Abstract
Nontuberculous mycobacteria (NTM) are environmentally acquired opportunistic pathogens that can cause chronic lung disease. Within the U.S., Hawai'i shows the highest prevalence rates of NTM lung infections. Here, we investigated a potential role for active volcanism at the Kīlauea Volcano located on Hawai'i Island in promoting NTM growth and diversity. We recovered NTM that are known to cause lung disease from plumbing biofilms and soils collected from the Kīlauea environment. We also discovered viable Mycobacterium avium, Mycobacterium abscessus, and Mycobacterium intracellulare subsp. chimaera on volcanic ash collected during the 2018 Kīlauea eruption. Analysis of soil samples showed that NTM prevalence is positively associated with bulk content of phosphorus, sulfur, and total organic carbon. In growth assays, we showed that phosphorus utilization is essential for proliferation of Kīlauea‐derived NTM, and demonstrate that NTM cultured with volcanic ash adhere to ash surfaces and remain viable. Ambient dust collected on O'ahu concurrent with the 2018 eruption contained abundant fresh volcanic glass, suggestive of inter‐island ash transport. Phylogenomic analyses using whole genome sequencing revealed that Kīlauea‐derived NTM are genetically similar to respiratory isolates identified on other Hawaiian Islands. Consequently, we posit that volcanic eruptions could redistribute environmental microorganisms over large scales. While additional studies are needed to confirm a direct role of ash in NTM dispersal, our results suggest that volcanic particulates harbor and can redistribute NTM and should therefore be studied as a fomite for these burgeoning, environmentally acquired respiratory infections.
Keywords: volcanic ash, nontuberculous mycobacteria, genomics, airborne, transmission, public health
Key Points
Long‐range transport of hitch‐hiking infectious agents has never been reported for volcanic eruptions globally
Ash recovered from the 2018 Kīlauea Volcano eruption harbors species of nontuberculous mycobacteria (NTM) known to cause lung disease
Genomic evaluation reveals Kīlauea‐derived NTM are genetically similar to respiratory isolates identified on other Hawaiian Islands
1. Introduction
Nontuberculous mycobacteria (NTM) inhabit natural and built environments globally, causing opportunistic lung disease (LD) (Honda et al., 2018). In the United States, Hawai'i shows the highest number of NTM LD cases per capita among Medicare beneficiaries and the highest age‐adjusted mortality rates (Adjemian et al., 2017; Adjemian, Olivier, Seitz, Falkinham, et al., 2012; Adjemian, Olivier, Seitz, Holland, et al., 2012; Mirsaeidi et al., 2014). Resident Asians are at significantly higher risk compared to other racial/ethnic groups (Adjemian et al., 2017; Adjemian, Olivier, Seitz, Falkinham, et al., 2012; Adjemian, Olivier, Seitz, Holland, et al., 2012; Mirsaeidi et al., 2014), suggesting a genetic vulnerability, but it is also likely that specific geographic and environmental factors contribute to disparate NTM infection rates.
Respiratory infections caused by NTM are thought to occur via inhalation of aerosolized bacteria from environmental sources (e.g., tap water or aerosolized soil) or with colonization of the oropharynx and subsequent microaspiration (Wallace, 1987). Numerous studies have demonstrated that pathogenic NTM reside within freshwater biofilms, dust, and soil (De Groote et al., 2006; Falkinham, 2011; Falkinham et al., 2008; Gebert et al., 2018; Honda et al., 2018). We reported respiratory relevant NTM, for example, Mycobacterium avium and Mycobacterium abscessus subsp. abscessus, from Hawai'i household water biofilms and garden soil with a predominance of Mycobacterium intracellulare subsp. chimaera (referred to as M. chimaera henceforth), whose common recovery mirrored Hawai'i respiratory specimens (Honda et al., 2016; Nelson et al., 2021; Virdi et al., 2021). We also demonstrated that the indoor environments of Hawai'i are more likely to harbor respiratory relevant NTM compared to outdoor environments, particularly for M. chimaera (Virdi et al., 2021). More broadly, NTM species diversity varied between outdoor and indoor environments and among Kaua'i, O'ahu, Maui, and Hawai'i Island.
Wetlands, specific soil components, and geospatial variables have all been correlated with NTM presence in outdoor niches of Hawai'i (Glickman et al., 2020; Nelson et al., 2021; Virdi et al., 2021). However, a unique environmental feature of Hawai'i is active volcanism, including the Kīlauea Volcano on Hawai'i Island. The areas surrounding active volcanoes have diverse ecosystems that support an array of bacteria, including the phylum Actinobacteria, to which NTM belong (Riquelme et al., 2015). Kīlauea is one of the world's most active volcanoes, transitioning between effusive and explosive behavior. It has erupted near‐continuously from 1983 to present, emitting tons of sulfur dioxide (SO2) per day (Elias et al., 2020; Kern et al., 2020) and resulting in an environment that is uniquely different from urban areas of Hawai'i. For example, reaction of SO2 with oxygen, water, and particles in the atmosphere form sulfuric acid droplets that acidify soil (Elias & Sutton, 2017); correspondingly, Hawai'i Island soil is more acidic (pH 5.5) than the other Hawaiian Islands (average, pH 7) (Glickman et al., 2020). Kīlauea's summit erupted explosively in 2018 for the first time since 1924, which produced multiple gaseous plumes that deposited lithic and juvenile ash in surrounding areas (Neal et al., 2019). Inhalation of volcanic particulates can aggravate pre‐existing respiratory conditions and poses a respiratory hazard for sensitive individuals (Stewart et al., 2022). However, whether volcanic particulates have a role in the environmental distribution or acquisition of pathogenic mycobacteria has not been reported.
Based on our prior discoveries of environmental NTM in Hawai'i, we hypothesized that the active volcanic environment of Kīlauea is enriched for NTM. In the present study, we combine environmental sampling and compositional characterization with microbiological culture techniques, partial and whole bacterial genome sequencing, and in vitro assays to evaluate NTM diversity at Kīlauea. The 2018 eruption afforded a unique opportunity to determine whether particulate matter derived from the Kīlauea environment can carry opportunistic LD‐causing NTM species, and to explore volcanic particulate matter as a possible environmental fomite for NTM within and between islands in Hawai'i.
2. Materials and Methods
2.1. Soil and Plumbing Biofilm Sampling
The sampling strategy used for Kīlauea environmental sampling was constrained due to limited site access within the Kīlauea caldera (now Kaluapele). Soil samples and plumbing biofilm swabs were collected within Hawai'i Volcanoes National Park as previously described (Honda et al., 2016; Virdi et al., 2021) under permit #HAVO‐2017‐SCI‐0058 (U.S. Department of the Interior, National Park Service). Biofilm samples were collected using sterile synthetic flock swabs from seven households and five non‐household buildings. To collect soil, a small area of the ground's surface was cleared of any organic litter to expose the mineral soil, then the top ∼13 cm (5 inches) of soil was sampled into a sterile 50 ml conical screw cap tube. Soil was collected from seven household and 35 environmental sites. All samples were collected between November 2017 and January 2018. A sampling map (Figure 1a) was constructed using ArcMap 10.5.1 software (World Geodetic System 84) with raster and shapefiles obtained from the State of Hawai'i GIS portal (http://geoportal.hawaii.gov/).
Figure 1.
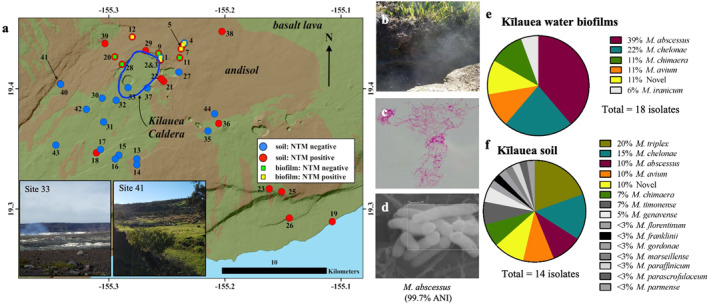
Environmental sampling sites and identification of nontuberculous mycobacteria (NTM) from the Kīlauea environment. (a) Map of soil (circles) and biofilm (squares) sampling locations, Kīlauea Volcano, Hawai'i Island. The pre‐2018 summit caldera is encircled in blue. Square symbols inside of a circle indicate locations where both soil and biofilm samples were collected. Inset: Photographs of NTM culture negative site 33 and NTM culture positive test site 41. (b) Photograph of a NTM culture positive steam vent, site 29. (c) An isolate with smooth colonies was cultured from soil collected at site 29 and is acid‐fast stain positive (1,000× total magnification). (d) The isolate was imaged using scanning electron microscopy (230,00× total magnification) and identified as M. abscessus by rpoB gene sequencing. NTM species diversity of (e) 18 isolates recovered from Kīlauea biofilms and (f) 41 isolates recovered from Kīlauea soil are shown.
