of the low-pH-triggered influenza virus fusion protein (HA), HA1, has a topology similar to that of the HIV SU (67).
The ability to easily trigger fusion in vitro has allowed extensive study of the low-pH fusion pathway. For HA, receptor binding anchors the virus to the target cell surface but does not induce significant conformational changes in HA and does not trigger the fusion reaction (60). Triggering occurs upon a decrease in the local pH during endocytosis. Titration of charged residues along the interface between HA1 (analogous to SU) and HA2 (analogous to TM) alters the forces between the HA1 subunits of the trimer, causing them to separate (34). This head group separation releases the clamp on HA2, triggering fusion (28, 38). The addition of protons and separation of the head groups are accomplished without significantly altering the conformation of HA1 (9, 58).
HIV env is the best-studied receptor-activated fusion protein (reviewed in reference 25). Binding of the primary receptor, CD4, induces conformational changes in gp120 that stabilize its core and alter the orientation of the V1/V2 and V3 loops, thereby exposing the binding site for a second receptor, a chemokine receptor. This process also exposes the TM subunit to fusion-inhibiting antibodies and peptides, suggesting that the fusion process has been partially triggered. Binding of the second receptor allows increased exposure of the TM subunit and its refolding into a hairpin structure, which drives the fusion process.
Very little is known about the molecular mechanism of the hybrid two-step fusion activation process. Our hypothesis is that receptor binding induces a conformational change in the SU subunit that is transmitted to the TM subunit by a conformational coupling mechanism to trigger the first steps of fusion. Low pH may allow or stabilize a conformation required to complete fusion. We are using avian sarcoma/leukosis virus subtype A (ASLV-A) as a model system for studying two-step fusion activation because a single viral glycoprotein (EnvA) and a single receptor (Tva), at temperatures of ≥22°C, are sufficient to initiate fusion activation (16, 21, 27, 33, 52). Furthermore, a soluble 47-residue domain of the receptor (sTva) (64) is sufficient for these fusion-triggering properties. The ASLV-A env protein, Env-A, exists in a typical retroviral trimer of SU-TM heterodimer subunits format. Examination of the SU-A sequence and mapping of the variable regions (11) reveal that the variable regions are spread throughout SU-A (Fig. 1A) and that there is no proline-rich hinge region. Thus, the SU-A structure is likely to be organized more like that of gp120 and HA1 than that of the murine leukemia virus SU.
FIG. 1.
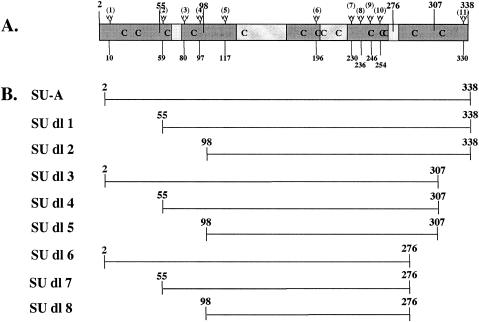
SU-A subunit and deletion (SU-A dl) mutant proteins. (A) Diagram of the SU subunit of the ASLV-A envelope protein. The two major variable regions are the central gray boxes; the minor variable regions are the smaller gray boxes. “Branches” mark the 11 glycosylation sites, and their residue numbers are given below. Cysteines are designated “C.” The larger numbers at the top of the figure designate the sites of the deletions for the mutant SU-As. (B) Depiction of the SU-A domain segment included in each deletion mutant protein.
The ASLV-A receptor, Tva, is a member of the low-density lipoprotein receptor (LDLR) family. LDLR family members were originally recognized for their ligand uptake capabilities (35). LDLR family members also serve as receptors for a variety of pathogens (1, 4, 30). It is becoming clear that LDLR family members also have important roles as transducers of extracellular signals in development (reviewed in reference 61) and in maintenance of the nervous system (reviewed in reference 13). The physiological role of Tva has yet to be determined.
All LDLR family members bind their ligands through their ligand binding repeat (LBR) domains. These LBR domains consist of a 40-residue repeat with six invariant Cys residues that form three invariant disulfide bonds. In addition, these LBRs contain a cluster of acidic residues near their C termini, four of which participate in the chelation of a Ca2+ ion. Variable arrangements of multiple LBR domains provide specificity to the various members of the LDLR family. Tva is the simplest known family member, having only one LBR domain. sTva is a soluble form of this single LBR domain.
As a first step in elucidating the mechanisms by which information is transmitted from the receptor binding subunit to the fusion subunit to trigger fusion and how low pH completes the process, we sought to examine the effect of receptor binding and subsequent low-pH exposure on an isolated SU subunit or a receptor binding fragment of SU-A. We therefore prepared a plasmid for the expression of SU-A and a series of nested-deletion mutant proteins. We found that only the full-length SU-A subunit was capable of binding sTva with high affinity. We purified this protein and characterized its interactions with sTva and the subsequent effects of low pH on the complex.
MATERIALS AND METHODS
sTva.
The pMal/sTva 47 plasmid, which encodes a fusion protein consisting of maltose binding protein and a 47-residue protein encompassing the single LBR of Tva, has been described previously (33, 64). sTva was expressed and purified, refolded, and cleaved as previously described (64). The lyophilized sTva fractions from the final high-pressure liquid chromatography purification step were stored as a powder at room temperature. Aliquots were dissolved in appropriate buffers as needed. The purified sTva is a monomeric protein whose theoretical molecular mass is 5,114 Da.
SU-A and SU-A deletion mutant proteins.
cDNAs encoding SU-A and a series of proteins with nested deletion mutations were prepared and inserted into pMTBipC (Invitrogen) for expression in Schneider S2 cells (Invitrogen) as follows. An SnaBI site was generated at the codon for V2, V55, or V98, and a BstEII site was generated at G216, G307, or the 3′ end of SU-A in the pCB6/WTA plasmid (27) by quick-change mutagenesis (Stratagene) in accordance with the manufacturer's instructions. The SnaBI-BstEII fragments were excised and inserted into the SmaI-BstEII sites in pMTBiPC. The resulting plasmids encode secreted proteins (via the drosophila BiP secretion signal) under the control of the inducible metallothionein promoter. The cloning strategy leaves a five-residue extension (RSPWP) N terminal toV2, V55, or V98 and both a V5 epitope tag and six His residues at the C termini. The inserts were verified by sequencing, and stable cell lines were prepared in accordance with the manufacturer's instructions.
Expression and purification of SU-A.
Cell lines were maintained in Sigma serum-free insect medium I supplemented with 10% heat-inactivated fetal bovine serum, 1× penicillin-streptomycin, 1× l-glutamine, 1× pyruvate, and 300 μg of hygromycin per ml (all from Gibco-BRL). For expression of the various SU-A proteins, cells were removed from the dishes, washed two times with phosphate-buffered saline (PBS) containing calcium and magnesium (Cellgro), and plated at 1 × 106 to 2 × 106/ml in Sigma serum-free insect medium I supplemented with 1× penicillin-streptomycin, 1× l-glutamine, 1× pyruvate, and 1% Ex-Cyte (Serologicals Corp.). Two days later, CuSO4 was added to a final concentration of 1 mM. Cells were fed with additional medium as needed to keep them from reaching stationary phase and harvested 5 days postinduction. Culture supernatants were collected and cleared of cell debris, and the supernatant was centrifuged at 31,400 × g for 20 min. The supernatant from this second centrifugation was diluted by adding an equal volume of 2× NTB (1× NTB is 50 mM HEPES, 400 mM NaCl, 15% glycerol, and 5 mM imidazole, pH 7.0) and then an additional one-third volume of 1× NTB. The pH of this mixture was readjusted to 7.0, and the solution was allowed to bind pre-equilibrated TALON (BD Biosciences) by gently rotating the mixture for 3 h at 4°C. This resin was chosen because proteins can be eluted from the column with lower concentrations of imidazole than from the more common divalent cation chelating resin Ni-nitrilotriacetic acid agarose (Ni-NTA) (QIAGEN); the higher concentrations of imidazole required for Ni-NTA elution precipitated SU-A on the column. The TALON was collected, washed copiously by filtration, including a stringent wash with NTB containing 400 mM NaCl and 2% Tween 20, and re-equilibrated with 1× NTB. The washed TALON was then loaded into a column, and bound protein was eluted with NTB containing 50 mM imidazole and 4 mM CaCl2. Fractions containing SU-A were combined and diluted 1:1 with SPB (25 mM HEPES, 10% glycerol, 0.02% sodium azide, pH 6.8) containing 60 mM NaCl, and the pH was adjusted to 6.8. SP-Sepharose (Amersham) was precharged with 500 mM NaCl in SPB and then equilibrated with SPB containing 110 mM NaCl. The SU-A solution was allowed to interact with the SP-Sepharose during 1 to 2 h of gentle rotation at room temperature, loaded into a column, washed with 20 column volumes of SPB-110 mM NaCl, and then the SU-A was eluted with SPB containing 250 mM NaCl. SU-A-containing fractions were combined, concentrated to approximately 1 mg/ml, and stored at 4°C until used.
