Abstract
Kaposi's sarcoma-associated herpesvirus (also called human herpesvirus type 8 [HHV8]) latently infects a number of cell types. Reactivation of latent virus can occur by treatment with the phorbol ester tetradecanoyl phorbol acetate (TPA) or with the transfection of plasmids expressing the lytic switch activator protein K-Rta, the gene product of ORF50. K-Rta expression is sufficient for the activation of the entire lytic cycle and the transactivation of viral genes necessary for DNA replication. In addition, recent evidence has suggested that K-Rta may participate directly in the initiation of lytic DNA synthesis. We have now generated a recombinant HHV8 bacterial artificial chromosome (BAC) with a large deletion within the ORF50 locus. This BAC, BAC36Δ50, failed to produce infectious virus upon treatment with TPA and was defective for DNA synthesis. Expression of K-Rta in trans in BAC36Δ50-containing cells was able to abolish both defects. Real-time PCR revealed that K-bZIP, ORF40/41, and K8.1 were not expressed when BAC36Δ50-containing cells were induced with TPA. However, the mRNA levels of ORF57 were over fivefold higher in TPA-treated BAC36Δ50-containing cells than those observed in similarly treated wild-type BAC-containing cells. In addition, immunohistochemical analysis showed that while the latency-associated nuclear antigen (LANA) was expressed in the mutant BAC-containing cells, ORF59 and K8.1 expression was not detected in TPA-induced BAC36Δ50-containing cells. These results showed that K-Rta is essential for lytic viral reactivation and transactivation of viral genes contributing to DNA replication.
Kaposi's sarcoma-associated herpesvirus, also called human herpesvirus type 8 (HHV8), primarily latently infects B cells and is typically characterized by the expression of the latency-associated nuclear antigen (LANA) (8, 10, 11). The initiation of lytic DNA replication is mediated by the lytic switch protein K-Rta, which is encoded by the ORF50 locus (6, 16, 26, 29). K-Rta transactivates a number of viral genes and autoregulates its own expression via various DNA elements within promoter regions (15, 21, 25, 29, 30). In addition, K-Rta interacts with the CCAAT/enhancer binding protein alpha (C/EBPα) to upregulate K-Rta expression by an interaction with the K-Rta promoter (31, 32). K-Rta also interacts with the early virally encoded protein encoded by K8, K-bZIP (7, 13). The interaction of K-bZIP with K-Rta appears to selectively downregulate the ability of K-Rta to activate certain viral promoters in transient assays. In addition, it was recently suggested that maintenance of viral latency involves an interaction between LANA and K-Rta (12).
Recently, it was demonstrated that K-Rta and K-bZIP are required for amplification of the HHV8 lytic origin in a cotransfection-replication assay (1, 33). Evidence suggests that the role of K-Rta in DNA replication may be to transactivate a K-Rta-responsive promoter within the oriLyt region. This observation implicates K-Rta as having a direct role in lytic DNA replication as well as activating the viral genes required for viral growth.
Although many studies have focused on the results of exogenous expression of K-Rta in HHV8-infected cells, it has not been demonstrated that the expression of K-Rta is required for progression to the lytic replication cycle in the context of the viral genome. Studies from a closely related virus, gammaherpesvirus 68, demonstrated that the expression of gene 50 from the viral genome, a homolog of HHV8 ORF50, was essential for virus replication (19). This was shown by the construction of a recombinant virus in which the gene 50 open reading frame (ORF) was deleted. Zhu et al. were able to reduce the level of K-Rta expression by 90% with oligonucleotides that recruit cellular RNase P, which resulted in over a 100-fold decrease in virus replication (36). These data suggest that the expression of K-Rta in HHV8 is essential for virus replication and growth; however, the generation of a viral mutant that is defective for K-Rta expression will serve to answer the question of the dependence of K-Rta for the progression of the virus into the lytic cycle.
The recent subcloning of the HHV8 genome into a bacterial artificial chromosome (BAC) vector allowed the generation of viral deletion mutants targeting specific open reading frames (ORFs) within the virus (17, 35). In this report, we generated a recombinant BAC with a deletion in the ORF50 locus (BAC36Δ50) and show that K-Rta is required for lytic DNA replication and viral growth. Real-time PCR analysis revealed that all representative early and late kinetic classes of viral transcripts are not expressed in BAC36Δ50-containing cells induced with TPA. However, there was an apparent increase in the accumulation of mRNA encoding ORF57, a viral transactivator. BAC36Δ50 was efficiently rescued, with respect to production of infectious virus and DNA replication, upon the expression of K-Rta in trans in BAC36Δ50-containing cells. These data indicated that K-Rta is essential for lytic viral reactivation and transactivation of viral genes contributing to DNA replication.
MATERIALS AND METHODS
Cells.
293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. BAC36-infected 293 cells obtained from S. Gao (University of Texas) (35) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1 mg of hygromycin per ml. BCBL-1 cells infected with HHV8 were cultured with RPMI 1640 medium supplemented with 20% heat-inactivated fetal bovine serum. To induce HHV8 lytic replication, the cultured cells were treated with 25 ng of 12-O-tetradecanoyl phorbol-13-acetate (TPA; Sigma) per ml and 1,000 units of alpha interferon per ml for 2 to 4 days.
Plasmids.
BAC DNA containing the HHV8 genome (BAC36) was purified from BAC36-infected 293 cells with an alkaline lysis procedure (27) and transformed into Escherichia coli strain DH10B by electroporation. The shuttle plasmid pST76K_SR was obtained from M. Messerle, Max con Pettenkofer Institute (3). ORF50 and its flanking sequence (nucleotides 69405 to 77640) was PCR amplified with HHV8 genome DNA as the template and the primer set forward primer, 5′-GTTGAACTTATTTTCCCTTTTGACCTGCGT-3′, and reverse primer, 5′-CTTTATTTGTGGCCTCGATACTAGGTCACT-3′. The 8.2-kb PCR product was ligated into the pGEM-T easy Vector (Promega, catalog number A1360) according to the manufacturer's protocol to generate construct pGEM-ORF50flank. A 7.6-kb fragment containing ORF50 and its flanking sequence (nucleotides 69405 to 77003) was released by cleaving pGEM-ORF50flank with NotI, treated with E. coli DNA polymerase Klenow fragment, and ligated into pST76K_SR cleaved with SmaI. The resultant construct, pKSR-ORF50flank, was then cleaved with ApaI and upon removal of 1.21 kb of DNA sequence within ORF50, the plasmid was religated to make the construct pKSR-ORF50mut. This final construct was then used as a shuttle plasmid to generate an HHV8 BAC recombinant with a deletion within the ORF50 gene locus.