2.2. Ambient Dust and Volcanic Ash Collection
Passive dust collectors were deployed in four urban and residential settings in central and southeastern O'ahu between January 2018 and January 2019 and the collected dust was recovered between August 2018 and December 2019. The May‐September 2018 Kīlauea eruption was an unanticipated event that commenced ∼5 months after the environmental soil and plumbing sampling was performed. The collectors would have sampled material during this entire period whenever winds and eruption dynamics were favorable.
To create dust collectors, newly purchased Bundt‐cake pans, marbles, and 0.5 cm metal mesh were thoroughly cleaned with 70% ethanol. After filling disinfected pans with marbles, they were covered with the mesh and secured on using zip ties (Figure S1 in Supporting Information S1) (Reheis & Kihl, 1995; Reheis et al., 1995). To simulate dust fluxes and ensure that the material in the collectors was dominated by dry deposition, collectors were placed under residential eaves or roofs. To recover samples, the metal mesh was removed, and the marbles poured into a plastic mesh fishing net, rinsed with deionized water into a Ziploc bag, and the volume transferred to a plastic bottle for storage, shipping, and drying at 60°C.
From May through September 2018, lava erupted through fissures in Kīlauea's lower East Rift Zone in conjunction with recurring explosion‐collapse events at Kīlauea's summit, each event resulted in the production of ash‐containing plumes (Neal et al., 2019). A volcanic ash sample collected on 28 May was used for this study. The sample was collected as it fell in a clean, five‐gallon high density polyethylene bucket deployed eight km downwind from the Kīlauea summit, in the Ka'ū Desert. Investigation by scanning electron microscopy (SEM) with energy‐dispersive X‐ray spectroscopy (EDX) confirmed that the sample contained an abundance of fragmented older lavas derived from fragmentation of the Kīlauea edifice, and was devoid of soil (Table S2 in Supporting Information S1). An SEM image of Kīlauea ash is shown in Figure 2a. The sample was previously characterized for soluble compounds (Damby et al., 2018; Tomasek et al., 2021) following established volcanic ash leaching methods (Stewart et al., 2020). Two size fractions were isolated from the bulk sample for analysis: a fine (<32 μm) fraction, obtained by sieving through a series of mesh sieves of decreasing size, and a respirable (<4 μm) fraction, obtained by aerodynamic separation as previously described (Tomasek et al., 2016). This was done to determine whether there is a microbial difference between respirable and coarser material.
Figure 2.
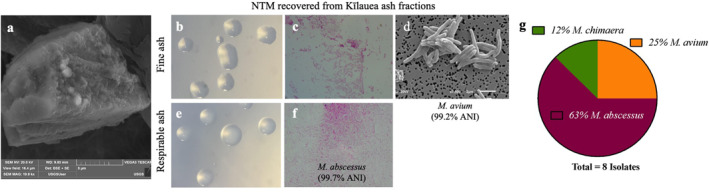
Recovery of viable nontuberculous mycobacteria (NTM) from Kīlauea fine and respirable ash fractions. (a) scanning electron microscopy (SEM) image of Kīlauea ash (198,00× total magnification). (b), (e) Two NTM‐like colonies were cultured from fine (<32 μm) and respirable (<4 μm) ash fractions respectively and visualized on a dissecting microscope (80× total magnification). (c), (f) Acid‐fast bacilli staining of the smooth colonies (1,000× total magnification). (d) The Kīlauea ash isolate shown in b and c was identified as M. avium by rpoB gene sequencing and imaged using SEM (7,500× total magnification). The Kīlauea ash isolate shown in e and f was identified as M. abscessus by rpoB gene sequencing. (g) Species diversity of the eight isolates microbiologically cultured from Kīlauea ash and identified by partial rpoB gene sequencing.
2.3. Soil Bulk Compositional Analyses
Soil pH was measured by an ISFET pH probe attached to an IQ Scientific Instruments IQ170 pH and conductivity meter inserted into ∼15 ml of soil dampened with Milli‐Q water.
For bulk chemical characterization, soils were first air dried, crushed, and homogenized. Soils with ample material were crushed using a tungsten carbide SPEX ShatterBox. A SPEX tungsten carbide grinding vial set was used to crush soil with less than 70 g of material.
Loss on ignition (LOI) measurements were performed to quantify what was lost to the atmosphere as H2O vapor and CO2. Crucibles used for analyses were heated to 1,000°C and cooled in a desiccator for 30 min prior to addition of 2 g of soil. Crucibles with soil sample were placed in a 400°C oven to remove organic material, cooled in a desiccator, and weighed. Following this calcination step, crucibles were placed in a 1,000°C oven to remove any remaining water and carbonate material, cooled, and weighed. Sample mass was tabulated using a Mettler AE 50‐S scale with ±0.0001 g precision.
Major element abundances were determined using a Rigaku ZSX Primus II X‐ray fluorescence spectrometer (Nelson et al., 2013). Reported abundances were recalculated to account for LOI. Pyrolysis was performed using a Wildcat Technologies HAWK Workstation. Approximately 50 mg of soil was placed into a crucible at 100°C, heated to 650°C to measure organic carbon content, and finally heated to 850°C to measure inorganic carbon content.
2.4. Particle Imaging and Chemistry
O'ahu dust was examined by SEM and optical microscopy. SEM imaging and energy‐dispersive X‐ray analysis were conducted to verify that apparent glass fragments were volcanic in origin. A small amount of dust was suspended in deionized water and allowed to evaporate on a silicon wafer affixed to a standard SEM stub. Analyses were conducted with a FEI Apreo SEM under low‐vacuum conditions. Glass fragments were identified as bright grains using back‐scatter electrons (the Fe‐rich basaltic glass appears bright against the silicon wafer) and EDX spectra were obtained. Fragments were basaltic ash when Si, Fe, Mg, Al, Na, Ca, and O peaks were present.
To verify the basaltic fragments in the sample as glass, we used polarized‐light optical microscopy, where the illumination of glass is extinct under cross‐polarized light. Dust was rinsed three times in deionized water to remove soluble salts and loaded from an aqueous suspension by transferring ∼1 ml to a 22 mm glass cover slip and allowed to dry. The cover slip was inverted and placed on a standard microscope slide over a small drop of Zrax optical resin (refractive index ∼1.7) and cured on a hot plate.
2.5. NTM Microbiological Culture
Environmental plumbing biofilm samples were processed as published (Honda et al., 2016; Virdi et al., 2021). For soil, 1 g was weighed into a sterile 50 ml conical vial, resuspended in 20 ml of sterile Milli‐Q water and mixed on an end‐to‐end shaker at room temperature (22°C) for two hours. After shaking, the sample was placed upright and allowed to settle for 30 min. For disinfection, 50 μl of 1% cetylpyridinium chloride was added to 450 μl of sample suspension, vortexed, incubated at room temperature for 30 min, and 100 μl was spread onto duplicate Middlebrook 7H10‐OADC and 7H10‐OADC with malachite green agar plates and incubated at 30°C or 37°C for 21 days.
To culture NTM from volcanic ash, 1 mg of fine or respirable ash fractions were resuspended in 7 ml of Middlebrook 7H9 broth, briefly vortexed, then incubated at 30°C or 37°C. After 3, 7, and 14 days of incubation, samples were briefly vortexed and 100 μl of broth was spread onto duplicate Middlebrook 7H10‐OADC and 7H10‐OADC with malachite green agar plates and incubated at 30°C or 37°C for 21 days. NTM colonies typified by smooth or rough morphologies that appeared on solid media after 3 days were picked and cultured into Middlebrook 7H9 broth and stocks were made.
2.6. DNA Extraction and Gene Sequencing
Genomic DNA was extracted from isolate cell pellets of both fine and respirable volcanic ash fractions according to a protocol tailored for lysing mycobacterial cells with bead‐beating (Epperson & Strong, 2020). NTM were identified by sequencing a 711bp RNA polymerase beta subunit (rpoB) gene region and a 16S gene region (515F, 926R) using Sanger sequencing (Quintara Biosciences, CA) (Adekambi et al., 2003; Walters et al., 2016). Sequencing results were trimmed for quality control and then compared to rpoB and 16S reference sequences in the National Center for Biotechnology Information (NCBI) GenBank using their BLAST algorithm. If an isolate had a >90% BLAST identity and BLAST query coverage match to any Mycobacterium, but no BLAST identity above 97%, it was categorized as a putatively “novel” NTM.
2.7. Mycobacterial Microbiome Profiling
Genomic DNA extracted directly from volcanic ash served as template for rpoB amplification using primers adapted for Illumina indexing and sequenced on the MiSeq using 2 × 300 v3 chemistry (Adekambi et al., 2003). Resulting forward and reverse reads were trimmed and joined with 10 ambiguous N bases. The concatenated reads were processed using QIIME2 for quality, sorting, and annotated with a custom rpoB data set derived from a query of the NCBI non‐redundant database (Accessed on 6/18/2018, gap extension penalty set to 0) (Bolyen et al., 2019).