Biotinylation of sTva.
sTva was conjugated to EZ-Link Sulfo-NHS-Biotin (Pierce) dissolved in PBS supplemented with 1 mM MgCl2. The reaction was allowed to proceed for a minimum of 20 h at 4°C to inactivate any remaining Sulfo-NHS-Biotin before using the biotinylated sTva. In some cases, the biotinylated sTva was separated from unconjugated Sulfo-NHS-Biotin on a Superdex G-75 column (Amersham).
Coprecipitation assays.
To coprecipitate sTva with SU-A, SU-A was bound via the V5 tag to anti-V5 (Invitrogen) that had been prebound to protein G-agarose beads (Roche). After extensive washing, biotinylated sTva was added in 100 μl of buffer and allowed to bind for 1 h with rotation at 4°C. The beads were then washed extensively, boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, resolved by SDS-PAGE, transferred to nitrocellulose, and visualized with horseradish peroxidase-conjugated streptavidin. SU-A was coprecipitated with sTva in a similar manner, except that biotinylated sTva was bound to avidin-agarose beads, SU-A was bound for 1 h in 100 μl of immunoprecipitation buffer (20 mM HEPES [pH 7.4], 130 mM NaCl, 1% NP-40), and the blot was probed with horseradish peroxidase-conjugated anti-V5.
Surface plasmon resonance.
Surface plasmon resonance biosensor data were collected on a BiaCore 3000 optical biosensor (BiaCore AB, Uppsala, Sweden). To orient sTva on the biosensor chip, the chip was first conjugated with streptavidin. This was accomplished with an amine coupling kit (BiaCore AB) in accordance with the manufacturer's instructions. Briefly, the chip was activated with N-ethyl-N′-(dimethylaminopropyl)carbodiimide in buffer HSB-P (10 mM HEPES [pH 7.4], 150 mM NaCl, 0.005% polysorbate 20 [P20, Biacore no. BR-1000-54]) for 7 min. Streptavidin (100 mg/ml in 50 mM sodium acetate) was allowed to bind for 7 min, and the remaining free carboxyl groups were quenched with 1 M ethanolamine HCl, pH 8.5, for an additional 7 min. Fast protein liquid chromatography-purified, biotin-labeled sTva diluted in HSB-P (HSB-P alone for the control chip) was manually injected over the streptavidin-conjugated chip to precisely control the surface density to 15 resonance units. This low conjugation density was necessary to obtain the high-affinity data measured here. Conjugation at higher densities resulted in apparent affinities that were significantly lower. This is most likely due to depletion of SU-A from the buffer layer adjacent to the chip because of a binding rate that exceeds the rate of diffusional replenishment from the flowing solution (32). Serial 1:1 dilutions of samples with HBS-P were prepared. Kinetic studies were performed on duplicate injections at 25°C with a flow rate of 50 μl/min. Samples were injected for 60 s and dissociated for 300 s. In some cases, dissociation was measured for 1,000 s. The chip was regenerated by injection of 30 μl of 50 mM NaOH-1 M NaCl. The binding kinetics were calculated by subtracting binding to the blank streptavidin chip from binding to the sTva-conjugated streptavidin chip, and the data were fitted with a 1:1 Langmuir binding model with the Biaevaluation software package, version 3.1.
CD.
The circular dichroism (CD) spectra of SU-A were measured on an AVIV 215 CD spectrophotometer scanning at 0.5-nm intervals with an averaging time of 0.5 s/datum point. The temperature was maintained at 24°C by a circulating water bath. The values from three scans were averaged, and the resulting spectrum was deconvoluted with software supplied by the manufacturer. The contributions of the buffer were subtracted from all spectra.
Protease digestion.
SU-A was mixed either with sTva in a 1:4 molar ratio or with an equal volume of PBS. The sample was allowed to equilibrate at 4°C for 30 min and then incubated at the indicated temperature for an additional 30 min. For thermolysin digestion, trypsin (Sigma) was added to a final concentration of 1.5 mg/ml and the reaction was allowed to proceed at 4°C for 20 min before being quenched with 50 mM EDTA. For trypsin digestion, 2,000 U of sequencing grade trypsin (Promega) was added and the digestion was carried out for 30 min at 4°C before being quenched with 2 mg of soybean trypsin inhibitor (Sigma) per ml. For digestion with GluC, samples were incubated with 2 mg of GluC per ml for 2 h at 4°C and then stopped with 70 mg of Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK; Sigma) per ml. Samples were resolved by SDS-PAGE and visualized by silver staining.
Fluorescence spectroscopy.
All fluorescence measurements were taken with a Jobin Yvon Fluorolog 3 spectrofluorimeter equipped with an F-3004 Peltier sample cooler controlled by a Wavelength Electronic LFI-3751 temperature controller. The excitation slits were set to a 1-nm band pass, while the emission slits were set to 3 nm. All samples were prepared at 5 μM, and measurements were made with 4-mm path length quartz cuvettes. All spectra were measured at 22°C. For intrinsic tryptophan fluorescence (ITF) measurements, the samples were excited at 295 nm and spectra were taken from 305 to 405 nm by three-scan averaging. For bis-ANS (4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid) fluorescence measurements, samples were excited at 395 nm and scanned from 410 to 580 nm. To determine the fluorescence resonance energy transfer (FRET) between tryptophans and bis-ANS, samples were excited at 295 nm and the emission was scanned from 305 to 580 nm. For measurements of maximum bis-ANS and FRET, bis-ANS was added to the sample for a final bis-ANS concentration of 50 μM and mixed with the solution by pipetting up and down. The sample was brought to 22°C and allowed to equilibrate for at least 5 min before spectra were taken. To determine the titer of bis-ANS binding to SU-A, sTva, and their complex, the sample was allowed to equilibrate in the 22°C sample chamber for at least 5 min in the absence of bis-ANS and a spectrum was taken. Two-nanomole (roughly 5 μM) increments of bis-ANS were then added directly to the sample until the sample appeared to reach its saturation point. Spectra were taken at each concentration. The spectrum of each respective buffer under the respective condition was subtracted before plotting the data.
RESULTS
Generation of a soluble SU-A subunit and soluble SU-A deletion mutant proteins.
Our goal was to obtain a pure, soluble form of SU-A and/or a minimal receptor binding fragment for biochemical and biophysical studies of the conformational changes in SU-A induced by receptor binding. Because SU-A contains 11 glycosylation sites (Fig. 1A), most of which are required for proper folding and receptor binding (17) and because the protein also contains 14 cysteines, we were unable to recover functional protein after expression in Escherichia coli (S. Delos, unpublished results). To produce large quantities of protein, we therefore turned to a Drosophila Expression System (Invitrogen). Secreted proteins harboring a C-terminal V5 tag and six His residues at their C termini can be expressed with the pMTBiP/V5-His vector. We prepared stable S2 cell lines for the expression of secreted, C-terminally tagged SU-A and a nested set of N- and/or C-terminal deletion mutants (Fig. 1B). We induced these cells and examined culture supernatants 4 days later for the expression of secreted, V5-tagged proteins. In each case, a soluble, secreted protein was produced that could be observed as a discrete band on an immunoblot (Fig. 2A). Most deletion mutant proteins migrated at a position somewhat higher than the predicted molecular weight (Fig. 2A and Table 1), as is common for glycosylated proteins.
FIG. 2.
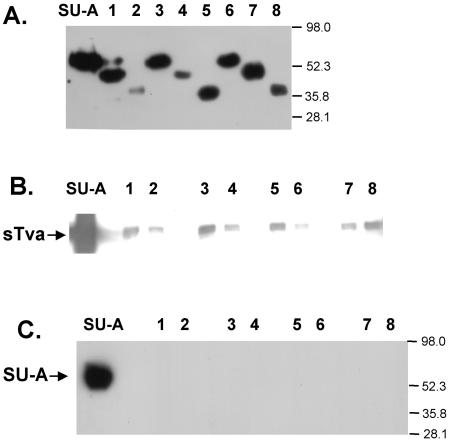
Expression and receptor binding of SU-A and SU-A deletion mutant proteins. (A) Expression of secreted SU-A and SU-A deletion mutant proteins. Culture supernatants were harvested 4 days postinduction, and samples were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with an anti-V5 antibody. Coprecipitation of sTva with SU-A (B) or SU-A with sTva (C) was performed as described in Materials and Methods. Lane numbers refer to the deletion mutant proteins depicted in Fig. 1B. The values on the right are molecular sizes in kilodaltons.
