Abstract
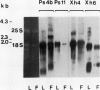
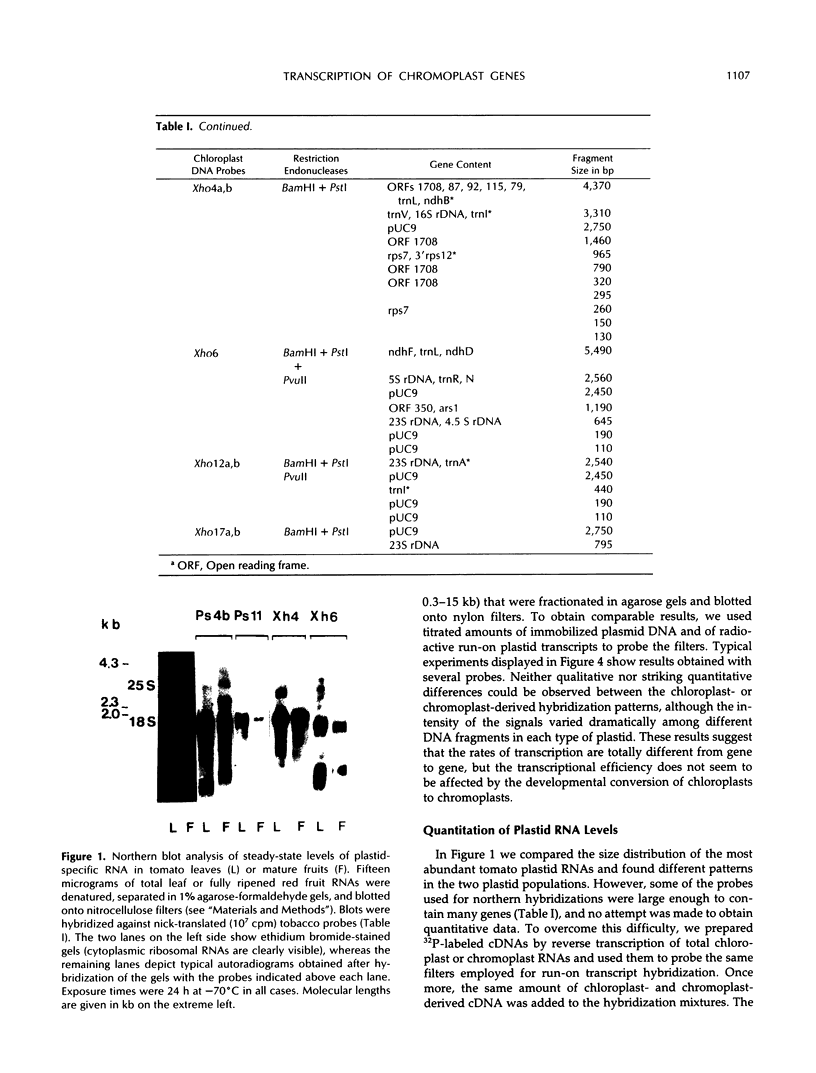
The size distribution of plastid transcripts during chromoplast differentiation in ripening tomato (Lycopersicon esculentum L.) fruits was determined using northern blot analysis. Hybridization of total cellular RNA from leaves and fruits with several tobacco chloroplast DNA probes showed distinct transcript patterns in chloroplasts and chromoplasts. We also compared transcriptional rates by probing immobilized DNA fragments of small size (representing about 85% of the plastid genome) with run-on transcripts from tomato plastids. The relative rates of transcription of the various DNA regions were very similar in chloro- and chromoplasts. Parallel determination of the steady-state levels of plastid RNA showed no strict correlation between synthesis rate and RNA accumulation. Differences in the relative abundance of transcripts between chloro- and chromoplasts were not very pronounced and were limited to a small number of genes. The results indicate that the conversion of chloroplasts to chromoplasts at the onset of tomato fruit ripening proceeds with no important variations in the relative transcription rates and with only moderate changes in the relative stability of plastid-encoded transcripts.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988 Sep;7(9):2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambl R. Mitochondrial biogenesis during fungal spore germination. Development of cytochrome c oxidase activity. Arch Biochem Biophys. 1977 Jul;182(1):273–281. doi: 10.1016/0003-9861(77)90308-3. [DOI] [PubMed] [Google Scholar]
- Carrillo N., Bogorad L. Chloroplast DNA replication in vitro: site-specific initiation from preferred templates. Nucleic Acids Res. 1988 Jun 24;16(12):5603–5620. doi: 10.1093/nar/16.12.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. W., Gruissem W. Constitutive transcription and regulation of gene expression in non-photosynthetic plastids of higher plants. EMBO J. 1988 Nov;7(11):3301–3308. doi: 10.1002/j.1460-2075.1988.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. W., Gruissem W. Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell. 1987 May 8;49(3):379–387. doi: 10.1016/0092-8674(87)90290-x. [DOI] [PubMed] [Google Scholar]
- Deng X. W., Stern D. B., Tonkyn J. C., Gruissem W. Plastid run-on transcription. Application to determine the transcriptional regulation of spinach plastid genes. J Biol Chem. 1987 Jul 15;262(20):9641–9648. [PubMed] [Google Scholar]