2.8. Microbiology Imaging
NTM colony morphologies were imaged on a dissecting microscope at total magnification of 80X (Ken‐A‐Vision). Acid‐fast isolates were identified with Ziehl‐Neelsen staining and imaged by light microscopy at total magnification of 1,000× using a Laxco SeBa Pro 4 microscope.
SEM was performed on Kīlauea‐derived M. abscessus and Mycobacterium chelonae that were cultured with volcanic ash. NTM and ash cultures were prepared by inoculating 1 × 106 NTM cells into 1 ml 7H9 broth containing 1 mg of fine ash and incubated on a rotary shaker at 37°C for 48 hr. The NTM‐ash cultures were deposited onto Whatman No. 5 filter paper (2.5 μm) and fixed by submersion in 2.5% glutaraldehyde diluted with 0.1 M cacodylate buffer. Imaging and chemical analyses were conducted on a Tescan VEGA3 Variable Pressure SEM equipped with two Oxford Instruments XmaxN 150 mm2 silicon drift detectors for EDX.
2.9. NTM Viability Assay
To test whether NTM remain viable in the presence of Kīlauea ash, triphenyl tetrazolium chloride (TTC) assays were performed. 1 × 104 cells were inoculated into 7H9 broth or broth containing 0.5 mg of fine ash. Broth alone and broth containing 0.5 mg fine ash without NTM were used as negative controls. Cultures were incubated on a rotary shaker at 37°C for 48 hr. Following incubation, particulates were allowed to settle, supernatants discarded, and replaced with fresh broth. 125 μl of TTC reagent (Sigma, T8877) (0.5 mg/ml) was added and incubated at 37°C for 48 hr. Viability was indicated by solution color change from clear to red.
2.10. Phosphate Utilization in Culture
Informed by observed correlations of NTM prevalence with sample chemistry, we tested whether phosphate utilization is essential for colony formation. Kīlauea‐derived NTM cultures were streaked onto Pikovskayas agar plates (HiMedia Laboratories) and incubated at 30°C or 37°C for 7–21 days. Images of NTM cultured on Pikovskayas agar plates were taken using the iBright imaging system (Thermo Fisher). Determination of the production of a clear zone indicative of phosphorus solubilization was made in comparison to a positive control isolate, Pseudomonas aeruginosa.
2.11. Respiratory NTM Isolates
The rpoB gene sequencing protocol was used to identify NTM among a collection of 595 sputum samples submitted to the Diagnostic Laboratory Services, Inc (Aiea, HI) in 2016 for mycobacterial testing and culture. Samples were from de‐identified Hawaiian and other Pacific Islanders with suspected tuberculosis, similar to our prior work (Honda et al., 2016; Virdi et al., 2021). From this initial collection, 11 isolates of M. avium and 41 isolates of M. abscessus were subjected to whole genome sequencing (WGS) due to their importance as opportunistic LD pathogens and were chosen to represent the genomic and geographic diversity of NTM in Hawai'i. At the time, obtaining specific clinical details about these patients was beyond the scope of this project; thus, it is not known whether these isolates were from individuals who met diagnostic criteria for NTM‐LD set forth by the American Thoracic Society.
2.12. Phylogenomic Analyses
To investigate Kīlauea ash as a potential fomite for NTM, the rpoB sequences of the M. avium, M. abscessus, and M. chimaera isolates derived from the Kīlauea environment and its ash were compared against M. avium, M. abscessus, and M. chimaera isolates recovered from other non‐Kīlauea environments of Hawai'i as well as respiratory isolates from individuals with NTM living on other Pacific Islands. rpoB sequences were aligned using MUSCLE (Edgar, 2004) and compared using observed single nucleotide polymorphisms (SNPs) in complete sites with no missing data in MEGA (Stecher et al., 2020). Isolates were determined to be similar if their rpoB sequence showed 0 SNP differences (100% sequence identity) to another isolate of the same species.
WGS was conducted for ash and respiratory isolates with similar rpoB sequences and representative samples collected in Hawai'i. Nextera XT DNA or DNA FLEX sample preparation was used to prepare WGS libraries and libraries were sequenced using the Illumina HiSeq 2500. Sequence reads were trimmed of adapters and base calls with quality scores less than Q20 using Skewer (Jiang et al., 2014). Trimmed reads were assembled into scaffolds using Unicycler (Wick et al., 2017). Genome assemblies were compared against a collection of reference genomes to estimate average nucleotide identity (ANI) and assign a species call to each isolate (Goris et al., 2007; Richter & Rosselló‐Móra, 2009). A cutoff ANI of ≥ 95% indicated the isolate and reference genome belonged to the same species. Based upon taxonomic assignment using the highest ANI score above 95% for each genome, trimmed sequence reads were mapped to respective reference genomes, for example, M. avium H87 and M. chimaera CDC 2015‐22‐71 (Hasan et al., 2017; Zhao et al., 2017), using Bowtie2 (Langmead & Salzberg, 2012) and SNPs identified as previously described (Hasan et al., 2017). SNPs among isolates were extracted and concatenated to reconstruct a species‐specific phylogeny using Maximum Likelihood with 500 bootstrap replicates in RAxML‐NG using nucleotide general time‐reversible model. Phylogenies were visualized using ggtree (Yu et al., 2017).
To identify a SNP threshold for genetically similar isolates, we classify that isolates with a shared recent ancestry have a pairwise genetic distance less than or equal to 36 SNPs as published (Bryant et al., 2016). Isolates with a pairwise distance of greater than 36 SNPs were defined as different strains.
3. Results
3.1. NTM in the Kīlauea Environment
Between November 2017 and January 2018, soil and plumbing biofilms were collected from the surrounding areas of the Kīlauea summit (Figure 1a). In total, 44 sites were sampled, 10 with plumbing biofilm and soil, two where only plumbing biofilms were collected, and 32 sites where only soil was collected (Table S1 in Supporting Information S1). Of all 44 sampling sites, 26 (59%) were NTM culture positive. As an example, we show a Kīlauea steam vent location (Figure 1b) where soil was collected and from which we recovered viable smooth colonies of acid‐fast positive bacilli (Figure 1c) identified as M. abscessus and imaged by SEM (Figure 1d).
Of the 12 Kīlauea plumbing biofilm sites sampled, eight (67%) were NTM culture positive. From these eight positive sites, 44 plumbing biofilm bacterial isolates were recovered, of which 18/44 (41%) were identified as NTM using Sanger amplicon sequencing of the rpoB gene. The diversity of the plumbing biofilm NTM isolates is evidenced by the recovery of M. abscessus (7/18%, 39%), M. chelonae (4/18%, 22%), M. chimaera (2/18%, 11%), M. avium (2/18%, 11%), potentially novel NTM species (2/18%, 11%), and M. iranicum (1/18%, 6%) (Figure 1e).
Of the 42 sites where soil was collected, 23 (55%) were NTM culture positive. In all, 124 soil bacterial isolates underwent rpoB sequencing, of which 41 (33%) were identified as NTM, primarily M. triplex (8/41%, 20%), M. chelonae (6/41%, 15%), M. abscessus (4/41%, 10%), M. avium (4/41%, 10%), M. chimaera (3/41%, 7%), and others (Figure 1f).
Both plumbing biofilms and soil contained potentially novel species of NTM, demonstrating sequence similarity to the Genus Mycobacteria but diverging in rpoB sequence identity from reference NTM species. These isolates would need to be further investigated to determine whether they represent new species of NTM. From the same plumbing biofilm and soil samples, non‐NTM organisms were also identified by partial rpoB and 16S gene sequencing (Figures S2a and S2b in Supporting Information S1).
3.2. Recovery of Viable NTM From Kīlauea Volcanic Ash
The two size fractions (fine and respirable) of volcanic ash were both microbiologically cultured to determine whether there is a microbial difference between respirable and comparatively coarser material. In total, eight isolates were recovered from the volcanic ash fractions. These were identified as M. abscessus (5/8%; 63%), M. avium (2/8%; 25%), and M. chimaera (1/8%; 12%) by rpoB sequencing (Figure 2g). Of the eight isolates, three (1 M. avium, 2 M. abscessus) were derived from the fine ash fraction, and the five others (1 M. avium, 3 M. abscessus, 1 M. chimaera) from the respirable ash fraction. We also performed NTM‐specific microbiome amplification and sequencing by targeting a region of the rpoB gene to further characterize the mycobacterial taxa associated with the ash fractions, revealing the predominance of M. avium in both fine ash (94%) and respirable ash (87%) fractions (Figures S3a and S3b in Supporting Information S1). Of note, it is not uncommon to recover different ratios of NTM by the molecular versus culture methods.
We highlight two examples of NTM recovered from the two ash sized fractions that were inoculated into pure culture, producing smooth colonies (Figures 2b and 2e) of acid‐fast bacilli (Figures 2c and 2f). Sanger rpoB sequencing identified the exemplar fine ash isolate viewed by SEM (Figure 2d) as M. avium and the respirable ash isolate as M. abscessus.
