Abstract
Homologs of the UL51 protein of herpes simplex virus have been identified in all herpesvirus subfamilies, but until now, no function has been assigned to any of them. To investigate function of the UL51 gene product of the alphaherpesvirus pseudorabies virus (PrV), we isolated and analyzed a mutant lacking the major part of the open reading frame, PrV-ΔUL51F, and a rescuant. One-step growth analysis of PrV-ΔUL51F revealed only slightly reduced titers, but plaque size was notably diminished and reached only approximately 30% the plaque size of wild-type PrV. Ultrastructurally, intracytoplasmic capsids were found in large numbers either without envelope or in different stages of envelopment, indicating that secondary envelopment in the cytoplasm was less efficient. However, neuroinvasion in the mouse trigeminal pathway after intranasal infection was only slightly delayed. A PrV UL11 mutant also showed a defect in secondary envelopment (M. Kopp, H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter, J. Virol. 77:5339-5351, 2003). Since both proteins are part of the viral tegument and are predicted to be membrane associated, they may serve similar, possibly redundant functions during viral morphogenesis. Therefore, we also isolated a mutant simultaneously lacking UL51 and UL11. This mutant exhibited further reduced plaque size compared to the single-deletion mutants, but viral titers were comparable to those for the UL11 mutant. In electron microscopic analyses, the observed defect in secondary envelopment was similar to that found in the UL11 single-deletion mutant. In conclusion, both conserved tegument proteins, either singly or in combination, are involved in virion morphogenesis in the cytoplasm but are not essential for viral replication in vitro and in vivo.
Herpesviruses are large, enveloped DNA viruses, and the genomes of the subfamily Alphaherpesvirinae contain between 70 and 80 protein-coding genes (39). Approximately half of the encoded gene products are part of the mature herpes virion and have to be assembled during viral morphogenesis. The mechanism of how this assembly process is accomplished is only incompletely understood (reviewed in reference 33).
The genome of the alphaherpesvirus pseudorabies virus (PrV), the causative agent of Aujeszky's disease (32), is similar in gene content and arrangement to the genomes of other alphaherpesviruses (22). It carries a core set of approximately 40 genes which are conserved in the alpha- (31), beta- (9), and gammaherpesvirus subfamilies (2). The core gene products comprise 8 capsid or capsid-associated proteins (UL6, UL18, UL19, UL25, UL26, UL26.5, UL35, UL38), 10 proteins involved in DNA replication (UL2, UL5, UL8, UL12, UL29, UL30, UL39, UL42, UL50, UL52), 5 proteins participating in DNA cleavage and/or encapsidation (UL15, UL17, UL28, UL32, UL33), 1 activator of viral transcription (UL54), 2 proteins necessary for nuclear egress (UL31, UL34), one protein kinase (UL13), 5 tegument proteins engaged in secondary envelopment in the cytoplasm (UL11, UL16, UL21, UL36, UL37), 5 envelope (glyco)proteins (UL1, UL10, UL22, UL27, UL49.5), and 4 gene products with unassigned function in the replication cycle (UL7, UL14, UL24, UL51) (38).
The mature herpesvirus particle consists of at least 30 different proteins which have to be assembled during virion formation (42). While the capsid is assembled in the nucleus, the majority of viral tegument and envelope proteins are collected in the cytoplasm. Recent data indicate that tegumentation and secondary envelopment are the result of an intricate network of protein-protein interactions with an extensive functional redundancy (reviewed in references 33 and 34). Tegumentation might start at two different sites, at the nucleocapsid after nuclear egress and at vesicles derived from the Golgi apparatus which contain viral glycoproteins embedded in the future viral envelope. The capsid-apposed tegument is thought to consist of the UL36, UL37, and US3 gene products, whereas other tegument proteins, like UL49 and UL11, physically and/or functionally interact with viral glycoproteins to connect tegument and envelope (reviewed in references 33 and 34). However, no complete picture has so far been obtained of the molecular composition of the tegument and of the roles individual tegument proteins play in virion formation.
Among the conserved herpesvirus proteins that have not yet been assigned a specific function are the homologs of the herpes simplex virus type 1 (HSV-1) UL51 protein. A mutant HSV-1 which lacked the UL51 gene could be isolated on noncomplementing cells, indicating that, despite the high conservation, the protein product is dispensable for viral replication in cell culture. The mutant replicated with only slightly delayed kinetics but formed very small plaques (5). The HSV-1 UL51 gene products were found as 27-, 29-, and 30-kDa phosphorylated proteins in infected cells as well as in virions (11). In transfected cells, HSV-1 UL51 localized at the cytoplasmic face of Golgi membranes, and for this targeting, the N-terminal 15 amino acids were sufficient. In infected cells, UL51-specific fluorescence was found predominantly on the inner side of cytoplasmic vesicles and/or the viral envelope (11, 36). It has been suggested that palmitoylation at the N-terminal cysteine, which is conserved in all UL51 homologs, is required for Golgi localization (36). Thus, the HSV-1 UL51 protein may be involved in targeting capsids to the final envelopment site and/or in virus particle trafficking associated with the Golgi apparatus (36).
