Abstract
We characterized the regions involved in replication and mobilization of the 40-kb plasmid pNZ4000, encoding exopolysaccharide (EPS) production in Lactococcus lactis NIZO B40. The plasmid contains four highly conserved replication regions with homologous rep genes (repB1, repB2, repB3, and repB4) that belong to the lactococcal theta replicon family. Subcloning of each replicon individually showed that all are functional and compatible in L. lactis. Plasmid pNZ4000 and genetically labeled derivatives could be transferred to different L. lactis strains by conjugation, and pNZ4000 was shown to be a mobilization plasmid. Two regions involved in mobilization were identified near two of the replicons; both included an oriT sequence rich in inverted repeats. Conjugative mobilization of the nonmobilizable plasmid pNZ124 was promoted by either one of these oriT sequences, demonstrating their functionality. One oriT sequence was followed by a mobA gene, coding for a trans-acting protein, which increased the frequency of conjugative transfer 100-fold. The predicted MobA protein and the oriT sequences show protein and nucleotide similarity, respectively, with the relaxase and with the inverted repeat and nic site of the oriT from the Escherichia coli plasmid R64. The presence on pNZ4000 of four functional replicons, two oriT sequences, and several insertion sequence-like elements strongly suggests that this EPS plasmid is a naturally occurring cointegrate.
Lactococci are known to harbor conjugative plasmids that are used for industrial strain improvement since they encode important metabolic traits such as lactose fermentation, protease activity, bacteriophage resistance, or production of exopolysaccharide (EPS). Therefore, these plasmids are studied for their functional properties as well as for their mode of replication and transfer capacities. Two different mechanisms of replication are known to operate in Lactococcus lactis: rolling circle and theta replication. Rolling circle replication seems to be restricted to relatively small lactococcal plasmids with cryptic functions (19). Two of these, the related promiscuous plasmids pWV01 and pSH71, have been developed into widely used cloning and expression vectors (6). The replication regions of several theta replicating lactococcal plasmids that encode metabolic functions have been analyzed, and all are members of a family of highly related, compatible theta replicons as first identified for plasmid pCI305 (17, 35). They all contain a homologous repB gene encoding the replication protein. The conserved region upstream of repB is likely to include the origin of replication and also contains 22-bp repeats which have a replicon-specific regulatory role in plasmid replication and an inverted repeat overlapping the repB promoter which is a RepB binding site (7).
The capacity for conjugal transfer is an important characteristic of some lactococcal plasmids. Self-transmissible conjugative plasmids have the ability to form effective cell-to-cell contact, while mobilization plasmids are able only to prepare their DNA for transfer (36). The conjugation process in gram-negative bacteria is initiated at the origin of transfer (oriT) by the formation of a relaxosome, usually containing a relaxase and accessory DNA binding proteins. The relaxase catalyzes the cleavage of a specific phosphodiester bond at the nic site in the oriT, after which it is covalently linked to the 5′ end of the cleaved strand through a tyrosyl residue. Single-stranded DNA is transferred to the recipient cell and subsequently ligated through the cleaving-joining activity of the relaxase, resembling the process of leading-strand replication by rolling circle replication (20). To date, very little is known about genes required for conjugation in lactococci and other gram-positive bacteria (12). The chromosomally encoded sex factor and the homologous conjugative element pRS01 of L. lactis 712 and ML3, respectively, can mediate a high-frequency transfer of nonconjugative lactose plasmids and confer a cell aggregation (Clu) phenotype (1, 10). The sex factor cluA gene encodes a protein that is involved in cell aggregation during conjugation (14). On the bacteriophage resistance plasmid pCI528, a 2-kb region involved in conjugative mobilization has been identified. It contains a putative oriT and a mobA gene which is predicted to encode a protein involved in mobilization (24).