We next investigated the capacity of Kīlauea‐derived NTM to bind to Kīlauea ash in vitro by SEM imaging. After co‐culture for 48 hr, Kīlauea‐derived M. abscessus and M. chelonae were found adhered to Kīlauea ash (Figures 3a and 3b). TTC viability assays demonstrate NTM remain viable in the presence of Kīlauea ash (Figure 3c).
Figure 3.
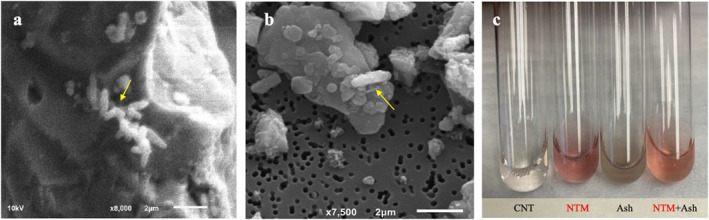
Kīlauea‐derived nontuberculous mycobacteria (NTM) bind to Kīlauea ash and remains viable. Scanning electron microscopy imaging of (a) M. abscessus and (b) M. chelonae isolates (yellow arrows) recovered from Kīlauea soil cultured with 1 mg of Kīlauea fine ash, 48 hr post inoculation. (c) Triphenyl tetrazolium chloride (TTC) viability assay indicates that NTM (M. abscessus shown) remain viable after culture with Kīlauea ash for 48 hr (tube labeled “NTM + Ash”) as indicated by the red color change (“CNT” control tube is control for no ash or NTM, “NTM” tube is NTM only, “Ash” tube is ash only).
3.3. Kīlauea Environmental Factors Increase NTM
Kīlauea NTM were variably recovered from a broad spectrum of soil pH, ranging from 3.5 to 7.9. The soil samples themselves were predominantly acidic, and a majority (78%) were recovered from acidic soil, pH 4.0–5.99 (Figure 4a) but NTM were also detected in some soils with pH > 7. To reveal other factors pertaining to Kīlauea soil associated with NTM, a suite of soil chemical features was examined (Table S3 in Supporting Information S1). After logistic modeling, bulk sample sulfur, pH, total organic carbon (TOC), and phosphorus (reported as P2O5) were soil chemical features that positively associated with the NTM in Kīlauea soil (Figure 4b).
Figure 4.
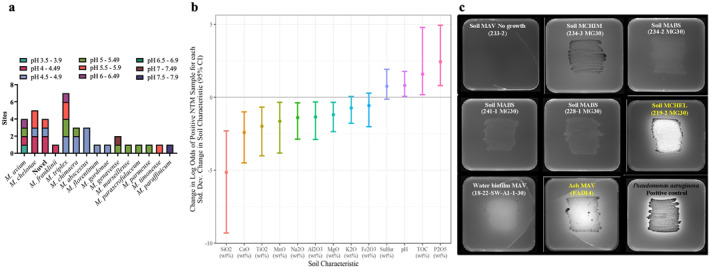
Kīlauea nontuberculous mycobacteria (NTM) prefer acidic soil and phosphate. (a) NTM cultured from Kīlauea soil in relation to the pH at soil collection sites. Soil pH measurements were grouped at intervals of pH 0.5. (b) Log odds ratios of NTM detection for a 1‐standard deviation increase in each Kīlauea soil variable. Soil chemistry is reported as major and minor element oxides, except sulfur. TOC = total organic carbon. (c) Detection of phosphate solubilizing NTM from Kīlauea soil and ash using Pikovskayas agar. Yellow text identifies isolates that solubilize and use phosphate. Pseudomonas aeruginosa was used as a positive control for phosphate solubilization.
Because NTM presence correlated strongly with soil P2O5, Kīlauea‐derived M. abscessus, M. chelonae, M. chimaera, and M. avium isolates were screened for phosphate utilization using selective Pikovskayas microbiological media. M. avium recovered from ash and M. chelonae derived from Kīlauea soil produced halos around the bacteria at the site of inoculation, indicating phosphate solubilization (Figure 4c).
3.4. Discovery of Volcanic Glass Fragments in O'ahu Dust
Samples collected by the O'ahu dust collectors were imaged to investigate the potential of NTM transportation with ash and particulate clouds across the ∼330 km separating Kīlauea on Hawai'i Island and O'ahu. SEM‐EDX analysis and optical microscopy verified the presence of volcanic glass particles in the recovered samples. By SEM, numerous fragments with a composition compatible with basalt (Si‐, Fe‐rich) were found. In plane‐polarized light, numerous observable glass fragments were noted (Figures 5a–5f) that were all extinct under crossed nicols. Similar to platy fragments illustrated in published identification guides (Marsaglia et al., 2013; Shapley, 2022), platy fragments with raised ridges that meet in triple junctions were observed, representing the interface of growing glass bubbles (Figure 5a). Glass fragments ranged from brown to clear, had conspicuous sharp edges, and exhibited conchoidal fractures typical of volcanic glass particles.
Figure 5.
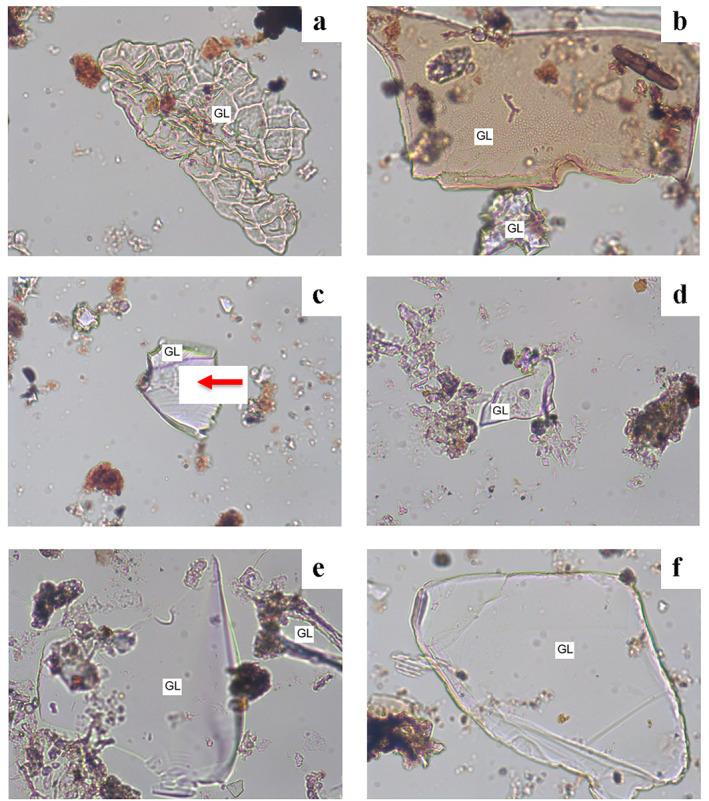
Examples of glass (GL) found in O'ahu dust collectors deployed concurrent with the 2018 Kīlauea eruption. (a) A platy fragment with raised ridges separating adjacent vesicles. (b) Brown platy and smaller clear glass fragments. (c) Clear glass fragment exhibiting conchoidal fracture (red arrow). (d) Glass fragment with concave and convex edges. (e) Thin platy glass fragment and adjacent small ash particle. (f) Platy glass fragment. The presence of glass was verified by extinction under cross polarized light. Width of each photomicrograph is 300 μm.
3.5. Genomic Analysis of Environment and Respiratory NTM Isolates
Finally, we explored volcanic ash as a possible fomite for NTM transmission using comparative rpoB gene similarity and phylogeographic analyses comparing core‐genome SNPs and geospatial relationships of the NTM isolates recovered from Kīlauea ash.
Two M. avium isolates derived from Kīlauea ash were identical to or showed one SNP difference with clinical isolates from Maui, O'ahu, and Guam, two Kīlauea environmental and three O'ahu environmental M. avium isolates as assessed by rpoB sequencing (Figure S4a in Supporting Information S1). The M. abscessus isolates derived from Kīlauea ash showed high rpoB similarity (<5 SNPs) to clinical M. abscessus isolates from O'ahu, Hawai'i Island, and Guam (Figure S4b in Supporting Information S1). The single M. chimaera isolate recovered from Kīlauea ash did not show rpoB similarity to any other environmental or clinical M. chimaera isolate examined in this study (>5 SNPs). However, M. chimaera from the Kīlauea environment were identical to 39 respiratory M. chimaera isolates from O'ahu and Maui, yet different from reference M. chimaera isolates from Canada, Switzerland, and Italy (Figure S4c in Supporting Information S1).