TABLE 1.
Characteristics of SU-A deletion mutant proteins
| Protein | Base MWa | No. of glycosylated sites | MWthb | MWappc | Binds sTvad | Bound by sTvad |
|---|---|---|---|---|---|---|
| SU-A | 40,700 | 11 | 52,129 | 58,393 | +++ | +++ |
| SU dl 1 | 34,870 | 10 | 45,260 | 45,945 | + | − |
| SU dl 2 | 30,140 | 7 | 37,413 | 37,625 | ± | − |
| SU dl 3 | 37,290 | 10 | 47,680 | 52,842 | + | − |
| SU dl 4 | 31,450 | 9 | 40,801 | 42,416 | ± | − |
| SU dl 5 | 26,730 | 6 | 32,964 | 33,375 | + | − |
| SU dl 6 | 33,880 | 10 | 44,270 | 50,772 | − | − |
| SU dl 7 | 28,050 | 9 | 37,401 | 41,578 | ± | − |
| SU dl 8 | 23,320 | 6 | 29,554 | 32,067 | + | − |
Theoretical molecular weight based upon amino acid sequence.
Calculated on the basis of the normal S2 glycan unit weighing 1,039 Da (26).
Calculated from the mobility on an SDS-10% PAGE gel as visualized on an immunoblot probed with anti-V5.
+++, very high; +, moderate; ±, low; −, negligible binding based on visual inspection of the gels shown in Fig. 2B and C.
Our criterion for a functional protein is its ability to bind its receptor. We therefore determined the ability of SU-A and each deletion mutant protein to interact with a soluble receptor fragment, sTva (64), in reciprocal coprecipitation assays. As shown in lanes 1 of Fig. 2B and C, SU-A and sTva were readily coprecipitated by each other. The results obtained with the deletion mutant proteins were more variable. When we asked whether the SU-A species could coprecipitate sTva (Fig. 2B), a small amount of binding was observed for deletion mutant proteins 1, 3, 5, and 8. Less binding was observed for mutant proteins 2, 4, and 7. Deletion mutant protein 6 exhibited only a background level of binding. The amount of sTva that could be bound by even the most effective mutant proteins was minimal compared to the amount coprecipitated by SU-A. When we performed the experiment in the opposite direction, we were unable to coprecipitate any of the deletion mutant proteins with sTva (Fig. 2C). No signal was observed, even when the immunoblots were exposed for long periods of time (data not shown). These results suggest that residues near the N and C termini of the SU-A subunit are required for proper folding of SU-A into a receptor binding-competent module. Therefore, only full-length SU-A was used in further studies.
SU-A is a monomeric, globular protein.
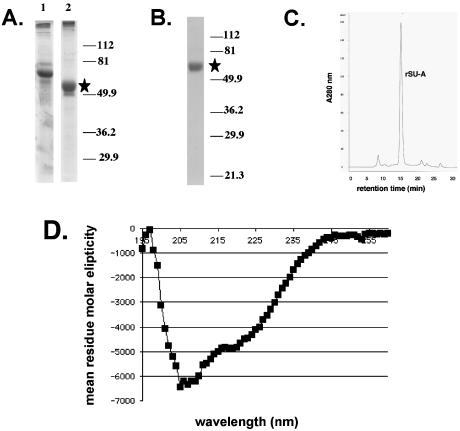
A two-step protocol was developed to purify SU-A. In the first step, SU-A was concentrated and partially purified from culture supernatants on TALON, a cobalt-chelating resin (compare lane 2 with lane 1 of Fig. 3A). A subsequent pass over SP-Sepharose, as described in Materials and Methods, yielded pure protein (Fig. 3B). SU-A migrates on a sizing column as a monomer with an apparent molecular mass of 52.4 kDa (Fig. 3C). This is in good agreement with the theoretical molecular mass (Table 1), suggesting that the protein is globular. The narrow width of the SU-A peak and the lack of a leading or trailing shoulder further suggest that the folding and glycosylation of the protein are uniform. The overall structural characteristics of SU-A were determined by CD (Fig. 3D) as described in Materials and Methods. The protein has 21% α-helix, 25% β-sheet, and 43% random coil.
FIG. 3.
Purification and characterization of SU-A. Initial concentration and partial purification of SU-A on TALON beads (A) and purification to a single species on SP-Sepharose (B) were analyzed by resolving column fractions by SDS-PAGE and visualizing them by Coomassie staining. The position of the SU-A band is marked by a star. The values on the right of panels A and B are molecular sizes in kilodaltons. Lanes in panel A: 1, S2 culture supernatant; 2, SU-A fraction eluted with 50 mM imidazole. (C) Size exclusion chromatography analysis of the apparent molecular weight of purified SU-A. An aliquot of the purified SU-A solution was separated on a Superdex G-200 column (Amersham). The apparent molecular weight of the major peak was determined by comparison of the retention time of SU-A with those of a set of protein standards (Bio-Rad). (D) CD spectrum of SU-A. The CD spectrum of SU-A (4.75 μM) was measured on an AVIV 215 CD spectrophotometer as described in Materials and Methods.
SU-A binds sTva with high affinity.
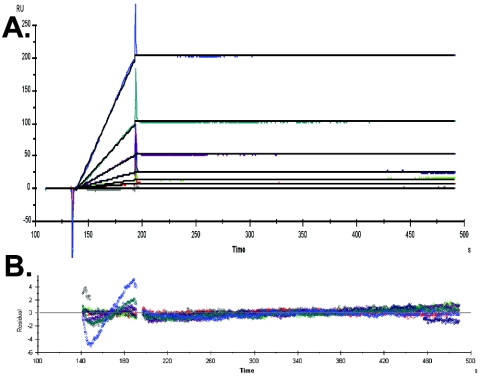
We have shown that SU-A associates with sTva in reciprocal coprecipitation assays (Fig. 2B and C). To further characterize the association, we used a biosensor assay to determine the binding kinetics of SU-A with sTva. As sTva does not have any lysine residues, it can be covalently labeled specifically at its N terminus with Sulfo-NHS-Biotin. This label does not interfere with the ability of sTva to bind SU-A (Fig. 2) or to trigger EnvA (33), and the biotinylated protein remains monomeric. As shown in Fig. 4A, SU-A and sTva associate with high affinity and this high affinity is characterized by a negligible off rate. The data, fitted to a 1:1 Langmuir binding model, yielded an apparent KD of 3.05 × 10−12 M (χ2 value = 0.53), with an on rate (ka) of 7.05 × 104 M−1 s−1 and an off rate (kd) of 2.15 × 10−7 s−1. Extending the measurement time for the off rate did not alter the apparent kd. The data presented are for a representative experiment. The residuals for this experiment are shown in Fig. 4B. Although the apparent kd for the binding of SU-A and sTva is at the lower limits of BiaCore sensitivity, we have reproduced these values with a variety of SU-A and sTva preparations, obtaining apparent KD values ranging between 2.5 × 10−12 and 5 × 10−12 M (Delos, unpublished). Moreover, we have detected differences between the off rates for the wild-type SU-A/sTva complex and those for SU-A and mutant sTvas (Delos, unpublished). In this regard, it is worth noting that to obtain linear data, BiaCore analysis of the kinetics of binding between SU-A and sTva required binding sTva to the chip at a very low density (see Materials and Methods). At higher conjugation densities, the binding was diffusion limited (32) and the measured affinities were 10- to 1,000-fold lower (Delos, unpublished). Because the affinity of SU-A for sTva is so high, association of 1:1 mixtures was assumed to be complete in the experiments described below.
FIG. 4.
Surface plasmon resonance determination of SU-A/sTva binding kinetics. Surface plasmon resonance biosensor data were collected on a BiaCore 3000 optical biosensor (BiaCore AB) as described in Materials and Methods. (A) BiaCore sensorgram with superimposed data fits. (B) Residuals for the traces shown in panel A. The concentrations of SU-A applied to the sTva-conjugated chip were, 30, 15, 7.5, 3.75, 1.88 0.94, 0.47, and 0.23 nM, respectively.
The SU-A/sTva binding interface includes hydrophobic surfaces.