A similar function has been proposed for another conserved viral tegument protein, the product of the UL11 gene (3, 7). The HSV-1 UL11 protein is a small, phosphorylated, myristoylated, and palmitoylated virion protein which is associated with membranes of infected cells and the viral envelope (4, 7, 28, 30). Membrane binding was found to be mediated by the two fatty acid groups, while an acidic cluster and a dileucine motif were identified as important for the recycling of UL11 from the plasma membrane to the Golgi apparatus (28). HSV-1 UL11 has been demonstrated to be involved in nucleocapsid envelopment and egress (3), and a PrV mutant lacking UL11 showed a defect in secondary envelopment in the cytoplasm (24). Nucleocapsids accumulated in the cytoplasm and were often found associated with tegument in aggregated structures, while intracytoplasmic membranes appeared distorted and tightly connected, indicating that the partly tegumented capsids are impaired in access to the budding site in the absence of UL11 (24). A role for UL11 in secondary envelopment was further highlighted by the phenotype of a mutant PrV that simultaneously lacked gM and UL11. In cells infected with this virus mutant, no infectious enveloped virions were produced but huge accumulations of capsids embedded in tegument were observed (25). Absence of the UL11 homolog in human cytomegalovirus blocked intracytoplasmic virion formation (40).
Thus, both proteins, UL11 and UL51, are predicted to share several features. (i) Both open reading frames (ORFs) are conserved in the three herpesvirus subfamilies. (ii) The gene products constitute small virion components which are most probably part of the envelope-associated tegument layer (11, 23, 27, 36). (iii) Membrane interaction for the HSV-1 UL11 and UL51 homologs was shown to rely on fatty acid modifications: while HSV-1 UL11 is myristoylated as well as palmitoylated, HSV-1 UL51 seems to constitute a type III palmitoylated protein (36, 37). (iv) Both proteins seem to contain specific Golgi-targeting signals, suggesting that both proteins might serve similar functions.
Previously, we sequenced the PrV UL51 gene (6) and identified its protein product as a 30-kDa tegument component (27). Since the antipeptide sera available at that time did not react in immunofluorescence analyses, we used fractionation of infected cells followed by immunoblotting and found reactivity primarily in the nuclear fraction (27). This was in marked contrast to results published for the UL51 homologs of HSV-1 (11, 36) and bovine herpesvirus 1 (BHV-1) (17). In cells infected by these viruses, the corresponding gene products were detected mainly in the cytoplasm. Moreover, colocalization with Golgi-specific proteins was observed (36).
To reinvestigate the subcellular location of the PrV UL51 protein, we produced new, potent, monospecific polyclonal antisera. For analysis of the functional role of PrV UL51, we constructed a mutant in which the major part of the UL51 ORF was deleted by mutating the PrV genome cloned as a bacterial artificial chromosome (BAC) (24, 41). This mutant, as well as a corresponding rescue mutant, was investigated. To assay for possible functional interaction between the UL51 and UL11 proteins, a UL11-UL51 double mutant was also constructed and analyzed.
MATERIALS AND METHODS
Viruses and cells.
All PrV mutants were derived from the wild-type laboratory strain Kaplan (PrV-Ka [19]). Viruses were grown in rabbit kidney (RK13) or porcine kidney (PSEK) cells in Eagle's minimum essential medium supplemented with 10 or 5% fetal calf serum, respectively. Cloning of PrV as a BAC, pPrV-ΔgB, has been described previously (24). For generation of constitutively UL51-expressing cells, pcDNA-UL51 was prepared after PCR amplification of the complete UL51 ORF (see Fig. 1B and C) by using primers UL51For (5′-CACAGAATTCACGCGGATCATGCTGGGCGG-3′; nucleotides [nt] 1097 through 1078, accession number X87246 [6]) and UL51Rev (5′-CACACTCGAGCGCCCGCCCTCTCCGGCTAC-3′; nt 362 through 381, accession number X87246 [6]) (UL51 start and stop codons were part of the primer sequences and are shown in bold), with Pfx DNA polymerase (Invitrogen, Karlsruhe, Germany) and cloned genomic BamHI fragment 5′ as template. EcoRI and XhoI restriction sites (shown in italics) were introduced for convenient cloning into appropriately cleaved vector pcDNA3 (Invitrogen). pcDNA-UL51 was transfected into RK13 cells by using Superfect transfection reagent (QIAGEN, Hilden, Germany), followed by selection with 0.5 mg of G418 (Invitrogen)/ml and immunofluorescence screening of single-cell clones with the monospecific anti-UL51 sera (see below). One cell clone, named RK13-UL51, was chosen for further experiments. UL51 expression in this cell clone was also verified by immunoblot analysis (data not shown).
FIG. 1.
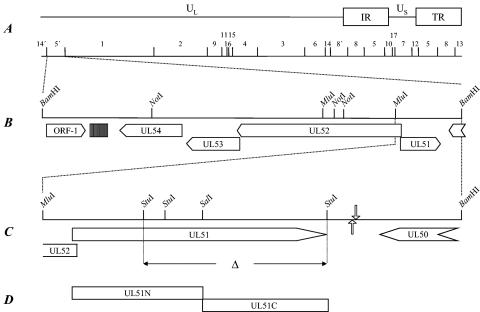
Construction of virus mutants. (A) A schematic map of the PrV genome showing the unique long (UL) and unique short (US) regions, the inverted repeat sequences (IR, TR), and the positions of the BamHI restriction fragments which are numbered according to their size. (B) Enlargement of the UL51 gene region. The UL51 gene is transcribed into a unique mRNA antiparallel to the neighboring genes UL50, coding for dUTPase, and UL52, encoding the putative primase subunit of the primase/helicase complex (6). A region of reiterated sequences located between ORF-1 and UL54 is shown by a hatched box. Relevant restriction sites are indicated. (C) Construction of PrV-ΔUL51F. Deletion of UL51-specific sequences between the StuI sites is indicated. Locations of the polyadenylation signals are shown by hollow arrows (pointing downward for the UL50 gene and pointing upward for the UL51 gene). (D) Amino-terminal (UL51N) and carboxy-terminal (UL51C) regions used for the generation of specific antisera.
Preparation of monospecific anti-UL51 sera.