To complement the species characterization by rpoB sequencing and to better understand isolate derivation, WGS was performed on Kīlauea ash‐derived M. avium and M. abscessus, and sequences were compared to the WGS of other M. avium and M. abscessus isolates recovered from other environments and respiratory isolates from Hawai'i (Table S4 in Supporting Information S1). These comparisons enabled us to assess isolate relatedness using phylogenomic and comparative genomic methods (Davidson et al., 2013). We postulated that high genomic similarity among the core genomes of Kīlauea ash‐derived NTM and clinical NTM from other Hawaiian Islands provides a plausible route of environmental exposure, via NTM‐laden ash traveling within Hawai'i Island and between neighboring islands. No similarity was observed between one Kīlauea ash‐derived and three clinical M. avium genomes (Figure S5 in Supporting Information S1). However, high genomic similarities (≤36 SNP differences among the core genome) were observed between the genomes of the three (3/3 = 100%) Kīlauea ash‐derived M. abscessus isolates, 15 (15/26 = 58%) other Hawai'i environmental M. abscessus isolates, and 6 (6/10 = 60%) clinical M. abscessus isolates (Figure 6a). The phylogenetic tree of isolate genomes (red box, Figure 6a) shows the percent genomic differences between isolates are <0.001% SNPs across the entire core genome comprising >4M nucleotide positions. After mapping these M. abscessus isolates onto their island of origin, Kīlauea ash‐derived isolates are highly genetically similar to environmental and respiratory M. abscessus isolates observed from other Hawai'i islands (Figure 6b).
Figure 6.
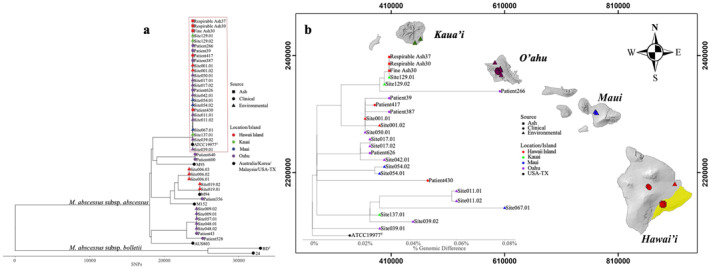
Whole genome sequence analyses reveal phylogenetically related M. abscessus in Kīlauea ash, the local environment, and respiratory isolates from individuals on neighboring islands. (a) Phylogenetic relationships between Kīlauea ash‐derived M. abscessus (red squares), environmental isolates (triangles), and respiratory isolates (circles) from individuals residing in Hawai'i. The red rectangle highlights genetically similar (>99.999%) isolates. (b) Phylogenetic tree of isolate genomes within the highlighted box in “A” show the % genomic differences between isolates are <0.001% single nucleotide polymorphisms (SNPs) (36 SNPs) cross the entire core genome comprising >4M nucleotide positions. These M. abscessus are mapped onto their island of origin. Volcanic ash isolates from Hawai'i Island are genetically identical to environmental and respiratory M. abscessus from other islands. The geographic area of the Kīlauea Volcano is highlighted in yellow. The Universal Transverse Mercator Coordinate System (zone 5) was used for the map; values in meters (m) are indicated. The superscript “T” designates that the isolate is a type strain for a given taxon.
4. Discussion and Conclusions
Island‐specific environmental factors are likely to impact NTM acquisition in Hawai'i. The predominance of M. abscessus and M. chelonae in Kīlauea household plumbing biofilm differed from what we previously reported for O'ahu, where M. chimaera was the dominant species (Honda et al., 2016; Virdi et al., 2021); this difference may be due to the smaller sample size studied in prior work. A large diversity of NTM species—including those that are respiratory relevant—were cultured from soil compared to plumbing biofilms, supplementing prior work showing predominance of NTM from indoor environments of Hawai'i (Honda et al., 2016). Geographically, there is a qualitative difference in general soil characteristics between NTM‐positive sampling locations (areas where soil development has occurred or on the coastal lava delta; i.e., andisols) and NTM‐negative locations (comprised predominantly of soil‐poor locations on the Ka'ū desert, downwind of the Kīlauea summit during normal trade‐wind conditions).
Previous work has identified iron oxide minerals as a key soil component that supports NTM in Hawai'i (Glickman et al., 2020; Parsons et al., 2022). Our observations that phosphorus, sulfur, and TOC in Kīlauea soil are positively associated with NTM presence highlight additional factors that may enrich for NTM, particularly within the state of Hawai'i and other volcanic environments. These findings illustrate the unique environment of Kīlauea compared to other regions and among the islands of Hawai'i. The successful solubilization of phosphate by ash‐derived M. avium may indicate the importance of a phosphorous‐bearing phase such as volcanic glass (Table S2 in Supporting Information S1) or apatite, which is present on the surfaces of 2018 Kīlauea ash (Emily Oborski & Damby, 2020) in supporting growth and provides promising research directions for identifying NTM‐harboring environmental substrates. Additionally, our findings underscore the need for further characterization of how these and potentially other volcanic factors contribute to the increased presence of NTM in the Kīlauea environment, and if these factors similarly influence LD‐causing NTM in active volcanic environments in other parts of the world.
A prominent finding from this study is the discovery of viable M. avium, M. abscessus, and M. chimaera from the Kīlauea environment, including on erupted summit volcanic ash. The presence of NTM would not be expected for ash generated from fragmentation of fresh magma as temperatures would be too high for NTM survival and the ash surfaces would be newly formed. The 2018 Kīlauea ash sample tested was predominantly comprised of fragmented edifice material produced from episodic collapse of the volcano summit, a mechanism that would favor incorporation of pre‐existing, NTM‐laden material (e.g., soils and older lava surfaces). As demonstrated by our environmental sampling, material in the Kīlauea environment was variably rich with diverse NTM. Consequently, we favor a scenario whereby older, NTM‐harboring edifice material was fragmented and entrained into the eruption column. Even still, NTM must have survived in the eruption column in order to be transported in a viable state down‐plume. Evidence from salt‐forming reactions within the volcanic plume suggests high temperatures were reached (King et al., 2023), on the order of >600°C in the core of the eruption column based on comparison to experimental data in (Ayris et al., 2014), though temperatures would have been substantially cooler (<<100°C) where the plume rapidly mixed with the atmosphere. Further investigation of eruptive plume temperatures—with implications for NTM survival and transport—would be a novel application of the plethora of monitoring data collected during the 2018 eruption.
Our recovery of viable NTM from Kīlauea ash and demonstration that NTM recovered from the Kīlauea environment bind to Kīlauea ash implicates volcanic ash as a novel substrate for environmental dispersal of NTM. While we acknowledge that only one ash sample was analyzed currently, we found variability in both NTM species and relative abundances in the two size fractions analyzed, as would be expected for a larger number of ash samples. The limited sample masses of pristine ash able to be collected during the eruption prevented more detailed analysis. However, given our above assessment that ash‐bound NTM derived from re‐distribution of environmental material and that volcanic ash produced throughout the 2018 summit eruption similarly comprised aged edifice material, we might expect that all summit ash samples contain NTM. Dispersion of microbiota by explosive volcanic eruptions has been reported (Van Eaton et al., 2013) and volcanic ash is an established substrate for bacterial colonization (Witt et al., 2017). However, we were unable to find documentation of a hitch‐hiking infectious agent related to volcanism as has been reported for long‐range transport of other environmental plumes, such as wildland fire smoke (Kobziar & Thompson, 2020) and Saharan dust events (Marone et al., 2020).
The transport of ultrafine volcanic ash during an eruption is poorly understood (Gouhier et al., 2019). However, the discovery of abundant individual volcanic glass fragments in dust from O'ahu provides suggestive evidence for inter‐island atmospheric transport of ash and, by extension, the plausibility for transport of attached microbes on materials lofted during ash‐dispersing events. The glass particles identified likely originated from Kīlauea and were transported and trapped during the time that the dust samplers were deployed on O'ahu. Although Kīlauea is ∼330 km from Honolulu, it is the closest source of fresh volcanic glass (Porder et al., 2007), and the timing is coincident. In support of this (Figures 6a–6d in Supporting Information S1) illustrates selected air‐trajectory models (HYSPLIT, 2022) run forward for 48 hr on days that major eruptive and ash‐dispersing events occurred at the Kīlauea Volcano (HYSPLIT, 2018). On many days, the dispersal of ash would be to the west‐southwest due to the northeast trade winds; however, during non‐trade wind conditions, ash would have been transported from Kīlauea toward O'ahu, especially material that was lofted high into the atmosphere (i.e., 2,000 m) (Neal et al., 2019). Similarly, during the 2018 eruption, operational ash transport modeling using Ash3D (Schwaiger et al., 2012) of an explosive eruption of Kīlauea showed ash reaching other islands under non‐trade wind conditions. Further, volcanic emissions such as vog (volcanic smog) emanating from Kīlauea are known to affect the air quality of other islands, including O'ahu (Kern et al., 2020). Between 2009 and 2014, Honolulu experienced 7–24 “vog days” per year (Halliday et al., 2019; Tofte et al., 2017) determined that wind from Hawai'i Island blows toward O'ahu 11% of the time. Of note, the presence of fresh/juvenile volcanic glass and size of the fragments up to 300 μm in length (Figure 5b) suggests that they derive from the eruptive fissures in the lower East Rift Zone rather than summit events; summit ash is predominantly older material, as mentioned, and is comparatively fine grained (unpublished data).