While EPS production by lactococci has long been known to be a plasmid-encoded trait, it was only recently established that structural genes involved in EPS biosynthesis are located on these plasmids (37, 38). The best-characterized EPS plasmid to date is the 40-kb pNZ4000 from L. lactis NIZO B40, which contains a 12-kb gene cluster encoding EPS biosynthesis (37). Furthermore, it contains multiple replicons, since we were able to separate pNZ4000 in two XhoI-SphI fragments that upon labeling with an erythromycin resistance (Eryr) marker could each replicate in L. lactis (37). In this study, we report the identification and characterization of the regions involved in plasmid replication and mobilization of this EPS plasmid. Plasmid pNZ4000 contains four functional replicons and two regions involved in mobilization; one codes for an active trans-acting mobilization protein, and both contain a cis-acting oriT region.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown in L-broth-based medium at 37°C (33). L. lactis was grown at 30°C in M17 broth (Difco Laboratories) supplemented with 0.5% glucose (GM17). If appropriate, the media contained chloramphenicol (10 μg/ml), erythromycin (10 μg/ml for L. lactis and 150 μg/ml for E. coli), rifampin (50 μg/ml), streptomycin (100 μg/ml), tetracycline (25 μg/ml), or ampicillin (100 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | 16 | |
| L. lactis NIZO B40 | Lac+ Eps+ multiplasmid strain harboring pNZ4000 | 37 |
| L. lactis MG1363 | Plasmid free | 11 |
| L. lactis MG1614 | Rifr Strr, plasmid free | 11 |
| L. lactis IL1403 | Plasmid free | 3 |
| L. lactis NZ4010 | Rifr Strr Eps+, MG1614 harboring pNZ4000 | 37 |
| Plasmids | ||
| pCI182 | Tetr, 8.0-kb pBR322 derivative carrying the Tn919 tetM gene | 18 |
| pUC19Ery | Eryr, 3.8-kb pUC19 carrying the Eryr gene of pIL253 | 37 |
| pUC18Ery | Eryr, 3.8-kb pUC18 carrying the Eryr gene of pIL253 | 37 |
| pNZ4000 | 40-kb plasmid encoding EPS production | 37 |
| pNZ4001 | Eryr, 7.1-kb derivative of pUC19Ery carrying repB1 | This study |
| pNZ4002 | Eryr, 7.2-kb derivative of pUC19Ery carrying repB2 | This study |
| pNZ4003 | Eryr, 7.5-kb derivative of pUC18Ery carrying repB3 | This study |
| pNZ4004 | Eryr, 6.7-kb derivative of pUC19Ery carrying repB4 | This study |
| pNZ4006 | Eryr, 5.0-kb derivative of pUC19Ery carrying a 1.2-kb EcoRI-XbaI fragment of pNZ4000 with orfD1 | This study |
| pNZ4007 | Eryr, 7.9-kb derivative of pUC19Ery carrying a 4.1-kb EcoRI-XbaI fragment of pNZ4000 with orfD2 | This study |
| pNZ4010 | Eryr, 45-kb, pNZ4000 containing an integrated copy of pNZ4006 | This study |
| pNZ4017 | Eryr, 48-kb, pNZ4000 containing an integrated copy of pNZ4007 | This study |
| pNZ124 | Cmr, 2.8-kb pSH71 replicon | 28 |
| pNZ4021 | Cmr, 3.6-kb derivative of pNZ124 carrying oriT1 | This study |
| pNZ4022 | Cmr, 3.4-kb derivative of pNZ124 carrying oriT2 | This study |
| pNZ4023 | Cmr, 5.8-kb derivative of pNZ124 carrying oriT1 and mobA | This study |
| pNZ4025 | Tetr, 10.2-kb derivative of pUC18 carrying repB1 and tetM | This study |
| pNZ4026 | Tetr, 11.4-kb derivative of pCI182 carrying repB2 | This study |
| pNZ4027 | Tetr, 10.9-kb derivative of pCI182 carrying repB4 | This study |
Lac+, lactose fermenting; Eps+, EPS producing; Rifr, rifampin resistant; Strr, streptomycin resistant; Tetr, tetracycline resistant; Eryr, erythromycin resistant; Cmr, chloramphenicol resistant.
DNA isolation, manipulation, and transfer.
Isolation of E. coli plasmid DNA and standard recombinant DNA techniques were performed as described by Sambrook et al. (33). Large-scale isolation of E. coli plasmid DNA for nucleotide sequence analysis was performed with Qiagen columns as instructed by the manufacturer. Isolation and transformation of L. lactis plasmid DNA were performed as previously described (5). For whole-cell lysates of L. lactis, 1.5 ml of a late-log-phase culture was harvested and suspended in 100 μl of a buffer containing 30 mM Tris-HCl (pH 8.0), 3 mM MgCl2, 25% sucrose, 10 μg of lysozyme ml−1, and 0.1 mg of RNase ml−1. This suspension was incubated at 37°C for 30 min. Lysis was achieved by addition of 100 μl of 2% sodium dodecyl sulfate and vortexing at top speed for 1 min, after which the lysate was treated with 20 μg of proteinase K ml−1 at 37°C for 30 min. Conjugation was performed by filter matings as described before (37). The ratio of donor and recipient was 2:1.
Nucleotide sequence analysis.
Automatic double-stranded DNA sequence analysis was performed on both strands with an ALF DNA sequencer (Pharmacia Biotech). Sequencing reactions, performed with an AutoRead sequencing kit, were initiated by using fluorescein-labeled universal and reverse primers and continued with synthetic primers in combination with fluorescein-15-dATP, following the instructions of the manufacturer (Pharmacia Biotech). Sequence data were assembled and analyzed using the PC/GENE program (version 6.70; IntelliGenetics). The GenBank database (February 1998 release) was screened for homologies by using TFASTA.
Construction of plasmids.