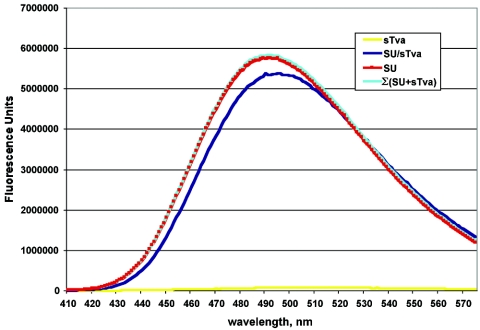
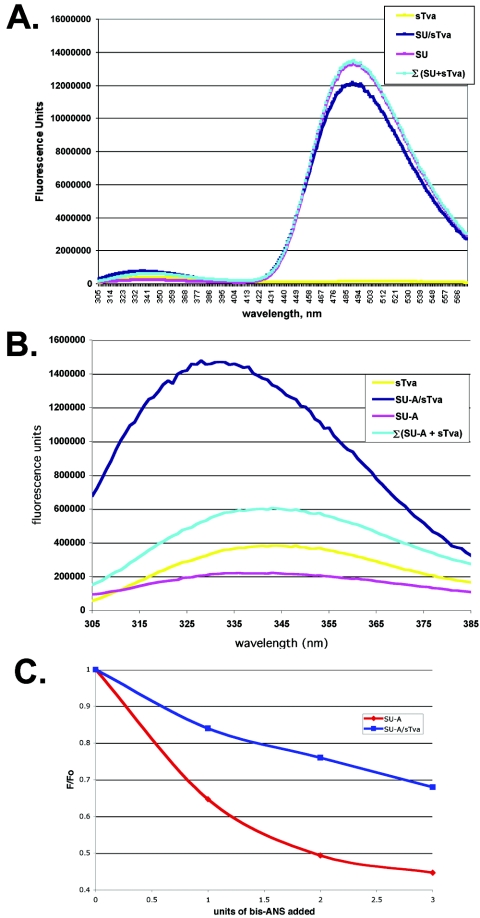
Charged residues have been implicated in SU-A/sTva binding (56). Indeed, charged residues have been identified as critical for association of many LDLR family members with their ligands (2, 18, 55, 65). However, the high affinity of the SU-A/sTva association suggested that hydrophobic interactions are also important. We therefore assessed the ability of bis-ANS to bind to SU-A, sTva, and the SU-A/sTva complex. Bis-ANS is soluble in aqueous solutions but readily binds to hydrophobic surfaces of proteins. Binding is accompanied by dequenching of the bis-ANS fluorescence. The amount of dequenching is proportional to the amount of bis-ANS bound, which is proportional the accessible hydrophobic surface. Binding of bis-ANS has been used to measure changes in exposed hydrophobic surfaces during virion assembly (12, 14, 62, 63) and fusion triggering (7, 14, 37, 40, 41). We therefore asked whether we could observe a change in the accessible hydrophobic surface area on SU-A and sTva before and after complex formation. We first determined the amount of bis-ANS required to saturate a 5 μM solution (2 nmol of total protein) of each protein (data not shown) and then determined the relative abilities of saturating solutions of bis-ANS to bind SU-A and sTva before and after complex formation. As shown in Fig. 5, bis-ANS binding to sTva is minimal. However, SU-A binds large amounts of bis-ANS. Less bis-ANS is able to bind to the SU-A/sTva complex than to SU-A, and the wavelength of the fluorescence is red shifted. These results suggest that the hydrophobic surface area of SU-A has decreased and the environment of the bound bis-ANS has been altered.
FIG. 5.
Comparison of bis-ANS binding to SU-A, sTva, and the SU-A/sTva complex. A saturating concentration (50 μM) of bis-ANS was added to 5 μM solutions of SU-A (red) and sTva (yellow) and a pre-equilibrated equimolar mixture of SU-A and sTva (blue), and the resulting fluorescence spectrum was measured as described in Materials and Methods. The data presented here are averages from two or more experiments. In each case, the spectrum of bis-ANS in buffer was subtracted. The Σ(SU-A+sTva) (cyan) curve is the sum of the theoretical contribution of each component to an equimolar mixture in which no interaction occurs.
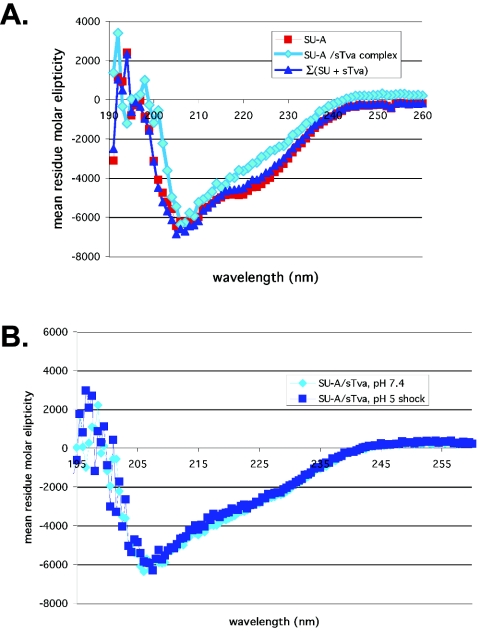
sTva binding changes the CD spectrum of SU-A.
Upon binding to CD4, gp120 undergoes significant conformational changes that stabilize several structural elements (43, 51). These changes are reflected by changes in the CD spectrum (51) and by altered protease and antibody sensitivity of exposed loops (reviewed in reference 25). We therefore first asked whether we could observe changes in the CD spectrum of SU-A upon receptor binding. As shown in Fig. 6A, association of sTva with SU-A results in a loss of α-helical content. Because sTva has only 47 residues whereas SU-A has 367, the bulk of the contribution to the spectrum is from SU-A (notice the minimal change in the value of the theoretical sum of the SU-A and sTva contributions [dark blue trace] from that of SU-A [red trace]). Furthermore, it has been reported that LBRs do not alter their structure upon ligand binding (18). For these reasons we expect that the observed loss of α-helical character reflects changes in SU-A structure induced by sTva binding.
FIG. 6.
Comparison of CD spectra of SU-A and the SU-A/sTva complex. (A) CD spectra of SU-A (red) and a 1:1 molar mixture of SU-A and sTva (cyan) that had been incubated at 37°C for 30 min were determined as described in the legend to Fig. 3D. The theoretical sum of the mean residue contribution of an equimolar mixture of nonbinding SU-A and sTva at neutral pH (dark blue) is provided for comparison. (B) Comparison of the spectrum of the SU-A/sTva complex in panel A (cyan) with that obtained for the complex that had been subjected to 10 min at pH 5 at 37°C and then reneutralized (dark blue).
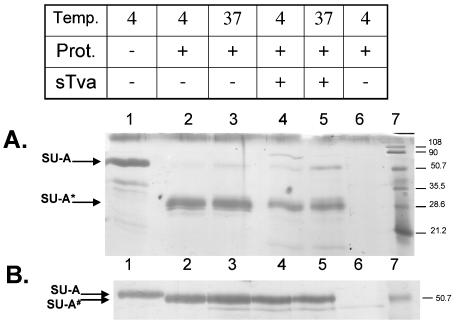
Protease sensitivity of SU-A is neither receptor nor temperature dependent.
We next asked if we could detect changes in the conformation of SU-A upon receptor binding by assessing changes in the susceptibility of SU-A to proteases. We examined the protease sensitivity of SU-A in the presence and absence of sTva at both 4 and 37°C. The results for digestion by thermolysin and trypsin are presented in Fig. 7A and B, respectively. As shown in Fig. 7A, a thermolysin-sensitive site(s) is present in the isolated SU-A monomer, resulting in a resistant fragment of approximately 30 kDa. This same fragment is obtained whether the reaction is performed at 4 or 37°C, in the absence or presence of soluble receptor. Similarly, digestion with trypsin gave an ∼51-kDa fragment at both 4 and 37°C, whether or not the receptor had been prebound (Fig. 7B). Digestion with GluC yielded two bands that were also both temperature and receptor independent (data not shown).
FIG. 7.
Protease (Prot.) sensitivity of SU-A and the SU-A/sTva complex. (A) Sensitivity to thermolysin. (B) Sensitivity to trypsin. Purified SU-A was incubated on ice for 30 min in the absence (lanes 1 to 3) or presence (lanes 4 to 5) of sTva and heated to 37°C for 30 min (lanes 3 and 5) or kept on ice (lanes 1, 2, 4, and 6). Samples were then incubated with thermolysin (1.5 mg/ml; panel A, lanes 2 to 6) or trypsin (2,000 U; panel B, lanes 2 to 6) for 30 min on ice and quenched with EDTA (55 mM) (A) or soybean trypsin inhibitor (1 mg/ml) (B). Untreated SU-A is shown in lanes 1. Protease and inhibitor only are shown in lanes 6. sTva runs off the bottom of these gels. SU-A*, thermolysin-resistant fragment; SU-A#, trypsin-resistant fragment. The values on the right are molecular sizes in kilodaltons.