While it would be extremely challenging to show definitely in humans (short of an actual challenge) that NTM is acquired from respirable volcanic ash, our current unique findings indicate Kīlauea ash is an environmental source of respiratory relevant NTM. Indeed, the 2018 eruption post‐dates our 2016 respiratory NTM isolates. However, in analyzing the WGS of Kīlauea ash‐derived M. avium and M. abscessus as well as other environmental and respiratory isolates from neighboring islands, including O'ahu, we identified appreciable genetic similarity between these isolates. The level of genomic similarity observed between ash‐derived M. abscessus isolates and isolates from individuals on O'ahu, Maui, and Kaua'i falls within epidemiologically established thresholds of genomic similarity to imply possible transmission and shared recent common ancestry. These matches ranged from 15 to 36 SNPs among the core genomes. Moreover, considerable similarity among M. abscessus isolates found in ash and other environmental isolates from Hawai'i Island, Kaua'i, Maui and O'ahu and individuals from Hawai'i Island and O'ahu were noted. While genomic matches between ash‐derived M. avium and clinical isolates were not observed, we did find genomic matches to environmental isolates from Hawai'i Island and O'ahu. In this study, M. chimaera isolates recovered from both the Kīlauea environment and clinical respiratory samples were identical at the rpoB gene level; however, they differed from the single Kīlauea ash‐derived M. chimaera isolate, suggesting distinct provenances.
Of consequence, ash and other volcanic particulates can remain in the environment for decades, with continuous resuspension and remobilization by wind and human activities, resulting in the potential for both prolonged exposure (Hansell et al., 2006) and movement between the islands of Hawai'i. While geologic and historical records at Kīlauea identified numerous ash‐forming eruptions (e.g., 1790 and 1924) (Donald A. Swanson et al., 2014), we recognize that large ash‐generating eruptions are infrequent at Kīlauea Volcano over relevant timescales, and pre‐existing volcanic particulates (i.e., historic deposits or volcanic soil) may be more frequent volcanic fomites in intra‐ and inter‐island transport in Hawai'i. Even still, other mineral particles, such as desert dust (Kellogg & Griffin, 2006) are known to transport microbes over multiple spatial scales; this raises the question of a similar propensity for volcanic ash, which is an established agent in the Earth system (D. B. Dingwell et al., 2012). Using WGS and a global collection of clinical isolates, Bryant et al. (2016) surmised that the majority of M. abscessus infections belong to a dominant circulating clone likely acquired through transmission via fomites and aerosols that have spread globally. Our work suggests that volcanic particulate matter may be one such fomite.
While these data focus on understanding the unique environmental drivers that may contribute to high NTM prevalence in Hawai'i, they provide a framework for additional studies to explore how different volcanic soil and ash, or other environmental plumes such as wildland fire smoke (Kobziar & Thompson, 2020; Moore et al., 2021)—may disperse NTM and help drive the emergence of NTM lung infections on a global scale. Our findings also lend some credence that natural disasters that invariably disperse water and soil increase exposure to NTM (Honda et al., 2015). Future studies could investigate the environmental origins and survival of NTM in the context of dust transport. As extreme weather events continue to increase in frequency respiratory and infectious disease communities should remain vigilant of opportunistic environmental pathogens such as NTM.
Conflict of Interest
The authors have no conflicts of interest to declare.
Supporting information
Supporting Information S1
Acknowledgments
We thank Morgan Nasholds, Shauna Bladt, Gail Ferguson, Mike Pearson, Howard Hoshide, and Tina Neal for assistance with sample collection and acknowledge the Hawai'i Volcanoes National Park for sampling permit HAVO‐2017‐SCI‐0058, Michael Galloway for reagents, and Eric Wartchow for SEM expertise. We also acknowledge Diagnostic Laboratory Services, Inc. for their assistance with Hawai'i NTM clinical isolates and Rachel Wilsey for project assistance. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. Support for this work was provided by the National Science Foundation Grant 1743587 Division of Environmental Biology as part of the Evolution of Infectious Diseases program (JRH, MS, STN, JLC, EDC); Padosi Foundation (JRH); University of Colorado Anschutz Medical Campus Graduate Experiences for Multicultural Students GEMS) program NIH #HL103286 (JM). Funders were not involved in study design, data interpretation, writing, or in the decision to submit this work for publication.
Dawrs, S. N. , Virdi, R. , Norton, G. J. , Elias, T. , Hasan, N. A. , Robinson, S. , et al. (2024). Hawaiian volcanic ash, an airborne fomite for nontuberculous mycobacteria. GeoHealth, 8, e2023GH000889. 10.1029/2023GH000889
Data Availability Statement
The shapefiles used for sample map generation in ArcMap are available from the Hawaii Statewide GIS Program's open geospatial data porta (GIS). Chemical data for Kīlauea soils additional to those provided in the Supplementary Information, including LOI and pH, used for correlating NTM presence with environmental factors are available in Robinson (2019). The WGS data that support the findings of this study have been deposited in the National Center for Biotechnology Information (NCBI) database with accession codes under BioProject PRJNA718117 (NCBI BioProject Database, 2021). All reference isolates are contained within the NCBI non‐redundant BLAST database. The HYSPLIT model used for air trajectory simulations is freely available through a web‐based interface from NOAA Air Resources Laboratory (Stein et al., 2015) with no registration requirement. The MUSCLE (Edgar, 2004) and MEGA (Stecher et al., 2020) software used for rpoB sequence alignments and comparisons, respectively, are freely available on their respective webpages. The R package ggtree used for creating phylogenetic trees is available from the developer on Github (Yu et al., 2017).
References
- Adekambi, T. , Colson, P. , & Drancourt, M. (2003). rpoB‐based identification of nonpigmented and late‐pigmenting rapidly growing mycobacteria. Journal of Clinical Microbiology, 41(12), 5699–5708. 10.1128/jcm.41.12.5699-5708.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjemian, J. , Frankland, T. B. , Daida, Y. G. , Honda, J. R. , Olivier, K. N. , Zelazny, A. , et al. (2017). Epidemiology of nontuberculous mycobacterial lung disease and tuberculosis, Hawaii, USA. Emerging Infectious Diseases, 23(3), 439–447. 10.3201/eid2303.161827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjemian, J. , Olivier, K. N. , Seitz, A. E. , Falkinham, J. O., 3rd , Holland, S. M. , & Prevots, D. R. (2012). Spatial clusters of nontuberculous mycobacterial lung disease in the United States. American Journal of Respiratory and Critical Care Medicine, 186(6), 553–558. 10.1164/rccm.201205-0913OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjemian, J. , Olivier, K. N. , Seitz, A. E. , Holland, S. M. , & Prevots, D. R. (2012). Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. American Journal of Respiratory and Critical Care Medicine, 185(8), 881–886. 10.1164/rccm.201111-2016OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayris, P. M. , Delmelle, P. , Cimarelli, C. , Maters, E. C. , & Suzuki, Y. (2014). HCl uptake by volcanic ash in the high temperature eruption plume: Mechanistic insights. Geochimica et Cosmochimica Acta, 144, 188–201. 10.1016/j.gca.2014.08.028 [DOI] [Google Scholar]
- Bolyen, E. , Rideout, J. R. , Dillon, M. R. , Bokulich, N. A. , Abnet, C. C. , et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37(8), 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, J. M. , Grogono, D. M. , Rodriguez‐Rincon, D. , Everall, I. , Brown, K. P. , Moreno, P. , et al. (2016). Emergence and spread of a human‐transmissible multidrug‐resistant nontuberculous mycobacterium. Science, 354(6313), 751–757. 10.1126/science.aaf8156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damby, D. E. , Peek, S. , Lerner, A. H. , & Elias, T. (2018). Volcanic ash leachate and rainwater chemistry from increased 2018 activity of Kīlauea Volcano, Hawaii. Retrieved from https://www.sciencebase.gov/catalog/item/5b69cbe6e4b006a11f775784