We recently described the generation of sera raised against four UL51-specific peptides which detected the protein in immunoblot analysis but not in immunofluorescence assays (27). To generate more potent antisera, we expressed the UL51 ORF in bacteria and used the purified expression product as antigen for immunization. Since the complete UL51 ORF could not be expressed in bacteria, the 5′ and 3′ parts of the coding region were cloned and expressed separately (see Fig. 1D). For expression of the N-terminal part, the coding region was amplified using primers ML-98 (5′-GGGATCCGATGCTGGGCGGCATCTTCTCCGG-3′; nt 1088 through 1066, with the start codon shown in bold, accession number X87246 in reference 6, with the BamHI site shown in italics) and ML-99 (5′-GTCCGCGTCGACGGCGCCGACCGAGAG-3′, nt 720 through 746, accession number X87246, with the SalI site shown in italics). The resulting 375-bp PCR product which encompassed codons 1 to 121 of PrV UL51 was cloned into vector pET-23b (Novagen, Schwalbach, Germany) as a BamHI-SalI fragment by using the introduced BamHI site and the internal SalI site (see Fig. 1). For expression of the C-terminal part of the UL51 protein, PCR was performed using primers ML-100 (5′-GGCGCCGTCGACGCGGACACGGACGAC-3′; nt 737 through 711, accession number X87246, with the SalI site shown in italics) and ML-101 (5′-GGCTCGAGCATCTTGCAGGCCTCGGC-3′; nt 381 through 398, accession number X87246, with the XhoI site shown in italics), resulting in a 365-bp PCR product containing codons 119 to 236 which was cloned as SalI-XhoI fragment into pET-23b by using again the internal SalI site (see Fig. 1) and the introduced XhoI restriction site. pET-UL51N and pET-UL51C were expressed in Escherichia coli BL21 pLYS S cells as fusion proteins with a C-terminal histidine tag. Purification of the expression products was performed according to the manufacturer's instructions on nickel-nitrilotriacetic acid-agarose (QIAGEN) under denaturing conditions. Immunization of rabbits was done as previously described (24).
Isolation of PrV-ΔUL51F and PrV-ΔUL51F/11G.
For generation of an UL51-negative PrV mutant, the 1.2-kbp MluI-BamHI subfragment of the BamHI fragment 5′ containing the complete UL51 gene region (Fig. 1C) (6) was cloned into BamHI-SmaI-cleaved vector pUC19. To delete the major part of the UL51 ORF without interfering with the transcription of neighboring genes, two StuI fragments encompassing 493 bp of the UL51 ORF (see Fig. 1C) representing codons 69 to 233 were deleted and substituted with a kanamycin resistance gene flanked by Flp recombinase recognition target sites that was derived from plasmid pKD13 as a 1.258-bp BstBI fragment (12), giving rise to plasmid pUC-ΔUL51KF. The 2-kb insert of this plasmid was amplified by PCR with vector-specific primers M13 (−47) und M13 (−48) (New England Biolabs, Frankfurt/Main, Germany) and with Pfx DNA polymerase (Invitrogen). The PCR product was excised after gel electrophoresis and used for the mutagenesis of pPRV-ΔgB (23) in E. coli by using the Red recombinase of bacteriophage λ (12). PrV-ΔUL51F could be isolated after excision of the kanamycin resistance gene by using Flp recombinase (10) and cotransfection into RK13 cells by calcium-phosphate coprecipitation (13) together with pUC-B1BclI (23), which contains the authentic gB gene of PrV-Ka and restores the gB defect. The introduction of the correct deletion was confirmed by PCR and sequence analysis (data not shown). For isolation of a UL51 rescue mutant, genomic DNA of PrV-ΔUL51F was cotransfected with a plasmid containing a 2.1-kb NotI-BamHI subfragment of BamHI 5′ (see Fig. 1B). Transfection progeny was tested for UL51 expression by immunofluorescence using the monospecific anti-UL51 sera. One single plaque isolate, PrV-ΔUL51FR, was further analyzed.
For isolation of a mutant unable to express the UL51 and UL11 proteins, genomic DNA of PrV-ΔUL51F was cotransfected with plasmid pΔUL11GFP into RK13-UL11 cells (23). In this plasmid, codons 16 to 32 of the UL11 ORF had been deleted and substituted with a green fluorescent protein expression cassette. Recombinants were titrated on RK13-UL11 cells (23) and analyzed for autofluorescence. One single-plaque isolate, PrV-ΔUL51F/11G, was further tested.
The deletion and rescue mutants were characterized by restriction typing and Southern blot analysis (data not shown).
Virus purification and immunoblotting.
For virus purification, PSEK cells were infected with PrV-Ka, PrV-ΔUL51F, PrV-ΔUL51FR, and PrV-ΔUL51F/11G, and virions were purified as described previously (23). Purified virions were separated by electrophoresis in sodium dodecyl sulfate-10 or 15% polyacrylamide gels (26), electrotransferred onto nitrocellulose membranes, and reacted with monospecific antisera against the UL51 (UL51N and/or UL51C; 1:10,000 [this study]), UL11 (1:10,000 [23]), UL37 (1:100,000 [23]), UL49 (1:100,000 [8]), or gH (1:50,000 [21]) proteins. After incubation with peroxidase-conjugated secondary antibody (Dianova, Hamburg, Germany), bound antibody was detected by chemiluminescence (Super Signal; Pierce, Bonn, Germany) recorded on X-ray film.
Indirect immunofluorescence and confocal laser scan microscopy.