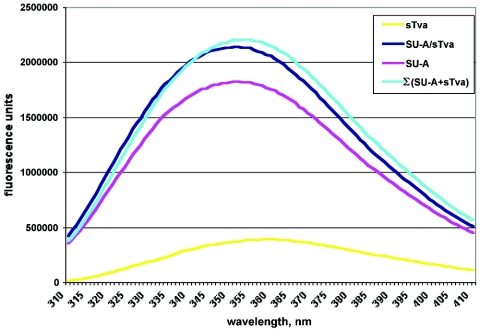
Tryptophan environment is altered by SU-A/sTva complex formation.
ITF has been used to monitor conformational changes in proteins under different conditions. We therefore asked whether ITF might be a useful tool for monitoring conformational changes in SU-A induced by sTva binding. A plot representing the averaged fluorescence values from three or more experiments is shown in Fig. 8. sTva has a fluorescence maximum of 358 nm, indicating that its two tryptophans are in a hydrophilic environment, consistent with published nuclear magnetic resonance structures (64, 66). The fluorescence maximum for SU-A is 351 nM, suggesting that most SU-A tryptophans are in a less hydrophilic environment than those of sTva. The ITF of the SU-A/sTva complex was slightly blue shifted and slightly quenched relative to the value expected for a noninteracting mixture of SU-A and sTva, suggesting that one or more tryptophans are involved at the SU-A/sTva interface.
FIG. 8.
ITF of SU-A, sTva, and the SU-A/sTva complex. (A) Neutral pH. Five-micromolar samples of SU-A (purple) and sTva (yellow) and an equimolar mixture of SU-A and sTva (dark blue) were incubated at 37°C for 30 min and then equilibrated to 22°C, and ITF was measured between 305 and 405 nm after excitation at 295 nm. The theoretical ITF of a mixture of noninteracting SU-A and sTva (cyan) is also shown. In all cases, the contribution of the buffer has been subtracted from the spectra.
To further assess the possible occlusion of tryptophans in SU-A upon complex formation, we performed FRET assays between tryptophan and bis-ANS for SU-A, sTva, and the SU-A/sTva complex (Fig. 9). Figure 9A shows the complete spectra for the FRET assays. Figure 9B is an expansion of the tryptophan fluorescence portion (305 to 385 nm) of these spectra. FRET is manifested as a decrease in tryptophan fluorescence (305 to 385 nm) and a concomitant increase in bis-ANS fluorescence (420 to 580 nm). As expected, since sTva does not bind bis-ANS to any measurable extent, there is minimal FRET between sTva and bis-ANS (Fig. 9A). This result is consistent with the tryptophans in sTva being near acidic residues (64, 66). In contrast, there is considerable FRET between tryptophans in isolated SU-A and bis-ANS (Fig. 9A, purple trace just below cyan trace). In fact, most of the tryptophan fluorescence in isolated SU-A is quenched (Fig. 9B, purple trace), suggesting that most of the tryptophans in isolated SU-A are near hydrophobic patches at the surface of SU-A. When the FRET for the SU-A/sTva complex (Fig. 9A, dark blue trace) was examined, the wavelength of the maximum fluorescence was slightly blue shifted (2 nm) and the amplitude of the bis-ANS fluorescence was decreased compared to that of isolated SU-A (Fig. 9A). Correspondingly, the residual tryptophan fluorescence for the complex was both larger and blue shifted (more than 10 nm) compared to that for isolated SU-A (Fig. 9B, dark blue trace versus purple trace). Thus, tryptophans accessible to bis-ANS in isolated SU-A are no longer accessible in the complex. A further indication that one or more tryptophans in SU-A are occluded by SU-A/sTva complex formation comes from a bis-ANS titration experiment. When we titrated the amount of bis-ANS required to saturate SU-A, we noticed that the fluorescence of one or more tryptophans in SU-A is rapidly quenched by small amounts of bis-ANS and that this rapid quenching was no longer observed after complex formation (Fig. 9C).
FIG. 9.
FRET between tryptophan and bis-ANS. (A) FRET between tryptophan and bis-ANS was measured by monitoring the emission between 305 and 580 nm after excitation at 295 nm. The theoretical FRET of a mixture of noninteracting SU-A and sTva is also shown (cyan). In all cases, the contribution of the buffer has been subtracted from the spectra. (B) Blow-up of the residual tryptophan fluorescence. Colors are the same as those in Fig. 8. (C) Titer of bis-ANS binding measured by loss of ITF. Increasing amounts of bis-ANS were added to SU-A or the SU-A/sTva complex, and ITF was measured. After subtraction of the contribution of the buffer, the spectra were normalized to the maximum fluorescence of the untreated sample at λmax (F0). A plot of the change in fluorescence (F/F0) in the presence of increasing amounts of bis-ANS is shown.
Exposure to low pH does not induce further detectable changes in the SU-A/sTva complex.
Because ASLV fusion requires the sequential application of receptor followed by low pH, we asked if we could observe additional effects of low pH on the SU-A/sTva complex. A hallmark of LDLR function is the ability to release its ligand in an early endosomal compartment. This appears to be triggered by a conformational change in the receptor resulting from the decreased pH of this compartment (29). We therefore asked if the application of low pH might be sufficient to dissociate SU-A and sTva in our BiaCore binding assays. No increase in the off rate was observed at any pH between 7.4 and 4.8 (data not shown).
We next asked if we might detect low-pH-induced conformational changes in the complex by CD. As shown in Fig. 6B, no significant changes in the CD spectrum of the SU-A/sTva complex were observed after application of a low-pH pulse at 37°C.
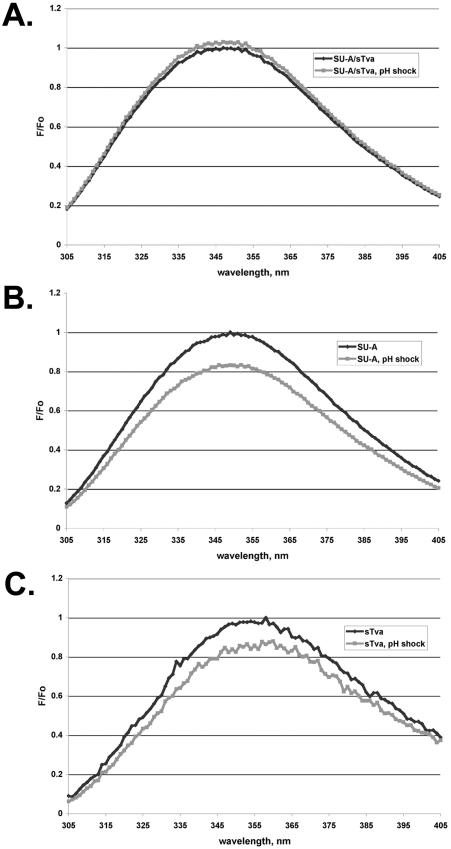
We also examined the effect of a low-pH pulse on the ITF of the SU-A/sTva complex. SU-A and sTva were premixed and “triggered” (incubated at 37°C for 10 min) as for Fig. 8, and an ITF spectrum was measured. The complex was then adjusted to pH 5 and incubated at 37°C for 10 min. After reneutralization, a second spectrum was measured. As shown in Fig. 10A, there was no change in the spectrum. Treatment of either SU-A or sTva with a low pH caused a decrease in their ITF spectra (Fig. 10B and C) indicative of a more flexible structure, likely due to partial unfolding of the isolated proteins. Thus, complex formation protects both SU-A and sTva from irreversible changes induced by exposure to a low pH.
FIG. 10.
pH dependence of tryptophan fluorescence. ITF was measured on the preformed SU-A/sTva complex (A), SU-A (B), or sTva (C) as described in the legend to Fig. 8. Samples were then treated with acid to pH 5, held for approximately 10 min, and then reneutralized with base, all at 22°C. The ITF of the reneutralized samples was then measured. The spectrum of the buffer alone was subtracted from each spectrum, and the spectrum of the low-pH-treated samples was corrected for the respective dilution factors. The resulting spectra were normalized to Fmax at the λmax (F0) of the non-pH-treated sample. Black trace, before low-pH treatment; grey trace, after low-pH treatment.
DISCUSSION
Search for a minimal RBD.