For replicon screening and plasmid integration, the E. coli plasmid pUC19Ery or pUC18Ery, carrying the Eryr gene, or pCI182, carrying the tetracycline resistance (Tetr) gene, was used. For plasmids pNZ4001, pNZ4002, and pNZ4004, a 3.3-kb EcoRI-XbaI fragment, a 3.4-kb EcoRI fragment, and a 2.9-kb Sau3AI fragment of pNZ4000 were cloned into pUC19Ery digested with EcoRI-XbaI, EcoRI, and BamHI, respectively. For plasmid pNZ4003, a 3.7-kb XhoI-HincII fragment of pNZ4000 was cloned into SalI-SmaI-digested pUC18Ery. To construct plasmid pNZ4025, a 3.3-kb EcoRI-XbaI fragment of pNZ4000 was cloned in pUC18 (41) digested with EcoRI-XbaI, and subsequently the pCI182 tetM gene was cloned on a 4.2-kb HincII fragment in the pUC18 HincII site. For plasmids pNZ4026 and pNZ4027, a 3.4-kb EcoRI and a 2.9-kb Sau3AI fragment of pNZ4000 were cloned in pCI182 digested with EcoRI or BglII, respectively.
To obtain Eryr derivatives of pNZ4000 (pNZ4010 and pNZ4017), plasmids pNZ4006 and pNZ4007 were constructed. Plasmids pNZ4006 and pNZ4007 are pUC19Ery derivatives carrying 1.2- and 4.1-kb EcoRI-XbaI fragments of pNZ4000, respectively. These plasmids were used for plasmid integration by a single crossover to form pNZ4010 and pNZ4017, respectively.
For functional analysis of the putative oriT regions, fragments containing the oriT1 or oriT2 sequence were cloned in plasmid pNZ124. Plasmid pNZ4021, carrying oriT1, was constructed by cloning a 1.8-kb BglII-XhoI fragment of pNZ4000 in BglII-XhoI-digested pNZ124. Plasmid pNZ4022, carrying oriT2, was constructed by cloning a Klenow enzyme-treated 0.64-kb NspV-NcoI fragment of pNZ4000 in pNZ124 linearized with ScaI. To study the functionality of mobA, plasmid pNZ4023, carrying both oriT1 and mobA, was constructed by cloning a 3.0-kb BglII-AccI fragment of pNZ4000 with a Klenow enzyme-treated AccI site in BglII-ScaI-digested pNZ124. All plasmids were constructed in E. coli.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the replication and mobilization regions are available under GenBank accession no. AF03685, AF03686, and AF03687.
RESULTS AND DISCUSSION
The EPS plasmid pNZ4000 contains four functional replicons.
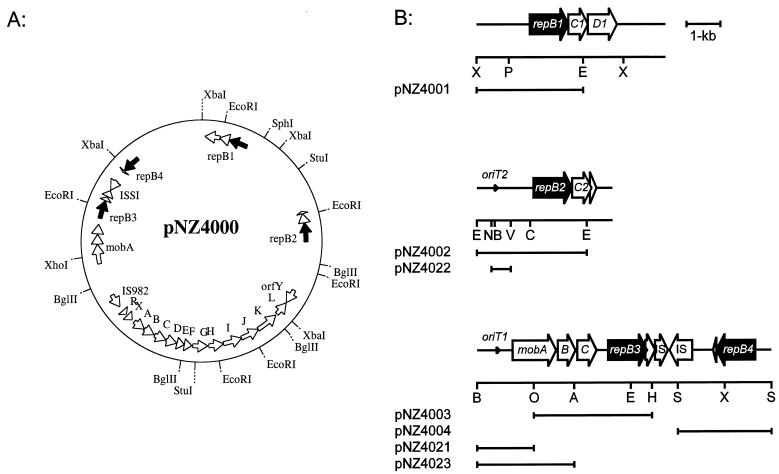
The 40-kb plasmid pNZ4000 is essential for EPS production in strain NIZO B40 and includes the 12-kb eps gene cluster involved in EPS biosynthesis (37). The nucleotide sequence of the EPS plasmid was determined, and analysis of the data revealed the unusual presence of four highly homologous replication regions that belong to a family of lactococcal theta replicons (35) which are located outside the eps gene cluster (Fig. 1). DNA fragments carrying these putative replicons were cloned into pUC19Ery or pUC18Ery, which can be used as replicon screening vectors in L. lactis. The resulting plasmids (pNZ4001, pNZ4002, pNZ4003, and pNZ4004) were transformed to L. lactis MG1363, and in all cases Eryr transformants that harbored plasmids with the expected configuration were obtained (results not shown). These results indicate that all four replicons are functional in L. lactis. Since plasmid replication requires only one of these replicons, pNZ4000 must have derived fragments of several plasmids, which might have formed cointegrates during conjugation processes. This conclusion is corroborated by the presence of complete and truncated copies of ISSI-like elements (Fig. 1), since it is known that ISSI mediates cointegration of the L. lactis ML3 lactose plasmid pSK08 with the conjugal plasmid pRS01 (29). In addition, a complete copy of an IS982-like element is present on pNZ4000 (Fig. 1).