- Davidson, R. M. , Hasan, N. A. , de Moura, V. C. , Duarte, R. S. , Jackson, M. , & Strong, M. (2013). Phylogenomics of Brazilian epidemic isolates of Mycobacterium abscessus subsp. bolletii reveals relationships of global outbreak strains. Infection, Genetics and Evolution, 20, 292–297. 10.1016/j.meegid.2013.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groote, M. A. , Pace, N. R. , Fulton, K. , & Falkinham, J. O. (2006). Relationships between Mycobacterium isolated from patients with pulmonary mycobacterial infection and potting soils. Applied and Environmental Microbiology, 72(12), 7602–7606. 10.1128/aem.00930-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwell, D. B. , Lavallée, Y. Y. , & Kueppers, U. (2012). Volcanic ash: A primary agent in the Earth system. Physics and Chemistry of the Earth, 45–46, 2–4. 10.1016/j.pce.2011.07.007 [DOI] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity [Software] [Comparative Study]. BMC Bioinformatics, 5(1), 113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias, T. , Kern, C. , Sutton, A. J. , & Horton, K. (2020). Sulfur dioxide emission rates from Kīlauea Volcano, Hawaii 2008–2013. 10.5066/P9K0EZII [DOI]
- Elias, T. , & Sutton, A. J. (2017). Volcanic air pollution hazards in Hawaii. U. S. G. Service; (pp. 1–4). [Google Scholar]
- Emily Oborski, P. L. K. , & Damby, D. E. (2020). Tracking the reactions of SO 2 and glass in the 2018 Kilauea ash. Goldschmidt2020, Virtual
- Epperson, L. E. , & Strong, M. (2020). A scalable, efficient, and safe method to prepare high quality DNA from mycobacteria and other challenging cells. Journal of Clinical Tuberculosis Other Mycobacterial Diseases, 19, 100150. 10.1016/j.jctube.2020.100150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkinham, J. O., 3rd. (2011). Nontuberculous mycobacteria from household plumbing of patients with nontuberculous mycobacteria disease. Emerging Infectious Diseases, 17(3), 419–424. 10.3201/eid1703.101510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkinham, J. O., 3rd , Iseman, M. D. , de Haas, P. , & van Soolingen, D. (2008). Mycobacterium avium in a shower linked to pulmonary disease. Journal of Water and Health, 6(2), 209–213. 10.2166/wh.2008.032 [DOI] [PubMed] [Google Scholar]
- Gebert, M. , Delgado‐Baquerizo, M. , Oliverio, A. , Webster, T. , Nichols, L. , Honda, J. , et al. (2018). Ecological analyses of mycobacteria in showerhead biofilms and their relevance to human health. MBio, 9(5), 10–1128. Retrieved from http://biorxiv.org/content/early/2018/07/10/366088.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, C. M. , Virdi, R. , Hasan, N. A. , Epperson, L. E. , Brown, L. , Dawrs, S. N. , et al. (2020). Assessment of soil features on the growth of environmental nontuberculous mycobacterial isolates from Hawai'i. Applied and Environmental Microbiology, 86(21). 10.1128/AEM.00121-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris, J. K. , Konstantinidis, K. T. , Klappenbach, J. A. , Coenye, T. , Vandamme, P. , & Tiedje, J. M. (2007). DNA‐DNA hybridization values and their relationship to whole‐genome sequence similarities. International Journal of Systematic and Evolutionary Microbiology, 57(1), 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Gouhier, M. , Eychenne, J. , Azzaoui, N. , Guillin, A. , Deslandes, M. , et al. (2019). Low efficiency of large volcanic eruptions in transporting very fine ash into the atmosphere. Scientific Reports, 5(9), 1–12. 10.1038/s41598-019-38595-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, T. J. , Lynham, J. , & De Paula, Á. (2019). Vog: Using volcanic eruptions to estimate the health costs of particulates. The Economic Journal, 129(620), 1782–1816. 10.1111/ecoj.12609 [DOI] [Google Scholar]
- Hansell, A. L. , Horwell, C. J. , & Oppenheimer, C. (2006). The health hazards of volcanoes and geothermal areas. Occupational and Environmental Medicine, 63(2), 149–156. 10.1136/oem.2005.022459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, N. A. , Warren, R. L. , Epperson, L. E. , Malecha, A. , Alexander, D. C. , Turenne, C. Y. , et al. (2017). Complete genome sequence of Mycobacterium chimaera SJ42, a Nonoutbreak strain from an immunocompromised patient with pulmonary disease. Genome Announcements, 5(37). 10.1128/genomeA.00963-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, J. R. , Bernhard, J. N. , & Chan, E. D. (2015). Natural disasters and nontuberculous mycobacteria: A recipe for increased disease? Chest, 147(2), 304–308. 10.1378/chest.14-0974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, J. R. , Hasan, N. A. , Davidson, R. M. , Williams, M. D. , Epperson, L. E. , Reynolds, P. R. , et al. (2016). Environmental nontuberculous mycobacteria in the Hawaiian islands. PLoS Neglected Tropical Diseases, 10(10), e0005068. 10.1371/journal.pntd.0005068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, J. R. , Virdi, R. , & Chan, E. D. (2018). Global environmental nontuberculous mycobacteria and their contemporaneous man‐made and natural niches. Frontiers in Microbiology, 9, 2029. 10.3389/fmicb.2018.02029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HYSPLIT (Hybrid Single‐Particle Lagrangian Integrated Trajectory model) . (2018). Retrieved from https://www.ssd.noaa.gov/VAAC/ARCH18/archive.html
- HYSPLIT (Hybrid Single‐Particle Lagrangian Integrated Trajectory model) . (2022). Retrieved from https://www.ready.noaa.gov/HYSPLIT.php
- Jiang, H. , Lei, L. , Ding, S. W. , & Zhu, S. (2014). Skewer: A fast and accurate adapter trimmer for next‐generation sequencing paired‐end reads. BMC Bioinformatics, 15(182), 1–12. 10.1186/1471-2105-15-s9-s1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, C. A. , & Griffin, D. (2006). Aerobiology and the global transport of desert dust. Trends in Ecology & Evolution, 21(11), 638–644. 10.1016/j.tree.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Kern, C. , Lerner, A. , Elias, T. , Nadeau, P. , Holland, L. , Kelly, P. , et al. (2020). Quantifying gas emissions associated with the 2018 rift eruption of Kilauea Volcano using ground‐based DOAS measurements. Bulletin of Volcanology, 82(55), 1–24. [Google Scholar]
- King, P. L. , Ávila, J. N. , Oborski, E. M. , Damby, D. E. , Patkar, A. , Williams, I. S. , et al. (2023). Sulfur isotopes in Kīlauea Volcano's 2018 summit ash: Tracing sulfur‐bearing reactions from the source to hydrothermal system and plume.
- Kobziar, L. N. , & Thompson, G. R. (2020). Wildfire smoke, a potential infectious agent. Science, 370(6523), 1408–1410. 10.1126/science.abe8116 [DOI] [PubMed] [Google Scholar]
- Langmead, B. , & Salzberg, S. L. (2012). Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9(4), 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone, A. , Touré Kane, C. , Mbengue, M. , Jenkins, G. S. , Niang, D. N. , Drame, M. S. , & Gernand, J. M. (2020). Dust events in Dakar, Senegal, West Africa. Geohealth, 4(6), 1–8. 10.1029/2019gh000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsaglia, K. M. , Milliken, K. , & Doran, L. (2013). Technical Note 1: Smear slides of marine mud for IODP Core Description: Part I: Methodology and atlas of siliciclastic and volcanogenic components: International Ocean Discovery Program (263 p.). 10.2204/iodp.tn.1.2013 [DOI]
- Mirsaeidi, M. , Machado, R. F. , Garcia, J. G. , & Schraufnagel, D. E. (2014). Nontuberculous mycobacterial disease mortality in the United States, 1999–2010: A population‐based comparative study. PLoS One, 9(3), e91879. 10.1371/journal.pone.0091879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R. A. , Bomar, C. , Kobziar, L. N. , & Christner, B. C. (2021). Wildland fire as an atmospheric source of viable microbial aerosols and biological ice nucleating particles. The ISME Journal, 15(2), 461–472. 10.1038/s41396-020-00788-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI BioProject Database . (2021). Mycobacterium intracellulare subsp. chimaera. Pacific island nontuberculous mycobacteria raw sequence reads [Dataset]. Retrieved from https://www.ncbi.nlm.nih.gov/bioproject/PRJNA718117
- Neal, C. A. , Brantley, S. R. , Antolik, L. , Babb, J. L. , Burgess, M. , Calles, K. , et al. (2019). The 2018 rift eruption and summit collapse of Kilauea Volcano. Science, 363(6425), 367–374. 10.1126/science.aav7046 [DOI] [PubMed] [Google Scholar]
- Nelson, S. T. , Robinson, S. , Rey, K. , Brown, L. , Jones, N. , Dawrs, S. N. , et al. (2021). Exposure pathways of nontuberculous mycobacteria through soil, streams, and groundwater, Hawai'i, USA. Geohealth, 5(4), e2020GH000350. 10.1029/2020GH000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, S. T. , Tingey, D. G. , & Selck, B. (2013). The denudation of ocean islands by ground and surface waters: The effects of climate, soil thickness, and water contact times on Oahu, Hawaii. Geochimica et Cosmochimica, 103, 276–294. 10.1016/j.gca.2012.09.046 [DOI] [Google Scholar]