The K-Rta expression plasmid pCMV RTA 5′Flag was a gift from Ren Sun (University of California-Los Angeles). pOPRSVI-50 which contains ORF50 (K-Rta) with the Flag epitope in the same reading frame context was subcloned from pCMV-RTA 5′Flag with PCR primers 5′-CCGCTCGAGTTGATCTACCATGGACTACAAAGACGATGACGA-3′ and 5′-TCCTCTAGAGATCTATCAGTCTCGGAAGTAATTACGCCAT-3′. Primers were designed such that XhoI and XbaI restriction endonuclease cleavage sites were placed on the ends of the oligonucleotides. After cleavage with XhoI and XbaI the resulting PCR product was ligated into XhoI- and XbaI-cleaved pOPRSVI/MCS (Stratagene) and used to generate the inducible ORF50 cell line.
BAC mutagenesis.
Mutagenesis of the HHV8 BAC plasmid was performed according to the protocol provided by M. Messerle (3). Briefly, shuttle plasmid pKSR-ORF50mut was electroporated into E. coli DH10B bacteria that already contained HHV8 BAC36. Transformants were selected at 30°C on Luria-Bertani (LB) agar plates containing chloramphenicol (15 μg/ml) and kanamycin (50 μg/ml). Clones containing cointegrates were identified by streaking the bacteria onto new LB plates containing chloramphenicol (15 μg/ml) and kanamycin (50 μg/ml) followed by incubation at 43°C. To allow resolution of the cointegrate clones were streaked onto LB plates containing chloramphenicol (15 μg/ml) and incubated at 30°C.
To select clones that had resolved the cointegrate and that contained the mutant BAC plasmid, bacteria were restreaked onto LB plates containing chloramphenicol (15 μg/ml) and 5% sucrose. Resolution of the cointegrate was confirmed by testing for the loss of the kanamycin marker encoded by the shuttle plasmid. BAC plasmid DNA was isolated from 10-ml overnight cultures by the alkaline lysis procedure and characterized by restriction enzyme analysis followed by Southern blot analysis. Large preparations of HHV8 BAC plasmids were obtained from 500-ml E. coli cultures with the Qiagen Large construct kit according to the instructions of the manufacturer.
Southern blot analysis.
The DNA probe used for Southern blot hybridizations was amplified as a 1.4-kb fragment (corresponding to the ORF50 locus) with the HHV8 genome as the template and primer set forward, 5′-ATGCAGCGGGGTGAGCCTGCCTCCAGCC-3′, and reverse, 5′-TTGCAGAATACTGGACAACAGCGCGTCG-3′. Purified HHV8 BAC plasmid DNA was cleaved with Hind III and separated by electrophoresis on 0.5% agarose gels in 1X Tris-borate-EDTA buffer for 14 to 18 h at 2.5 V/cm. DNA fragments were visualized by ethidium bromide staining, denatured, and transferred to Zeta-Probe GT genomic tested blotting membranes (Bio-Rad). DNA probes were radiolabeled with [α-32P]dCTP (Amersham) with the Rediprime II random prime labeling system (Amersham). Prehybridization was performed at 65°C for1 h in hybridization buffer (7% sodium dodecyl sulfate, 10% polyethylene glycol, 1.5X SSPE [1X SSPE is 0.18 M NaCl, 10 mM NaPO4, and 1 mM EDTA, pH 7.7]). DNA blots were hybridized with radiolabeled probes in the same solution at 65°C for about 16 h. Blots were washed twice for 15 min with 2X SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate and twice for 30 min with 0.1X SSC-0.1% sodium dodecyl sulfate at 65°C. Blots were exposed to X-ray film for 3 to 5 h at room temperature.
Establishment of stable BAC36Δ50-containing 293 cell lines.
293 cells were transfected with purified BAC36Δ50 DNA with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Stable cell lines harboring BAC36Δ50 were selected with 1 mg of hygromycin per ml.
Generation of an inducible ORF50 (K-Rta) 293 cell line, 293-LAC50.
293 cells were cotransfected with pOPRSVI-50 and pCMVLacI (Stratagene) with 293 TransIT (Mirus) transfection reagent. Cells were passed at 48 h posttransfection into hygromycin (250 μg/ml) and G418 (1.5 mg/ml) and further incubated for 1 to 2 weeks. Dual-drug-resistant colonies were picked and tested for isopropylthiogalactopyranoside (IPTG)-induced expression of K-Rta with anti-Flag antibody.
Detection of supernatant virus.
293 cells plated onto 12-well tissue culture dishes were infected with supernatant containing BAC36 or BAC36Δ50 virus plus Polybrene (5 μg/ml). Supernatants were obtained from BAC36- or BAC36Δ50-containing cells treated with TPA or from the inducible cell line 293-LAC50 transfected with BAC constructs induced with IPTG. Supernatants were centrifuged three times at 5,000 x g for 15 min to remove floating cells. Clarified supernatants were then subjected to one freeze-thaw cycle and incubated with fresh 293 cells. Green fluorescent protein (GFP) expression was used to monitor infection.
RNA purification.