FIG. 1.
(A) Physical and genetic map of plasmid pNZ4000. The eps gene cluster is located between IS982 and orfY. (B) Physical and genetic maps of the replication and mobilization regions of pNZ4000. The fragments used for functional analysis are depicted below. A, AccI; C, ClaI; E, EcoRI; H, HincII; N, NcoI; O, XhoI; P, SphI; S, Sau3AI; V, NspV; X, XbaI. For AccI, ClaI, HincII, NcoI, NspV, and Sau3AI, only sites relevant for subcloning are included. Sequences are available under GenBank accession no. AF036485, AF03686, and AF03687.
GenBank analysis of the proteins encoded by the repB genes of the four replicons on pNZ4000 showed them to be highly homologous to putative replication proteins of several other lactococcal plasmids which all carry a single replicon and belong to a family of lactococcal theta replicons (34), including pVS40 (86.0% identity with RepB1) (39), pWV04 (98.5% identity with RepB2) (34), pCI528 (99.8% identity with RepB3) (23), and pFV1201 (99.2% identity with RepB4) (GenBank accession no. X96949). The upstream regions of the repB genes of pNZ4000 were highly conserved and corresponded to those found in the other lactococcal replicons as first identified for pCI305 (17). They all contain an A/T-rich region that could be the recognition site for host-encoded functions involved in replication (35), a 22-bp sequence repeated 3.5 times which was shown to have a replicon-specific regulatory role in plasmid replication (7), and two inverted repeats, one of which overlapped the −35 region of the repB promoter (inverted repeat 1 [IR1]) and was found to be a RepB binding site (7). The upstream region of repB1 showed a slightly different architecture and contained only a 2.5-times-repeated 22-bp direct repeat.
All four replicons are compatible but show differences in organization.
The minimal replicons for repB1, repB2, and repB4 were labeled with the tetM gene to generate pNZ4025, pNZ4026, and pNZ4027, respectively. These plasmids were combined with either pNZ4001, pNZ4002, pNZ4003, or pNZ4004 and transformed to MG1363 to make six strains including all combinations of different replicons carrying a set of Eryr and Tetr genes (Table 2). All plasmids had comparable copy numbers, as judged from the intensity of ethidium bromide-stained plasmid DNA separated by agarose gel electrophoresis (results not shown). Stable transformants were obtained for all heteroplasmid combinations following selection for Eryr and Tetr, indicating that these replicons are compatible. The compatibility of the plasmids carrying different replicons was confirmed by determining the segregational stability after growth for 35 generations in medium containing no antibiotics (Table 2). Plasmids carrying the replicons with repB1, repB2, and repB4 formed highly stable heteroplasmid combinations. In contrast, the segregational stability of the repB3-containing replicon was significantly lower than that of the others. This was also observed when this replicon was present as a single replicon in MG1363. After 20 generations without selection pressure, 61% of the population was plasmid containing; after 40 generations 16%, and after 60 generations only 3%, of the population contained plasmids. The reason for the difference in stability between the repB3-containing replicon and the other three highly homologous replicons is unclear. It seems that there is interference with the maintenance functions of the repB3-containing replicon which are not directly involved in replication. The orfC genes located downstream of and partly overlapping the repB genes (Fig. 1) are not likely to be involved in this process. The predicted OrfC proteins are homologous to RepB287 (45 and 43% identity for OrfC1 and OrfC2, respectively) encoded by the Tetragenococcus halophilus theta-replicating plasmid pUCL287. RepB287 is not essential for replication, as is OrfC, but its presence reduces the copy number and the segregational stability (2). The N-terminal parts of the OrfC proteins are highly conserved and contain a helix-turn-helix motif which is probably involved in DNA binding. While repB1 and repB2 are followed by almost complete orfC genes, orfC3 and orfC4 encode only the N-terminal parts of OrfC-like proteins. If the role of the lactococcal OrfC was similar to that of RepB287, we would expect the stability of the replicons containing repB1 and repB2 to be lower than that of the replicons containing repB3 and repB4, which is not as we observed.
TABLE 2.
Segregational stability of the four replicons of pNZ4000
| Replicon
|
Fraction of plasmid-containing cellsa
|
|||
|---|---|---|---|---|
| Tetr | Eryr | Tetr | Eryr | Tetr + Eryr |
| repB1 | repB2 | 0.8 | 0.9 | 0.9 |
| repB1 | repB3 | 0.9 | 0.01 | 0.01 |
| repB4 | repB1 | 1.0 | 1.0 | 0.9 |
| repB2 | repB3 | 0.7 | 0.05 | 0.06 |
| repB2 | repB4 | 0.8 | 0.9 | 0.7 |
| repB4 | repB3 | 0.9 | 0.08 | 0.07 |
After 35 generations without selection pressure, cultures containing the Tetr and Eryr replicons were plated on medium containing tetracycline, erythromycin, both tetracycline and erythromycin, or no antibiotics, and the fraction of antibiotic-resistant colonies was determined.