To aid analysis of the interactions between SU-A and sTva, we attempted to define a minimal RBD. We therefore prepared a series of deletion mutant forms of SU-A. Because the regions in ASLV SU sequences that define receptor specificity occur toward the center of the sequence, we only deleted sequences at the N and C termini. Although all of the deletion mutant proteins were expressed, appeared to undergo normal glycosylation, and were secreted from Drosophila S2 cells, none of them retained significant receptor binding activity. One possibility is that the SU-A deletion mutant proteins form oligomers that prevent sTva binding. The mutant proteins are not grossly aggregated, however, because they remain in solution after 30 min of centrifugation at 31,000 × g and during prolonged storage. Another possibility is that residues in the N- and C-terminal regions of SU-A affect (formation of) important structural features of the RBD. Similar results have been observed for N- and C-terminal deletion mutant forms of gp120 (19, 68). Interestingly, residues in these segments of gp120 also appear to interact with the HIV TM (8, 36, 45, 54). If a similar relationship among the ASLV-A receptor binding site, the SU-A N and C termini, and TM-A exists, it lends support to the hypothesis that for receptor-triggered fusion proteins there is a direct allosteric conduit between the receptor binding site and the SU-TM interface.
Characteristics of SU-A.
As has been found for HIVgp120 (50) and simian immunodeficiency virus (SIV) gp120 (39), SU-A purified after expression in drosophila S2 cells is a stable, soluble, monomeric glycoprotein. Similarly, the HA “top” (HA1 residues 28 to 238), isolated by proteolytic cleavage of low-pH-treated HA, is monomeric (58). The ability to isolate and store these SU-A-like subunits as monomers suggests that the primary trimerization motifs in these fusion proteins are contained within the TM subunits, as previously suggested for EnvA (22).
We have shown previously that glycosylation of sites within SU-A is necessary for proper folding, secretion, and receptor binding in the context of full-length EnvA (17). It is well known that insect cells do not modify their glycans to the same extent as do mammalian cells (26). A concern was that this difference in glycan modification might affect the structure of SU-A and its ability to bind receptor. An added concern was that not all of the glycosylation sites required for proper folding and receptor binding would be used. However, the abilities to be secreted, to be purified and stored as a stable, monomeric protein, and to bind sTva with high affinity show that complex glycans are not necessary for SU-A function. It has been reported that, in the context of an SU(-A)-immunoglobulin G fusion protein, 10 of the 11 potential glycosylation sites in SU-A are utilized (42). The apparent molecular weight of SU-A, determined by size exclusion chromatography, suggests that most of the potential glycosylation sites within SU-A are also utilized by the Drosophila glycosylation machinery (Table 1).
The SU-A-sTva interface.
SU-A binds sTva with a remarkably high affinity (KD = 3.0 pM) that is characterized by a negligible off rate (Fig. 4). This association is accompanied by a loss of α-helix (Fig. 6A). Although dissociation constants of other isolated retroviral SU proteins for their receptors have been reported to be in the 1 to 100 nM range (39, 51, 70), this is the first report of a picomolar dissociation constant. It has been shown that isolated (monomeric) SIV gp120 binds its primary receptor, CD4, in a 1:1 complex with a KD of 60 nM, whereas when in the context of trimeric env gp120 binds CD4 in a 3:1 ratio with a KD of 190 to 210 nM (39). The increase in affinity of isolated SIV gp120 for CD4 is characterized by a slower off rate. The reported affinities between trimeric full-length EnvA and sTva vary between 0.5 and 20 nM (5, 31, 69, 71). As for the SIV system, this may reflect differences in the binding of sTva to the trimer due to steric factors or to alterations in SU-A structure when it is part of the full-length protein rather than an isolated monomer. It has been observed that sTva binding to full-length, trimeric EnvA is cooperative (15), suggesting that binding to one SU-A subunit within a trimer facilitates binding to additional subunits. The high affinity of the binding between SU-A and sTva may help explain the unusual stability of receptor-triggered EnvA and the low-pH requirement for complete fusion. When the affinity of HA for its receptor was increased, the ability of HA to mediate the transition from small to large fusion pores was inhibited (53).
It is well documented that basic residues within their ligands are essential for binding to the LBRs of various LDLR family members (55). These residues are thought to interact with acidic residues present in the C-terminal sequences of all LBRs. Four of the implicated residues are directly involved in calcium coordination (23). Interestingly, the Ca2+-coordinating residues also appear to be at the ligand interface of known LBR-ligand structures (18, 57, 65). There are also additional acidic residues in each LBR that can interact with some ligands. Basic residues within the hr-2 domain of SU-A have been implicated in sTva interactions (56). The bis-ANS binding data show that hydrophobic interactions are also involved in SU-A/sTva complex formation. Although bis-ANS does not bind well to sTva, the published nuclear magnetic resonance structures clearly show areas of hydrophobicity on the surface of sTva (64, 66). However, these areas are small and bounded by highly charged residues. Repulsion by the charged residues may prevent the binding of bis-ANS to sTva. In contrast, our titration data suggest that more than 20 mol of bis-ANS can bind each mole of SU-A (J. Godby and S. Delos, unpublished results). When the SU-A/sTva complex was treated with bis-ANS, approximately 10% of the total bis-ANS binding expected for a 1:1 mixture of unpaired SU-A and sTva was lost. Thus, our data suggest that hydrophobic interactions make a contribution to the interaction of SU-A with sTva. These results support the model of Prevost and Raussens (55), in which hydrophobic, as well as ionic, interactions are important at an LBR-ligand contact site.
Our ITF and FRET data show that the tryptophan environment is altered upon complex formation. We believe that most of this change is due to tryptophans in both SU-A and sTva at the binding interface. An aromatic residue at W48 of sTva is required for infectivity (71). Mutation of this residue to A decreases the affinity of SU-A/sTva interaction, severely impairs the ability of the resulting sTva to trigger the conformational changes in EnvA that allow it to bind membranes, and impairs the ability of sTva to mediate infection (33, 71). By analogy with the known or modeled structures of LBRs with their ligands (18, 55, 65) and the β-propeller interface with the LBR4 and LBR5 modules of the LDLR ectodomain at pH 5.5 (57), we predict that W33 of sTva is also at the ligand interface. There are a number of candidate tryptophans in hr-1 and hr-2 of SU-A for involvement in sTva interactions. For example, Melder et al. have identified two tryptophans in SU-A hr-1 that are mutated when ASLV-A is grown on chicken cells in the presence of a soluble immunoglobulin G-Tva (quail isoform) construct (48).
Activation of fusion proteins has often been monitored by changes in their susceptibility to protease digestion. Indeed, we have shown previously that a thermolysin-sensitive site is exposed in the SU-A domain of the full-length trimeric EnvA protein upon association with sTva at fusion-permissive temperatures (27). Alterations in the orientation of variable loops 1, 2, and 3 of gp120 upon CD4 binding have been observed by changes in protease susceptibility (59). However, we were unable to identify a protease that gave different digestion patterns for monomeric SU-A before and after association with sTva. This may be due to the protection of such sites by glycosylation and/or a lack of exposed mobile loops. We did, however, observe a prominent 30-kDa, thermolysin-resistant fragment of SU-A whose formation was independent of both sTva association and temperature. The size of this fragment is consistent with its being equivalent to the thermolysin-resistant fragment of SU-A formed in full-length trimeric EnvA upon association with Tva in a highly temperature-dependent manner. If this is the case, then the thermolysin-sensitive site in SU-A is likely either at the SU-A-SU-A or the SU-A-TM-A interface in the native trimer.
Effect of low pH on the SU-A/sTva complex.
Application of a low-pH pulse to the SU-A/sTva complex did not significantly alter its structure as measured by CD (Fig. 6B) or cause complex dissociation as measured by surface plasmon resonance. Furthermore, ITF experiments revealed that complex formation protected both sTva and SU-A from an acid-induced increase in flexibility (Fig. 10). Thus, as for CD4 interaction with gp120 (51), sTva interaction with SU-A appears to have stabilized the components of the protein complex. By three independent methods, we were unable to observe any irreversible low-pH effects on the SU-A/sTva complex. In contrast, we have identified specific low-pH-induced irreversible conformational changes in the TM subunit of sTva-triggered EnvA (47b). One hypothesis is that low pH affects TM-A directly rather than causing additional changes in SU-A that are relayed to TM-A. We cannot, however, rule out the possibility of pH-induced changes at the SU-A surface (at the SU-A-SU-A and/or SU-A-TM-A interface), not measured by any of the techniques used here, that are relevant to late stages of fusion.
LDLR family members release their natural ligands when they encounter the decreased pH of early endosomes. However, the SU-A/sTva complex did not dissociate upon exposure to a low pH. This may be due to the absence of the epidermal growth factor-like (EGF) domain of Tva in sTva. The crystal structure of the LDLR ectodomain at low pH revealed that the β-propeller of the EGF domain interacts with the LBR modules critical for ligand binding (57). This observation led to the prediction that the EGF domain displaces natural ligands as the pH decreases. Because the natural ligand(s) for Tva is not known, we are unable to test this prediction for our system. However, the suggestion that ASLV-A virions that have been internalized after association with the transmembrane form of Tva (which contains the EGF domain) can be recycled to the plasma membrane (47) may indicate that neither a low pH nor the Tva EGF domain is sufficient to induce SU-A/Tva dissociation. It has been reported that when trimeric EnvA is triggered by sTva to bind target membranes at neutral pH, sTva is released (16, 33, 49). However, the presence of liposomes did not alter the dissociation kinetics of the SU-A/sTva complex at neutral pH (Delos, unpublished). This may mean that association of the TM-A subunit of the EnvA trimer with the target membranes (through the fusion peptide) exerts a structural influence on the SU-A subunit that disrupts its association with sTva.