293 cells containing BAC36 or BAC36Δ50 were induced with 25 ng of TPA per ml and harvested at various times postinduction. For the 293-LAC50 cell line, cells were transfected with BAC36 or BAC36Δ50 and total RNA was harvested at various times post-IPTG treatment. Total RNA was isolated with the Absolutely RNA reverse transcription-PCR miniprep kit (Stratagene, catalog number 400800) according to the instructions of the manufacturer. Residual DNA contamination was eliminated with Turbo DNA-free DNase (Ambion, catalog number 1907). RNA quality was assessed by separation of RNA through a 1% formaldehyde denaturing agarose gel. Only samples with 260 nm/280 nm absorbance ratios of greater than 1.9 were used for subsequent experiments.
Quantitative real-time PCR analysis.
cDNA (20 μl) for real-time PCR was generated from 2 μg of total RNA with the SuperScript First-Strand Synthesis System (Invitrogen, catalog number11904-018), priming with 250 ng of random hexamers, according to the manufacturer's instructions. The probe and primer sequences for the real-time TaqMan PCR system were chosen and synthesized with Applied Biosystems's Assay-by-Design program (Applied Biosystems, 4331348) for ORF57, K8, K8.1, ORF40/41, and LANA genes, respectively (Table 1). The two unlabeled PCR primers and the FAM (6-carboxyfluorescein) dye-labeled TaqMan MGB probe for each sequence were formulated into a single-tube, ready-to-use mixture at a 20x concentration. The real-time PCRs were performed with the TaqMan Universal PCR Master Mix (Applied Biosystems, catalog number 4304437), which consists of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 5 mM MgCl2, 300 mM each dATP, dCTP, and dGTP, 600 mM dUTP, 0.625 U of AmpliTaq Gold DNA polymerase per reaction, and 0.25 U of uracil N-glycosylase per reaction. Reactions in a total volume of 25 μl consisted of 12.5 μl of Mastermix, appropriate amounts of cDNA (10% for detection of viral transcripts, 2% for detection of cellular transcripts), 900 nM each primer, and 200 nM TaqMan probe. Following 2 min at 50°C, the AmpliTaq Gold DNA polymerase was activated at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and at 60°C for 1 min. All reactions were carried out six times with a 7700 ABI Prism sequence detector (Applied Biosystems).
TABLE 1.
Primers and probes used for real-time PCR
| Gene | Primer and probe | Sequence (5′-3′) |
|---|---|---|
| ORF59 | Forward | TGGAAGCCGGTGGTAGGA |
| Reverse | GTAGGAATGCACCCGTTTTGC | |
| Probe | CCACCAGGCTTCTCCTCT | |
| ORF57 | Forward | CATCCTAGAGGACTCTGT |
| Reverse | TTGCTCGTCTTCCAGTGT | |
| Probe | TCTGAGTTTGACGAATC | |
| ORF40-41 | Forward | TTTGGAGCCTGAGCAATG |
| Reverse | GAGAATACAAGATCCCCCGAAG | |
| Probe | GGTTTACTCTGAAGGG | |
| K8 | Forward | CAAGAGGCGACTACATAGAAA |
| Reverse | GATCACATACTTCGGCCTTAAC | |
| Probe | AGGAACGCTTATGCAC | |
| K8.1 | Forward | AATATCAGCCTTTTCAGGATCA |
| Reverse | CACCACTATTTCTGCCGTTTTC | |
| Probe | AACCATCCAGGACCAC | |
| LANA | Forward | CGCTCCCGCAACACCTTTA |
| Reverse | GGGAGGAAGACGTGGTTACG | |
| Probe | CCTCCACCGGCACTCT |
RNA isolated from the BCBL-1 cells was used to generate a standard curve for each gene examined. The standard curve was used to calculate the relative amount of specific cDNA present in a sample. Data from each sample was normalized to human cyclophilin (Applied Biosystem, part 4326316E) levels for the equal amount of cDNA input in each real-time PCR. As an additional control, RNA from each sample without the reverse transcriptase step was analyzed by real-time PCR to ensure that samples were free of contaminating DNA. Each experiment was performed 12 times, and error bars are the standard deviations from the mean of the 12 experiments. Data are reported as the increase in mRNA accumulation compared to the untreated BAC36 mRNA levels. As a negative control, each plate contained a minimum of three wells lacking template.
Viral DNA accumulation analysis with real-time PCR.
Reactions were performed as described for cDNA real-time PCR with the exception that total cellular DNA was used from 293 (HHV8 BAC36) or 293 (BAC36Δ50) cells. Cells were harvested at various times after induction with DNA extraction buffer (2% sodium dodecyl sulfate, 100 mM Tris-HCl, pH 8, 10 mM EDTA, and 50 mg of proteinase K per ml). The cell lysates was incubated at 60°C for 1 h, followed by phenol-chloroform extraction and then chloroform extraction. DNA was precipitated with 100% ethanol. DNA was resuspended in TE (10 mM Tris, pH 8, 1 mM EDTA) and the concentration was determined with a spectrophotometer. Equal amounts (100 ng) of the total DNA were used in a real-time PCR with primers and probe specific for the ORF59 locus (Table 1). Data are reported as the increase in DNA accumulation compared to the untreated BAC36 DNA level. Each experiment was done in triplicate. A standard curve was generated with BAC36-purified DNA with the ORF59 primers and probe.
Immunofluorescence assay.
Cells were plated on glass coverslips and fixed at 96 h postinduction with 1% paraformaldehyde in phosphate-buffered saline for 10 min at 20°C and then permeabilized with 0.2% Triton X-100 in phosphate-buffered saline for 20 min at 4°C. Cells were rinsed with phosphate-buffered saline, incubated with 10% goat serum for 20 min, and then rinsed with phosphate-buffered saline. Coverslips were incubated for 1 h at 37°C in a humidified tray with 2% goat serum and a 1:2,500 dilution of primary antibody. After incubation, coverslips were washed in phosphate-buffered saline three times for 5 min each and incubated with a 1:600 dilution of Alexa Fluor 555 goat anti-mouse or anti-rat immunoglobulin antibody (Molecular Probes, Inc.) for 45 min at 37°C. Prior to the addition of mounting solution, cells were once again washed for 5 min in phosphate-buffered saline three times.