Downstream of orfC1 and orfC2, we found a partly overlapping third ORF (orfD1 and orfD2, respectively [Fig. 1]). The predicted gene product OrfD1 shows considerable homology (47% identity) to the product of an hsdS-like gene from the lactococcal plasmid pIL2614, which encodes the specificity subunit of a type IC restriction-modification system (34). The hsdS-like gene is the last of a putative operon of five genes, the first two of which are replication genes homologous to repB and orfC. These are followed by three genes coding for the endonuclease, methylase, and specificity subunits, respectively, of a type I replication-modification system (34). This finding indicates that pNZ4000 and pIL2614 contain similarly organized and homologous operons, the one in pNZ4000 lacking the genes encoding the endonuclease and methylase subunits.
The EPS plasmid pNZ4000 is a mobilization plasmid.
We have previously shown that plasmid pNZ4000 can be conjugally transferred together with the lactose plasmid from the L. lactis NIZO B40 to the recipient strain MG1614 (37). To study the intraspecific conjugative transfer of pNZ4000 in more detail, the Eryr derivatives pNZ4010 and pNZ4017 were used. These plasmids were transformed to the plasmid-free strain MG1363, and the resulting strains were used as donors in filter matings with strain MG1614. Conjugative transfer of either of these plasmids between these isogenic L. lactis subsp. cremoris strains occurred at a frequency of 10−6 per donor. Plasmid pNZ4017 was also transformed to the plasmid-free L. lactis subsp. lactis strain IL1403, from which it could be transferred to L. lactis subsp. cremoris MG1614 at a frequency of 10−8 per donor. These results demonstrate that pNZ4000 can be mobilized from strains MG1363 and IL1403. It is likely that differences in chromosomal conjugation functions account for the differences in transfer efficiency of the pNZ4000 derivatives from both L. lactis subspecies, which are known to share approximately 70 to 80% sequence identity in characterized genes and differ by the presence of a large chromosomal inversion of about half of the genome (13, 21). Furthermore, MG1363 harbors the sex factor that encodes conjugative functions (12), which may play a role in mobilization of pNZ4000.
pNZ4000 contains two functional oriT sites.
Mobilization involves a cis-acting oriT region and a trans-acting gene encoding a relaxase (20). Nucleotide sequence analysis of pNZ4000 revealed the presence of a region upstream of repB3 (Fig. 1), which is almost identical (98.3% identity) to a 2.0-kb fragment involved in mobilization of the lactococcal plasmid pCI528 (24). It contains a mobA gene encoding a putative mobilization protein. The upstream region of the mobA gene contains three inverted repeats and a direct repeat and has been postulated to be the oriT region (24). We tested the functionality of the putative oriT sequence (oriT1) by cloning it in the nonconjugative plasmid pNZ124 and transforming the resulting plasmid pNZ4021 to strain MG1363. This strain was mated with MG1614 and chloramphenicol-resistant transconjugants were selected (Table 3). The 1.8-kb region containing the oriT1 sequence was sufficient to achieve conjugal transfer of the nonconjugative plasmid pNZ124, showing that the cloned fragment contains a functional oriT.
TABLE 3.
Transfer frequencies of pNZ124 derivatives from MG1363 to MG1614
| Plasmid(s) | Genotype
|
Transfer frequencya | |||
|---|---|---|---|---|---|
| oriT1 | oriT2 | mobA in cis | mobA in trans | ||
| pNZ124 | − | − | − | − | <10−10 |
| pNZ4021 | + | − | − | − | 10−7 |
| pNZ4021, pNZ4017 | + | − | − | + | 10−5 |
| pNZ4022 | − | + | − | − | 10−7 |
| pNZ4022, pNZ4017 | − | + | − | + | 10−5 |
| pNZ4023 | + | − | + | − | 10−5 |
Number of transconjugants per donor (average of two independent experiments).
A second oriT region sharing 96.6% identity in 417 nucleotides with oriT1 was found upstream of repB2. It was cloned as a 0.64-kb fragment in pNZ124, and the resulting plasmid, pNZ4022, had the same transfer frequency as pNZ4021 (Table 3), indicating the presence of two functional oriT sequences on pNZ4000, one upstream of mobA (oriT1) and one upstream of repB2 (oriT2) (Fig. 1), oriented in opposite directions.