RK13 cells grown on coverslips were infected for 16 h with PrV-Ka or PrV-ΔUL51F or transfected with pcDNA-UL51. Thereafter, cells were fixed for 20 min with 3% paraformaldehyde, followed by 10-min incubation with 3% paraformaldehyde containing 0.3% Triton X-100. Cells were then incubated in phosphate-buffered saline (PBS) supplemented with 10% fetal calf serum for 30 min at room temperature. After repeated washing with PBS, cells were incubated with monospecific sera raised against the N- and/or C-terminal part of UL51 (UL51N, UL51C; 1:400 [this study]) or monoclonal antibodies against gB (A20-c26 [35]), gC (B16-c8 [21]), γ-Adaptin (clone 100/3; Sigma-Aldrich, Taufkirchen, Germany), or Golgi-specific P3-b4 (16), followed by incubation with Alexa 488- or Alexa 594-conjugated secondary antibodies (Molecular Probes, Leiden, The Netherlands). Slides were washed repeatedly after each step. Fluorescence was preserved in a 9:1 mixture of glycerol and PBS containing 25 mg of 1,4-diazabicyclooctane per ml and 1 μg of propidium iodide per ml for chromatin counterstaining, if indicated. Slides were analyzed in a confocal laser scan microscope (LSM510; Zeiss, Göttingen, Germany)
In vitro replication properties.
RK13, RK13-UL51 (this study), or RK13-UL11 (23) cells were infected at a multiplicity of infection (MOI) of 10 with PrV-Ka, PrV-ΔUL51F, PrV-ΔUL11 (23), or PrV-ΔUL51F/11G and incubated on ice for 1 h. Cells were then overlaid with prewarmed medium and incubated at 37°C for 1 h. Remaining extracellular virus was inactivated by low-pH treatment, and cells were scraped into the medium immediately as well as after 4, 8, 12, 24, and 36 h of incubation at 37°C. Cells were lysed by freezing (−70°C) and thawing (37°C), and titers of progeny virus were determined on RK13-UL51 cells. Mean values from three independent experiments and standard deviations were calculated. Plaque diameters on RK13 or complementing cells were analyzed as described previously (23). For each virus mutant, 30 plaques were measured microscopically, and the relative average plaque diameters compared to those of PrV-Ka, which were set at 100%, were determined. Average values from two independent experiments and standard deviations were calculated.
Electron microscopy.
For ultrathin sectioning, RK13, RK13-UL51, and RK13-UL11 cells were infected at an MOI of 1 with PrV-ΔUL51F or PrV-ΔUL51F/11G and fixed 14 h postinfection (p.i.). Fixation, dehydration, and epoxy embedding were performed as described previously (14, 15). The counterstained ultrathin sections were analyzed with a transmission electron microscope (Technai 12; Philips, Eindhoven, The Netherlands).
Animal experiments.
For determination of mean survival times, 10 6- to 8-week-old CD1 mice for PrV-Ka and PrV-ΔUL51F and three animals for PrV-ΔUL51FR were anesthetized and infected by bilateral intranasal instillation of a 5-μl suspension containing 106 PFU. Animals were observed three times a day and euthanized when severe pruritus with excitations and self-mutilation (“mad-itch” syndrome) occurred (20). To analyze the kinetics of viral spread in the trigeminal circuit, mice were euthanized sequentially every 24 h after inoculation, and tissues were prepared and analyzed as described previously (20). Virus-infected cells were identified by incubation with monospecific antiserum against the major capsid protein of PrV (1:500 in Tris-buffered saline [20]), followed by biotinylated goat anti-rabbit immunoglobulin G1 (1:200; Vector, Burlingame, Calif.). Antibody binding was visualized with the avidin-biotin complex method (1:10 in Tris-buffered saline; Vector) (18) using AEC substrate (Dako, Hamburg, Germany) as chromogen.
RESULTS
Subcellular localization of PrV UL51.
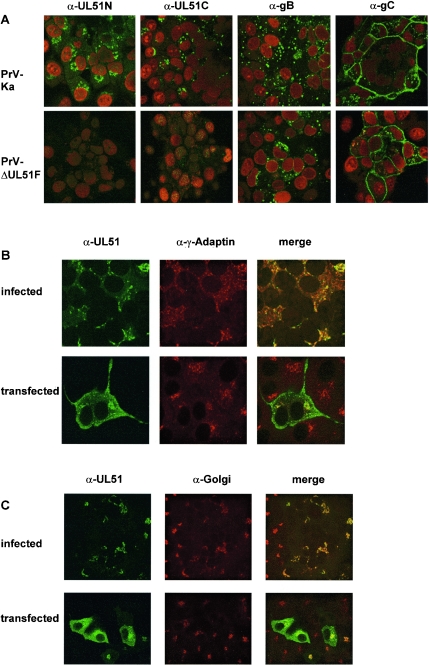
The inadequacy of our PrV UL51 antipeptide sera prompted us to generate more potent sera by using bacterially expressed PrV UL51 for rabbit immunization. Since the complete UL51 ORF could not be expressed successfully, it was cloned in two parts, and rabbits were immunized with expression products corresponding to the N- or C-terminal regions of the protein (Fig. 1D). Both antisera reacted in indirect immunofluorescence assays with PrV-Ka-infected RK13 cells but not with noninfected cells or cells infected with PrV-ΔUL51F, verifying the specificity of the generated sera (Fig. 2). In contrast to previous results from cell fractionation (27), UL51-specific fluorescence was detected in PrV-Ka-infected cells with both antisera predominantly in large speckles in the cytoplasm in close proximity to the nucleus (Fig. 2A through C) but not within the nucleus, thus paralleling results found for the homologous proteins in BHV-1 and HSV-1 (17, 36). Fluorescence partially colocalized with γ-Adaptin, which is specific for the Golgi adaptor complex AP-1 and used as marker for the trans-Golgi network (Fig. 2B), or with a monoclonal antibody against an unspecified Golgi antigen (Fig. 2C) (16). It is also evident that the Golgi apparatus is partially redistributed in infected as well as transfected cells compared to the noninfected or nontransfected cells in the vicinity (Fig. 2C), as has been described to occur in HSV-1 infection (1). Antibodies against PrV gB and gC were used as controls (Fig. 2A).