- Gold B., Carrillo N., Tewari K. K., Bogorad L. Nucleotide sequence of a preferred maize chloroplast genome template for in vitro DNA synthesis. Proc Natl Acad Sci U S A. 1987 Jan;84(1):194–198. doi: 10.1073/pnas.84.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W. Chloroplast gene expression: how plants turn their plastids on. Cell. 1989 Jan 27;56(2):161–170. doi: 10.1016/0092-8674(89)90889-1. [DOI] [PubMed] [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C. R., Meng B. Y. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989 Jun;217(2-3):185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Ngernprasirtsiri J., Akazawa T. Transcriptional regulation and DNA methylation in plastids during transitional conversion of chloroplasts to chromoplasts. EMBO J. 1990 Feb;9(2):307–313. doi: 10.1002/j.1460-2075.1990.tb08112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J., Schell J., Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987 May 15;163(1):16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Marano M. R., Carrillo N. Chromoplast formation during tomato fruit ripening. No evidence for plastid DNA methylation. Plant Mol Biol. 1991 Jan;16(1):11–19. doi: 10.1007/BF00017913. [DOI] [PubMed] [Google Scholar]
- Mullet J. E., Klein R. R. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987 Jun;6(6):1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngernprasirtsiri J., Kobayashi H., Akazawa T. DNA Methylation Occurred around Lowly Expressed Genes of Plastid DNA during Tomato Fruit Development. Plant Physiol. 1988 Sep;88(1):16–20. doi: 10.1104/pp.88.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson G. L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977 Dec;83(2):346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Rodermel S. R., Bogorad L. Maize plastid photogenes: mapping and photoregulation of transcript levels during light-induced development. J Cell Biol. 1985 Feb;100(2):463–476. doi: 10.1083/jcb.100.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G., Gruissem W. Chloroplast mRNA 3' end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 1991 Jun;10(6):1493–1502. doi: 10.1002/j.1460-2075.1991.tb07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra E. C., Carrillo N. DNA polymerase activity of tomato fruit chromoplasts. FEBS Lett. 1990 Nov 26;275(1-2):102–106. doi: 10.1016/0014-5793(90)81449-x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Stirdivant S. M., Crossland L. D., Bogorad L. DNA supercoiling affects in vitro transcription of two maize chloroplast genes differently. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4886–4890. doi: 10.1073/pnas.82.15.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullis R. H., Rubin H. Calcium protects DNase I from proteinase K: a new method for the removal of contaminating RNase from DNase I. Anal Biochem. 1980 Sep 1;107(1):260–264. doi: 10.1016/0003-2697(80)90519-9. [DOI] [PubMed] [Google Scholar]
- Wu M., Lou J. K., Chang D. Y., Chang C. H., Nie Z. Q. Structure and function of a chloroplast DNA replication origin of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6761–6765. doi: 10.1073/pnas.83.18.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]