The oriT site of the streptococcal plasmid pMV158 is homologous to sequences of several plasmids from gram-positive hosts (15). However, no significant homology between the oriT regions of pNZ4000 and these sequences could be detected. In contrast, the pNZ4000 oriT sequences contain an inverted repeat (IR3) which is highly homologous to that of the oriT from IncI1 plasmid R64 (Fig. 2). This includes the R64 mobilization protein NikA binding site (9). Moreover, the homology between the pNZ4000 oriT sequences and that of R64 also includes the sequence next to the repeat containing the nic site (Fig. 2). In the absence of experimental evidence, we therefore postulate that these sequences may contain the pNZ4000 nic sites. The streptococcal plasmid pIP501 and the staphylococcal plasmid pGO1 oriT regions are homologous to oriT sequences of several gram-negative plasmids. They all contain a conserved sequence with the nic site next to a nonconserved inverted repeat centered around the nucleotide sequence 5′-GAA-3′ (4, 40). Although no significant homology between these oriT regions and those of pNZ4000 could be detected, the IR3 sequence of each of the pNZ4000 oriT regions is also situated around a 5′-GAA-3′ nucleotide sequence.
FIG. 2.
DNA sequence alignment of the IR3 segments of the oriT regions found on pNZ4000 (oriT1 and oriT2) and the sequence of the IncI1 plasmid R64 oriT. For R64 oriT, the inverted repeat is underlined, the NikA binding site is indicated in boldface, and the nic site is indicated with an arrowhead (9).
mobA encodes a product trans-acting on oriT-carrying plasmids.
The involvement of mobA in mobilization was studied by comparing the transfer frequencies of plasmids carrying only oriT sequences or carrying oriT and mobA either in cis or in trans (Table 3). When mobA was provided in trans on pNZ4017, the transfer frequencies of pNZ4021 and pNZ4022 increased significantly. The same effect was achieved by when mobA was present in cis as on plasmid pNZ4023, containing oriT1 and mobA. These results indicate that mobA encodes a trans-acting element involved in mobilization.
To verify the relaxation activity of the mobA gene product (25), whole-cell lysates of MG1363 harboring oriT1- or oriT2-carrying plasmids with or without mobA (in cis or in trans) were separated by agarose gel electrophoresis. The plasmid profiles of pNZ4021 and pNZ4022 showed a significant increase in open circular plasmid DNA only when pNZ4017 was present (approximately half of the oriT-carrying plasmids were in the open circular form). Moreover, pNZ4023 carrying oriT1 and mobA showed a similar high degree of open circular DNA (data not shown). These results indicate that the plasmids carrying oriT fragments are relaxed by the trans-acting mobA gene product.
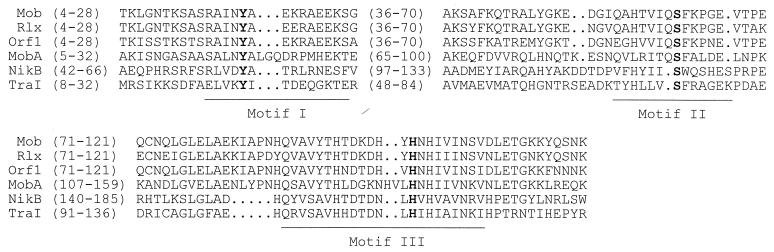
The predicted MobA protein reveals significant homologies (approximately 30% identity) with three mobilization proteins found on antibiotic resistance plasmids of Staphylococcus aureus (30–32) and moderate homology (23% identity in 388 amino acids) with the N-terminal part of TraI from the E. coli IncPα plasmid RP4. TraI is a relaxase and forms together with TraJ the relaxosome at oriT (26). TraI contains three conserved regions found in several relaxases (27). Motifs I and III are involved in catalyzing the cleaving-joining reaction. Motif I contains a conserved tyrosine residue which after nicking is covalently attached to the 5′ end of the cleaved DNA. Motif III contains a conserved histidine residue that is likely to activate the tyrosine of motif I by proton extraction. Motif II contains a conserved serine and is thought to be involved in DNA recognition (27). Multiple sequence alignment of MobA, the four homologous proteins, and the E. coli plasmid R64 relaxase NikB, which is homologous to TraI (8), showed that the three conserved domains and the tyrosine, serine, and histidine residues needed for relaxase activity are present in MobA (Fig. 3). This conservation strongly suggests that the lactococcal MobA is a relaxase which is involved in nicking the nic sites of the oriT sequences (Fig. 2), which is corroborated by the formation of open circular DNA of plasmids carrying an oriT sequence when mobA is present (see above).
FIG. 3.
Amino acid sequence comparison of the three conserved regions involved in relaxase activity as determined for TraI (27) for the relaxases MobA (Mob), Rlx, and Orf1 from S. aureus plasmids pC221, pS194, and pC223, respectively, MobA (MobA) from pNZ4000, NikB from E. coli plasmid R64 (8), and TraI from the E. coli plasmid RP4. The tyrosine (motif I) and histidine (motif III) residues involved in cleaving-joining reaction, and the serine residue (motif II) involved in DNA binding, are indicated in boldface.