Images were captured by confocal microscopy (Nikon E800 epifluorescence microscope). Detection of HHV8 LANA was carried out with the anti-LANA rat monoclonal antibody (Advanced Biotechnologies Inc., catalog number 13-210-100). Detection of HHV8 ORF59 protein (PF-8) was carried out with anti-HHV8 ORF59 mouse monoclonal antibody (Advanced Biotechnologies Inc., catalog number 13-211-100). Detection of HHV8 ORF K8.1 protein was carried out with anti-K8.1A monoclonal antibody (Advanced Biotechnologies Inc., catalog number 13-213-100).
RESULTS
Generation of an HHV8 recombinant BAC with a large deletion within the ORF50 locus.
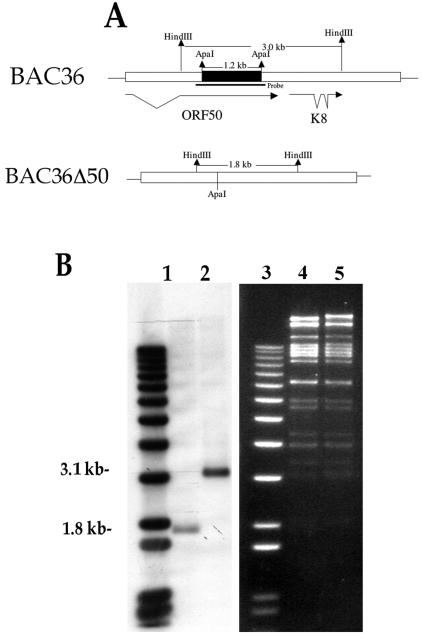
In order to generate a recombinant BAC, we initially subcloned a 7.6-kb fragment corresponding to the ORF50 region into the pST76K-SR vector (3). Subsequently, a 1,210-nucleotide segment (nucleotides 73139 to 74348) of DNA between the ApaI restriction sites was removed from within the ORF50 coding region (Fig. 1A). This deletion removed approximately 400 amino acids from the K-Rta protein. Recombination experiments were performed with BAC36 as the template, and the resultant HHV8 BAC mutant, BAC36Δ50, was purified and analyzed by Southern blot. The recombinant BAC36Δ50 DNA along with BAC36 DNA was cleaved with HindIII and hybridized with a probe specific for the ORF50 locus (Fig. 1A). A 1.8-kb band was detected in the BAC36Δ50 DNA sample compared to the nondeleted 3.1-kb band observed in the BAC36 DNA lane (Fig. 1B, compare lanes 1 and 2). In addition, DNA sequencing confirmed the deletion in the recombinant BAC. Also shown is the ethidium bromide-stained gel of BAC36Δ50 (Fig. 1, lane 4) and BAC36 (Fig. 1, lane 5) DNA cleaved with HindIII.
FIG. 1.
Construction of a recombinant HHV8 BAC with a large deletion in ORF50. (A) Schematic showing the region deleted from within ORF50 and the resultant configuration of the genome after the deletion. Also depicted are the HindIII sites used to confirm the proper BAC construction and the position of the hybridization probe. (B) Southern blot of HindIII-cleaved BAC36 and BAC36Δ50 DNAs hybridized with a probe specific for the ORF50 region. Lanes: 1, BAC36Δ50; 2, BAC36. Also shown is an ethidium bromide-stained gel of BAC36Δ50 and BAC36 DNA cleaved with HindIII. Lanes: 3, molecular size markers; 4, BAC36Δ50 DNA; 5, BAC36 DNA.
BAC36Δ50-transfected cells fail to produce supernatant virus and accumulate viral DNA in the presence of TPA.
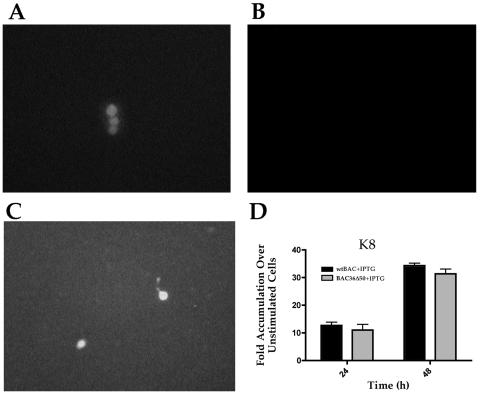
We first evaluated the ability of BAC36Δ50 to produce supernatant virus upon treatment with TPA, an inducer of the viral lytic cycle. BAC36 and BAC36Δ50 were transfected into 293 cells, and cell lines were generated with hygromycin selection. Cell lines were treated with TPA, and supernatants were collected and used to infect fresh 293 cells. The wild-type (BAC36) virus-containing cell line was capable of producing infectious virus, as demonstrated by the appearance of green cells upon inoculation of fresh 293 cells (Fig. 2A). The cell line harboring BAC36Δ50 failed to produce green cells upon inoculation of fresh 293 cells with supernatants from BAC36Δ50-containing cells treated with TPA (Fig. 2B). However, when BAC36Δ50 was transfected into the inducible K-Rta cell line 293-LAC50, which was subsequently treated with IPTG to induce the expression of K-Rta, infectious virus was produced, as demonstrated by the appearance of green cells on freshly inoculated 293 cells (Fig. 2C). This indicated that the ORF50 mutant virus was capable of producing infectious virus when K-Rta was supplied in trans and that the mutation within the HHV8 BAC genome affected only the ORF50 locus.
FIG. 2.
Complementation of BAC36Δ50 virus production by expression of K-Rta. For complementation, BAC-containing 293 cell lines were treated with TPA or 293-LAC50 cells were transfected with BAC36Δ50 and induced to express K-Rta by treatment with IPTG. Supernatants from each treatment were harvested 4 days postinduction and used to inoculate fresh 293 cells. (A) Cells were inoculated with supernatant from BAC36-containing 293 cells treated with TPA. (B) Cells were inoculated with supernatant from BAC36Δ50-containing 293 cells treated with TPA. (C) Cells were inoculated with supernatant from BAC36Δ50-transfected 293-Lac50 cells treated with IPTG. (D) IPTG induction of ORF50 complements the defect in BAC36Δ50. 293-LAC50 cells were transfected with BAC36 or BAC36Δ50 and treated with IPTG. Real-time PCR was performed, and the mRNA accumulation for K8 was determined.