- Parsons, A. W. , Dawrs, S. N. , Nelson, S. T. , Norton, G. J. , Virdi, R. , Hasan, N. A. , et al. (2022). Soil properties and moisture synergistically influence nontuberculous mycobacterial prevalence in natural environments of Hawai'i. Applied and Environmental Microbiology, 88(9), e0001822. 10.1128/aem.00018-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porder, S. , Hilley, G. E. , & Chadwick, O. A. (2007). Chemical weathering, mass loss, and dust inputs across a climate by time matrix in the Hawaiian Islands. Earth and Planetary Science Letters, 258(3–4), 414–427. 10.1016/j.epsl.2007.03.047 [DOI] [Google Scholar]
- Reheis, M. C. , Goodmacher, J. C. , Harden, J. W. , McFadden, L. D. , Rockwell, T. K. , Shroba, R. R. , et al. (1995). Quaternary soils and dust deposition in southern Nevada and California. The Geological Society of America Bulletin, 107(9), 1003–1022. [DOI] [Google Scholar]
- Reheis, M. C. , & Kihl, R. (1995). Dust deposition in southern Nevada and California, 1984–1989: Relations to climate, source area, and source lithology. Journal of Geophysical Research, 100, D5–D8918. 10.1029/94jd03245 [DOI] [Google Scholar]
- Richter, M. , & Rosselló‐Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences of the U S A, 10(106), 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme, C. , Marshall Hathaway, J. J. , Enes Dapkevicius Mde, L. , Miller, A. Z. , Kooser, A. , Northup, D. E. , et al. (2015). Actinobacterial diversity in volcanic caves and associated geomicrobiological interactions. Frontiers in Microbiology, 6, 1342. 10.3389/fmicb.2015.01342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, S. M. (2019). Geological and geochemical controls on non‐tuberculous mycobacterium transmission: Examples from Hawai'i [Dataset]. Brigham Young University. Retrieved from https://scholarsarchive.byu.edu/etd/8558/
- Schwaiger, H. , Denlinger, R. , & Mastin, L. G. (2012). Ash3d: A finite‐volume, conservative numerical model for ash transport and tephra deposition. Journal of Geophysical Research, 117(B04204). 10.1029/2011jb008968 [DOI] [Google Scholar]
- Shapley, M. (2022). Volcanic glass. University of Minnesota. Retrieved from https://tmi.csd.umn.edu/uniqueIdentification/show/27 [Google Scholar]
- Stecher, G. , Tamura, K. , & Kumar, S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS [Software]. Molecular Biology and Evolution, 37(4), 1237–1239. 10.1093/molbev/msz312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, A. F. , Draxler, R. R. , Rolph, G. D. , Stunder, B. J. B. , Cohen, M. D. , & Ngan, F. (2015). NOAA's HYSPLIT atmospheric transport and dispersion modeling system [Software]. https://www.ready.noaa.gov/HYSPLIT.php
- Stewart, C. , Damby, D. E. , Horwell, C. J. , Elias, T. , Ilyinskaya, E. , Tomašek, I. , et al. (2022). Volcanic air pollution and human health: Recent advances and future directions. Bulletin of Volcanology, 84(11), 1–25. 10.1007/s00445-021-01513-9 [DOI] [Google Scholar]
- Stewart, C. , Damby, E. D. , Tomašek, I. , Horwell, C. J. , Plumlee, G. S. , Armienta, M. A. , Ruiz Hinojosa, M. G. , et al. (2020). Assessment of leachable elements in volcanic ashfall: A review and evaluation of a standardized protocol for ash hazard characterization. Journal of Volcanology and Geothermal Research, 392, 1–22. 10.1016/j.jvolgeores.2019.106756 [DOI] [Google Scholar]
- Swanson, D. A. , Rose, T. R. , Mucek, A. E. , Garcia, M. O. , Fiske, R. S. , & Mastin, L. G. (2014). Cycles of explosive and effusive eruptions at Kīlauea Volcano, Hawai'i. Geology, 42(7), 631–634. 10.1130/g35701.1 [DOI] [Google Scholar]
- Tofte, K. , Chu, P. S. , & Barnes, G. M. (2017). Large‐scale weather patterns favorable for volcanic smog occurrences on O'ahu, Hawai'i. Air Quality, Atmosphere & Health, 10(10), 1163–1180. 10.1007/s11869-017-0502-z [DOI] [Google Scholar]
- Tomasek, I. , Damby, D. E. , Stewart, C. , Horwell, C. J. , Plumlee, G. , Ottley, C. J. , et al. (2021). Development of a simulated lung fluid leaching method to assess the release of potentially toxic elements from volcanic ash. Chemosphere, 278, 130303. 10.1016/j.chemosphere.2021.130303 [DOI] [PubMed] [Google Scholar]
- Tomasek, I. , Horwell, C. J. , Damby, D. E. , Barosova, H. , Geers, C. , Petri‐Fink, A. , et al. (2016). Combined exposure of diesel exhaust particles and respirable Soufriere Hills volcanic ash causes a (pro‐) inflammatory response in an in vitro multicellular epithelial tissue barrier model. Particle and Fibre Toxicology, 13(1), 67. 10.1186/s12989-016-0178-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eaton, A. R. , Harper, M. A. , & Wilson, C. J. N. (2013). High‐flying diatoms: Widespread dispersal of microorganisms in an explosive volcanic eruption. Geology, 41(11), 1187–1190. 10.1130/g34829.1 [DOI] [Google Scholar]
- Virdi, R. , Lowe, M. E. , Norton, G. J. , Dawrs, S. N. , Hasan, N. A. , Epperson, L. E. , et al. (2021). Lower recovery of nontuberculous mycobacteria from outdoor Hawai'i environmental water biofilms compared to indoor samples. Microorganisms, 9(2), 224. 10.3390/microorganisms9020224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, R. J., Jr. (1987). Nontuberculous mycobacteria and water: A love affair with increasing clinical importance. Infectious Disease Clinics of North America, 1(3), 677–686. 10.1016/s0891-5520(20)30139-2 [DOI] [PubMed] [Google Scholar]
- Walters, W. , Hyde, E. R. , Berg‐Lyons, D. , Ackermann, G. , Humphrey, G. , Parada, A. , et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4‐5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems, 1(1). 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, R. R. , Judd, L. M. , Gorrie, C. L. , & Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Computational Biology, 13(6), 1–22. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt, V. , Ayris, P. M. , Damby, D. E. , Cimarelli, C. , Kueppers, U. , Dingwell, D. B. , & Worheide, G. (2017). Volcanic ash supports a diverse bacterial community in a marine mesocosm. Geobiology, 15(3), 453–463. 10.1111/gbi.12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Smith, D. K. , Zhu, H. , Guan, Y. , & Lam, T. T. Y. (2017). GGTREE: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution, 8(1), 28–36. 10.1111/2041-210x.12628 [DOI] [Google Scholar]
- Zhao, X. , Epperson, L. E. , Hasan, N. A. , Honda, J. R. , Chan, E. D. , Strong, M. , et al. (2017). Complete genome sequence of Mycobacterium avium subsp. hominissuis strain H87 isolated from an indoor water sample. Genome Announcements, 5(16). 10.1128/genomeA.00189-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI BioProject Database . (2021). Mycobacterium intracellulare subsp. chimaera. Pacific island nontuberculous mycobacteria raw sequence reads [Dataset]. Retrieved from https://www.ncbi.nlm.nih.gov/bioproject/PRJNA718117
- Robinson, S. M. (2019). Geological and geochemical controls on non‐tuberculous mycobacterium transmission: Examples from Hawai'i [Dataset]. Brigham Young University. Retrieved from https://scholarsarchive.byu.edu/etd/8558/
- Stein, A. F. , Draxler, R. R. , Rolph, G. D. , Stunder, B. J. B. , Cohen, M. D. , & Ngan, F. (2015). NOAA's HYSPLIT atmospheric transport and dispersion modeling system [Software]. https://www.ready.noaa.gov/HYSPLIT.php
Supplementary Materials
Supporting Information S1
Data Availability Statement
The shapefiles used for sample map generation in ArcMap are available from the Hawaii Statewide GIS Program's open geospatial data porta (GIS). Chemical data for Kīlauea soils additional to those provided in the Supplementary Information, including LOI and pH, used for correlating NTM presence with environmental factors are available in Robinson (2019). The WGS data that support the findings of this study have been deposited in the National Center for Biotechnology Information (NCBI) database with accession codes under BioProject PRJNA718117 (NCBI BioProject Database, 2021). All reference isolates are contained within the NCBI non‐redundant BLAST database. The HYSPLIT model used for air trajectory simulations is freely available through a web‐based interface from NOAA Air Resources Laboratory (Stein et al., 2015) with no registration requirement. The MUSCLE (Edgar, 2004) and MEGA (Stecher et al., 2020) software used for rpoB sequence alignments and comparisons, respectively, are freely available on their respective webpages. The R package ggtree used for creating phylogenetic trees is available from the developer on Github (Yu et al., 2017).