On pNZ4000, a second ORF, here designated mobB, was found downstream of mobA, the putative start codon of which overlaps the stop codon of mobA. This configuration resembles that of the S. aureus plasmid pC223, which contains two overlapping mobilization genes orf1 and orf2 (30, 32). In addition to the homologous Orf1 and MobA proteins, the predicted MobB protein shares moderate homology (24% identity) with Orf2 of pC223. A third ORF, designated mobC, was detected 16 bp downstream of the stop codon of mobB. Its gene product showed no homology to any protein in the GenBank database, and the involvement of mobC in the conjugation process remains to be established.
The region on pNZ4000 containing the mobilization genes and the third replicon has a high degree of homology with the same regions on pCI528. Plasmid pCI528 is a 46-kb plasmid encoding the production of a hydrophilic polymer containing glucose and rhamnose that reduces phage adsorption to its lactococcal host (22). Although pCI528 does not encode EPS production whereas pNZ4000 does, there may be a close relationship between the two plasmids or their ancestors.
In summary, we demonstrated that plasmid pNZ4000 contains four homologous and active replicons that are compatible with each other. It contains two functional oriT sequences. One oriT is followed by the mobA gene coding for a trans-acting protein. The predicted MobA protein and the oriT sequences are homologous to the R64 relaxase and the oriT. The R64 relaxase is known to nick a site which is also conserved in the oriT sequences of pNZ4000.
ACKNOWLEDGMENTS
This work was partly supported by European Community research grant 1116/92 1.6.
We thank Norwin Willem and Sónia Mendes for technical assistance in the sequencing of the replicons. We are grateful to Joey Marugg for advice at the initial stages of this work. We acknowledge Michiel Kleerebezem and Roland Siezen for critically reading the manuscript.
REFERENCES
- 1.Anderson D G, McKay L L. Genetic and physical characterization of recombinant plasmids associated with cell aggregation an high-frequency conjugal transfer in Streptococcus lactis ML3. J Bacteriol. 1984;158:954–962. doi: 10.1128/jb.158.3.954-962.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benachour A, Frère J, Novel G, Auffray Y. Molecular analysis of the replication region of the theta-replicating plasmid pUCL287 from Tetragenococcus (Pediococcus) halophilus ATCC33315. Mol Gen Genet. 1997;255:504–513. doi: 10.1007/s004380050523. [DOI] [PubMed] [Google Scholar]
- 3.Chopin A, Chopin M C, Moillo-Batt A, Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984;11:260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- 4.Climo M W, Sharma V K, Archer G L. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol. 1996;178:4975–4983. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vos W M, Vos P, de Haard H, Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989;85:169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- 6.de Vos W M, Simons G F M. Gene cloning and expression systems in Lactococci. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, England: Chapman and Hall; 1994. pp. 52–105. [Google Scholar]
- 7.Foley S, Bron S, Venema G, Daly C, Fitzgerald G F. Molecular analysis of the replication origin of the Lactococcus lactis plasmid pCI305. Plasmid. 1996;36:125–141. doi: 10.1006/plas.1996.0040. [DOI] [PubMed] [Google Scholar]
- 8.Furuya N, Nisioka T, Komano T. Nucleotide sequence and functions of the oriT operon in IncI1 plasmid R64. J Bacteriol. 1991;173:2231–2237. doi: 10.1128/jb.173.7.2231-2237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuya N, Komano T. Mutational analysis of the R64 oriT region: requirement for precise location of the NikA-binding sequence. J Bacteriol. 1997;179:7291–7297. doi: 10.1128/jb.179.23.7291-7297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasson M J, Davies F L. Conjugal transfer of the drug resistance plasmid pAMβ1 in the lactic streptococci. FEMS Microbiol Lett. 1980;7:51–53. [Google Scholar]
- 11.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasson M J, Godon J-J, Pillidge C J, Eaton T J, Jury K, Shearman C A. Characterization and exploitation of conjugation in Lactococcus lactis. Int Dairy J. 1995;5:757–762. [Google Scholar]
- 13.Godon J-J, Delorme C, Ehrlich S D, Renault P. Divergence of genomic sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1992;58:4045–4047. doi: 10.1128/aem.58.12.4045-4047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godon J-J, Jury K, Shearman C A, Gasson M J. The Lactococcus lactis sex-factor aggregation gene cluA. Mol Microbiol. 1994;12:655–663. doi: 10.1111/j.1365-2958.1994.tb01053.x. [DOI] [PubMed] [Google Scholar]