Since the level of virus produced from TPA induction of 293 cells was very low, we examined K8 mRNA levels from 293-LAC50 cells transfected with BAC36 and BAC36Δ50. This was done to show that the expression of ORF50 could complement the mutation in BAC36Δ50. When cells were treated with IPTG, we observed a similar level of K8 mRNA accumulation between BAC36- and BAC36Δ50-containing cells (Fig. 2D). This demonstrated that the defect in BAC36Δ50 could be fully complemented, with respect to gene expression, by supplying ORF50 in trans.
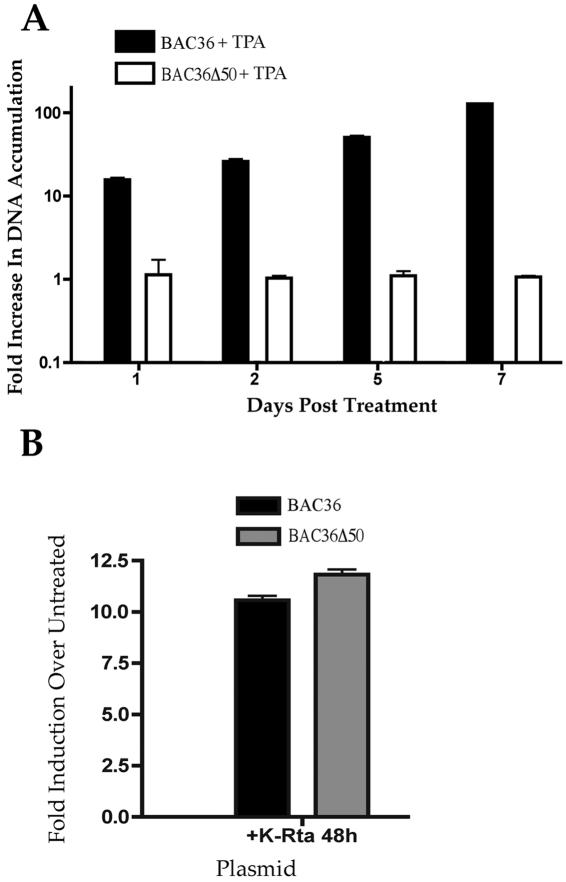
We next examined the ability of BAC36Δ50 to accumulate viral DNA when induced with TPA. Since transient expression of K-Rta has been shown to activate viral gene expression, we sought to evaluate the level of viral DNA accumulation in the absence of K-Rta. We anticipated that the lack of K-Rta expression would result in failure of the virus to enter the lytic phase of DNA replication.
293 cells harboring either BAC36Δ50 or BAC36 latent viral DNA were treated with TPA or transfected with a K-Rta expression plasmid, pCMV-RTA 5′-Flag. Total cellular DNA was harvested at various days post-TPA treatment and transfection and viral DNA accumulation was evaluated by real-time PCR with primers and probes specific for the ORF59 gene locus (Table 1). Cells harboring BAC36Δ50 DNA failed to accumulate viral DNA in the presence of TPA for up to 7 days postinduction, whereas BAC36-transfected cells showed a significant increase in viral DNA accumulation (Fig. 3A). The transfection of a K-Rta expression plasmid, pCMV RTA 5′Flag, had a similar effect to that of TPA treatment on BAC36 DNA accumulation (Fig. 3B). In addition, the introduction of the K-Rta expression plasmid into cells harboring BAC36Δ50 resulted in an increase in mutant BAC viral DNA similar to that observed from BAC36-containing cells (Fig. 3B). These results indicated that BAC36Δ50 was defective for viral DNA synthesis and this defect was abolished upon transfection of a K-Rta expression construct. Taken together, these series of experiments revealed that BAC36Δ50 was unable to respond to TPA treatment but could be complemented by supplying K-Rta in trans.
FIG. 3.
BAC36Δ50 is defective for DNA replication. (A) 293 cells containing BAC36Δ50 DNA fail to accumulate viral DNA when induced with TPA. BAC36- and BAC36Δ50-containing cells were treated with TPA, and total cellular DNA was harvested, and real-time PCR was performed to determine viral DNA accumulation. Error bars are the standard deviations from three experiments. (B) BAC36Δ50 DNA replication defect can be overcome when K-Rta is expressed in trans. BAC36- and BAC36Δ50-containing cells were transfected with a K-Rta expression vector, and total cellular DNA was harvested 48 h posttransfection, and real-time PCR was performed to determine viral DNA accumulation. Error bars are the standard deviations from 4 experiments.
Early and late viral gene expression is defective in BAC36Δ50-containing cells.
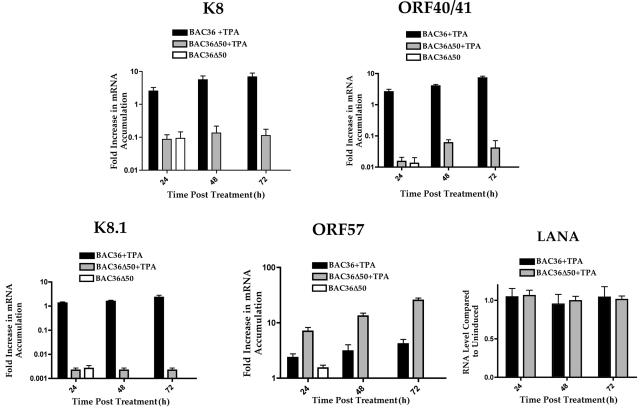
Since we were unable to detect an increase in accumulation of viral DNA or production of supernatant virus from BAC36Δ50-containing cells induced with TPA, we wanted to survey key mRNAs corresponding to viral transcripts from each kinetic class with real-time PCR. We evaluated various viral mRNA levels in cells containing either BAC36Δ50 or BAC36 DNA upon treatment with TPA. TPA treatment normally activates the expression of K-Rta resulting in the expression of viral early and late genes. One goal of the examination of viral early and late gene expression is to determine if these mRNA levels are affected by the absence of K-Rta expression in cell containing BAC36Δ50 DNA. We chose to evaluate the level of gene expression, with real-time PCR, from two early or delayed-early ORFs, K8 (K-bZIP) and ORF40/41, and the late gene K8.1. For these evaluations, the probes and primers used spanned the known spliced regions for these three transcripts (Table 1).