FIG. 2.
Subcellular localization of PrV UL51. (A) RK13 cells were infected with PrV-Ka or PrV-ΔUL51F, fixed 16 h p.i., and incubated with UL51-specific antisera (α-UL51N, α-UL51C) or monoclonal antibodies against gB (A20-c26) or gC (B16-c8). Fluorescence of Alexa 488-conjugated anti-rabbit or anti-mouse antibodies (green) and of propidium iodide-stained chromatin (red) was analyzed in a confocal laser scan microscope. (B and C) RK13 cells were infected with PrV-Ka or transfected with plasmid pcDNA-UL51 and then incubated with the anti-UL51 serum (green fluorescence) and a monoclonal antibody against γ-Adaptin (panel B, red fluorescence) or an unspecified Golgi marker (panel C, red fluorescence) (16). Columns labeled “merge” depict the merger of the green and red fluorescence.
Deletion of the UL51 gene from the PrV genome.
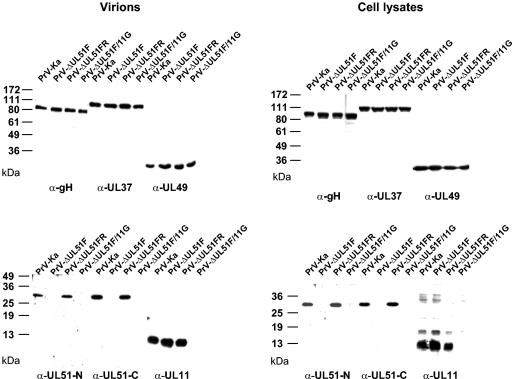
PrV-ΔUL51F, which lacks the major part of the UL51 ORF, was isolated after BAC mutagenesis. Since the UL51 ORF partly overlaps in antiparallel orientation with the UL52 ORF (6) which codes for the essential primase subunit of the primase/helicase complex, the UL51 coding sequence could not be completely deleted. The introduced partial deletion ensures that most of the UL51 gene is gone but that neither the UL52 ORF nor any regulatory elements immediately upstream are affected. PrV-ΔUL51F could be isolated on noncomplementing cells, indicating that UL51 is dispensable for viral replication in cell culture. As expected, in immunoblot analysis of purified virions (Fig. 3, left panels) or infected cell lysates (Fig. 3, right panels), UL51-specific reactivity was absent from PrV-ΔUL51F- and PrV-ΔUL51F/11G-infected cells and purified virions but restored in the rescue mutant, PrV-ΔUL51FR. In the generated mutants, a polypeptide composed of the 69 N-terminal amino acids of UL51, 11 amino acids derived from the remaining Flp recombinase recognition target site, and the C-terminal 4 amino acids of UL51 could theoretically be expressed. However, a corresponding signal was not observed either in indirect immunofluorescence (Fig. 2) or in immunoblot analysis (Fig. 3). In contrast, both UL51-specific antisera reacted with the full-length UL51 protein in PrV-Ka- and PrV-ΔUL51FR-infected cells (Fig. 3). Envelope glycoprotein gH and tegument proteins UL37 and UL49 were present in all virus preparations and infected cell lysates in similar amounts, whereas the UL11 gene product, as expected, was absent from PrV-ΔUL51F/11G-infected cells and virions.
FIG. 3.
Immunoblots of mutant viruses. Purified virions (left panels) or lysates of RK13 cells (right panels) infected with PrV-Ka, PrV-ΔUL51F, PrV-ΔUL51FR, or PrV-ΔUL51F/11G were separated by electrophoresis in sodium dodecyl sulfate gels containing 10% (for detection of gH, UL37, and UL49 proteins) or 15% (for detection of the UL51 and UL11 proteins) polyacrylamide. After transfer to nitrocellulose filters, blots were probed with monospecific sera against UL51N, UL51C, UL11, UL37, UL49, and gH. Locations of molecular mass markers are indicated to the left of each panel.
PrV UL51 is not essential for viral replication in cell culture.
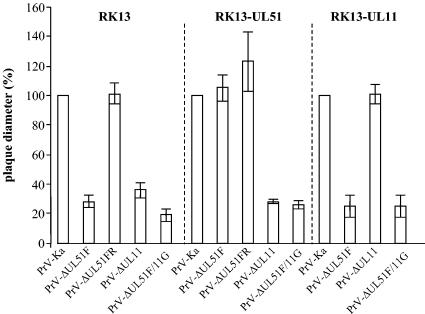
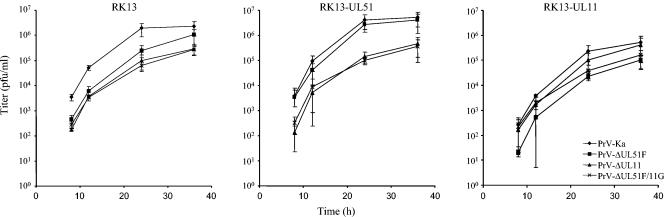
To analyze growth properties in more detail, RK13 and RK13-UL51 cells were infected with PrV-Ka, PrV-ΔUL51F, and PrV-ΔUL51FR under plaque assay conditions, and plaques were measured microscopically 2 days p.i. While plaques formed by PrV-ΔUL51FR were comparable in size and morphology to those formed by PrV-Ka on RK13 cells, the UL51 deletion mutant induced only small plaques with diameters approximately 30% the size of those formed by PrV-Ka. A large plaque phenotype was restored on UL51-expressing cells but not on UL11-expressing cells (Fig. 4). In one-step growth analyses, PrV-ΔUL51F exhibited decreased viral titers at all time points tested, with titers at 36 h p.i. reduced approximately fivefold compared to those of PrV-Ka (Fig. 5) and the rescue mutant PrV-ΔUL51FR (data not shown). The replication defect was complemented on RK13-UL51 cells (Fig. 5), indicating that no further unwanted mutations contribute to the observed phenotype.