SU-A, a hybrid receptor binding subunit.
In summary, we have shown that the SU-A subunit is a stable monomer that binds its receptor with a KD of 3.0 pM. This association induces a conformational change in SU-A resulting in loss of α-helical structure, occlusion of a hydrophobic surface(s), and occlusion of tryptophan residues as measured by loss of FRET between tryptophan and bis-ANS. We did not detect any irreversible changes in the SU-A/sTva complex following its exposure to a low pH. Nonetheless, these results are consistent with a two-step mechanism for ASLV-A virion-membrane fusion in which, in step 1, receptor binding induces conformational changes in the SU-A subunit of EnvA that, in turn, allow exposure of the fusion peptide in TM-A, its interaction with target membranes, and, under some experimental conditions, hemifusion of the membranes. In step 2, protonation of residues in TM-A, at the SU-A-TM-A interface, or at the SU-A-SU-A interface, causes conformational changes needed to complete fusion. Such two-step mechanisms may, in fact, be more common than is currently appreciated. Association of HIV env with its primary receptor induces conformational changes in the HIV SU that increase exposure of TM; association with a second receptor is required to complete the fusion reaction (25). For HA, protons are needed for the initial separation of the HA1 subunits; more protons (lower pH) are needed to complete the fusion reaction (10).
Acknowledgments
We thank Carolyn Teschke and Robert Nakamoto for helpful discussions. We thank Deyu Wang of the University of Virginia Biomolecular Core Facility for performing the BiaCore studies and preparing the graphs shown in Fig. 4. We gratefully acknowledge the expert technical assistance of Kurt Becker and Susana Contreras-Alcantara.
This work was supported by NIH grant AI22470 to J.M.W. and American Heart Association, Mid-Atlantic Division, grant 0365322U, a University of Virginia School of Medicine R&D Award, and a SEED Grant from American Cancer Society Institutional Research Grant IRG-81-001-20 (to the University of Virginia) to S.E.D.
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, O. M., L. L. Christensen, P. A. Christensen, E. S. Sorensen, C. Jacobsen, S. K. Moestrup, M. Etzerodt, and H. C. Thogersen. 2000. Identification of the minimal functional unit in the low density lipoprotein receptor-related protein for binding the receptor-associated protein (RAP). A conserved acidic residue in the complement-type repeats is important for recognition of RAP. J. Biol. Chem. 275:21017-21024. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 4.Avramoglu, R. K., J. Nimpf, R. S. McLeod, K. W. Ko, Y. Wang, D. FitzGerald, and Z. Yao. 1998. Functional expression of the chicken low density lipoprotein receptor-related protein in a mutant Chinese hamster ovary cell line restores toxicity of Pseudomonas exotoxin A and degradation of α2-macroglobulin. J. Biol. Chem. 273:6057-6065. [DOI] [PubMed] [Google Scholar]
- 5.Balliet, J. W., J. Berson, C. M. D'Cruz, J. Huang, J. Crane, J. M. Gilbert, and P. Bates. 1999. Production and characterization of a soluble, active form of Tva, the subgroup A avian sarcoma and leukosis virus receptor. J. Virol. 73:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett, A. L., and J. M. Cunningham. 2001. Receptor binding transforms the surface subunit of the mammalian C-type retrovirus envelope protein from an inhibitor to an activator of fusion. J. Virol. 75:9096-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bethell, R., N. Gray, and C. Penn. 1995. The kinetics of the acid-induced conformational change of influenza virus haemagglutinin can be followed using 1,1′-bis(4-anilino-5-naphthalenesulphonic acid). Biochem. Biophys. Res. Commun. 206:355-361. [DOI] [PubMed] [Google Scholar]
- 8.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bizebard, T., B. Gigant, P. Rigolet, B. Rasmussen, O. Diat, P. Bosecke, S. A. Wharton, J. J. Skehel, and M. Knossow. 1995. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 376:92-94. [DOI] [PubMed] [Google Scholar]
- 10.Bottcher, C., K. Ludwig, A. Herrmann, M. van Heel, and H. Stark. 1999. Structure of influenza haemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett. 463:255-259. [DOI] [PubMed] [Google Scholar]
- 11.Bova, C. A., J. C. Olsen, and R. Swanstrom. 1988. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J. Virol. 62:75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein micro 1 mediates membrane disruption. J. Virol. 76:9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper, J. A., and B. W. Howell. 1999. Lipoprotein receptors: signaling functions in the brain? Cell 97:671-674. [DOI] [PubMed] [Google Scholar]
- 14.Daenke, S., and S. Booth. 2000. HTLV-1-induced cell fusion is limited at two distinct steps in the fusion pathway after receptor binding. J. Cell Sci. 113:37-44. [DOI] [PubMed] [Google Scholar]
- 15.Damico, R., and P. Bates. 2000. Soluble receptor-induced retroviral infection of receptor-deficient cells. J. Virol. 74:6469-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damico, R. L., J. Crane, and P. Bates. 1998. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc. Natl. Acad. Sci. USA 95:2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delos, S. E., M. J. Burdick, and J. M. White. 2002. A single glycosylation site within the receptor binding domain of the avian sarcoma/leukosis virus glycoprotein is critical for receptor binding. Virology 294:354-363. [DOI] [PubMed] [Google Scholar]
- 18.Dolmer, K., W. Huang, and P. G. Gettins. 2000. NMR solution structure of complement-like repeat CR3 from the low density lipoprotein receptor-related protein. Evidence for specific binding to the receptor binding domain of human α2-macroglobulin. J. Biol. Chem. 275:3264-3269. [DOI] [PubMed] [Google Scholar]
- 19.Dowbenko, D., G. Nakamura, C. Fennie, C. Shimasaki, L. Riddle, R. Harris, T. Gregory, and L. Lasky. 1988. Epitope mapping of the human immunodeficiency virus type 1 gp120 with monoclonal antibodies. J. Virol. 62:4703-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earp, J., S. E. Delos, H. E. Park, and J. M. White. 2004. The many mechanisms of viral membrane fusion proteins, p. 25-66. In M. Marsh (ed.), Membrane trafficking in viral replication. Springer Verlag, New York, N.Y.
- 21.Earp, L. J., S. E. Delos, R. C. Netter, P. Bates, and J. M. White. 2003. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 77:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Einfeld, D. A., and E. Hunter. 1997. Mutational analysis of the oligomer assembly domain in the transmembrane subunit of the Rous sarcoma virus glycoprotein. J. Virol. 71:2383-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fass, D., S. Blacklow, P. S. Kim, and J. M. Berger. 1997. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388:691-693. [DOI] [PubMed] [Google Scholar]
- 24.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 Å resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 25.Gallo, S. A., C. M. Finnegan, M. Viard, Y. Raviv, A. Dimitrov, S. S. Rawat, A. Puri, S. Durell, and R. Blumenthal. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36-50. [DOI] [PubMed] [Google Scholar]
- 26.Gardsvoll, H., F. Werner, L. Sondergaard, K. Dano, and M. Ploug. 2004. Characterization of low-glycosylated forms of soluble human urokinase receptor expressed in Drosophila Schneider 2 cells after deletion of glycosylation-sites. Protein Expr. Purif. 34:284-295. [DOI] [PubMed] [Google Scholar]
- 27.Gilbert, J. M., L. D. Hernandez, J. W. Balliet, P. Bates, and J. M. White. 1995. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J. Virol. 69:7410-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godley, L., J. Pfeifer, D. Steinhauer, B. Ely, G. Shaw, R. Kaufmann, E. Suchanek, C. Pabo, J. J. Skehel, D. C. Wiley, and S. Wharton. 1992. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68:635-645. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein, J. L., H. H. Hobbs, and M. S. Brown. 1995. Familial hypercholesterolemia, p. 1981-2030. In C. R. Schriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular bases of inherited disease, 7th ed. McGraw Hill, New York, N.Y.