ORF40/41 is one of the components of the putative helicase-primase complex. This gene is normally expressed under conditions associated with lytic gene expression and participates in the enzymatic synthesis of HHV8 DNA (1, 2, 34). K-bZIP is the proposed origin binding protein and is also, along with ORF40/41, required for oriLyt-dependent DNA replication (1). Although the exact role of K-bZIP in DNA replication is unknown, it has been shown to be upregulated by K-Rta (4). K8.1 encodes a viral structural protein that displays true late-gene kinetics (17, 20, 28).
We compared mRNA levels from BAC36Δ50-containing cells treated with TPA to those obtained from cells containing the wild-type BAC36 when treated with TPA. mRNA accumulation is reported as the increase in accumulation over mRNA levels from the untreated BAC36-containing cell lines. K8 (K-bZIP) mRNA accumulation was almost 90-fold lower in BAC36Δ50-containing cells treated with TPA at 72 h postinduction and was comparable to that in untreated cells (Fig. 4, K8 graph). As expected, an increase in K8 mRNA accumulation was observed from 24 to 72 h post-TPA treatment in BAC36 samples.
FIG. 4.
Real-time PCR evaluation of early and late gene expression from BAC36Δ50-containing cells treated with TPA. Total cellular RNA was harvested from either BAC36- or BAC36Δ50-containing cells incubated with TPA. Real-time PCR was performed at various times posttreatment. ORF40/41, K8, K8.1, LANA, and ORF57 mRNA accumulation was determined and compared to those of untreated BAC36-containing cells. Error bars are the standard deviations from the mean of 12 separate experiments except for the data for LANA, which are from three separate experiments.
When BAC36Δ50-containing cells were treated with TPA, mRNA accumulation for the early transcript encoding ORF40/41 remained at levels similar to those observed from untreated cells (Fig. 4, ORF40/41 graph). No significant increase in ORF40/41 mRNA accumulation was observed at any time point tested (Fig. 4, ORF40/41 graph). The late gene transcript encoding the viral structural protein K8.1 was also unaffected in TPA-treated BAC36Δ50-containing cells and remained at mRNA accumulation levels similar to those observed from untreated samples (Fig. 4, graph K8.1). Although mRNA accumulation in TPA-treated BAC36-containing cells was only about fourfold higher than those observed from untreated cells at 72 h posttreatment, mRNA accumulation in BAC36Δ50-containing cells was almost undetectable (Fig. 4, K8.1 graph). These results indicated that early and late viral mRNA accumulation in BAC36Δ50-containing cells was defective and strongly suggested that the lack of K-Rta expression has a profound effect on mRNA induction for these genes.
As a control, we examined the mRNA levels for the latency-associated transcript LANA. Since BAC36- and BAC36Δ50-containing cells should accumulate equal amounts of LANA RNA, this should serve as a good internal control for overall gene expression patterns between the two cell lines. As can be seen in Fig. 4, the levels of LANA mRNA accumulation were similar between the cell lines throughout TPA treatment.
ORF57 gene expression is upregulated in the absence of K-Rta expression.
Since we demonstrated that the absence of K-Rta expression had a deleterious effect on the mRNA levels of two early genes, we next wanted to evaluate the mRNA levels of an immediate-early gene. We selected ORF57 since it is the homolog of Epstein-Barr virus Mta, an immediate-early gene involved in reactivation and DNA replication. However, transient HHV8 assays reveled that the promoter for ORF57 was transactivated by K-Rta, and Northern analysis reveled that ORF57 mRNA was detected at about 4 h post-TPA treatment a short time after ORF50 expression (16). Neither of these observations gives a clear indication that K-Rta is required for the expression of ORF57 in the context of the viral genome.
Consequently, we examined relative mRNA accumulation of ORF57 in BAC36Δ50-containing cells verses accumulation in BAC36-containing cells. Cells were treated with TPA, and RNA was harvested at various times posttreatment, and real-time PCR was performed with a probe specific for ORF57 (spanning an intron, see Table 1). Interestingly, mRNA accumulation of ORF57 was approximately fivefold higher at 72 h post-TPA treatment in BAC36Δ50-containing cells compared to TPA-treated BAC36-containing cells (Fig. 4, ORF57 graph). ORF57 mRNA accumulation was over 20-fold higher than that observed in uninduced cells (Fig. 4, ORF57 graph). This result suggests that, in contrast to observations from transient promoter assays where ORF50 upregulated ORF57 expression, the lack of K-Rta in the virus genome resulted in an increase, or dysregulation, of ORF57 expression, resulting in higher mRNA accumulations than in BAC36-containing cells under similar conditions.
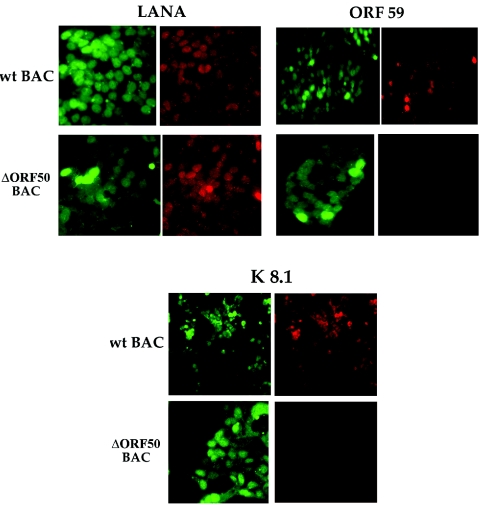
Evaluation of LANA, ORF59, and K8.1 protein expression in BAC36- and BAC36Δ50-containing 293 cells.