FIG. 4.
Plaque size of mutant viruses on noncomplementing and complementing cells. RK13, RK15-UL51, or RK13-UL11 cells were infected with the indicated viruses under plaque assay conditions. Plaques were microscopically measured 2 days p.i. Relative plaque sizes were calculated and compared to those of PrV-Ka which were set as 100%. Average values and standard deviations from at least two independent experiments are shown.
FIG. 5.
One-step growth analysis. RK13, RK13-UL51, or RK13-UL11 cells were infected with PrV-Ka or the respective mutants, harvested at the indicated times after infection, and titrated. Average values and standard deviations from three independent experiments are shown.
Absence of UL51 impairs secondary envelopment in the cytoplasm.
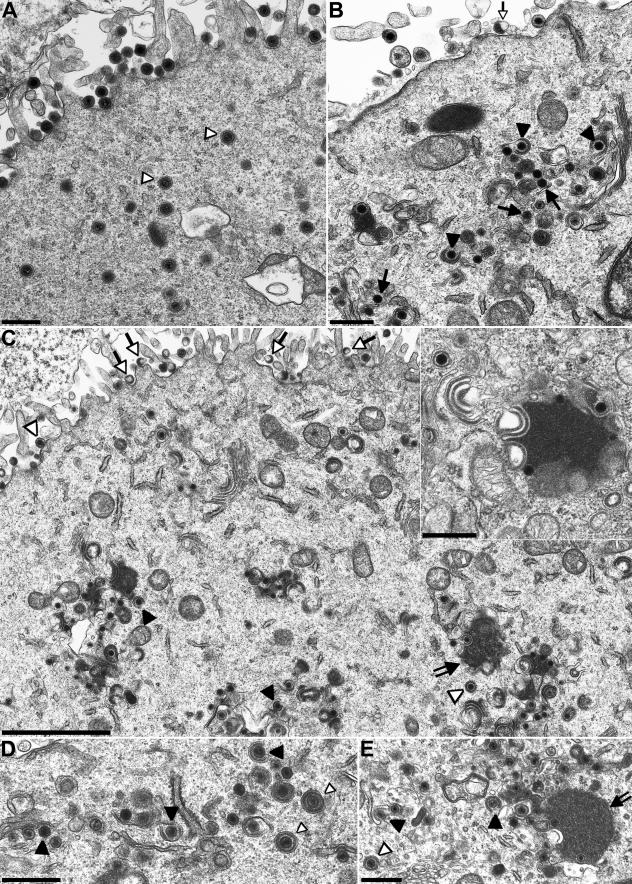
To investigate the role of PrV UL51 in viral replication, RK13 cells were infected with PrV-Ka (Fig. 6A) or PrV-ΔUL51F (Fig. 6B), fixed 14 h p.i., and processed for transmission electron microscopy. In PrV-Ka-infected cells, numerous extracellular virions were found lining the plasma membrane (Fig. 6A). In contrast, only a few intracytoplasmic nonenveloped nucleocapsids or nucleocapsids undergoing secondary envelopment were observed (Fig. 6A). In PrV-ΔUL51F-infected cells, mature extracellular virus particles were also observed at the plasma membrane, although less numerous, indicating that virus morphogenesis proceeds in the absence of PrV UL51. This parallels the results of the one-step growth kinetics (Fig. 5). In addition, an unusually large number of unenveloped nucleocapsids or nucleocapsids in the process of secondary envelopment were obvious (Fig. 6B) which were observed in PrV-Ka-infected cells significantly less frequently (Fig. 6A) (14). Formation of capsidless L-particles was also observed (Fig. 6B). These findings indicate that in the absence of UL51, secondary envelopment in the cytoplasm is less efficient or proceeds slower, resulting in the observed accumulation of intracytoplasmic capsid maturation stages.
FIG. 6.
Transmission electron microscopy of infected cells. RK13 (A through C), RK13-UL11 (D), or RK13-UL51 (E) cells were infected with PrV-Ka (A), PrV-ΔUL51F (B), or PrV-ΔUL51F/11G (C through E) at an MOI of 1 and fixed 14 h p.i. Arrows point to intracytoplasmic nucleocapsids, closed triangles point to nucleocapsids undergoing secondary envelopment, and open triangles point to enveloped virions. Open arrows indicate L-particles. In PrV-ΔUL51F/11G-infected RK13 or RK13-UL51 cells, intracytoplasmic nucleocapsids were sometimes observed in close contact with tegument and distorted intracytoplasmic membranes (panels C and E, double-shafted arrows; also panel C, inset) as previously described for a PrV UL11 deletion mutant (23, 24), whereas infected RK13-UL11 cells exhibited an increased number of nucleocapsids in the process of secondary envelopment as observed in RK13 cells infected by PrV-ΔUL51F. Bars, 500 nm (A, B, C inset, D, and E) and 2 μm (C).
Kinetics of viral spread in the trigeminal circuit.