- 30.Gruenberger, M., R. Wandl, J. Nimpf, T. Hiesberger, W. J. Schneider, E. Kuechler, and D. Blaas. 1995. Avian homologs of the mammalian low-density lipoprotein receptor family bind minor receptor group human rhinovirus. J. Virol. 69:7244-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo, Y., X. Yu, K. Rihani, Q. Y. Wang, and L. Rong. 2004. The role of a conserved acidic residue in calcium-dependent protein folding for a low density lipoprotein (LDL)-A module: implications in structure and function for the LDL receptor superfamily. J. Biol. Chem. 279:16629-16637. [DOI] [PubMed] [Google Scholar]
- 32.Hall, D. R., J. R. Cann, and D. J. Winzor. 1996. Demonstration of an upper limit to the range of association rate constants amenable to study by biosensor technology based on surface plasmon resonance. Anal. Biochem. 235:175-184. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez, L. D., R. R. Peters, S. E. Delos, J. A. T. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 139:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, Q., R. P. Sivaramakrishna, K. Ludwig, T. Korte, C. Bottcher, and A. Herrmann. 2003. Early steps of the conformational change of influenza virus hemagglutinin to a fusion active state: stability and energetics of the hemagglutinin. Biochim. Biophys. Acta 1614:3-13. [DOI] [PubMed] [Google Scholar]
- 35.Hussain, M. M., D. K. Strickland, and A. Bakillah. 1999. The mammalian low-density lipoprotein receptor family. Annu. Rev. Nutr. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 36.Ivey-Hoyle, M., R. K. Clark, and M. Rosenberg. 1991. The N-terminal 31 amino acids of human immunodeficiency virus type 1 envelope protein gp120 contain a potential gp41 contact site. J. Virol. 65:2682-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones, P. L., T. Korte, and R. Blumenthal. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273:404-409. [DOI] [PubMed] [Google Scholar]
- 38.Kemble, G. W., D. L. Bodian, J. Rosé, I. A. Wilson, and J. M. White. 1992. Intermonomer disulfide bonds impair the fusion activity of influenza virus hemagglutinin. J. Virol. 66:4940-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, M., B. Chen, R. E. Hussey, Y. Chishti, D. Montefiori, J. A. Hoxie, O. Byron, G. Campbell, S. C. Harrison, and E. L. Reinherz. 2001. The stoichiometry of trimeric SIV glycoprotein interaction with CD4 differs from that of anti-envelope antibody Fab fragments. J. Biol. Chem. 276:42667-42676. [DOI] [PubMed] [Google Scholar]
- 40.Korte, T., and A. Herrmann. 1994. pH-dependent binding of the fluorophore bis-ANS to influenza virus reflects the conformational change of hemagglutinin. Eur. Biophys. J. 23:105-113. [DOI] [PubMed] [Google Scholar]
- 41.Korte, T., K. Ludig, F. P. Booy, R. Blumenthal, and A. Herrmann. 1999. Conformational intermediates and fusion activity of influenza virus hemagglutinin. J. Virol. 73:4567-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kvaratskhelia, M., P. K. Clark, S. Hess, D. C. Melder, M. J. Federspiel, and S. H. Hughes. 2004. Identification of glycosylation sites in the SU component of the avian sarcoma/leukosis virus envelope glycoprotein (subgroup A) by mass spectrometry. Virology 326:171-181. [DOI] [PubMed] [Google Scholar]
- 43.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with CD4 receptor and a neutralizing human antibody. Nature 393:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavillette, D., B. Boson, S. J. Russell, and F. L. Cosset. 2001. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J. Virol. 75:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leavitt, M., E. J. Park, I. A. Sidorov, D. S. Dimitrov, and G. V. Quinnan, Jr. 2003. Concordant modulation of neutralization resistance and high infectivity of the primary human immunodeficiency virus type 1 MN strain and definition of a potential gp41 binding site in gp120. J. Virol. 77:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 47.Lim, K. I., S. Narayan, J. A. Young, and J. Yin. 2004. Effects of lipid rafts on dynamics of retroviral entry and trafficking: quantitative analysis. Biotechnol. Bioeng. 86:650-660. [DOI] [PubMed] [Google Scholar]
- 47b.Matsuyama, S., S. E. Delos, and J. M. White. 2004. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 78:8201-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melder, D. C., V. S. Pankratz, and M. J. Federspiel. 2003. Evolutionary pressure of a receptor competitor selects different subgroup A avian leukosis virus escape variants with altered receptor interactions. J. Virol. 77:10504-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melikyan, G. B., R. J. Barnard, R. M. Markosyan, J. A. Young, and F. S. Cohen. 2004. Low pH is required for avian sarcoma and leukosis virus Env-induced hemifusion and fusion pore formation but not for pore growth. J. Virol. 78:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139-1142. [DOI] [PubMed] [Google Scholar]
- 51.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Netter, R. C., S. M. Amberg, J. W. Balliet, M. J. Biscone, A. Vermeulen, L. J. Earp, J. M. White, and P. Bates. 2004. Heptad repeat 2-based peptides inhibit avian sarcoma and leukosis virus subgroup A infection and identify a fusion intermediate. J. Virol. 78:13430-13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohuchi, M., R. Ohuchi, T. Sakai, and A. Matsumoto. 2002. Tight binding of influenza virus hemagglutinin to its receptor interferes with fusion pore dilation. J. Virol. 76:12405-12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poumbourios, P., A. L. Maerz, and H. E. Drummer. 2003. Functional evolution of the HIV-1 envelope glycoprotein 120 association site of glycoprotein 41. J. Biol. Chem. 278:42149-42160. [DOI] [PubMed] [Google Scholar]
- 55.Prevost, M., and V. Raussens. 2004. Apolipoprotein E-low density lipoprotein receptor binding: study of protein-protein interaction in rationally selected docked complexes. Proteins 55:874-884. [DOI] [PubMed] [Google Scholar]
- 56.Rong, L., A. Edinger, and P. Bates. 1997. Role of basic residues in the subgroup-determining region of the subgroup A avian sarcoma and leukosis virus envelope in receptor binding and infection. J. Virol. 71:3458-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rudenko, G., L. Henry, K. Henderson, K. Ichtchenko, M. S. Brown, J. L. Goldstein, and J. Deisenhofer. 2002. Structure of the LDL receptor extracellular domain at endosomal pH. Science 298:2353-2358. [DOI] [PubMed] [Google Scholar]
- 58.Ruigrok, R. W. H., A. Aitken, L. J. Calder, S. R. Martin, J. J. Skehel, S. A. Wharton, W. Weis, and D. C. Wiley. 1988. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 69:2785-2795. [DOI] [PubMed] [Google Scholar]
- 59.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 61.Strickland, D. K., S. L. Gonias, and W. S. Argraves. 2002. Diverse roles for the LDL receptor family. Trends Endocrinol. Metab. 13:66-74. [DOI] [PubMed] [Google Scholar]
- 62.Teschke, C. M., and J. King. 1993. Folding of the phage P22 coat protein in vitro. Biochemistry 32:10839-10847. [DOI] [PubMed] [Google Scholar]
- 63.Teschke, C. M., J. King, and P. E. Prevelige, Jr. 1993. Inhibition of viral capsid assembly by 1,1′-bi(4-anilinonaphthalene-5-sulfonic acid). Biochemistry 32:10658-10665. [DOI] [PubMed] [Google Scholar]
- 64.Tonelli, M., R. J. Peters, T. L. James, and D. A. Agard. 2001. The solution structure of the viral binding domain of Tva, the cellular receptor for subgroup A avian leukosis and sarcoma virus. FEBS Lett. 509:161-168. [DOI] [PubMed] [Google Scholar]
- 65.Verdaguer, N., I. Fita, M. Reithmayer, R. Moser, and D. Blaas. 2004. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11:429-434. [DOI] [PubMed] [Google Scholar]
- 66.Wang, Q. Y., W. Huang, K. Dolmer, P. G. Gettins, and L. Rong. 2002. Solution structure of the viral receptor domain of Tva and its implications in viral entry. J. Virol. 76:2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366-372. [DOI] [PubMed] [Google Scholar]
- 68.Wyatt, R., E. Desjardin, U. Olshevsky, C. Nixon, J. Binley, V. Olshevsky, and J. Sodroski. 1997. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J. Virol. 71:9722-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, X., Q. Y. Wang, Y. Guo, K. Dolmer, J. A. Young, P. G. Gettins, and L. Rong. 2003. Kinetic analysis of binding interaction between the subgroup A Rous sarcoma virus glycoprotein SU and its cognate receptor Tva: calcium is not required for ligand binding. J. Virol. 77:7517-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, W., G. Canziani, C. Plugariu, R. Wyatt, J. Sodroski, R. Sweet, P. Kwong, W. Hendrickson, and I. Chaiken. 1999. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry 38:9405-9416. [DOI] [PubMed] [Google Scholar]
- 71.Zingler, K., and J. A. T. Young. 1996. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J. Virol. 70:7510-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]