Since we observed aberrant transcript levels for selected early and late HHV8 genes, we next sought to evaluate the protein expression of representative latent, early,and late genes by immunofluorescence staining of TPA-induced BAC36- and BAC36Δ50-containing cells. 293 cells containing either BAC36 or BAC36Δ50 were treated with TPA and reacted with antibodies specific for LANA, ORF59, or K8.1. Both BAC36- and BAC36Δ50-containing cells expressed the latency-specific protein LANA, indicating that the mutant BAC was capable of establishing a latent state within 293 cells (Fig. 5, LANA panel). However, when the cells were evaluated for the expression of ORF59, which encodes the polymerase accessory protein, only wild-type-BAC-containing cells reacted positively with the antibody specific for this protein (Fig. 5, ORF59 panel). In addition, when cells were reacted with the antibody specific for K8.1, no positive BAC36Δ50-containing cells were observed (Fig. 5, K8.1 panel). These results confirmed that the expression of K8.1 was aberrant in BAC36Δ50-containing cells and also established that the replication protein ORF59 was not expressed in TPA-treated mutant BAC-containing cells. However, both wild-type and mutant BAC viral DNAs were capable of establishing a latent environment within 293 cells, as was evident by the efficient expression of LANA.
FIG. 5.
BAC36Δ50 failed to express ORF59 and K8.1 proteins. 293 cells containing BAC36 (wild-type BAC) or BAC36Δ50 were treated with TPA, fixed, and reacted with monoclonal antibodies specific for LANA, ORF59, or K8.1 viral proteins. Cells containing BAC constructs are positive for green fluorescent protein. Cells positively reacting with antibodies are shown as red-fluorescing cells.
DISCUSSION
HHV8 has many features in common with the closely related gammaherpesvirus Epstein-Barr virus. However, some noted differences include the apparent mechanism involved in the initiation of lytic reactivation and DNA replication. For HHV8, K-Rta, the gene product of ORF50, the homolog of Epstein-Barr virus Rta, is sufficient for lytic reactivation when introduced into latently infected cells (6, 16). K-Rta has strong transactivation properties and evidently activates HHV8 early and late genes upon transfection into latently infected cells (4, 6, 15, 16, 21, 24, 29). For Epstein-Barr virus, it is Zta, the homolog of HHV8 K8 (K-bZIP), that drives the lytic cycle. In HHV8, the gene product of K8 has no transactivation function and appears to be regulated by ORF50 (K-Rta). Also, K-Rta and K8 (K-bZIP) are required for origin-dependent DNA replication (1). K-Rta may have a direct role in DNA replication by interacting with a K-Rta-responsive element within oriLyt (1). This scenario is again in contrast to Epstein-Barr virus, where it is Zta that apparently performs essential transactivation and DNA replication functions with Epstein-Barr virus oriLyt (22).
ORF50 is the product of a spliced transcript that can be detected as early as 1 h post-TPA treatment (16). The ORF50 gene product, K-Rta, is a highly phosphorylated protein that was initially shown to be necessary for lytic reactivation. In addition, K-Rta upregulates its own expression through a mechanism involving the binding of the transcription factor Oct-I to sequences within the promoter region (21). K-Rta appears to activate gene expression by directly interacting with elements within promoters for several early genes and the immediate-early gene encoding ORF57 (15). Although in some cases these activation elements share some sequence homology, there appear to be many distinct K-Rta activation elements controlling the expression of various viral proteins (4, 15, 24). In addition to direct binding of K-Rta to promoter elements, cellular factors also contribute to the transactivation of viral gene expression (30). However, it is the repression of K-Rta expression that maintains the latent state of the virus. This control of K-Rta expression may be due to histone modification and chromatin remodeling, resulting in the hindrance of transcriptional activation of the ORF50 gene (14).
We have now demonstrated that the lack of K-Rta expression in the context of the viral genome results in a null phenotype with respect to early gene expression, virus production, and DNA synthesis. No increase in the accumulation of viral BAC36Δ50 DNA was observed in the presence of TPA, indicating that the normal process of lytic cycle induction was defective when K-Rta expression was abolished. For real-time PCR analysis of viral mRNAs, we chose to evaluate only spliced mRNAs (except for LANA). All primers and probes were designed so that they spanned an intron. This eliminated the possibility that the observed signal was from the viral genome. Although in most cases the accumulation of viral mRNAs from TPA-treated BAC36-containing cells was modest, the mutant BAC consistently showed much lower accumulation levels and was similar to those obtained from untreated cells.
mRNA accumulation for early and late gene expression from BAC36Δ50-containing cells treated with TPA was consistent with a viral phenotype with a defect in transcriptional activation. Although transient transfection experiments show that K-bZIP and ORF57 are upregulated by K-Rta, there are no data concerning the activation of ORF40/41 by K-Rta. We assume, however, that ORF40/41 mRNA production is also influenced by K-Rta and the onset of viral DNA synthesis. It is for this reason that we believe that the ORF40/41 mRNA accumulation pattern is similar to that of K8 in TPA-induced mutant virus-containing cells. Indeed, our data support the hypothesis that ORF40/41, and probably all other replication genes, is dependent on the expression of K-Rta.
Our results also suggest that there is an apparent dysregulation of the expression of another viral transactivator, ORF57, in the absence of K-Rta expression. This is in contrast to findings from transient assays where K-Rta transactivated ORF57 expression (15, 24, 29). ORF57 protein was found to act synergistically with ORF50 (K-Rta) to upregulate the expression of some HHV8 gene-specific promoters and to have an effect on the accumulation of several viral mRNAs (9). Recently, it was demonstrated that ORF57 interacts with K-Rta and this protein complex upregulated K-Rta expression in transient assays (18). The ORF57 gene product is the predicted homolog of Epstein-Barr virus BMLF1 (Mta) (5, 23). Mta is one of three major transacting factors encoded by Epstein-Barr virus and was shown to bind to specific RNAs and shuttle them between the nucleus and cytoplasm (23).