The first symptoms after infection with PrV-ΔUL51F (depression, anorexia, cowering in a hunched position) were observed 2 days p.i., which is approximately 24 h later than that observed in PrV-Ka-infected animals (Table 1). Subsequently, the animals showed increasing excitations and convulsions, heavy dyspnea, and extensive scratching of the facial and nasal skin, thereby causing severe hemorrhagic dermal erosions (mad-itch syndrome). PrV-ΔUL51F-infected mice died at an average time of 64 h p.i. compared to 50 h p.i. with PrV-Ka. In mice infected with either virus strain, detection of the PrV major capsid protein by immunohistochemistry revealed infection of cells in the nasal mucosa from 24 h p.i. on (Table 1). Whereas in PrV-Ka-infected mice, few infected neurons in the trigeminal ganglion were already detectable at 24 h p.i., first infection of trigeminal ganglion neurons by PrV-ΔUL51F was observed at 48 h p.i. Subsequently, the number of ganglionic neurons infected by either virus strain strongly increased until the time of death. Infected cells were also detected in the second-order spinal trigeminal nuclei in PrV-Ka-infected animals at 48 h p.i. and in PrV-ΔUL51F-infected mice at 62 h p.i. but not in third-order neurons of the ectorhinal cortex (Table 1).
TABLE 1.
Mean time to death, clinical symptoms, and appearance of infected cells in mice inoculated experimentally with PrV-Ka and PrV-ΔUL51F
| Parameter | Virus
|
|
|---|---|---|
| PrV-Ka | PrV-ΔUL51F | |
| Mean time to death (h)a | 50.0 (1.9) | 64 (2.1) |
| Clinical symptoms (days p.i.)b | ||
| 1 | 0 | 0 |
| 2 | +++ | + |
| 3 | † | +++ |
| Immunohistochemistry (days p.i.)c | ||
| Nasal cavity | 1 | 1 |
| First-order neurons | 1 | 2 |
| Second-order neurons | 2 | 3 |
| Third-order neurons | −d | − |
Averages calculated for 10 animals each. Standard deviations are indicated in parentheses.
Clinical symptoms were measured as follows: 0, clinically normal mice; +, slight depression, hunched position, ruffled hair coat; ++, apathy, anorexia, moderate dyspnea, slight facial pruritus; +++, severe attacks of excitation, self-mutilation, skin erosions, heavy dyspnea; †, animals moribund or dead.
Immunohistochemistry was measured as the time in days p.i. of first antigen detection at the following different neuronal levels of the trigeminal pathway: nasal cavity in respiratory mucosal epithelium, first-order neurons in the trigeminal ganglion, second-order neurons in the spinal trigeminal nucleus (Sp5), and third-order neurons in the ectorhinal cortex.
−, no infected cells detectable.
Isolation and characterization of PrV-ΔUL51F/11G.
Since the UL11 and UL51 proteins of PrV share several features (see the introduction) and may exert similar, possibly redundant functions, we simultaneously deleted the UL51 and UL11 genes from the PrV genome. Replication properties of the double mutant in cultured cells were investigated by plaque assays, one-step growth kinetics, and transmission electron microscopy. PrV-ΔUL51F/11G formed only very small plaques on noncomplementing RK13 cells with diameters reaching only approximately 20% of those of wild-type plaques (Fig. 4), while plaque formation was restored to levels of those of the UL11 single-deletion mutant, PrV-ΔUL11 (23), on RK-UL51 cells. On RK13-UL11 cells, diameters were comparable to those of the UL51 deletion mutant on RK13 cells. In one-step growth kinetics, PrV-ΔUL51F/11G replicated on noncomplementing cells in a manner similar to that of the UL11 deletion mutant, with reduced titers at all time points, and with approximately 10-fold-lower final titers (Fig. 5). In electron microscopic analyses of PrV-ΔUL51F/11G-infected RK13 cells, the phenotype of the UL11 deletion mutant was predominant with distorted intracytoplasmic membranes and nucleocapsids accumulating in the cytoplasm associated with tegument in aggregated structures (Fig. 6C). However, intracytoplasmic and extracellular enveloped virions as well as extracellular L-particles were also observed (Fig. 6C). The phenotype of the double mutant was similar on UL51-expressing cells (Fig. 6E) while on RK13-UL11 cells infected with PrV-ΔUL51F/11G (Fig. 6D), the phenotype of the single UL51 deletion mutant was observed.
DISCUSSION
Homologs of the nonessential HSV-1 UL51 tegument protein are conserved throughout the herpesvirus family, but no function has been assigned to them yet. Previously, the PrV UL51 protein has been identified as a virion component, most likely residing in the tegument (27). In Western blot analyses on fractionated infected cells, PrV UL51 was detected mainly in the nuclear fraction. This was in marked contrast to the homologous proteins of HSV-1 and BHV-1, which were detected exclusively or dominantly in the cytoplasm (11, 17, 36). Since the PrV UL51 antipeptide sera used in the previous study (27) did not detect the protein in indirect immunofluorescence assays, we generated antisera by using bacterially expressed UL51 polypeptides. Using these novel sera in immunofluorescence analyses, UL51 was found predominantly in the cytoplasm colocalizing with Golgi markers. Thus, these results are in line with those for the HSV-1 and BHV-1 homologs. The differences from our earlier results are probably due to the inadequacy of the cell fractionation method. In summary, the HSV-1, BHV-1, and PrV UL51 proteins share a cytoplasmic location in infected cells. This finding correlates with the notion that tegument assembly occurs primarily in the cytoplasm (33, 34).
Even in the absence of other viral proteins, the HSV-1 and PrV UL51 proteins partly colocalized with various Golgi proteins (36; this study), although the appearance of the Golgi apparatus in transfected cells was altered compared to that in adjacent nontransfected cells (Fig. 2C). For this intrinsic Golgi targeting, palmitoylation of the N-terminal cysteine at position 9 of the HSV-1 protein was required (36). In contrast to N-myristoylation, palmitoylation is transient and may serve to regulate membrane association (37). Therefore, it was suggested that UL51 might function in targeting viral nucleocapsids to the site of secondary envelopment (36). Amino acid sequence alignment of alphaherpesvirus UL51 homologs, including PrV, demonstrates that this cysteine residue is conserved (data not shown), indicating that palmitoylation might be a general feature for UL51 proteins. Studies on fatty acid modifications of PrV UL51 are under way.