- 15.Guzmán L M, Espinosa M. The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J Mol Biol. 1997;266:688–702. doi: 10.1006/jmbi.1996.0824. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Hayes F, Vos P, Fitzgerald G F, de Vos W M, Daly C. Molecular organization of the minimal replicon of novel narrow-host-range, lactococcal plasmid pCI305. Plasmid. 1991;25:16–26. doi: 10.1016/0147-619x(91)90003-f. [DOI] [PubMed] [Google Scholar]
- 18.Hill C, Venema G, Daly C, Fitzgerald G F. Cloning and characterization of the tetracycline resistance determinant of and several promoters from within the conjugative transposon Tn919. Appl Environ Microbiol. 1988;54:1230–1236. doi: 10.1128/aem.54.5.1230-1236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan S A. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 21.Le Bourgeois P, Lautier M, van den Berghe L, Gasson M J, Ritzenthaler P. Physical and genetic map of the Lactococcus lactis subsp. cremoris MG1363 chromosome: comparison with that of Lactococcus lactis subsp. lactis IL1403 reveals a large chromosomal inversion. J Bacteriol. 1995;177:2840–2850. doi: 10.1128/jb.177.10.2840-2850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucey M, Daly C, Fitzgerald G F. Cell surface characteristics of Lactococcus lactis harbouring pCI528, a 46 kb plasmid encoding inhibition of bacteriophage adsorption. J Gen Microbiol. 1992;138:2137–2143. [Google Scholar]
- 23.Lucey M, Daly C, Fitzgerald G F. Identification and sequence analysis of the replication region of the phage resistance plasmid pCI528 from Lactococcus lactis subsp. cremoris UC503. FEMS Microbiol Lett. 1993;10:249–256. doi: 10.1111/j.1574-6968.1993.tb06330.x. [DOI] [PubMed] [Google Scholar]
- 24.Lucey M, Daly C, Fitzgerald G. Analysis of a region from the bacteriophage resistance plasmid pCI528 involved in its conjugative mobilization between Lactococcus strains. J Bacteriol. 1993;175:6002–6009. doi: 10.1128/jb.175.18.6002-6009.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novick R. Plasmid-protein relaxation complexes in Staphylococcus aureus. J Bacteriol. 1976;127:1177–1187. doi: 10.1128/jb.127.3.1177-1187.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pansegrau W, Ziegelin G, Lanka E. Covalent association of the traI gene product of plasmid RP4 with the 5′-terminal nucleotide at the relaxation nick site. J Biol Chem. 1990;265:10637–10644. [PubMed] [Google Scholar]
- 27.Pansegrau W, Schröder W, Lanka E. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J Biol Chem. 1994;269:2782–2789. [PubMed] [Google Scholar]
- 28.Platteeuw C, Simons G, de Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1993;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polzin K M, Shimizu-Kadota M. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J Bacteriol. 1987;169:5481–5488. doi: 10.1128/jb.169.12.5481-5488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Projan S J, Novick R. Comparative analysis of five related staphylococcal plasmids. Plasmid. 1988;19:203–221. doi: 10.1016/0147-619x(88)90039-x. [DOI] [PubMed] [Google Scholar]
- 31.Projan S, Moghazeh J, Novick R P. Nucleotide sequence of pS194, a streptomycin-resistance plasmid from Staphylococcus aureus. Nucleic Acids Res. 1988;16:2179–2187. doi: 10.1093/nar/16.5.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Projan S J, Archer G L. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol. 1989;171:1841–1845. doi: 10.1128/jb.171.4.1841-1845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Schouler C, Clier F, Lerayer A L, Ehrlich S D, Chopin M-C. A type IC restriction-modification system in Lactococcus lactis. J Bacteriol. 1998;180:407–411. doi: 10.1128/jb.180.2.407-411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seegers J F M, Bron S, Franke C M, Venema G, Kiewiet R. The majority of lactococcal plasmids carry a highly related replicon. Microbiology. 1994;140:1291–1300. doi: 10.1099/00221287-140-6-1291. [DOI] [PubMed] [Google Scholar]
- 36.Steele J L, McKay L L. Conjugal transfer of genetic material in lactococci: a review. J Dairy Sci. 1989;72:3388–3397. [Google Scholar]
- 37.van Kranenburg R, Marugg J D, Willem N J, van Swam I I, de Vos W M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide production in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 38.van Kranenburg, R., H. R. Vos, I. I. van Swam, M. Kleerebezem, and W. M. de Vos. 1998. Unpublished results.
- 39.Von Wright A, Raty K. The nucleotide sequence for the replication region of pVS40, a lactococcal food grade cloning vector. Lett Appl Microbiol. 1993;17:25–28. doi: 10.1111/j.1472-765x.1993.tb01427.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang A, Macrina F L. Streptococcal plasmid pIP501 has a functional oriT site. J Bacteriol. 1995;177:4199–4206. doi: 10.1128/jb.177.15.4199-4206.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]