For HHV8, ORF57 is characterized as a lytic gene, and expression can be seen between 2 and 4 h post-TPA treatment in BCLB-1 cells (16). ORF57 itself is the product of a spliced transcript, and the protein product is localized to the cell nucleus (9). One reason for the increase in mRNA accumulation of ORF57 in cells containing BAC36Δ50 could be that K-Rta may serve to downregulate the expression of ORF57 at very early times after lytic cycle induction. When this regulation is removed, as in the case of a mutant virus unable to express K-Rta, then the levels of ORF57 mRNA may increase. Another possibility is that K-Rta may destabilize ORF57 mRNA in lytically infected cells and thereby regulate the effects of ORF57 posttranscriptionally.
We used an inducible cell line to complement viral growth and DNA synthesis of BAC36Δ50. We developed this inducible K-Rta system because of the apparent toxicity of constitutive expression of K-Rta (data not shown). This inducible cell line provides an efficient method for controlled expression of K-Rta in trans. We were also able to induce lytic replication from wild-type BAC (BAC36) upon transfection and subsequent IPTG induction of the K-Rta-containing 293 cell line. As observed in other studies, we also had a low level of production of infectious virus from BAC36-containing 293 cells upon induction with TPA (17). Nevertheless, the numbers of green cells observed upon inoculation of fresh 293 cells with supernatants from either TPA-treated BAC36 or complemented BAC36Δ50-containing cells were similar. This indicated that expression of K-Rta in trans was sufficient to supply all the functions necessary for progression to the lytic cycle.
Acknowledgments
This work was supported by PHS grant CA085164.
We thank S. Gao for the BAC36 construct and Ren Sun for the K-Rta expression construct.
REFERENCES
- 1.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. [DOI] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., and G. S. Pari. 2002. The human herpesvirus-8 (Kaposi's sarcoma-associated herpesvirus) ORF 40/41 region encodes two distinct transcripts. J. Gen. Virol. 83:189-193. [DOI] [PubMed] [Google Scholar]
- 3.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byun, H., Y. Gwack, S. Hwang, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame (ORF) 50 transactivates K8 and ORF57 promoters via heterogeneous response elements. Mol. Cell 14:185-191. [PubMed] [Google Scholar]
- 5.Chen, L., G. Liao, M. Fujimuro, O. J. Semmes, and S. D. Hayward. 2001. Properties of two EBV Mta nuclear export signal sequences. Virology 288:119-128. [DOI] [PubMed] [Google Scholar]
- 6.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komatsu, T., M. E. Ballestas, A. J. Barbera, and K. M. Kaye. 2002. The KSHV latency-associated nuclear antigen: a multifunctional protein. Front. Biosci. 7:d726-d730. [DOI] [PubMed] [Google Scholar]
- 11.Komatsu, T., A. J. Barbera, M. E. Ballestas, and K. M. Kaye. 2001. The Kaposi' s sarcoma-associated herpesvirus latency-associated nuclear antigen. Viral Immunol. 14:311-317. [DOI] [PubMed] [Google Scholar]
- 12.Lan, K., D. A. Kuppers, S. C. Verma, and E. S. Robertson. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 78:6585-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu, F., J. Zhou, A. Wiedmer, K. Madden, Y. Yuan, and P. M. Lieberman. 2003. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J. Virol. 77:11425-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luna, R. E., F. Zhou, A. Baghian, V. Chouljenko, B. Forghani, S. J. Gao, and K. G. Kousoulas. 2004. Kaposi's sarcoma-associated herpesvirus glycoprotein k8.1 is dispensable for virus entry. J. Virol. 78:6389-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik, P., D. J. Blackbourn, M. F. Cheng, G. S. Hayward, and J. B. Clements. 2004. Functional co-operation between the Kaposi's sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J. Gen. Virol. 85:2155-2166. [DOI] [PubMed] [Google Scholar]
- 19.Pavlova, I. V., H. W. t. Virgin, and S. H. Speck. 2003. Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication. J. Virol. 77:5731-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raab, M. S., J. C. Albrecht, A. Birkmann, S. Yaguboglu, D. Lang, B. Fleckenstein, and F. Neipel. 1998. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J. Virol. 72:6725-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semmes, O. J., L. Chen, R. T. Sarisky, Z. Gao, L. Zhong, and S. D. Hayward. 1998. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J. Virol. 72:9526-9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song, M. J., H. Deng, and R. Sun. 2003. Comparative study of regulation of rta-rEsponsive genes in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 77:9451-9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the Promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, T. Q., D. A. Fernstermacher, and J. M. Vos. 1994. Human artificial episomal chromosomes for cloning large DNA fragments in human cells. Nat Genet. 8:33-41. [DOI] [PubMed] [Google Scholar]
- 28.Tang, S., and Z. M. Zheng. 2002. Kaposi's sarcoma-associated herpesvirus K8 exon 3 contains three 5′-splice sites and harbors a K8.1 transcription start site. J. Biol. Chem. 277:14547-14556. [DOI] [PubMed] [Google Scholar]
- 29.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 146:1415-1426. [DOI] [PubMed] [Google Scholar]
- 30.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, S. E., F. Y. Wu, M. Fujimuro, J. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of CCAAT/enhancer-binding protein alpha (C/EBPalpha) in activation of the Kaposi's Sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding Protein-alpha is induced during the early stages of Kaposi's Sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 77:9590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, Y., H. Li, M. Y. Chan, F. X. Zhu, D. M. Lukac, and Y. Yuan. 2004. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent dNA Replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J. Virol. 78:8615-8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's Sarcoma-Associated Herpesvirus DNA Replication Compartments in Transient Cotransfection Assays and Association with the ORF-K8 Protein and Cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a ercombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, J., P. Trang, K. Kim, T. Zhou, H. Deng, and F. Liu. 2004. Effective inhibition of Rta expression and lytic replication of Kaposi's sarcoma-associated herpesvirus by human RNase P. Proc. Natl. Acad. Sci. USA 101:9073-9078. [DOI] [PMC free article] [PubMed] [Google Scholar]