Despite its conservation, the UL51 protein is not essential for the replication of HSV-1 (5, 29) or PrV (this study). Unfortunately, to our knowledge, no corresponding mutants have been investigated for members of other herpesvirus subfamilies. In HSV-1 and PrV, UL51 deletion mutants formed only small plaques, which in PrV were 70% reduced compared to those in PrV-Ka on noncomplementing cells, and viral titers were reduced approximately fivefold. In electron microscopic examinations, the presence of numerous particles in the process of secondary envelopment, which were not observed in this abundancy in wild-type PrV-infected cells (14), indicated that virion formation is delayed. Thus, UL51 might be necessary for an efficient envelopment process.
A similar phenotype has recently been described for a PrV mutant lacking UL11 (23). Since both proteins are small, membrane-associated conserved tegument proteins, it was conceivable that deletion of either one does not produce a robust phenotype due to functional redundancy between the two. Thus, we generated a mutant simultaneously lacking UL11 and UL51. However, PrV-ΔUL51F/11G showed only a moderate additive effect on the plaque size of the double deletion but replicated with kinetics similar to those of PrV-ΔUL11. In electron microscopic examinations, the UL11 phenotype was dominant, indicating that there is no functional redundancy between the UL11 and UL51 proteins of PrV.
The UL51 protein of PrV is not required for productive replication in RK13 cells and is also not needed for neuroinvasion after intranasal infection of mice. The mean time to death of PrV-ΔUL51F-infected animals of 64 h is similar to that of PrV-ΔUL11-infected mice (67 h) (20). Moreover, a similar kinetics of neuroinvasion was observed, with minor symptoms becoming apparent at day 2 after infection and with the full-blown disease appearing only on day 3. In contrast, after PrV-Ka infection, animals exhibit strong symptoms already after 2 days and are dead by day 3. Nevertheless, transneuronal infection occurred and PrV-ΔUL51F infection could easily be observed in second-order neurons in Sp5, indicating that the UL51 protein is not required for neuroinvasion and transneuronal spread of PrV in the mouse trigeminal system.
In conclusion, although deletion of the conserved UL51 gene from the PrV genome impaired replication in cell culture and in the mouse model to a limited extent, no robust phenotype could be observed. As part of the tegument, a viral structure with an abundance of protein-protein interactions, it is conceivable that functional redundancy, which has previously been observed with nonessential tegument components and viral glycoproteins (reviewed in reference 33), also applies to the function of UL51. Thus, multiple deletions will have to be introduced into the viral genome to uncover the specific function of single viral proteins.
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (Me854/5-2).
We thank Jens Teifke for advice on animal experiments and histopathology and Uta Hartwig, Mandy Jörn, Petra Meyer, Diana Werner, and Elke Zorn for expert technical and photographical assistance.
REFERENCES
- 1.Avitabile, E., S. Di Gaeta, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 310:207-211. [DOI] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, D. E., and B. Roizman. 1990. Identification of three genes nonessential for growth in cell culture near the right terminus of the unique sequences of long component of herpes simplex virus 1. Virology 177:684-691. [DOI] [PubMed] [Google Scholar]
- 6.Baumeister, J., B. G. Klupp, and T. C. Mettenleiter. 1995. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J. Virol. 69:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowzard, J. B., R. J. Visalli, C. B. Wilson, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L.W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C.M. Brown, R. Cerny, T. Horsnell, C. A. Hutchinson III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 10.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 11.Daikoku, T., K. Ikenoya, H. Yamada, F. Goshima, and Y. Nishiyama. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J. Gen. Virol. 79:3027-3031. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 14.Granzow, H., F. Weiland, A. Jöns, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossmann, A., F. Weiland, and E. Weiland. 1989. Autoimmunity induced by lactate dehydrogenase-elevating virus: monoclonal autoantibodies against Golgi antigens and other subcellular elements. Autoimmunity 2:201-211. [DOI] [PubMed] [Google Scholar]
- 17.Hamel, F., H. Boucher, and C. Simard. 2002. Transcriptional and translational expression kinetics of the bovine herpesvirus 1 UL51 homologue gene. Virus Res. 84:125-134. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, S. M., L. Raine, and H. Fanger. 1981. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J. Histochem. Cytochem. 29:577-580. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 20.Klopfleisch, R., J. P. Teifke, W. Fuchs, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2004. Influence of tegument proteins of pseudorabies virus on neuroinvasion and transneuronal spread in the nervous system of adult mice after intranasal inoculation. J. Virol. 78:2956-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopp, M., H. Granzow, W. Fuchs, B. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lenk, M., N. Visser, and T. C. Mettenleiter. 1997. The pseudorabies virus UL51 gene product is a 30-kilodalton virion component. J. Virol. 71:5635-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 30.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70:3147-3157. [DOI] [PubMed] [Google Scholar]
- 31.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region of the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 32.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 33.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 35.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effect of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nozawa, N., T. Daikoku, T. Koshizuka, Y. Yamauchi, T. Yoshikawa, and Y. Nishiyama. 2003. Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J. Virol. 77:3204-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451:1-16. [DOI] [PubMed] [Google Scholar]
- 38.Roizman, B., and D. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields' virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 39.Roizman, B., and P. Pellet. 2001. The family herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields' virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 40.Silva, M., Q.-C. Yu, L. W. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-coded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.